Abstract
Genetic regulation of gene expression is dynamic, as transcription can change during cell differentiation and across cell types. We mapped expression quantitative trait loci (eQTLs) throughout differentiation to elucidate the dynamics of genetic effects on cell type specific gene expression. We generated time-series RNA-sequencing data, capturing 16 time points from induced pluripotent stem cells to cardiomyocytes, in 19 human cell lines. We identified hundreds of dynamic eQTLs that change over time, with enrichment in enhancers of relevant cell types. We also found nonlinear dynamic eQTLs, which affect only intermediate stages of differentiation, and cannot be found by using data from mature tissues. These fleeting genetic associations with gene regulation may represent a new mechanism to explain complex traits and disease. We highlight one example of a nonlinear eQTL that is associated with body mass index.
One Sentence Summary:
We provide a high-resolution analysis of temporal dynamics of genetic effects on gene expression throughout cardiomyocyte differentiation.
Genetic variants that alter gene regulation play an essential role in the genetics of human disease and other complex phenotypes (1, 2). Large studies have identified thousands of genetic loci associated with complex diseases, most of which are in non-coding regions of the genome and therefore are putatively involved in gene regulation (2). Expression quantitative trait locus (eQTL) analysis has shown that many disease-associated loci influence the regulation of nearby genes (3, 4) but still, a substantial fraction of disease-associated loci remain unexplained (5, 6).
Much effort has been dedicated to map and identify eQTLs across tissues and cell types, as regulatory impact of disease-associated loci may be most evident in cell types relevant to each disease. Regulatory genetic effects can be also timepoint-specific or environment-dependent (7, 8), and may influence temporal programs of gene regulation. Yet, almost all studies of the genetics of gene regulation, including the multi-tissue GTEx project (7), involve data collected at a single time point, usually from adult individuals. Dynamic gene expression data can add another dimension to eQTL analysis, allowing identification of genetic variants with transient effects that may not have been found in analysis of static data.
We took advantage of a panel of induced pluripotent stem cell (iPSC) lines from 19 individuals to investigate high-resolution temporal genetic effects on gene regulation over time during cardiomyocyte differentiation. Specifically, we collected gene expression data throughout the differentiation from iPSCs to cardiomyocytes in 19 well-characterized, human Yoruba HapMap cell lines (9). For each cell line, RNA was extracted and sequenced every 24 hours for 16 days, to capture the entire differentiation process; in total, we sequenced 297 RNA samples (Figs. S1–S2). Combined with available whole genome sequences and genotype data for each cell line, these data provide a resource with which to investigate how gene expression and genetic regulation change throughout cardiomyocyte differentiation with high temporal resolution.
During iPSC culturing, differentiation, RNA extraction, and processing for sequencing, we recorded extensive metadata on each sample (Table S1). Quality controls and filtering yielded 16,319 genes for downstream analysis (10). Following standardization and normalization of the RNA sequencing data (10), we evaluated the contribution of potential confounders to overall variation in our data, confirming that our study design was effective (Fig. S3). We also used replicates from an independent differentiation to confirm that the gene expression patterns we observed in our iPSCs and iPSC-derived cardiomyocytes are robust with respect to variance that may be associated with the differentiation procedure (Fig. S4) (9, 10).
We evaluated the efficiency of our differentiation by FACS (Table S2), and by considering the time course expression of known cell type specific marker genes (11, 12) (Fig. S5). As expected (12), cardiomyocyte purity and the expression of lineage marker genes are variable across our samples. This variability between cell lines was observed across the entire time course, though the effect of differentiation time is the primary source of variation in the data (Figs. 1A, S3, S6).
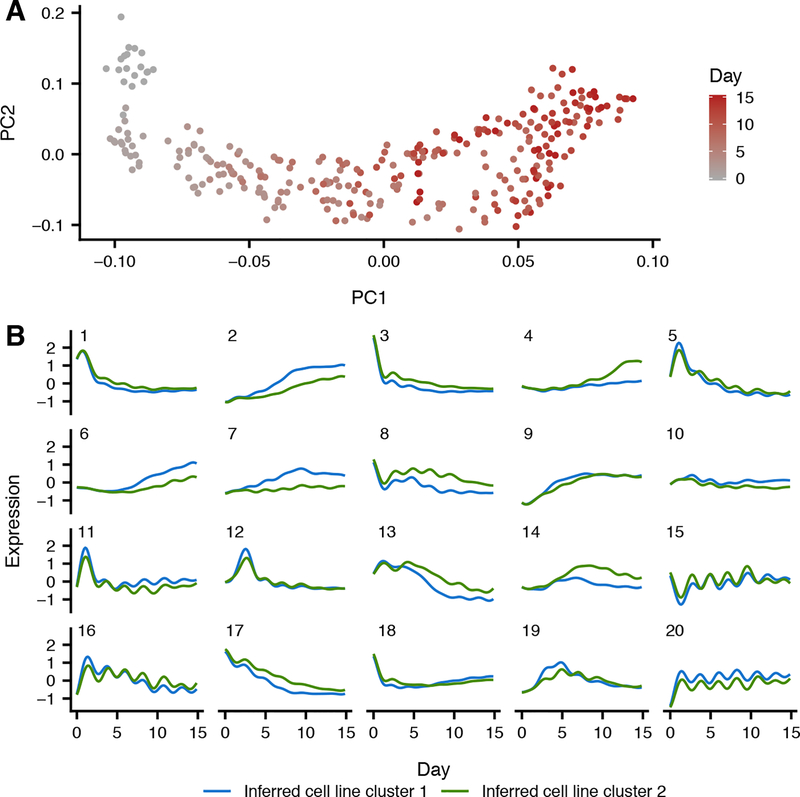
Fig. 1. Gene expression trends throughout cardiomyocyte differentiation.
(A) The first two gene expression principal component loadings for all 297 RNA-seq samples across cell lines, where each sample is colored by day of collection. (B) Predicted cell line cluster expression trajectories for 20 gene clusters according to split-GPM. Many gene clusters (8, 11, 15, 16, and 20) exhibit periodic expression trajectories that correspond with cell culture media changes.
We characterized global patterns of gene expression across time by applying split-GPM, an unsupervised probabilistic model that infers time course trajectories of gene expression using Gaussian processes, while simultaneously performing clustering of genes and cell lines (10). Using this approach, we identified two clusters of cell lines that displayed broad differences in the expression patterns of multiple clusters of genes; within each gene cluster, genes exhibit shared expression changes over time. The assignment of cell lines to clusters is robust with respect to the parameters we tested, such as the number of gene clusters we infer (Fig. S7).
The two cell line clusters we identified differ in the efficiency of cardiomyocyte differentiation. Cell lines in the first (larger) cluster display greater Troponin expression levels in the final six timepoints of differentiation (p=.014, Wilcoxon rank-sum test). The expression of a group of genes enriched for myogenesis also increases by a greater magnitude over time in cell lines in the first cluster (Bonferroni p=9.29e-14; gene cluster 2 in Fig. 1B) (13). Cell lines in the second, smaller cluster, show high expression of genes related to KRAS activation (Bonferroni p=0.005; gene cluster 4 in Fig. 1B), which is associated with increased self-renewal of undifferentiated iPSCs and decreased neuronal differentiation propensity (14). Other gene clusters illuminate broad changes in gene expression over time such as a transient rise in MYC and E2F target genes in the early days of differentiation (gene cluster 13 in Fig. 1B; Table S3). Together, this analysis documents patterns of gene expression trajectories over time and captures differences among our cell lines that are not obvious from the individual time point data alone.
Next, we evaluated the impact of genetic variation on gene regulation in our system. We used WASP (15) to identify cis-eQTLs in the data from each time point, independently (10). To control for latent confounders in the independent analysis of data from each time point, we included the first three expression PCs using data from samples of the corresponding time point as covariates (Figs. S8, S9A, S9B). At an empirical false discovery rate (eFDR) of 5%, we identified a median of 111 genes (range 71 – 231) with at least one eQTL in each time point (Figs. S9C, S10). As expected, the eQTLs we identified early in the time course replicated in data from iPSCs, whereas eQTLs from later time points were better supported by data from iPSC-derived cardiomyocytes (both p < 0.001, linear regression; Fig. 2A) (9).
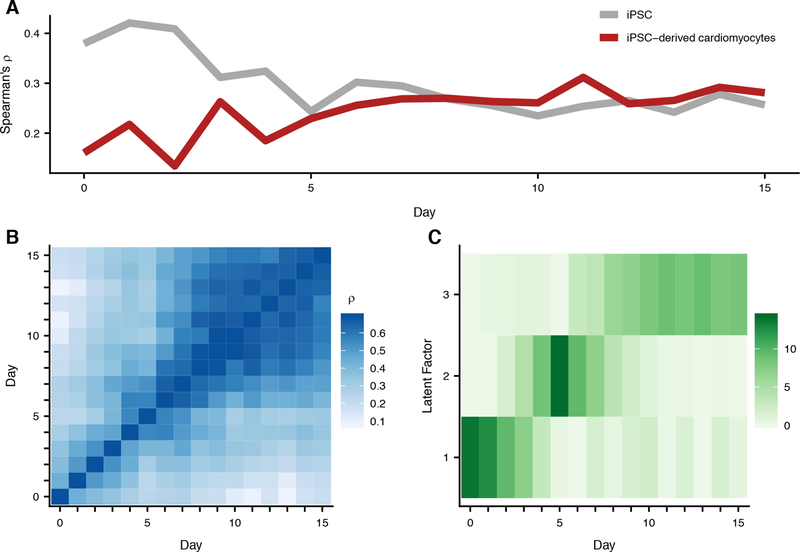
Fig. 2. eQTL patterns during cardiomyocyte differentiation.
We limit to genes with at least one significant eQTL (WASP combined haplotype test; eFDR <= .05) across time points. If a gene has more than one significant eQTL, we select a single variant for that gene with the smallest geometric mean p-value across all 16 time points. (A) Spearman correlation of p-values between eQTLs from each day (x-axis) and existing iPSC (grey) and iPSC-derived cardiomyocyte (red) eQTLs. (B) Spearman correlation of eQTL p-values for each pair of days. (C). Factors identified via sparse matrix factorization of eQTL -log10 p-values using 3 latent factors and a L1 penalty of .5.
We computed the correlation of the significant eQTL summary statistics for each pair of time points (Fig. 2B). We observed that correlation between eQTL summary statistics increases as the distance between time points decreases (p <= 2e-16, linear regression). Though this observation is intuitive, it indicates that the dynamic impact of genetic variation on gene regulation in our data is not random, and is related to the temporal process of cardiomyocyte differentiation.
To more formally quantify the temporal structure of genetic regulation throughout differentiation, we performed sparse non-negative matrix factorization on the matrix of significant eQTL summary statistics from all time points (10). The learned factors capture genetic signal that is largely specific to a subset of differentiation time (Fig. 2C), a pattern that is robust with respect to the number of latent factors or sparse prior choice (Fig. S11).
Our analysis indicates that temporal structure dominates the patterns of genetic association with gene expression in our data. However, the observation that most significant non-dynamic eQTLs can be identified in only a few time points (median of 2; Fig. S12) is most likely explained by incomplete power to identify eQTLs in each time point independently. To robustly identify dynamic eQTLs whose effect varies significantly over time, leveraging power across all time points (Fig. 3A), we used a Gaussian linear model applied jointly to data from the entire experiment. Specifically, we quantified the effect of interactions between genotype and differentiation time on gene expression, controlling for linear effects of both differentiation time and genotype. In addition, we accounted for the systematic differences in differentiation trajectories identified between cell lines (Figs. 1B, S13–S16, Table S4) (10), which would otherwise lead to false positives in our analysis. Using this approach, we identified 550 genes with a significant dynamic eQTL (eFDR <= .05; Figs. S17–S20, Table S5).
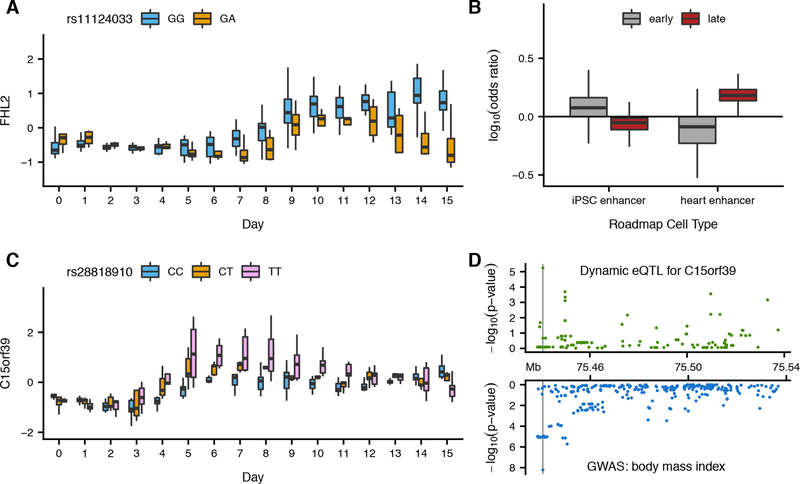
Fig. 3. Dynamic eQTLs detect genetic regulatory changes caused by cardiomyocyte differentiation.
(A) Linear interaction association between genotype (color) of rs11124033 and time point (x-axis) on residual gene expression (cell line effects regressed on expression) of FHL2 (y-axis). (B) Enrichment of dynamic eQTLs within cell type specific chromHMM enhancer elements relative to 1000 sets of randomly selected matched background variants. Dynamic eQTLs were classified as early or late (C) Nonlinear interaction association between genotype (color) of rs28818910 and time point (x-axis) on residual gene expression of C15orf39 (y-axis). (D) Nonlinear interaction association significance of all variants tested within 50 KB of the C15orf39 transcription start site with expression of C15orf39 (green) and GWAS significance for BMI of variants in the same window (blue). Vertical line depicts genomic location of the most significant nonlinear dynamic eQTL (rs28818910) for C15orf39.
We classified the 550 dynamic eQTL as early (eQTL effect size decreasing over time), late (eQTL effect size increasing over time), or switch (eQTL effect size exhibiting different directions of effect over time; Fig. S21) (10). We found that the early dynamic eQTLs are enriched for chromHMM enhancer elements annotated in iPSC Roadmap cell types but not in heart-related cell types (16, 17). In turn, late dynamic eQTLs are enriched for chromHMM enhancer elements annotated in heart-related Roadmap cell types but not in iPSCs (Figs. 3B, S22). These observations indicate that dynamic eQTL mapping can capture temporal changes in cellular gene regulation reflecting changes in regulatory element activity as the cell cultures differentiate.
The observation that we are able to capture the function of cell-type-specific regulatory elements prompted us to consider dynamic eQTLs in other contexts. We found that dynamic eQTLs are enriched for genes with roles in myogenesis (Bonferroni p = .0019, Fisher’s exact; Table S6) (13), and also show significant enrichment for genes related to dilated cardiomyopathy (p = .001, Fisher’s exact; Table S7) (10, 18). Two significant dynamic eQTLs in particular, rs7633988 and rs6599234 (in strong LD, R2 = 0.93), are GWAS variants for QRS duration and QT interval, respectively (Fig. S23) (19, 20). Both variants show an association with the expression levels of SCN5A, which is involved in the creation of sodium channels and is in the dilated cardiomyopathy gene set (21). Another dynamic eQTL, rs11124033, associated with the expression of FHL2 (Fig. 3A), is also associated with dilated cardiomyopathy. This variant lies in a Roadmap chromHMM promoter element annotated in heart-related cell types but not in iPSCs (16, 17). Interestingly, none of these examples were identified as eQTLs in the non-dynamic QTL analysis of each time point from our dataset or in the GTEx heart tissue data (7).
Finally, we sought to identify a wider range of dynamic regulatory patterns, including nonlinear associations such as when a genetic effect increases in magnitude in the middle of the time course before decreasing or disappearing. To identify nonlinear dynamic eQTLs we expanded our linear model using a second order polynomial basis function (10). We acknowledge that our study is underpowered to expand to a more general class of nonlinear dynamic eQTLs that do not assume a continuous effect of differentiation time (Fig. S24) (10).
We identified 693 genes with a nonlinear dynamic eQTL (eFDR <= .05; Figs. S17B, S19B, Table S8), 28 of which have their strongest genetic effect in the middle of the differentiation time course (middle dynamic eQTLs; Fig. S25) (10). It is worth noting that 25 of these middle dynamic eQTL genes and their strongest associated variant are not identified as eQTLs in our non-dynamic QTL analysis in either iPSCs (day 0) or cardiomyocytes (day 15).
In one example of a non-linear dynamic eQTL, rs8107849 is associated with the expression of ZNF606 with a larger magnitude of effect during days 4 through 11 (Fig. S26). The rs8107849 locus does not lie in iPSC or heart-related chromHMM regulatory regions and was not identified in our analysis as a non-dynamic eQTL in any time point. While ZNF606 is known to have a role in differentiation of chondrocytes (22), it is possible this is a conserved process involved in the differentiation of additional cell types, including cardiomyocytes. Another nonlinear dynamic eQTL reveals an association between rs28818910 and C15orf39. The rs28818910 variant is also associated with BMI (p < 6.07 e-9, reported; Fig. 3C, 3D) (23) and weakly associated with red blood cell count (p < 1.48 e-6, reported) (24). This dynamic eQTL and both traits show similar patterns of association across the region (Fig. S27). The rs28818910 locus is associated with inter-individual differences in gene expression only during intermediate stages of differentiation; it does not lie in annotated regulatory elements of either iPSCs or cardiomyocytes and is not identified as an eQTL in iPSCs, mature cardiomyocytes, or either of the two GTEx heart tissues. Thus, this is an example of a temporary, dynamic regulatory effect that may have phenotypic consequences.
In summary, our time course study design allowed us to identify hundreds of dynamic eQTLs throughout the differentiation of human iPSCs to cardiomyocytes. Dynamic eQTLs, in particular those with nonlinear effects, may often be transient and will not be found in studies that only consider gene expression data from either stem cells or mature tissues and cell types. Many of our dynamic eQTLs lie in regions without known regulatory annotations, as functional studies have focused on static cell types. Thus, these loci are candidates for novel regulatory effects, which may be followed up with further functional validation in relevant intermediate time points. Dynamic genetic effects identified in our study, or in future time series genomic datasets, provide a novel resource for investigating mechanisms underlying disease associations that cannot be characterized based on studies of terminal cell types.
Supplementary Material
Acknowledgments:
We thank H. Kyung Im for help with GWAS analysis and J. K. Pritchard for providing comments on the manuscript.;
Funding: YG and AB were supported by NIH/NIGMS R01GM120167. RE was supported by the NIH MSTP Training Grant T32GM007281. KR was supported by NIH GRTG 5T32GM007197 and AHA Predoctoral Fellowship 18PRE34030197. The computational resources were provided by the University of Chicago Research Computing Center.;
Footnotes
Competing interests: Authors declare no competing interests;
Data and materials availability: The fastq files and RNA-seq counts are available at GEO under accession GSE122380. Files containing non-dynamic and dynamic eQTL summary statistics are available at https://zenodo.org/badge/latestdoi/175270219. The split-GLM package can be found at https://doi.org/10.5281/zenodo.2590826. Scripts used for this analysis can be found at https://zenodo.org/badge/latestdoi/156638838.
References and Notes:
- 1.Li YI et al. , RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert FW, Kruglyak L, The role of regulatory variation in complex traits and disease. Nat. Rev. Genet 16, 197–212 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Zhu Z et al. , Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Nicolae DL et al. , Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6, e1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joehanes R et al. , Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol 18, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen X, Pique-Regi R, Luca F, Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS Genet 13, e1006646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium GTEx et al. , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles DA et al. , Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods 14, 699–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banovich NE et al. , Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res 28, 122–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supplementary materials at the Science website.
- 11.Okita K, Ichisaka T, Yamanaka S, Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lian X et al. , Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U. S. A 109, E1848–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubara K et al. , Status of KRAS in iPSCs Impacts upon Self-Renewal and Differentiation Propensity. Stem Cell Reports 11, 380–394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Geijn B, McVicker G, Gilad Y, Pritchard JK, WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 12, 1061–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst J, Kellis M, Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roadmap Epigenomics Consortium et al. , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke MA, Cook SA, Seidman JG, Seidman CE, Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. J. Am. Coll. Cardiol 68, 2871–2886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong K-W et al. , Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum. Mol. Genet 23, 6659–6667 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Arking DE et al. , Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet 46, 826–836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rook MB, Evers MM, Vos MA, Bierhuizen MFA, Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc. Res 93, 12–23 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z et al. , ZNF606 interacts with Sox9 to regulate chondrocyte differentiation. Biochem. Biophys. Res. Commun 479, 920–926 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Abbott L et al. , Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank-- Neale Lab (2017), (available at http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank). [Google Scholar]
- 24.Astle WJ et al. , The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burridge PW et al. , Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degner JF et al. , DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lázaro-Gredilla M, Van Vaerenbergh S, Lawrence ND, Overlapping Mixtures of Gaussian Processes for the data association problem. Pattern Recognit 45, 1386–1395 (2012). [Google Scholar]
- 28.Hensman J, de AG Matthews G, Ghahramani Z, Scalable Variational Gaussian Process Classification. J. Mach. Learn. Res (2015). [Google Scholar]
- 29.Gamazon ER, Huang RS, Dolan ME, Cox NJ, Im HK, Integrative genomics: quantifying significance of phenotype-genotype relationships from multiple sources of high-throughput data. Front. Genet 3, 202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedregosa F et al. , Learning scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830 (2011). [Google Scholar]
- 31.Abbott L et al. , UK Biobank -- Neale lab (2018), (available at http://www.nealelab.is/uk-biobank/). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.