Abstract
Advances in mouse neural circuit genetics, brain atlases, and behavioral assays provide a powerful system for modeling the genetic basis of cognition and psychiatric disease. However, a critical limitation of this approach is how to achieve concordance of mouse neurobiology with the ultimate goal of understanding the human brain. Previously, the common marmoset has shown promise as a genetic model system toward the linking of mouse and human studies. However, the advent of marmoset transgenic approaches will require an understanding of developmental principles in marmoset compared to mouse. In this study, we used gene expression analysis in marmoset brain to pose a series of fundamental questions on cortical development and evolution for direct comparison to existing mouse brain atlas expression data. Most genes showed reliable conservation of expression between marmoset and mouse. However, certain markers had strikingly divergent expression patterns. The lateral geniculate nucleus and pulvinar in the thalamus showed diversification of genetic organization between marmoset and mouse, suggesting they share some similarity. In contrast, gene expression patterns in early visual cortical areas showed marmoset-specific expression. In prefrontal cortex, some markers labeled architectonic areas and layers distinct between mouse and marmoset. Core hippocampus was conserved, while afferent areas showed divergence. Together, these results indicate that existing cortical areas are genetically conserved between marmoset and mouse, while differences in areal parcellation, afferent diversification, and layer complexity are associated with specific genes. Collectively, we propose that gene expression patterns in marmoset brain reveal important clues to the principles underlying the molecular evolution of cortical and cognitive expansion.
Introduction
During primate evolution, the cerebral cortex dramatically increased in the size and number of functional areas underlying the acquisition of higher cognitive functions including language, episodic memory, and executive control (Kaas, 2008). However, the genetic mechanism by which the cortex developed new areas remains a question of fundamental importance toward understanding higher brain functions and diseases.
The most commonly used “primate model species” for studying brain anatomy and behavior are Old World monkeys, typically rhesus monkeys (Macaca mulatta), and long-tailed (Macaca fascicularis) macaques because of resource availability and historical descriptions of these species (Rakic, 1988; Takahata et al., 2006, 2009; Watakabe et al., 2006, 2007). However, the disadvantages of using such monkeys for experimental manipulations include their large body size, inefficient fertility, and slow development to sexual maturity. Previously, the common marmoset (Callithrix jacchus), a small New World monkey species, has emerged as a promising model for scientific studies including the investigation of neurological disorders, basic and behavioral neuroscience, neuroimaging, stem cell research, drug toxicology, immunity and autoimmune diseases, and reproductive biology (Poswillo et al., 1972; Mansfield, 2003; Iwanami et al., 2005a,b; Sasaki et al., 2005; Leuner et al., 2007; Tomioka et al., 2010; Hikishima et al., 2011; Yamazaki et al., 2011). The marmoset has a number of advantageous characteristics as an experimental animal, including a small body size, high fertility, and early sexual maturity (they reach adulthood at 1.5–2 years). The marmoset cortex is relatively smooth, but the gyrencephalic and cortical sheet is divided into functionally distinct cortical areas, as in Old World monkeys (Rosa and Tweedale, 2005) and is routinely used in studies of higher cognitive functions and social communication (Bonnin et al., 2007; Eliades and Wang, 2008). Therefore, marmosets are considered to be a good experimental model animal to understand the evolution of brain development and function. Moreover, transgenic marmosets have been generated previously, demonstrating the possibility of gene manipulation in this species (Sasaki et al., 2009). However, if the marmoset is to be used as a productive genetic model for bridging rodent and primate neurobiology, the understanding of the relationship of brain genomic development between the two species is critically needed. Moreover, a comparison of marmoset genome information with recent information from gene expression studies in the developing human cortex will provide strong tools to understand primate-specific brain evolution (Johnson et al., 2009; Kang et al., 2011).
In this study, we compared molecules expressed in specific cortical areas, layers, and thalamic nuclei in mouse brain with their expression profiles in marmoset brain regions that are important for cognitive functions, including the prefrontal cortex (PFC) and visual cortex, and highly connected regions, including the dorsal lateral geniculate nucleus (dLGN), pulvinar, and hippocampus. These results represent the first detailed study of gene expression patterns in marmoset brain and anatomical comparison to mouse brain.
Materials and Methods
Animals.
Experiments were performed in 10 (five male, five female) neonatal common marmosets [postnatal day (P0)] and four embryonic marmosets (3 months old) derived from a breeding colony at the RIKEN Brain Science Institute. Neonatal marmosets were anesthetized with a lethal dose of pentobarbitone (100 mg/kg), and after three failed attempts to elicit a foot withdrawal reflex, the animals were transcardially perfused with 4% paraformaldehyde (PFA) in PBS. Adult pregnant common marmosets were anesthetized, and embryos were collected by cesarean section. The brains were removed, fixed for 3 d in 30% sucrose/4% PFA, and sectioned in the coronal plane on a Leica sledge microtome at 20 μm for postnatal brain and 28 μm for embryonic brain. Sections were individually mounted on slides and processed for in situ hybridization (ISH) to visualize expression of various genes. The use of the marmosets was in accord with the guidelines approved by the RIKEN Institutional Animal Care and Use Committee.
Probes.
Primers for PCR targeting common marmoset genes were designed with software (Primer3, version 0.4.0) based on the common marmoset genome sequence (Table 1). Total RNAs were isolated from frozen embryonic and postnatal marmoset brain, collected in Trizol reagent (Invitrogen). cDNA was generated from total RNA using the prime script first-strand cDNA synthesis kit (Takara) according to the manufacturer's recommended procedure.
Table 1.
Primer sequences and the position of the genes used as probes
| Gene | Encoding protein product | Probe length | Forward primer | Reverse primer | NCBI gene ID |
|---|---|---|---|---|---|
| Btbd3 | BTB (POZ) domain containing 3 | 767 bp | TTCACTCGATTAGGGAGAGAGC | AGGAAGATGTCGTTGGTCTCAT | XM_002747367 |
| Cdh6 | Cadherin 6 | 528 bp | GGACAGCCCTATTTTTCAGTTG | CTCATCTACATCCTCCACCACA | XM_002745091 |
| Cdh8 | Cadherin 8 | 637 bp | TGCATGAATTTCCAAGTGGTAG | TAAGCAAATCACCACGAAAATG | XM_002761039 |
| Cplx3 | Complexin3 | 691 bp | TACGGAATTCAGAACGAGACAGATGAGAGCCAG | ACGTGGATCCCTACAGGAACAGAACAGAAATTG | XM_002753343 |
| CTGF | Connective tissue growth factor | 694 bp | AGCAGCTGCAAGTACCAGTGTA | TGTCTCCGTACATCTTCCTGTG | XM_002806723 |
| EphA4 | Eph receptor A4 | 704 bp | TTTACCTGGCTTTTCAGGATGT | AGCCCAGATTTCAAAGGTGTAA | XM_002749827 |
| EphA5 | Eph receptor A5 | 700 bp | CCCCTATCCACACATACCAAGT | TAACTGTGAGGTGGACATTTGC | XM_002745799 |
| EphA6 | Eph receptor A6 | 743 bp | ACGAGAGTTTTACCCAGATGGA | TATCACGGAACTGTTGATCAGG | XM_002761301 |
| EphA7 | Eph receptor A7 | 558 bp | CCACAGAACCTCATTTTCAACA | AGCAGTAAAAGCTCGAATCTGG | XM_002746832 |
| EFNA5 | Ephrin A5 | 538 bp | CAGATTCCAGAGGGGTGACTAC | GAACAGTAGGATTGCCAAAAGG | XM_002744672 |
| ER81 | Ets variant gene 1 | 767 bp | GTGTTCTGGTGGAGGACTTTTC | CATACAGCCTTCTGTTCTGCTG | XM_002751560 |
| Foxp2 | Forkhead box P2 | 794 bp | CTTAGCCTGCACAAGTGTTTTG | TGAACCCTTAAGAGCCACATTT | XM_002751710 |
| GFRalpha1 | Glial cell line derived neurotrophic factor family receptor alpha 1 | 932 bp | CTGAAGCAGAAGTCGCTCTACA | GTGGGAATTTCATTCTCAGACC | XM_002756616 |
| Kitl | Kit ligand | 656 bp | CTCCAGAACAGCTAAACGGAGT | CTGTCATTCCTAAGGGAGTTGG | XM_002752832 |
| Lhx9 | LIM homeobox protein 9 | 652 bp | CCACTTGCAACAAGACTCTGAC | TTAGAGGTCACGGCTGTCACTA | XM_002760614 |
| NR1D1 | Nuclear receptor subfamily 1, group D, member 1 | 638 bp | TGATTCCCCCTACACACTCTCT | GACACTTCTTGAAGCGACACTG | XM_002806862 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 611 bp | TCGCTGTCAGTACTGTCGATTT | CGTGTCTCTCTGTGACCATAGC | XM_002749430 |
| Ntng2 | Netrin G2 | 627 bp | GAATGTTAGGCTTCCACCAAAG | CTTGGGATACTCCTTCCAAGTG | XM_002751158 |
| Relin | Reelin | 650 bp | AGTTTCCAAGGTGATGATTGCT | TCTTCATGGGTATCGCCTAAGT | XM_002751684 |
| RORalpha | RAR-related orphan receptor alpha | 633 bp | GACAGCTTGTATGCAGAAGTGC | CACCACCTCTAGAGAACCTGCT | XM_002753196 |
| Satb2 | Special AT-rich sequence binding protein 2 | 657 bp | CCACCACAGTACTCCAATCAGA | ATCTCCTGTTGGATCTCGTCAT | XM_002749606 |
| Sema6A | Sema domain, transmembrane domain (TM), and cytoplasmic domain (semaphorin) 6A | 671 bp | GTCATTTACCGGAGTCTTGGAG | CGGTATCTGACCATTGTTCTCA | XM_002744676 |
| Tbr1 | T-box brain gene 1 | 560 bp | TAGCGGCTACTACGGCTTCTAC | ATTTCACAGGCTTTGTTCCACT | XM_002749394 |
| Tcf7l2 | Transcription factor 7-like 2, T-cell specific, HMG box | 654 bp | CTCTCCACCTTTTGTCAATTCC | TTTGCCATGATACTGTCTGTCC | XM_002757270 |
| Zic1 | Zinc finger protein of the cerebellum 1 | 348 bp | TCCACGAATCCTCCTCGCAGGG | CCAATAGACCCAGGACGAGTAA | NM_003412 |
| Zic4 | Zinc finger protein of the cerebellum 4 | 633 bp | CTTTCAGGAAAGGGAGGAGCAC | TCCTAACAACGCTGATGAAACG | NM_001168378 |
BTB, BR-C, ttk, and bab; POZ, Pox virus and Zinc finger; HMG, High Mobility Group; RAR, retinoic acid receptor.
ISH.
Single-color nonradioactive ISH was performed as described previously, with some modifications (Grove et al., 1998). Sections on glass slides were fixed in 4% PFA in PBS and treated with proteinase K (Roche). Sections were then fixed again and hybridized with digoxigenin (DIG)-labeled probes (Roche) at 70°C overnight in hybridization solution (50% formamide, 5× SSC, 1% SDS, 500 μg/ml yeast tRNA, 50 μg/ml heparin, and 200 μg/ml acetylated BSA). Excess probe was washed out, and sections were blocked with lamb serum and incubated with solution containing Alkaline phosphatase conjugated to DIG antibody (Roche). Color was developed with a combination of the chromagens nitroblue tetrazolium (Nacalai; 350 mg/ml) and 5-bromo-4-chloro-3-indolylphosphate for blue staining.
Images and illustrations.
Images were taken with an Olympus VS-100 virtual slide system with a 10× objective lens. Contrast, brightness, and sharpness of the entire image were adjusted with Adobe Photoshop CS5.
Results
Marker gene isolation in common marmoset brain and comparison with mouse brain
To obtain precise information about the gene expression pattern in neonatal marmoset brain, we used ISH, as reported previously, with some modifications (Grove et al., 1998). The average gestation period of the marmoset is 148 d, and its rate of development is different from mouse embryos. Based on a previous report (Lebrand et al., 2006) examining the degree of cortical laminae maturation, level of serotonin expression, its receptor expression, and serotonin transporter expression in brain at birth, we determined that the equivalent stage of a P0 marmoset brain is approximately P14 to P20 in mouse. Thus, we considered mouse P14 brain (or later) for gene expression comparison with neonatal marmoset brain. For anatomical terminology of the common marmoset brain, we used the Digital Brain Atlas of the Common Marmoset and previous publications (Tokuno et al., 2009a,b). All primer sequences and positions of the genes used as probes are listed in Table 1.
Gene expression in afferent regions of the PFC–pulvinar
In rodents, connectivity suggests that the lateral posterior (LP) nucleus complex is equivalent to the pulvinar (Baldwin et al., 2011). However, if further studies validate this hypothesis, it may be premature to conclude there is no pulvinar in rodents, as suggested in the literature (Ogren and Racic, 1981; Fosse and Fonnum, 1987; Jones, 2007; Bystron et al., 2008; Clowry et al., 2010). This controversy highlights the points that direct anatomical comparisons between rodent pulvinar/LP and primate pulvinar is not an easy task. Therefore, to facilitate the resolution of this issue with a genetic approach, we decided to study molecular markers expressed in marmoset pulvinar showing areal identities. The pulvinar has strong connections with the cortex and can be divided anatomically into medial (PuM), lateral (PuL), inferior (PuI), and anterior (PuA) regions (Romanski et al., 1997) (Fig. 1A). However, these cytoarchitectonically defined divisions do not correspond well to divisions based on connectivity, neurochemistry, or electrophysiological properties (Gutierrez et al., 1995; Stepniewska and Kaas, 1997; Adams et al., 2000). The PuL and PuI have widespread connections with visual cortical association areas (Romanski et al., 1997). Moreover, the PuI receives input from the superior colliculus, which seems to function as a second route of visual input to the cortex (Stepniewska and Kaas, 1997; Weller et al., 2002). Injection of neuronal tracers demonstrated that the PuM has widespread connections with layers 3 and 4 of the PFC (Romanski et al., 1997). Thalamic lesion studies of pulvinar in humans and monkeys result in attention deficits such as visuospatial hemineglect, a syndrome associated with the failure to direct attention to the contralesional space (Petersen et al., 1987; Karnath et al., 2002). Therefore, revealing the gene expression pattern in this unique complex of nuclei in primate thalamus is important for understanding the evolutionary mechanisms of circuit development.
Figure 1.
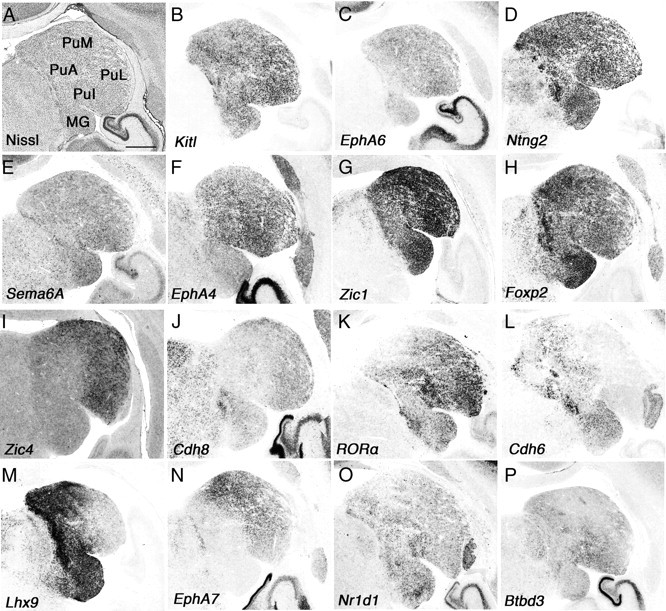
Gene expression in connecting regions to the prefrontal cortex–pulvinar. A–P, Coronal section of neonate marmoset brain for Nissl staining (A) and ISH for Kitl (B), EphA6 (C), Ntng2 (D), Sema6A (E), EphA4 (F), Zic1 (G), Foxp2 (H), Zic4 (I), Cdh8 (J), RORA (K), Cdh6 (L), Lhx9 (M), EphA7 (N), Nr1d1 (O), and Btbd3 (P). Scale bar: (in A) A–P, 1 mm.
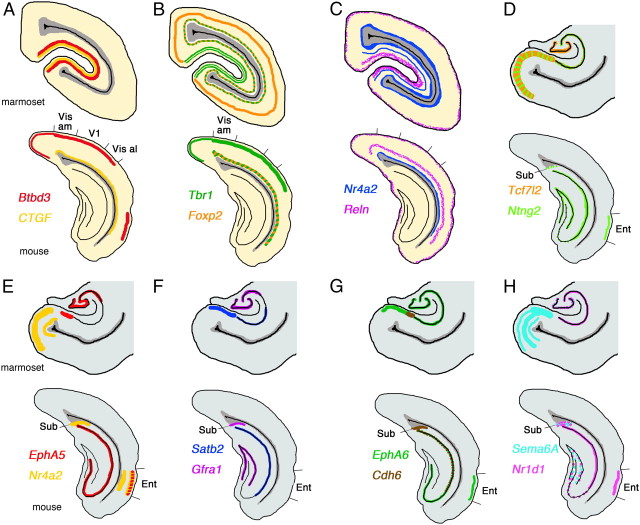
First, we demonstrated expression of various genes (Kitl, EphA6, Ntng2, Sema6a, Reln, Zic1, and Foxp2) in the entire marmoset pulvinar (Fig. 1), whose expression has been reported in mouse thalamus (Suzuki-Hirano et al., 2011; Yuge et al., 2011). Kitl, Ntng2, and Zic1 showed strong expression in the pulvinar and medial geniculate body (MG) (Fig. 1B,D,G), whereas EphA6, Sema6a, and Reln showed scattered expression throughout the entire pulvinar (Fig. 1C,E) (data not shown). Next, we examined molecules expressed in a region-specific manner and found that Zic4 and Cdh8 are expressed in the PuM and PuL (Fig. 1I,J). RORA showed strong expression in the PuL and PuI, and its expression became weaker toward the PuM (Fig. 1K). Cdh6 and Lhx9 showed counter gradients (PuM was strong, and PuL was weak) and were also expressed in the PuI (Fig. 1L,M). We also observed strong PuM and weak PuL gradients of EphA7, but there was no expression in the PuI (Fig. 1N). Nr1d1 and Btbd3 were expressed relatively strongly in the PuI, but it was hard to define a border for each subdivision (Fig. 1O,P). Although the functions of these molecules expressed within the pulvinar remain unknown, they may control afferent and efferent projections. Together, we demonstrated that molecular markers can distinguish anatomically characterized subdivisions in the pulvinar. Further study is required to understand the function of these molecules to reveal the molecular mechanisms that specify pulvinar development; however, it provides evolutionarily and biomedically valuable information between marmoset and human (Rakic, 2009).
Marmoset gene expression in the dLGN
As a consequence of evolving visual cortex complexity in the marmoset, the dLGN in the thalamus also requires the development of complexity, including regional identities, different cell types, and unique gene expression. We used the same set of marker genes characterized above to reveal the dynamic expression patterns in the dLGN. From Nissl staining, we observed that the neonatal marmoset dLGN is already segregated into layers with three different cell types (magnocellular, parvocellular, and koniocellular) as reported previously (Goodchild and Martin, 1998). These cell layers are labeled in Figure 2A as the external magnocellular layer (ME), internal magnocellular layer (MI), external parvocellular layer (PE), internal parvocellular layer (PI), and koniocellular layers (K1–K4). Generally, the PI and MI receive input from the ipsilateral eye, and the PE and ME receive input from the contralateral eye (Jones, 2007). Axons leaving the magnocellular layers and the parvocellular layers reach layer 4 in primary visual cortex (V1) (Jones, 2007). The koniocellular layers also send their axons to layer 4 in V1, and one report showed that K1 and K2 project directly to the middle temporal area (MT) in the cortex (Bourne et al., 2007). Studies involving blindsight have suggested that projections from the LGN not only travel to V1, but also to the higher cortical areas including secondary (V2) and tertiary (V3) visual cortex (Schmid et al., 2010). Together, unlike the relatively simple mouse visual pathway, the marmoset visual pathway is more complex anatomically. Therefore, examining molecular markers that delineate each cell type and identify layers, as well as axon guidance molecules, is important for understanding the developmental mechanisms operating within this area.
Figure 2.
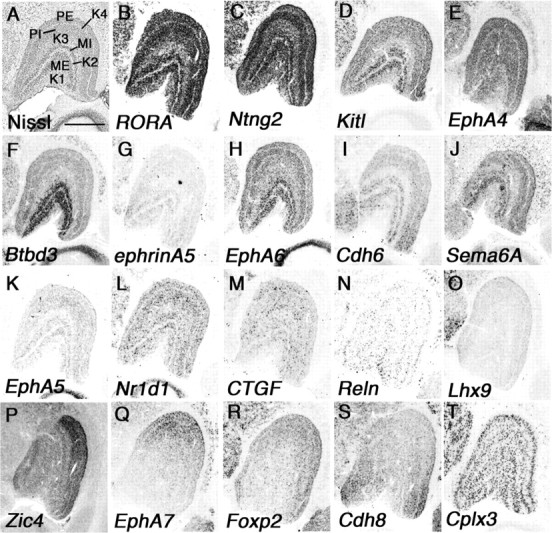
Marker genes show regional identities in marmoset dLGN. A–T, Coronal section of neonate marmoset brain for Nissl staining (A) and ISH for related orphan receptor alpha (RORA) (B), Ntng2 (C), Kit ligand (Kitl) (D), EphA4 (E), Btbd3 (F), ephrinA5 (G), EphA6 (H), Cdh6 (I), Sema6A (J), EphA5 (K), Nr1d1 (L), CTGF (M), reelin (Reln) (N), Lhx9 (O), Zic4 (P), EphA7 (Q), Foxp2 (R), Cdh8 (S), and Cplx3 (T). Scale bar: (in A) A–T, 1 mm.
First, we examined genes expressed only in specific layers in the dLGN and found that RORA, Ntng2, Kitl, EphA4, and Cplx3 were expressed only in magnocellular and parvocellular layers but not in koniocellular layers (Fig. 2B–E,T). Next we examined genes expressed in more specific layers and found that Btbd3 and ephrinA5 were specifically expressed in the ME and MI (Fig. 2F,G). EphA6, Cdh6, and Sema6A were strongly expressed in the MI and ME, and additional weak expression in parvocellular layers was seen (Fig. 2H–J). EphA5, Nr1d1, connective tissue growth factor (CTGF), and Reln were seen in all layers in a scattered expression pattern, but expression in koniocellular layers was always weaker than in other layers (Fig. 2K–N). In rodents, gradient expression of axon guidance molecules in the dLGN has important roles in afferent and efferent targeting (Huberman et al., 2005; Torii and Levitt, 2005). In neonatal marmoset, Lhx9, Zic4, EphA7, Foxp2, and Cdh8 also showed gradient expression in the dLGN, but these expression patterns were quite different from those in rodents, which show no obvious area identity (Fig. 2O–S).
Gene expression differences in primary and secondary visual cortex
One of the more remarkable features of the primate visual cortex is the evolution of visual association areas. There is a clear boundary between V1 and V2 that is easily distinguished cytoarchitectonically. The visual cortex in mouse brain also has a V2-like area in the lateral medial region of visual cortex suggested from its connectivity; however, morphologically there is no clear border between V1 and V2 (Wang and Burkhalter, 2007). We examined genes that delineate the border of these two areas in the occipital cortex to explore their differences at the molecular level in marmoset brain. First, we noticed that Btbd3 and CTGF labeled primary visual cortex exclusively (Fig. 3B,C). Btbd3 was expressed in layers 4a, 4b, and 4c in V1 (Fig. 4C). Expression of Btbd3 was also detected in mouse visual cortex, but no clear evidence for specific labeling of V1 in mouse cortex was observed (see Fig. 8A). The expression of CTGF was restricted to the subplate and layer 5 in the entire mouse cortex, but surprisingly expression in neonatal marmoset in V1 was restricted to layers 4a and 4b (Fig. 4Q). We compared the position and sequence of the probe we used for CTGF ISH to those used for mouse and human ISH in the Allen Brain Atlas (http://www.brain-map.org/), and confirmed that 494 bp of marmoset probe overlap with mouse and human probes with >90% similarity (data not shown). Next, to test whether neonate marmoset brain has a subplate, we further tested for expression of complexin3 (Cplx3) which labels the subplate in mouse cortex (Hoerder-Suabedissen et al., 2009). As seen in mouse cortex, Cplx3 clearly labeled subplate neurons in marmoset visual cortex (Figs. 3S, 4S). Therefore, the different gene expression of CTGF is specific to marmoset brain but not because the lack of a subplate structure in visual cortex. Next, we observed that expression of Tbr1 could be detected in layers 4a and 4c in V1, but not in V2, whereas expression in layer 6 was uniform in V1 and V2 (Figs. 3D, 4N). In mouse visual cortex, Tbr1 expression in layer 4 was ubiquitous and no specificity to V1 (see Fig. 8B). In marmoset, RORA and Nr1d1 were broadly expressed in layer 4 of V1 (Fig. 3E,F). Expression then became very narrow and was restricted to a single stripe in the V2 region (Fig. 4D,E). Foxp2 was expressed in layers 5 and 6 but not seen in layer 4 of V1, but expression appeared in layer 4 in V2 (Figs. 3G, 4I). In mouse cortex, Foxp2 was also expressed in layers 5 and 6, but no expression was observed in layer 4 (see Fig. 8B). EphA6, EphA7, EphA5, Cdh8, and Sema6A were not expressed in layers 4a and 4b of V1, but expression was seen in layer 4 in V2 of marmoset visual cortex (Figs. 3H–L, 4F–H,J,K). Although the expression level of these marker genes were different in mouse visual cortex, expression patterns were similar to marmoset visual cortex. Expression of Nr4a2 in layer 5 in marmoset V1 disappeared in V2, but expression in layer 6 was uniform in marmoset V1 and V2 (Figs. 3M, 4P). In contrast, Nr4a2 was expressed in mouse subplate, but no expression was observed in layer 5 in visual cortex (see Fig. 8C). In mouse cortex, ephrinA5 was expressed widely in visual cortex; however, expression in layers 4a–4c of marmoset V1 became a narrow band in layer 4 of V2, which made it possible to distinguish the border of V1 and V2 (Figs. 3N, 4O). Scattered expression of Reln in layer 1 was observed in the entire mouse and marmoset visual cortex; however, expression in other layers was different in both species. In marmoset brain, Reln expression in layer 2 was restricted in V1, but in mouse brain, there was expression of Reln in layer 5 in entire cortex (Figs. 3O, 4B, 8C). ER81 (layer 5) and Satb2 (multiple layers) in V1 were conserved in V2 (Fig. 3P,Q, 4L,M,). Here we examined marker genes that can define cortical layers and areas in neonatal marmoset visual cortex. However, marmoset visual cortex further develops in the first few months of postnatal life depending on experience, and, therefore, the genes we examined here may dynamically change their expression pattern in later postnatal stages, an idea that may be tested in the future (Bourne and Rosa, 2006).
Figure 3.
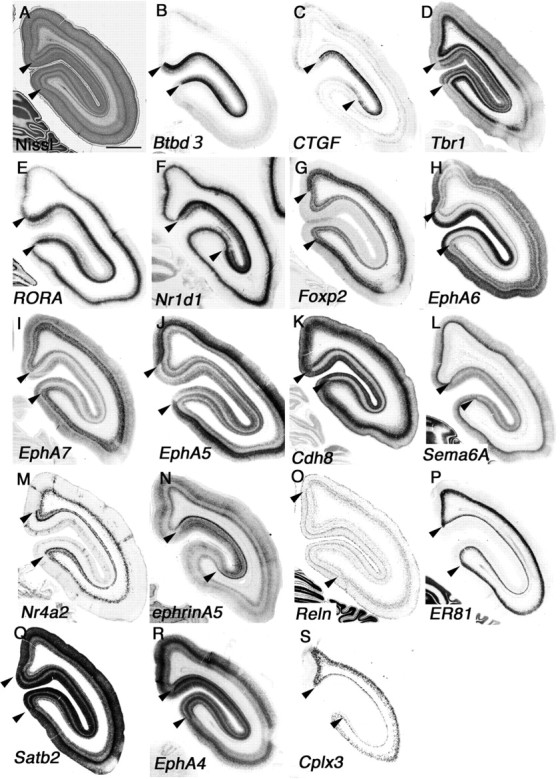
Regional identities in neonatal marmoset occipital cortex. A–S, Coronal section of neonate marmoset occipital region for Nissl staining (A) and ISH for Btbd3 (B), CTGF (C), Tbr1 (D), RORA (E), Nr1d1 (F), Foxp2 (G), EphA6 (H), EphA7 (I), EphA5 (J), Cdh8 (K), Sema6A (L), Nr4a2 (M), ephrinA5 (N), Reln (O), ER81 (P), Satb2 (Q), EphA4 (R), and Cplx3 (S). Borders of V1 are indicated by arrowheads. Scale bar: (in A) A–S, 1 mm.
Figure 4.
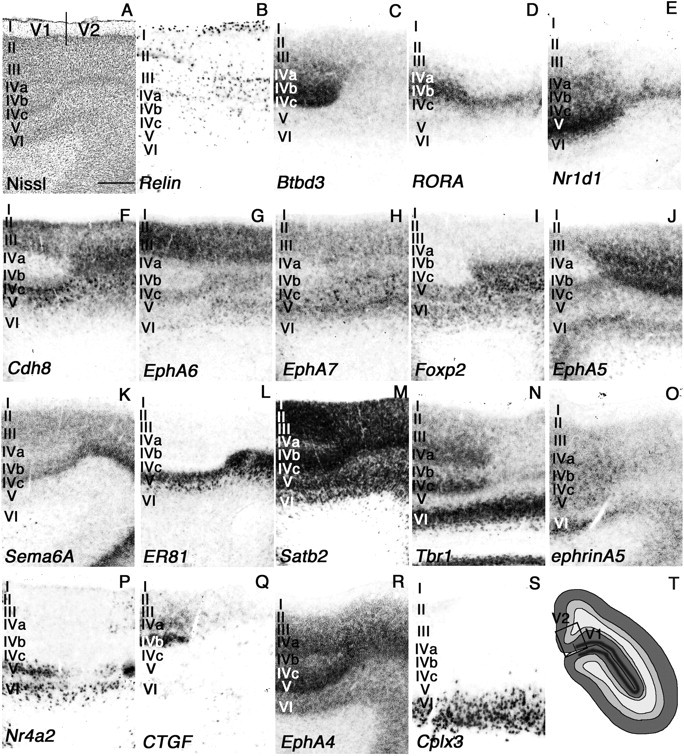
Layer specificity in genes expression in neonatal marmoset V1/V2 cortex. A higher magnification of V1 and V2 regions shown in Figure 4 is shown. A–S, Nissl staining (A) and ISH for Reln (B), Btbd3 (C), RORA (D), Nr1d1 (E), Cdh8 (F), EphA6 (G), EphA7 (H), Foxp2 (I), EphA5 (J), Sema6A (K), ER81 (L), Satb2 (M), Tbr1 (N), ephrinA5 (O), Nr4a2 (P), CTGF (Q), EphA4 (R), and Cplx3 (S). T, Diagram summary for approximate region taken for picture in visual cortex is indicated. The borders of V1 and V2 are indicted in A. The relative position in visual cortex is indicated by a box in T. Scale bar, 400 μm.
Figure 8.
Diagram summary of differently expressed marker genes. A–H, Summarized gene expression difference in mouse and marmoset visual cortex (A–C) and hippocampal area, subiculum, and entorhinal cortex (D–H). Vis al, Visual cortex anterolateral.
Gene expression in the MT, a unique area in the marmoset cortex
The primate visual cortex is one of the most distinct areas of cortex when compared to mouse visual cortex. As its connectivity is more complex, the number of visual areas are also different. Especially V5 (also known as the middle temporal area) is connected to a wide array of cortical and subcortical brain areas, such as the visual cortical areas V1, V2, and dorsal V3, the koniocellular regions of the LGN, and the inferior pulvinar (Ungerleider and Desimone, 1986). Generally, V1 is the major input to MT; however, several studies have demonstrated that neurons in MT can process visual information, even after V1 has been destroyed or inactivated (Rodman et al., 1989). Hence, MT can be considered one of the important visual areas, and to test how similar the MT and V1 are, we tested expression of the V1-specific marker genes, Btbd3 and CTGF, in marmoset MT. We observed clear expression of Btbd3 and CTGF in marmoset MT, but the cortical layers where these genes were expressed slightly different than that in V1 (Fig. 5A,B,F,G). Especially CTGF expression in subplate was weak but evident in MT, suggesting dynamic expression of CTGF in the subplate during development in the marmoset brain (Fig. 5B,G, arrows). Furthermore, we examined additional genes expressed in the MT and found that ephrinA5 and Cdh6 were expressed in layers 2–4 and 6 in the MT (Fig. 5C,D). Expression of ephrinA5 was seen in V1 and V2 (Figs. 3N, 4O). In contrast, Cdh6 was not expressed in V1 or V2 (data not shown). Therefore, this MT-specific gene expression pattern may make this region functionally different from V1. Next, we examined CTGF expression in embryonic brain to test whether the MT develops earlier than other cortical areas, which would suggest that the MT has a similar character to the primary visual cortical area (Bourne and Rosa, 2006). Embryonic brains were collected at approximately embryonic day 90 (E90). In the embryonic presumptive MT area, detectable CTGF expression was observed, whereas CTGF expression in the PFC was still immature (Fig. 5I). Expression of CTGF and EphA6 in the PFC was still weak, and the expression patterns were quite different from those in neonatal cortex (Fig. 5J,K). Therefore, from the gene expression analysis, the MT area appears to develop relatively earlier and may be considered a primary cortical visual area. However, to test whether MT functions as a seed for an intracortical signaling center will require further analysis to discover signaling molecules that control development of visual cortical areas in marmoset.
Figure 5.
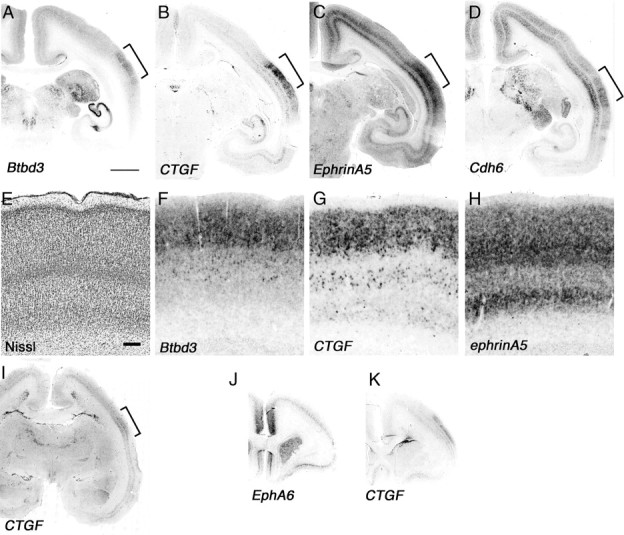
MT is already developed in embryonic marmoset brain. Sections include the MT region (brackets) of the coronal section of neonate marmoset brain for ISH for Btbd3 (A), CTGF (B), ephrinA5 (C), and Cdh6 (D). E–H, Higher magnification of MT regions in A–C (brackets) and Nissl staining (E). Arrows indicate position of subplate (B, G). I–K, ISH of E90 marmoset brain at presumptive MT (I, bracket) and PFC regions (J, K). Scale bars: A (for A–D, I–K), 2 mm; E (for E–H), 200 μm.
Dynamic gene expression in efferent regions of the PFC–hippocampus
Many primary sensory areas change their connectivity, morphology, and function during early postnatal life in a process called activity-dependent plasticity (Wiesel and Hubel, 1963; Woolsey and Wann, 1976). Previously, this type of plasticity was reported in the developing PFC, which facilitates the morphological and functional changes necessary to refine behaviorally relevant communication derived from hippocampal oscillatory activity (Brockmann et al., 2011). Although gene expression studies have been reported for neonatal marmoset hippocampus using marmoset-specific DNA microarray (Datson et al., 2009), we examined molecules whose expression has not been reported in wider regions such as the entorhinal (Ent) cortex. The Ent cortex receives inputs from the cortex, especially the PFC, and projects to the hippocampus to play an important role in memory formation and consolidation (Wall and Messier, 2001). As reported previously, there are no significant gene expression differences between the mouse and marmoset CA fields and dentate gyrus (DG) (Datson et al., 2009). However, we found many differences in gene expression in the Ent cortex and subiculum (Sub) (Fig. 6A). EphA5 was strongly expressed in marmoset prosubiculum (ProS) (Fig. 6B), and this expression is not detectable in mouse brain (see Fig. 8). In mouse brain, EphA6 and Satb2 are not expressed in the Sub, but these genes are clearly expressed in marmoset (Fig. 6C,D). In contrast, EphA6 is strongly expressed in mouse Ent (see Fig. 8G), but no signal was observed in marmoset Ent (Fig. 6C). Next, we analyzed expression of Nr4a2, which is restricted to the marmoset Ent (Fig. 6E). In mouse, expression of Nr4a2 was also evident in the Ent, but the expression was also observed in the Sub (see Fig. 8E). Sema6A was expressed in multiple layers in the marmoset Ent and Sub (Fig. 6F), but expression was not detected in mouse Ent (see Fig. 8H). We observed identical expression of Ntng2 in the Ent, Sub, and CA field in mouse and marmoset brain, but expression in DG was seen only in mouse (Figs. 6G, 8D). In hippocampal CA and DG fields, mouse cells did not express CTGF (see Fig. 8A), but we observed strong expression in marmoset CA2 and scattered expression in the DG (Fig. 6H). No Cdh6 expression in CA fields was detected in marmoset hippocampus (Fig. 6I), but mouse hippocampus strongly expressed Cdh6 in the CA1 (see Fig. 8G). Gfrα1 was strongly expressed in mouse Sub, but not in marmoset Sub. Moreover, expression in CA3 was detected only in marmoset hippocampus, while expression in DG was observed in both species (Figs. 6J, 8F). We observed a high level of expression of Nr1d1 in mouse Sub and Ent, but no expression in marmoset (Figs. 6K, 8H). Finally, the expression of Tcf7l2 in marmoset was observed in the CA field, DG, Sub, and Ent (Fig. 6L), whereas it is not expressed in any of these structures in mouse brain (see Fig. 8D). In addition to these differentially expressed genes, we also examined Btbd3, Cdh8, Tbr1, ER81, EphA7, Reln, RORA, and Foxp2 and observed similar expression patterns in marmoset and mouse hippocampus (Figs. 6M–T, 8). Together, our results suggest gene expression in marmoset hippocampus, especially in areas that function as a gateway from cortex to the hippocampus such as the Ent, is distinct from that in mouse.
Figure 6.
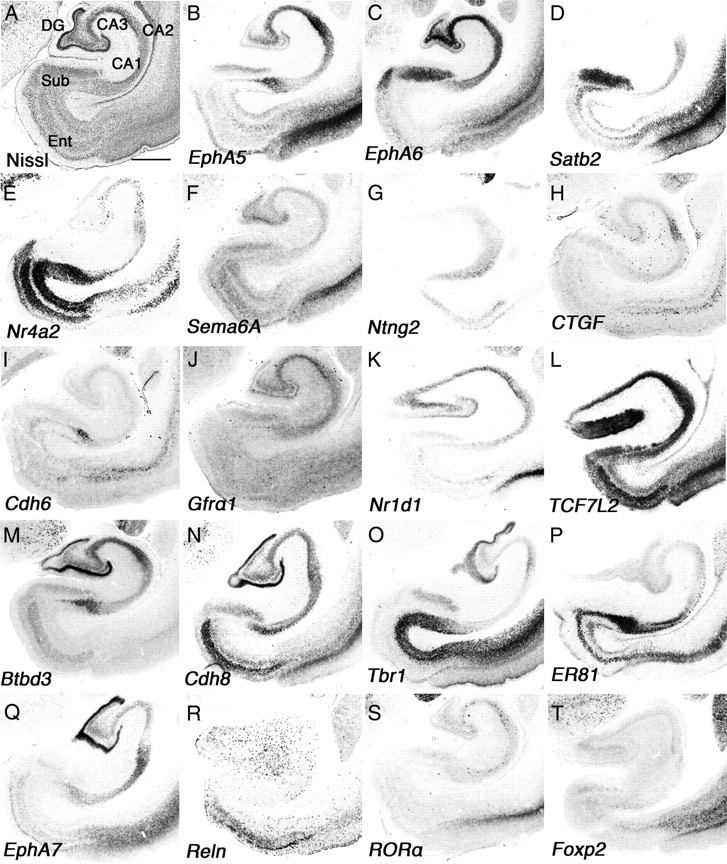
Divergent gene expression in neonatal marmoset hippocampus. A–T, Coronal section of neonate marmoset hippocampal region for Nissl staining (A) and ISH for EphA5 (B), EphA6 (C), Satb2 (D), Nr4a2 (E), Sema6A (F), Ntng2 (G), CTGF (H), Cdh6 (I), Gfrα1 (J), Nr1d1 (K), Tcf7l2 (L), Btbd3 (M), Cdh8 (N), Tbr1 (O), ER81 (P), EphA7 (Q), Reln (R), RORA (S), and Foxp2 (T). CA1–CA3, Hippocampal CA field. Scale bar: (in A) A–T, 1 mm.
Primate-specific gene expression in the PFC
The PFC has been implicated in planning complex cognitive behavior, personality expression, decision making, and moderating correct social behavior (Barbey et al., 2009). It can be divided into several subdomains based on connections to other parts of the brain. In our study, we divided the PFC into three areas: the cingulate gyrus (CG), frontal cortex (CxF), and insular cortex (CxI) (Fig. 7A). A study of the efferent fiber connections of the rat's mediodorsal nucleus of the thalamus projection to cortex demonstrated some similarities to those of monkey prefrontal cortex. However, the rat's medial dorsal nucleus-projection cortex differs from that in the monkey in that it lacks a granular layer and appears to have no prominent direct associations with temporal and juxtahippocampal areas (Leonard, 1969). To compare these differences with molecular markers, we first examined genes expressed in mouse anterior cortex (Tbr1, EphA4, EphA5, EphA6, EphA7, Cdh6, Cdh8, and RORA) to test whether they share similar expression patterns in neonatal marmoset PFC. All of these marker genes were expressed in common marmoset PFC, but with some differences from the mouse expression pattern. As in mouse brain, Tbr1 was expressed in layer 6 of the entire PFC, but the strongest expression was seen in CxI (Fig. 7B, arrow). Tbr1 was also expressed in layers 4 and 5 of mouse brain, in addition to layer 6 in frontal cortex, a pattern that was not evident in common marmoset PFC (Fig. 7B). There was no expression of Tbr1 in layers 4 and 5 of the neonatal marmoset in more caudal regions such as parietal cortex and occipital cortex, besides primary visual cortex (data not shown). The gene expression patterns of EphrinA-EphA family members have been thoroughly studied in both mouse and Old World monkeys (Donoghue and Rakic, 1999; Yun et al., 2003). In both mouse and rhesus monkeys, the embryonic brain shows uniform expression of EphA4 in the PFC, a pattern that was also seen in neonatal marmoset PFC (Fig. 7C). EphA5 expression in the rhesus monkey shows strong anterior and weak posterior gradients (Donoghue and Rakic, 1999), which were also evident in neonatal marmoset brain (Fig. 7D). The gradient expression was detected until the parietal cortex (data not shown). As in P14 mouse brain, EphA6 in neonatal marmoset brain showed uniform expression throughout the entire PFC, but weak expression in layer 4 (Fig. 7E). However, this expression was different from E80 macaque monkey brain, which shows expression only in the posterior half of the neocortex (Donoghue and Rakic, 1999). Therefore, we also tested EphA6 expression in E90 marmoset brain (Fig. 5L) (data not shown). In embryonic marmoset, expression of EphA6 shows a weak anterior and strong posterior gradient as in the macaque embryonic brain, but the expression is quite different from mouse (Yun et al., 2003). This different expression pattern of EphA6 during embryogenesis may contribute to formation of the primate-specific neocortex. Expression of EphA7 in neonatal marmoset PFC was seen both in layers 6 and 4, which is different from the weak expression in mouse cortex. Moreover, EphA7 expression in the PFC was strong in the CG, and no expression was seen in the ventrolateral region (predicting monkey area 47) (Fig. 7F, bracket). Similar expression patterns were seen for EphA5, EphA6, Cdh6, Foxp2, Satb2, and ephrinA5 (Fig. 7D,E,G,L,M,R, brackets). This region exhibits the architectonic characteristics of area 45 in human brain (Petrides and Pandya, 2002), the role of which is still controversial. However, some evidence suggests that the area functions to subserve semantic retrieval or semantic working memory processes (Badre and Wagner, 2007). In mouse cortex, Cdh6 was strongly expressed in layers 4 and 6, but in neonatal marmoset PFC, both genes were strongly expressed in layers 2/3 and 5 (Fig. 7G). Cdh8 was strongly expressed in layers 2/3–5 in mouse cortex, but its expression in neonatal marmoset was strongest in layer 4 (Fig. 7H). Cdh8 also determines area identity in the PFC in marmoset, which is not evident in mouse. Cdh6 showed the strongest expression in the CG, whereas Cdh8 showed the strongest expression in dorsolateral, CxI, and ventromedial regions of the PFC (Fig. 7G,H). In mouse cortex, RORA is expressed both in primary motor and primary sensory cortex, but not in cingulate cortex (Nakagawa and O'Leary, 2003). However, in neonatal marmoset, RORA expression was uniform throughout the entire cortex including the PFC (Fig. 7I) (data not shown). Even in later stages in mouse brain (P54) when RORA was more widely expressed, it was still not expressed in the medial PFC in marmoset. Therefore, we concluded that the expression differences in the neonatal marmoset were not the only differences in this developmental stage between the two species. To further test for differences in cortical layer markers, we next examined the expression of Reln (layer 1 in mouse), ER81 (layer 5 in mouse), Foxp2 (layer 6 in mouse), and Satb2 (determines upper layer neuron specification in the mouse neocortex) in neonatal marmoset. Reln and ER81 expression in the neonatal marmoset was similar to the expression pattern in mouse and other Old World monkeys in layers 1 and 4 and layer 5, respectively (Fig. 7J,K) (Rodriguez et al., 2000; Yoneshima et al., 2006). However, both Foxp2 and CTGF showed different expression patterns in neonatal marmoset brain. High-level expression of Foxp2 expression in mouse cortex in layer 6b and some additional staining was observed in somatosensory layer 4 (Yuge et al., 2011). In neonatal marmoset PFC, we observed strong expression of Foxp2 in layer 6, but in more caudal regions, the expression in layer 6 became weaker, and expression in layers 5 and 4 became much stronger (data not shown). Satb2 showed strong expression in the entire PFC and weak expression in ventrolateral regions (Fig. 7M, bracket).
Figure 7.
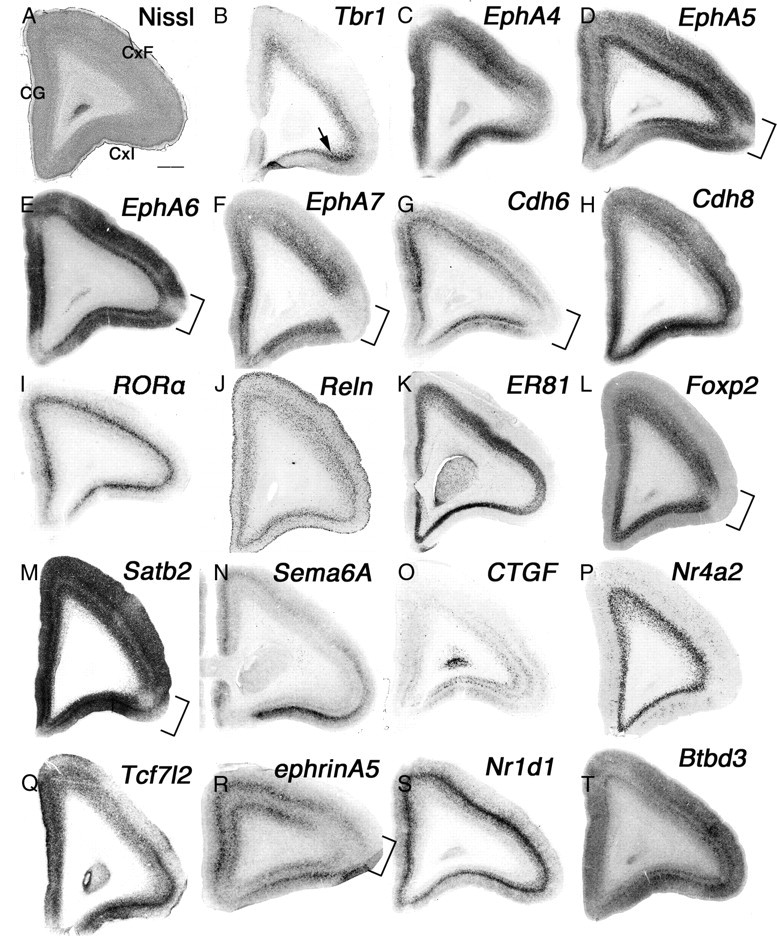
Primate-specific gene expression in the PFC. A–T, Coronal section of neonate marmoset PFC Nissl staining (A) and ISH for Tbr1 (B), EphA4 (C), EphA5 (D), EphA6 (E), EphA7 (F), Cdh6 (G), Cdh8 (H), RORA (I), Reln (J), ER81 (K), Foxp2 (L), Satb2 (M), Sema6A (N), CTGF (O), Nr4a2 (P), Tcf7l2 (Q), ephrinA5 (R), Nr1d1 (S), and Btbd3 (T). The arrow in A indicates strong expression of EphA7 in CxI, and brackets in D–G, L, and M indicate position of the ventrolateral region. Scale bar: (in A) A–T, 1 mm.
We next examined genes that show more regional identities in the PFC and found that expression of both Sema6A and CTGF was strong in CxI (Fig. 7N,O). Moreover, CTGF expression in mouse is strong in subplate neurons (Hoerder-Suabedissen et al., 2009), but in neonatal marmoset, expression in the cortex was quite different. First, expression in the subplate was weak, but strong expression in upper layers was observed (data not shown), which further shows area identities such as the MT region and primary visual cortex (Figs. 3, 5). We examined expression of Nr4a2, which is restricted to the claustrum and subplate in mouse cortex (Hoerder-Suabedissen et al., 2009). Surprisingly, Nr4a2 was expressed in the entire layer 6 of the PFC (Fig. 7P) and the claustrum (data not shown) in neonatal marmoset. Moreover, this layer 6 expression of Nr4a2 was evident in the entire cortex and was quite different from the restricted expression in mouse subplate (Hoerder-Suabedissen et al., 2009). We examined more genes expressed in the PFC but in different layers compared to their expression in mouse. First, Tcf7L2, which is expressed in layer 5 of the PFC in mouse, was expressed in marmoset in layers 4–6 (Fig. 7Q). In mouse, ephrinA5 expression is strong in primary somatosensory layer 4, but there is no expression in the PFC (Yun et al., 2003). Expression of ephrinA5 in neonatal marmoset was split between layers 4 and 6 in the PFC, and its expression in dorsolateral regions was weak (Fig. 7R, bracket). Nr1d1 expression in mouse brain was strong in upper layers (layers 2/3–4), but the expression of Nr1d1 in neonatal marmoset was restricted to layer 4 (Fig. 7S). Finally, Btbd3 shows strong expression in early postnatal mouse primary somatosensory cortex (T. Shimogori, unpublished data), and its expression in the entire PFC becomes stronger in later postnatal stages (http://www.brain-map.org). In neonatal marmoset PFC, Btbd3 showed gradient expression, which was strong in lateral regions and became weaker in the dorsal and medial PFC (Fig. 7T). Together, we observed many differences in gene expression levels in the neonatal marmoset PFC, not only within area identities, but also in specific layers. This suggests functional and connectivity differences in the PFC that are specific to primates.
Discussion
Mouse genetics can be a powerful tool to address many aspects of brain development, but faces obvious limitations for investigating the development of circuitry underlying higher cognitive functions. There is thus a pressing need to develop a new animal model that can make use of insights gained from studying mouse brain development to directly investigate the evolution and development of the primate neocortex. Here, we used in situ hybridization analysis in marmoset brain to identify similarities and differences in the expression pattern of genes found previously to regulate mouse brain development. In this study, we revealed that these genes show similar broad regional patterns of expression in both species. However, when examined in detail, the expression patterns of these genes are quite different, especially in brain areas that have connections to PFC and are presumably involved in higher cognitive functions.
Comparative analysis of mouse and monkey brain
Comparative gene expression studies in embryonic mouse and monkey brains showed small differences in expression related to area identities (Donoghue and Rakic, 1999; Yun et al., 2003; Jones and Rubenstein, 2004). Another comparative gene expression analysis in adult mouse and marmoset brain using microarray analysis did not report major differences (Datson et al., 2009). However, studies comparing gene expression in postnatal monkey and mouse brain identified multiple expression differences in gene expression within cortical layers and cell populations (Takahata et al., 2006, 2009; Watakabe et al., 2006, 2007). Therefore, to find species-specific gene expression, it is important to analyze gene expression at the cellular level in specific regions for higher cognitive functions with high resolution in situ hybridization approaches. To do this, the marmoset is an ideal animal because of many benefits of its nature (a small body size, high fertility, and early sexual maturity) and relatively simple brain structure but complicated behavior (relatively smooth cortex but the gyrencephalic has higher cognitive functions and social communication).
Differential gene expression in developing cognitive circuits
We identified several genes expressed similarly between mouse and marmoset PFC but also many that were expressed differentially, including some identifying areas within the PFC. Segregation of each subregion within the PFC remains difficult with current marker genes. Therefore, the P0 marmoset PFC may not be fully developed, or we may have only examined molecules that do not identify specific areas. However, it was remarkable to find genes specifically expressed in the subplate in mouse brain that exhibited expression in different layers in the marmoset PFC (CTGF and Nr4a2). There are marked differences in the thickness of the subplate zone in rodents, carnivores, humans, and nonhuman primates. A thicker subplate is observed in relation to the cortical plate in larger-brained mammals (Kostovic and Rakic, 1990), suggesting that the evolutionary expansion of the neocortex and the expansion of the subplate zone are linked. In addition, it is hypothesized that the evolutionary appearance of the subplate zone led to a change in the axonal ingrowth pattern from above to below the cortical plate (Aboitiz, 1999; Supèr and Uylings, 2001). The authors suggested that this change was crucial for the radial expansion of the dorsal cortex and the development of the six-layered mammalian neocortex. Therefore, the gene expression differences in mouse subplate markers in the PFC suggest dynamic thalamocortical axon growth in the marmoset PFC. Furthermore, CTGF expression is dynamic among different species such as mouse, rat, chick, turtle, and ferret, suggesting that CTGF plays an important role in species-specific thalamus–cortex connectivity. Moreover, Nr4a2 knock-out mice show a phenotype similar to schizophrenia and depression (Xing et al., 2006; Rojas et al., 2007, 2010), suggesting that the expanded expression of Nr4a2 in marmoset may play a central role in models of severe mental disorders in these animals.
Tbr1 is a putative transcription factor that is highly expressed in glutamatergic early-born cortical neurons. In Tbr1-deficient mice, these early-born neurons show molecular and functional defects. Cajal–Retzius cells express decreased levels of Reln, resulting in a reeler-like cortical migration disorder. Impaired subplate differentiation is associated with ectopic projection of thalamocortical fibers into the basal telencephalon. Layer 6 defects contribute to errors in thalamocortical, corticothalamic, and callosal projections. Collectively, these results show that Tbr1 is a neonatal genetic determinant of the differentiation of early-born glutamatergic neocortical neurons and provides insights into the functions of these neurons as regulators of cortical development. In marmoset PFC, Tbr1 expression was restricted to layer 6, suggesting that it specifically functions in the development of layer 6 characteristics.
Many reports have been described on the participation of hippocampus–PFC pathways in higher-order functions including selective attention, decision making, and goal-directed behaviors. The hippocampus is an old structure in cortex, and its architectonic structure and gene expression patterns are generally conserved among species (Datson et al., 2009). Surprisingly, detailed gene expression in neonatal marmoset revealed many different gene expression patterns in the subiculum and entorhinal cortex (Figs. 6, 8D–H). We do not know the significance of these different gene expression patterns for neonatal marmoset brain function, but evidence of coupled oscillations between the hippocampus and PFC for plasticity in mouse suggests that these gene expression differences may contribute to developmental diversity of marmoset PFC. Moreover, it will be interesting to examine gene expression patterns in later stages of marmoset development after exposing the animals to different environments to identify experience-dependent gene expression.
Evolution of the cortical area map
The view that the cortical primordium is initially patterned by gradients of signaling molecules and transcription factors in similar ways as the rest of the embryo is widely accepted (Grove and Fukuchi-Shimogori, 2003). Our gene expression study provides insights into current hypotheses on the mechanism of cortical area map evolution. First, we found that some axon guidance molecules were expressed only in layer 4 of V2 (EphA6, EphA7, EphA5, Sema6A, and ephrinA5), suggesting different axon projections into V2 compared to layer 4 of the V1 (Figs. 3, 4). This may result from segregation of thalamic input that projected into the visual cortex from the dLGN and pulvinar (Krubitzer, 2007). Our finding of common gene expression in V1 and MT suggests that MT functions as a “primary” visual area, supporting the idea of duplicating and adding new areas to older modules (Fig. 5) (Rosa and Tweedale, 2005). To support this idea, it is shown that the difference in development and organization between areas 17 and 18 in Old World primates derives from the cell fate decision that originated already in the proliferative ventricular zone before neurons began to migrate to their areal and laminar positions (Kornack and Rakic, 1998). From this study, we found that gene expression in neonatal marmoset brain is distinct from that in mouse, and therefore we can test how these different gene expression patterns contribute to cortical evolution not only by using genetically modified mice, but also in the future by genetically modifying marmosets. Moreover, a population of interneurons specific to human, which are generated in cortical ventricular zone, was reported previously (Letinic et al., 2002), and marmoset-specific gene expression may have a contribution to primate-specific neurogenesis, a hypothesis that cannot be tested in mouse.
Another question raised here is how did an additional region in the thalamus such as the pulvinar evolve? In rodents, connectivity suggests that the LP nucleus complex is equivalent to the pulvinar (Baldwin et al., 2011). However, based on its connection to the visual cortex, it is also reasonable to hypothesize that it shares some origins with the dLGN. Therefore, it may be premature to conclude that there is no pulvinar in rodents, as suggested in the literature (Fosse and Fonnum, 1987; Jones, 2007; Bystron et al., 2008; Clowry et al., 2010). This controversy highlights the point that direct anatomical comparisons or gene expression comparison between rodent pulvinar/LP and primate pulvinar is not an easy task. Our study shows that many genes are similarly expressed in the dLGN and PuL, which have dense projections to the visual cortex. To further test this idea, gene expression analysis of early embryonic stages in the pulvinar will be required. The earliest stage we examined was E90, when the pulvinar is already formed and gene expression is already similar to that observed at P0.
Mechanism of marmoset specific gene expression
Two obvious questions raised by the current findings are how is differential gene expression controlled in marmoset? and what is the function of these differentially expressed genes? Recent observations that may help to answer these questions indicate that noncoding genes can control the area and timing of coding gene expression (Ohshima and Okada, 2005; Pollard et al., 2006; Goodier and Kazazian, 2008; Britten, 2010) of signaling molecules to regulate patterning, neuronal growth rate, and DNA damage repair (Goodier and Kazazian, 2008; Sasaki et al., 2008; Lambert et al., 2011). The generation of transgenic marmosets for enhancer activity assays will be a very powerful tool to test this hypothesis (Sasaki et al., 2009). Furthermore, direct gene manipulation will allow testing the function of these differently expressed molecules in marmoset brain. For this, the combination of common marmoset and recent technical improvements in viral vectors (Callaway, 2008; Luo et al., 2008) and in utero electroporation (Shimogori and Ogawa, 2008) will provide direct evidence of how genetic divergence triggered cognitive neuronal circuit development during evolution.
Notes
Supplemental material for this article is available at http://mmtd.brain.riken.jp/P0marmoset/. It includes all data of ISH for neonate marmoset brain and a table that contains references for mouse gene expression. This material has not been peer reviewed.
Footnotes
This work was supported by the RIKEN Brain Science Institute (T.S.), the MEXT Strategic Research Program for Brain Science (T.S., H.O.), and the Funding Program for World-Leading Innovative R&D on Science and Technology (T.S., H.O.). We thank Dr. Yokoyama, Dr. Blackshaw, and Dr. Matsui for critical reading.
The authors declare no competing financial interests.
References
- Aboitiz F. Feature article evolution of isocortical organization. A tentative scenario including roles of reelin, p35/cdk5 and the subplate zone. Cereb Cortex. 1999;9:655–661. doi: 10.1093/cercor/9.7.655. [DOI] [PubMed] [Google Scholar]
- Adams MM, Hof PR, Gattass R, Webster MJ, Ungerleider LG. Visual cortical projections and chemoarchitecture of macaque monkey pulvinar. J Comp Neurol. 2000;419:377–393. doi: 10.1002/(sici)1096-9861(20000410)419:3<377::aid-cne9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Badre D, Wagner AD. Left ventrolateral prefrontal cortex and the cognitive control of memory. Neuropsychologia. 2007;45:2883–2901. doi: 10.1016/j.neuropsychologia.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Baldwin MK, Wong P, Reed JL, Kaas JH. Superior colliculus connections with visual thalamus in gray squirrels (Sciurus carolinensis): evidence for four subdivisions within the pulvinar complex. J Comp Neurol. 2011;519:1071–1094. doi: 10.1002/cne.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey AK, Krueger F, Grafman J. An evolutionarily adaptive neural architecture for social reasoning. Trends Neurosci. 2009;32:603–610. doi: 10.1016/j.tins.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin A, Torii M, Wang L, Rakic P, Levitt P. Serotonin modulates the response of embryonic thalamocortical axons to netrin-1. Nat Neurosci. 2007;10:588–597. doi: 10.1038/nn1896. [DOI] [PubMed] [Google Scholar]
- Bourne JA, Rosa MG. Hierarchical development of the primate visual cortex, as revealed by neurofilament immunoreactivity: early maturation of the middle temporal area (MT) Cereb Cortex. 2006;16:405–414. doi: 10.1093/cercor/bhi119. [DOI] [PubMed] [Google Scholar]
- Bourne JA, Warner CE, Upton DJ, Rosa MG. Chemoarchitecture of the middle temporal visual area in the marmoset monkey (Callithrix jacchus): laminar distribution of calcium-binding proteins (calbindin, parvalbumin) and nonphosphorylated neurofilament. J Comp Neurol. 2007;500:832–849. doi: 10.1002/cne.21190. [DOI] [PubMed] [Google Scholar]
- Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann MD, Pöeschel B, Cichon N, Hanganu-Opatz IL. Coupled oscillations mediate directed interactions between prefrontal cortex and hippocampus of the neonatal rat. Neuron. 2011;71:332–347. doi: 10.1016/j.neuron.2011.05.041. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowry G, Molnar Z, Rakic P. Renewed focus on the developing human neocortex. J Anat. 2010;217:276–288. doi: 10.1111/j.1469-7580.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson NA, Morsink MC, Steenbergen PJ, Aubert Y, Schlumbohm C, Fuchs E, de Kloet ER. A molecular blueprint of gene expression in hippocampal subregions CA1, CA3, and DG is conserved in the brain of the common marmoset. Hippocampus. 2009;19:739–752. doi: 10.1002/hipo.20555. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Rakic P. Molecular gradients and compartments in the embryonic primate cerebral cortex. Cereb Cortex. 1999;9:586–600. doi: 10.1093/cercor/9.6.586. [DOI] [PubMed] [Google Scholar]
- Eliades SJ, Wang X. Neural substrates of vocalization feedback monitoring in primate auditory cortex. Nature. 2008;453:1102–1106. doi: 10.1038/nature06910. [DOI] [PubMed] [Google Scholar]
- Fosse VM, Fonnum F. Biochemical-evidence for glutamate and/or aspartate as neurotransmitters in fibers from the visual-cortex to the lateral posterior thalamic nucleus (pulvinar) in rats. Brain Res. 1987;400:219–224. doi: 10.1016/0006-8993(87)90620-2. [DOI] [PubMed] [Google Scholar]
- Goodchild AK, Martin PR. The distribution of calcium-binding proteins in the lateral geniculate nucleus and visual cortex of a New World monkey, the marmoset, Callithrix jacchus. Vis Neurosci. 1998;15:625–642. doi: 10.1017/s0952523898154044. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Grove EA, Fukuchi-Shimogori T. Generating the cerebral cortical area map. Annu Rev Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Gutierrez C, Yaun A, Cusick CG. Neurochemical subdivisions of the inferior pulvinar in macaque monkeys. J Comp Neurol. 1995;363:545–562. doi: 10.1002/cne.903630404. [DOI] [PubMed] [Google Scholar]
- Hikishima K, Quallo MM, Komaki Y, Yamada M, Kawai K, Momoshima S, Okano HJ, Sasaki E, Tamaoki N, Lemon RN, Iriki A, Okano H. Population-averaged standard template brain atlas for the common marmoset (Callithrix jacchus) Neuroimage. 2011;54:2741–2749. doi: 10.1016/j.neuroimage.2010.10.061. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Wang WZ, Lee S, Davies KE, Goffinet AM, Rakić S, Parnavelas J, Reim K, Nicolić M, Paulsen O, Molnár Z. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb Cortex. 2009;19:1738–1750. doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Murray KD, Warland DK, Feldheim DA, Chapman B. Ephrin-As mediate targeting of eye-specific projections to the lateral geniculate nucleus. Nature Neurosci. 2005;8:1013–1021. doi: 10.1038/nn1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami A, Yamane J, Katoh H, Nakamura M, Momoshima S, Ishii H, Tanioka Y, Tamaoki N, Nomura T, Toyama Y, Okano H. Establishment of graded spinal cord injury model in a nonhuman primate: the common marmoset. J Neurosci Res. 2005a;80:172–181. doi: 10.1002/jnr.20435. [DOI] [PubMed] [Google Scholar]
- Iwanami A, Kaneko S, Nakamura M, Kanemura Y, Mori H, Kobayashi S, Yamasaki M, Momoshima S, Ishii H, Ando K, Tanioka Y, Tamaoki N, Nomura T, Toyama Y, Okano H. Transplantation of human neural stem cells for spinal cord injury in primates. J Neurosci Res. 2005b;80:182–190. doi: 10.1002/jnr.20436. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG. The thalamus. 2nd edition. Cambridge: Cambridge UP; 2007. [Google Scholar]
- Jones EG, Rubenstein JL. Expression of regulatory genes during differentiation of thalamic nuclei in mouse and monkey. J Comp Neurol. 2004;477:55–80. doi: 10.1002/cne.20234. [DOI] [PubMed] [Google Scholar]
- Kaas JH. The evolution of the complex sensory and motor systems of the human brain. Brain Res Bull. 2008;75:384–390. doi: 10.1016/j.brainresbull.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnath HO, Himmelbach M, Rorden C. The subcortical anatomy of human spatial neglect: putamen, caudate nucleus and pulvinar. Brain. 2002;125:350–360. doi: 10.1093/brain/awf032. [DOI] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci U S A. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 1990;297:441–470. doi: 10.1002/cne.902970309. [DOI] [PubMed] [Google Scholar]
- Krubitzer L. The magnificent compromise: cortical field evolution in mammals. Neuron. 2007;56:201–208. doi: 10.1016/j.neuron.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Lambert N, Lambot M-A, Bilheu A, Albert V, Englert Y, Libert F, Noel J-C, Sotiriou C, Holloway AK, Pollard KS, Detours V, Vanderhaeghen P. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. Plos One. 2011;6:e17753. doi: 10.1371/journal.pone.0017753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C, Gaspar P, Nicolas D, Hornung JP. Transitory uptake of serotonin in the developing sensory pathways of the common marmoset. J Comp Neurol. 2006;499:677–689. doi: 10.1002/cne.21137. [DOI] [PubMed] [Google Scholar]
- Leonard CM. Prefrontal cortex of rat. i. Cortical projection of mediodorsal nucleus. 2. Efferent connections. Brain Res. 1969;12:321–343. doi: 10.1016/0006-8993(69)90003-1. [DOI] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Leuner B, Kozorovitskiy Y, Gross CG, Gould E. Diminished adult neurogenesis in the marmoset brain precedes old age. Proc Natl Acad Sci U S A. 2007;104:17169–17173. doi: 10.1073/pnas.0708228104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield K. Marmoset models commonly used in biomedical research. Comp Med. 2003;53:383–392. [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Dynamic patterned expression of orphan nuclear receptor genes ROR alpha and ROR beta in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Ogren MP, Racic P. The prenatal development of the pulvinar in the monkey—thymidine-h-3 autoradiographic and morphometric analyses. Anat Embryol. 1981;162:1–20. doi: 10.1007/BF00318090. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- Petersen SE, Robinson DL, Morris JD. Contributions of the pulvinar to visual spatial attention. Neuropsychologia. 1987;25:97–105. doi: 10.1016/0028-3932(87)90046-7. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and corticocortical connection patterns in the monkey. European J Neurosci. 2002;16:291–310. doi: 10.1046/j.1460-9568.2001.02090.x. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Poswillo DE, Hamilton WJ, Sopher D. Marmoset as an animal model for teratological research. Nature. 1972;239:460–462. doi: 10.1038/239460a0. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman HR, Gross CG, Albright TD. Afferent basis of visual response properties in area mt of the macaque. i. Effects of striate cortex removal. J Neurosci. 1989;9:2033–2050. doi: 10.1523/JNEUROSCI.09-06-02033.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MA, Pesold C, Liu WS, Kriho V, Guidotti A, Pappas GD, Costa E. Colocalization of integrin receptors and reelin in dendritic spine postsynaptic densities of adult nonhuman primate cortex. Proc Natl Acad Sci U S A. 2000;97:3550–3555. doi: 10.1073/pnas.050589797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas P, Joodmardi E, Hong Y, Perlmann T, Ogren SO. Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol Psychiatry. 2007;12:756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- Rojas P, Joodmardi E, Perlmann T, Ogren SO. Rapid increase of Nurr1 mRNA expression in limbic and cortical brain structures related to coping with depression-like behavior in mice. J Neurosci Res. 2010;88:2284–2293. doi: 10.1002/jnr.22377. [DOI] [PubMed] [Google Scholar]
- Romanski LM, Giguere M, Bates JF, Goldman-Rakic PS. Topographic organization of medial pulvinar connections with the prefrontal cortex in the rhesus monkey. J Comp Neurol. 1997;379:313–332. [PubMed] [Google Scholar]
- Rosa MGP, Tweedale R. Brain maps, great and small: lessons from comparative studies of primate visual cortical organization. Philos Trans Royal Soc B Biol Sci. 2005;360:665–691. doi: 10.1098/rstb.2005.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Hanazawa K, Kurita R, Akatsuka A, Yoshizaki T, Ishii H, Tanioka Y, Ohnishi Y, Suemizu H, Sugawara A, Tamaoki N, Izawa K, Nakazaki Y, Hamada H, Suemori H, Asano S, Nakatsuji N, Okano H, Tani K. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus) Stem Cells. 2005;23:1304–1313. doi: 10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, Kokubo N, Kimura-Yoshida C, Matsuo I, Sumiyama K, Saitou N, Shimogori T, Okada N. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci U S A. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- Schmid MC, Mrowka SW, Turchi J, Saunders RC, Wilke M, Peters AJ, Ye FQ, Leopold DA. Blindsight depends on the lateral geniculate nucleus. Nature. 2010;466:373–377. doi: 10.1038/nature09179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Ogawa M. Gene application with in utero electroporation in mouse embryonic brain. Dev Growth Differ. 2008;50:499–506. doi: 10.1111/j.1440-169X.2008.01045.x. [DOI] [PubMed] [Google Scholar]
- Stepniewska I, Kaas JH. Architectonic subdivisions of the inferior pulvinar in New World and Old World monkeys. Vis Neurosci. 1997;14:1043–1060. doi: 10.1017/s0952523800011767. [DOI] [PubMed] [Google Scholar]
- Supèr H, Uylings HB. The early differentiation of the neocortex: a hypothesis on neocortical evolution. Cereb Cortex. 2001;11:1101–1109. doi: 10.1093/cercor/11.12.1101. [DOI] [PubMed] [Google Scholar]
- Suzuki-Hirano A, Ogawa M, Kataoka A, Yoshida AC, Itoh D, Ueno M, Blackshaw S, Shimogori T. Dynamic spatiotemporal gene expression in embryonic mouse thalamus. J Comp Neurol. 2011;519:528–543. doi: 10.1002/cne.22531. [DOI] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Activity-dependent expression of occ1 in excitatory neurons is a characteristic feature of the primate visual cortex. Cereb Cortex. 2006;16:929–940. doi: 10.1093/cercor/bhj034. [DOI] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Differential expression patterns of occ1-related genes in adult monkey visual cortex. Cereb Cortex. 2009;19:1937–1951. doi: 10.1093/cercor/bhn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuno H, Tanaka I, Umitsu Y, Akazawa T, Nakamura Y. Digital brain atlas of the common marmoset 2.0. Neurosci Res. 2009a;65:S225–S225. doi: 10.1016/j.neures.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Tokuno H, Tanaka I, Umitsu Y, Akazawa T, Nakamura Y. Web-accessible digital brain atlas of the common marmoset (Callithrix jacchus) Neurosci Res. 2009b;64:128–131. doi: 10.1016/j.neures.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Tomioka I, Maeda T, Shimada H, Kawai K, Okada Y, Igarashi H, Oiwa R, Iwasaki T, Aoki M, Kimura T, Shiozawa S, Shinohara H, Suemizu H, Sasaki E, Okano H. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells. 2010;15:959–969. doi: 10.1111/j.1365-2443.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii M, Levitt P. Dissociation of corticothalamic and thalamocortical axon targeting by an EphA7-mediated mechanism. Neuron. 2005;48:563–575. doi: 10.1016/j.neuron.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Ungerleider LG, Desimone R. Cortical connections of visual area mt in the macaque. J Comp Neurol. 1986;248:190–222. doi: 10.1002/cne.902480204. [DOI] [PubMed] [Google Scholar]
- Wall PM, Messier C. The hippocampal formation—orbitomedial prefrontal cortex circuit in the attentional control of active memory. Behav Brain Res. 2001;127:99–117. doi: 10.1016/s0166-4328(01)00355-2. [DOI] [PubMed] [Google Scholar]
- Wang Q, Burkhalter A. Area map of mouse visual cortex. J Comp Neurol. 2007;502:339–357. doi: 10.1002/cne.21286. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Ohsawa S, Hashikawa T, Yamamori T. Binding and complementary expression patterns of semaphorin 3E and plexin D1 in the mature neocortices of mice and monkeys. J Comp Neurol. 2006;499:258–273. doi: 10.1002/cne.21106. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Ichinohe N, Ohsawa S, Hashikawa T, Komatsu Y, Rockland KS, Yamamori T. Comparative analysis of layer-specific genes in mammalian neocortex. Cereb Cortex. 2007;17:1918–1933. doi: 10.1093/cercor/bhl102. [DOI] [PubMed] [Google Scholar]
- Weller RE, Steele GE, Kaas JH. Pulvinar and other subcortical connections of dorsolateral visual cortex in monkeys. J Comp Neurol. 2002;450:215–240. doi: 10.1002/cne.10298. [DOI] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH. Single-cell responses in striate cortex of kittens deprived of vision in 1 eye. J Neurophysiol. 1963;26:1003–1017. doi: 10.1152/jn.1963.26.6.1003. [DOI] [PubMed] [Google Scholar]
- Woolsey TA, Wann JR. Areal changes in mouse cortical barrels following vibrissal damage at different postnatal ages. J Comp Neurol. 1976;170:53–66. doi: 10.1002/cne.901700105. [DOI] [PubMed] [Google Scholar]
- Xing GQ, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res. 2006;84:36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Echigo C, Saiki M, Inada M, Watanabe S, Iriki A. Tool-use learning by common marmosets (Callithrix jacchus) Exp Brain Res. 2011;213:63–71. doi: 10.1007/s00221-011-2778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshima H, Yamasaki S, Voelker CCJ, Molnar Z, Christophe E, Audinat E, Takemoto M, Nishiwaki M, Tsuji S, Fujita I, Yamamoto N. ER81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137:401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]
- Yuge K, Kataoka A, Yoshida AC, Itoh D, Aggarwal M, Mori S, Blackshaw S, Shimogori T. Region-specific gene expression in early postnatal mouse thalamus. J Comp Neurol. 2011;519:544–561. doi: 10.1002/cne.22532. [DOI] [PubMed] [Google Scholar]
- Yun ME, Johnson RR, Antic A, Donoghue MJ. EphA family gene expression in the developing mouse neocortex: Regional patterns reveal intrinsic programs and extrinsic influence. J Comp Neurol. 2003;456:203–216. doi: 10.1002/cne.10498. [DOI] [PubMed] [Google Scholar]