Abstract
The phosphotyrosine interacting domain-containing protein 1 (PID1) serves as a cytosolic adaptor protein of the LDL receptor-related protein 1 (LRP1). By regulating its intracellular trafficking, PID1 controls the hepatic, LRP1-dependent clearance of pro-atherogenic lipoproteins. In adipose and muscle tissues, LRP1 is present in endosomal storage vesicles containing the insulin-responsive glucose transporter 4 (GLUT4). This prompted us to investigate whether PID1 modulates GLUT4 translocation and function via its interaction with the LRP1 cytosolic domain. We initially evaluated this in primary brown adipocytes as we observed an inverse correlation between brown adipose tissue glucose uptake and expression of LRP1 and PID1. Insulin stimulation in wild type brown adipocytes induced LRP1 and GLUT4 translocation from endosomal storage vesicles to the cell surface. Loss of PID1 expression in brown adipocytes prompted LRP1 and GLUT4 sorting to the plasma membrane independent of insulin signalling. When placed on a diabetogenic high fat diet, systemic and adipocyte-specific PID1-deficient mice presented with improved hyperglycemia and glucose tolerance as well as reduced basal plasma insulin levels compared to wild type control mice. Moreover, the improvements in glucose parameters associated with increased glucose uptake in adipose and muscle tissues from PID1-deficient mice. The data provide evidence that PID1 serves as an insulin-regulated retention adaptor protein controlling translocation of LRP1 in conjunction with GLUT4 to the plasma membrane of adipocytes. Notably, loss of PID1 corrects for insulin resistance-associated hyperglycemia emphasising its pivotal role and therapeutic potential in the regulation of glucose homeostasis.
Keywords: Adaptor proteins, type 2 diabetes, GLUT4, insulin, glucose homeostasis, adipose tissues
1. Introduction
Adipose tissues contribute to systemic glucose and lipid homeostasis under both anabolic and catabolic conditions [1]. In white adipose tissue (WAT), the uptake and processing of dietary nutrients is stimulated by insulin in the postprandial state, enabling efficient storage of excess energy as triglycerides within lipid droplets. In the fasted state, lack of insulin signaling facilitates lipolysis by intracellular lipases to release fatty acids for e.g. VLDL production in the liver and ATP generation in muscles [1, 2]. In contrast to WAT, energy substrates internalized by brown adipose tissue (BAT) are used for heat production upon cold-induced non-shivering thermogenesis, mediated primarily by uncoupling protein 1 (UCP1) [3]. Both thermogenic activation by cold-exposure as well as anabolic insulin signaling promote BAT uptake of glucose and lipids [4–7]. Accordingly, systemic insulin resistance triggered by obesity leads to impaired glucose and lipid disposal into adipose tissues. These metabolic dysfunctions contribute to hyperglycemia and dyslipidemia and promote the development of type 2 diabetes and atherosclerosis that are associated with obesity [8, 9]. BAT activation can correct these metabolic disturbances, and consequently the obesity-associated comorbidities [5, 10–12].
The capacity for glucose uptake in adipocytes and skeletal muscle cells is regulated by insulin-dependent redistribution of glucose transporter 4 (GLUT4) from intracellular endosomal vesicles to the plasma membrane. Mice with adipose tissue-specific GLUT4 deficiency or overexpression presented with impaired or enhanced glucose tolerance, respectively, underscoring the relevance of GLUT4 in adipocytes for systemic glucose homeostasis [13, 14]. The insulin signaling cascade that initiates GLUT4 translocation to the cell surface depends on activation of the protein kinase B (AKT) pathway. Subsequently, GLUT4-containing vesicles are targeted to the plasma membrane in a process dependent on numerous proteins involved in signaling (e.g. the TBC1 domain family member 4 also known as AS160 [15]), protein recycling (e.g. syntaxin 6 and tumor suppressor candidate 5 [16, 17]), vesicle tethering (e.g. tether, containing a UBX domain, for GLUT4 [18]) as well as vesicle fusion (e.g. vesicle-associated membrane proteins [19, 20]). The same GLUT4-storage vesicles also contain low-density lipoprotein receptor related protein 1 (LRP1) [21], for which insulin-dependent transport to the plasma membrane has been described in hepatocytes and adipocytes [22–24]. Insulin-mediated LRP1 translocation in the liver is dependent on the phosphorylation of its distal NPxYxxL motif [23, 25]. Notably, by binding to the unphosphorylated form of this motif, the protein phosphotyrosine interacting domain containing 1 (PID1) serves as an intracellular adaptor for LRP1 [23, 26, 27]. In addition, PID1 has been implicated to regulate glucose uptake via modulation of insulin signaling in adipocytes and muscle cells [28–31]. We recently demonstrated that insulin signaling in hepatocytes disrupts PID1 binding to LRP1, which results in the translocation of LRP1 to the cell surface and thus the efficient lipoprotein receptor-mediated endocytosis in the postprandial state [23]. Based on these findings and the high co-expression of PID1 and LRP1 in adipose tissues [23], we propose that PID1 has a fundamental role in systemic glucose homeostasis by regulating sorting of GLUT4 vesicles via insulin-dependent trafficking of LRP1.
We here provide in vitro and in vivo evidence that PID1 serves as a negative regulator of glucose uptake into adipose tissues and muscles and that its inactivation improves glucose uptake, hyperglycemia and hyperinsulinemia under diabetogenic conditions. This suggests that PID1 is not only an important regulator for hepatic lipoprotein disposal but also for systemic glucose metabolism emphasizing the potential for PID1 as a novel therapeutic target for the treatment of obesity-associated metabolic abnormalities.
2. Materials and Methods
2.1. Mice and animal housing
Pid1−/− and Pid1fl/fl mice were generated as described before [23]. For generation of adipose tissue-specific Pid1 knockout mice, Pid1fl/fl mice were crossed with mice expressing Cre recombinase under control of the adiponectin promotor (B6;FVB-Tg(Adipoq-cre)1Evdr/J mice from Jackson) to generate Pid1fl/fl-AdipoqCre− mice and Pid1fl/fl-AdipoqCre+ mice. All mice were on C57BL6/J background. Male mice kept at 22°C on a 12 hour/12 hour light dark cycle with ad libitum access to food and water were used for metabolic studies. Chow-fed mice were routinely aged 12–18 weeks. For high-fat diet (HFD) feeding, mice received an obesogenic HFD (Bio-Serv F3282, 60% calories from fat) ad libitum for 18–20 weeks beginning at 4–6 weeks of age. For the temperature acclimation studies, mice were housed for 1 week at either 6°C, 22°C or 30°C in temperature-controlled cabinets (Memmert). All procedures were performed with approval from the animal care committees of the University Medical Center Hamburg-Eppendorf and the Behörde für Gesundheit und Verbraucherschutz Hamburg.
2.2. Experimental procedures and organ harvest
Insulin tolerance tests were performed by intraperitoneal injection of insulin into 4 hour fasted mice (Insuman Rapid, Aventis, 1 U/kg body weight). Oral glucose tolerance tests were performed by gavage of glucose (2 g/kg body weight) after a 4 hour fasting period. Blood glucose concentrations were measured using commercially available AccuCheck Aviva sticks (Roche). Plasma insulin was measured using a rat/mouse insulin assay kit (Chrystal Chem).
For radioactive uptake studies, the glucose gavage was traced with 0.79 kBq 2-deoxy-D-[1,2-3H(N)]-glucose/g body weight. Mice were anaesthetized with injection of 300 mg/kg ketamin and 30 mg/kg rompun, and transcardially perfused with PBS containing 10 U/ml heparin. Organs were solubilized in 10x (v/w) solvable (Perkin Elmer) over night at 60°C and counted using Aquasafe 300 Plus scintillation liquid (Zinsser Analytic) in a liquid scintillation counter (Perkin Elmer Tricarb). For in vivo insulin signaling studies, fasted mice were injected intraperitoneally with 2 U/kg insulin (Insuman Rapid, Aventis) or sterile saline. Ten minutes after injection, mice were sacrificed and organs were harvested for further analysis. Organs harvested for Western blot or gene expression analysis were weighed and then immediately snap-frozen in liquid nitrogen.
2.3. mRNA expression analysis
Total RNA was isolated from organs using TRIzol (Invitrogen) and a NucleoSpin RNAII kit (Macherey & Nagel) according to manufacturer’s instructions. After DNase treatment (RNase-free DNase set, Qiagen), 400 ng RNA was used for cDNA preparation (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems) according to manufacturer’s instructions. Quantitative real-time RT-PCR was performed using assays-on-demand primer/probe sets provided by Applied Biosystems (Assay IDs: mAcaca, Mm01304285_m1; mCd36, Mm00432403_m1; mDio2, Mm00515664_m1; mElovl3, Mm00468164_m1; mFasn, Mm00662319_m1; mGlut4 = mSlc2a4, Mm01245502_m1; mLpl, Mm00434764_m1; mLrp1, Mm00464608_m1; mPid1, Mm01545237_m1; mPpargc1a, Mm00447183_m1; mTbp, Mm00446973_m1; mUcp1, Mm00494069_m1. Relative mRNA expression was calculated by the ΔΔCT method und normalization to the housekeeper (Tbp) mRNA expression as described previously [32].
2.4. Cell culture
For the isolation of primary brown adipocytes, WT or Pid1−/− mice (aged 3–5 weeks) were sacrificed, interscapular brown adipose tissue was removed and digested in PBS containing collagenase II (Biochrom). Pre-adipocytes were cultured for 10 days in DMEM containing 10% FCS and 1% PenStrep and were differentiated through addition of 20 nM insulin (Sigma), 1 nM T3 (trijodothyronine-sodium, Sigma), 0.5 mM IBMX (3-isobutyl-1-methylxanthine, Sigma), 1 μM Dexa (Dexamethasone, Sigma) and 1 μM Rosiglitazone (Sigma) to obtain mature brown adipocytes. For the generation of LRP1-deficient primary brown adipocytes, pre-adipocytes were isolated from Lrp1fl/fl mice [22] and cultured as described above. Two days after seeding, cells were infected with Ad-Cre adenovirus (Cat. No. ADV-005, Cellbiolabs), yielding infection rates of ~50%. Afterwards, cells were differentiated and mature brown adipocytes were analysed as described below.
2.5. Antibodies
Antibodies used in the study were: sheep polyclonal anti-LRP1 (made in-house, IF 1:250; IP 1:200 [33]), rabbit monoclonal anti-LRP1 (abgent, AJ1448a; WB 1:10000), rabbit polyclonal anti-PID1 (Sigma-Aldrich, HPA36103; WB 1:200), rabbit polyclonal anti-GLUT4 (kindly provided by A. Schürmann, DIfE, Potsdam-Rehbrücke, Germany; WB 1:1000, IF 1:500), rabbit monoclonal anti-IRAP (Cell Signaling 6918, IF 1:250), mouse monoclonal anti-GLUT1 (Abcam, ab40084, IF 1:100), rabbit polyclonal anti-AS160 (Millipore 07–741, WB 1:500), mouse monoclonal anti-GST (Santa Cruz sc-138, WB 1:500), rabbit polyclonal anti-AKT (Cell Signaling, 9272, WB 1:1000) and rabbit polyclonal anti-p-AKT (Ser473) (Cell Signaling, 9271, WB 1:1000).
All following secondary antibodies were purchased from Jackson Immuno Research: goat anti-rabbit HRP (#111-035-144, WB 1:5000); goat anti-mouse HRP (#115-035-003, WB 1:5000); Cy2-donkey-anti-sheep (#713-225-147, IF 1:250), Cy2-donkey-anti-sheep (#713-166-147, IF 1:500), Cy2-donkey-anti-rabbit (#711-225-152, IF 1:250), Cy3-donkey-anti-rabbit (#711-165-152; IF 1:500), Alexa488-donkey-anti-mouse (#715-486-150, IF 1:250).
2.6. Protein extraction, SDS-PAGE and Western blotting
Western blots were performed using standard procedures [34]. Cells were lysed in lysis buffer (2 mM CaCl2, 80 mM NaCl, 1% (v/v) TritonX-100, 50 mM Tris/HCl, pH 8.0) supplemented with cOmplete Mini inhibitor cocktail (Roche). Snap-frozen tissue samples were lysed in 10x (v/w) RIPA buffer (50 mM Tris-HCl pH7.4, 5 mM EDTA, 150 mM NaCl, 1 mM Na-Pyrophosphate, 1 mM NaF, 1 mM Na-Vanadate, 1% (v/v) NP-40) supplemented with cOmplete Mini inhibitor cocktail (Roche) and 0.1% SDS using a TissueLyser (Qiagen). Protein concentrations were determined using the Lowry method and subsequently, proteins were separated on NuPAGE Bis-Tris 4–12 % gradient gels (Invitrogen) or 10% Tris-glycine gels. For Western blotting, proteins were transferred to nitrocellulose membranes, blocked for 1h in 5% (w/v) milk in TBS-T (20 mM Tris, 150 mM NaCl, 0.1% (v/v) Tween 20) and incubated overnight at 4°C in the respective primary antibodies, diluted in 5% (w/v) BSA in TBS-T. After washing in TBS-T, the membranes were incubated with respective secondary horseradish peroxidase-labelled antibodies. Signals were detected with enhanced chemiluminescence (ECL) using Amersham Hyperfilm (GE Healthcare) or Amersham Imager 600 (GE), and densitometric quantification was carried out using LICOR image studio lite.
2.7. Immunoprecipitation
To analyse the interaction between LRP1 and PID1, BAT and muscle was harvested from fasted wild type mice and immediately placed in ice-cold RIPA buffer supplemented with phosphatase and protease inhibitors. Tissue lysates were generated as described above and 1 mg of total protein (in 300 μl) were pre-cleared using sepharose beads (25 μl Protein G Sepharose, GE, washed 2 times with RIPA buffer). Then, the lysates were incubated with anti-LRP1-antibody for 4 hours on a rotation wheel at 4°C. Protein G-coupled magnetic beads (50 μl dynabeads, Invitrogen) were washed 2 times with RIPA buffer and incubated with the antibody-containing lysates for additional 2 hours. Afterwards, the beads were washed 3 times with 500 μL RIPA, and elution was performed using NuPage SDS sample buffer (Invitrogen) for 10 min at 60°C. LRP1 and PID1 levels in input as well as pulldown fractions were analysed by Western blotting. WT and Pid1−/− lysates were used as controls.
2.8. GST-pulldown experiments
For pulldown analysis, muscle lysates (150 μg) were incubated overnight at 4°C with a GSTPID1 fusion protein [23], which prior was incubated with glutathione sepharose. Subsequently, pulldown and unbound fractions were separated by centrifugation for 3 min at 9000 rpm. Supernatants were removed and the sepharose pellets were washed six times with PBS. Pellets were re-suspended in NuPAGE reducing sample buffer, heated at 60°C for 10 min and analysed by Western blot.
2.9. Membrane preparation
For the isolation of plasma membranes, muscle samples were harvested from WT and Pid1−/− mice and homogenized in 6x (v/w) homogenization buffer (20 mM Tris-HCl, 2 mM MgCl2 0.25 M sucrose, pH 7.4), supplemented with protease inhibitors (Roche). Lysates were centrifuged for 15 min at 800 g and 4°C, supernatants were taken, and centrifugation steps were repeated. The resulting supernatants were centrifuged at 100,000 g for 1 hour at 4°C. The pellets were re-suspended in 200 μl buffer (50 mM Tris-HCl, 2 mM CaCl2, 80 mM NaCl, 1% (v/v) Triton X-100, pH 8.8) supplemented with protease inhibitors, and centrifuged at 100,000 g for 1 hour at 4°C. The supernatants, containing membrane proteins, were used for Western blot analysis.
2.10. Immunofluorescence
To analyse the localization of GLUT4, LRP1, IRAP and GLUT1 before and after insulin stimulation, primary wild type and Pid1−/− brown adipocytes were generated as described above and seeded onto glass coverslips. After differentiation, the cells were starved for 2 hours and then incubated without or with 10 nM Insulin in DMEM supplemented with 10% FCS for 10 min at 37°C. For the analysis of LRP1fl/fl cells, fully differentiated brown adipocytes, infected with pAd-Cre, were starved for 2 hours. In both cases, cells were then washed with PBS, fixed with 4% PFA and analysed by indirect immunofluorescence using specific antibodies. Co-localization analysis was performed using a Nikon A1 confocal laser scanning microscope.
2.11. Statistical analyses and data processing
Data were processed using Microsoft Excel and GraphPad Prism 6. Two-tailed, unpaired Student’s t-test or One-Way ANOVA with Dunnett correction for multiple comparisons were performed to calculate statistical significance. P ≤ 0.05 was considered to be statistically significant.
3. Results
3.1. Inverse regulation of PID1 and LRP1 with GLUT4 levels and glucose uptake in response to cold-induced activation of brown adipose tissue
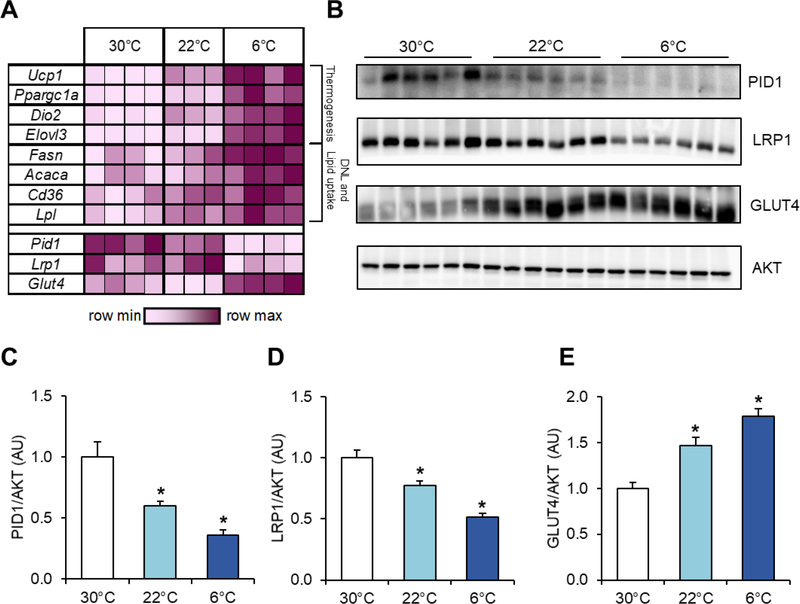
Adaptive thermogenesis and energy uptake by BAT are tightly controlled by the ambient temperature [3, 9]. At thermoneutrality (around 30–32°C for mice), metabolic activity of brown adipocytes is minimal, whereas decreasing ambient temperature stimulates sympathetic activation of BAT-mediated heat production and nutrient uptake [35, 36]. To study the effects of ambient temperature on gene expression of Pid1 and Lrp1 in relation to the expression of Glut4 and thermogenic marker genes, BAT samples of wild type mice acclimated to different ambient temperatures were analysed (Figure 1A). As expected, the expression of the established BAT activation markers uncoupling protein 1 (Ucp1), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Ppargc1a), deiodinase 2 (Dio2) and elongation of very long chain fatty acids 3 (Elovl3) as well as the critical lipid metabolism genes fatty acid synthase (Fasn), acetyl-CoA carboxylase (Acaca), cluster of differentiation 36 (Cd36), and lipoprotein lipase (Lpl) was increased in response to cold exposure. Notably, while the expression of the insulin-responsive glucose transporter 4 (Glut4) was up-regulated in activated BAT, we detected decreased expression of the low-density lipoprotein receptor-related protein 1 (Lrp1) and the phosphotyrosine interacting domain containing 1 (Pid1). Western blot analysis (Figure 1B) and densitometric quantification (Figures 1C–E) showed cold-induced reduction in PID1 and LRP1 proteins while levels of GLUT4 were increased.
Figure 1.
Inverse effects of environmental temperature on PID1 and LRP1 compared to GLUT4 expression levels in BAT. Male C57BL/6J wild type mice were acclimated to 30°C, 22°C or 6°C for 1 week. For gene expression analysis, BAT samples were harvested after a 4 hour fasting period while protein levels were determined two hours after an oral gavage of a glucose solution containing tracer amounts of 3H-deoxyglucose. (A) Expression of genes important for thermogenesis, de novo lipogenesis (DNL) and lipid uptake is presented in a heatmap as fold change over the mean BAT expression levels from 30°C acclimated mice (n=3–4 per group). (B) Western blot analysis of PID1, LRP1, GLUT4 and AKT in BAT tissue extracts (n=6 per group). (C) PID, (D) LRP1 and (E) GLUT4 protein levels normalized to AKT (n=6 per group). Data are presented as means ± SEM. Statistical significance was calculated using One-Way ANOVA with Dunnett correction for multiple comparisons. *p ≤ 0.05 for 30°C vs. 22°C or 6°C, respectively.
Ucp1: uncoupling protein 1; Ppargc1a: peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Dio2: deiodinase 2; Elovl3: elongation of very long chain fatty acids 3; Fasn: fatty acid synthase; Acaca: acetyl-CoA carboxylase; Cd36: cluster of differentiation 36; Lpl: lipoprotein lipase; Glut4: glucose transporter 4; Lrp1: low-density lipoprotein receptor-related protein 1; Pid1: phosphotyrosine interacting domain containing 1.
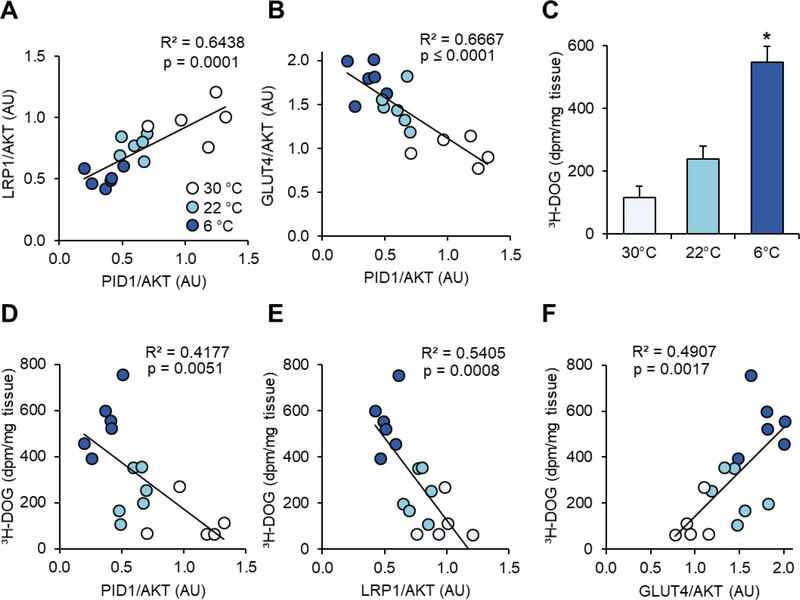
Correlation analysis revealed an inverse association between PID1 and LRP1 expression in relation to GLUT4 expression (Figure 2A–B). In line with previous observations we detected an incremental increase in glucose disposal with increasing BAT activation (Figure 2C). Moreover, expression of both PID1 and LRP1 correlated negatively, while GLUT4 showed a positive association with BAT glucose uptake (Figures 2D–F).
Figure 2.
Correlation of PID1, LRP1 and GLUT4 expression with BAT glucose uptake in differentially cold-activated mice. Protein levels shown in Figure 1 were used for correlation analysis. Pearson correlation analysis of PID1 expression with (A) LRP1 and (B) GLUT4. (C) Uptake of 3H-deoxyglucose (3H-DOG) into BAT of mice described in Figure 1 is calculated as dpm/mg tissue and presented as means ± SEM. (D) PID1, (E) LRP1 and (F) GLUT4 protein levels were related to radioactive glucose uptake using Pearson correlation analysis. For C, statistical significance was calculated using One-Way ANOVA with Dunnett correction for multiple comparisons. n=5–6 per group, *p ≤ 0.05 for 30°C vs. 6°C.
In summary, these data show that the higher metabolic activity of BAT at lower ambient temperatures is associated with reduced expression of LRP1 and its adaptor protein PID1.
3.2. PID1 controls insulin-dependent LRP1 and GLUT4 trafficking in brown adipocytes
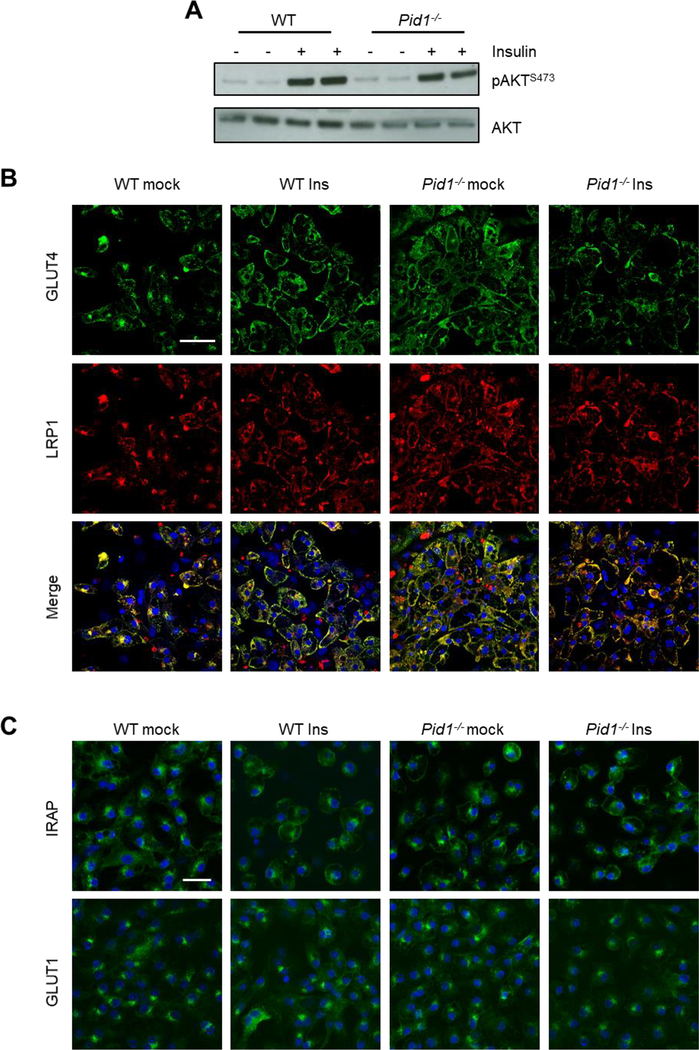
To elucidate the functional role of PID1 for LRP1 in adipose tissues, we first determined insulin-dependent phosphorylation of AKT in primary brown adipocytes isolated from wild type and PID1-deficient (Pid1−/−) mice (Figure 3A). Similar levels of phosphorylated AKT were observed between wild type and Pid1−/− adipocytes at baseline and after insulin stimulation. In vivo insulin administration in wild type and Pid1−/− mice led to comparable levels of phosphorylated AKT in BAT as well as inguinal and epididymal WAT depots (Supplementary figures S1A–F). Similarly, insulin administration induced equal glucose lowering in wild type and Pid1−/− mice fed either a chow or a high fat diet (HFD; Supplementary figures S1G,H). These findings indicate that PID1 does not directly influence insulin receptor-dependent signaling in vitro and in vivo. Next we investigated the impact of PID1 on intracellular sorting of LRP1 and GLUT4 in primary brown adipocytes using indirect immunofluorescence. In wild type cells, LRP1 and GLUT4 co-localized in perinuclear compartments under basal conditions and at the cell surface after insulin stimulation (Figure 3B). In contrast, LRP1 and GLUT4 were detected primarily at the plasma membrane irrespective of the presence of insulin in Pid1−/− primary brown adipocytes (Figure 3B). To determine whether this effect of PID1-deficiency was specific to GLUT4, we analysed the intracellular localization of another component of GLUT4-storage vesicles, the insulin-responsive aminopeptidase IRAP [37]. In WT cells under basal conditions, the majority of the IRAP signal was detected in perinuclear compartments (Figure 3C). Similar to GLUT4, insulin treatment resulted in increased plasma membrane abundance of IRAP in WT cells. In Pid1−/− adipocytes, IRAP was already found at higher levels at the cell surface in the basal state, and insulin treatment of Pid1−/− adipocytes did not further alter the intracellular localization of IRAP (Figure 3C). To exclude general alterations in intracellular sorting in PID1-deficient adipocytes, we also analysed the localization of the insulin-independent glucose transporter GLUT1 [38]. We found GLUT1 to be present in both intracellular compartments as well as at the plasma membrane in WT cells under basal conditions. However, and in contrast to LRP1, GLUT4 and IRAP, neither insulin treatment nor PID1-deficiency altered this distribution (Figure 3C). Altogether, these data suggest that PID1 directly controls the cell surface abundance of proteins within GLUT4-storage vesicles, possibly via the insulin-dependent vesicular trafficking of LRP1.
Figure 3.
Effect of PID1-deficiency on insulin signaling and localization of LRP1 and GLUT4 in primary brown adipocytes. (A) Insulin signaling was determined by Western blot analysis of AKT phosphorylation at position Ser473. For this purpose, differentiated brown adipocytes prepared from wild type (WT) and Pid1−/− mice were analysed under basal and insulin-stimulated conditions. (B) Immunofluorescence analysis of GLUT4 (green) and LRP1 (red) in WT and Pid1−/− primary brown adipocytes under basal and insulin-stimulated conditions. Yellow fluorescence indicates co-localization. (C) Immunofluorescence analysis of IRAP and GLUT1 in WT and Pid1−/− primary brown adipocytes under basal and insulin-stimulated conditions. Nuclei are stained with DAPI (blue; scale bar in (B) = 20 μm; scale bar in (C) = 25 μm).
3.3. PID1 interacts via LRP1 with GLUT4-containing vesicles in adipose tissue and muscle
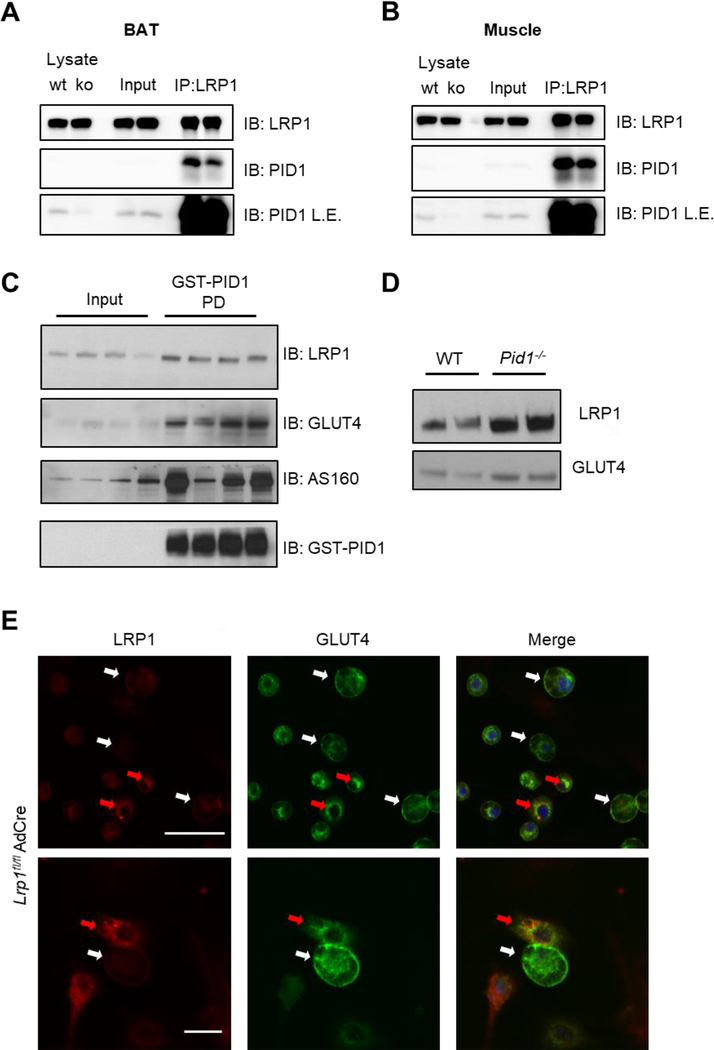
As we previously demonstrated the role of PID1 as a retention adaptor protein for LRP1 in hepatocytes [23], we studied the interaction of PID1 and LRP1 by co-immunoprecipitation experiments using lysates of brown adipose tissue (Figure 4A) and muscle (Figure 4B). An high enrichment of PID1 in the LRP1 pulldown fractions was observed in both tissues (Figure 4A,B), confirming the binding of PID1 to LRP1 also in adipose tissue and muscle. Given the presence of LRP1 in GSVs [21], we investigated whether PID1 interacts with GLUT4-storage vesicles via LRP1. Using GST-PID1 we pulled down LRP1, GLUT4 and AS160 from detergent-free muscle lysates (Figure 4C), indicating interaction of PID1 with LRP1 and thus GSVs. To study whether PID1-deficiency influence GSV trafficking in vivo, we isolated plasma membranes from muscle tissue of WT and Pid1−/− mice. Compared to controls, Western blot analysis revealed higher levels of GLUT4 and LRP1 in plasma membranes isolated from PID1-deficient muscles (Figure 4D). (Figure 4D). To study whether the effects on GLUT4 trafficking observed in the absence of PID1 can be mimicked by LRP1-deficiency, we infected primary brown adipocytes isolated from Lrp1fl/fl mice with an adenovirus containing a CRE expression cassette. This resulted in a mixed population of LRP1-containing and LRP1-deficient brown adipocytes. Notably, LRP1-deficient adipocytes (white arrows) are characterized by higher cell surface abundance of GLUT4, while in LRP1-positive adipocytes (red arrows) GLUT4 was found predominantly in perinuclear compartments (Figure 4E).
Figure 4.
Relevance of LRP1 and PID1 interaction for GLUT4 trafficking in brown adipose tissue and muscle. Immunoprecipitation of LRP1 was performed in brown adipose tissue (A) as well as muscle (B), and LRP1 as well as PID1 were detected in input and IP fractions by Western blotting. To confirm the specificity of the PID1 antibody, total lysates from WT and Pid1−/− muscle and brown adipose tissue were used as controls. To show the presence of PID1 in lysates and inputs, blots with longer exposure times (L.E.) are shown. (C) Muscle lysates from WT mice were incubated with GST-tagged recombinant PID1, and pulldown experiments were performed. LRP1, GLUT4, AS160, as well as GST-PID1 were detected in lysates and pulldown-fractions (PD) by Western blotting. (D) Plasma membrane preparation from muscles of WT and Pid1−/− mice. GLUT4 and LRP1 levels were detected by Western blot. (E) Immunofluorescent staining against GLUT4 (green) and LRP1 (red) in primary brown LRP1fl/fl adipocytes infected with an adenovirus containing a CRE expression cassette. Nuclei are stained with DAPI (blue; scale bar upper picture 50 μm and lower picture 25 μm). Red arrows indicate LRP1-positive cells and white arrows indicate LRP1-negative cells.
3.4. PID1-deficiency improves hyperglycemia by stimulating glucose uptake into adipose tissues and muscles under diabetogenic conditions.
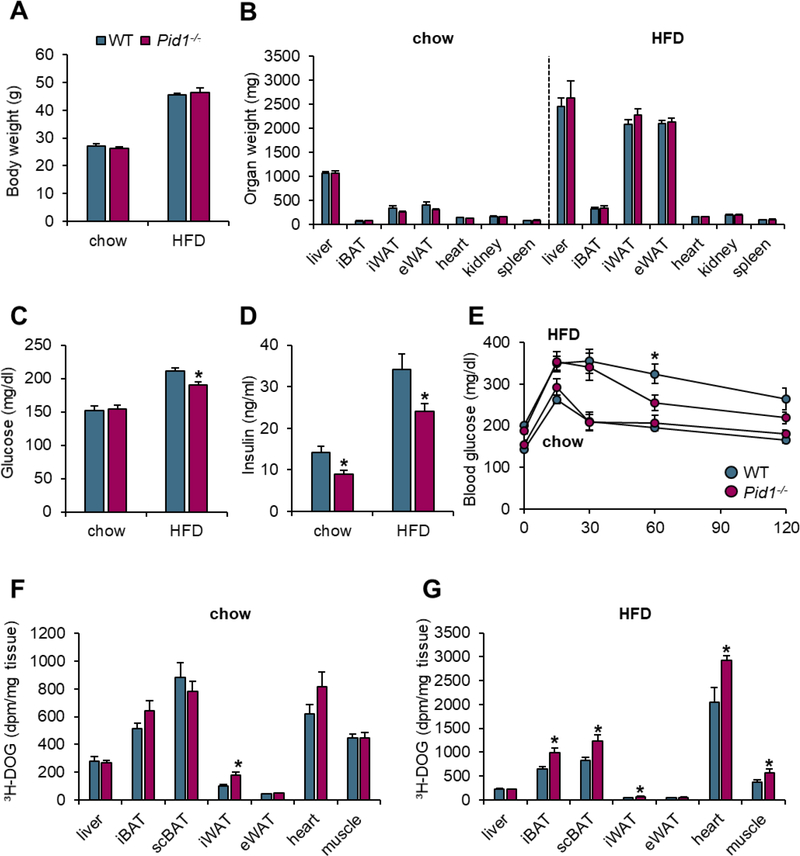
To study the functional relevance of PID1 under healthy conditions and obesity-associated insulin resistance, wild type and Pid1−/− mice were fed a chow diet or a diabetogenic HFD. Opposite to the down-regulation of Pid1 observed in catabolic cold-activated BAT (Figure 1A), we detected increased Pid1 expression in WAT under anabolic HFD-feeding conditions (Supplementary figures S2A,B). On both diets, wild type mice and Pid1−/− littermates had similar body and organ weights (Figures 5A,B). Furthermore, Pid1−/− mice on chow diet did not show altered glucose plasma levels, while under diabetogenic conditions, PID1-deficiency resulted in reduced plasma glucose levels compared to wild type controls (Figure 5C). Both in the healthy and in the insulin-resistant state, reduced insulin levels were observed in Pid1−/− mice (Figure 5D).
Figure 5.
Body and organ weights, glucose tolerance and uptake in wild type and Pid1−/− mice fed a chow or obesogenic high-fat diet (HFD). (A) Body and (B) organ weights as well as (C) fasting glucose and (D) insulin levels of chow- and HFD-fed WT and Pid1−/− mice (n=7 for chow and n=21–23 for HFD per group) (E) For assessment of glucose tolerance, fasted chow- and HFD-fed WT and Pid1−/− mice received an oral gavage of a glucose solution containing a tracer dose of 3H-deoxyglucose (3H-DOG). Plasma glucose levels were determined at indicated time points. (F, G) Uptake of 3H-DOG into metabolically active tissues was determined as dpm/mg tissue two hours after oral gavage in (F) chow- and (G) HFD-fed WT and Pid1−/− mice (n=7–8 per group). Data are shown as means ± SEM. Statistical significance was calculated using Student’s t test. *p ≤ 0.05 for WT vs. Pid1−/− mice.
iBAT - interscapular brown adipose tissue; scBAT - subscapular brown adipose tissue; iWAT - inguinal white adipose tissue; eWAT - epididymal white adipose tissue.
In chow-fed mice that are characterized by normal insulin sensitivity, wild type and Pid1−/− mice displayed a similar oral glucose tolerance (Figure 5E), and a comparable organ uptake of the simultaneously administered glucose tracer 3H-deoxyglucose (Figure 5F). On the other hand, a significantly improved glucose tolerance was detected in insulin-resistant Pid1−/− mice when compared to insulin-resistant controls (Figure 5E). This was accompanied by increased 3H-deoxyglucose disposal into PID1, LRP1 and GLUT4 expressing tissues, such as brown and white adipose tissues, heart and skeletal muscles of Pid1−/− mice (Figure 5G). In conclusion, these data support that the higher cell surface abundance of GLUT4 in PID1-deficient mice confers beneficial effects on glucose homeostasis particularly in the insulin-resistant state.
3.5. Adipocyte-specific PID1-deficiency improves glucose tolerance in the insulin-resistant state.
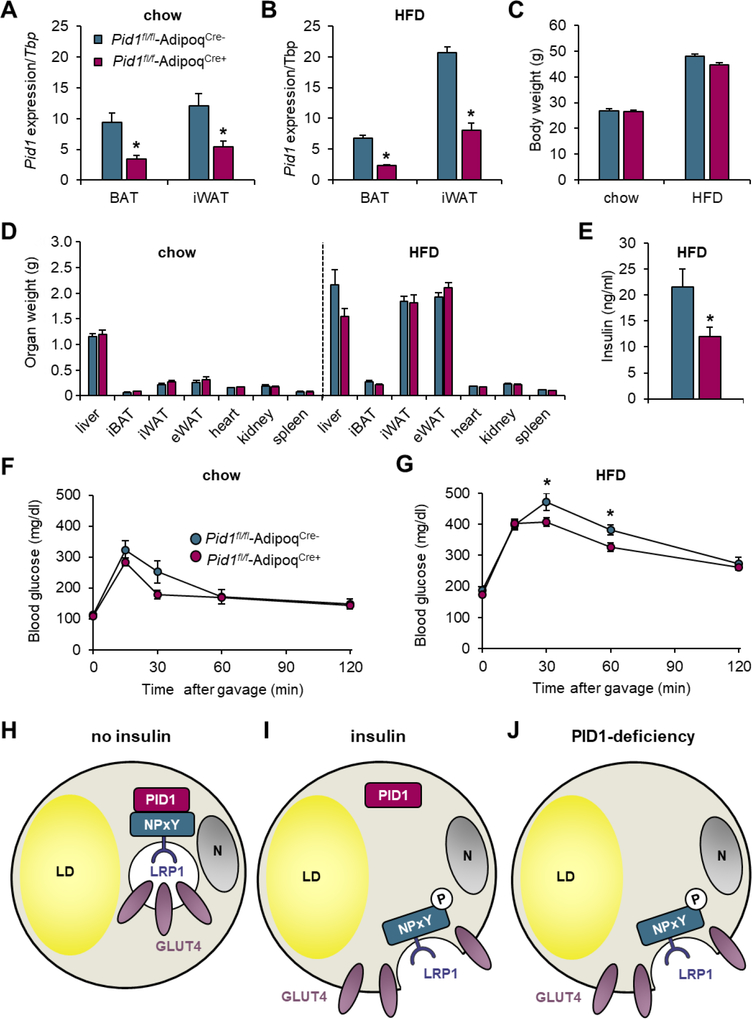
To exclude systemic effects of the total PID1-knockout and to specifically investigate the relevance of PID1 in adipose tissue depots, we generated adipocyte-specific PID1-deficient mice by crossing Pid1fl/fl mice [23] with mice expressing Cre recombinase under control of the adiponectin promoter (AdipoqCre+). These Pid1fl/fl-AdipoqCre+ mice and littermate controls (Pid1fl/fl-AdipoqCre-) were fed a chow or a diabetogenic HFD. The expression of Pid1 in white and brown adipose tissues of Pid1fl/fl-AdipoqCre+ mice was significantly reduced on both chow and HFD (Figures 6A,B). Residual Pid1 expression may be explained by expression in the stroma vascular cells, such as pre-adipocytes, macrophages and endothelial cells. Body and organ weights were not affected by adipocyte-specific loss of Pid1 (Figures 6C,D). In line with the effects of systemic PID1-deficiency, we observed lower fasting insulin levels in HFD-fed Pid1fl/fl-AdipoqCre+ mice compared to respective controls (Figure 6E). In an oral glucose tolerance test, we observed comparable glucose tolerance in chow-fed Pid1fl/fl-AdipoqCre− and Pid1fl/fl-AdipoqCre+ mice (Figure 6F). In the insulin-resistant state, Pid1fl/fl-AdipoqCre+ mice displayed an improved glucose tolerance (Figure 6G), demonstrating the regulatory role of adipocyte PID1 for adipocyte-dependent control of systemic glucose metabolism.
Figure 6.
Effects of adipocyte-specific PID1-deficiency on body weight, organ weights, gene expression and glucose metabolism. Adipocyte-specific PID1-deficient mice were generated by crossing Pid1-floxed mice with mice expressing Cre recombinase under the control of the adiponectin promoter (Pid1fl/fl-AdipoqCre+). Cre-negative littermates were used as controls (Pid1fl/fl-AdipoqCre−). Gene expression analysis of Pid1 normalized to Tbp in BAT and iWAT of (A) chow- and (B) HFD-fed mice (n=5–9 per group). (C) Body and (D) organ weights in chow- and HFD-fed mice (n=5–11 per group). (E) Insulin levels in HFD-fed mice after a 4 hour fasting period (n=7–9 per group). Oral glucose tolerance tests in (F) chow- and (G) HFD-fed mice (n=5–9 per group). Data are shown as means ± SEM. Statistical significance was calculated using Student’s t test. *p ≤ 0.05 for Pid1fl/fl-AdipoqCre− vs. Pid1fl/fl-AdipoqCre+. (H–J) Model of the proposed role of PID1 in the regulation of GLUT4-mediated glucose uptake into adipocytes under (H) basal and (I) insulin stimulated conditions. (J) PID1-deficiency results in the perpetual presence of LRP1 and GLUT4 at the cell surface.
4. Discussion
PID1 is an intracellular adaptor protein for LRP1 with higher expression in adipose tissues of obese compared to lean subjects [26, 39]. Recently, we have shown that PID1 controls clearance of pro-atherogenic lipoproteins into the liver by regulating cell surface abundance of LRP1 [23]. In addition to its role in hepatic lipoprotein metabolism, elevated PID1 expression was previously found to be associated with reduced glucose uptake in adipocytes and myotubes [31, 40]. In the current study, we show that PID1 determines the cellular localization of both LRP1 and GLUT4 that are found in perinuclear compartments of primary adipocytes under non-stimulated, fasting conditions. As expected, insulin stimulation leads to the translocation of both proteins to the plasma membrane [21, 24]. In Pid1−/− adipocytes and muscle, LRP1 and GLUT4 are targeted to the cell surface irrespective of insulin signaling. Notably, LRP1-deficiency also resulted in elevated cell surface levels of GLUT4 in primary brown adipocytes, supporting the concept that PID1-LRP1 interaction is involved in the intracellular trafficking of GSVs. Furthermore, we show the in vivo relevance of PID1 for glucose metabolism in global as well as adipocyte-specific PID1-deficient mice. The lack of PID1 did not alter insulin signaling and responsiveness in vivo, but resulted in an improved glucose tolerance and increased glucose disposal into GLUT4-expressing tissues despite insulin resistance. These data support the model (Figure 6H–J) that in the basal state, PID1 interacts with the non-phosphorylated distal NPxY motif within the intracellular domain of LRP1 [23, 41, 42] to retain GLUT4-containing storage vesicles in perinuclear compartments of adipocytes (Figure 6H). Insulin-mediated phosphorylation of the distal NPxY motif of LRP1 promotes the disintegration of the PID1-LRP1 complex. This process ultimately leads to the insulin-triggered targeting of GLUT4 to the cell surface to facilitate efficient glucose uptake in the postprandial situation (Figure 6I). Consequently, the lack of PID1 resulted in the perpetual cell surface presence of GLUT4, stimulating glucose uptake even in insulin-resistant, diabetogenic conditions (Figure 6J). The proposed mechanism is in line with our recent findings demonstrating that PID1 is involved in the hepatic clearance of pro-atherogenic lipoproteins [23], a process dependent on the insulin-mediated translocation of LRP1 to the cell surface [22]. Mechanistically, we showed that PID1 functions as an adaptor protein for LRP1 retaining the non-phosphorylated form of this receptor in intracellular storage compartments. In the postprandial phase, insulin signaling triggers the phosphorylation of the LRP1 - 60NPxYxxL66 motif, thereby disrupting the LRP1-PID1 complex ultimately leading to the vesicular transport of LRP1 to the plasma membrane [23].
Here, we provide evidence that in a similar manner PID1 regulates LRP1-mediated GLUT4 transport and glucose uptake in adipose and muscle tissues. In the current view, the GTPase-activating proteins (GAP) AS160 and TBC1D1 inhibit the RAB proteins RAB8A, RAB10, RAB13 and RAB14, which coordinate the translocation of GLUT4 to the plasma membrane [43]. Insulin receptor activation stimulates the AKT2-dependent inactivation of AS160 and TBC1D1, allowing RAB-dependent exocytosis of GLUT4-containing vesicles [43]. It is noteworthy that PID1 possesses only a phospho-tyrosine binding domain but it lacks GAP activity or additional motifs that could mediate the association of PID1 with the endosomal sorting machinery. Hence, unlike AS160 and TBC1D1, it is conceivable that PID1 directly competes with the endosomal sorting machinery that mediates vesicular trafficking of LRP1 and thus indirectly GLUT4. The retromer complex and associated adaptor proteins are good candidates to mediate cell surface trafficking of LRP1 by replacing the retention adaptor PID1. The retromer is important for retrograde transport of cargo proteins from endosomes back to the Golgi apparatus and consists of vesicular protein sorting proteins (Vps35, Vps29, Vps26) and sorting nexins (SNX1, SNX2 or SNX5, SNX6) [44]. Recently, it was shown that in conjunction with other SNX proteins, this complex also mediates endosome to plasma membrane transport [45]. Moreover, recycling of SNX17 cargoes, such as LRP1, is dependent on a retromer-like complex termed the retriever [46]. Furthermore, phosphorylation of LRP1-Y 63 has been implicated in SNX17-facilitated endosome to basolateral surface recycling of this receptor [42, 47]. Thus, it seems likely that PID1 is the insulin-dependent regulator that controls the SNX17-mediated LRP1 transport, and, accordingly the GLUT4-driven glucose uptake into adipose and muscle tissues. It remains to be determined which kinases are responsible for the insulin-mediated LRP1 phosphorylation, and whether this novel PID1 pathway involves additional, unidentified vesicular compartments and/or sorting factors.
In addition to providing insight in PID1 cell biology, the current study emphasizes the potential role of adipose PID1 as a therapeutic target for maintaining glucose homeostasis in insulin-resistant diabetic individuals. It remains to be evaluated if PID1 inactivation can also improve glucose metabolism in type 1 diabetes models. In line with the data presented here, adipose-specific LRP1-deficient mice displayed improved glucose tolerance [48], which could be explained by diminished intracellular vesicular retention, and thus higher abundance of GLUT4 at the cell surface. However, the lack of LRP1 results in a delayed postprandial lipoprotein clearance, limiting the value of this receptor for pharmacological targeting. In contrast, PID1-deficiency resulted in an accelerated LRP1- and LDL receptor-mediated endocytosis of pro-atherogenic lipoproteins into the liver [23]. Here, we show that next to this beneficial lipid-lowering effect, PID1-deficiency also improves glucose tolerance by stimulating glucose uptake into GLUT4-expressing tissues despite diet-induced insulin resistance. These findings have important consequences for type 2 diabetes, a disease characterized by hyperglycemia and dyslipidemia causing cardiovascular disease [8, 49, 50]. It is encouraging that in insulin-sensitive models PID1 inactivation does not result in hypoglycemia suggesting that PID1 targeting as a therapy for type 2 diabetes would likely not result in episodes of hypoglycemia often seen with insulin or sulfonylurea therapy [49, 51].
Taken together this study highlights the physiologic and pathophysiologic relevance as well as the therapeutic potential of PID1 for glucose metabolism. We show that PID1 is a negative regulator of glucose metabolism and adipose tissue metabolic activity. PID1, presumably via its interaction partner LRP1, serves as a retention factor to keep GLUT4 in a perinuclear storage compartment under fasting conditions. Global deficiency for PID1 improves glucose tolerance and peripheral glucose disposal in states of insulin resistance. This is due to increased GLUT4 abundance at the plasma membrane, an effect not crucial in chow-fed animals, since these mice display functional insulin signaling. Adipose-tissue-specific deletion of PID1 alone also improves systemic glucose metabolism, highlighting the importance of adipose tissue for regulation of glucose homeostasis. Hence, in addition to a lipid-lowering effect via stimulation of hepatic TRL-remnant and LDL-clearance, PID1 inhibition could serve as a potential target to improve glucose clearance in insulin-resistant states, thereby reducing metabolic burden in diabetic patients.
Supplementary Material
Highlights for.
Metabolic (in)activity determines PID1 expression in brown and white adipose depots
PID1 is an adaptor protein regulating insulin-dependent GLUT4 sorting in adipocytes
PID1 impairs glucose disposal into GLUT4-expressing adipose tissues and muscles
PID1-deficiency ameliorates hyperglycemia in insulin-resistant mice
Acknowledgement
We thank Anette Rosche, Sandra Ehret and Birgit Henkel for excellent technical assistance.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (research training group GRK1459 and Heisenberg Professorship HE3645/10–1) and an NIH UCSD/UCLA DRC Pilot and Feasibility grant to P.L.S.M.G. (P30 DK063491). A.W.F. was supported by a fellowship from the German National Academic foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No potential conflicts of interest were disclosed.
References
- [1].Scheja L, Heeren J, Metabolic interplay between white, beige, brown adipocytes and the liver, J. Hepatol, 64 (2016) 1176–1186. [DOI] [PubMed] [Google Scholar]
- [2].Young SG, Zechner R, Biochemistry and pathophysiology of intravascular and intracellular lipolysis, Genes Dev, 27 (2013) 459–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cannon B, Nedergaard J, Brown adipose tissue: function and physiological significance, Physiol. Rev, 84 (2004) 277–359. [DOI] [PubMed] [Google Scholar]
- [4].Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, Scheinin M, Taittonen M, Niemi T, Enerback S, Virtanen KA, Different metabolic responses of human brown adipose tissue to activation by cold and insulin, Cell Metab, 14 (2011) 272–279. [DOI] [PubMed] [Google Scholar]
- [5].Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmuller A, Gordts PL, Rinninger F, Bruegelmann K, Freund B, Nielsen P, Merkel M, Heeren J, Brown adipose tissue activity controls triglyceride clearance, Nat. Med, 17 (2011) 200–205. [DOI] [PubMed] [Google Scholar]
- [6].Heine M, Fischer AW, Schlein C, Jung C, Straub LG, Gottschling K, Mangels N, Yuan Y, Nilsson SK, Liebscher G, Chen O, Schreiber R, Zechner R, Scheja L, Heeren J, Lipolysis Triggers a Systemic Insulin Response Essential for Efficient Energy Replenishment of Activated Brown Adipose Tissue in Mice, Cell Metab, (2018). [DOI] [PubMed] [Google Scholar]
- [7].Roberts-Toler C, O’Neill BT, Cypess AM, Diet-induced obesity causes insulin resistance in mouse brown adipose tissue, Obesity (Silver Spring, Md.), 23 (2015) 1765–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Petersen MC, Shulman GI, Mechanisms of Insulin Action and Insulin Resistance, Physiol. Rev, 98 (2018) 2133–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Heeren J, Scheja L, Brown adipose tissue and lipid metabolism, Curr. Opin. Lipidol, 29 (2018) 180–185. [DOI] [PubMed] [Google Scholar]
- [10].Berbee JF, Boon MR, Khedoe PP, Bartelt A, Schlein C, Worthmann A, Kooijman S, Hoeke G, Mol IM, John C, Jung C, Vazirpanah N, Brouwers LP, Gordts PL, Esko JD, Hiemstra PS, Havekes LM, Scheja L, Heeren J, Rensen PC, Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development, Nat. Commun, 6 (2015) 6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chondronikola M, Volpi E, Borsheim E, Porter C, Saraf MK, Annamalai P, Yfanti C, Chao T, Wong D, Shinoda K, Labbe SM, Hurren NM, Cesani F, Kajimura S, Sidossis LS, Brown Adipose Tissue Activation Is Linked to Distinct Systemic Effects on Lipid Metabolism in Humans, Cell Metab., 23 (2016) 1200–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanssen MJ, Hoeks J, Brans B, van der Lans AA, Schaart G, van den Driessche JJ, Jorgensen JA, Boekschoten MV, Hesselink MK, Havekes B, Kersten S, Mottaghy FM, van Marken Lichtenbelt WD, Schrauwen P, Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus, Nat. Med, 21 (2015) 863–865. [DOI] [PubMed] [Google Scholar]
- [13].Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB, Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue, J. Biol. Chem, 268 (1993) 22243–22246. [PubMed] [Google Scholar]
- [14].Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB, Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver, Nature, 409 (2001) 729–733. [DOI] [PubMed] [Google Scholar]
- [15].Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE, Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation, J. Biol. Chem, 278 (2003) 14599–14602. [DOI] [PubMed] [Google Scholar]
- [16].Perera HK, Clarke M, Morris NJ, Hong W, Chamberlain LH, Gould GW, Syntaxin 6 regulates Glut4 trafficking in 3T3-L1 adipocytes, Mol. Biol. Cell, 14 (2003) 2946–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Beaton N, Rudigier C, Moest H, Muller S, Mrosek N, Roder E, Rudofsky G, Rulicke T, Ukropec J, Ukropcova B, Augustin R, Neubauer H, Wolfrum C, TUSC5 regulates insulin-mediated adipose tissue glucose uptake by modulation of GLUT4 recycling, Molecular metabolism, 4 (2015) 795–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bogan JS, Hendon N, McKee AE, Tsao TS, Lodish HF, Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking, Nature, 425 (2003) 727–733. [DOI] [PubMed] [Google Scholar]
- [19].Bryant NJ, Gould GW, SNARE proteins underpin insulin-regulated GLUT4 traffic, Traffic, 12 (2011) 657–664. [DOI] [PubMed] [Google Scholar]
- [20].Randhawa VK, Bilan PJ, Khayat ZA, Daneman N, Liu Z, Ramlal T, Volchuk A, Peng XR, Coppola T, Regazzi R, Trimble WS, Klip A, VAMP2, but not VAMP3/cellubrevin, mediates insulin-dependent incorporation of GLUT4 into the plasma membrane of L6 myoblasts, Mol. Biol. Cell, 11 (2000) 2403–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jedrychowski MP, Gartner CA, Gygi SP, Zhou L, Herz J, Kandror KV, Pilch PF, Proteomic analysis of GLUT4 storage vesicles reveals LRP1 to be an important vesicle component and target of insulin signaling, J. Biol. Chem, 285 (2010) 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Laatsch A, Merkel M, Talmud PJ, Grewal T, Beisiegel U, Heeren J, Insulin stimulates hepatic low density lipoprotein receptor-related protein 1 (LRP1) to increase postprandial lipoprotein clearance, Atherosclerosis, 204 (2009) 105–111. [DOI] [PubMed] [Google Scholar]
- [23].Fischer AW, Albers K, Krott LM, Hoffzimmer B, Heine M, Schmale H, Scheja L, Gordts P, Heeren J, The adaptor protein PID1 regulates receptor-dependent endocytosis of postprandial triglyceride-rich lipoproteins, Mol. Metab, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Descamps O, Bilheimer D, Herz J, Insulin stimulates receptor-mediated uptake of apoE-enriched lipoproteins and activated alpha 2-macroglobulin in adipocytes, J. Biol. Chem, 268 (1993) 974–981. [PubMed] [Google Scholar]
- [25].Gordts PL, Bartelt A, Nilsson SK, Annaert W, Christoffersen C, Nielsen LB, Heeren J, Roebroek AJ, Impaired LDL receptor-related protein 1 translocation correlates with improved dyslipidemia and atherosclerosis in apoE-deficient mice, PLoS ONE, 7 (2012) e38330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Caratu G, Allegra D, Bimonte M, Schiattarella GG, D’Ambrosio C, Scaloni A, Napolitano M, Russo T, Zambrano N, Identification of the ligands of protein interaction domains through a functional approach, Mol. Cell. Proteomics, 6 (2007) 333–345. [DOI] [PubMed] [Google Scholar]
- [27].Kajiwara Y, Franciosi S, Takahashi N, Krug L, Schmeidler J, Taddei K, Haroutunian V, Fried U, Ehrlich M, Martins RN, Gandy S, Buxbaum JD, Extensive proteomic screening identifies the obesity-related NYGGF4 protein as a novel LRP1-interactor, showing reduced expression in early Alzheimer’s disease, Mol. Neurodegener, 5 (2010) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zeng XQ, Zhang CM, Tong ML, Chi X, Li XL, Ji CB, Zhang R, Guo XR, Knockdown of NYGGF4 increases glucose transport in C2C12 mice skeletal myocytes by activation IRS-1/PI3K/AKT insulin pathway, J. Bioenerg. Biomembr, 44 (2012) 351–355. [DOI] [PubMed] [Google Scholar]
- [29].Zhang CM, Chen XH, Wang B, Liu F, Chi X, Tong ML, Ni YH, Chen RH, Guo XR, Over-expression of NYGGF4 inhibits glucose transport in 3T3-L1 adipocytes via attenuated phosphorylation of IRS-1 and Akt, Acta Pharmacol. Sin., 30 (2009) 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang CM, Zeng XQ, Zhang R, Ji CB, Tong ML, Chi X, Li XL, Dai JZ, Zhang M, Cui Y, Guo XR, Effects of NYGGF4 knockdown on insulin sensitivity and mitochondrial function in 3T3-L1 adipocytes, J. Bioenerg. Biomembr, 42 (2010) 433–439. [DOI] [PubMed] [Google Scholar]
- [31].Chen L, Wang X-Y, Zhu J-G, You L-H, Wang X, Cui X-W, Shi C-M, Huang F-Y, Zhou Y-H, Yang L, Pang L-X, Gao Y, Ji C-B, Guo X-R, PID1 in adipocytes modulates whole-body glucose homeostasis, Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1861 (2018) 125–132. [DOI] [PubMed] [Google Scholar]
- [32].Schlein C, Talukdar S, Heine M, Fischer AW, Krott LM, Nilsson SK, Brenner MB, Heeren J, Scheja L, FGF21 Lowers Plasma Triglycerides by Accelerating Lipoprotein Catabolism in White and Brown Adipose Tissues, Cell Metab, 23 (2016) 441–453. [DOI] [PubMed] [Google Scholar]
- [33].Heeren J, Grewal T, Jackle S, Beisiegel U, Recycling of apolipoprotein E and lipoprotein lipase through endosomal compartments in vivo, J. Biol. Chem, 276 (2001) 42333–42338. [DOI] [PubMed] [Google Scholar]
- [34].Fischer AW, Schlein C, Cannon B, Heeren J, Nedergaard J, Intact innervation is essential for diet-induced recruitment of brown adipose tissue, Am. J. Physiol. Endocrinol. Metab, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fischer AW, Cannon B, Nedergaard J, Optimal housing temperatures for mice to mimic the thermal environment of humans: An experimental study, Mol. Metab, 7 (2018) 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fischer AW, Csikasz RI, von Essen G, Cannon B, Nedergaard J, No insulating effect of obesity, Am. J. Physiol. Endocrinol. Metab, 311 (2016) 202–213. [DOI] [PubMed] [Google Scholar]
- [37].Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW, Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase, Biochem. J, 402 (2007) 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ebeling P, Koistinen HA, Koivisto VA, Insulin-independent glucose transport regulates insulin sensitivity, FEBS Lett, 436 (1998) 301–303. [DOI] [PubMed] [Google Scholar]
- [39].Wang B, Zhang M, Ni YH, Liu F, Fan HQ, Fei L, Pan XQ, Guo M, Chen RH, Guo XR, Identification and characterization of NYGGF4, a novel gene containing a phosphotyrosine-binding (PTB) domain that stimulates 3T3-L1 preadipocytes proliferation, Gene, 379 (2006) 132–140. [DOI] [PubMed] [Google Scholar]
- [40].Wu WL, Gan WH, Tong ML, Li XL, Dai JZ, Zhang CM, Guo XR, Over-expression of NYGGF4 (PID1) inhibits glucose transport in skeletal myotubes by blocking the IRS1/PI3K/AKT insulin pathway, Mol. Genet. Metab, 102 (2011) 374–377. [DOI] [PubMed] [Google Scholar]
- [41].Gordts PL, Reekmans S, Lauwers A, Van Dongen A, Verbeek L, Roebroek AJ, Inactivation of the LRP1 intracellular NPxYxxL motif in LDLR-deficient mice enhances postprandial dyslipidemia and atherosclerosis, Arterioscler. Thromb. Vasc. Biol, 29 (2009) 1258–1264. [DOI] [PubMed] [Google Scholar]
- [42].Farfan P, Lee J, Larios J, Sotelo P, Bu G, Marzolo MP, A sorting nexin 17-binding domain within the LRP1 cytoplasmic tail mediates receptor recycling through the basolateral sorting endosome, Traffic, 14 (2013) 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jaldin-Fincati JR, Pavarotti M, Frendo-Cumbo S, Bilan PJ, Klip A, Update on GLUT4 Vesicle Traffic: A Cornerstone of Insulin Action, Trends Endocrinol. Metab, 28 (2017) 597–611. [DOI] [PubMed] [Google Scholar]
- [44].Seaman MN, McCaffery JM, Emr SD, A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast, J. Cell Biol, 142 (1998) 665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M, SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors, Nat. Cell Biol, 13 (2011) 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD, Endosomal receptor trafficking: Retromer and beyond, Traffic, 19 (2018) 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Donoso M, Cancino J, Lee J, van Kerkhof P, Retamal C, Bu G, Gonzalez A, Caceres A, Marzolo MP, Polarized traffic of LRP1 involves AP1B and SNX17 operating on Y-dependent sorting motifs in different pathways, Mol. Biol. Cell, 20 (2009) 481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hofmann SM, Zhou L, Perez-Tilve D, Greer T, Grant E, Wancata L, Thomas A, Pfluger PT, Basford JE, Gilham D, Herz J, Tschop MH, Hui DY, Adipocyte LDL receptor-related protein-1 expression modulates postprandial lipid transport and glucose homeostasis in mice, J. Clin. Invest, 117 (2007) 3271–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fox CS, Golden SH, Anderson C, Bray GA, Burke LE, de Boer IH, Deedwania P, Eckel RH, Ershow AG, Fradkin J, Inzucchi SE, Kosiborod M, Nelson RG, Patel MJ, Pignone M, Quinn L, Schauer PR, Selvin E, Vafiadis DK, American L Heart Association Diabetes Committee of the Council on, C.o.C.C.C.o.C. Cardiometabolic Health, C.o.C.S. Stroke Nursing, C.o.Q.o.C. Anesthesia, R. Outcomes, A. the American Diabetes, Update on Prevention of Cardiovascular Disease in Adults With Type 2 Diabetes Mellitus in Light of Recent Evidence: A Scientific Statement From the American Heart Association and the American Diabetes Association, Diabetes Care, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Prentki M, Nolan CJ, Islet beta cell failure in type 2 diabetes, J. Clin. Invest, 116 (2006) 1802–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sola D, Rossi L, Schianca GP, Maffioli P, Bigliocca M, Mella R, Corliano F, Fra GP, Bartoli E, Derosa G, Sulfonylureas and their use in clinical practice, Arch. Med. Sci, 11 (2015) 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.