Abstract
The retinoblastoma protein (Rb) family members are essential regulators of cell cycle progression, principally through regulation of the E2f transcription factors. Growing evidence indicates that abnormal cell cycle signals can participate in neuronal death. In this regard, the role of Rb (p105) itself has been controversial. Germline Rb deletion leads to massive neuronal loss, but initial reports argue that death is non-cell autonomous. To more definitively resolve this question, we generated acute murine knock-out models of Rb in terminally differentiated neurons in vitro and in vivo. Surprisingly, we report that acute inactivation of Rb in postmitotic neurons results in ectopic cell cycle protein expression and neuronal loss without concurrent induction of classical E2f-mediated apoptotic genes, such as Apaf1 or Puma. These results suggest that terminally differentiated neurons require Rb for continuous cell cycle repression and survival.
Introduction
The re-expression of cell cycle proteins in neurons, or cell cycle re-entry, has been observed in neurodegenerative conditions and injury such as stroke (Greene et al., 2007; Herrup and Yang, 2007; Rashidian et al., 2007). Recent experimental models implicate essential cell cycle regulators as critical upstream events in disease progression (Park et al., 1997b; Rashidian et al., 2005). For example, cyclin-dependent kinases (CDKs), key proteins involved in cell cycle progression, have been shown to regulate upstream events during neuronal loss in both in vitro and in vivo systems (Park et al., 1997b; Rashidian et al., 2005). In these cases, the pathogenic mechanism relating to reactivation of the cell cycle machinery has yet to be fully determined. The paradoxical case of cell cycle reactivation in a seemingly terminally quiescent neuronal population suggests the need for constant repressive action on cell cycle components.
The retinoblastoma protein (Rb), the first tumor suppressor identified, is best characterized for its role in regulating cell proliferation by the repression of E2f transcription factors (Burkhart and Sage, 2008). Upon proliferation-inducing stimulus, Rb is phosphorylated by CDK–cyclin complexes that disassociate the Rb/E2f complex allowing for the transcription of genes that initiate S-phase of the cell cycle (Burkhart and Sage, 2008). Rb has also been shown to regulate apoptosis by both direct and indirect mechanisms (Polager and Ginsberg, 2009). Rb targets include genes regulating cell cycle progression (CyclinE1, CyclinA2) (Burkhart and Sage, 2008), apoptosis (Apaf1, Puma, SIVA) (Polager and Ginsberg, 2009), and nonclassical functions (Neogenin, Bnip3) (Tracy et al., 2007; Andrusiak et al., 2011). Previous studies examining the role of cell cycle regulators in neuronal cell death observed that CDK inhibition, as well as Rb overexpression, are protective against apoptotic stimuli (Park et al., 1997a, 2000). Additionally, Rb is phosphorylated in a number of neuronal injury and degeneration models (Park et al., 2000; Biswas et al., 2007; Yu et al., 2012). These studies initially suggested that CDK-mediated inactivation of Rb may be critical in mediating neuronal loss; however, several pieces of evidence questioned this interpretation. First, while Rb germline knock-out mice showed massive neuronal loss, this was due to placental defects rather than neuron-specific pathways (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992; de Bruin et al., 2003). Consistent with this, conditional Rb-deficient neural precursors divided ectopically, but were able to survive and differentiate into neurons (Ferguson et al., 2002; MacPherson et al., 2003). Whether this was due to compensation by the Rb homologs p107 and p130 is unknown. Second, previous studies implicated p130 and not Rb as the main mediator of neuronal death particularly in models of NGF withdrawal (Liu et al., 2005). Together, it poses the important question of whether Rb itself plays any significant role in neuronal cell cycle-mediated death.
Here, we sought to define the specific role for Rb in terminally differentiated postmitotic neurons. We show that Rb is essential to maintain survival of postmitotic neurons throughout adulthood. Rb loss induces expression of cell cycle regulatory genes; however, death following Rb loss appears to occur independently of transcriptional upregulation of apoptotic genes. This evidence demonstrates a crucial role for Rb in maintaining quiescence and survival of postmitotic neurons.
Materials and Methods
Animals.
All experiments were approved by the Animal Care Ethics Committee of the University of Ottawa and adhered to the Guidelines of the Canadian Council on Animal Care. Rbflox (Marino et al., 2000) and CamKCreERT2 (EMMA ID: 02125) (Erdmann et al., 2007) mice were maintained on FVBN and C57BL/6 backgrounds, respectively. Animals were genotyped according to standard protocols with previously published primers. All CamKCreERT2 animals used were heterozygotes for Cre expression. CamKCreERT2;Rbflox/+ mice were crossed with Rbflox/flox mice to generate experimental animals, which at 5–6 weeks of age were given tamoxifen (TAM; Sigma) (180 mg/kg/d, i.p., for 5 d) and killed 1 or 4 weeks after the final injection. In all experiments, both female and male animals were used.
Primary cortical neurons.
Embryonic cortical neurons were isolated by standard procedures (Fortin et al., 2001). Neurons were infected at the time of plating with a pWPXLD lentiviral vector expressing control GFP, GFP-tagged Cre recombinase, or dominant-negative DP1Δ103–126 at a multiplicity of infection of 2. For immunofluorescence, cells were grown on coverslips and treated as indicated in the figure legends. Cells were then fixed with 4% PFA, and stained for Tuj1 (Covance), Ki67 (Cell Marque), γ-H2AX (Millipore), and DAPI. Alexa Fluor 488/594 secondary antibodies were used (Invitrogen). Statistical differences were determined using a one-way ANOVA, where p < 0.05 was considered statistically significant.
Western blots.
Protein was isolated from cultured cortical neurons or cortical tissue, and Western blot analyses were performed as previously described (Ferguson et al., 2002) with antibodies directed toward activated caspase-3 (Cell Signaling Technology), Rb (PharMingen), CyclinA2 (Abcam), γ-H2AX (Millipore), and β-actin (Sigma).
Real-time PCR.
Quantitative real-time PCR (qRT-PCR) assays were performed on primary cortical neurons using a Rotor-Gene RG-3000 (Corbett Research). Total RNA was isolated using Trizol method (Invitrogen) at 6 d in vitro (DIV). The SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) was used to amplify indicated target genes. All expression values were normalized to GAPDH. Primer sequences are available upon request. Expression values were obtained from three independent cultures, and significance was determined by a two-tailed Student's t test, where p < 0.05 was considered statistically significant.
Microarray.
Rbflox/flox neurons were treated with GFP or Cre-GFP lentivirus and harvested after 6 DIV from three independent dissections. Total RNA was purified using Trizol method (Invitrogen). Samples were hybridized to Mouse Gene 1.0ST array (Affymetrix) at the Ottawa Hospital Research Institute Stemcore facility (Ontario, Canada). Data were normalized using the Robust Multichip Average expression measure (Irizarry et al., 2003), and statistically significant gene changes were determined using significance analysis of microrarrays (Tusher et al., 2001) in the TM4 MultiExperiment Viewer software package (Saeed et al., 2003). Significant changes in gene expression were computed using a fold increase of ≥1.5 and a false discovery rate (FDR) of <5%. Gene functional classification was performed using DAVID Bioinformatics (http://david.abcc.ncifcrf.gov/).
Tissue processing, immunohistochemistry, and cell quantification.
Brains were perfused and fixed as previously described (Fortin et al., 2001). Sections were collected as 14 μm coronal cryosections on slides. For immunohistochemistry, sections underwent antigen retrieval in Target Retrieval Solution (Dako) and were incubated overnight at 4°C with the following primary antibodies: NeuN (Millipore), Ki67 (Cell Marque), and γ-H2AX (Millipore). Sections were incubated in blocking solution containing donkey anti-rabbit Alexa Fluor488 (Invitrogen) or donkey anti-mouse Cy3 (Jackson Immunoresearch) and DAPI. All images were acquired using a Zeiss 510 metaconfocal microscope under a 40× objective. For cell quantification, a minimum of three sections containing the frontal cortex was analyzed per brain. For each section, two fields in layer 2–4 and two fields in layer 5–6 were picked in the primary motor cortex, and the percentage of NeuN+ cells among the total DAPI+ cells, Ki67+ cells among NeuN+ cells or γ-H2AX+ cells among Ki67+ cells were quantified. Statistical analysis was determined by a two-tailed Student's t test, where p < 0.05 was considered statistically significant.
Results
Rb loss triggers apoptosis in primary cortical neurons
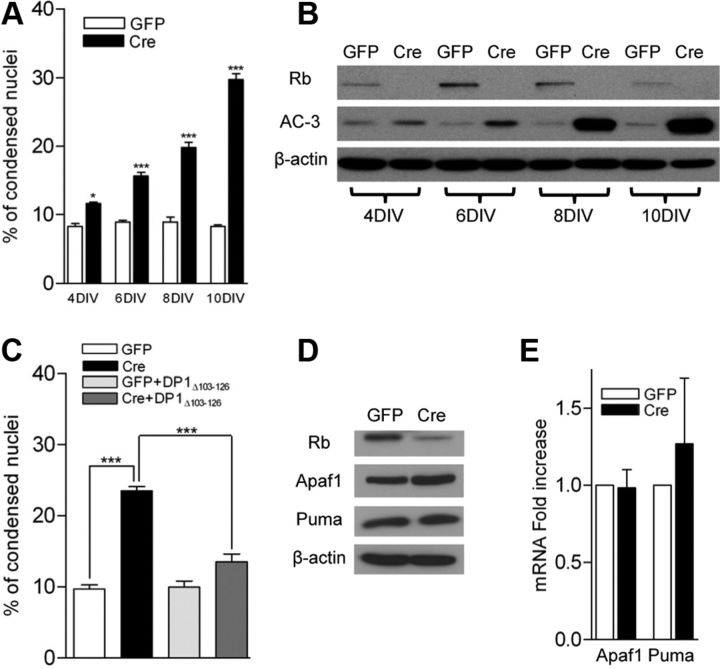
Previous evidence, as stated above, suggested that Rb does not regulate cell death in immature neural precursors. We were interested to see whether Rb may play a role in fully differentiated neurons. To accomplish this, we first established an acute paradigm in which we treated primary cortical neurons from animals harboring a conditional Rb allele (Rbflox/flox) with lentiviruses expressing control-GFP (GFP) or Cre-GFP (Cre). Lentivirus carrying Cre was tested and had no toxic effects in wild-type neurons (data not shown). This system ablates Rb in cells infected with Cre virus (Fig. 1B). We examined neurons for induction of apoptosis by both microscopic assessment of condensed nuclei and Western blot analysis for activated caspase-3 (Fig. 1A,B). Rb loss was observed as early as 4 d after viral infection of Cre. Analysis of condensed nuclei by DAPI staining showed very little effect in Cre-infected neurons at 4 DIV; however, robust death was observed by 10 DIV (Fig. 1A). Activation of caspase-3 by Western blot analysis revealed an identical trend with caspase-3 activation peaking slightly earlier at 8 DIV (Fig. 1B). This evidence supports the notion that Rb plays an essential role in the survival of postmitotic neurons. To address whether the neuronal apoptosis we observe is E2f dependent, we broadly inhibited E2f activity using a dominant-negative DP1Δ103–126 construct (Wu et al., 1996). The inhibition of E2f activity was able to rescue the appearance of condensed nuclei upon Rb deletion, and had no deleterious effect on control neurons (Fig. 1C).
Figure 1.
Acute Rb removal results in neuronal apoptosis independent of classical E2f-regulated apoptotic genes. A, Cortical neurons were fixed at the indicated DIV and condensed nuclei were examined by DAPI staining (n = 3). B, Western blot analysis of total protein extracted from control and Rb-deficient cortical neurons at the indicated DIV. C, Cortical neurons were infected with the indicated constructs, and condensed nuclei were examined by DAPI staining at 8 DIV (n = 3). D, Western blots on total protein extracted at 6 DIV. E, qRT-PCR on RNA extracted at 6 DIV. RNA levels were normalized to GAPDH (n = 3). *p < 0.05,***p < 0.001. Error bars indicate SEM.
Rb loss does not induce classical E2f-regulated apoptotic genes
The Rb/E2f axis has been implicated in the direct transcriptional regulation of apoptotic machinery in a number of models (Polager and Ginsberg, 2009). The most well characterized gene targets consist of Apaf1 and the BH3-only protein Puma (Guo et al., 2001; Hershko and Ginsberg, 2004). We therefore asked the question of whether loss of Rb de-represses these apoptotic genes, thereby triggering a cell death cascade. We examined levels of the E2f-regulated apoptotic genes Apaf1 and Puma by Western blot and qRT-PCR (Fig. 1D,E). We did not observe any significant upregulation of either gene at the level of protein or transcript, suggesting an alternative pathway of death.
Acute removal of Rb results in upregulation of cell cycle machinery
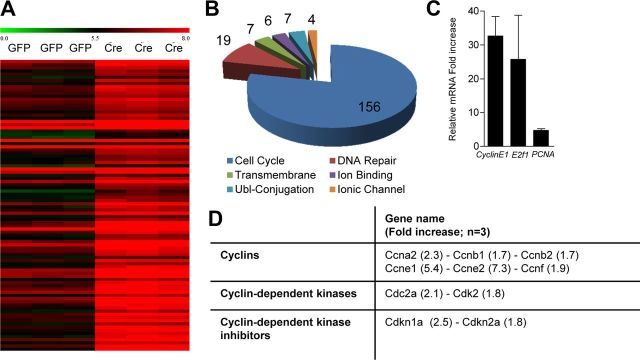
We next sought to identify the specific genes de-repressed upon loss of Rb in our primary cortical neurons. We used microarray to examine broad gene expression changes in response to Rb deletion. We examined neurons at 6 DIV as they do not yet display extensive apoptosis, which may result in indirect transcript dysregulation. Genes were deemed significant by significance analysis of microarray increase of ≥1.5-fold and a FDR of ≤5% (n = 3). We identified 377 increased transcripts upon Rb loss (Fig. 2A). Using DAVID Bioinformatics (http://david.abcc.ncifcrf.gov/), we used a functional classification of genes (Huang da W, et al., 2009) (Fig. 2B). Gene functional classification uses a modular enrichment analysis to assess term-to-term relationships to minimize redundancy in GO classification terms. Of the 377 genes identified as upregulated in our Rb-deficient neurons, 368 possessed DAVID IDs. Gene functional classification identified 14 gene clusters representing 216 genes. These clusters were manually reduced to Cell Cycle (156), DNA Repair (19), Transmembrane (7), Ion Binding (6), Ubl-conjugation (7), and Ionic Channel (4) (Fig. 2B). This over-representation of cell cycle genes suggests a restricted role for Rb in postmitotic neurons in the maintenance of the quiescent state. Several genes including PCNA and CyclinE1 were chosen for validation by qRT-PCR with upregulation on the array confirmed (Fig. 2C). Upregulated cell cycle genes were not confined to the G1/S transition, as factors associated with various facets of the cell cycle were induced (Fig. 2D). Our microarray data suggest that Rb plays a role in the regulation of specific gene classes that does not include classical E2f-regulated apoptotic genes in terminally differentiated neurons.
Figure 2.
Loss of Rb upregulates genes associated with cell cycle function. A, Heat map representing top 200 genes significantly upregulated by microarray upon acute Rb deletion (fold increase of ≥1.5, FDR <5%, n = 3). B, Gene functional classification of upregulated transcripts following Rb loss. C, qRT-PCR validation of targets identified by microarray analysis. D, Analysis of distribution of cell cycle regulatory genes induced upon Rb deletion. Error bars indicate SEM.
Acute removal of Rb triggers ectopic cell cycle protein expression and DNA damage
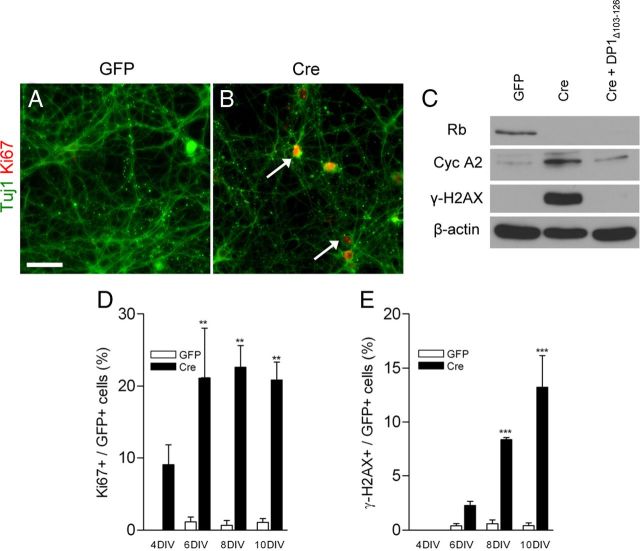
Our microarray analysis revealed a very specific cohort of genes related to cell cycle dynamics upregulated in the absence of Rb. We then asked whether upregulation of these genes would manifest itself as ectopic expression of cell cycle proteins. Rbflox/flox neurons infected with control and Cre lentivirus were fixed and stained for Tuj1 (neuronal marker) and Ki67 (proliferative marker). We observed Tuj1/Ki67+ very prominently in Rb-deficient neurons, which were rarely visible in control neurons (Fig. 3A,B). Neurons also exhibited an increase in protein levels of the known cell cycle regulatory protein CyclinA2 (Fig. 3C). Finally, we examined neurons for increased expression of γ-H2AX. γ-H2AX is the phosphorylated form of H2AX; this phosphorylation event is dependent upon induction of double strand breaks (DSBs) (Rogakou et al., 1998). We observed a significant increase in this mark in Rb-deficient neurons (Fig. 3C), suggesting that Rb loss triggers ectopic induction of cell cycle machinery and a DNA damage response. The induction of γ-H2AX and CyclinA2 is E2f dependent as introduction of DP1Δ103–126 was able to rescue to control levels (Fig. 3C). To examine the temporal relationship between cell cycle re-entry and DNA damage, we immunostained for Ki67 and γ-H2AX at 4, 6, 8, and 10 DIV. We observed an increase in the number of Ki67+ cells upon Rb deletion as early as 4 DIV (Fig. 3D). The appearance of γ-H2AX was not apparent until 6 DIV, and increased at 8 and 10 DIV (Fig. 3E). These data suggest that double strand breaks occur downstream of cell cycle re-entry and that both of these processes are mediated by E2f activation.
Figure 3.
Rb-deficient neurons display cell cycle and DNA damage markers. A, B, Cortical neurons were fixed at 8 DIV and stained for Ki67. C, Western blots were performed on total protein extracted at 8 DIV. D, E, Cortical neurons were fixed at the indicated DIV and quantified for the percentage of double labeling (Ki67+/GFP+ over GFP+, γ-H2AX/GFP+ over GFP+) (n = 3 or 4). **p < 0.01,***p < 0.001. Scale bar, 50 μm. Error bars indicate SEM.
Acute in vivo loss of Rb results in neurodegeneration
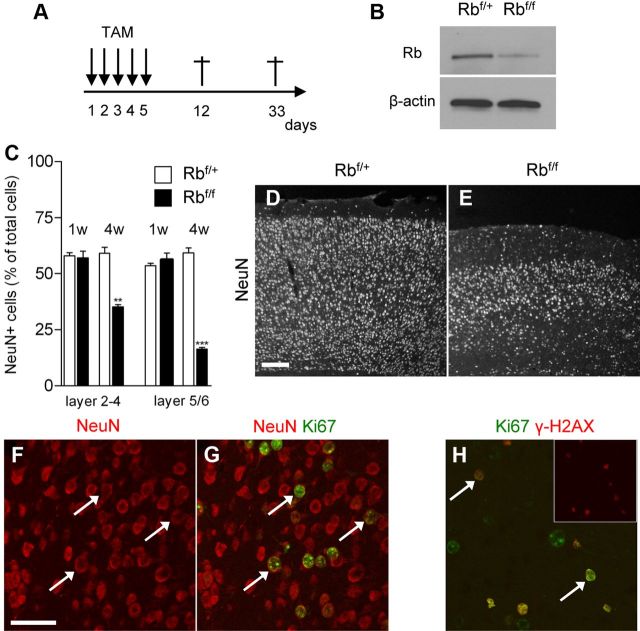
We finally asked whether conditional Rb loss would result in neurodegeneration of adult neurons in vivo. To this end, we used a TAM-inducible CamKCreERT2 mouse model to acutely delete Rb in adult forebrain neurons. Adult CamKCreERT2; Rbflox/+ (Ctrl) and CamKCreERT2; Rbflox/flox mice (5–6 weeks of age) were injected with TAM and killed 1 or 4 weeks after the last injection (Fig. 4A). Recombination efficiency was validated by Western blot (Fig. 4B). At 1 week postinjection, we did not detect any significant neuronal loss in the cerebral cortex of CamKCreERT2; Rbflox/flox animals (Fig. 4C). However, neuronal density was dramatically reduced in the cortex of knock-out animals 4 weeks after TAM induction (Fig. 4C–E). To determine whether Rb-deficient neurons ectopically express cell cycle proteins in vivo, we colabeled the neuronal marker NeuN with Ki67 at 1 week postinjection, before neuronal loss. We did not observe NeuN/Ki67+ neurons in Ctrl animals; however, 22.5 ± 16.2% and 16.9 ± 3.9% (n = 3) of NeuN+ neurons expressed Ki67 in the superficial (layers 2–4) and deep (layers 5–6) layers of Rb-deficient animals, respectively (Fig. 4F,G). Finally, a significant proportion of ectopic Ki67+ cells in the cortex of Rb-deficient animals also exhibit γ-H2AX immunoreactivity (layer 2–4: 81.2 ± 7.3%; layer 5–6: 54.9 ± 5.1%; n = 3), indicating that Rb loss induces cell cycle proteins and DSB, which may contribute to neuronal loss in vivo (Fig. 4H).
Figure 4.
Acute Rb removal in adult neurons triggers neuronal loss. A, TAM injection paradigm. B, Western blot of cortex lysates confirming the decrease of Rb levels in CamKCreERT2; Rbflox/floxmice (Rbf/f) 1 week after the last TAM injection. C, Rb loss results in neuronal loss in the cerebral cortex 4 weeks following TAM treatment (n = 3). D, E, Representative pictures of NeuN immunofluorescence displaying loss of neurons in the cortex of CamKCreERT2; Rbflox/floxmice (E) compared with CamKCreERT2; Rbflox/+mice (D). F, G, Ectopic expression of the proliferative marker Ki67 in NeuN+ neurons (arrows) of CamKCreERT2; Rbflox/flox mice 1 week after the last TAM injection. H, Ectopically proliferating Ki67+ cells exhibit γ-H2AX immunoreactivity (arrows) following Rb deficiency. **p < 0.01,***p < 0.001. Scale bars: D, E, 100 μm; F–H, 50 μm. Error bars indicate SEM.
Discussion
Our studies demonstrate several significant conclusions regarding neuronal survival. We have shown that acute loss of the cell cycle regulatory gene Rb in postmitotic neurons results in apoptosis. Critically, we show by microarray and candidate gene approaches that this neuronal loss is not due to de-repression of classical Rb/E2f-regulated apoptotic machinery. Instead, we observe the induction of mainly cell cycle- and DNA repair-related proteins. Finally, we provide in vivo evidence that acute Rb deletion is sufficient to induce neuronal loss. Together, these results highlight a specific role for Rb in the repression of cell cycle genes and survival in postmitotic neurons.
Importantly, our results highlight a differential dependence on Rb in proliferating neural precursors and terminally differentiated neurons. Germline Rb loss models displayed nonautonomous neuronal apoptosis, and in vitro differentiation of germline Rb-deficient precursors showcased the dispensable role for Rb at this stage of development (Callaghan et al., 1999). In more defined precursor-specific conditional deletion models, neuronal differentiation also proceeded in a largely normal fashion (Ferguson et al., 2002; MacPherson et al., 2003). In contrast, our present observations indicate that in terminally differentiated neurons, Rb is essential to maintain survival.
Our findings serve to further expand the role of the Rb family in the regulation of neuronal survival. In both steady-state and stress-induced paradigms, the Rb-related protein p130 was shown to be essential in the regulation of neuronal survival (Liu et al., 2005). Our data implicate Rb in a similar manner. It is important to note the latencies to cell death observed with either p130 or Rb loss. Upon siRNA-mediated p130 depletion, apoptosis was observed within 48 h (Liu et al., 2005), whereas, in our system Rb deletion results in similar levels of cell death after 120 h. This suggests that while both Rb and p130 may be essential in the maintenance of neuronal survival, the mechanisms used by each may differ. In neurons, p130 was shown to mediate gene repression via HDAC1-mediated interaction with Suv39h1-p130-E2f4 (Liu et al., 2005). The direct mechanisms that Rb employs to repress gene expression in neurons have yet to be fully described.
We and others have previously demonstrated a tight link between neuronal cell cycle re-entry and cell death. Importantly, our data emphasize that high E2f activity and induction of E2f target genes in the absence of Rb is deleterious for postmitotic neurons. However, the direct link between cell cycle re-entry and cell death remains correlative at this point, and future experiments will be required to determine exactly how cell cycle re-entry and DNA damage repair intersect to result in cell death in a postmitotic neuron. To our surprise, induction of cell death upon Rb deletion occurs independently of E2f-dependent transcriptional upregulation of classical Rb/E2f-mediated apoptotic factors such as Apaf1 and Puma. In this regard, comparison to other neuronal death paradigms involving abnormal cell cycle activation is informative. DNA damage-induced CDK activation, Rb phosphorylation, and subsequent cell loss occurs within hours of insult (Park et al., 2000). However, in this paradigm, independent activations of both CDK/Rb/E2f and p53 signals occur and work in concert to promote apoptosis (Morris et al., 2001). In contrast, our present data indicate that Rb deficiency alone results in a significantly protracted timing of death compared with DNA damage. This suggests that the efficiency and speed of death is regulated by the presence of multiple signals that act together to regulate apoptosis. In this regard, classical apoptotic targets like Apaf1 and Puma are known to be regulated by both E2fs and p53. Finally, it is interesting to note that while Rb loss itself induces markers of DNA damage and induction of DNA repair enzymes, the classical p53 response does not appear to be initially activated.
In conclusion, our results reveal an essential role for Rb in the maintenance of neuronal survival. Rb is able to actively repress genes that drive the cell cycle to prevent neurons from entering a proliferative state. Unlike other tissue systems, Rb loss does not trigger concurrent induction of classical E2f-regulated apoptotic targets. These findings reveal the need for constitutive Rb-mediated repression to prevent ectopic expression of cell cycle proteins and maintain survival in a terminally differentiated postmitotic neuron. It also highlights the differential role Rb plays in precursors compared with terminally differentiated neurons.
Footnotes
This work was supported by Canadian Institutes of Health Research grants to R.S.S. and D.S.P. M.G.A. is supported by awards from Ontario Graduate Scholarship and Heart and Stroke Foundation of Ontario. R.V. is supported by fellowships from the Alzheimer Society of Canada and the Vision 2010 strategic plan of the University of Ottawa, and by a travel award from the Léon Fredericq Funds (University of Liège, Liège, Belgium). We thank Jason G. MacLaurin and Linda Jui for excellent technical assistance. We thank Lisa Julian and Jacob Wong for critical review of the manuscript. We thank G. Schütz (Deutsches Krebsforschungszentrum, Heidelberg, Germany) for providing CamKCreERT2 animals.
References
- Andrusiak MG, McClellan KA, Dugal-Tessier D, Julian LM, Rodrigues SP, Park DS, Kennedy TE, Slack RS. Rb/E2F regulates expression of neogenin during neuronal migration. Mol Cell Biol. 2011;31:238–247. doi: 10.1128/MCB.00378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SC, Shi Y, Vonsattel JP, Leung CL, Troy CM, Greene LA. Bim is elevated in Alzheimer's disease neurons and is required for β-amyloid-induced neuronal apoptosis. J Neurosci. 2007;27:893–900. doi: 10.1523/JNEUROSCI.3524-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan DA, Dong L, Callaghan SM, Hou YX, Dagnino L, Slack RS. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev Biol. 1999;207:257–270. doi: 10.1006/dbio.1998.9162. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann G, Schütz G, Berger S. Inducible gene inactivation in neurons of the adult mouse forebrain. BMC Neurosci. 2007;8:63. doi: 10.1186/1471-2202-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KL, Vanderluit JL, Hébert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, Hakim A, Albert PR, Cecconi F, Helin K, Park DS, Slack RS. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol. 2001;155:207–216. doi: 10.1083/jcb.200105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Liu DX, Troy CM, Biswas SC. Cell cycle molecules define a pathway required for neuron death in development and disease. Biochim Biophys Acta. 2007;1772:392–401. doi: 10.1016/j.bbadis.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Yikang S, Yoshida H, Mak TW, Zacksenhaus E. Inactivation of the retinoblastoma tumor suppressor induces apoptosis protease-activating factor-1 dependent and independent apoptotic pathways during embryogenesis. Cancer Res. 2001;61:8395–8400. [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–8634. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19:719–732. doi: 10.1101/gad.1296405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Keramaris E, Rideout HJ, Slack RS, Dyson NJ, Stefanis L, Park DS. Cyclin-dependent kinases and P53 pathways are activated independently and mediate Bax activation in neurons after DNA damage. J Neurosci. 2001;21:5017–5026. doi: 10.1523/JNEUROSCI.21-14-05017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Greene LA, Geller HM. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J Neurosci. 1997a;17:1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Levine B, Ferrari G, Greene LA. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. J Neurosci. 1997b;17:8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Bremner R, Keramaris E, Padmanabhan J, Rosenbaum M, Shelanski ML, Geller HM, Greene LA. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons evoked by DNA damage. J Neurosci. 2000;20:3104–3114. doi: 10.1523/JNEUROSCI.20-09-03104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Rashidian J, Iyirhiaro G, Aleyasin H, Rios M, Vincent I, Callaghan S, Bland RJ, Slack RS, During MJ, Park DS. Multiple cyclin-dependent kinases signals are critical mediators of ischemia/hypoxic neuronal death in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:14080–14085. doi: 10.1073/pnas.0500099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidian J, Iyirhiaro GO, Park DS. Cell cycle machinery and stroke. Biochim Biophys Acta. 2007;1772:484–493. doi: 10.1016/j.bbadis.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borsovsky I, Liu Z, Vinsavich A, Trush V, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ren QG, Zhang ZH, Zhou K, Yu ZY, Luo X, Wang W. Phospho-Rb mediating cell cycle reentry induces early apoptosis following oxygen-glucose deprivation in rat cortical neurons. Neurochem Res. 2012;37:503–511. doi: 10.1007/s11064-011-0636-6. [DOI] [PubMed] [Google Scholar]