Abstract
Mutations in VAPB have been identified in a familial form of amyotrophic lateral sclerosis (ALS), and reduced VAPB levels have been found in patients with sporadic ALS. Vap protein family members from different species and cell types have been implicated in a number of cellular functions, but how Vap dysfunction in neurons and/or muscles contributes to motor neuron degeneration and death is poorly understood. Using Drosophila as a model organism, we show that Vap physically interacts with and affects the axonal functions of the Down syndrome cell adhesion molecule (Dscam). Dscam is a cell-surface receptor involved in axon and dendritic patterning and neuron self-recognition and avoidance. Alternative splicing of the Dscam transcript leads to the production of Dscam isoforms that contain one of two possible transmembrane (TM) domain and flanking sequences that either restrict the isoform to dendrites and cell bodies (TM1) or target the isoform to axon processes (TM2). We find that Vap specifically interacts with Dscam isoforms that contain the TM2 cytoplasmic juxtamembrane flanking sequences. Using loss-of-function genetics, we further show that Vap is required for localization of Dscam isoforms containing TM2 to axons and that Vap loss suppresses Dscam gain-of-function axon phenotypes. We propose that Vap function is required in neurons to selectively traffic proteins to axons, and disruption of this function may contribute to the pathology of ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that affects motor neurons. While most cases of ALS have an unknown etiology, ∼10% have a hereditary component and mutations in several genes have been implicated in these familial forms of ALS (FALS) (Pasinelli and Brown, 2006). Understanding how perturbations of FALS genes contribute to neuron dysfunction may lead to insights for treatment of sporadic ALS. A point mutation (P56S) in ALS8 is responsible for FALS cases that were first discovered in a Brazilian family (Nishimura et al., 2004). ALS8 encodes VAPB, an evolutionarily conserved type II membrane protein that is associated with the endoplasmic reticulum and Golgi network (Soussan et al., 1999; Skehel et al., 2000; Kaiser et al., 2005). Overexpression of VAPB carrying the P56S point mutation (VAPBP56S) in model organisms suggests that VAPBP56S functions as a dominant-negative by causing the formation of cytoplasmic inclusions containing ubiquitinated forms of both VAPBP56S and wild-type VAPB, which lowers the cellular levels of functional nonaggregated VAPB (Teuling et al., 2007; Tsuda et al., 2008). However, motor neurons differentiated from induced pluripotent stem cells generated from ALS8 patient fibroblasts exhibited reduced levels of VAPB protein in the absence of inclusions, suggesting that reduction of VAPB levels is important for ALS8 pathogenesis (Mitne-Neto et al., 2011). Indeed, VAPB mRNA levels are reduced in spinal cords of sporadic ALS patients (Anagnostou et al., 2010).
VAPB has been implicated in a number of cell-autonomous functions, including lipid trafficking (Loewen et al., 2003), regulation of size and number of neuromuscular junction boutons (Pennetta et al., 2002), and mitochondria transport (Mórotz et al., 2012). Recent studies in flies and worms have uncovered a cell nonautonomous role for VAPB proteins (Tsuda et al., 2008; Han et al., 2012). The cytoplasmic portion of VAPB proteins contains a major sperm protein (MSP) domain that can be released from the membrane-spanning segment by proteolysis and secreted by neurons. Secreted MSP domains interact with a variety of axon guidance receptors including EPH receptors, Robo receptors, and Lar-like protein-tyrosine phosphatases to stabilize mitochondria networks at postsynaptic neuromuscular junctions (Tsuda et al., 2008; Han et al., 2012). Cleaved VAPB is also present in human serum (Tsuda et al., 2008), suggesting that human VAPB also has a cellular nonautonomous function mediated thorough axon guidance receptors.
Here we report that the Drosophila Down syndrome cell adhesion molecule (Dscam) interacts with Vap-33, the fly ortholog of vertebrate VAPB (hereafter referred to as Vap). We determine that Vap does not interact with the Dscam extracellular region, but rather specifically and directly interacts with the cytoplasmic domain of Dscam isoforms that localize to axonal processes. We further show that Vap is required for the axonal localization of Dscam isoforms, but is not required for Dscam expression or localization of Dscam isoforms to dendrites. Our findings support a model in which Vap functions to selectively traffic proteins to axonal processes.
Materials and Methods
Eukaryotic expression plasmids
Dscam1.30.30.1Juxta and Dscam1.30.30.2Juxta pUAST constructs were produced by the ligation of a tandem affinity purification (TAP) tag encoding DNA duplex sequence (TTTCCCTTGTCATCGTCATCCTTGTAGTCTCCCTTGTCATCGTCATCCTTGTAGTCATGGTGATGGTGATGGTGGCCTTGGAAGTACAGATTCTCC) into the PshAI site of Dscam1.30.30.1 and Dscam1.30.30.2 pUAST constructs, respectively. Dscam1.30.30.1C-term and Dscam1.30.30.2C-term pUAST were produced by annealing HFF-5 (CTAGTCACCATCACCATCACCATGATTACAAGGATGACGACGATAAGGATTACAAGGATGACGACGATAAGA) and HFF-3 (CTAGTCTTATCGTCGTCATCCTTGTAATCCTTATCGTCGTCATCCTTGTAATCATGGTGATGGTGATGGTGA) and ligation of the resulting duplex into the engineered XbaI site of Dscam1.30.30.1 and Dscam1.30.30.2 pUAST constructs, respectively (Wojtowicz et al., 2004). The DscamΔ2–7C-term pUAST construct was produced by releasing the sequences encoding Ig domains 2–7 by digestion with SmaI (partial) and XhoI, fill in of the XhoI overhang, and religation.
Dscam1.30.30.1C-term and Dscam1.30.30.2C-term pIZGM constructs were produced by releasing the TAP-tagged Dscam sequences from pBluescript (Stratagene) plasmids and religating them into the XbaI site of the pIZGM vector (Millard et al., 2007). Dscam1.30.30.1Juxta and Dscam1.30.30.2Juxta pIZGM constructs were produced by ligating the TAP tag duplex sequences into the PshAI site of Dscam1.30.30.1 and Dscam1.30.30.2 pIZGM constructs.
Construction of Dscam chimeras
Generation of the Dscam-rpsL-neo and Dscam-8Ig-rpsL-neo intermediate plasmids.
Dscam chimera constructs were generated using recombineering strategies and the counterselection BAC modification kit (Gene Bridges) with the modifications described below for use on high copy plasmids. For the FL Dscam ectodomain constructs, rpsL-neo counterselection cassette was PCR amplified using 17-rpsL-F (TGGCGGCTCTGCTCGTCATAATCATAGCGATAATTGTTATTTGTATACTGggcctggtgatgatggcgggatcg) and 24-rpsL-R (AAACACTAAGCTAAAATGCCACGCCCACCGCCGCAGAGGGCGCTTACAAAtcagaagaactcgtcaagaaggcg). For the truncated Dscam ectodomain construct, rpsL-neo counterselection cassette was PCR amplified using 11-rpsL-F (GCGTTCAGGCCCCTCCAGAGTTCACGGAGAAGCTGCGCAACCAGACCGCCggcctggtgatgatggcgggatcg) and 24-rpsL-R. Primers 17-rpsL-F and 24-rpsL-R contain 50 bp sequences (uppercase letters) from Dscam exon 17 and 24, respectively, and 24 bp sequences from the rpsL-neo counterselection cassette (lower case letters). Primer 11-rpsl-F contains 50 bp of sequence from Dscam exon 11. The plasmid, prpsL-neo, supplied in the counterselection BAC modification kit, was used as a template for amplifying the rpsL-neo fragment. The 1420 bp amplified fragment was purified using High Pure PCR purification kit (Roche Applied Science), mixed with pUAST-Dscam1.30.30.1, and coelectroporated into Escherichia coli DY380 cells containing the viral proteins encoded by the plasmid pSC101-BAD-gbaA-tet. Electroporation was performed at 1750 V, 200 ohms, 25 μF using an electroporation cuvette with a 1 mm gap. After electroporation, cells were allowed to recover at 30°C for 3 h in 500 μl of LB media. Two hundred microliters of this culture was plated on LB plates containing ampicillin, tetracycline, and kanamycin. Recombineered constructs, Dscam-rpsL-neo and Dscam-8Ig-rpsL-neo, were obtained from positive colonies.
Generation of the Dscam-neurexinTM-CTD chimera.
The Neurexin TM-CTD fragment was amplified using 74 bp primers 16-NrxTMCTD-F (GGCTTCACTGTGGCCGAATACGACTTTGCCACCTTAACCGTTACCGGAGGCtctgtgctgttggcatgtctgcttg) and 24-NrxTMCTD-R (AAACACTAAGCTAAAATGCCACGCCCACCGCCGCAGAGGGCGCTTACAAAttagataaagatctctgttcgcttc). These primers contained 50 bp sequences (uppercase letters) from Dscam exon 16 and 24, respectively, and 24 bp sequences (lower case letters) from the Neurexin cDNA clone RE18634 from Berkeley Drosophila Gene Collection (BDGC). This clone was used as a template for the amplification of the transmembrane (TM) and C-terminal domains (CTD) of Neurexin. The amplified fragment was purified and coelectroporated with Dscam-rpsL-neo using the conditions mentioned above and recombineered constructs were selected on plates containing ampicillin and streptomycin.
Generation of the Dscam-neuroliginTM-CTD chimera.
The Neuroligin TM-CTD fragment was amplified using 74 bp primers 16-Nl2TMCTD-F (GGCTTCACTGTGGCCGAATACGACTTTGCCACCTTAACCGTTACCGGAGGCtatacgacagccctgatagccacc) and 24-Nl2TMCTD-R (AAACACTAAGCTAAAATGCCACGCCCACCGCCGCAGAGGGCGCTTACAAActagactgagatttcctgaatatg). These primers contained 50 bp sequences (uppercase letters) from Dscam exon 16 and 24, respectively, and 24 bp sequences (lower case letters) from the Neuroligin cDNA clone RH63339 from BDGC. This clone was used as a template for the amplification of the TM and CTD of Neuroligin. Electroporation conditions, recovery, and selection were as mentioned above.
Generation of the Dscam-8Ig-neurexinTM-CTD chimera.
The Neurexin TM-CTD fragment was amplified using 74 bp primers 11-NrxTMCTD-F (GCGTTCAGGCCCCTCCAGAGTTCACGGAGAAGCTGCGCAACCAGACCGCCtctgtgctgttggcatgtctgcttg) and 24-NrxTMCTD-R. These two primers contained 50 bp sequences (uppercase letters) from Dscam exon 11 and 24, respectively, and 24 bp sequences (lower case letters) from the Neurexin cDNA clone RE18634 from BDGC. This clone was used as a template for the amplification of the TM and CTD of Neurexin. Electroporation conditions, recovery, and selection were as mentioned above.
Cell culture
Drosophila Schneider 2 (S2) cells were grown in Schneider's media (Life Technologies) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 1% pen-strep (Life Technologies). Effectene (Qiagen) was used for all plasmid transfections. For mass spectrometry (MS) experiments and Dscam TM preference studies, stably transfected cells were obtained by Zeocin selection (75 μg/ml; Life Technologies) for 4 weeks. For Dscam chimeric studies, S2 cells were transiently cotransfected with chimeric Dscam pUAST constructs and pRmHa-GAL4 in a 1:1 ratio using Effectene. After transfection, the cells were allowed to recover for 2 d before assay. For initial TM domain preference determination, stably transfected cell lines expressing tagged Dscam constructs were used. Dscam expression was induced by treating the cultures with 700 μm CuSO4 for 24 h.
Dscam expression and MS
Both control S2 cells and Dscam-expressing cells were subjected to the following expression and purification procedure. Cells were grown in a 1 L spinner flask with 100 rpm spinning. After cell density reached 1 × 106 cell/ml, the cells were induced with 0.7 mm CuSO4 for 1 d to induce Dscam expression. The cells were harvested and suspended in radioimmunoprecipitation assay (RIPA) buffer (150 mm NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0, 0.2 mm sodium orthovanadate, 10 mm NaF, 0.4 mm EDTA, and 10% glycerol). The cells were lysed by sonication and the soluble fraction was collected by sequential centrifugation for 10 min at 16,100 × g followed by 30 min at 100,000 × g in a tabletop ultracentrifuge. Anti-FLAG M2 resin (Sigma) was added to the lysate and incubated for 2 h at 4°C. The resin was washed with RIPA buffer and specifically bound proteins were eluted by competition with 100 μg/ml 3xFLAG peptide (Sigma). Anti phospho-tyrosine resin (protein A with cross-linked 4G10 antibody (Millipore)) was added to the eluent to capture phosphorylated Dscam. The resin was washed with RIPA buffer followed by 100 mm triethanolamine (TEA) buffer, pH 8.0. The final sample was eluted with 100 mm TEA, pH 11.5, and evaporated to dryness using a SpeedVac.
To prepare samples for MS, purified proteins were resuspended in 50 mm ammonium bicarbonate with 0.2% RapiGest (Waters). Samples were reduced with 5 mm tris (2-carboxyethyl) phosphine at 56°C for 30 min, alkylated with 15 mm iodoacetamide for 1 h in the dark with vortexing, and then digested with 1 μg of trypsin overnight at 37°C. Finally, 0.5 μl of concentrated HCl was added and the samples were centrifuged to remove the RapiGest and dried using a SpeedVac. Subsequently, samples were resuspended in 5% acetonitrile, 0.1% trifluoroacetic acid before liquid chromatography and electrospray ionization-MS/MS analysis.
An Agilent 1100 nano-HPLC system was used to separate peptides using Zorbax C18 trap and 75 μm × 150 mm capillary columns. Peptides were eluted with a gradient of increasing acetonitrile in 0.1% formic acid at a flow of 300 nl/min and injected into a Thermo LTQ-Orbitrap XL using a nanoelectrospray source. The Orbitrap was operated in data-dependent MS/MS mode with one MS scan followed by 3 MS/MS scans and a dynamic exclusion window of 2 min. Data were searched against a database of Drosophila melanogaster protein sequences using both Sorcerer (Sage-N Research) and Mascot (MatrixScience) search software. Similar results were obtained with both. Database searches were set with a mass tolerance of 25 ppm, full tryptic cleavage, one allowed miscleavage, and carbamidomethyl cysteine modification. Proteins identified in experimental samples with Mascot protein scores >100 that were undetected in control samples were considered specific Dscam binding partners.
Immunoprecipitation
S2 cells or S2 cells expressing exogenous Dscam were lysed in RIPA buffer by sonication. Cleared lysate was achieved from a sequential centrifugation procedure consisting of 16,100 × g for 10 min at 4°C in a micro centrifuge followed by 100,000 × g at 4°C for 30 min in a tabletop ultracentrifuge. The lysates were incubated with anti-Dscam or anti-Vap antisera (2 μl of each) at 4°C for 1 h before adding 50 μl slurry of immobilized protein A resin (G-Biosciences) for 1 h at 4°C with continuous rotation. For FLAG immunoprecipitation (IP), 50 μl of anti-FLAG M2 resin (Sigma) was added directly to the lysate and incubated at 4°C for 1 h. The samples were washed three times with RIPA buffer and eluted with SDS loading buffer.
For whole fly extracts, five male and five female 1-d-old adult Drosophila were collected, pooled, and killed by freezing. Lysates were prepared in RIPA buffer by mechanical disruption using a micro centrifuge tube pestle attached to a handheld motor followed by sonication. Cleared lysates and IP were performed as described above.
Western analysis
Samples in SDS-PAGE sample buffer were subjected to SDS-PAGE. The protein was transferred to an Immobilon-P membrane (Millipore) and blocked for 1 h with 5% nonfat dry milk. Dscam was detected using anti-Dscam(cyto) antibody (1:1000, 1 h) or anti-FLAG M2 antibody (1:1000, 1 h) (Sigma) and Vap was detected using anti-Vap (1:500, 1 h) antibodies followed with horseradish peroxidase–conjugated secondary antibodies (mouse/rabbit 1:10,000) for 1 h and detected with ECL detection reagent (Thermo Scientific). For Figure 3, the integrated density values (IDV) of the Vap and Dscam immunoreactive bands were measured using the Fluorchem 5500 image system (Alpha Innotech).
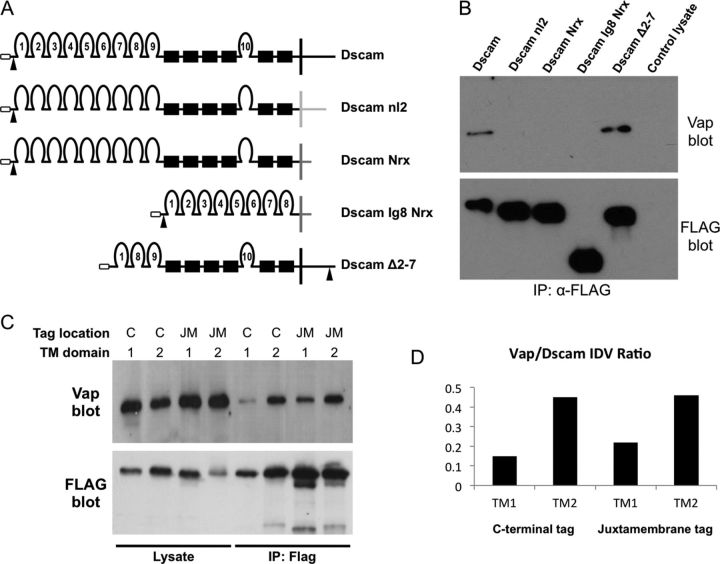
Figure 3.
Vap interacts with the Dscam cytodomain. A, Schematic representations of the Dscam chimeras used to map the Vap interaction region of Dscam. Dscam nl2 contains the Dscam ectodomain and the TM and cytodomain of nl2 (light gray). Dscam Nrx contains the Dscam ectodomain and the TM and cytodomain of Nrx (dark gray). Dscam Ig8 Nrx contains Dscam Ig domains 1 through 8 fused to the TM and cytodomain of Nrx (dark gray). Dscam Δ2–7 is a Dscam construct in which Ig domains 2–7 are deleted. Each Dscam construct includes a FLAG tag that is incorporated at the indicated location (arrowheads). B, Western analysis of FLAG IPs from lysates of S2 cells expressing the constructs in A. The control lysate was prepared from nontransfected S2 cells. All of the Dscam constructs were successfully expressed and purified as evidenced by the anti-FLAG blot (bottom); however, Vap immunoreactivity was only present in IPs of constructs containing the Dscam cytodomain. C, Western analysis of lysates and IPs of S2 cell culture-expressing FLAG-tagged Dscam isoforms. The tagged isoforms contained TM domain 1 or 2. The FLAG tag is located in either the C-terminal (C) or juxtamembrane (JM) location. The FLAG blot (bottom) indicates that each construct was successfully expressed and immunoprecipitated. The Vap blot (top) shows that approximately equal amounts of Vap are present in each lysate, but more Vap coprecipitates with Dscam isoforms that contain TM2. D, A graph of Vap to Dscam integrated density value ratios from the data obtained in A (top).
Bacterial expression of Dscam and Vap fusion proteins
The DNA sequences encoding the cytodomain of both TM1 Dscam and TM2 Dscam were PCR amplified and subcloned into the pGSTag2 vector. The PCR primers used were GATCCATATGCGTAGCAAGGGCAATCACC (TM1 forward) and GATCGAATTCTTACACTGCCATAGTATCGTAG (TM1 reverse) and GATCCATATGCGGAGGCGAGCGGACGA (TM2 forward) and GATCGAATTCTTACACTGCCATAGTATCGTAG (TM2 reverse). The amplified sequences were digested with NdeI and EcoRI and inserted into the corresponding sites of the pGSTag2 vector.
The cytodomain of Vap was PCR amplified and subcloned into the pT7-LOH vector. The primers used were GATCCATATGAGCAAATCACTCTTTGATC (forward) and GATCGAATTACTTCTCAGCCAGGACTGGG (reverse). The amplified Vap cytodomain sequence was digested with NdeI and EcoRI and inserted into the corresponding sites of the pT7-LOH vector.
In vitro Dscam binding assay
Both Dscam and Vap constructs were introduced into Novablue (DE3) cells (Novagen) and expressed. One liter of LB broth was inoculated with a single colony and incubated at 30°C until OD600 = 0.8. IPTG (0.4 mm) was added (Sigma) to induce expression of fusion proteins and the cultures were incubated at 18°C overnight. Cells were collected by centrifugation and lysed using a French press. Lysate containing 6xHisVap was incubated with Talon affinity resin (Clontech) to produce Vap-coated resin. Lysates containing the GST-Dscam TM1 or TM2 truncations were allowed to flow through the Vap coated on untreated Talon resin columns. After washing with lysis buffer, proteins bound to all columns were eluted with SDS loading buffer.
Fly lines
Standard Drosophila genetic technique was used for all crosses and transgenesis. All flies were raised at 25°C with 70% relative humidity. Fly lines used are as follows: UAS-VAP and VapΔ20/FM7,Kr-GFP (a gift from Hugo Bellen, Baylor College of Medicine, Texas); ppk-Gal4 (a gift from Wesley Grueber, Columbia University, New York); UAS-Dscam3.36.25.1Δ19-GFP and UAS-Dscam3.36.25.1Δ19-GFP (gifts from Tzumin Lee, Howard Hughes Medical Institute); 72OK-GAL4, UAS-syb-GFP, and UAS-Dscam1.30.30.2 (S.L. Zipursky, University of California Los Angeles); yw and UAS-CD8-mCherry (Bloomington Stock Center, IN). UAS-Dscam3.36.25.1Δ19-GFP was recombined with VapΔ20 and ppk-Gal4 was recombined with UAS-CD8-mCherry to aid in the crosses.
Fluorescence microscopy
For antenna lobe imaging, Drosophila brains were dissected from adult male flies expressing Dscam1.30.30.2 and synaptobrevin-GFP by the 72OK-GAL4 driver in Vap wild-type and mutant backgrounds 1–3 d after eclosion and fixed with 2% paraformaldehyde (PFA) in phosphate-buffered lysine for 1 h. They were then blocked with 10% normal goat serum in PBS and incubated with rabbit anti-GFP (1:1000, Invitrogen) and NC82 (1:100; Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biology, Iowa City, IA) for 3 h at room temperature, followed by a 1 h incubation with anti-rabbit Alexa Fluor 488 (1:1000; Invitrogen) and anti-mouse Alexa Fluor 568 (1:1000; Invitrogen). Brains were mounted in VECTASHIELD (Vector Laboratories) and imaged using an LSM 710 confocal microscope (Zeiss).
For Dscam optic lobe immunostaining, adult male flies were collected and dissected in PBS buffer, fixed with 4% PFA in PBS for 30 min. A mouse monoclonal antibody against an amino acid sequence common to all Dscam isoforms (ab 18; Shi et al., 2007) was used to detect Dscam. A secondary antibody conjugated to Cy3 (Jackson ImmunoResearch; Invitrogen) was used at 1:250. Images were captured using an LSM 510 confocal microscope (Zeiss).
For Dscam ventral nerve cord immunostaining, embryo preparation and fixation of either sex was performed as described previously (Patel, 1994). Rabbit anti-Dscam (Schmucker et al., 2000) was used at 1:1000, and detected by anti-rabbit Alexa Fluor 488 (1:1000). Embryos were mounted in VECTASHIELD and imaged using an LSM 710 confocal microscope.
For Dscam-GFP localization studies, male third instar larva was collected and dissected in PBS buffer, fixed with 4% PFA in PBS for 30 min and mounted in VECTASHIELD. Preparations were imaged immediately after mounting. Images were captured using an LSM 710 confocal microscope.
Results
Isolation of Dscam interacting proteins
In both vertebrates and invertebrates, the extracellular domains of Dscam axon guidance receptors participate in homophilic isoform-specific binding, which results in repulsion of interacting neurites for branch segregation or maintenance of neuronal spacing (Wojtowicz et al., 2004; Zhan et al., 2004; Matthews et al., 2007; Millard et al., 2007; Fuerst et al., 2008). The Dscam extracellular domain also participates in heterophilic interactions with draxin (Ahmed et al., 2011) and netrins (Andrews et al., 2008; Ly et al., 2008; Liu et al., 2009), secreted proteins that serve as guidance cues. In Drosophila, mutually exclusive cassette-like alternative splicing of the sequences encoding Ig domains 2, 3, and 7 allows the Dscam gene to produce ∼19,000 structurally related protein isoforms, each having an extracellular domain with a unique binding specificity (Schmucker et al., 2000; Wojtowicz et al., 2004, Wojtowicz et al., 2007; Sawaya et al., 2008). Mutually exclusive cassette-like alternative splicing also occurs in the sequences encoding the TM domain that results in restriction of Dscam isoforms to the cell body and dendrites if TM1 is present or localization to axons if TM2 is present (Wang et al., 2004; Shi et al., 2007). Finally, non-mutually exclusive alternative splicing within sequences encoding the cytoplasmic domain further contribute to Dscam isoform diversity (Wang et al., 2004; Yu et al., 2009).
Dscam cell culture expression constructs were generated to identify proteins that physically interact with Dscam receptor isoforms. TAP tags comprising 6xHis sequence and two copies of the FLAG sequence were incorporated in either a cytoplasmic juxtamembrane location or a location near the C terminus (Fig. 1A). The locations of these tag sites are not within any identified sequence motifs that may serve as docking sites for other proteins. A stable cell line expressing Dscam1-30-30-1 with the C-terminal affinity tag was established and grown in spinner flasks. Lysates from these cells as well as control nontransfected S2 cells were prepared and subjected to identical purification procedures.
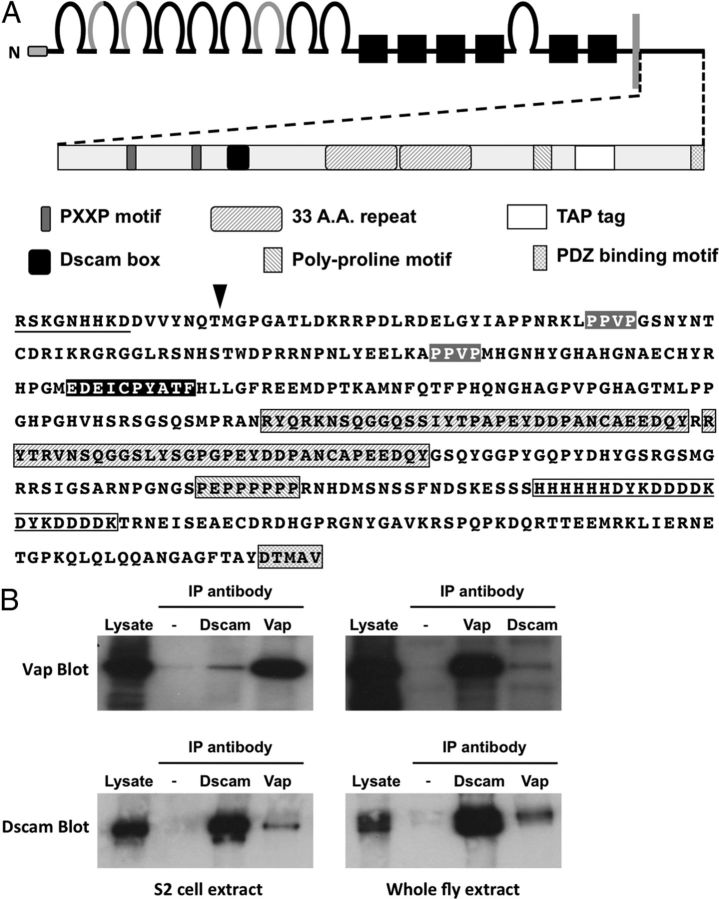
Figure 1.
Dscam and Vap form a complex in vivo. A, Top, Diagram of the overall Dscam domain structure. The extracellular domain comprises an N-terminal (N) signal peptide (open box), 10 Ig domains (loops), and 6 fibronectin type 3 domains (filled boxes). The regions of Ig domains 2, 3, and 7 that are varied by alternative splicing are gray. The cytoplasmic domain is separated from the extracellular domain by a single membrane-spanning segment (vertical gray bar), which is also varied by alternative splicing. A diagram detail of the cytoplasmic domain is shown below the overall structure. Identified sequence motifs present in the cytoplasmic domain are represented by boxes in the diagram and highlighted regions in the amino acids sequence (bottom). These motifs include SH3 domain binding PXXP motifs (gray), the Dscam box present in all Drosophila Dscam genes (black), a 33 aa repeated sequence (upward diagonal pattern), a poly-proline motif (downward diagonal pattern), and a C-terminal sequence proposed to bind PDZ domains (checked pattern). The juxtamembrane sequence unique to TM domain 1 variants is underlined. The location and composition of the C-terminal TAP tag is boxed and the location of the juxtamembrane TAP-tag insertion site is indicated (arrowhead). B, Western analysis of endogenous Vap and Dscam IP experiments from extracts prepared from S2 cell culture (left) or whole flies (right). Vap immunoreactivity is detected at levels above control precipitates that lack primary antibody (−) in both Dscam and Vap IPs (top), while Dscam immunoreactivity is similarly detected in both Dscam and Vap precipitates (bottom).
Affinity-tagged Dscam was purified by the sequential binding to and elution from anti-FLAG and anti-phosphotyrosine resins (see Materials and Methods). The 6×His tag was ultimately not used in these purifications due to poor purification yields. anti-phosphotyrosine antibodies were used instead because Dscam is known to be heavily tyrosine phosphorylated. Proteins copurifying with Dscam were identified by tandem MS after cleavage with trypsin. A large fraction of identified peptides in the experimental samples were derived from Dscam itself, which is likely due to the large size of the Dscam protein (>2000 aa) and the level of expression. Three additional proteins were identified that had a highly significant Mascot score > 100 and were not present in the control purification (Table 1). Dreadlocks is an SH2/SH3 adapter protein that has been previously shown to interact with Dscam (Schmucker et al., 2000). Vap and Ubiquitin-63E have not been previously described as Dscam interacting proteins.
Table 1.
Dscam interacting proteins identified by MS
| Protein name | Unique identified peptides | Mascot scorea | HGNC ortholog symbol | Molecular function |
|---|---|---|---|---|
| Dreadlocks | 7 | 345 | NCK | Adapter protein |
| Vap-33 | 3 | 113 | VAPB | Neuron apoptosis |
| Ubiquitin-63-E | 2 | 114 | UBC | Protein degradation |
aMascot score = −10log(P) where P is the probability that the match is incorrect. HGNC, HUGO Gene Nomenclature Committee.
We chose to focus on Vap in the rest of this study because of its role in synaptic modulation and association with ALS. To determine whether Vap associates with endogenous Dscam we conducted IP experiments using cleared lysates of nontransfected S2 cells. Immune precipitation of Dscam from S2 cells coprecipitates Vap (Fig. 1B, top left), while IP of Vap is able to coprecipitate Dscam (Fig. 1B, bottom left). We further find that endogenous Vap and Dscam coprecipitate from lysates prepared from whole flies (Fig. 1B, right).
Vap influences Dscam function and expression patterns
Loss of Vap protein has a widespread effect on morphology and patterning of neurons, including neuromuscular junctions and the mushroom body (Pennetta et al., 2002; Tsuda et al., 2008). Therefore, we examined if Vap influences Dscam function in axon targeting or patterning. The 72OK enhancer trap can be used to express proteins in two olfactory receptor neuron (ORN) subclasses that target the VM1 and VM4 glomeruli in the antenna lobe (Fig. 2A). Exogenous Dscam expression using 72OK induces repulsion of axon terminals in the region of the VM1 and VM4 glomeruli and also enhances mistargeting of axons to the dorsal region of the antenna lobes (Fig. 2B). Gene dosage reduction of Vap results in suppression of the Dscam-induced dorsal ectopic targeting phenotype, but has little effect on Dscam-induced repulsion in the VM1 and VM4 glomeruli (Fig. 2C), suggesting that Vap dosage reduction can modify Dscam-induced targeting phenotypes. Total loss of Vap protein is largely pupa lethal with only a few adult escapers that emerge from pupa cases. These animals are severely uncoordinated and die shortly after eclosion. Analysis of 72OK-positive ORNs expressing Dscam in Vap-null escapers reveals ectopic targeting of these ORNs within and outside of the antenna lobe and a general increase in disorder of the antenna lobe glomerular structure and surrounding brain regions (Fig. 2D). These data indicate that Vap dosage reduction can modify Dscam-induced targeting phenotypes while preserving gross brain structure, whereas complete loss of Vap protein has a widespread effect on patterning in the adult nervous system.
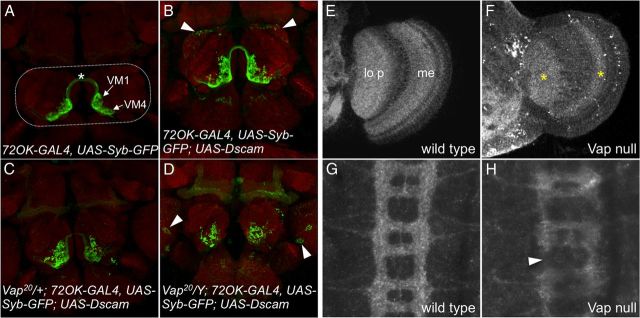
Figure 2.
Vap loss of function affects Dscam-induced phenotypes and localization. A–D, Axons of ORNs terminate in the antenna lobes (indicated by dotted rounded rectangle in A) of the Drosophila brain and form glomerular structures that can be visualized with the neuropil marker NC82 (red). 72OK-GAL4 was used to drive expression of synaptobrevin-GFP (green) in two subsets of ORNs that terminate in the VM1 and VM4 glomeruli of the antenna lobe (labeled in A) and extend contralateral projections (*). A, Wt 72OK expressed syb-GFP pattern (n = 25). B, Expression of a Dscam transgene induces homophilic repulsion of 72OK axons resulting in disorganization of the VM1 and VM4 glomeruli (100% penetrance, n = 27) and the ectopic targeting of some of these axons to the dorsal region of the antenna lobe (arrowheads; 75% penetrance, n = 27). C, Single copy loss of Vap eliminates the dorsal ectopic targeting (100% show no dorsal ectopic targeting, n = 21) but VM1 and VM4 disorganization is still present (100% penetrance, n = 21). D, Complete loss of Vap in hemizygous mutant flies results in general disorganization of antenna lobe glomerular structure, which enhances VM1 and VM4 disorganization, disrupts commissural projections, and induces ectopic targeting outside of the antenna lobe (arrowheads; similar phenotypes in all animals, n = 10). E–H, Dscam expression and localization in the adult optic lobe (E, F) and embryo ventral nerve cord (G, H) are assessed by Dscam immunofluorescence. E, Wild-type optic lobe has an even but layered Dscam expression pattern, with the highest labels of Dscam seen in the lobula plate (lo p) and the medulla layers (me). F, Vap-null optic lobes show disruption of Dscam expression patterns in most regions; however, there are regions in the lobula plate and medulla where Dscam localization appears to be less affected (*). G, Dscam localizes to the ladder-like pattern of the axon scaffold in the embryo ventral nerve cord and is evenly distributed in the commissural axons and the longitudinal tracts. F, Dscam localization is disrupted in Vap mutant embryos with greatly reduced levels of Dscam immunoreactivity in general and breaks in the longitudinal tracts (arrowhead).
Examination of endogenous Dscam expression in the adult optic lobe reveals an even distribution of Dscam in distinct layers with the highest expression in the medulla and lobula plate (Fig. 2E). This pattern is disrupted in Vap-null mutants, with puncta of intense Dscam immunofluorescence in the medulla and loss of immunofluorescence in the medulla and lobula plate (Fig. 2F). Interestingly, there are regions in the lobula plate and inner medulla that appear to have Dscam immunofluorescence patterns that are less affected by Vap loss. In the embryo ventral nerve cord, Dscam can be seen broadly expressed in the longitudinal tracts and commissures of the CNS (Fig. 2G). This pattern is disrupted in Vap-null animals, which exhibit less Dscam immunofluorescence in general with breaks in the longitudinal tracts and a reduction in Dscam levels in the commissures (Fig. 2H). Based on these results, we conclude that Vap is an attractive Dscam interacting protein candidate that may be important for aspects of Dscam function and localization.
Vap does not interact with the Dscam extracellular domain
Since Vap can be found associated with the endoplasmic reticulum and Golgi as well as proteolytically processed and secreted, it is plausible that Vap interacts with the Dscam extracellular domain, the Dscam cytoplasmic domain, or both. To address this we generated a series of FLAG-tagged Dscam transgenes that contains deletions of different Dscam extracellular regions or replacement of the TM and cytoplasmic domain with the corresponding domains of Neurexin 1 (Nrx) or neuroligin (nl2) (Fig. 3A). We found that endogenous Vap coprecipitated with experimental Dscam transgenes that contained the Dscam TM and cytoplasmic domains, even when the variable region of Dscam is removed, but did not copurify with Dscam constructs that had the TM and cytodomains replaced by Nrx or nl2 sequences (Fig. 3B). These data demonstrate that Vap interacts preferentially with the Dscam TM or cytoplasmic domain and is unlikely to function as a secreted ligand that binds to the Dscam ectodomain.
Vap preferentially interacts with Dscam isoforms containing alternative TM domain 2
Alternative splicing of the Dscam transcript produces receptor isoforms that contain one of two possible TM domains (TM1 or TM2), which are encoded by alternative exons 17.1 and 17.2. These alternative exon 17s also encode unique intracellular and extracellular juxtamembrane flanking sequences. To determine whether Vap shows any binding preference for TM1-containing versus TM2-containing Dscam isoforms, we transfected S2 cells with expression constructs encoding TAP-tagged Dscam TM1 or TM2 isoform variants. The TAP tags are located near the C terminus or in the juxtamembrane location 7 aa downstream of the exon 17–18 junction (Fig. 1). Endogenous Vap coimmunoprecipitates with each of these tagged Dscam isoforms, regardless of TAP-tag location or TM sequence present (Fig. 3C). However, more Vap protein copurifies with TM2-containing isoforms than with TM1-containing isoforms (Fig. 3C,D). This suggests that Vap may have a higher affinity for Dscam isoforms that contain TM2.
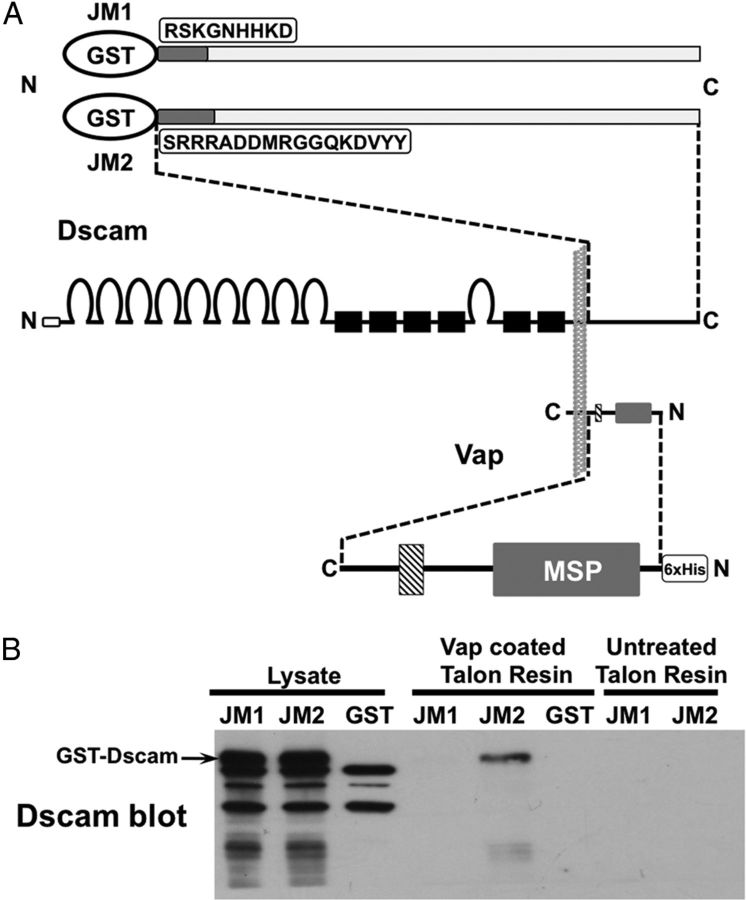
Dscam isoforms are known to engage in cis-interactions (within the same cellular membrane) in an isoform-independent manner, which is mediated by the cytodomains (J.C. Clemens, J.J. Flanagan, and S. L. Zipursky, unpublished observations). Therefore it is possible that the Vap that copurifies with TAP-tagged TM1 isoforms in our experiments is actually bound to endogenous Dscam TM2 isoforms that are cis-associated with the tagged protein. To address this, glutathione S-transferase (GST) fusions of Dscam cytodomains containing the TM1 or TM2 juxtamembrane flanking sequences (JM1 or JM2, respectively) were created (Fig. 4A). The only difference between the JM1 and JM2 GST fusions is the unique cytoplasmic juxtamembrane sequences that were fused to GST: RSKGNHHKD (JM1) and SRRRADDMRGGQKDVYY (JM2). A 6xHis-Vap fusion construct was also created that contained all of the Vap cytoplasmic domain sequence, but not the TM or C-terminal sequences (Fig. 4A). We find that GST-Dscam fusion proteins containing the JM2 sequence were retained on Talon resin precoated with 6xHis-Vap, but not on untreated Talon resin (Fig. 4B). GST-Dscam JM1 fusion protein did not bind to Vap coated or uncoated Talon resin. These data indicate that Vap specifically binds to Dscam TM2-containing isoforms and that the cytoplasmic juxtamembrane sequence (JM2) encoded by the TM2 exon is sufficient for Vap interaction.
Figure 4.
Vap preferentially associates with Dscam isoforms containing TM domain 2. A, Schematic of Dscam and Vap in a biological membrane (vertical gray structure) and the bacterially expressed fusion protein constructs derived from these proteins. The N termini (N) and C termini (C) of the constructs are indicated. GST-Dscam fusion proteins contain N-terminal GST fused to the Dscam cytodomain of TM1-containing or TM2-containing isoforms. The Dscam cytodomain sequences are identical (long light gray rectangle) except for the juxtamembrane (JM) sequences (listed) that are encoded by alternatively spliced exons 17.1 for JM1 and 17.2 for JM2 (dark gray rectangles in schematic). The 6xHis-Vap fusion construct contains an N-terminal 6xHis tag fused to the cytodomain of Vap, which contains the MSP domain (labeled) and a coiled coil domain (downward diagonal pattern). B, The presence of GST-Dscam fusion proteins is detected by Western analysis. Full-length GST-Dscam immunoreactivity (indicated on blot) is present in cleared bacterial lysates containing GST-DscamJM1 (JM1) and GST-DscamJM2 (JM2), but not GST alone. Three bands are visible in all three lysate lanes that correspond to bacterial proteins that react with antibodies in the Dscam antiserum. Additional lower molecular weight Dscam immunoreactivity is present in the JM1 and JM2 lanes, which likely arises from breakdown of full-length GST-Dscam. The GST lysates were applied to Talon resin that was precoated with 6xHis-Vap or untreated. GST-DscamJM2 is retained on Vap-coated resin but not on untreated Talon resin. GST-DscamJM1 was not retained on either resin.
Vap is required for the localization of Dscam isoforms containing TM domain 2 to axons
Vap mutant flies die in pupation and those few that eclose are uncoordinated and quickly die. The nervous system of these animals is disorganized and Dscam protein distribution is disrupted; however, there are regions in the optic lobe where Dscam protein distribution appears relatively normal (Fig. 2F). Since Vap specifically interacts with Dscam isoforms containing TM domain 2, it is possible that Vap loss specifically affects TM2-containing isoforms and not those containing TM1. Therefore, the regions in the optic lobe where Dscam distribution appears relatively normal, the inner medulla and the lobula plate, might be regions where TM1 isoforms of Dscam are localized.
To determine whether Vap loss of function is required specifically for the function of Dscam TM2 containing isoforms, we expressed TM1- or TM2-containing GFP-tagged Dscam isoforms using ppk-GAL4 in dendritic arborization (DA) neurons. The dendrites and cell bodies of DA neurons reside in the body wall, and they extend axons to the longitudinal tracts and commissures of the ventral nerve cord. These structures can be visualized by expressing CD8-mCherry, which generally labels the membranes of these neurons (Fig. 5A,B). Simultaneous expression of DscamTM1-GFP and CD8-mCherry showed Dscam labeling of cell bodies and puncta in dendrites, but no DscamTM1-GFP fluorescence was detected in the ventral nerve cord of any of the animals examined (N = 15, Fig. 5C,D). The pattern of DscamTM1-GFP was not changed in Vap-null animals (N = 12, Fig. 5E,F). Expression of DscamTM2-GFP and CD8-mCherry showed that TM2-bearing isoforms localize to axons, dendrites, and cell bodies in all animals examined (N = 15; Fig. 5G,H) as had been previously described (Shi et al., 2007) and leads to disorder in the axon scaffold of all 15 animals in the ventral nerve cord through the induction of Dscam-mediated homophilic repulsion of DA axons that form synapses in this region (Fig. 5H′). DscamTM2 is absent from axons in a Vap-null background even though it is still present in the cell bodies and dendrites in 100% of animals examined (Fig. 5I,J), and the loss of expressed DscamTM2 in axons restores order to the axon scaffold in the ventral nerve cord (12 of 12; Fig. 5J′). Coexpression of Vap and DscamTM2 in an otherwise Vap-null background (Fig. 5K,L) results in restoration of DscamTM2 localization to axons (Fig. 5L) as well as induction of Dscam-mediated repulsion in the ventral nerve cord in all animals examined (N = 5; Fig. 5L′). These studies demonstrate that Vap protein is required for the axonal localization of Dscam isoforms containing TM domain 2 and mediates Dscam function in axon patterning.
Figure 5.
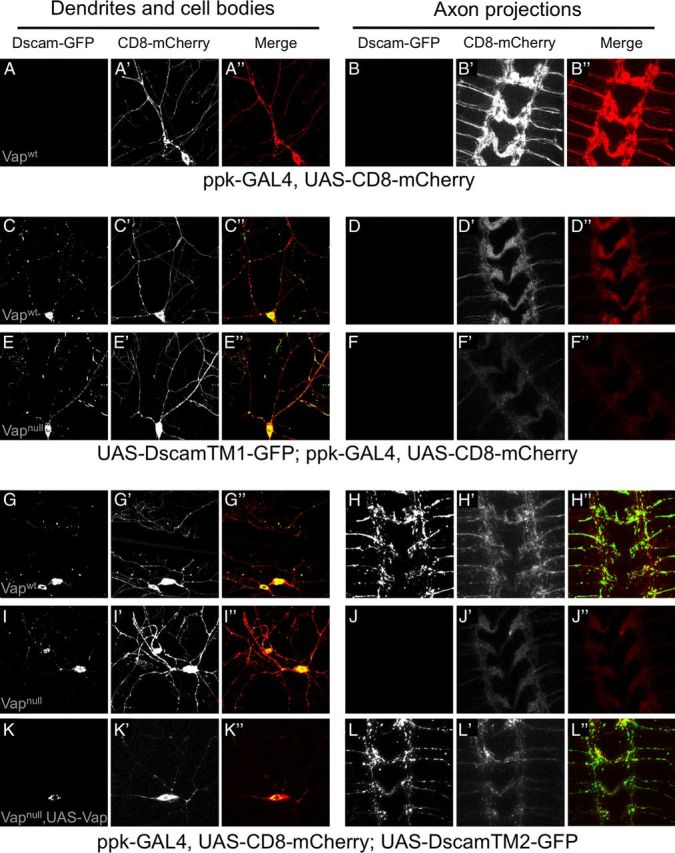
Vap is required for axonal localization of Dscam isoforms containing TM domain 2. Fluorescent micrographs of Dscam-GFP isoform localization (A–L), CD8-mCherry localization (A′–L′), or merged images (A″–L″) in the cell bodies and dendrites (A, C, E, G, I, K) or axon projections in the ventral nerve cord (B, D, F, H, J, L) of DA neurons in Vap wild-type (Vapwt) or Vap-null (Vapnull) backgrounds. Fifteen animals were observed for the Vapwt backgrounds, 12 animals for the Vapnull background, and 5 animals for the Vap expression rescue background. All images were taken with identical confocal settings and are representative of all individuals within the given genotype. A, B, CD8-mCherry expressed by the ppk-GAL4 driver localizes to dendrites and cell bodies (A) as well as axons (B). C–F, Coexpression of DscamTM1-GFP and CD8-mCherry in a Vapwt (C, D) and Vapnull (E, F) background shows that DscamTM1 is present in cell bodies and dendrites, but restricted from axons. G–H, Coexpression of DscamTM2-GFP and CD8-mCherry reveals that DscamTM2 is not only present in the cell bodies and dendrites (G) but also trafficks to axons (H) in a Vapwt background and induces disorganization of the axon scaffold. I–J, In a Vapnull background, expressed DscamTM2-GFP is no longer present in axons and the axon scaffold appears normal (J), but DscamTM2-GFP is still present in cell bodies and dendrites (I). K–L, Simultaneous expression of Vap and DscamTM2-GFP by the ppk-GAL4 driver in an otherwise Vapnull background does not affect dendritic and cell body localization (K), but restores DscamTM2 localization to axons and disorganization of the axon scaffold (L).
Discussion
We have identified three proteins that copurify with tagged Dscam expressed in Drosophila S2 cells using tryptic digestion followed by HPLC and tandem MS. Dreadlocks is a known Dscam-interacting protein and was in fact the protein that was originally used to purify and identify Drosophila Dscam (Schmucker et al., 2000). Ubiquitin and Vap had not been previously described as Dscam interacting proteins. There certainly must be other Dscam interacting proteins, but we were unable to identify them with any degree of confidence in our study. It is possible that the harsh conditions required to extract Dscam from the cell membrane may disrupt protein interactions. Alternatively, the sheer number and intensity of Dscam peptides may have suppressed peptide signals from interacting proteins. Therefore, the three proteins that we identify here likely represent proteins that are more strongly associated with Dscam or are simply more abundant.
Given the involvement of Vap in synaptic connectivity and the causative role of VAPB mutations in a subset of familial ALS cases, we further characterized the interaction between Vap and Dscam and assessed the requirement of Vap for Dscam function. Vap proteins have membrane-spanning sequences and are found primarily on the cytoplasmic surface of the endoplasmic reticulum and Golgi network. Proteolytic processing can free the cytoplasmic portion of Vap, including the MSP domain, from the membrane-spanning sequence so that it can be secreted by an unknown mechanism and bind to cell surface receptors involved in axon guidance. Our studies show that Vap does not interact with the extracellular domain of Dscam, but rather preferentially binds to the cytoplasmic domain of Dscam isoforms containing alternative TM domain 2. The juxtamembrane amino acid sequence SRRRADDMRGGQKDVYY is the only sequence that is unique to the cytodomain of TM2-containing isoforms and likely contains the Vap binding site. Previous studies have found that the Vap MSP domain binds to FFAT motifs (two phenylalanines (FF) in an acidic tract (AT)) (Loewen et al., 2003), which does not appear to be present in the TM2 juxtamembrane sequence. A detailed binding analysis and structural studies will have to be performed to determine the precise sites of interaction for Dscam and Vap.
Alternative splicing of the Dscam TM domain creates isoforms that differentially localize: TM1-containing isoforms are restricted from axons and localize to dendrites and cell bodies, while TM2-containing isoforms show robust axonal localization in addition to dendrite and cell body localization (Wang et al., 2004; Shi et al., 2007). Previous work found that point mutations in the unique TM2 cytoplasmic juxtamembrane sequence interfere with axonal localization of these isoforms (Yang, 2008). Our in vitro binding study is consistent with Vap binding directly to this sequence, and our Vap loss-of-function studies demonstrate that Vap is necessary for the localization of TM2-containing isoforms to axons, while dendritic localization of TM1-containing isoforms is not affected. Expression of DscamTM2-GFP using the DA neuron driver ppk-GAL4 results in the appearance of robust GFP fluorescence in the axons of these neurons within the ventral nerve cord (Fig. 5H). Similar expression of DscamTM2-GFP in a Vap-null background results in loss of both axonal GFP fluorescence in the ventral nerve cord and Dscam homophilic repulsion-induced disorganization of the axon scaffold (Fig. 5J). Vap is not required for expression of TM2-containing isoforms, as DscamTM2-GFP fluorescence is present in cell bodies and weakly in dendrites in both wild-type and Vap-null backgrounds. Vap does not appear to have a general role in axonal trafficking of all cell-surface receptors as CD8-mCherry still appears in the axons of DA neurons within the ventral nerve cord, although the levels of CD8-mCherry do appear to be reduced in the Vap-null background (compare Fig. 5D′,F′,H′,J′). This notion is further supported by Pennetta et al. (2002) who found that Vap function is not required for the axonal localization of Fasciclin II.
Given the requirement of Vap for DscamTM2 localization to axons, it should be the case that Dscam axon loss-of-function phenotypes can be phenocopied by Vap loss-of-function, and Dscam gain-of-function axon phenotypes can be suppressed by Vap loss. Dscam serves as a molecular basis for self-recognition and self-avoidance in Drosophila and ensures that dendritic and axonal branches segregate from one another. In the mushroom body, Dscam mutants fail to segregate sister axon branches, leading to the loss of one of the two mushroom body lobes (Wang et al., 2002, Wang et al., 2004; Zhan et al., 2004). This is also seen in Vap mutant animals at high frequency suggesting that a failure to localize DscamTM2 variants contributes to this phenotype (Tsuda et al., 2008). Expression of a single DscamTM2 isoform in neighboring neurons induces homophilic repulsion between neurons leading to disorganization of axon patterning (Zhan et al., 2004). In this study, expression of DscamTM2 in DA neurons results in disorganization of the axon scaffold in the ventral nerve cord, which can be completely suppressed by Vap loss (Fig. 5H′,J′). Single copy loss of Vap can also partially suppress dominant DscamTM2 expression phenotypes in ORNs (Fig. 2B,C). Our studies suggest that Vap mutants may be useful for specifically studying the requirement of Dscam function in axons.
Vaps have been implicated in a number of cellular functions, including lipid and membrane transport, mainly due to the functions of Vap-associated proteins from a number of different cell types (Lev et al., 2008). Because of the association of VAPB mutants with ALS, overexpression of the ALS8 mutant form of VAPB has implicated Vap dysfunction in numerous processes including formation of inclusions, induction of the unfolded protein response, and interfering with anterograde axon transport of mitochondria. Although the defects caused by ALS8 mutation are still under investigation, the mutation likely produces loss-of-function or dominant-negative effects. In addition, a recent study has found that VAPB protein levels are reduced and cytoplasmic inclusions are absent in ALS8 motor neurons derived from differentiation of ALS8-induced pluripotent stem cells (Mitne-Neto et al., 2011). Also, patients with sporadic ALS have reduced spinal cord expression of VAPB (Anagnostou et al., 2010), which is consistent with VAPB loss playing an important role in ALS pathology. Here, using biochemistry and genetic loss of function, we define a new neuronal role for Drosophila VAPB in the direct binding and trafficking of DscamTM2 cell surface receptors to axons. It is highly likely that VAPB is required for the axon trafficking of other cell-surface proteins in addition to Dscam and we propose that impairment of this function in humans may contribute to ALS pathology.
Footnotes
This work is supported by a grant from the Klingenstein Foundation (J.C.C.). H.T. was supported by grants from the ALS Association and the Canadian Institutes of Health Research (MOP-115153). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interests.
References
- Ahmed G, Shinmyo Y, Ohta K, Islam SM, Hossain M, Naser IB, Riyadh MA, Su Y, Zhang S, Tessier-Lavigne M, Tanaka H. Draxin inhibits axonal outgrowth through the netrin receptor DCC. J Neurosci. 2011;31:14018–14023. doi: 10.1523/JNEUROSCI.0943-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostou G, Akbar MT, Paul P, Angelinetta C, Steiner TJ, de Belleroche J. Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol Aging. 2010;31:969–985. doi: 10.1016/j.neurobiolaging.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Andrews GL, Tanglao S, Farmer WT, Morin S, Brotman S, Berberoglu MA, Price H, Fernandez GC, Mastick GS, Charron F, Kidd T. Dscam guides embryonic axons by Netrin-dependent and -independent functions. Development. 2008;135:3839–3848. doi: 10.1242/dev.023739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SM, Tsuda H, Yang Y, Vibbert J, Cottee P, Lee SJ, Winek J, Haueter C, Bellen HJ, Miller MA. Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev Cell. 2012;22:348–362. doi: 10.1016/j.devcel.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci U S A. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Millard SS, Flanagan JJ, Pappu KS, Wu W, Zipursky SL. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature. 2007;447:720–724. doi: 10.1038/nature05855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitne-Neto M, Machado-Costa M, Marchetto MC, Bengtson MH, Joazeiro CA, Tsuda H, Bellen HJ, Silva HC, Oliveira AS, Lazar M, Muotri AR, Zatz M. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum Mol Genet. 2011;20:3642–3652. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mórotz GM, De Vos KJ, Vagnoni A, Ackerley S, Shaw CE, Miller CC. Amyotrophic lateral sclerosis-associated mutant VAPBP56S perturbs calcium homeostasis to disrupt axonal transport of mitochondria. Hum Mol Genet. 2012;21:1979–1988. doi: 10.1093/hmg/dds011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Pennetta G, Hiesinger PR, Fabian-Fine R, Meinertzhagen IA, Bellen HJ. Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron. 2002;35:291–306. doi: 10.1016/s0896-6273(02)00769-9. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Wojtowicz WM, Andre I, Qian B, Wu W, Baker D, Eisenberg D, Zipursky SL. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. 2008;134:1007–1018. doi: 10.1016/j.cell.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Shi L, Yu HH, Yang JS, Lee T. Specific Drosophila Dscam juxtamembrane variants control dendritic elaboration and axonal arborization. J Neurosci. 2007;27:6723–6728. doi: 10.1523/JNEUROSCI.1517-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel PA, Fabian-Fine R, Kandel ER. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc Natl Acad Sci U S A. 2000;97:1101–1106. doi: 10.1073/pnas.97.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussan L, Burakov D, Daniels MP, Toister-Achituv M, Porat A, Yarden Y, Elazar Z. ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J Cell Biol. 1999;146:301–311. doi: 10.1083/jcb.146.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuling E, Ahmed S, Haasdijk E, Demmers J, Steinmetz MO, Akhmanova A, Jaarsma D, Hoogenraad CC. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Han SM, Yang Y, Tong C, Lin YQ, Mohan K, Haueter C, Zoghbi A, Harati Y, Kwan J, Miller MA, Bellen HJ. The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell. 2008;133:963–977. doi: 10.1016/j.cell.2008.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. TM/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ. The molecular mechanisms underlying the polarized distribution of Drosophila Dscam in neurons: a dissertation. Worcester: University of Massachusetts; 2008. [Google Scholar]
- Yu HH, Yang JS, Wang J, Huang Y, Lee T. Endodomain diversity in the Drosophila Dscam and its roles in neuronal morphogenesis. J Neurosci. 2009;29:1904–1914. doi: 10.1523/JNEUROSCI.5743-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]