The universal triple-nucleotide genetic code is often viewed as a given, randomly selected through evolution. However, as summarized in this article, many observations and deductions within structural and thermodynamic frameworks help to explain the forces that must have shaped the code during the early evolution of life on Earth.
KEYWORD: Genetic code
ABSTRACT
The universal triple-nucleotide genetic code is often viewed as a given, randomly selected through evolution. However, as summarized in this article, many observations and deductions within structural and thermodynamic frameworks help to explain the forces that must have shaped the code during the early evolution of life on Earth.
INTRODUCTION
The universal triple-nucleotide genetic code, allowing DNA-encoded mRNA to be translated into the amino acid sequences of proteins using transfer RNAs (tRNAs) and many accessory and modification factors, is essentially common to all living organisms on Earth (1–3). Thousands of studies have focused on various aspects of the genetic code, revealing aspects of the basis for its structure and evolution (4–6). And no wonder, since the code provides a molecular explanation for the transmission of information from DNA to mRNA to protein (the central dogma of biology). All of genetics and molecular biology depend on the forces and factors that determine how the nucleotide triplet code translates into amino acid sequences.
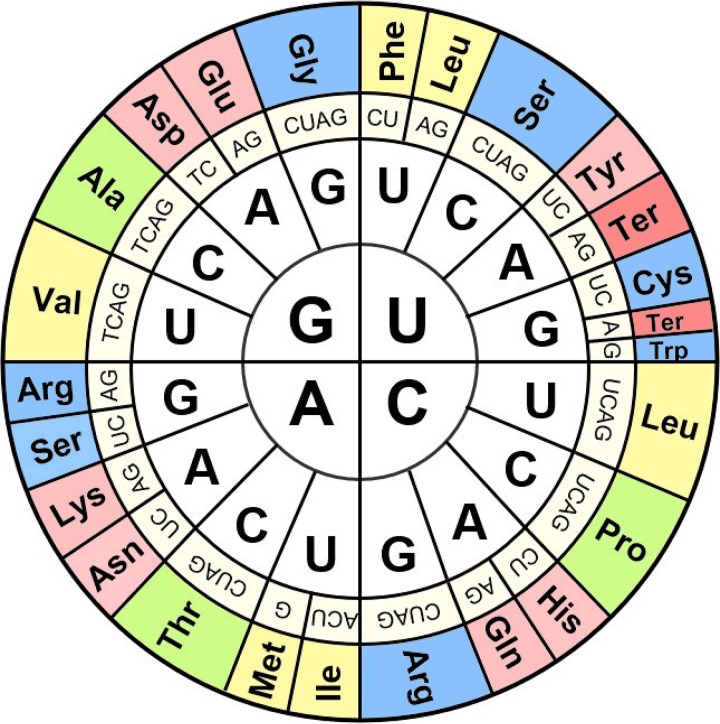
The codon wheel, used in virtually all textbooks and websites, has the nucleotide at position 1 determining the quadrant, with thymine (T, DNA) or uracil (U, RNA) in the 1st quadrant and cytosine (C), adenine (A), and guanine (G) in the 2nd, 3rd, and 4th quadrants, respectively (Fig. 1), where T, U, and C are small bases (pyrimidines) while G and A are large bases (purines). This convention is technically correct but may not be optimally helpful for conceptualization of the forces that dictate the code. Instead, the second nucleotide position should be emphasized as it is the one determining the nature of the amino acids encoded. How was this first deduced?
FIG 1.
The codon wheel as it appears in textbooks and websites. It allows any user to identify the amino acid encoded by any DNA/RNA codon. Codon position 1 is in the center of the wheel, codon position 2 is in the middle of the wheel, and codon position 3 is near the periphery of the wheel, next to the three-letter amino acid designation at the outermost part of the wheel. While technically correct, this wheel does not facilitate learning the essential features determining the rules that make sense of the code. TER, a polypeptide chain termination codon.
RELATIVE IMPORTANCE OF THE THREE CODON POSITIONS
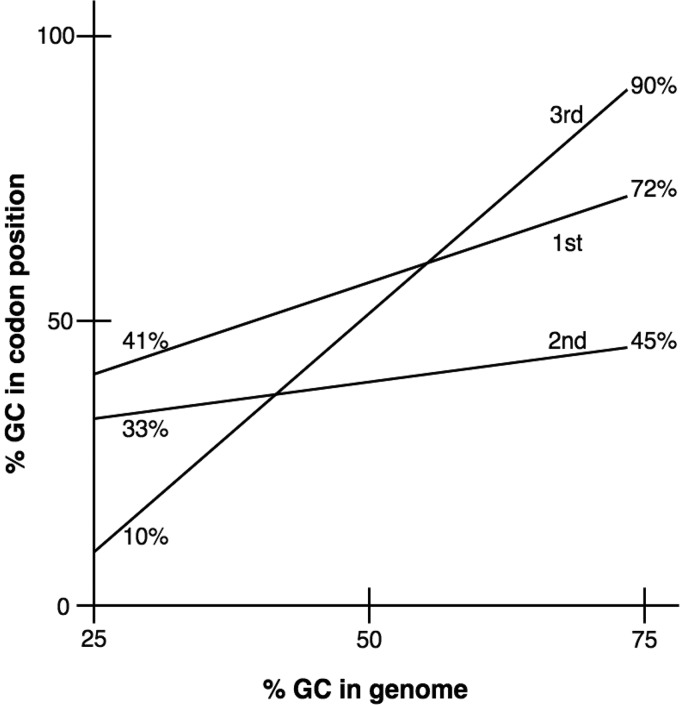
Living organisms have DNA guanine/cytosine (GC) contents that range from about 20% GC to 80% GC or 80% AT to 20% AT, respectively. When variations in the GC contents of the three codon positions, P1, P2, and P3, are plotted versus the GC contents of many genomes (Fig. 2), position 1 varies from 41% GC to 72% GC, a change of 31%. In contrast, position 2 varies from 33% to 45%, a change of only 12%. Position 3 varies from 10% to 90%, a whopping 80% change (7, 8). How did these differences arise during evolutionary history? Since point mutations normally arise randomly, with the advantageous ones being selected for while the deleterious ones are selected against, it can be assumed that these differences reflect the constraints imposed on mutations arising in these three codon positions. These constraints are apparently greatest for codon position 2 (P2) and least for codon position 3 (P3) (9). As we shall see, this is because P2 specifies the type of amino acid, codon position 1 (P1) usually specifies the specific amino acid, and P3 is highly redundant as several bases specify a particular amino acid. The different evolutionary rates of divergence can best be explained by the “negative selection principle,” i.e., functionally less important parts evolve (change) more rapidly than more important parts (10, 11). Thus, it would appear that P2 in codons is most important, P1 is of intermediate importance, and P3 is least important for specifying the amino acids in proteins (7).
FIG 2.
Correlation of G+C (GC) contents of the total genomic DNA of various organisms with the GC contents of the three codon positions. The first, second, and third positions of the three nucleotides in the mRNA codons, specifying amino acids in proteins, are labeled as such. (Modified from reference 7.)
AN ALTERNATIVE CODON WHEEL
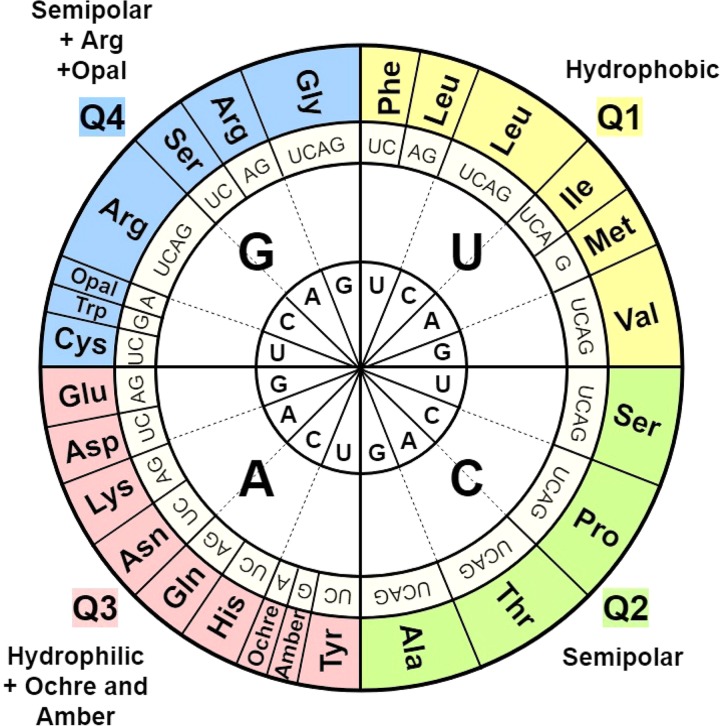
The relative importance of the three codon positions can be better understood if the helical wheel is plotted as shown in Fig. 3 (12, 13). With T/U in position 2 (quadrant 1, upper right), all amino acids are strongly hydrophobic without exception, but with A in position 2 (quadrant 3; lower left), all amino acids are strongly hydrophilic, also without exception. With C or G in position 2, most codons code for semipolar amino acids. Thus, when C is in position 2 (quadrant 2 in Fig. 3), there is no exception, but with G in position 2 (quadrant 4 in Fig. 3), there are two exceptions. Arginine, a strongly hydrophilic residue, and opal (UGA), a chain termination codon, are found within this quadrant (13). Interestingly, however, UGA can also code for amino acids: l-selenocysteine (14, 15), l-tryptophan (16), and glycine (17), all semipolar residues (18). One can imagine that the primordial code specified three types of amino acids, one hydrophobic, one hydrophilic, and two semipolar.
FIG 3.
Wheel representation of codon usage emphasizing the primary importance of the central codon position (position 2) in determining the type of amino acid, the secondary role of position 1 in determining the specific amino acid, and the relatively minor role of the third (wobble) position for amino acid specification. As in Fig. 1, the three-letter abbreviations of the amino acids are used. The three chain termination codons are indicated by name (UAA, ochre; UAG, amber; and UGA, opal). Quadrants 1 to 4 (Q1 to Q4, respectively) are indicated.
RELATED CODONS OFTEN SPECIFY RELATED AMINO ACIDS
Amino acids that exhibit similar properties are often encoded by codons that differ only in one position, P1, P2, or P3. For example, Asp and Glu are the two strongly acidic amino acids in proteins, and they are encoded by GAPy and GAPu (Py, pyrimidine; Pu, purine), respectively, differing only in P3. Moreover, Asn and Gln are derived from Asp and Glu by amidation, and their codons are AAPy (Asn) and CAPu (Gln), differing from those of their parental acidic amino acids only in P1. The two aliphatic hydroxy amino acids, Ser and Thr, are encoded by UCN and ACN (N, any nucleotide), respectively, differing only in P1. The two strongly basic amino acids, Lys and Arg, are encoded by AAPu and AGPu, respectively, differing only in P2, although Arg is also encoded by CGN. The two closely related aromatic residues, Phe and Tyr, are encoded by UUPy and UAPy, respectively, also differing only in P2. Finally, the aliphatic hydrophobic amino acids are all encoded by codons with U in position 2 as noted above, and many such codons differ from each other only in a single position.
THE WOBBLE POSITION: WHAT IS IMPORTANT FOR AMINO ACID SPECIFICATION IN P3?
What quality of the position 3 nucleotide influences amino acid selection? Examination of the codon wheel shown in Fig. 3 reveals that when P3 is important, it is only important whether the base in P3 is a purine (A or G) or a pyrimidine (U or C). Thus, only the type of the base at position 3 is important (12) (see next section). However, there are two exceptions: Ile/Met and Trp/opal (Fig. 3). Three codons specify isoleucine (AUU, AUC, and AUA) with only one codon (AUG) specifying methionine, while one each specifies tryptophan (Trp; UGG) and chain termination (opal; UGA). Interestingly, though, as noted above, some organisms and organelles, including mitochondria, use both codons (UGG and UGA) to specify Trp, and then UGA is not a stop codon (12). Similarly, when UGA specifies selenocysteine or glycine, it does not terminate extension of the growing polypeptide chain. In all other cases where P3 is important, only the type of base is important as noted above.
THE WOBBLE POSITION: WHEN IS P3 IMPORTANT?
Referring to Fig. 3 again, it can be seen that when P2 is C, P3 is never important. When P2 is an A, P3 is always important, determined only by whether it is a purine or pyrimidine but not by which of the two purines or pyrimidines it is. However, when P2 is a G or U, P3 is sometimes important. Thus, P2 primarily determines when P3 plays a role in specifying an amino acid.
IF P2 IS A G OR U, WHEN IS P3 IMPORTANT?
When P2 is a G or U, the wobble position is important if and only if P1 is an A or U, not when P1 is a G or C. Since an A-U base pair has two H bonds while a G-C base pair has three, this suggests that H-bond strength plays a dominant role although base shape complementarity may also play a role (19). In other words, with P2 as a G or U, the type of base pair at P1 (A-U versus G-C) determines the importance of P3. The H-bond strength of P2 plus P1 likely is a determinative factor, but, clearly, this does not provide a full explanation. We need to further refine our understanding of the specifications that determine the importance of P3.
THE H-BOND STRENGTH OF A-U (mRNA-tRNA) MAY NOT BE THE SAME AS U-A (mRNA-tRNA)
Careful consideration of Fig. 3 suggests that A-U (mRNA-tRNA) is not equivalent to U-A and that G-C is not equivalent to C-G. In fact, U-A probably forms stronger bonds than A-U, and C-G probably forms stronger bonds than G-C. In other words, the H bonds may be stronger when the pyrimidine is in the mRNA and the purine is in the tRNA. This explains why the wobble position is never important when C is in P2 of the mRNA although it is sometimes important when G is in P2 of the mRNA (that is, when an A or U is in P1). Similarly, the wobble position is sometimes important when U is in P2 (that is, when A or U is in P1) but always important when A is in P2. These differences in H-bond strength between U-A and A-U or between C-G and G-C may have to do with the established fact that straight H bonds are the strongest (20), suggesting that both the numbers and configurations of the H bonds dictate their thermodynamic consequences. In this case, the curvature of the anticodon on the tRNA may be responsible. It has been argued that discrimination between tRNAs is dependent on steric (shape) complementarity of the bases (9, 21) and that base modification of the tRNAs could play a role (22).
DEPENDENCY OF TRANSLATION ON tRNA MODIFICATIONS
A new frontier in understanding the details of the central dogma of biology involves the effects of posttranscriptional tRNA modifications, some of which may be nearly universal across phyla while others are phylum specific (23). More than 100 such tRNA modifications have been identified, a major fraction in their RNA anticodon loops (24). Modifications include deamination of adenosine to inosine, introduction of the modified nucleoside, queuosine, thiolation, methylation, isopentenylation, 5-methoxycarbonyl methylation, threonyl carbamoylation, and others (25–28). These modifications are necessary for the speed and fidelity of translation. Hypomodification can inhibit translation and thereby inhibit growth (29, 30). Changes in tRNA modifications have been shown to be involved in diseases in humans as well as bacterial pathogenesis.
Particularly relevant to this minireview, these modifications favor specific codon-anticodon affinities by stabilizing specific base pairs, thus fine-tuning protein synthesis (31). Codon bias promotes preferential utilization of certain synonymous codons that differ only in P3 of the codon (32). Moreover, modification-dependent tRNA cleavage can facilitate downregulation of protein synthesis in response to stress signals (31). To make matters even more complicated, one tRNA modification may influence the activity of an enzyme catalyzing another modification reaction (33). From these observations, it is clear that numerous posttranscriptional modifications of tRNAs play important roles in the efficiency and accuracy of translation.
CHAIN INITIATION CODONS
Initiation codons, acting with an initiation tRNA, usually encode formyl methionine (fMet) in bacteria, chloroplasts, and mitochondria or methionine (Met) in archaea and the cytosol of eukaryotes (34–36). The codon wobble position is P1, where the order of usage for prokaryotes is usually AUG > GUG > UUG > CUG. However, in high-GC-content organisms, the frequencies of GUG relative to those of AUG increase, and in many eukaryotes, the order of initiation codon usage is AUG > CUG > GUG > UUG (37, 38). While many codons can be used to initiate translation at low frequencies (39), the initiation factors and mechanisms of chain initiation are complex but similar in different organisms (40), and either fMet or Met is used as the initiating amino acid, depending on conditions, regardless of the codon used (41). It should be reemphasized that Met codon discrimination depends on anticodon modifications and is often species specific (42, 43).
CHAIN TERMINATION (STOP OR NONSENSE) CODONS
UAA (ochre) is the best and most frequently used chain termination (stop or nonsense) codon, particularly in low- or moderate-GC-content organisms (44). It virtually never codes for anything other than stop. UAG (amber), used in smaller amounts but almost invariant with respect to GC content, can also code for pyrrolysine, which is an active-site residue in some methyltransferases (45). This amino acid is found most frequently in archaea but occasionally in bacteria (46). Of the three stop codons, UGA (opal) is used for chain termination primarily in high-GC-content organisms, but the actual frequency depends also on the organismal type (44). These three codons are recognized by release factors (RFs): RF1 (which recognizes UAA and UAG), RF2 (which recognizes UAA and UGA), and RF3 (which functions to recycle RF1 and RF2 in Escherichia coli). These release factors may have coevolved with the stop codons (47–49). Thus, in most organismal phyla, UAA is used more frequently than UAG or UGA (44). The importance of the UAA stop codon is illustrated by the observation that highly expressed genes predominantly end with UAA (44).
It is interesting that all of the common nonsense codons use U in position 1 which is invariant, with two purines in positions 2 and 3. Since AU base pairs have two hydrogen (H) bonds while GC has three, the best stop codon (UAA) potentially would have only six H bonds (2 per codon position) if it were to pair with its complementary sequence in a tRNA, while the other two would have seven (20). Codons, in general, have between six and nine H bonds, depending on their AU versus GC contents, suggesting that weaker hydrogen bonding potentially may have played a role in the selection of the chain termination codon(s) early in the formulation of the code.
AMINO ACIDS IN THE PREBIOTIC PRIMORDIAL SOUP
It seems probable that the earliest evolving microorganisms had to survive on compounds that were present in the prebiotic primordial soup (50, 51). Stanley Miller’s atmospheric spark discharge experiments and subsequent studies showed that 10 of the 20 common, naturally occurring amino acids in proteins could be generated abiotically by using simulated primordial Earth conditions (52). Moreover, these compounds corresponded roughly in relative abundance to those in meteorites (53). These 10 abiotic amino acids, in order of their relative abundances, were Gly > Ala > Asp > Glu > Val > Ser > Ile > Leu > Pro > Thr (54). This order proved to correlate with the free energies of their syntheses, suggesting that thermodynamics determined their relative amounts. In more recent experiments, not only amino acids but also nucleic acid bases and fatty acids could be made from inorganic sources of hydrogen, carbon, nitrogen, and sulfur in the presence of UV radiation under plausible prebiotic conditions (55). These observations further strengthen the argument that prebiotic conditions led to the synthesis of molecules that facilitated the evolution of simple life forms from preexisting compounds. This argument is applicable regardless of whether life arose here on Earth or came here from some other source in outer space.
The eight top amino acids, listed in Table 1, fall into three groups: the semipolar amino acids (Gly, Ala, and Ser), the acidic hydrophilic amino acids (Asp and Glu), and the aliphatic hydrophobic amino acids (Val, Ile, and Leu). And as discussed above, when the second base in a codon (P2) is G or C, semipolar residues are usually encoded. In the primordial code, if this were true, what semipolar amino acid would have been preferred? Examining the codon wheels in Fig. 1 and 3, we find that when C is in P2, Ala, Ser, Thr, and Pro are encoded, but when G is in P2, Gly and Ser are encoded. Thus, if we were to select a single primordial amino acid, the most abundant one, Gly, is the preferred choice with G in P2, but Ala is the preferred choice with C in P2. Examining the codon wheels further, we note that if G is in P1, regardless of which base is at P2, Gly, Glu/Asp, Ala, and Val are encoded, which prove to be the five most abundant amino acids predicted for the primordial soup (Table 1). Thus, if we are to propose a primitive code involving specific amino acids, we might suggest only four or five amino acids encoded by four codons: GGN encoding Gly, GAN encoding Glu/Asp, GCN encoding Ala, and GUN encoding Val (where N is any base). Thus, it is possible that G (with three H bonds) in P1 yielded the four original codons, coding for the four or five most prevalent amino acids in the prebiotic soup.
TABLE 1.
Properties of the 8 amino acids believed to be present in greatest amounts in the prebiotic primordial soup
| Amino acid | Rank in the soupa | Rank in proteinsb | Polarityc | Hydrophobicityd | Vol (Å)e | Surface area (Å2)f |
|---|---|---|---|---|---|---|
| Glycine | 1 | 5 | 0 | −0.4 | 48 | 85 |
| Alanine | 2 | 2 | 0 | +0.8 | 67 | 113 |
| Serine | 6 | 7 | 0.1 | +0.8 | 73 | 122 |
| Aspartate | 3 | 10 | 50 | −3.5 | 91 | 151 |
| Glutamate | 4 | 6 | 50 | −3.5 | 109 | 183 |
| Valine | 5 | 3 | 0.1 | +4.2 | 105 | 160 |
| Isoleucine | 7 | 4 | 0.1 | +3.8 | 124 | 182 |
| Leucine | 8 | 1 | 0.1 | +4.5 | 124 | 180 |
The identities and relative concentrations of the 8 most abundant amino acids on the prebiotic Earth (and elsewhere in the universe) based on two lines of evidence: first, by the relative amounts of these amino acids produced in prebiotic chemistry experiments (52, 83, 88), and, second, by the concentrations of these compounds in meteorites (54). While these two lines of evidence lead to the same relative values, they also correlate with the free energies of the amino acid syntheses; the amino acids synthesized with expenditure of the least amount of energy are present in the largest amounts (54, 56, 85).
Derived from analyses of present-day protein compositions (56).
Polarity is a measure of the electric field strength around the molecule (89).
Relative hydrophobicity is based on the values reported by Kyte and Doolittle (90).
Surface area represents the area accessible to water in an unfolded peptide (91).
Data in Table 1 tabulate properties of the common amino acids: polarity, hydrophobicity (a positive [+] value) versus hydrophilicity (a negative [−] value), molecular volume, and surface area (see also the footnotes to Table 1) (56). The three groups of amino acids (semipolar, polar, and nonpolar) are clearly delineated on the basis of these properties, suggesting means by which the types of amino acids could have been distinguished by an evolving coding system. Of course, later stepwise evolutionary events presumably involved expansion of the code to include eventually all 20 common protein amino acids. Thus, expansion would result from the subdivision of codon blocks in which some of the similar codons assigned to an early amino acid were reassigned to a late amino acid. These subdivisions would usually involve the introduction of related amino acids so as to minimize the consequences of mutations and translational errors. The current code would thus be a relic of the early code (56).
WHY IS THE GENETIC CODE SO WELL CONSERVED?
The standard extant genetic code includes a number of minor organismal differences, particularly in eukaryotic organelles as well as in parasitic and symbiotic prokaryotes with small genomes. However, the standard code is essentially universal (54). Several scientists have suggested why the code should be so well conserved, and the consensus is that there is probably more than one reason. One is referred to the “frozen accident.” By this, it is suggested that a codon reassignment gives rise to harmful effects on translation, decreasing the robustness of the standard, nonrandom code, which appears to be designed, in part, to minimize the deleterious consequences of mutations and errors in translation (56). This argument assumes that the code was optimized long ago, so it is now almost perfect.
Whether this is true or not is controversial, but at least we can claim that the code is good enough and hard to change. Another argument suggests that codon variation among organisms would inhibit the occurrence of lateral (horizontal) transfer of genetic materials between organisms. This would be detrimental as adaptation to environmental changes often depends on interorganismal genetic exchange mechanisms, of which we currently recognize several (13). Lateral transfer is most common in microbes that live in changing environments and that need to adapt quickly to survive. Conditions that existed early during the evolution of the genetic code and early life were, of course, very different from those we experience today, including anaerobiosis and high dissolved Fe2+ concentrations. However, regardless of conditions, horizontal gene transfer was probably more important then than it is now (57).
BENEFITS OF A REDUNDANT GENETIC CODE
As noted above, the genetic code is redundant, with as many as six synonymous codons specifying a single amino acid. Synonymous rare codons are now known to have diverse functions, including regulation of cotranslational protein folding, facilitation of covalent protein modifications during or after synthesis, and co- or posttranslational secretion (58). It has also been argued that the redundant code decreases the deleterious consequences of random point mutations (9, 59–61). This is currently an active field of research, and new advances are continuously being made.
Exchanging synonymous codons can cause diseases in humans and other organisms (62, 63), an observation that is not surprising when it is considered that translational pausing is programmed, allowing, for example, coordinated folding of the nascent proteins (64). Synonymous codon selection may also play a role in epigenetic modifications (65). Current studies indicate that there are additional benefits as noted above.
CODON FREQUENCIES VERSUS GENE EXPRESSION LEVELS
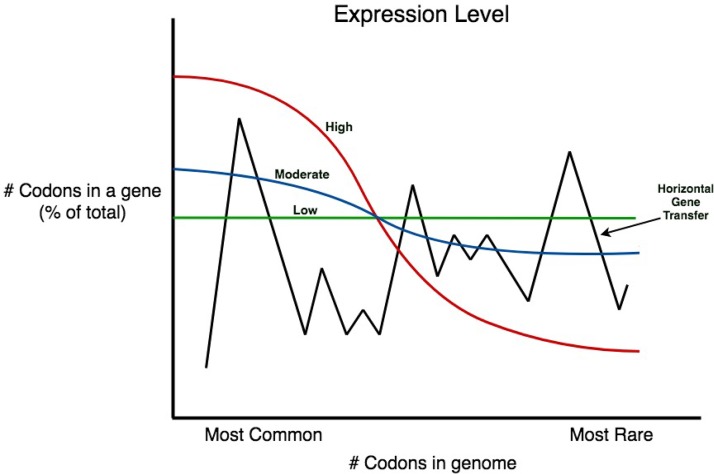
For any organism, some codons specifying an amino acid are used frequently while others are infrequently used (rare codons) although the set of preferred codons differs for phylogenetically distant organisms. This is a hot topic of investigation as ∼100 papers are published over a single year on this subject alone. Figure 4 shows a schematic view of the use of the most common codons versus rare codons for genes expressed at different levels in a range of organisms. If expressed at high levels (e.g., ribosomal proteins), the common codons are used with high frequency while rare codons are seldom used (Fig. 4, red line) (58, 66). If a gene is expressed at very low levels (e.g., the gene for the E. coli lactose repressor lacI (Fig. 4, green line), there is little preference for common codons. As expected, moderately expressed genes, or highly expressed genes (e.g., the lactose operon) induced under rare conditions (Fig. 4, blue line), use common codons with intermediate frequencies (67, 68). The presumption is that the use of common codons, corresponding to the most prevalent tRNAs, favors rapid and accurate translation and therefore increases the level of the gene product (67, 69). This is expected since a higher rate of translation should result if the cytoplasmic concentrations of the tRNAs used are high. Furthermore, it has been shown that the use of suboptimal codons leads to misincorporation of amino acids by the ribosome (70–72). This is particularly detrimental for proteins needed in large amounts but of little importance for proteins for which only a few copies are required (72).
FIG 4.
Schematized correlation between the level of gene expression and the frequency of common versus rare codons used in the coding region of the corresponding gene. The red line represents the codon usage pattern for highly expressed genes, the blue line shows the same for genes expressed at a moderate level or those that are induced to high levels only under certain conditions, and the green line represents the codon usage pattern for genes that are expressed at very low levels. Finally, the black line reveals the pattern for a gene with little or no correlation of its codon usage with the frequency of codons used in the organism. Such a gene was presumably obtained by horizontal (lateral) gene transfer from an organism with a very different set of codon usage frequencies. Note that codon frequencies roughly correlate with the levels of the corresponding tRNAs in the cytoplasm of the organism in which that gene evolved (68, 70, 92), and the levels of the tRNAs in the cell determine the benefit for highly expressed genes using the commonly used codons. Genes expressed at low levels do not prefer common codons because low rates of translation of these genes are not deleterious.
Horizontally transferred genes, obtained from another organism (which often has different codon preferences [73]), will not show a correlation with the codon preferences of the recipient organism (Fig. 4, black line). Studies have shown that it takes hundreds of millions of years for such a gene to come to equilibrium with the codon usage pattern of the recipient (74). For this reason, computer programs could be designed to estimate not only what type of organism the gene came from but also when in evolutionary history the transfer event occurred (75–81). But additional benefits that result from the coding options chosen include maximizing recombinant gene expression, controlling protein folding, and attenuating viruses.
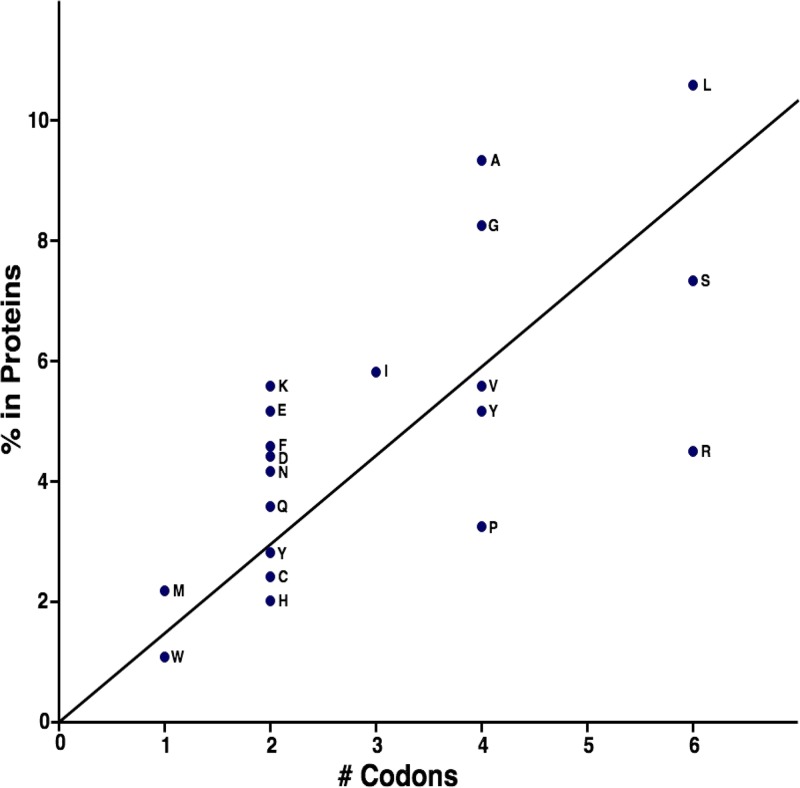
FREQUENCIES OF AMINO ACIDS IN PROTEINS AS A FUNCTION OF THE NUMBERS OF ENCODING CODONS
Examination of Fig. 3 reveals that some amino acids (Trp and Met) have only one codon, while others (Leu, Ser, and Arg) have six codons each. All others have two, three, or four codons. In Fig. 5, the percentage of a particular amino acid in an array of randomly selected proteins is plotted versus the number of codons that specify that amino acid. Trp and Met are the rarest amino acids in proteins, and, as noted above, each is encoded by only one codon. A quick perusal of Fig. 5 shows that while there is a rough correlation between percent occurrence in proteins and numbers of encoding codons, there is considerable scatter from a straight line. A similar plot with less scatter was obtained by King and Jukes when a set of proteins exclusively of mammalian origin was examined (82). Thus, codon numbers correlate roughly with relative amino acid frequencies in proteins. The availability of certain thermodynamically stable amino acids in the primordial soup may have played a role in the selection of the amino acids first to be incorporated into proteins (83, 84). This is because these are the amino acids that predominated before amino acid biosynthetic pathways evolved (see “Amino Acids in the Prebiotic Primordial Soup” above) (85).
FIG 5.
Plot of amino acid frequency in proteins versus the numbers of codons specifying these amino acids. The one-letter abbreviations of the amino acids are adjacent to the points representing the positions corresponding to their relative abundances, expressed as a percentage of the total in proteins on the y axis. The numbers of codons that specify the amino acids are plotted on the x axis. The amino acid frequencies in randomly selected representative proteins from all domains of living organisms were taken from Saier (13). (Republished from reference 13 with permission of the publisher.)
Which came first, the need for a greater amount of a particular amino acid or an increased number of codons? Possibly the former was the driving force that was responsible for the differing numbers of codons used to specify the different amino acids. However, the correlation observed in Fig. 5 leads to additional unanswered questions. Why does this correlation exist, and what does it tell us? While we can guess at the answers, further research will be needed to provide definitive answers.
TWELVE RULES SUMMARIZING THE FORCES THAT DETERMINE THE GENETIC CODE
Simple observations noted in this article correlate with and may provide an explanation for some of the factors influencing the specification of amino acids by codons within the genetic code. These are summarized here. (i) Position 2 (P2) is most important of the three nucleotide codon positions because it specifies the type of amino acid, while position 1 (P1) determines the specific amino acid, sometimes with the aid of P3, the wobble position. (ii) The frequency of an amino acid in proteins roughly correlates with the number of codons that specify it. (iii) Initiation codons, acting with an initiation tRNA, encode formyl methionine or methionine, but the codon wobble position is P1 where the order of usage is AUG > GUG > UUG > CUG in many organisms and organelles. (iv) Chain termination codons (UAA > UAG or UGA) have an invariant U in position 1 with two purines in P2 and P3; weak hydrogen (H) bonding may have influenced their evolution. (v) Highly expressed genes use the most common codons in an organism while genes expressed at low levels use rare codons with higher frequencies, but horizontally (laterally) transferred genes may show no correlation. (vi) When P3 is important for amino acid specification, it is important only whether P3 is a purine (A or G) or a pyrimidine (U or C) with just a couple of exceptions. (vii) Whether or not P3 is important is determined by the nucleotide at P2: when P2 is a C, P3 is never important; when P2 is an A, P3 is always important; when P2 is a U or G, P3 is sometimes important. (viii) When P2 is a U or G, P3 is important only when P1 is an A or U but not when P1 is a G or C, so the numbers of H bonds in P2 plus P1 determine the importance of P3. (ix) It makes a difference if an A or U is in the mRNA or the tRNA to the H-bond strength. U-A (mRNA-tRNA) is stronger than A-U, and C-G is stronger than G-C. Thus, a pyrimidine in the mRNA forms stronger H bonds with the tRNA than when the corresponding H-bonded purine is in the mRNA. (x) Related amino acids are often encoded by similar codons, differing in a single position, suggesting that one derived from the other. (xi) Rare synonymous codons can be programmed for translational pausing, promoting cotranslational protein folding, covalent modification, and secretion. (xii) The most common amino acids in proteins are often, but not always, the thermodynamically most stable ones.
These observations allow thermodynamic rationalization of many aspects of the genetic code and lead to postulates about how the code may have evolved, first from four types of amino acids, then with the specification of certain specific amino acids, and then by expansion with the specification of additional related amino acids.
CONCLUSIONS
Science strives to reveal the laws of nature, and critical to an understanding of all of biology is the central dogma, the basic framework whereby genetic information flows from DNA to RNA to protein. Conceptually, the RNA polymerase-mediated transcription of DNA to RNA is relatively straightforward, but the translation of RNA into proteins is much more complicated. It is this last subject, involving the triplet genetic code, that is the focus of this minireview. Based on our knowledge that C and U are pyrimidines, very different in structure from purines (G and A), and that A-U pairs form two hydrogen bonds while G-C pairs form three, we have been able to come to important suggestions regarding the thermodynamic basis for amino acid specification in proteins by the nucleotide codons in mRNAs. We are also able to formulate hypotheses, based on sound principles and compelling experimental evidence, as to how this code arose. The appearance of the code, dictated by thermodynamic principles, probably followed a logical sequence of events in which a limited number of readily available amino acids, present in the primordial soup, and a simple nucleotide code to specify as few as 4 amino acids but as many as 8 or 10 amino acids gradually expanded as additional amino acids became available due to evolving anabolic pathways. This would have involved the use of an increasing number of smaller blocks of codons specifying a correspondingly increased number of amino acids (54, 57, 85–87). The next step would be to experimentally examine these observations to test the hypotheses put forth and to generate a better understanding of the fine details by which the nearly universal genetic code specifies the 22 encoded amino acids in proteins.
ACKNOWLEDGMENTS
I thank professors Steven Baird, Russ Doolittle, Adam Hockenberry, Jack Kyte, William Margolin, Arturo Medrano-Soto, Mauricio Montal, Sheila Podell, Ralf Rabus, Jack Trevors, and Chris Wills for helpful comments on the contents of this article.
The work reported has been used for teaching purposes at UCSD and was supported by grant GM077402 from the U.S. National Institutes of Health.
Biography
Milton H. Saier, Jr., is a professor of molecular biology at the University of California at San Diego. His current research focuses on molecular evolution involving several aspects of the central dogma of molecular biology. These include (i) membrane protein evolution, (ii) transport and metabolic regulation, and (iii) transposon-mediated directed mutation. He has taught many graduate and undergraduate courses over the years, most recently including microbial biochemistry, microbial genetics, and microbial physiology as well as human impact on the environment. He is a long-standing member of the ASM and the AAAS as well as an honorary member of La Société Française de Microbiologie (SFM) and the Alexander von Humboldt Stiftung of Germany. His wife, Jeanne, and he have performed chamber music throughout most of their lives. They have three adult children, Hans, Anila, and Amanda, and six grandchildren.
REFERENCES
- 1.Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. 1961. General nature of the genetic code for proteins. Nature 192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 2.Kubyshkin V, Acevedo-Rocha CG, Budisa N. 2018. On universal coding events in protein biogenesis. Biosystems 164:16–25. doi: 10.1016/j.biosystems.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Tamura K. 2015. Origins and early evolution of the tRNA molecule. Life (Basel) 5:1687–1699. doi: 10.3390/life5041687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanofsky C. 2007. Establishing the triplet nature of the genetic code. Cell 128:815–818. doi: 10.1016/j.cell.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 5.Khorana HG. 1979. Total synthesis of a gene. Science 203:614–625. doi: 10.1126/science.366749. [DOI] [PubMed] [Google Scholar]
- 6.Nirenberg MW, Matthaei JH. 1961. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci U S A 47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muto A, Osawa S. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A 84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krawiec S, Riley M. 1990. Organization of the bacterial chromosome. Microbiol Rev 54:502–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeland SJ, Hurst LD. 1998. The genetic code is one in a million. J Mol Evol 47:238–248. doi: 10.1007/PL00006381. [DOI] [PubMed] [Google Scholar]
- 10.Anderson WW. 1989. Selection in natural and experimental populations of Drosophila pseudoobscura. Genome 31:239–245. doi: 10.1139/g89-041. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M, Ohta T. 1974. On some principles governing molecular evolution. Proc Natl Acad Sci U S A 71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagerkvist U. 1981. Unorthodox codon reading and the evolution of the genetic code. Cell 23:305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- 13.Saier MH., Jr. 2008. The bacterial chromosome. Crit Rev Biochem Mol Biol 43:89–134. doi: 10.1080/10409230801921262. [DOI] [PubMed] [Google Scholar]
- 14.Zinoni F, Birkmann A, Leinfelder W, Bock A. 1987. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci U S A 84:3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Flores JN, Shetty SP, Dubey A, Copeland PR. 2013. The molecular biology of selenocysteine. Biomol Concepts 4:349–365. doi: 10.1515/bmc-2013-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osawa S, Muto A, Ohama T, Andachi Y, Tanaka R, Yamao F. 1990. Prokaryotic genetic code. Experientia 46:1097–1106. doi: 10.1007/BF01936919. [DOI] [PubMed] [Google Scholar]
- 17.Hanke A, Hamann E, Sharma R, Geelhoed JS, Hargesheimer T, Kraft B, Meyer V, Lenk S, Osmers H, Wu R, Makinwa K, Hettich RL, Banfield JF, Tegetmeyer HE, Strous M. 2014. Recoding of the stop codon UGA to glycine by a BD1-5/SN-2 bacterium and niche partitioning between Alpha- and Gammaproteobacteria in a tidal sediment microbial community naturally selected in a laboratory chemostat. Front Microbiol 5:231. doi: 10.3389/fmicb.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saier MH., Jr. 1987. Enzymes in metabolic pathways: a comparative study of mechanism, structure, evolution and control. Harper & Row, Publishers, Inc, New York, NY. [Google Scholar]
- 19.Dzantiev L, Alekseyev YO, Morales JC, Kool ET, Romano LJ. 2001. Significance of nucleobase shape complementarity and hydrogen bonding in the formation and stability of the closed polymerase-DNA complex. Biochemistry 40:3215–3221. doi: 10.1021/bi002569i. [DOI] [PubMed] [Google Scholar]
- 20.Pauling L. 1960. The nature of the chemical bond, 3rd ed Cornell University Press, Ithaca, NY. [Google Scholar]
- 21.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016. New structural insights into translational miscoding. Trends Biochem Sci 41:798–814. doi: 10.1016/j.tibs.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Pan T. 2018. Modifications and functional genomics of human transfer RNA. Cell Res 28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Percival HG. 1989. Initial continence testing of sleeved monolayer colonic anastomoses in sheep: a comparative bench study. Dis Colon Rectum 32:21–25. doi: 10.1007/BF02554719. [DOI] [PubMed] [Google Scholar]
- 24.Bednarova A, Hanna M, Durham I, VanCleave T, England A, Chaudhuri A, Krishnan N. 2017. Lost in translation: defects in transfer RNA modifications and neurological disorders. Front Mol Neurosci 10:135. doi: 10.3389/fnmol.2017.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuorto F, Lyko F. 2016. Genome recoding by tRNA modifications. Open Biol 6:160287. doi: 10.1098/rsob.160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai Y, Nakai M, Yano T. 2017. Sulfur modifications of the wobble U34 in tRNAs and their intracellular localization in eukaryotic cells. Biomolecules 7:17. doi: 10.3390/biom7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer U, Bohleber S, Fradejas-Villar N. 2017. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol 14:1197–1208. doi: 10.1080/15476286.2017.1294309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agris PF, Eruysal ER, Narendran A, Vare VYP, Vangaveti S, Ranganathan SV. 2018. Celebrating wobble decoding: half a century and still much is new. RNA Biol 15:537–553. doi: 10.1080/15476286.2017.1356562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNally KP, Panzica MT, Kim T, Cortes DB, McNally FJ. 2016. A novel chromosome segregation mechanism during female meiosis. Mol Biol Cell 27:2576–2589. doi: 10.1091/mbc.e16-05-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjan N, Rodnina MV. 2017. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc 139:5857–5864. doi: 10.1021/jacs.7b00727. [DOI] [PubMed] [Google Scholar]
- 31.Duechler M, Leszczyńska G, Sochacka E, Nawrot B. 2016. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci 73:3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrenhofer-Murray AE. 2017. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules 7:E14. doi: 10.3390/biom7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy B, Liu Q, Shoji S, Fredrick K. 2018. IF2 and unique features of initiator tRNA(fMet) help establish the translational reading frame. RNA Biol 15:604–613. doi: 10.1080/15476286.2017.1379636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramesh V, Kohrer C, RajBhandary UL. 2002. Expression of Escherichia coli methionyl-tRNA formyltransferase in Saccharomyces cerevisiae leads to formylation of the cytoplasmic initiator tRNA and possibly to initiation of protein synthesis with formylmethionine. Mol Cell Biol 22:5434–5442. doi: 10.1128/MCB.22.15.5434-5442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharyya S, Varshney U. 2016. Evolution of initiator tRNAs and selection of methionine as the initiating amino acid. RNA Biol 13:810–819. doi: 10.1080/15476286.2016.1195943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinnebusch AG. 2017. structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42:589–611. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31:1717–1731. doi: 10.1101/gad.305250.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng X, Hsu SJ, Kasbek C, Chaiken M, Price CM. 2017. CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res 45:4281–4293. doi: 10.1093/nar/gkx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gualerzi CO, Pon CL. 2015. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell Mol Life Sci 72:4341–4367. doi: 10.1007/s00018-015-2010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JM, Seok OH, Ju S, Heo JE, Yeom J, Kim DS, Yoo JY, Varshavsky A, Lee C, Hwang CS. 2018. Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway. Science 362:eaat0174. doi: 10.1126/science.aat0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones TE, Brown CL, Geslain R, Alexander RW, Ribas de Pouplana L. 2008. An operational RNA code for faithful assignment of AUG triplets to methionine. Mol Cell 29:401–407. doi: 10.1016/j.molcel.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 43.Jones TE, Ribas de Pouplana L, Alexander RW. 2013. Evidence for late resolution of the aux codon box in evolution. J Biol Chem 288:19625–19632. doi: 10.1074/jbc.M112.449249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korkmaz G, Holm M, Wiens T, Sanyal S. 2014. Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance. J Biol Chem 289:30334–30342. doi: 10.1074/jbc.M114.606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibba M, Soll D. 2002. Genetic code: introducing pyrrolysine. Curr Biol 12:R464–R466. doi: 10.1016/S0960-9822(02)00947-8. [DOI] [PubMed] [Google Scholar]
- 46.Crnkovic A, Suzuki T, Soll D, Reynolds NM. 2016. Pyrrolysyl-tRNA synthetase, an aminoacyl-tRNA synthetase for genetic code expansion. Croat Chem Acta 89:163–174. doi: 10.5562/cca2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi X, Joseph S. 2016. Mechanism of translation termination: RF1 dissociation follows dissociation of RF3 from the ribosome. Biochemistry 55:6344–6354. doi: 10.1021/acs.biochem.6b00921. [DOI] [PubMed] [Google Scholar]
- 48.Wei Y, Wang J, Xia X. 2016. Coevolution between stop codon usage and release factors in bacterial species. Mol Biol Evol 33:2357–2367. doi: 10.1093/molbev/msw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baggett NE, Zhang Y, Gross CA. 2017. Global analysis of translation termination in E. coli. PLoS Genet 13:e1006676. doi: 10.1371/journal.pgen.1006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oberbeck VR, Fogleman G. 1990. Impact constraints on the environment for chemical evolution and the continuity of life. Orig Life Evol Biosph 20:181–195. doi: 10.1007/BF01808281. [DOI] [PubMed] [Google Scholar]
- 51.Melendez-Hevia E, Montero-Gomez N, Montero F. 2008. From prebiotic chemistry to cellular metabolism–the chemical evolution of metabolism before Darwinian natural selection. J Theor Biol 252:505–519. doi: 10.1016/j.jtbi.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 52.Ring D, Wolman Y, Friedmann N, Miller SL. 1972. Prebiotic synthesis of hydrophobic and protein amino acids. Proc Natl Acad Sci U S A 69:765–768. doi: 10.1073/pnas.69.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bada JL. 2013. New insights into prebiotic chemistry from Stanley Miller's spark discharge experiments. Chem Soc Rev 42:2186–2196. doi: 10.1039/c3cs35433d. [DOI] [PubMed] [Google Scholar]
- 54.Koonin EV, Novozhilov AS. 2017. Origin and evolution of the universal genetic code. Annu Rev Genet 51:45–62. doi: 10.1146/annurev-genet-120116-024713. [DOI] [PubMed] [Google Scholar]
- 55.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. 2015. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nature Chem 7:301–307. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higgs PG. 2009. A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16. doi: 10.1186/1745-6150-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barge LM. 2018. Considering planetary environments in origin of life studies. Nat Commun 9:5170. doi: 10.1038/s41467-018-07493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaney JL, Clark PL. 2015. Roles for synonymous codon usage in protein biogenesis. Annu Rev Biophys 44:143–166. doi: 10.1146/annurev-biophys-060414-034333. [DOI] [PubMed] [Google Scholar]
- 59.Freeland SJ, Knight RD, Landweber LF, Hurst LD. 2000. Early fixation of an optimal genetic code. Mol Biol Evol 17:511–518. doi: 10.1093/oxfordjournals.molbev.a026331. [DOI] [PubMed] [Google Scholar]
- 60.Dufton MJ. 1985. Genetic code redundancy and the evolutionary stability of protein secondary structure. J Theor Biol 116:343–348. doi: 10.1016/S0022-5193(85)80272-1. [DOI] [PubMed] [Google Scholar]
- 61.Dufton MJ. 1983. The significance of redundancy in the genetic code. J Theor Biol 102:521–526. doi: 10.1016/0022-5193(83)90388-0. [DOI] [PubMed] [Google Scholar]
- 62.Maraia RJ, Iben JR. 2014. Different types of secondary information in the genetic code. RNA 20:977–984. doi: 10.1261/rna.044115.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM. 2013. Rare codons regulate KRas oncogenesis. Curr Biol 23:70–75. doi: 10.1016/j.cub.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Onofrio DJ, Abel DL. 2014. Redundancy of the genetic code enables translational pausing. Front Genet 5:140. doi: 10.3389/fgene.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maleszka R, Mason PH, Barron AB. 2014. Epigenomics and the concept of degeneracy in biological systems. Brief Funct Genomics 13:191–202. doi: 10.1093/bfgp/elt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutman GA, Hatfield GW. 1989. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A 86:3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trotta E. 2011. The 3-base periodicity and codon usage of coding sequences are correlated with gene expression at the level of transcription elongation. PLoS One 6:e21590. doi: 10.1371/journal.pone.0021590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Supek F, Vlahovicek K. 2005. Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinformatics 6:182. doi: 10.1186/1471-2105-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 70.Gingold H, Pilpel Y. 2011. Determinants of translation efficiency and accuracy. Mol Syst Biol 7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quax TE, Claassens NJ, Soll D, van der Oost J. 2015. Codon bias as a means to fine-tune gene expression. Mol Cell 59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Athey J, Alexaki A, Osipova E, Rostovtsev A, Santana-Quintero LV, Katneni U, Simonyan V, Kimchi-Sarfaty C. 2017. A new and updated resource for codon usage tables. BMC Bioinformatics 18:391. doi: 10.1186/s12859-017-1793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence JG, Ochman H. 1997. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol 44:383–397. doi: 10.1007/PL00006158. [DOI] [PubMed] [Google Scholar]
- 75.Blanco E, Abril JF. 2009. Computational gene annotation in new genome assemblies using GeneID. Methods Mol Biol 537:243–261. doi: 10.1007/978-1-59745-251-9_12. [DOI] [PubMed] [Google Scholar]
- 76.Jani M, Sengupta S, Hu K, Azad RK. 2017. Deciphering pathogenicity and antibiotic resistance islands in methicillin-resistant Staphylococcus aureus genomes. Open Biol 7:170094. doi: 10.1098/rsob.170094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu QH, Guo ZG, Ren JH. 2012. Phylogenetic application and analysis of horizontal transfer based on the prokaryote eno gene. Yi Chuan 34:907–918. (In Chinese.) doi: 10.3724/SP.J.1005.2012.00907. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen M, Ekstrom A, Li X, Yin Y. 2015. HGT-Finder: a new tool for horizontal gene transfer finding and application to Aspergillus genomes. Toxins (Basel) 7:4035–4053. doi: 10.3390/toxins7104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Podell S, Gaasterland T. 2007. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol 8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schonknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Brautigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilkerson C, Rensing SA, Gagneul D, Dickenson NE, Oesterhelt C, Lercher MJ, Weber AP. 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 81.Tuller T, Girshovich Y, Sella Y, Kreimer A, Freilich S, Kupiec M, Gophna U, Ruppin E. 2011. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res 39:4743–4755. doi: 10.1093/nar/gkr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.King JL, Jukes TH. 1969. Non-Darwinian evolution. Science 164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 83.Miller SL. 1974. The atmosphere of the primitive earth and the prebiotic synthesis of amino acids. Orig Life Evol Biosph 5:139–151. doi: 10.1007/BF00927019. [DOI] [PubMed] [Google Scholar]
- 84.Friedmann N, Miller SL. 1969. Phenylalanine and tyrosine synthesis under primitive earth conditions. Science 166:766–767. doi: 10.1126/science.166.3906.766. [DOI] [PubMed] [Google Scholar]
- 85.Higgs PG, Pudritz RE. 2009. A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 9:483–490. doi: 10.1089/ast.2008.0280. [DOI] [PubMed] [Google Scholar]
- 86.Miller SL. 1986. Current status of the prebiotic synthesis of small molecules. Chem Scr 26B:5–11. [PubMed] [Google Scholar]
- 87.Wu M, Higgs PG. 2009. Origin of self-replicating biopolymers: autocatalytic feedback can jump-start the RNA world. J Mol Evol 69:541–554. doi: 10.1007/s00239-009-9276-8. [DOI] [PubMed] [Google Scholar]
- 88.Miller SL, Schlesinger G. 1983. The atmosphere of the primitive earth and the prebiotic synthesis of organic compounds. Adv Space Res 3:47–53. doi: 10.1016/0273-1177(83)90040-6. [DOI] [PubMed] [Google Scholar]
- 89.Zimmerman JM, Eliezer N, Simha R. 1968. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol 21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 90.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 91.Miller S, Janin J, Lesk AM, Chothia C. 1987. Interior and surface of monomeric proteins. J Mol Biol 196:641–656. [DOI] [PubMed] [Google Scholar]
- 92.Neurath H, Hill RL (ed). 1979. The proteins, 3rd ed Academic Press, New York, NY. [Google Scholar]