Abstract
Next Generation Sequencing allows for testing and typing of entire genes of the HLA region. A better and comprehensive sequence assessment can be achieved by the inclusion of full gene sequences of all the common alleles at a given locus. The common alleles of DRB5 are under-characterized with the full exon-intron sequence of two alleles available. In the present study the DRB5 genes from 18 subjects alleles were cloned and sequenced; haplotype analysis showed that 17 of them had a single copy of DRB5 and one consanguineous subject was homozygous at all HLA loci. Methodological approaches including robust and efficient long-range PCR amplification, molecular cloning, nucleotide sequencing and de novo sequence assembly were combined to characterize DRB5 alleles. DRB5 sequences covering from 5’UTR to the end of intron 5 were obtained for DRB5*01:01, 01:02 and 02:02; partial coverage including a segment spanning exon 2 to exon 6 was obtained for DRB5*01:03, 01:08N and 02:03. Phylogenetic analysis of the generated sequences showed that the DRB5 alleles group together and have distinctive differences with other DRB loci. Novel intron variants of DRB5*01:01:01, 01:02 and 02:02 were identified. The newly characterized DRB5 intron variants of each DRB5 allele were found in subjects harboring distinct associations with alleles of DRB1, B and/or ethnicity. The new information provided by this study provides reference sequences for HLA typing methodologies. Extending sequence coverage may lead to identify the disease susceptibility factors of DRB5 containing haplotypes while the unexpected intron variations may shed light on understanding of the evolution of the DRB region.
Keywords: Next Generation Sequencing, de novo assembly, HLA-DRB5, Single Tandem Repeats, Gene conversion
1. Introduction
The major histocompatibility complex (MHC) was initially identified because differences in proteins from different individuals that are encoded in this genetic system play a major role in the rejection of tissues and organs. The class I and II MHC genes encode cell-surface heterodimers that play central roles in antigen presentation, tolerance, and self/non-self recognition [1–3]. The human histocompatibility class II genes encode for three cell-surface isotypes, designated HLA-DR, HLA-DQ, and HLA-DP. Each functional HLA class molecule is a heterodimer formed by an alpha and a beta subunit [4]. Molecular studies of the DR sub-region show one DRA gene, encoding the alpha chain, and multiple DRB genes, encoding the beta chains, on different haplotypes, which display, in addition, copy number variation. The DRA gene in humans is highly conserved while nine different HLA-DRB genes have been described. HLA-DRB1, -B3, -B4 and -B5 encode functional gene products, whereas -B2, -B6, -B7, -B8, and -B9 represent pseudogenes as manifested by various insertions/deletions (indels) and deleterious mutations [5]. Among the expressed HLA-DRB genes, DRB1 is the most polymorphic locus while DRB3, -B4 and -B5 have significantly less alleles as reported to the IMGT/HLA database [6]. The most common DRB5 alleles include DRB5*01:01:01, 01:02, 01:03, 01:08N and 02:02 [7–8]. Within the human population, five major region haplotype configurations have been described associated to serotypes DR1, DR51, DR52, DR8, and DR53, that are characterized by the presence of a unique combinations of DRB genes/pseudogenes [9–10]. For the chimpanzee (Pan troglodytes) and some macaque species 9 to more than 30 different DRB haplotypes have been described [11–12].
In the human MHC, the DRB5 locus is unique to the haplotypes bearing the DR51 serotype; DRB5 is adjacent to the DRB6 pseudogene. The DRB5 locus appears is likely to derive from ancestral from DRBl alleles, and was generated more than 20 million yr; DRB5 is present in chimpanzees and gorillas [13–14]. The DRB gene organization with duplications and the extensive allelic polymorphism of expressed DR-beta molecules are striking features. These findings suggest that the DRB genes and the corresponding functional molecules are under distinct selective pressures that resulted from episodic evolution involving both, population expansions and contractions with functional adaptations [14].
The evolutionary history of the HLA-DRB1 locus has been delineated thoroughly by the analysis of genomic full-length alleles (10–15 kb) of human and non-human primates [15]. In contrast, the full evolutionary history of the second expressed DRB loci of different haplotypes such as those bearing DRB5 has not been assessed because of incomplete intron sequence information. Significant information about the evolution and biological functions of DRB loci can be obtained through complete sequencing analysis in the second expressed DRB genes. These should include evaluation of both coding and non-coding regions such as the microsatellite (GT)x(GA)x repeats adjacent to exon 2 region [16]. This microsatellite in the MHC- DRB1 genes is interesting since not only in the exon/intron architecture is exactly conserved among all studied vertebrates but also has revealed that the exceptional polymorphism of exon 2 correlates with the variability of this microsatellite locus [17].
HLA matching for transplantation and mapping disease susceptibility and resistance factors, accurate and highly informative HLA allele assignment is desired. The application of whole gene next-generation sequencing (NGS) to the study of highly polymorphic and structurally complex regions of the human genome increases the throughput, accuracy, and resolution of genetic analysis by several orders of magnitude, presenting an opportunity to better understand the biological mechanisms underlying HLA disease associations [18]. For the evaluation of sequencing data, a comprehensive sequence reference database is needed in order to obtain accurate HLA assignments. Genomic references for some HLA allele lineages and loci are missing from the HLA sequences compiled by IMGT [6]. To address these limitations, we embarked in the characterization of genomic sequences of less studied or overlooked loci. In the present study we focused in the characterization of all common alleles of HLA-DRB5.
2. Materials and methods
2.1. Samples and DNA preparation
Samples from the international workshop cell lines (Research Cell Bank, Fred Hutchinson Cancer Research Center, Seattle, Washington) and from selected individuals previously typed by NGS were selected for this study. Genomic DNA was obtained using the QIAamp 96 DNA blood kit (Qiagen, Valencia, CA). Table 1 shows the samples and their HLA alleles. With exception of PGF, a cell line that is consanguineous and homozygous at all HLA loci [19], all the cell lines or subjects included in this study are heterozygous in DRB1 and carried one copy of DRB5.
Table 1:
HLA genotypes obtained by Next Generation Sequencing and reference sequences of 18 subjects and cell lines carrying DRB5.
| CLASS II ALLELES | CLASS I ALLELES | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| International Workshop Cell line # |
Local Sample ID |
Ethnic Origin |
Accession # | IMGT Submission # |
DRB5 | DRB1 | DQA1 | DQB1 | DPA1 | DPB1 | HLA-A | HLA-B | HLA-C |
| IHW0 9228 | JS | European - North America | AL713966 | DRB5*01:01:01 DRB3*02:02:01:01 |
DRB1*15:01:01 DRB1*03:01:01:01 |
DQA1*01:02:01:01 DQA1*05:01:01:01 |
DQB1*06:02:01 | DPA1*01:03:01:05 DPA1*01:03:01:01 |
DPB1*04:02:01:02 DPB1*02:02 |
A*03:01:01:01 A*30:03 |
B*07:02:01 B*18:01:01:01 |
C*07:02:01:03 C*05:01:01:01 |
|
| IHW0 9318 | PGF | European - England, Europe | AL713966 | DRB5*01:01:01 | DRB1*15:01:01 | DQA1*01:02:01 | DQB1*06:02:01 | DPA1*01:03:01:02 | DPB1*04:01:01:01 | A*03:01:01:01 | B*07:02:01 | C*07:02:01:03 | |
| IHW0 9123 | HAY, BD | Australian Aboriginal |
KU593577 | HWS10025809 | DRB5*01:01:01 STR1a DRB3*02:02:01:02 |
DRB1*15:01:01 DRB1*14:07:01 |
DQA1*01:02:01:01 DQA1*01:04:01 |
DQB1*06:02:01 DQB1*05:03:01:01 |
DPA1*01:03:01:02 DPA1*02:02:02 |
DPB1*04:01:01:01 DPB1*05:01:01 |
A*24:02:01:01 | B*07:02:01 B*40:02:01 |
C*07:02:01:03 C*15:02:01:01 |
| IHW0 9394 | BPOT | KU593577 | HWS10025809 | DRB5*01:01:01_STR1a | DRB1*15:01:01 DRB1*13:01:01 |
DQA1*01:02:01:01 DQA1*01:03:01:02 |
DQB1*06:02:01 DQB1*06:03:01 |
DPA1*01:03:01:04 | DPB1*04:01:01:01 | A*02:01:01:01 | B*07:03 B*15:01:01:01 |
C*03:04:01:01 C*03:03:01 |
|
| STA1001 | Asian | KU593580 | HWS10025817 | DRB5*01:01:01v1b | DRB1*15:01:01 DRB1*08:03:02 |
DQA1*01:02:01:03 DQA1*01:03:01 |
DQB1*06:02:01 DQB1*06:01:01 |
DPA1*01:03:01:01 DPA1*02:02:02 |
DPB1*05:01:01 | A*02:01:01:01 A*32:01:01 |
B*54:01:01 B*52:01:01:02 |
C*01:02:01 C*12:02:02 |
|
| STA1029 | Asian | KU593580 | HWS10025817 | DRB5*01:01:01v1b DRB4*01:03:01:02N |
DRB1*15:01:01 DRB1*07:01:01 |
DQA1*01:02:01:03 DQA1*02:01 |
DQB1*06:02:01 DQB1*03:03:02:01 |
DPA1*02:01:01 | DPB1*13:01:01 | A*02:06:01:01 A*03:01:01:01 |
B*35:01:01:02 B*35:03:01 |
C*03:03:01 C*12:03:01:01 |
|
| IHW0 9368 | Asian - Unknown | KU593576 | HWS10025819 | DRB5*01:01:01v1_STR1c | DRB1*15:01:01 DRB1*09:01:02 |
DQA1*01:02:01:01 DQA1*03:02 |
DQB1*06:02:01 DQB1*03:03:02:02 |
DPA1*01:03:01:01 DPA1*02:02:02 |
DPB1*02:01:02 DPB1*05:01:01 |
A*24:02:01:01 A*26:04 |
B*38:02:01 B*39:01:03 |
C*07:02:01:01 | |
| IHW0 9327 | THA | European - India, Asia | KU593571 | HWS10025805 | DRB5*01:02e1d DRB3*02:02:01 |
DRB1*15:02:02 DRB1*13:08 |
DQA1*01:03:01:01 DQA1*01:04:01 |
DQB1*06:01:01 DQB1*05:03:01 |
DPA1*01:03:01:04 DPA1*01:03:01:02 |
DPB1*04:01:01 | A*31:01:02:01 A*03:01:01:01 |
B*35:03:01 B*15:18:01 |
C*04:01:01:01 C*07:04:01 |
| STA1005 | Asian | KU593573 | HWS10025807 | DRB5*01:02e1 STR1e | DRB1*15:02:01 DRB1*10:01:01 |
DQA1*01:01:01 DQA1*01:05:01 |
DQB1*05:01:24 DQB1*05:01:01:02 |
DPA1*01:03:01:01 DPA1*02:02:02 |
DPB1*02:01:02 DPB1*13:01:01 |
A*11:01:01:01 A*29:01:01:01 |
B*38:02:01 B*07:05:01 |
C*15:05:02 C*07:02:01:01 |
|
| STA1007 | Asian | KU593574 | HWS10025813 | DRB5*01:03e1f DRB3*03:01:03 |
DRB1*15:02:01 DRB1*12:02:01 |
DQA1*01:01:01 DQA1*06:01:01 |
DQB1*05:02:01 DQB1*03:01:01:01 |
DPA1*02:02:02 DPA1*01:03:01:03 |
DPB1*05:01:01 DPB1*2 1:01 |
A*02:03:01 A*24:02:01:01 |
B*40:02:01 B*15:02:01 |
C*03:04:01:01 C*08:01:01 |
|
| STA1010 | Asian | KU593575 | HWS10025815 | DRB5*01:08Ne1g | DRB1*15:02:01 DRB1*10:01:01 |
DQA1*01:01:01 DQA1*01:05:01 |
DQB1*05:01:24 DQB1*05:01:01:02 |
DPA1*01:03:01:03 DPA1*02:02:02 |
DPB1*21:01 DPB1*05:01:01 |
A*11:01:01:01 A*29:01:01:01 |
B*38:02:01 B*07:05:01 |
C*07:02:01:01 C*15:05:02 |
|
| IHW0 9277 | HS67 | Asian - Japan | KU593572,KX687265 | HWS10025811 | DRB5*02:02e1h | DRB1*16:02:01v2k DRB1*08:03:02 |
DQA1*01:02:02 DQA1*01:03:01 |
DQB1*05:02:01 DQB1*06:01:01 |
DPA1*02:02:02 | DPB1*05:01:01 | A*11:01:01:01 A*24:02:01:01 |
B*67:01:01 B*48:01:01 |
C*07:02:01:01 C*08:03:01 |
| IHW0 9258 | DAN72 3 | American Indian - North America | KU593572,KX687282 | HWS10025811 | DRB5*02:02e1h DRB3*01:01:02 |
DRB1*16:02:01v1l DRB1*14:02:01 |
DQA1*05:05:01:03 DQA1*05:03 |
DQB1*03:01:01:01 DQB1*03:03:02:02 |
DPA1*02:01:01 DPA1*02:02:02 |
DPB1*14:01:01 DPB1*05:01:01 |
A*31:01:02:01 | B*15:08:01 B*40:02:01 |
C*01:02:01 C*03:04:01:02 |
| IHW0 9317 | FORE | European - France | KU593572,KX687283 | HWS10025811,IHW09317 | DRB5*02:02e1h DRB4*01:03:01:03 |
DRB1*16:04 DRB1*04:04:01 |
DQA1*01:02:02 DQA1*03:01:01 |
DQB1*05:02:01 DQB1*03:02:01 |
DPA1*01:03:01:03 | DPB1*03:01:01 DPB1*06:01 |
A*02:01:01:01 A*24:02:01:01 |
B*51:01:01:01 B*15:01:01:01 |
C*14:02:01 C*03:03:01 |
| IHW0 9365 | GRC-138 | American Indian - Brazil | KU593572 | HWS10025811 | DRB5*02:02e1h DRB3*01:01:02 |
DRB1*16:02:01 DRB1*14:13 |
DQA1*05:05:01 DQA1*05:03 |
DQB1*03:01:01:01 | DPA1*01:03:01:05 | DPB1*04:02:01:02 | A*02:01:01:01 A*02:11:01 |
B*40:03 B*15:04:01 |
C*03:04:01:02 C*03:03:01 |
| STA1016 | European | KU593572 | HWS10025811 | DRB5*02:02e1h DRB3*01:01:02 |
DRB1*16:02:01 DRB1*13:03 |
DQA1*01:02:02 DQA1*05:05:01:01 |
DQB1*03:01:01:03 DQB1*03:01:01:01 |
DPA1*01:03:01:05 DPA1*01:03:01:02 |
DPB1*04:02:01:02 DPB1*104:01 |
A*24:02:01:01 A*02:01:01:01 |
B*44:05:01 B*41:02:01 |
C*02:02:02:01 C*17:03 |
|
| IHW0 9112 | CHA, AJ | European | KU593578,KX687264 | HWS100 258 21,HWS10 026687 | DRB5*02:02e1_STR1i DRB3*02:02:01:01 |
DRB1*16:01:01e1 DRB1*03:01:01:02 |
DQA1*01:02:02 DQA1*05:01:01 |
DQB1*05:02:01 DQB1*02:01:01 |
DPA1*01:03:01:04 | DPB1*04:01:01 | A*03:01:01:01 A*24:03:01 |
B*39:06:02 B*50:01:01 |
C*12:03:01:01 C*15:04:01 |
| STA1020 | Asian | KU593579,KX687282 | HWS10025823 | DRB5*02:03e1j DRB3*01:01:02 |
DRB1*16:02:01v1l DRB1*13:02:01 |
DQA1*01:02:02 DQA1*01:03:01:02 |
DQB1*05:02:01 DQB1*06:03:01 |
DPA1*02:02:02 | DPB1*02:02 DPB1*05:01:01 |
A*02:03:01 A*33:03:01 |
B*38:02:01 B*58:01:01:01 |
C*07:02:01:01 C*03:02:02:01 |
|
DRB5*01:01:01_STR1 is an intronic variant of DRB5*01:01:01 with copy number variations in the intron 2 STR region
DRB5*01:01:01v1 is an intronic variant of DRB5*01:01:01
DRB5*01:01:01v1_STR1 is an intronic variant of DRB5*01:01:01 with copy number variations in the intron 2 STR region
DRB5*01:02e1 is an extended genomic sequence of DRB5*01:02
DRB5*01:02e1_STR1 is an intronic variant of DRB5*01:02 with copy numper variations in the intron 2 STR region
DRB5*01:03e1 is an extended genomic sequence of DRB5*01:03
DRB5*01:08Ne1 is an extended genomic sequence of DRB5*01:08N
DRB5*02:02e1 is an extended genomic sequence of DRB5*02:02
DRB5*02:02e1_STR1 is an extended genomic sequence of DRB5*02:02
DRB5*02:03e1 is an extended genomic sequence of DRB5*02:03
DRB1*16:02:01v2 is an intronic variant of DRB1*16:02:01
DRB1*16:02:01v1 is an intronic variant of DRB1*16:02:01
2.2. HLA Database Construction Strategy
2.2.1. PCR Amplification
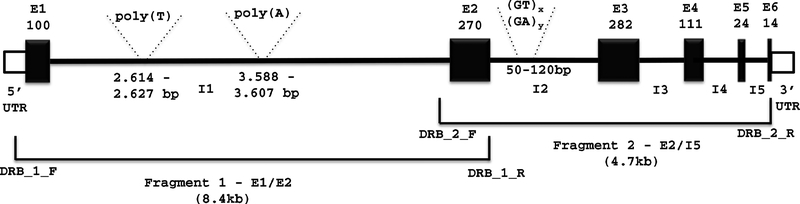
DRB alleles were amplified by long-range PCR (Long AMP polymerase, New England Biolabs) of genomic DNA. In order to determine the nucleotide sequences, two fragments overlapping in the highly polymorphic exon 2 region were amplified (Fig. 1). The primers were designed on the basis of examination of DRB sequences available in the IMGT database [6] and Ensembl [20] with the aid of Integrated DNA Technologies OligoAnalyzer v3.1 software. 0.2 M Trehalose was introduced into PCR reaction to obtain reliable and efficient amplification [21].
Figure 1:
Schematic illustration of amplified fragments of DRB5. The DRB5*01:01:01 is used as a reference for sequence annotation. Exons are represented by black boxes. E1–E6 denotes exons 1–6. I1–I5 denotes introns 1–5, 5’3’UTR, homopolymer (Poly Ts/As) and STRs locations. The numbers represent the exon length in base pairs. The two amplified fragments are showed at the bottom; they overlap in Exon 2 and flanking sequences. Primers DRB_1_F and DRB_1_R were used to amplify fragment 1 and primers DRB_2_F and DRB_2_R were utilized to amplify fragment 2.
2.2.2. Molecular Cloning of PCR products
The two PCR products were gel purified and cloned into the pCR-XL-TOPO vector using the TOPO® XL PCR Cloning Kit (ThermoFisher Scientific). The positively identified clones confirmed via gel agarose electrophoresis were further examined by Sanger sequencing (Elim Biopharmaceuticals, Inc).
2.2.3. Nucleotide Sequencing by NGS
The library construction protocol of the allelic clones for NGS was performed exactly as described by Wang et al. [22]. Sequencing was performed in a MiSeq sequencer using 250 bp paired-end reads run according to the manufacturer’s instructions (Illumina, San Diego).
2.2.4. Processing of the NGS data and de-novo Assembly
The fastx_barcode_splitter.pl from Hannon/CSHL [23] was used to demultiplex of the raw fastq data generated by NGS sequencer. Read trimming was performed with bwa -q 20 [24]. The fastq file produced for each sample was converted to a corresponding fasta, which was subsequently used as the input to a developed de novo assembly process. The developed algorithm was a blast based assembler [25] and performed the sequence assembly for each cloned amplicon. For the accurate determination of the STR and homopolymer lengths a localized sequence assembly, micro- assembly, of the reads mapping around these locations analyzed directly.
The exact HLA Database construction Strategy with evaluation and comparison with other available algorithms is included in the Supplementary Materials and Methods.
2.3. Phylogenetic Analysis
The full-length generated DRB sequences were deposited into the NIH [6] and IMGT HLA genetic database [26] (Table 1). The non-human primates DRB5 sequences of Pan troglodytes part-DRB5*01:02 and Macaca mulatta mamu-DRB5*03:01 were retrieved from the IPD-MHC-NHP database [27,28].
Pairwise comparisons of the DRB sequences were performed with the EMBOSS needle program using the Needleman-Wunsch algorithm [29]. Multiple sequence alignments executed by the Clustal Omega program [29]. The software MEGA 6.0 [30] was employed to calculate nucleotide diversity and to construct phylogenetic trees. The phylogenetic trees were reconstructed using the maximum likelihood method with Jukes-Cantor model [31]. The phylograms were a consensus of 500 boostraps replicates.
3. Results
3.1.
3.1.1. Description of HLA-DRB5 alleles
This report provides additional information regarding sequence variation at both, exons and introns of the most common DRB5 alleles. Out of the 87 samples genotyped, 18 samples harbored the DRB5 alleles (Table 1); the resulting consensus sequences were analyzed and compared between them and with other alleles of the DRB gene families as well as DRB5 alleles of non-human primates.
In the present study we identified intron variants resulting from SNP or STR variation. We developed a local naming convention where the e suffix indicated sequence extension, the suffix v specified intron variants while the suffix _STR indicated differences in the STR length (Table 1).
The sequence analyses showed that seven individuals carried DRB5*01:01:01, six DRB5*02:02, two DRB5*01:02, and one of each, DRB5*01:03, DRB5*01:08N and DRB5*02:03. The sequence of DRB5*01:01:01 was confirmed in two samples (JS and PGF, Table 1). The CDS present in IMGT [6] for DRB5*01:02 and DRB5*02:02 was confirmed and extended from the 5’UTR up to the end of intron 5 including all intervening introns and exons with the total length of 12,681 and 12,638 bp respectively (Supplementary Tables 1 and 2). For DRB5*02:03, DRB5*01:03 and DRB5*01:08N alleles the cloning data extended the previously known sequence from the beginning of exon 2 to the end of intron 5 with lengths of 4,658, 4,693 and 4,672 bp respectively (Supplementary Table 2).
3.1.2. 5’ UTR sequences (PCR Fragment 1)
A segment of 173 bp upstream of exon 1 was identical for DRB5*01:01:01, 01:02 and 02:02. Examination of sequences from three alleles in which fragment 1 (5’UTR to exon 2) was analyzed showed that exon 1 of DRB5*01:02 is identical to that of DRB5*01:01:01; these alleles differ from DRB5*02:02 by two nucleotide substitutions resulting in a one amino acid change in codon −16 (K to V substitution) (Table 2). There is higher sequence divergence between DRB5*01 and DRB5*02 alleles in intron 1 (Fig. 2A,B, Supplementary Tables 1,2). The nucleotide differences between DRB5*01:01:01, DRB5*01:02 and DRB5*02:02 are shown in Figure 2A,B and Supplementary Tables 1,2. The location of two intron 1 homopolymers in reference to the first nucleotide of DRB5*01:01:01 in intron 1 is shown in Figures 2A,B. length variation (Table 2) and the sequences of these (T)x (Fig. 2A) and (A)x (Fig. 2B) homopolymers are distinctive among the described alleles. An estimation example of the length of the (A)x and (T)x intron 1 homopolymers is be shown in Figures 3A and 3B.
Table 2:
SNPs outside of exon 2 and STRs variations in all DRB5 alleles examined.
| Exon 1 | Intron1 Homopolymers | Intron2 STR Structure | Intron 2 SNPs | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Codon16(aa) | poly(T) | poly(A) | 498 | 548 | 565 | 837 | 1480 | 1550 | 1670 | 1804 | 1890 | 122 | 391 | 412 | |||||||||
| DRB5*01:01:01 | AAG(K) | 14 | 20 | (GT)21 | (GA)5 | (GGAA) | (GA)4 | CA | T | G | T | T | G | T | A | T | C | A | A | C | ||||
| DRB5*01:01:01 STR1a | (GT)19 | (GA)5 | (GGAA) | (GA)4 | CA | T | G | T | T | G | T | A | T | C | A | A | C | |||||||
| DRB5*01:01:01v1b | (GT)18 | (GA)8 | (GGAA) | (GA)4 | CA | T | G | T | T | G | T | G | T | C | A | A | C | |||||||
| DRB5*01:01:01v1 STR1c | (GT)20 | (GA)8 | (GGAA) | (GA)4 | CA | T | G | T | T | G | T | G | T | C | A | A | C | |||||||
| DRB5*01:02e1d | AAG(K) | 16 | 19 | (GT)22 | (GA)8 | (GGAA) | (GA)4 | CA | (GA)7 | (GGAA) | (GA)4 | CA | T | G | T | T | G | G | G | C | C | A | A | C |
| DRB5*01:02e1 STR1e | (GT)21 | (GA)9 | (GGAA) | (GA)4 | CA | (GA)7 | (GGAA) | (GA)4 | CA | T | G | T | T | G | G | G | C | C | A | A | C | |||
| DRB5*01:03e1f | (GT)21 | (GA)9 | (GGAA) | (GA)4 | CA | (GA)7 | (GGAA) | (GA)4 | CA | T | G | T | T | G | G | G | C | C | A | A | C | |||
| DRB5*01:08Ne1g | (GT)20 | (GA)9 | (GGAA) | (GA)4 | CA | (GA)7 | (GGAA) | (GA)4 | CA | T | G | T | T | G | G | G | C | C | A | A | C | |||
| DRB5*02:02e1h | GTG(V) | 12 | 16 | (GT) | (GA)11 | (GGAA) | (GA)4 | CA | G | A | C | A | C | G | G | C | T | G | C | T | ||||
| DRB5*02:02e1 STR1i | (GT)15 | (GA)11 | (GGAA) | (GA)4 | CA | G | A | C | A | C | G | G | C | T | G | C | T | |||||||
| DRB5*02:03e1j | (GT)15 | (GA)11 | (GGAA) | (GA)4 | CA | G | A | C | A | C | G | G | C | T | G | C | T | |||||||
| patr-DRB5*01:02 | GTG(V) | 13 | 18 | (GT)4 GA(GT)7 | (GA)7 | (GCAA) | (GA)4 | CA | (GA)3 | T | G | C | A | C | G | G | C | C | A | A | C | |||
| mamu-DRB5*03:01 | del | 10 | (GT)2 | (GA)2 AAGAAA(GA)2 | AAGAAA | (GA)4 | (GC)4 | T | G | C | A | C | G | G | T | C | A | C | C | |||||
| DRB1*16:01:01:01 | (GT)18 | (GA)8 | (GGAA) | (GA)6 | ||||||||||||||||||||
| DRB1*16:02:01:01 | (GT)18 | (GA)8 | (GGAA) | (GA)6 | ||||||||||||||||||||
| DRB1*15:01:01:01 | (GT)22 | (GA)5 | CA | (GA)4 | CA | (GA)3 | (GGAA) | (GA)4 | ||||||||||||||||
| DRB1*09:01:02 | (GT)10 | (GA)9 | (GGAA) | (GA)4 | CA | GAAAGAGGGA | ||||||||||||||||||
DRB5*01:01:01_STR1 is an intronic variant of DRB5*01:01:01 with copy number variations in the intron 2 STR region
DRB5*01:01:01v1 is an intronic variant of DRB5*01:01:01
DRB5*01:01:01v1_STR1 is an intronic variant of DRB5*01:01:01 with copy number variations in the intron 2 STR region
DRB5*01:02e1 is an extended genomic sequence of DRB5*01:02
DRB5*01:02e1_STR1 is an intronic variant of DRB5*01:02 with copy numper variations in the intron 2 STR region
DRB5*01:03e1 is an extended genomic sequence of DRB5*01:03
DRB5*01:08Ne1 is an extended genomic sequence of DRB5*01:08N
DRB5*02:02e1 is an extended genomic sequence of DRB5*02:02
DRB5*02:02e1_STR1 is an extended genomic sequence of DRB5*02:02
DRB5*02:03e1 is an extended genomic sequence of DRB5*02:03
DRB1*16:02:01v2 is an intronic variant of DRB1*16:02:01
DRB1*16:02:01v1 is an intronic variant of DRB1*16:02:01
Figure 2.
A: CLUSTALW partial alignment of three de novo assembled DRB5 alleles within intron 1 shows the length variation of the poly(T) repeats. Numbering on the right indicates the relative nucleotide position at this intron for the corresponding allele. B: CLUSTALW partial alignment of three de novo assembled DRB5 alleles within intron 1 exhibits the length variation of the poly(A) repeats. Numbering on the right indicates the relative nucleotide position at this intron for the corresponding allele.
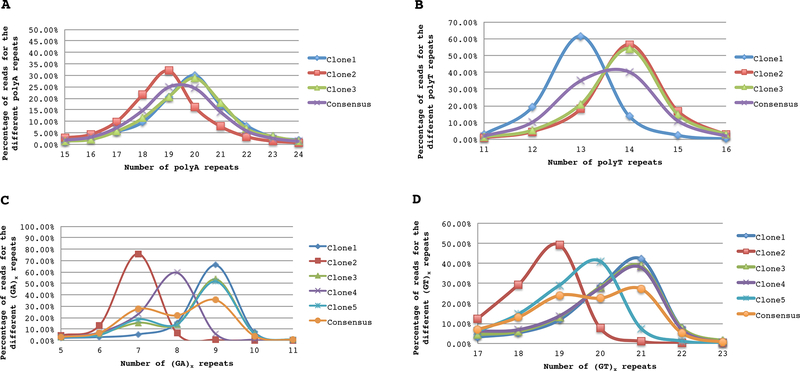
Figure 3.
A: Estimation of the length of intron 1 poly(A) in DRB5*01:01:01 from three clones isolated from cell line JS. The most representative STR length was selected for each clone according to percentages of reads; the length found in more clones was assigned as the final length for the tandem repeat. B: Estimation of the length of intron 1 poly(T) in DRB5*01:01:01 from three clones isolated from the cell line JS. The most representative STR length was selected for each clone according to percentages of reads; the length found in more clones was assigned as the final length for the tandem repeat. C: Estimation of the length of intron 2 (GA)x STR in DRB5*01:03 found in cell 119990 from five clones of cell 119990. The most representative STR length was selected for each clone according to percentages of reads; the length found in more clones was assigned as the final length for the tandem repeat. D: Estimation of the length of intron 2 (GT)x STR in DRB5*01:03 found in cell 119990 from five clones of cell 119990. The most representative STR length was selected for each clone according to percentages of reads; the length found in more clones was assigned as the final length for the tandem repeat.
3.1.3. Extended sequence coverage (PCR Fragment 2)
Fragment 2 (exon 2 to intron 5) was cloned and analyzed for all 18 samples bearing DRB5. In these samples, the length of this fragment ranged from 4,657 bp (DRB5*01:01:01v1) to 4,693 bp (DRB5*01:02_STR1) (Fig. 1). Interestingly, almost all the length variation between these alleles resulted from differences in the number of two dinucleotide STRs, (GT)x and (GA)x of intron 2 (50bp after exon 2) (Table 2).
As previously described [32–33], additional length variation results from the deletion of 2 and 19 nucleotides in exon 2 and exon 3, respectively of DRB5*01:10N and DRB5*01:08N (Supplementary Tables 1,2).
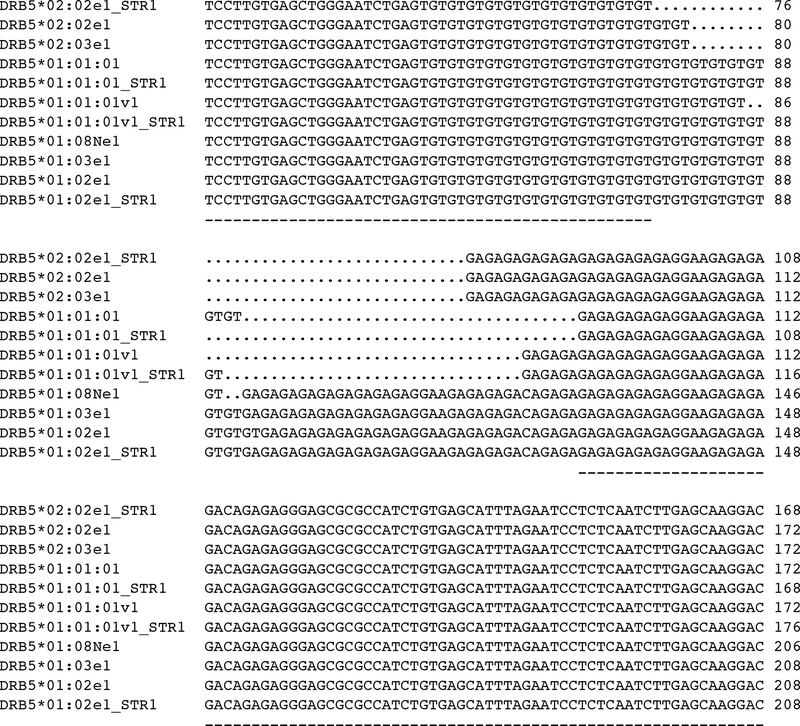
Three additional intron variants were identified among 7 subjects carrying DRB5*01:01:01; these are defined by differences in STR length in intron 2 (DRB5*01:01:01_STR1, DRB5*01:01:01v1, DRB5*01:01:01v1_STR1) and an intron 2 SNP variation (9990A/G) (DRB5*01:01:01v1, DRB5*01:01:01v1_STR1) (Table 2). Figure 1 shows the location of two contiguous STRs located at the beginning of intron 2 in reference to DRB5*01:01:01. The sequences of both STRs of all DRB5 alleles described in the present study are shown in Figure 4; their length variation is shown in Table 2. Based on the analyses of SNPs in introns 2 and 3 (Table 2) three DRB5 lineages have been identified, namely DRB5*01:01:01, DRB5*01:02 (including also 01:03 and 01:08N) and DRB5*02. Table 1 shows the cell lines or subjects that carry the four variants of DRB5*01:01:01 also carry DRB1*15:01:01; these DRB5*01:01:01 intron variants are found in distinct haplotypes defined either by variations in the non-coding regions of the DRB1*15:01:01, in HLA-B or by distinguished by ethnic differences.
Figure 4:
CLUSTALW partial alignment of 11 de novo assembled DRB5 alleles within the intron 2 region depicts the length variation of (GA)x and (GT)x repeats. Numbering on the right indicates the relative nucleotide position at this intron for the corresponding allele.
Two intron 2 STR variants of DRB5*01:02 were described in the present study (DRB5*01:02e1 and DRB5*01:02_STR1). These alleles were found in cells carrying DRB1*15:02:02 for DRB5*01:02e1 and DRB1*15:02:01 for DRB5*01:02e1_STR1 and they differ also in DQA1, DQB1 or at both DQ loci (Table 1).
The DRB5*01:02_STR1 and DRB5*01:03 alleles differ only by one nucleotide substitution in exon 2 and were identical in the gene segment spanning exon 2 to 5 (Table 2). The estimation of the STR lengths in DRB5*01:03 found in cell 119990 are shown in Figures 3C (GA)x and 3D (GT)x. The alleles DRB5*01:02_STR1 and DRB5*01:03 had high similarity with DRB5*01:08N in the assembled genomic region.
In this report two STR variants of DRB5*02:02 were identified (DRB5*02:02e1 and DRB5*02:02e1_STR1) (Fig. 4 and Table 2) and the genomic sequence of DRB5*02:03e1 was extended. The variants of DRB5*02:02 were found in haplotypes carrying different subtypes of DRB1*16. DRB5*02:02e1 and DRB5*02:03e1 were found in cells carrying DRB1*16:02:01 and DRB1*16:04 while the allele DRB5*02:02e1_STR1 was found in the cell line CHA, AJ that carries DRB1*16:01:01(Table 1).
3.2.
3.2.1. Comparison with other DRB genes
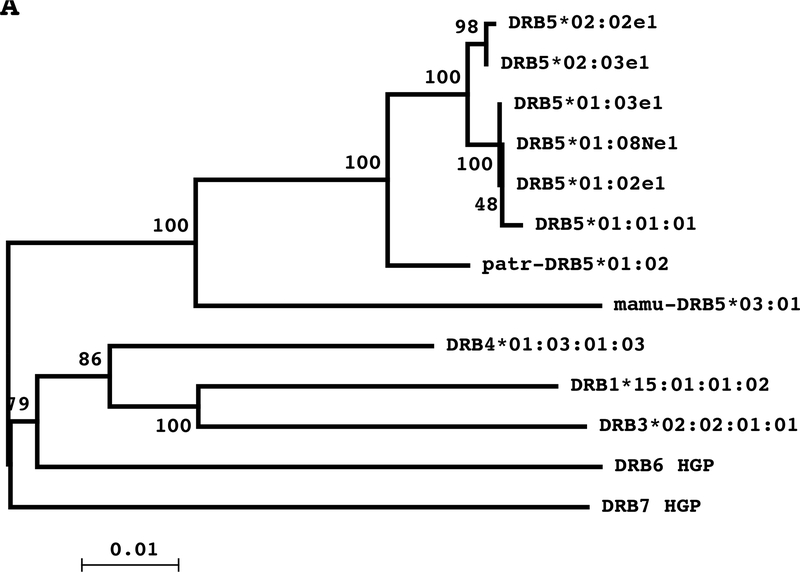
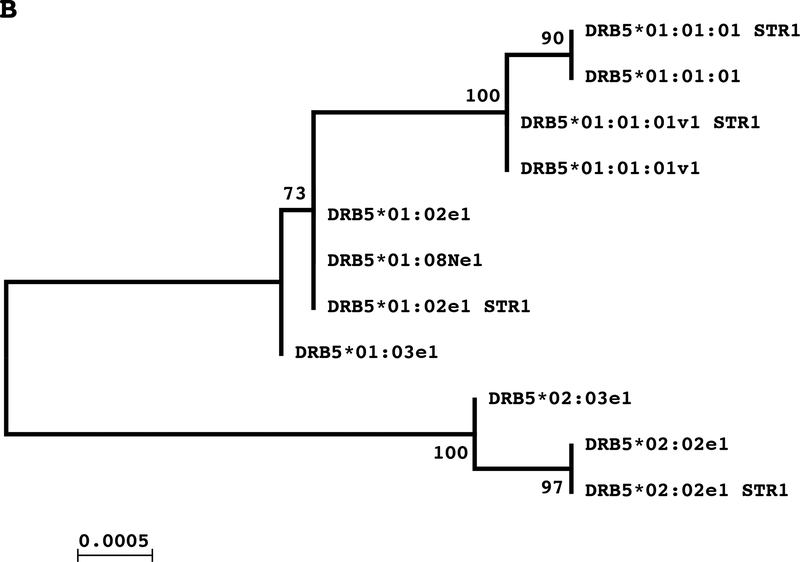
A global alignment (EMBOSS Needle) [29] and a phylogenetic tree was employed to determine fragment 2 homologies and phylogenetic relationships between DRB5*01:01:01 with other DRB5 alleles and DRB genes (DRB1/3/4/6/7). DRB5 alleles had very high similarity scores with distinctive differences with other DRB loci (Fig. 5A). The intra pairwise alignment displayed lower similarity scores between DRB5*01:01:01 and other DRB genes; the DRB5 gene had higher similarity with DRB4 followed by DRB3 and DRB1*15:01. The alignments between DRB5*01:01:01 and DRB pseudogenes (DRB6 and DRB7) showed the highest genetic dissimilarity (Supplementary Table 3). The pairwise comparisons between DRB5 alleles indicated that DRB5*01:02, DRB5*01:03 and DRB5*01:08N are highly homologous and as a group present the lowest distances with DRB5*01:01:01, DRB5*02:02 and DRB5*02:03 alleles (Supplementary Table 4). The phylogenetic analysis identified three clusters of alleles with the DRB5*01:02 group being in the middle and closer to the DRB5*01:01 group than to the DRB5*02 group (Fig. 5B).
Figure 5.
A: Phylogenetic tree of DRB sequences spanning from exon 2 to exon 5 obtained by the Maximum Likelihood method based on the Jukes-Cantor model. Numbers on the branches indicate the bootstrap values for 500 repeats. B: Phylogenetic tree of DRB5 sequences spanning from exon 2 to exon 5 obtained by the Maximum Likelihood method based on the Jukes-Cantor model. Numbers on the branches indicate the bootstrap values for 500 repeats.
3.2.2. Comparison with Non-Human primates DRB5 alleles
Phylogenetic analysis of exon 2 to intron 5 sequences and intron 2 only for human and non-human primate DRB genes and DRB5 alleles were performed. The results indicated that the hominid DRB5 alleles (Homo sapiens and Pan troglodytes) and the old world monkey mamu-DRB5*03:01 allele of Macaca mulatta form a separate clade in comparison to the other DRB genes (Fig. 5A, Supplementary Figure 1 ). In addition, a global alignment was conducted, using the Needleman-Wunsch algorithm [29], among HLA-DRB5 alleles, the common chimpanzee patr-DRB5*01:02 and Rhesus macaque mamu-DRB5*03:01. The results showed high homology of 92.6% between HLA-DRB5*01:01:01 and patr-DRB5*01:02 but lower with the other human DRB5 alleles. The mamu-DRB5*03:01 and HLADRB5*01:01:01 sequences had 91.5% similarity which was higher than the alignment among other DRB genes (Supplementary Table 3). Further global pairwise alignment analysis was performed for separate introns between human and non-human primates of DRB5 alleles and can be found in Supplementary Table 5. Interestingly, SNPs that are informative and separate the human DRB5 families can be found also in non-human species (Table 2).
3.3. STR/Homopolymer Analysis of DRB5 alleles
In this study, the variations of simple repeat stretches with the basic structures poly(A), poly(T) and (GT)x(GA)x in the different intronic areas of the HLA-DRB5 alleles were extensively investigated (Table 2, Fig. 3A–D). The NGS data showed an inverse correlation between the accuracy in the determination of STR length and the length of the repeats which is concordant with previous studies [34–37]. The short repeats T(6,9) in intron 1 of DRB5*01:01:01 were determined with high confidence because more than 90% of the reads in this area for each clone indicated the same poly(T) length. On the contrary, for longer homopolymers (T)14, (A)20 in intron 1 of DRB5*01:01:01 and STRs GA(9), GT(21) in intron 2 of DRB5*01:03e1, the reads showed a broader Gaussian like distribution; the highest frequency of the most representative reads for this area per clone were ranging from 22% to 60% (Fig. 3A–D). This broader distribution results in more difficult determination of the length of these STRs; more than 3 clones were required to be sequenced separately (instead of pooling them) for achieving accurate analysis.
The length variation and the basic structure of intron 2 STRs for all the DRB alleles for human and primates that has been examined in this report are be shown in Table 2.
The (GT)x variations associate with specific haplotypes and ethnic background as shown in Table 1 while the intron (GA)x variants correlate with the three DRB5 lineages namely DRB5*01:01:01, DRB5*01:02 (including also 01:03 and 01:08N) and DRB5*02 (Table 2).
Comparing the human DRB5 intron 2 microsatellite structures with other DRB genes shows only similarity with DRB1*15/16 and DRB1*09 alleles (Table 2). The intron data indicates that DRB1*16:01:01 was generated through a gene conversion event involving DRB1*15:01:01:01 (recipient allele) and DRB5*01:01:01v1 (donor allele) alleles that are found on the same haplotype (Table 1); the exchange segment is shown in Figure 6.
Figure 6:
Nucleotide sequences of a putative segmental exchange involved in the generation of DRB1*16:01:01. Boxes show the segments shared by DRB1*15:01 and DRB5*01:01 defining the boundaries of the segmental exchange.
Numbering on the right indicates the relative nucleotide position from the beginning of exon 2 for the corresponding allele.
Only three DRB5 alleles were examined in the first segment spanning intron 1; The poly(A) and poly(T) exhibited high variation in the number of repeats in both human and other primates DRB5 alleles (Table 2). More sequencing data is required to further characterize and determine the diversity and possible evolution of this region.
4. Discussion
In the present study, robust and improved methodological approaches were applied to characterize the gene structure and diversity of the second DRB locus expressed in haplotypes bearing DRB1*15 and DRB1*16 alleles. The novel strategy for full coverage and cost-effective allele sequencing took advantage of a modified long range PCR, highly efficient molecular cloning, clonal pooling, Illumina’s NGS platform and a novel assembly algorithm specific to HLA genes [Supplementary Materials and Methods] [21, 38–39]. Long read sequencing technologies were not used in this study since may present major drawbacks such as PCR-chimera formation and biased reference alignment, which need to be considered when attempting to phase variants [38].
Because of the noise inherent to NGS protocols, cloning errors and sequence complexity in the HLA alleles, de-novo assemblers such as Velvet (v1.2.1) and SSAKE (v3.8.2) [40–41] could not generate the full length and error-free sequence of each selected allele. These limitations led us to design an in-house assembly algorithm [Supplementary Materials and Methods]. The addition of a local de novo assembly and the evaluation of isolated clones was able to resolve the low complexity and high error areas such as homopolymers and STR regions (Fig. 3A–D) [42–44]. This approach provided significant and valuable information that could have not been obtained otherwise. The novel variants reported here were not deemed to receive an official name by WHO Nomenclature Committee for Factors of the HLA System. Nevertheless, the information and publication of these sequences provides significant value for the application of NGS based methods to HLA typing and further delineate evolutionary relations of HLA alleles and haplotypes [45,46].
The SNP analysis of the most common DRB alleles shows that all the variation in DRB5 locus resides in the 5’ side of the gene including exon 1 up to intron 3, whereas the region comprises exon 4 to intron 5 is completely conserved. Virtually all SNPs observed in introns 2 and 3 and the intron 2 (GA)x correlate exactly with three DRB5 lineages (Fig 5B) also defined previously by exon analyses only (Table 2). The intron 2 (GT)x appears to define recently generated variants in specific haplotypes and ethnic background as shown in Table 1. At the DRB5 locus, the intron 2 (GT)x appears to evolve more rapidly in comparison to the intron 2 (GA) x; the evolution rate of these STR is not similar at other DRB genes. This observation may be explained by: 1) different mutation rates among the DRB allele families and loci; and/or 2) different selective pressures in each DRB families. In the present study all alleles of the same DRB5 subfamily contain the same length (GA)x in intron 2 while they may differ in the length of the (GT)x. Both STRs are long; therefore, the mutation rate observed for these STRs indicates that the variation does not necessarily results from the repeat length. Interruptions in the perfect (GA)x repeats may decrease the mutation rate; in addition differentially acting selective pressures such convergent evolution may decrease the exon variability to that of the intron microsatellites [47,48].
The observed intron variability of the DRB5 STRs could result in possible biological and functional effects that need to be further evaluated. Permanent coevolution with exons suggests a possible biological role of these composite intron microsatellites [49–53]. Despite the repeat length variations of the STRs, the basic structure of (GT)x(GA)x is highly conserved between different families of DRB5 gene (Table 2). Interestingly, similar structures can be seen in DRB1*09/15/16 alleles (Table 2) which led us to confirm a previous hypothesis regarding a reciprocal intergenic exchange between DRB loci [54]. This report shows that the DRB1*16:01:01 allele may have arisen by reciprocal intergenic exchange between DRB1*15:01:01:01 (recipient allele) and DRB5*01:01:01v1 (donor allele) in the DR51 haplotype and the postulated recombination sites are shown in Figure 6.
The phylogenetic analysis showed that the hominid DRB5 alleles (Homo sapiens and Pan troglodytes) and the old world monkey (OWM) mamu-DRB5*03:01 allele of Macaca mulatta form a separate clade in comparison to the other DRB genes (Fig. 5A). There were two major diversification events in the evolution of the HLA-DRB genes approximately 50 million years (my) ago. A DRB1*04 and an ancestor of the DRBl*03 cluster (DRBl*03, DRBl*15, and DRB3) diverged from each other approximately 50 million years (my) ago, and DRB5, DRB7, DRB8, and an ancestor of the DRB2 cluster (DRB2, DRB4, and DRB6) emerged by gene duplication [55]. These data confirm that the DRB5 locus is common to the Catarrhini’s DRB region and the DRB5 gene originated before the OWM–HOM deviation (~25 My ago) as has been reported in other studies [27, 56].
The second expressed DRB loci (DRB3, DRB4, and DRB5) exhibit only limited allelic polymorphism in humans [57]. HLA typing of DRB5 alleles examining exon sequences shows only a few common alleles [6]. The structural protein sequence conservation in different ethnic groups is remarkable in spite of the rich haplotype variation identified when examining DRB1-DQA1-DQB1 haplotype blocks containing DRB5 (Table 1) [58,59]. The present study identifies additional non-coding variation that is haplotype specific and appears to indicate that there may be constraints for further diversification at the protein level compared to variations in the non-coding regions. It is proposed that DRB5 alleles may exert specific functions and may complement with molecules present in DRB1*15/16 and in the corresponding DQA1-DQB1 heterodimers [60–62]. Further work is needed to examine what immune responses are principally determined or restricted by the DRB5 alleles.
In all world populations that include haplotypes bearing in cis the genes encoding for DQA1*01:01 and DQB1*05:01, it has been observed that the DRB5 locus is absent in spite of the presence of the DRB6 pseudogene. These haplotypes include predominantly DRB1*01 alleles [59,63]. In addition to these haplotypes, Asian populations present frequently non-DRB1*01 haplotypes that include the DQ genes for the same DQ heterodimer (DQA1*01:01:01:01-DQB1*05:01:24); these haplotypes include the allele DRB1*15:02:01:03 and carry either DRB5*01:02 or DRB5*01:08N [32,64]. We speculate that the non-expressed DRB5*01:08N allele may have arisen recently in haplotypes bearing the latter DRB1-DQ alleles that also include DRB5*01:02. The putative mutational event with a deletion of 19 nucleotides in exon 3 of DRB5*01:02e1_STR1 may have resulted in the generation of DRB5*01:08N. This deletion produces a truncated non-membrane bound protein [32]. Given the high frequency of the allele DRB5*01:08N, it can be speculated that the haplotypes bearing this allele may have been positively selected in Asian populations. A plausible explanation is that because the DRB expressed alleles may determine negative selection of some T cells in the thymus, the non-expression of a specific allele may allow for the generation of some T-cell clones that could be effective in responding pathogen antigens via presentation by other HLA class II molecules. The various haplotypes (bearing DRB1*01:01, DRB1*01:02, DRB1*15:02) that include genes in cis encoding DQA1*01:01 and DQB1*05:01 include alleles with identical protein sequences that differ by exon silent substitutions or intron variations; these observations suggest more distant origins with positive pressure for structural DQ conservation that may be related to absence of DRB5 expression.
The molecules encoded by the second expressed DRB genes DRB3, DRB4 and DRB5 appear to have lower expression than DRB1 [65–70]. It should be noted that DRB4*01:03:01:02N is another common null allele that is found frequently in subjects with European and Asian ancestry [59]. In Africans, approximately 8 percent of the haplotypes bearing DRB1*15:03 lack DRB5 gene [59]. Therefore, DRB5 alleles, as other DRB low expression alleles may play a role in both providing specific responses to pathogens as well as determining the size of T-cell repertoire of an individual.
The present study provides a novel strategy for generating extended allele sequences of high quality that are reliable sources for HLA genomic references. Additionally, the characterization of the most common DRB5 alleles may improve the NGS based HLA typing by providing valuable phasing information that can subsequently lead to greater precision in matching donor organs to transplant recipients.
Supplementary Material
Acknowledgements
We acknowledge sincerely the contribution made by investigators at the Research Cell Bank, Fred Hutchinson Cancer Research Center, Seattle, Washington. We thank Eleni Koukou and Dr. Despoina Alexandraki from University of Crete, Greece for their insightful discussions and comments.
Funding
This work was supported by grant U19NS095774 (KB, MFV) from the U.S. National Institutes of Health (NIH).
Abbreviations:
- HLA
Human Leukocyte Antigens
- MHC
Major Histocompatibility Complex
- NGS
Next-Generation Sequencing
- PCR
Polymerase Chain Reaction
- STR
Single tandem repeats
- UTR
Untranslated Region
- indel
insertion or deletion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
none
Appendix A. Supplementary Data
Supplementary Materials and Methods: The HLA Database Construction protocol with evaluation and comparison with other algorithms.
Supplementary Table 1: Genomic sequences of cloned 5’ fragment of DRB5 alleles aligned in reference to DRB5*01:01:01.
Supplementary Table 2: Genomic sequences of cloned 3’ fragment of DRB5 alleles aligned in reference to DRB5*01:01:01.
Supplementary Table 3: Exon 2 to Intron 5 pairwise sequence alignment similarities between DRB5*01:01:01 with other DRB5 alleles and DRB1, DRB3, DRB4, DRB6 and DRB7 gene using Global alignment EMBOSS Needle.
Supplementary Table 4: Estimates of Evolutionary Divergence between DRB genes. The number of base substitutions per site from between sequences are shown. Analyses were conducted using the Jukes-Cantor model.
Supplementary Table 5: Global pairwise alignment analysis for each intron including human and non-human primates DRB5 genes
Supplementary Figure 1: Molecular Phylogenetic analysis of DRB5 intron 2 sequences obtained by the Maximum Likelihood method based on the Jukes-Cantor model.
Legend: Numbers on the branches indicate the bootstrap values for 500 repeats.
References
- [1].Davis MM, Bjorkman PJ, A model for T cell receptor and MHC/peptide interaction, Adv. Exp. Med. Biol 254 (1989) 13–16. 10.1007/978-1-4757-5803-0_1 [DOI] [PubMed] [Google Scholar]
- [2].Marrack P, Kappler J, The antigen specific, major histocompatibility complex restricted receptor on T cells, Adv. Immunol 38 (1986) 1–30. 10.1016/S0065-2776(08)60005-X [DOI] [PubMed] [Google Scholar]
- [3].Marrack P, Bender J, Jordan M, Rees W, Robertson J, Schaefer BC, Kappler J, Major histocompatibility complex proteins and TCRs: do they really go together like a horse and carriage?, J. Immunol 167 (2001) 617–621. 10.4049/jimmunol.167.2.617 [DOI] [PubMed] [Google Scholar]
- [4].Koch N, McLellan AD, Neumann J, A revised model for invariant chain-mediated assembly of MHC class II peptide receptors, Trends in biochemical sciences 32 12 (2007) 532–537. 10.1016/j.tibs.2007.09.007 [DOI] [PubMed] [Google Scholar]
- [5].Marsh SG, HLA class II region sequences, 1998, Tissue Antigens 51 (1998) 467–507. 10.1111/j.1399-0039.1998.tb02984.x [DOI] [PubMed] [Google Scholar]
- [6].Anthony Nolan Research Institute, The IPD-IMGT/HLA Database https://www.ebi.ac.uk/ipd/imgt/hla/stats.html, 2019. (accessed 28 January 2019).
- [7].Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF, Fleischhauer K, Goodridge D, Klitz W, Little AM, Maiers M, Marsh SG, Muller CR, Noreen H, Rozemuller EH, Sanchez-Mazas A, Senitzer D, Trachtenberg E, Ferandez-Vina M, Common and well-documented HLA alleles: 2012 update to the CWD catalogue, Tissue Antigens 81 4 (2013) 194–203. 10.1111/tan.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cano P, Klitz W, Mack SJ, Maiers M, Marsh SGE, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernández-Viña M, Common and Well-Documented HLA Alleles: Report of the Ad-Hoc Committee of the American Society for Histocompatiblity and Immunogenetics, Hum. Immunol 68 (2007) 392–417. 10.1016/j.humimm.2007.01.014 [DOI] [PubMed] [Google Scholar]
- [9].Bodmer JG, Marsh SG, Albert ED, Bodmer WE, Dupont B, Erlich HA, Mach B, Mayr WR, Parham R, Sasazuki T, Schreuder GMT, Strominger JL, Svejgaard A, Terasaki EI, Nomenclature for factors of the HLA system, 1994., Tissue Antigens 44 (1994) 1–18. 10.1111/j.1399-0039.1994.tb02351.x [DOI] [PubMed] [Google Scholar]
- [10].Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Charron D, Dupont B, Erlich HA, Fauchet R, Mach B, Mayr WR, Parham P, Sasazuki T, Schreuder GMT, Strominger JL, Svejgaard A, Terasaki PI, Nomenclature for factors of the HLA system, 1996, Eur. J. Immunogenet 24 (1997) 105–151. 10.1046/j.1365-2370.1997.00265.x [DOI] [PubMed] [Google Scholar]
- [11].Antunes SG, de Groot NG, Brok H, Doxiadis G, Menezes AAL, Otting N, Bontrop RE, The common marmoset: A new world primate species with limited Mhc class II variability., Proc. Natl. Acad. Sci 95 (1998) 11745–11750. 10.1073/pnas.95.20.11745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].de Groot N, Doxiadis GGM, de Vos-Rouweler AJM, de Groot NG, Verschoor EJ, Bontrop RE, Comparative genetics of a highly divergent DRB microsatellite in different macaque species, Immunogenetics. 60 (2008) 737–748. 10.1007/s00251-008-0333-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gyllensten U, Sundvall M, Ezcurra I, Erlich HA, Genetic diversity at class II DRB loci of the primate MHC., J. Immunol 146 (1991) 4368–4376. [PubMed] [Google Scholar]
- [14].Andersson G, Evolution of the human HLA-DR region., Front. Biosci 3 (1998) 739–745. [DOI] [PubMed] [Google Scholar]
- [15].von Salomé J, Gyllensten U, Bergström TF, Full-length sequence analysis of the HLA-DRB1 locus suggests a recent origin of alleles, Immunogenetics 59 (2007) 261–271. 10.1007/s00251-007-0196-8 [DOI] [PubMed] [Google Scholar]
- [16].Riess O, Kammerbauer C, Roewer L, Steimle V, Andreas A, Albert E, Nagai T, Epplen JT, Hypervariability of intronic simple (gt)n(ga)m repeats in HLA-DRB genes., Immunogenetics 32 (1990) 110–116. 10.1007/BF00210448 [DOI] [PubMed] [Google Scholar]
- [17].Schwaiger FW, Epplen J, Exonic MHC-DRB polymorphisms and intronic simple repeat sequences: Janus’ faces of DNA sequence evolution., Immunological reviews. 143 (1995) 199–224. 10.1111/j.1600-065X.1995.tb00676.x [DOI] [PubMed] [Google Scholar]
- [18].Caillier SJ, Briggs F, Cree BAC, Baranzini SE, Fernández-Viña M, Ramsay PP, Khan O, Royal W, Hauser SL, Barcellos LF, Oksenberg JR, Uncoupling the Roles of HLA-DRB1 and HLADRB5 Genes in Multiple Sclerosis, J. Immunol 181 (2008) 5473–5480 10.4049/jimmunol.181.8.5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stewart CA, Horton R, Allcock RJN, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JMM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S, Complete MHC Haplotype Sequencing for Common Disease Gene Mapping, Genome Res. 14 (2004) 1176–1187. 10.1101/gr.2188104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Frankish A, Vullo A, Zadissa A, Yates A, Thormann A, Parker A, Gall A, Moore B, Walts B, Aken BL, Cummins C, Girón CG, Ong CK, Sheppard D, Staines DM, Murphy DN, Zerbino DR, Ogeh D, Perry E, Haskell E, Martin FJ, Cunningham F, Riat HS, Schuilenburg H, Sparrow H, Lavidas I, Loveland JE, To JK, Mudge J, Bhai J, Taylor K, Billis K, Gil L, Haggerty L, Gordon L, Amode MR, Ruffier M, Patricio M, Laird MR, Muffato M, Nuhn M, Kostadima M, Langridge N, Izuogu OG, Achuthan P, Hunt SE, Janacek SH, Trevanion SJ, Hourlier T, Juettemann T, Maurel T, Newman V, Akanni W, McLaren W, Liu Z, Barrell D, Flicek P, Ensembl 2018, Nucleic Acids Res. 46 (2017) D754–D761. 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Spiess AN, Mueller N, Ivell R, Trehalose Is a Potent PCR Enhancer: Lowering of DNA Melting Temperature and Thermal Stabilization of Taq Polymerase by the Disaccharide Trehalose, Clin. Chem 50 (2004) 1256–1259. 10.1373/clinchem.2004.031336 [DOI] [PubMed] [Google Scholar]
- [22].Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, Levinson D, Fernandez-Viña MA, Davis RW, Davis MM, Mindrinos M, High-throughput, high-fidelity HLA genotyping with deep sequencing, Proc. Natl. Acad. Sci 109 (2012) 8676–8681. 10.1073/pnas.1206614109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cold Spring Harbor Laboratory, Hannon Lab. http://hannonlab.cshl.edu/fastx_toolkit/links.html, 2016. (accessed 13 January 2016).
- [24].Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25 14 (2009) 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Z, Schwartz S, Wagner L, Miller W, A greedy algorithm for aligning DNA sequences, J Comput Biol, 7(2000) 203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- [26].National Center for Biotechnology Information, Genbank. https://www.ncbi.nlm.nih.gov/genbank/,2015. (accessed November 28 2015).
- [27].Doxiadis GGM, Hoof I, De Groot N, Bontrop RE, Evolution of HLA-DRB genes, Mol. Biol. Evol 29 (2012) 3843–3853. 10.1093/molbev/mss186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Anthony Nolan Research Institute, The IPD-MHC-NHP Database, https://www.ebi.ac.uk/ipd/mhc/ 2019. (accessed 28 January 2019).
- [29].McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R, Analysis Tool Web Services from the EMBL-EBI, Nucleic Acids Res. 41 (2013) 597–600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, MEGA6: Molecular evolutionary genetics analysis version 6.0, Mol. Biol. Evol 30 (2013) 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thomas RH, Molecular Evolution and Phylogenetics, in: Nei M, Kumar S, Oxford University Press, Oxford, 2000, pp. 333 10.1046/j.1365-2540.2001.0923a.x [DOI] [Google Scholar]
- [32].Voorter CEM, Roeffaers HET, du Toit ED, van den Berg-Loonen EM, The absence of DR51 in a DRB5-positive individual DR2ES is caused by a null allele (DRB5*0108N), Tissue Antigens. 50 (1997) 326–333. 10.1111/j.1399-0039.1997.tb02882.x [DOI] [PubMed] [Google Scholar]
- [33].Balas A, Ocon P, Vicario JL, Alonso A, HLA-DR51 expression failure caused by a two-base deletion at exon 2 of a DRB5 null allele (DRB5*0110N) in a Spanish gypsy family, Tissue Antigens. 55 (2000) 467–469. 10.1034/j.1399-0039.2000.550513.x [DOI] [PubMed] [Google Scholar]
- [34].Zavodna M, Bagshaw A, Brauning R, Gemmell NJ, The accuracy, feasibility and challenges of sequencing short tandem repeats using next-generation sequencing platforms, PLoS One. 9 (2014) e113862 10.1371/journal.pone.0113862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wall JD, Tang LF, Zerbe B, Kvale MN, Kwok P-Y, Schaefer C, Risch N, Estimating genotype error rates from high-coverage next-generation sequence data, Genome Res. 24 (2014) 1734–1739. 10.1101/gr.168393.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shin S, Park J, Characterization of sequence-specific errors in various next-generation sequencing systems, Mol. Biosyst 12 (2016) 914–922. 10.1039/C5MB00750J [DOI] [PubMed] [Google Scholar]
- [37].Chen G, Mosier S, Gocke CD, Lin M-T, Eshleman JR, Cytosine Deamination Is a Major Cause of Baseline Noise in Next-Generation Sequencing, Mol. Diagn. Ther 18 (2014) 587–593. 10.1007/s40291-014-0115-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Laver TW, Caswell RC, Moore KA, Poschmann J, Johnson MB, Owens MM, Ellard S, Paszkiewicz KH, Weedon MN, Pitfalls of haplotype phasing from amplicon-based long-read sequencing, Sci. Rep 6 (2016) 21746 10.1038/srep21746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lind C, Ferriola D, Mackiewicz K, Papazoglou A, Sasson A, Monos D, Filling the gaps - The generation of full genomic sequences for 15 common and well-documented HLA class I alleles using next-generation sequencing technology, Hum. Immunol 74 (2013) 325–329. 10.1016/j.humimm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- [40].Zerbino DR, Using the Velvet de novo assembler for short-read sequencing technologies, Curr. Protoc. Bioinforma Chapter 11 (2010) Unit-11.5. 10.1002/0471250953.bi1105s31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Warren RL, Holt RA, Jones SJM, Sutton GG, Assembling millions of short DNA sequences using SSAKE, Bioinformatics. 23 (2006) 500–501. 10.1093/bioinformatics/btl629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Narzisi G, Schatz MC, The Challenge of Small-Scale Repeats for Indel Discovery, Front. Bioeng. Biotechnol 3 (2015) 8 10.3389/fbioe.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ballantyne KN, Goedbloed M, Fang R, Schaap O, Lao O, Wollstein A, Choi Y, van Duijn K, Vermeulen M, Brauer S, Decorte R, Poetsch M, von Wurmb-Schwark N, de Knijff P, Labuda D, Vézina H, Knoblauch H, Lessig R, Roewer L, Ploski R, Dobosz T, Henke L, Henke J, Furtado MR, Kayser M, Mutability of Y-Chromosomal Microsatellites: Rates, Characteristics, Molecular Bases, and Forensic Implications, Am. J. Hum. Genet 87 (2010) 341–353. 10.1016/j.ajhg.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Treangen TJ, Salzberg SL, Repetitive DNA and next-generation sequencing: computational challenges and solutions, Nat. Rev. Genet 13 (2011) 36 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Carapito R, Radosavljevic M, Bahram S, Next-Generation Sequencing of the HLA locus: Methods and impacts on HLA typing, population genetics and disease association studies, Hum. Immunol 77 (2016) 1016–1023. 10.1016/j.humimm.2016.04.002 [DOI] [PubMed] [Google Scholar]
- [46].Clark PM, Duke JL, Ferriola D, Bravo-Egana V, Vago T, Hassan A, Papazoglou A, Monos D, Generation of full-length class i human leukocyte antigen gene consensus sequences for novel allele characterization, Clin. Chem 62 (2016) 1630–1638. 10.1373/clinchem.2016.260661 [DOI] [PubMed] [Google Scholar]
- [47].Epplen C, Santos EJM, Guerreiro JF, Van Helden P, Epplen JT, Coding versus intron variability: Extremely polymorphic HLA-DRB1 exons are flanked by specific composite microsatellites, even in distant populations, Hum. Genet 99 (1997) 399–406. 10.1007/s004390050379 [DOI] [PubMed] [Google Scholar]
- [48].Bergström TF, Engkvist H, Erlandsson R, Josefsson a, Mack SJ, Erlich a, Gyllensten U Tracing the origin of HLA-DRB1 alleles by microsatellite polymorphism., Am. J. Hum. Genet 64 (1999) 1709–1718. 10.1086/302401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hamada H, Seidman M, Howard BH, Gorman CM, Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence., Mol. Cell. Biol 4 (1984) 2622–2630. 10.1128/MCB.4.12.2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hamada H, Petrino MG, Kakunaga T, Seidman M, Stollar BD, Characterization of genomic poly(dT-dG).poly(dC-dA) sequences: structure, organization, and conformation., Mol. Cell. Biol 4 (1984) 2610–2621. 10.1128/MCB.4.12.2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mäueler W, Bassili G, Arnold R, Renkawitz R, Epplen JT, The (gt)(n)(ga)(m) containing intron 2 of HLA-DRB alleles binds a zinc- dependent protein and forms non B-DNA structures, Gene. 226 (1999) 9–23. 10.1016/S0378-1119(98)00573-3 [DOI] [PubMed] [Google Scholar]
- [52].Kobori JA, Strauss E, Minard K, Hood L, Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex, Science. 234 (1986) 173–179. 10.1126/science.3018929 [DOI] [PubMed] [Google Scholar]
- [53].Arnold R, Mäueler W, Bassili G, Lutz M, Burke L, Epplen TJ, Renkawitz R, The insulator protein CTCF represses transcription on binding to the (gt)22(ga)15 microsatellite in intron 2 of the HLADRB1*0401 gene, Gene. 253 (2000) 209–214. 10.1016/S0378-1119(00)00271-7 [DOI] [PubMed] [Google Scholar]
- [54].Wu S, Saunders TL, Bach FH, Polymorphism of human Ia antigens generated by reciprocal intergenic exchange between two DR beta loci, Nature. 324 (1986) 676–679. 10.1038/324676a0 [DOI] [PubMed] [Google Scholar]
- [55].Satta Y, Mayer WE, Klein J, Evolutionary relationship of HLA-DRB genes inferred from intron sequences, J. Mol. Evol 42 (1996) 648–657. 10.1007/BF02338798 [DOI] [PubMed] [Google Scholar]
- [56].Doxiadis GGM, de Groot N, de Groot NG, Doxiadis IIN, Bontrop RE, Reshuffling of ancient peptide binding motifs between HLA-DRB multigene family members: Old wine served in new skins, Mol. Immunol 45 (2008) 2743–2751. 10.1016/j.molimm.2008.02.017 [DOI] [PubMed] [Google Scholar]
- [57].Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SG, The IPD and IMGT/HLA database: allele variant databases, Nucleic Acids Research. 43 (2015) 423–431. 10.1093/nar/gku1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Moraes ME, Fernandez-Viña M, Stastny P, DNA typing for class II HLA antigens with allele-specific or group-specific amplification. IV. Typing for alleles of the HLA-DR2 group, Hum. Immunol 31 (1991) 139–144. 10.1016/0198-8859(91)90017-4 [DOI] [PubMed] [Google Scholar]
- [59].Fernandez-Viña MA, Gao X, Moraes ME, Moraes JR, Salatiel I, Miller S, Tsai J, Sun Y, An J, Layrisse Z, Gazit E, Brautbar C, Stastny P, Alleles at four HLA class II loci determined by oligonucleotide hybridization and their associations in five ethnic groups, Immunogenetics. 34 (2004) 299–312. https://doi-org.stanford.idm.oclc.org/10.1007/BF00211994 [DOI] [PubMed] [Google Scholar]
- [60].Prat E, Tomaru U, Sabater L, Park DM, Granger R, Kruse N, Ohayon JM, Bettinotti MP, Martin R, HLA-DRB5*0101 and -DRB1*1501 expression in the multiple sclerosis-associated HLA-DR15 haplotype, J. Neuroimmunol 167 (2005) 108–119. 10.1016/j.jneuroim.2005.04.027 [DOI] [PubMed] [Google Scholar]
- [61].Sospedra M, Muraro PA, Stefanová I, Zhao Y, Chung K, Li Y, Giulianotti M, Simon R, Mariuzza R, Pinilla C, Martin R, Redundancy in Antigen-Presenting Function of the HLA-DR and -DQ Molecules in the Multiple Sclerosis-Associated HLA-DR2 Haplotype, J. Immunol 176 (2006) 1951–1961. 10.4049/jimmunol.176.3.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, Cardon LR, Bell JI, Fugger L, Functional epistasis on a common MHC haplotype associated with multiple sclerosis, Nature. 443 (2006) 574 10.1038/nature05133 [DOI] [PubMed] [Google Scholar]
- [63].Creary LE, Gangavarapu S, Mallempati KC, Montero-Martín G, Caillier SJ, Santaniello A, Hollenbach JA, Oksenberg JR, Fernández-Viňa MA, Next-Generation Sequencing reveals new information about HLA genomic and haplotype diversity in a large European American population, Manuscript submitted to Human Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Baldassarre LA, Steiner NK, Jones P, Tang T, Slack R, Ng J, Hartzman RJ, Hurley CK, Limited diversity of HLA-DRB1*02 alleles and DRB1-DRB5 haplotype associations in four United States population groups, Tissue Antigens. 61 (2003) 249–252. 10.1034/j.1399-0039.2003.00018.x [DOI] [PubMed] [Google Scholar]
- [65].Núñez G, Ball EJ, Myers L, & Stastny P (1985). Allostimulating cells in man. Quantitative variation in the expression of HLA-DR and HLA-DQ molecules influences T-cell activation. Immunogenetics, 22, 85–91. 10.1007/BF00430597 [DOI] [PubMed] [Google Scholar]
- [66].Shackelford DA, Mann DL, van Rood JJ, Ferrara GB, Strominger JL, Human B-cell alloantigens DC1, MT1, and LB12 are identical to each other but distinct from the HLA-DR antigen, Proc. Natl. Acad. Sci 78 (1981) 4566 LP–4570. 10.1073/pnas.78.7.4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kavathas P, DeMars R, Bach FH, Shaw S, SB: a new HLA-linked human histocompatibility gene defined using HLA-mutant cell lines, Nature. 293 (1981) 747–749. 10.1038/293747a0 [DOI] [PubMed] [Google Scholar]
- [68].Tanigaki N, Tosi R, Duquesnoy RJ, Ferrara GB, Three Ia species with different structures and alloantigenic determinants in an HLA-homozygous cell line., J. Exp. Med 157 (1983) 231 LP–247. 10.1084/jem.157.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Long EO, Gorski J, Mach B, Structural relationship of the SB β-chain gene to HLA-D-region genes and murine I-region genes, Nature. 310 (1984) 233–235. 10.1038/310233a0 [DOI] [PubMed] [Google Scholar]
- [70].Nuñez G, Giles RC, Ball EJ, Hurley CK, Capra JD, Stastny P, Expression of HLA-DR, MB, MT and SB antigens on human mononuclear cells: identification of two phenotypically distinct monocyte populations, J. Immunol 133 (1984) 1300–1306 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.