Abstract
Background
Verticillium wilt caused by the fungus Verticillium dahliae race 1 is among the top disease concerns for lettuce in the Salinas and Pajaro Valleys of coastal central California. Resistance of lettuce against V. dahliae race 1 was previously mapped to the single dominant Verticillium resistance 1 (Vr1) locus. Lines of tomato resistant to race 1 are known to contain the closely linked Ve1 and Ve2 genes that encode receptor-like proteins with extracellular leucine-rich repeats; the Ve1 and Ve2 proteins act antagonistically to provide resistance against V. dahliae race 1. The Vr1 locus in lettuce contains a cluster of several genes with sequence similarity to the tomato Ve genes. We used genome sequencing and/or PCR screening along with pathogenicity assays of 152 accessions of lettuce to investigate allelic diversity and its relationship to race 1 resistance in lettuce.
Results
This approach identified a total of four Ve genes: LsVe1, LsVe2, LsVe3, and LsVe4. The majority of accessions, however, contained a combination of only three of these LsVe genes clustered on chromosomal linkage group 9 (within ~ 25 kb in the resistant cultivar La Brillante and within ~ 127 kb in the susceptible cultivar Salinas).
Conclusions
A single allele, LsVe1L, was present in all resistant accessions and absent in all susceptible accessions. This allele can be used as a molecular marker for V. dahliae race 1 resistance in lettuce. A PCR assay for rapid detection of race 1 resistance in lettuce was designed based on nucleotide polymorphisms. Application of this assay allows identification of resistant genotypes in early stages of plant development or at seed-level without time- and labor-intensive testing in the field.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1905-9) contains supplementary material, which is available to authorized users.
Keywords: Lactuca sativa, Genomics, Marker-assisted selection, Plant breeding, Wilt resistance
Background
The Salinas and Pajaro Valleys of coastal central California are among the most important lettuce-producing regions in the United States [1]. One of the top disease concerns for lettuce in the area is Verticillium wilt caused by the fungus Verticillium dahliae [2, 3], which is a soilborne pathogen with a wide host range that also includes artichoke, cotton, eggplant, hops, potato, sunflower, tobacco, and tomato [4, 5]. Two races of V. dahliae occur in coastal central California based on their differential virulence on cultivar La Brillante [6]; however, race 1 is more prevalent and economically important than race 2 [7]. In tomato, race 1 of V. dahliae carries Ave1 that is recognized by Ve1 in resistant genotypes [8]. Ve genes encode receptor-like proteins (RLPs) with extracellular leucine-rich repeats [9, 10]; such RLPs are widespread in land plants [11]. In addition to Ve1, tomato also contains the closely linked paralog Ve2; their encoded RLPs work antagonistically to confer resistance to V. dahliae race 1 [12]. Several Ve paralogs also confer resistance in otherwise V. dahliae-susceptible species including cotton [13], potato [14, 15], hops, and wild eggplant [11], but it is unknown whether they function analogously to the tomato Ve genes in conferring V. dahliae race 1 resistance. In lettuce, resistance to V. dahliae race 1 was originally identified in the Batavia-type cultivar, La Brillante, as conferred by a single dominant locus (Verticillium resistance 1, Vr1) located on chromosomal linkage group 9 [16]. The lettuce Vr1 locus contains several genes with sequence similarity to the Ve genes of tomato; it is very likely that one or more of these LsVe homologs are functional resistance genes.
The goals of this study were to identify the lettuce Ve allele(s) that play a role in resistance to V. dahliae race 1 and to develop PCR-based assays for marker-assisted selection. For this purpose, we analyzed the genome sequences of cultivars La Brillante (resistant to V. dahliae race 1) and the previously published Salinas [17] (iceberg type, susceptible to V. dahliae race 1). Subsequently, we sequenced and/or used allele-specific PCR screens of 150 additional lettuce accessions to identify the allele(s) of the LsVe genes that are exclusively present in resistant phenotypes.
Results
Phenotypic evaluation of resistance in field tests
One hundred and fifty accessions from ten horticultural types and L. serriola were evaluated in four field experiments. Twenty accessions (13.3%) showed no disease symptoms and were considered resistant. The proportion of disease incidence in susceptible accessions ranged from 0.07 to 1.00, with a mean disease incidence of 0.43 (± 0.02). There was a substantial difference in the distribution of resistant phenotypes across horticultural types. Among horticultural types with at least five tested accessions, the largest frequency of resistant accessions was found in Latin (6/7 = 85.7%), followed by Batavia (2/6 = 33.3%), red leaf (4/15 = 26.7%), and butterhead (3/14 = 21.4%; Table 1) types. In contrast, the lowest frequencies of resistant accessions were found in iceberg (0/46 = 0%), romaine (2/36 = 5.6%), and green leaf (2/18 = 11.1%) types. Oil (0/4 = 0%), stem (1/3 = 33.3%) types, and L. serriola (0/1 = 0%) had fewer than five tested accessions each. All oil type accessions were susceptible to the disease and had a very high disease incidence (0.98 in one accession, 1.00 in all others). Statistical analysis indicated that the frequency of resistant accessions was significantly (p < 0.01) higher than the overall frequency of 13.3% in Latin types, while it was significantly lower in iceberg types; however, the statistical power to detect significant differences for horticultural types with a small number of tested accessions is limited.
Table 1.
Difference among horticultural types in their phenotypic reaction to V. dahliae race 1 tested in field trials
| Horticultural type or species | No. of tested accessions | No. of accessions with disease incidence | Frequency of resistant accessions | Chi-square test a | ||||
|---|---|---|---|---|---|---|---|---|
| 0.00 (resistant) | 0.01–0.25 | 0.26–0.50 | 0.51–0.75 | 0.76–1.00 | ||||
| Batavia | 6 | 2 | 4 | 0.33 | 1.6 | |||
| Butterhead | 14 | 3 | 9 | 1 | 1 | 0.21 | 0.8 | |
| Green leaf | 18 | 2 | 2 | 10 | 2 | 2 | 0.11 | 0.1 |
| Iceberg | 46 | 10 | 23 | 10 | 3 | 0.00 | 12.8* | |
| Latin | 7 | 6 | 1 | 0.86 | 19.0* | |||
| Oil | 4 | 4 | 0.00 | 1.1 | ||||
| Red leaf | 15 | 4 | 3 | 4 | 4 | 0.27 | 2.0 | |
| Romaine | 36 | 2 | 9 | 20 | 4 | 1 | 0.06 | 2.2 |
| Stem | 3 | 1 | 2 | 0.33 | 0.8 | |||
| Lactuca serriola | 1 | 1 | 0.00 | 0.3 | ||||
| Total | 150 | 20 | 31 | 63 | 21 | 15 | 0.13 | |
a Values of two-sided χ2 test for the frequency of resistant accessions. Asterisk (*) indicates horticultural types with the frequency of resistant accessions significantly different at experiment-wise p < 0.01 from the overall frequency (0.13) observed for all tested types. Note that the statistical test has a low to absent detection power for significant results for horticultural types with a very few tested accessions
Lettuce genome assemblies
Genome assemblies were generated for 61 accessions of cultivated lettuce (Table 2). The assembly of cultivar La Brillante consisted of 41,939 scaffolds with a total length of 2.04 Gb and had an L50 of 90.84 kb. The remaining 60 draft de novo assemblies consisted of 1.0 to 3.2 M contigs (average 2.78 M) with a total length of 2.08 to 2.44 Gb (average 2.20 Gb) and an L50 of 1.22 to 3.66 kb (average 1.6 kb). Reads have been submitted to GenBank (BioProject PRJNA478460).
Table 2.
Accessions used in this study sorted by horticultural type and accession name
| Type | Accession | Identifier | Genome | Race 1 resistance | LsVe1L | LsVe3L | LsVe4L | LsVe1S | LsVe2S | LsVe3S | No. of tested plants | Proportion of symptomatic plants k |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batavia | Anuenuea, b | L2 | + | – | – | – | – | – | + | + | 90 | 0.26 (0.18–0.35) |
| Batavia | Batavia Reine des Glacesc, b | L90 | – | – | – | NDi | – | ND | ND | ND | 90 | 0.44 (0.35–0.55) |
| Batavia | Icebergc, b | L115 | – | – | – | ND | – | ND | ND | ND | 30 | 0.50 (0.33–0.67) |
| Batavia | La Brillanted, e | 10G364–1 | + | + | + | + | + | – | – | – | NTj | NT |
| Batavia | La Brillantec, b | L119 | – | + | + | ND | + | ND | ND | ND | 180 | 0.00 (0.00–0.02) |
| Batavia | Laurad, b | L43 | + | + | + | + | + | – | – | – | 60 | 0.00 (0.00–0.06) |
| Batavia | Reines des Glacesd, b | L53 | + | – | – | – | – | + | + | + | 30 | 0.40 (0.25–0.58) |
| Butterhead | Allegiancea, b | L85 | – | – | – | ND | + | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Butterhead | Ancoraa, f | L240 | – | + | + | ND | + | ND | ND | ND | 60 | 0.00 (0.00–0.06) |
| Butterhead | Bennetta, b | L93 | – | – | – | ND | + | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Butterhead | Bibbd, b | L26 | + | – | – | – | – | + | + | + | 30 | 0.07 (0.02–0.21) |
| Butterhead | Cobham Greena, b | L6 | + | – | – | + | + | + | + | – | 30 | 0.80 (0.63–0.91) |
| Butterhead | Dark Green Boston-Cd, b | L71 | + | – | – | – | + | + | + | – | 26 | 0.23 (0.11–0.42) |
| Butterhead | Grappaa, b | L106 | – | – | – | ND | – | ND | ND | ND | 30 | 0.07 (0.02–0.21) |
| Butterhead | Little Gem-Gd, b | L77 | + | + | + | + | + | – | – | – | 60 | 0.00 (0.00–0.06) |
| Butterhead | Margaritad, b | L45 | + | – | – | – | + | + | + | – | 30 | 0.30 (0.17–0.48) |
| Butterhead | Mariskad, b | L46 | + | – | – | – | + | + | + | – | 29 | 0.24 (0.12–0.42) |
| Butterhead | Martina, b | L124 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Butterhead | Ostinataa, b | L127 | – | – | – | ND | + | ND | ND | ND | 30 | 0.07 (0.02–0.21) |
| Butterhead | Taniaa, b | L149 | – | – | – | ND | + | ND | ND | ND | 30 | 0.07 (0.02–0.21) |
| Butterhead | Tintoa, b | L23 | + | + | + | + | + | – | – | – | 59 | 0.00 (0.00–0.06) |
| Green leaf | Alpinea, b | L86 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Green leaf | Franklina, b | L100 | – | – | – | ND | – | ND | ND | ND | 60 | 0.20 (0.12–0.32) |
| Green leaf | Genecorps Greena, b | L103 | – | – | – | ND | – | ND | ND | ND | 30 | 0.60 (0.42–0.75) |
| Green leaf | Grand Rapidsd, b | L37 | + | – | – | – | – | + | + | + | 60 | 0.45 (0.33–0.58) |
| Green leaf | Green Visiona, b | L109 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Green leaf | Haciendad, b | L40 | + | – | – | – | – | + | + | + | 30 | 0.97 (0.83–0.99) |
| Green leaf | Plymoutha, b | L19 | + | + | + | + | + | – | – | – | 120 | 0.00 (0.00–0.03) |
| Green leaf | Pybas Greena, b | L133 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Green leaf | Salad Bowla, f | L307 | – | – | – | ND | – | ND | ND | ND | 27 | 0.41 (0.25–0.59) |
| Green leaf | Shining Stara, b | L141 | – | – | – | ND | – | ND | ND | ND | 30 | 0.50 (0.33–0.67) |
| Green leaf | Sloboltd, b | L56 | + | – | – | – | – | + | + | + | 29 | 0.45 (0.28–0.62) |
| Green leaf | Tehamaa, b | L150 | – | – | – | ND | – | ND | ND | ND | 30 | 0.47 (0.30–0.64) |
| Green leaf | Tropicanad, b | L58 | + | – | – | – | – | + | + | + | 30 | 0.70 (0.52–0.83) |
| Green leaf | Two Stara, b | L25 | + | – | – | – | – | + | + | + | 150 | 0.39 (0.31–0.47) |
| Green leaf | Waldmann’s Greena, b | L160 | – | – | – | ND | – | ND | ND | ND | 30 | 0.50 (0.33–0.67) |
| Green leaf | Waldmans Green-Gd, b | L84 | + | – | – | – | – | + | + | + | 30 | 0.37 (0.22–0.54) |
| Green leaf | Western Greena, b | L161 | – | – | – | ND | – | ND | ND | ND | 30 | 0.90 (0.74–0.97) |
| Green leaf | Xenad, b | L63 | + | + | + | + | + | – | – | – | 59 | 0.00 (0.00–0.06) |
| Iceberg | Autumn Goldd, b | L66 | + | – | – | – | – | + | + | + | 28 | 0.54 (0.36–0.70) |
| Iceberg | Bayviewa, b | L92 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Big Bena, b | L94 | – | – | – | ND | – | ND | ND | ND | 30 | 0.27 (0.14–0.44) |
| Iceberg | Caliceld, b | L27 | + | – | – | – | – | + | + | + | 26 | 0.54 (0.35–0.71) |
| Iceberg | Calmara, b | L5 | + | – | – | – | – | + | + | + | 30 | 0.50 (0.33–0.67) |
| Iceberg | Cannery Rowa, b | L163 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Ciscod, b | L30 | + | – | – | – | – | + | + | + | 26 | 0.65 (0.46–0.81) |
| Iceberg | Coolguardd, b | L31 | + | – | – | – | – | + | + | + | 28 | 0.86 (0.69–0.94) |
| Iceberg | Coronaa, b | L169 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Diplomatd, b | L34 | + | – | – | – | – | + | + | + | 29 | 0.55 (0.38–0.72) |
| Iceberg | Durangoa, b | L173 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Early Birda, b | L35 | + | – | – | – | – | + | + | + | 30 | 0.40 (0.25–0.58) |
| Iceberg | El Doradoa, b | L174 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Empirea, b | L175 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Grand Slama, b | L105 | – | – | – | ND | – | ND | ND | ND | 30 | 0.27 (0.14–0.44) |
| Iceberg | Great Lakesa, b | L107 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Iceberg | Great Lakes 659-Gd, b | L73 | + | – | – | – | – | + | + | + | 30 | 0.37 (0.22–0.54) |
| Iceberg | Hallmarka, b | L111 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Home Runa, b | L182 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Iceberg | Icona, b | L116 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Ithaca ZAA-Cd, b | L74 | + | – | – | – | – | + | + | + | 30 | 0.83 (0.66–0.93) |
| Iceberg | Laguna Frescaa, b | L120 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Iceberg | Legenda, b | L121 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Libertya, b | L122 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Iceberg | Mesa 659-Cd, b | L78 | + | – | – | – | – | + | + | + | 30 | 0.80 (0.63–0.91) |
| Iceberg | Monumenta, b | L125 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Iceberg | Pacifica, b | L128 | – | – | – | ND | – | ND | ND | ND | 60 | 0.18 (0.11–0.30) |
| Iceberg | Primusa, b | L20 | + | – | – | – | – | + | + | + | 30 | 0.57 (0.39–0.73) |
| Iceberg | Salinasg | NAh | NA | – | – | – | – | + | + | + | NT | NT |
| Iceberg | Salinasc, b | L138 | – | – | – | ND | – | ND | ND | ND | 180 | 0.55 (0.48–0.62) |
| Iceberg | Salinas 88c, b | L139 | – | – | – | ND | – | ND | ND | ND | 120 | 0.48 (0.40–0.57) |
| Iceberg | Salinas 88-Gd, b | L81 | + | – | – | – | – | + | + | + | 30 | 0.40 (0.25–0.58) |
| Iceberg | Sharp Shootera, b | L140 | – | – | – | ND | – | ND | ND | ND | 30 | 0.60 (0.42–0.75) |
| Iceberg | Silveradoa, b | L143 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Snipera, b | L144 | – | – | – | ND | – | ND | ND | ND | 60 | 0.55 (0.42–0.67) |
| Iceberg | Sun Devila, b | L146 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Sure Shota, b | L148 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Telluridea, b | L151 | – | – | – | ND | – | ND | ND | ND | 90 | 0.34 (0.25–0.45) |
| Iceberg | Tibera, b | L152 | – | – | – | ND | – | ND | ND | ND | 60 | 0.53 (0.41–0.65) |
| Iceberg | Tributea, b | L153 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Iceberg | Trojana, b | L155 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Iceberg | Vandenberga, b | L156 | – | – | – | ND | – | ND | ND | ND | 30 | 0.47 (0.30–0.64) |
| Iceberg | Vanguarda, b | L157 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Iceberg | Vanguard-Cd, b | L82 | + | – | – | – | – | + | + | + | 30 | 0.30 (0.17–0.48) |
| Iceberg | Vanguard-Gd, b | L83 | + | – | – | – | – | + | + | + | 30 | 0.23 (0.12–0.41) |
| Iceberg | Venusa, b | L183 | – | – | – | ND | – | ND | ND | ND | 30 | 0.47 (0.30–0.64) |
| Iceberg | Winterhavend, b | L61 | + | – | – | – | – | + | + | + | 23 | 0.61 (0.41–0.78) |
| Latin | Barnwood Gema, b | L89 | – | + | + | ND | + | ND | ND | ND | 60 | 0.00 (0.00–0.06) |
| Latin | Brigadea, b | L97 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Latin | Gallegaa, b | L102 | – | + | + | ND | + | ND | ND | ND | 60 | 0.00 (0.00–0.06) |
| Latin | Eruptionc, b | L9 | + | + | + | + | + | – | – | – | 120 | 0.00 (0.00–0.03) |
| Latin | Little Gemc, b | L123 | – | + | + | ND | + | ND | ND | ND | 60 | 0.00 (0.00–0.06) |
| Latin | Little Gemd, b | L44 | + | + | + | + | + | – | – | – | 60 | 0.00 (0.00–0.06) |
| Latin | Pavanea, b | L16 | + | + | + | + | + | – | – | – | 90 | 0.00 (0.00–0.04) |
| Oil | PI 250020a, b | L17 | + | – | – | – | – | + | + | + | 30 | 1.00 (0.89–1.00) |
| Oil | PI 251245d, b | L50 | + | – | – | – | – | + | + | + | 30 | 1.00 (0.89–1.00) |
| Oil | PI 251245a, b | L18 | – | – | – | ND | – | ND | ND | ND | 30 | 1.00 (0.89–1.00) |
| Oil | PI 251246c, b | L131 | – | – | – | ND | – | ND | ND | ND | 60 | 0.98 (0.91–1.00) |
| Red leaf | Aragon Reda, b | L87 | – | – | – | ND | – | ND | ND | ND | 30 | 0.47 (0.30–0.64) |
| Red leaf | Battaliona, b | L91 | – | + | + | ND | + | ND | ND | ND | 60 | 0.00 (0.00–0.06) |
| Red leaf | Big Reda, b | L95 | – | – | – | ND | – | ND | ND | ND | 30 | 1.00 (0.89–1.00) |
| Red leaf | Deep Redd, b | L32 | + | – | – | – | – | + | + | + | 30 | 0.80 (0.63–0.91) |
| Red leaf | Lolla Rossaa, b | L14 | + | + | + | + | + | – | – | – | 60 | 0.00 (0.00–0.06) |
| Red leaf | Merlota, b | L15 | + | + | + | + | + | – | – | – | 90 | 0.00 (0.00–0.04) |
| Red leaf | New Reda, b | L126 | – | – | – | ND | – | ND | ND | ND | 30 | 0.97 (0.83–0.99) |
| Red leaf | Prizeheada, b | L132 | – | – | – | ND | – | ND | ND | ND | 30 | 0.70 (0.52–0.83) |
| Red leaf | Red Foxd, b | L51 | + | – | – | – | – | + | + | + | 30 | 0.70 (0.52–0.83) |
| Red leaf | Red Grenoblea, b | L134 | – | – | – | ND | + | ND | ND | ND | 60 | 0.30 (0.20–0.43) |
| Red leaf | Red Ragea, b | L135 | – | – | – | ND | – | ND | ND | ND | 30 | 0.97 (0.83–0.99) |
| Red leaf | Red Tidea, b | L136 | – | – | – | ND | – | ND | ND | ND | 30 | 0.57 (0.39–0.73) |
| Red leaf | Red Tided, b | L52 | + | – | – | – | – | + | + | + | 30 | 0.43 (0.27–0.61) |
| Red leaf | Sentrya, b | L21 | + | + | + | + | + | – | – | – | 180 | 0.00 (0.00–0.02) |
| Red leaf | Western Red Leafd, b | L60 | + | – | – | – | – | + | + | + | 30 | 0.53 (0.36–0.70) |
| Romaine | Annapolisc, b | L1 | + | + | + | + | + | – | – | – | 90 | 0.00 (0.00–0.04) |
| Romaine | Avalanchec, b | L88 | – | – | – | ND | – | ND | ND | ND | 90 | 0.22 (0.15–0.32) |
| Romaine | Blonde Lente a Monterc, b | L4 | + | – | – | – | – | + | + | + | 30 | 0.70 (0.52–0.83) |
| Romaine | Brave Heartc, b | L96 | – | – | – | ND | – | ND | ND | ND | 60 | 0.32 (0.21–0.44) |
| Romaine | Caesarc, b | L98 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Romaine | Camino Realc, b | L162 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Romaine | Clementec, b | L166 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Romaine | Coastal Starc, b | L167 | – | – | – | ND | – | ND | ND | ND | 30 | 0.50 (0.33–0.67) |
| Romaine | Conquistadorc, b | L168 | – | – | – | ND | – | ND | ND | ND | 30 | 0.27 (0.14–0.44) |
| Romaine | Costa Rica #4d, b | L70 | + | – | – | – | – | + | + | + | 30 | 0.43 (0.27–0.61) |
| Romaine | Costa Rica #4c, b | L170 | – | – | – | ND | – | ND | ND | ND | 30 | 0.33 (0.19–0.51) |
| Romaine | Darkland ELc, b | L171 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Romaine | Defenderc, b | L8 | + | + | + | + | + | – | – | – | 90 | 0.00 (0.00–0.04) |
| Romaine | EXP1752a, b | L99 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Romaine | Flashy Troutbackc, b | L10 | + | – | – | + | + | – | + | – | 30 | 0.30 (0.17–0.48) |
| Romaine | Fresh Heartc, b | L101 | – | – | – | ND | – | ND | ND | ND | 30 | 0.37 (0.22–0.54) |
| Romaine | Green Forestc, f | L189 | – | – | – | ND | – | ND | ND | ND | 28 | 0.46 (0.30–0.64) |
| Romaine | Green Towersc, b | L108 | – | – | – | ND | – | ND | ND | ND | 60 | 0.20 (0.10–0.37) |
| Romaine | Green Towersd, b | L39 | + | – | – | – | – | + | + | + | 30 | 0.23 (0.12–0.41) |
| Romaine | Hearts Delightc, b | L112 | – | – | – | ND | – | ND | ND | ND | 150 | 0.31 (0.24–0.38) |
| Romaine | Heavy Heartc, b | L113 | – | – | – | ND | – | ND | ND | ND | 30 | 0.30 (0.17–0.48) |
| Romaine | King Henryc, b | L117 | – | – | – | ND | – | ND | ND | ND | 30 | 0.20 (0.10–0.37) |
| Romaine | King Louie 2005d, b | L76 | + | – | – | – | – | + | + | + | 30 | 0.83 (0.66–0.93) |
| Romaine | Klamatha, b | L118 | – | – | – | ND | – | ND | ND | ND | 30 | 0.17 (0.07–0.34) |
| Romaine | Lee Talc, b | L12 | + | – | – | – | + | + | + | – | 59 | 0.17 (0.09–0.28) |
| Romaine | Lobjoits Cosc, b | L13 | + | – | – | – | – | + | + | + | 90 | 0.53 (0.43–0.63) |
| Romaine | Parris Island Cosc, b | L129 | – | – | – | ND | – | ND | ND | ND | 60 | 0.45 (0.33–0.58) |
| Romaine | Parris Island Cos 714 (PIC714)a, b | L130 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Romaine | Parris Island Cos-Gd, b | L80 | – | – | – | ND | – | ND | ND | ND | 30 | 0.43 (0.27–0.61) |
| Romaine | Passportd, b | L64 | + | – | – | – | – | + | + | + | 30 | 0.50 (0.33–0.67) |
| Romaine | PI 171674d, b | L49 | + | – | – | – | – | + | + | + | 90 | 0.32 (0.23–0.42) |
| Romaine | SM09Ac, b | L22 | + | – | – | – | – | + | + | + | 30 | 0.60 (0.42–0.75) |
| Romaine | Sunbeltc, b | L184 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Romaine | Triple Threatc, b | L24 | + | – | – | – | – | + | + | + | 60 | 0.58 (0.46–0.70) |
| Romaine | Tritonc, b | L154 | – | – | – | ND | – | ND | ND | ND | 30 | 0.47 (0.30–0.64) |
| Romaine | VJO3Ra, b | L159 | – | – | – | ND | – | ND | ND | ND | 30 | 0.27 (0.14–0.44) |
| Stem | Balady Banhac, b | L3 | + | + | + | + | + | – | – | – | 88 | 0.00 (0.00–0.04) |
| Stem | Celtucea, b | L165 | – | – | – | ND | – | ND | ND | ND | 30 | 0.40 (0.25–0.58) |
| Stem | Celtuce-Gd, b | L69 | + | – | – | – | – | + | + | + | 30 | 0.43 (0.27–0.61) |
| Lactuca serriola | 11-G99c, f | L185 | – | – | – | ND | + | ND | ND | ND | 30 | 0.70 (0.52–0.83) |
aGermplasm Collection of USDA-ARS Salinas, CA
bSeeds
cGenome Wide Association Mapping Collection (GBS) of USDA-ARS Salinas, CA
dUC Davis Collection
eDark-grown seedlings
fLeaf tissue from field
gReyes-Chin-Wo et al., 2017
hNot applicable (NA), because this genome was sequenced previously (Reyes-Chin-Wo et al., 2017)
iNot determined (ND)
jNot field tested in the present study (NT). Cultivars La Brillante and Salinas we previously confirmed to be resistant and susceptible, respectively, to V. dahliae race 1 (Hayes at al., 2011)
k95% confidence interval for the proportion of symptomatic plants is shown in parentheses
Ve genes and alleles of cultivars La Brillante, Salinas, and 60 other accessions
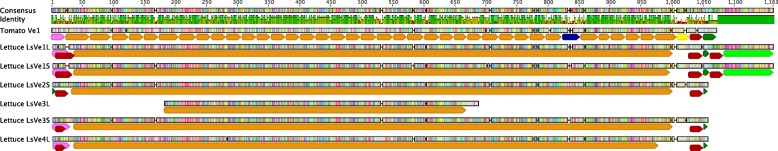
The expressed sequence tag marker QGD8I16.yg.ab1 at the Verticillium resistance 1 (Vr1) locus in lettuce [15] was used to query the genome assemblies of the lettuce cultivars La Brillante and Salinas using BLASTn. Three hits (e = 0.0) to scaffold linkage group 9 of the v8 reference assembly of the cultivar Salinas corresponded to three open reading frames (ORFs) that were named LsVe1S (because it had the highest sequence similarity of the three paralogs to Ve1 of tomato), LsVe2S, and LsVe3S (Fig. 1). The encoded proteins were comprised of 1133 aa, 1041 aa, and 1039 aa for LsVe1S, LsVe2S, and LsVe3S, respectively, and including a signal peptide, 37 extracellular leucine-rich repeats, a transmembrane domain, and a cytoplasmic region inferred from the N- to C-terminus (Fig. 2, Additional file 1). LsVe1S had an additional potential transmembrane domain and non-cytoplasmic region. Similarly, there were three hits (e = 0.0) on contig Lsat_LaBrillante_v1_g_2266 for La Brillante. The hits corresponded to three gene models that differed in sequence from the three Ve genes in cultivar Salinas. Phylogenetic analyses showed that two of the ORFs grouped with maximum support with the Ve1 and Ve3 alleles in Salinas (Fig. 3) and were therefore named LsVe1L and LsVe3L, respectively. The third gene sequence was sufficiently different from all three genes in cultivar Salinas and was therefore named LsVe4L (Fig. 3). LsVe1L, LsVe3L, and LsVe4L encode proteins measuring 1136, 503, and 1043 aa, respectively. The domains encoded by LsVe1L and LsVe4L were the same as for LsVe1S and LsVe3S, respectively; however, while the sequence of LsVe3L is similar to LsVe3S, premature stop codons result in a truncated protein encoded by LsVe3L (Fig. 2, Additional file 1).
Fig. 1.
Partial scaffolds of lettuce cultivars La Brillante (Lsat_LaBrillante_v1_g_2266) and Salinas (lg_9) containing LsVe genes
Fig. 2.
Alignment of tomato Ve1 and LsVe1, LsVe2, LsVe3, and LsVe4 alleles. Alleles are aligned in N to C orientation, residues are numbered across the top. Consensus and conserved residues across alleles are indicated in the top two tracks. Horizontal black boxes represent alleles, gray vertical lines represent substitutions, remaining colors inside black boxes represent different residues. Domains are indicated underneath alleles as colored boxes; the colors indicate the following: blue, non-LRR island domain (C2 domain); dark green, cytoplasmic domain; light green, non-cytoplasmic domain; orange, leucine-rich repeat region (individual repeats are indicated only for Ve1); pink, signal peptide; red, transmembrane domain; yellow, acidic domain. Domains follow [18] for Ve1 (GenBank accession ACR33105)
Fig. 3.
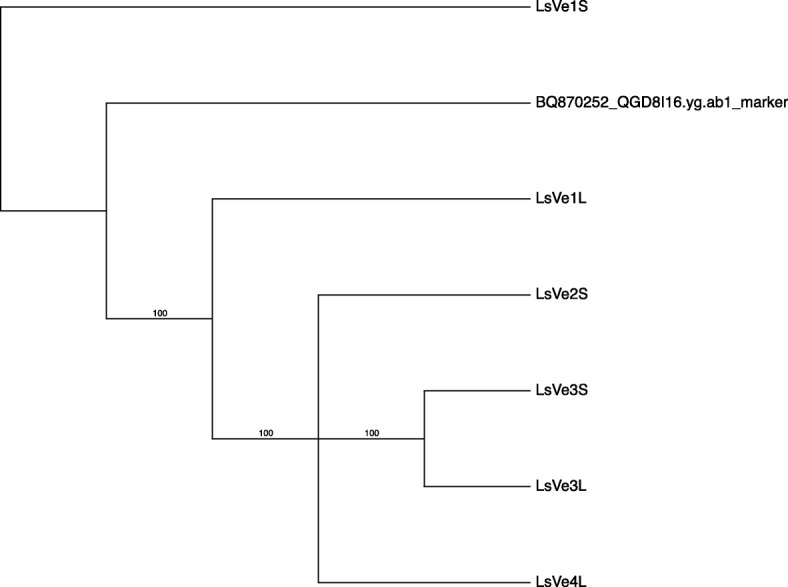
Unrooted parsimony bootstrap tree of cultivars La Brillante and Salinas Ve alleles. The topology shows that LsVe1 alleles plus BQ870252_QGD8I16.yg.ab1 marker (GenBank accession BQ870252) [16] group together with maximum statistical support; the LsVe3 alleles group together but LsVe4L and LsVe2S alleles do not. Bootstrap supports above 70% are indicated by the branches
The La Brillante and Salinas Ve alleles were then used as queries to identify homologs in diverse germplasm of cultivated lettuce. A total of 180 Ve sequences were extracted from genome assemblies of 60 lettuce cultivars (Additional file 2). The sequences represented 21 different alleles that were identical or similar to the Ve alleles from La Brillante and Salinas (Fig. 4). The LsVe1L clade contained a single allele and the remaining clades contained between two and six alleles (Fig. 4, Additional file 2). This analysis likely underestimated the total number of Ve alleles because only 47 of the 186 Ve sequences included in this study represented complete genes (Additional file 2). All cultivars contained three Ve genes, except cultivar Anuenue (susceptible), in which only two alleles were detected that clustered in the LsVe2 and LsVe3 clades, and cultivar Cobham Green (susceptible) that contained four Ve genes that clustered in the LsVs1S, LsVe2S, LsVe3L, and LsVe4L clades. For the remaining LsVe genotypes, see Table 2.
Fig. 4.
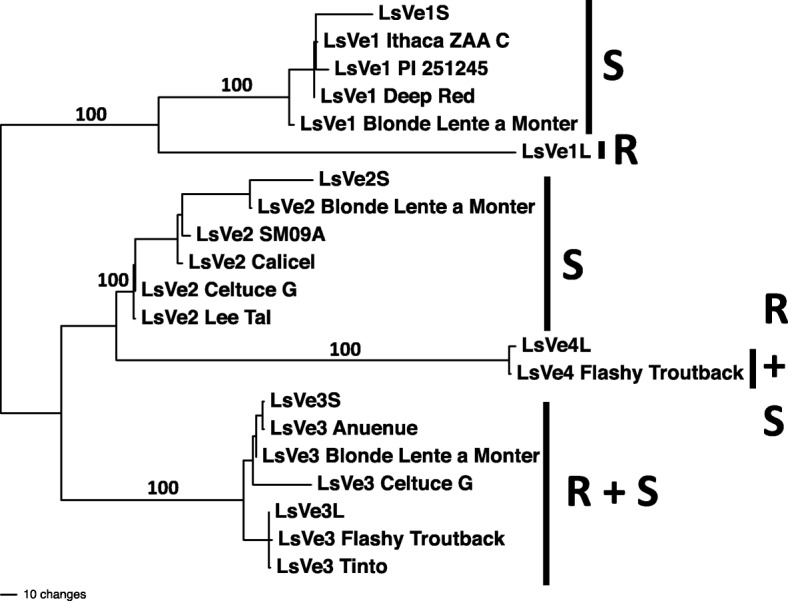
Phylogenetic tree of all LsVe alleles found in this study. Only one representative of each allele is included in the tree. Shown is one of 322 most parsimonious trees measuring 1120 steps; the tree is midpoint rooted. Taxon names consist of gene name followed by accession, except for cultivars La Brillante (L) and Salinas (S), where allele names are given. Numbers by the branches are bootstrap supports above 70%. Branch lengths are proportional to the number of changes occurring along the branches, the scale is given at the bottom. Association of alleles with resistance or susceptibility is indicated on the right side of vertical bars with R and S, respectively. All alleles are shown in Additional file 2
There were substantial differences in frequencies of LsVe alleles among lettuce horticultural types. For example, all tested Iceberg cultivars had the identical combination of three alleles, LsVe1S, LsVe2S, and LsVe3S, while none of the genotyped Latin accessions contained any of these alleles (Table 3). Only six combinations of LsVe alleles were detected in 62 accessions with sequenced genomes. The five combinations were found in susceptible accessions: 40 accessions with LsVe1S, LsVe2S, and LsVe3S; four accessions with LsVe4L, LsVe1S, and LsVe2S; one accession with LsVe3L, LsVe4L, and LsVe2S; one accession with LsVe3L, LsVe4L, LsVe1S, and LsVe2S; and one accession with LsVe2S and LsVe3S. In addition, one combination of alleles was found in all (15) resistant accessions LsVe1L, LsVe3L, and LsVe4L (Table 3).
Table 3.
LsVe allele frequencies in lettuce horticultural types included in this study. The number of accessions examined for the presence of the particular allele is given in parentheses
| Type | LsVe1L | LsVe3L | LsVe4L | LsVe1S | LsVe2S | LsVe3S |
|---|---|---|---|---|---|---|
| Batavia | 0.43 (7) | 0.50 (4) | 0.43 (7) | 0.25 (4) | 0.50 (4) | 0.50 (4) |
| Butterhead | 0.21 (14) | 0.43 (7) | 0.79 (14) | 0.71 (7) | 0.71 (7) | 0.14 (7) |
| Green leaf | 0.11 (18) | 0.25 (8) | 0.11 (18) | 0.75 (8) | 0.75 (8) | 0.75 (8) |
| Iceberg | 0 (47) | 0 (16) | 0 (47) | 1.00 (16) | 1.00 (16) | 1.00 (16) |
| Latin | 0.86 (7) | 1.00 (3) | 0.86 (7) | 0 (3) | 0 (3) | 0 (3) |
| Oil | 0 (4) | 0 (2) | 0 (4) | 1.00 (2) | 1.00 (2) | 1.00 (2) |
| Red leaf | 0.27 (15) | 0.43 (7) | 0.33 (15) | 0.57 (7) | 0.57 (7) | 0.57 (7) |
| Romaine | 0.06 (36) | 0.23 (13) | 0.11 (36) | 0.77 (13) | 0.85 (13) | 0.69 (13) |
| Stem | 0.33 (3) | 0.50 (2) | 0.33 (3) | 0.50 (2) | 0.50 (2) | 0.50 (2) |
| Lactuca serriola | 0 (1) | NDa | 1.00 (1) | ND | ND | ND |
aNot determined
Phylogenetic analyses of Ve-encoded amino acid sequences from cultivars La Brillante and Salinas with tomato Ve1 and homologs from other Asteraceae, Cannabaceae, Malvaceae, and Solanaceae species showed that these lettuce Ve alleles were monophyletic with 99% bootstrap support. Two equally parsimonious trees were obtained and the tree length was 3492 steps (Additional file 3).
PCR- based screening for LsVe1L and LsVe4L in 90 additional accessions
In order to determine the prevalence of candidate resistance alleles, 90 additional accessions were screened for the presence of LsVe1L and LsVe4L using allele-specific PCR (Additional file 4 and Additional file 5). LsVe1L-specific products were detected in six accessions and LsVe4L-specific products in 12 accessions. All accessions with LsVe1L also had LsVe4L (Table 2). LsVe3L screening was not performed because of the premature stop codons as mentioned above (Fig. 2).
Diagnostic PCR assays for race 1 resistance based on LsVe1L
LsVe1L and LsVe1S only share 89.5% sequence similarity differing by 358 single nucleotide polymorphisms (SNPs) and two indels. The overall ratio of non-synonymous (dN = 0.0754) to synonymous (dS = 0.2324) substitutions between the two alleles was 0.3246, providing no evidence for diversifying selection. These SNPs and indels provide multiple possibilities for allele-specific PCR-based assays. A PCR assay that selectively amplified LsVe1L was developed and validated as a marker for resistance to race 1 using selected accessions of lettuce with known Ve genotypes and resistance phenotypes. All PCR results were consistent with phenotypic observations and genome sequence data (Fig. 5). The LsVe1L allele was detected in 21 of the 152 tested accessions and all 21 were resistant to V. dahliae race 1 in field experiments (Table 2). Wilt symptoms were not observed on any of the 21 accessions with the exception of cultivar Plymouth, where two out of 30 plants showed root discoloration. However, the pathogen isolated from tap root tissue of cultivar Plymouth lacked the V. dahliae race 1 determinant Ave1 [8], thus revealing that the symptoms were not caused by V. dahliae race 1 (Additional file 6). LsVe4L was present in all resistant but also some susceptible accessions (Table 2). This is consistent with LsVe1L rather than LsVe4L conferring resistance to V. dahliae race 1.
Fig. 5.
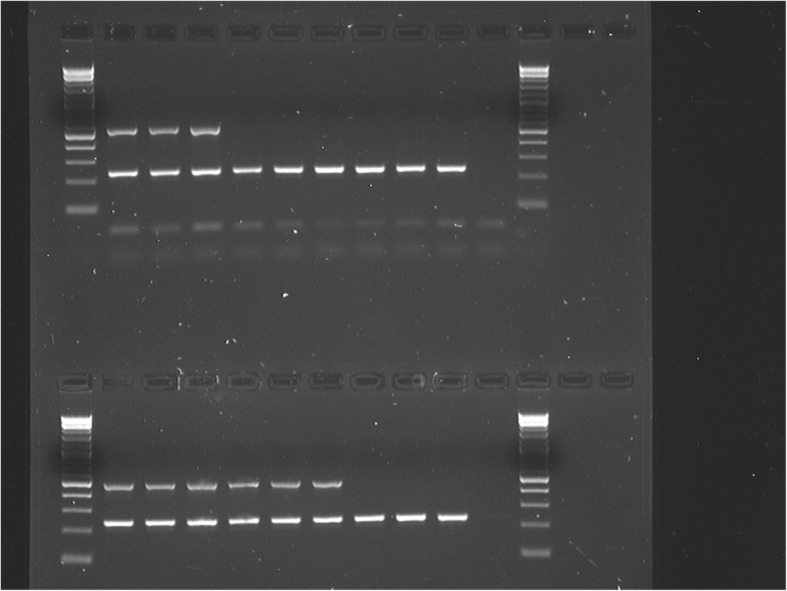
LsVe1L specific PCR assay is allele-specific. Shown are results of LsVe1L-specific PCR assays with selected lettuce accessions with known LsVe genotypes and resistance phenotypes. Resistance (R) and susceptibility (S) is indicated by capital letters for each accession. In all cases, the outcomes of the PCR assays were as expected from genome sequencing. Amplicon sizes are indicated by > and correspond to 200 and 500 bp. Lane numbers are: 1. 2-log ladder, 2. cultivar Balady Banha (Ve genotype: LBVe1, LBVe4, LBVe3), 3. cultivar Lolla Rossa (LBVe1, LBVe4, LBVe3), 4. cultivar Plymouth (LBVe1, LBVe4, LBVe3), 5. cultivar Cobham Green (LBVe4, LBVe3, SVe1, SVe4), 6. cultivar Lee Tal (LBVe4, SVe1, SVe2), 7. cultivar Margarita (LBVe4, SVe1, SVe2), 8. cultivar Anuenue (SVe2, SVe3), 9. cultivar Blonde Lente a Monter (SVe1, SVe2, SVe3), 10. cultivar Primus (SVe1, SVe2, SVe3), 11. negative control, and 12. 2-log ladder. PCR conditions are described in Table 4
Discussion
We tested 149 accessions of cultivated lettuce and a single accession of L. serriola in field experiments. Horticultural types with the greatest number of tested accessions were iceberg (46) and romaine (36) because they are the predominant types grown in the U.S. [1]. Despite the largest number of tested accessions, none of the iceberg cultivars were resistant to Verticillium wilt. This observation complements results from a previous study that tested accessions from multiple horticultural types for resistance to V. dahliae race 1 [19]. Therefore, development of modern iceberg-type cultivars with resistance to V. dahliae race 1 is one of the top priorities for public and private breeding efforts. USDA-ARS in Salinas released iceberg breeding lines [20–22] with their resistance derived from cultivar La Brillante. Cultivar La Brillante is a Batavia type lettuce with a small, round head that is less dense than those of modern iceberg cultivars. Because of the certain phenotypic similarities in the shape of heads, fewer backcrosses are usually needed to develop true to type iceberg cultivars when introgressing desirable genes from Batavia accessions than would be needed if those genes were introgressed from non-heading types of lettuces. Our current analyses showed that besides cultivar La Brillante, another Batavia cultivar (cultivar Laura) can also be used for a relatively rapid development of iceberg cultivars with resistance to V. dahliae race 1. Both of these cultivars contain the same combination of LsVe alleles (LsVe1L, LsVe3L, and LsVe4L).
Only two out of 36 romaine accessions were resistant to the disease in field experiments. One of the resistant accessions, cultivar Annapolis, is a dark red lettuce with a relatively small and light head that is usually grown for baby leaf production and is therefore harvested at early stages of development. The other resistant cultivar was Defender, which is green. Origin of resistance in this cultivar is unknown because it was developed through open pollination [23]. A high frequency (87.5%) of resistance to the disease was found in Latin type accessions that phenotypically resemble a small romaine lettuce with more pliable and oily leaves. Because of the phenotypic similarity between romaine and Latin types, Latin-type accessions may also be used for a relatively rapid development of romaine cultivars with resistance to V. dahliae race 1. Both romaine cultivars and three sequenced Latin cultivars (Eruption, Pavane, and Little Gem) that are resistant to the disease contain an identical combination of LsVe alleles (LsVe1L, LsVe3L, and LsVe4L).
Substantially different frequencies of LsVe1L alleles (Table 3) and resistant phenotypes (Table 1) in different horticultural types of lettuce are not unexpected considering that comparable differences were previously described for other monogenically inherited traits, such as resistance to lettuce dieback [24] and sensitivity to triforine [25]. Differences in the frequency of specific alleles among horticultural types are likely caused by the breeding approach that is used to develop lettuce cultivars. Only a few elite progenitors or founder lettuce cultivars have given rise to most of the modern commercial cultivars [26]. Each of these progenitors is frequently found in pedigrees of cultivars of the same horticultural type. Additionally, new cultivars are mainly developed by recurrent breeding within small pools of closely related germplasm of the same type [27]. Therefore, alleles present in an original progenitor(s) of a certain type are found in high frequency in cultivars of the same type, but may be absent or present in low frequency in cultivars of other horticultural types.
Our data are consistent with the LsVe1 gene identified in the cultivar La Brillante being involved in resistance to V. dahliae race 1 in lettuce. Among the 152 accessions included in this study, 21 were resistant to V. dahliae race 1 and all 21 contained the LsVe1L allele; this allele was not present in any of the susceptible accessions. The other La Brillante Ve alleles, LsVe3L and LsVe4L, were also present in all the resistant accessions, but they also occurred in two and twelve susceptible accessions, respectively. Therefore, LsVe1L is the strongest candidate as being required for resistance to V. dahliae race 1 in lettuce, although our data do not exclude LsVe3L or LsVe4L from also being involved similarly as in tomato [12]. Complementation and knock-out studies are still required to determine the functional basis of LsVe-mediated resistance to V. dahliae race 1.
The function and the significance of the differences between the LsVe1L and LsVe1S alleles (Additional file 7) remains to be investigated. The proteins encoded by LsVe1L and LsVe1S have the same domain organization, including the 37 extracellular, leucine-rich repeats separated by a short spacer region, as in previously characterized functional Ve proteins in other species [11, 18]. However, in addition to sequence diversity in the extracellular LRR domain, LsVe1L has an additional C-terminal transmembrane domain as compared to Ve1 and Ve2 in tomato, suggesting that maybe LsVe1L crosses the membrane three times and terminates with a non-cytoplasmic domain instead of a cytoplasmic domain.
The distribution of disease incidence in susceptible accessions (from 0.07 to 1.00) and across horticultural types (> 0.98 in stem types, but only 0.17 in a single susceptible Latin) indicates a possible presence of a modifying factor or factors that affect disease incidence. Our data do not exclude the possibility of interactions between two or more Ve genes, similar to those reported in tomato [12]. A more detailed study of accessions with different frequencies of disease incidence and allelic compositions is needed to elucidate the basis of variation in disease incidence.
Conclusions
There is a critical lack of iceberg and romaine type cultivars with resistance to V. dahliae race 1. Application of molecular markers can accelerate the lettuce breeding process while improving selection accuracy [28]. Therefore, the development of molecular marker assays for identification of desired genotypes is highly sought-after. The LsVe1L-specific PCR assay developed in this study can be used for the selection of lettuce genotypes with resistance to V. dahliae race 1. Application of this assay allows identification of resistant genotypes in early stages of plant development (or at a seed-level) without time- and labor-intensive testing of plants in the field. This molecular marker is a valuable addition to the tools available to breeders when developing improved cultivars of lettuce.
Methods
Lettuce accessions used for genome sequencing and PCR analysis
A total of 152 lettuce accessions representing all major types of cultivated lettuce (Batavia, butterhead, iceberg, Latin, leaf, oil, romaine, and stem) were analyzed (Table 2). The majority of accessions (111) were from the United States Department of Agriculture, Agricultural Research Service (USDA-ARS) lettuce collections at Salinas, California; the remaining accessions were from a variety of sources (Table 2), including Salinas, the previously sequenced cultivar [16]. When an accession was obtained from more than one source, each provenance was considered separately in the analyses.
Pathogenicity tests
Experiments were conducted in a field infested with V. dahliae race 1 [16] located at the USDA-ARS station in Salinas, California. One hundred and fifty accessions were direct-seeded in a randomized complete block design with three replications. The original seed batches of previously sequenced cultivars Salinas [17] and La Brillante (this publication) were not available for field tests; therefore, seed batches used in field tests are shown as separate entries (Table 2). Each plot was 7 m long and consisted of two seed lines on 1 m wide beds standard for lettuce production in coastal California. Plant spacing was approximately 28 cm between seed lines and 30 cm between plants within a seed line. All field experiments were maintained using standard cultural practices for coastal California lettuce production. Plants were evaluated for disease incidence after reaching harvest maturity. Unless indicated otherwise, ten plants from each plot were uprooted and visually evaluated. Disease incidence was assessed by cutting taproots longitudinally and recording the number of plants exhibiting the yellowish-brown discoloration of root vascular tissues that is typical of Verticillium wilt. Absence of V. dahliae race 1 in cultivars with race 1-resistant genotype was confirmed by plating surface-sterilized symptomatic root tissue on NP-10 semi-selective agar medium [29] and PCR screening any resulting isolates with Ave1-specific primers [8]. Three additional experiments were performed in the same field to confirm phenotypic observations. These experiments comprised only a subset of accessions that were either symptomless in the first experiment or were used as susceptible checks. Disease incidence values from all four experiments were combined and used for statistical analyses with JMP 14.2 (SAS Institute, Cary, NC, USA).
DNA extraction
DNA was extracted using FastDNA SPIN kit (MP Biomedicals, Solon, OH, USA) for most lettuce accessions and with the CTAB method [30] for La Brillante accession 10G364–1. For the FastDNA SPIN kit method, up to 100 mg seeds (~ 100 seeds) or freeze-dried leaf tissue was crushed in liquid nitrogen with a mortar and pestle, and DNA was extracted according to the manufacturer’s instructions for plant material. DNA quality was assessed using gel electrophoresis (0.7% agarose gel), a NanoDrop spectrophotometer, and a Qubit Fluorometer (both Thermo Fisher Scientific, Wilmington, DE, USA) as per the manufacturers’ instructions. DNA extraction for V. dahliae followed the same FastDNA SPIN kit protocol except that CLS-Y solution was used as suggested by the manufacturer.
Genome sequencing and assembly
For La Brillante accession 10G364–1, three genomic libraries were constructed, one with 180 bp insert size (with in-house protocols) and two Nextera (Illumina, San Diego, CA, USA) 2 and 7 kb mate-pair libraries. All libraries were sequenced in an Illumina Hi-Seq 2000 for 100 + 100 paired-end reads. Reads were directly imported into AllPaths-LG v49856 [31] and assembled using default parameters. Both mate-pair libraries were aligned to the AllPaths-LG assembly using BWA v0.7.4 [32] and these alignments were fed into SSpace v3.0 [33] together with the assembly to perform further scaffolding.
For the remaining accessions, DNA was sent to Novogene (Beijing, China) for library preparation (insert size 350 bp) and sequencing on Illumina HiSeq 4000 machines to generate ~ 800 M PE150 reads that provided approximately 25x whole genome coverage. Reads obtained from Novogene were further processed to remove low quality sequences using bbduk from the BBMap suite v33.65 [34]. This removed sequences with a quality score below 20 from both ends of the read and eliminated reads that had less than 50 bp after trimming. Genome assemblies were generated using MEGAHIT version 1.0.6 [35] or MaSuRCA version 2.3.2 [36]. MEGHIT was generally run with default settings and sometimes with meta-sensitive or bulk options in effect. MaSuRCA settings were insert size = 350 and standard deviation = 50, GRAPH_KMER_SIZE = 101, USE_LINKING_MATES = 0, LIMIT_JUMP_COVERAGE = 300, CA_PARAMETERS = cgwErrorRate = 0.15 ovlMemory = 4 GB, KMER_COUNT_THRESHOLD = 1, NUM_THREADS = 40, JF_SIZE = 10,000,000,000, and DO_HOMOPOLYMER_TRIM = 0. Assembly statistics were generated using the shell script stats.sh of BBMap version 37.68 [34].
Ve gene identification and naming
The expressed sequence tag marker QGD8I16.yg.ab1 at the Verticillium resistance 1 (Vr1) locus in lettuce [16] that has sequence similarity to the Ve genes of tomato was used to query the genome assemblies of V. dahliae race 1 susceptible cultivar Salinas [17] (assembly version 8, available at https://genomevolution.org/coge/GenomeInfo.pl?gid=28333) and race 1 resistant cultivar La Brillante, using local nucleotide BLAST v. 2.6 [37]. The LsVe sequences from cultivars La Brillante and Salinas were then used to query the remaining lettuce genome assemblies using BLASTn. Sequence alignments were generated with MAFFT version 7.309 [38, 39] using default settings. Phylogenetic analyses were performed with PAUP 4.0a (build 159) [40] using the maximum parsimony criterion, the heuristic search option, and 10 random addition replicates. Bootstrap branch support was based on 1000 random addition replicates. Default settings were used otherwise. Protein domains were annotated using the InterPro website (https://www.ebi.ac.uk/interpro/) [41]. Codon alignments were subjected to calculation of synonymous and non-synonymous substitution rates with PAL2NAL v. 14 [42].
PCR assays
La Brillante LsVe1L and LsVe4L-specific PCR assays were performed as follows. Each assay was a multiplex assay with two LsVeL-specific primers and a plant DNA control with two additional primers specific to the lettuce 4-hydroxyphenylpyruvate dioxygenase-encoding gene (HPPD), which has been used as the reference gene in real-time PCR assays [43]. PCR conditions and primer sequences are shown in Table 4.
Table 4.
PCR conditions and primer sequences used for LsVe1L and LsVe4L multiplex assays
| Target locusa | Forward primerb | Reverse primerb | Annealing temperaturec | Amplicon sized |
|---|---|---|---|---|
| LsVe1L | 5′-CAA GGG CTC TAT GTC ATT CCT CC | 5′-GAC CCA TGG AAG CTG TTG GAT CT | 60 °C | 569 bp |
| LsVe4L | 5′-CTT GTC CCA GAT AGA GTT GTC CAC C | 5′-CAG ACC CTG GAA ATC TTT GGT TTG A | 57 °C | 505 bp |
| HPPD 1 | 5′-TCC CAA CTC CTC ACA CTC CTT AAT C | 5′-GTA CGG AAC AAA GAG GAA GAG CC | 57 °C or 60 °C | 244 bp |
aThe lettuce HPPD was targeted as a DNA quality control in both the LsVe1L and LsVe4L multiplex assays
bEach 25 μL PCR reaction contained 1.25 μL of each of the four primers at 10 μM each to amplify HPPD plus LsVe1L or HPPD plus LsVe4L, 12.5 μL 2x GoTaq Colorless Master Mix (Promega Corp., Madison, WI), and 7.5 μL DNA template from a 1 ng/μL stock
cThe PCR program consisted of an initial denaturation at 94 °C for 2 min, followed by 32 cycles of denaturation at 94 °C for 10 s, 20 s at the assay-specific annealing temperature, extension at 72 °C for 1 min, and a final extension at 72 °C for 7 min. PCRs were set up on ice under sterile conditions and the thermocycler was preheated to 94 °C before adding the reactions
dPCR products (8 μL each) were run on a 1% agarose gel
Additional files
Alignment of tomato Ve1 and lettuce LsVe alleles. Domains are indicated; eLRR stands for extracellular leucine-rich repeat. Domain information for Ve1 is from [18]. (PDF 589 kb)
LsVe alleles found in this study. Provided are names of contigs or scaffolds, identical representatives included in Fig. 4, and completeness of sequencing coverage. (XLSX 15 kb)
Phylogenetic tree of cultivars La Brillante and Salinas Ve allele amino acid sequences and homologs from other plant families using maximum parsimony. One of two most parsimonious trees is shown measuring 3492 steps; the tree is midpoint rooted. Taxa names consist of species names followed by gene names. GenBank accession numbers are provided for sequences from other studies. Bootstrap supports above 60% are shown by the branches. Branch lengths are proportional to changes along the branches and the scale is provided. (PDF 16 kb)
Results of LsVe1L and LsVe4L PCR screening of 90 lettuce accessions that were not genome sequenced. The legend to lane numbers is in Additional file 5. For each accession, the top gel shows results of the LsVe1L screening, the bottom gel shows the results of the LsVe4L screening. Resistant accessions are marked with an R. Amplicon sizes are indicated by > and correspond to 200 and 500 bp. Size standard used is 2-log ladder. PCR conditions are described in Table 4. (PDF 1577 kb)
Legend to lane numbers in Additional file 4. For each lane, accession name, code, and PCR result are provided. (XLSX 12 kb)
PCR gel demonstrating that Verticillium dahliae strains isolated from symptomatic cultivar Plymouth tap roots did not contain Ave1, the specificity determinant of race 1, and were thus not race 1. Amplicon size marker indicated by > corresponds to 1000 bp. Lane numbers are: 1. 2-log ladder, 2. and 3. Verticillium dahliae strain isolated from symptomatic cultivar Plymouth tap root, 4. and 5. V. dahliae race 2 control strain Ls.17, 6. and 7. V. dahliae race 1 control strain Ls.16, and 8. negative control. (PDF 4236 kb)
Nucleotide sequences of six LsVe alleles from cultivars La Brillante (L) and Salinas (S). (DOCX 17 kb)
Acknowledgements
The authors would like to thank Richard Bostock, UC Davis, for providing lab space; Alex Kozik, UC Davis, for help with analyses; Jose Orozco, Rebecca Zhao, and Sharon Benzen, USDA Salinas, for the excellent technical assistance with field experiments. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Abbreviations
- HPPD
4-hydroxyphenylpyruvate dioxygenase-encoding gene
- LsVe1, LsVe2, LsVe3, and LsVe4
Lactuca sativa homologs of tomato Ve genes for resistance to Verticillium dahliae race 1
- ORF
Open reading frame
- RLP
Receptor-like proteins
- SNP
Single nucleotide polymorphisms
- Vr1
Verticillium resistance 1 locus
Authors’ contributions
PI, KVS, and IS conceived the study and obtained funding; PI carried out laboratory experiments and drafted the paper; PI, KVS, and IS carried out field experiments; PI and DL collected seeds and prepared samples for sequencing; PI, DL, and SRCW analyzed the sequenced data and generated the genome assemblies; PI and MC identified the Ve genes and developed the PCR-based assays; PI, MC, SRCW, RWM, KVS, and IS contributed to data interpretation and writing the paper. All authors read and approved the final manuscript.
Funding
External funding for this project was provided by the California Department of Food and Agriculture Specialty Crops Block Grants Program, Agreement Number SCB15048. The funding organization used internal and external reviewers to review the proposed study design, workplan, and budget before funding was provided. The funder had no role in the study design, data collection and analysis, or preparation of the manuscript.
Availability of data and materials
Data and results generated and analyzed during this study are included in this published article and its supplementary information files. Nucleotide sequences of six LsVe alleles from cultivars La Brillante and Salinas are provided in Additional file 7. Sequence data of 61 genomes generated and analyzed during the current study are available in GenBank at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA478460.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have approved the manuscript for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Patrik Inderbitzin and Marilena Christopoulou contributed equally to this work.
Contributor Information
Patrik Inderbitzin, Email: prin@ucdavis.edu.
Marilena Christopoulou, Email: marilena.christop@gmail.com.
Dean Lavelle, Email: dolavelle@ucdavis.edu.
Sebastian Reyes-Chin-Wo, Email: sreyeschinwo@ucdavis.edu.
Richard W. Michelmore, Email: rwmichelmore@ucdavis.edu
Krishna V. Subbarao, Email: kvsubbarao@ucdavis.edu
Ivan Simko, Email: Ivan.Simko@ars.usda.gov.
References
- 1.Simko I, Hayes RJ, Mou B, McCreight JD. Lettuce and spinach. In: Smith S, Diers B, Specht J, Carver B, editors. Yield gains in major US field crops. Madison: CSSA Special Publication; 2014. pp. 53–86. [Google Scholar]
- 2.Atallah ZK, Hayes RJ, Subbarao KV. Fifteen years of Verticillium wilt of lettuce in America's salad bowl: a tale of immigration, subjugation, and abatement. Plant Dis. 2011;95(7):784–792. doi: 10.1094/PDIS-01-11-0075. [DOI] [PubMed] [Google Scholar]
- 3.Inderbitzin P, Subbarao KV. Verticillium wilt. In: Subbarao KV, Davis RM, Gilbertson RL, Raid RN, editors. Compendium of lettuce diseases and pests. Second. St. Paul: APS Press; 2017. pp. 50–54. [Google Scholar]
- 4.Pegg GF, Brady BL. Verticillium wilts. Wallingford: CABI Publishing; 2002. [Google Scholar]
- 5.Inderbitzin P, Subbarao KV. Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology. 2014;104(6):564–574. doi: 10.1094/PHYTO-11-13-0315-IA. [DOI] [PubMed] [Google Scholar]
- 6.Vallad GE, Qin Q-M, Grube R, Hayes RJ, Subbarao KV. Characterization of race-specific interactions among isolates of Verticillium dahliae pathogenic on lettuce. Phytopathology. 2006;96(12):1380–1387. doi: 10.1094/PHYTO-96-1380. [DOI] [PubMed] [Google Scholar]
- 7.Gurung S, Short DPG, Atallah ZK, Subbarao KV. Clonal expansion of Verticillium dahliae in lettuce. Phytopathology. 2014;104(6):641–649. doi: 10.1094/PHYTO-10-13-0282-R. [DOI] [PubMed] [Google Scholar]
- 8.de Jonge R, van Esse PH, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci U S A. 2012;109(13):5110–5115. doi: 10.1073/pnas.1119623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci U S A. 2001;98(11):6511–6515. doi: 10.1073/pnas.091114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Fiers M, Ellendorff U, Wang Z, de Wit PJ, Angenent GC, et al. The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit Rev Plant Sci. 2010;29(5):285–299. doi: 10.1080/07352689.2010.502082. [DOI] [Google Scholar]
- 11.Song Y, Zhang Z, Seidl MF, Majer A, Jakse J, Javornik B, et al. Broad taxonomic characterization of Verticillium wilt resistance genes reveals an ancient origin of the tomato Ve1 immune receptor. Mol Plant Pathol. 2017;18(2):195–209. doi: 10.1111/mpp.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazar Ross N., Xu Xin, Kurosky Alexander, Robb Jane. Antagonistic function of the Ve R-genes in tomato. Plant Molecular Biology. 2018;98(1-2):67–79. doi: 10.1007/s11103-018-0764-3. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Li N, Ma X, Gupta VK, Zhang D, Li T, et al. The ectopic overexpression of the cotton Ve1 and Ve2-homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front Plant Sci. 2017;8:844. [DOI] [PMC free article] [PubMed]
- 14.Simko I, Costanzo S, Haynes KG, Christ BJ, Jones RW. Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor Appl Genet. 2004;108(2):217–224. doi: 10.1007/s00122-003-1431-9. [DOI] [PubMed] [Google Scholar]
- 15.Simko I, Haynes KG, Ewing EE, Costanzo S, Christ BJ, Jones RW. Mapping genes for resistance to Verticillium albo-atrum in tetraploid and diploid potato populations using haplotype association tests and genetic linkage analysis. Mol Gen Genomics. 2004;271(5):522–531. doi: 10.1007/s00438-004-1010-z. [DOI] [PubMed] [Google Scholar]
- 16.Hayes R, McHale L, Vallad G, Truco M, Michelmore R, Klosterman S, et al. The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. TAG Theor Appl Genet. 2011;123(4):509–517. doi: 10.1007/s00122-011-1603-y. [DOI] [PubMed] [Google Scholar]
- 17.Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun. 2017;8:14953. [DOI] [PMC free article] [PubMed]
- 18.Zhang Z, Song Y, Liu C-M, Thomma BPHJ. Mutational analysis of the Ve1 immune receptor that mediates Verticillium resistance in tomato. PLoS One. 2014;9(6):e99511. doi: 10.1371/journal.pone.0099511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes RJ, Vallad GE, Qin Q-M, Grube RC, Subbarao KV. Variation for resistance to Verticillium wilt in lettuce (Lactuca sativa L.) Plant Dis. 2007;91(4):439–445. doi: 10.1094/PDIS-91-4-0439. [DOI] [PubMed] [Google Scholar]
- 20.Hayes Ryan J., Sandoya German, Mou Beiquan, Simko Ivan, Subbarao Krishna V. Release of Three Iceberg Lettuce Populations with Combined Resistance to Two Soilborne Diseases. HortScience. 2018;53(2):247–250. doi: 10.21273/HORTSCI12559-17. [DOI] [Google Scholar]
- 21.Hayes Ryan J., Maruthachalam Karunakaran, Vallad Gary E., Klosterman Steven J., Simko Ivan, Luo Yaguang, Subbarao Krishna V. Iceberg Lettuce Breeding Lines with Resistance to Verticillium Wilt Caused by Race 1 Isolates of Verticillium dahliae. HortScience. 2011;46(3):501–504. doi: 10.21273/HORTSCI.46.3.501. [DOI] [PubMed] [Google Scholar]
- 22.Simko Ivan, Hayes Ryan J., Bull Carolee T., Mou Beiquan, Luo Yaguang, Trent Mark A., Atallah Amy J., Ryder Edward J., Sideman Rebecca G. Characterization and Performance of 16 New Inbred Lines of Lettuce. HortScience. 2014;49(5):679–687. doi: 10.21273/HORTSCI.49.5.679. [DOI] [Google Scholar]
- 23.Wehner Todd C. Vegetable Cultivar Descriptions for North America List 26 2002. HortScience. 2002;37(1):15–78. doi: 10.21273/HORTSCI.37.1.15. [DOI] [Google Scholar]
- 24.Simko I, Pechenick DA, McHale LK, Truco MJ, Ochoa OE, Michelmore RW, et al. Association mapping and marker-assisted selection of the lettuce dieback resistance gene Tvr1. BMC Plant Biol. 2009;9(1):135. doi: 10.1186/1471-2229-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simko I, Hayes RJ, Truco MJ, Michelmore RW. Mapping a dominant negative mutation for triforine sensitivity in lettuce and its use as a selectable marker for detecting hybrids. Euphytica. 2011;182(2):157–166. doi: 10.1007/s10681-011-0407-0. [DOI] [Google Scholar]
- 26.Mikel Mark A. Genealogy of Contemporary North American Lettuce. HortScience. 2007;42(3):489–493. doi: 10.21273/HORTSCI.42.3.489. [DOI] [Google Scholar]
- 27.Mikel MA. Genetic composition of contemporary proprietary US lettuce (Lactuca sativa L.) cultivars. Genet Resour Crop Evol. 2013;60(1):89–96. doi: 10.1007/s10722-012-9818-6. [DOI] [Google Scholar]
- 28.Simko I. Translational Genomics for Crop Breeding. Chichester, UK: John Wiley & Sons Ltd; 2013. Marker-Assisted Selection for Disease Resistance in Lettuce; pp. 267–289. [Google Scholar]
- 29.Kabir Z, Bhat RG, Subbarao KV. Comparison of media for recovery of Verticillium dahliae from soil. Plant Dis. 2004;88(1):49–55. doi: 10.1094/PDIS.2004.88.1.49. [DOI] [PubMed] [Google Scholar]
- 30.Doyle J, Doyle JL. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 1987;19(11):11–15. [Google Scholar]
- 31.Butler J., MacCallum I., Kleber M., Shlyakhter I. A., Belmonte M. K., Lander E. S., Nusbaum C., Jaffe D. B. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Research. 2008;18(5):810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate long-read alignment with burrows–wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2010;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 34.Bushnell B: BBMap. Retrieved from sourceforge.net/projects/bbmap/. 2016.
- 35.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 36.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Katoh K, Misawa K, Ki K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 41.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borowski JM, Galli V, da Silva Messias R, Perin EC, Buss JH, e Silva SDA, et al. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta. 2014;239(6):1187–1200. doi: 10.1007/s00425-014-2041-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of tomato Ve1 and lettuce LsVe alleles. Domains are indicated; eLRR stands for extracellular leucine-rich repeat. Domain information for Ve1 is from [18]. (PDF 589 kb)
LsVe alleles found in this study. Provided are names of contigs or scaffolds, identical representatives included in Fig. 4, and completeness of sequencing coverage. (XLSX 15 kb)
Phylogenetic tree of cultivars La Brillante and Salinas Ve allele amino acid sequences and homologs from other plant families using maximum parsimony. One of two most parsimonious trees is shown measuring 3492 steps; the tree is midpoint rooted. Taxa names consist of species names followed by gene names. GenBank accession numbers are provided for sequences from other studies. Bootstrap supports above 60% are shown by the branches. Branch lengths are proportional to changes along the branches and the scale is provided. (PDF 16 kb)
Results of LsVe1L and LsVe4L PCR screening of 90 lettuce accessions that were not genome sequenced. The legend to lane numbers is in Additional file 5. For each accession, the top gel shows results of the LsVe1L screening, the bottom gel shows the results of the LsVe4L screening. Resistant accessions are marked with an R. Amplicon sizes are indicated by > and correspond to 200 and 500 bp. Size standard used is 2-log ladder. PCR conditions are described in Table 4. (PDF 1577 kb)
Legend to lane numbers in Additional file 4. For each lane, accession name, code, and PCR result are provided. (XLSX 12 kb)
PCR gel demonstrating that Verticillium dahliae strains isolated from symptomatic cultivar Plymouth tap roots did not contain Ave1, the specificity determinant of race 1, and were thus not race 1. Amplicon size marker indicated by > corresponds to 1000 bp. Lane numbers are: 1. 2-log ladder, 2. and 3. Verticillium dahliae strain isolated from symptomatic cultivar Plymouth tap root, 4. and 5. V. dahliae race 2 control strain Ls.17, 6. and 7. V. dahliae race 1 control strain Ls.16, and 8. negative control. (PDF 4236 kb)
Nucleotide sequences of six LsVe alleles from cultivars La Brillante (L) and Salinas (S). (DOCX 17 kb)
Data Availability Statement
Data and results generated and analyzed during this study are included in this published article and its supplementary information files. Nucleotide sequences of six LsVe alleles from cultivars La Brillante and Salinas are provided in Additional file 7. Sequence data of 61 genomes generated and analyzed during the current study are available in GenBank at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA478460.