Abstract
Next-generation sequencing (NGS) hereditary pan-cancer panel testing can identify somatic variants, which exhibit lower allele frequencies than do germline variants and may confound hereditary cancer predisposition testing. This analysis examined the prevalence and characteristics of likely-somatic variants among 348,543 individuals tested using a clinical NGS hereditary pan-cancer panel. Variants showing allele frequencies between 10–30% were interpreted as likely somatic and identified in 753 (0.22%) individuals. They were most frequent in TP53, CHEK2 and ATM, commonly as C-to-T transitions. Among individuals who carried a likely-somatic variant and reported no personal cancer history, 54.2% (78/144) carried a variant in TP53, CHEK2 or ATM. With a reported cancer history, this percentage increased to 81.1% (494/609), predominantly in CHEK2 and TP53. Their presence was associated with age (OR=3.1, 95% CI 2.5, 3.7; p<0.001) and personal history of cancer (OR=3.3, 95% CI 2.7, 4.0; p<0.001), particularly ovarian cancer. Germline ATM pathogenic variant carriers showed significant enrichment of likely-somatic variants (OR=2.8, 95% CI 1.6, 4.9; p=0.005), regardless of cancer status. The appearance of likely-somatic variants is consistent with clonal hematopoiesis, possibly influenced by cancer treatment. These findings highlight the precision required of diagnostic laboratories to deliver accurate germline testing results.
Keywords: Next-generation sequencing, hereditary pan-cancer panel testing, somatic variant, clonal hematopoiesis
INTRODUCTION
Mosaicism is defined as the presence of two or more cell populations with unique genotypes in one individual. Low-level mosaicism is seen frequently in normal tissues, and particularly in those with higher turnover rates.[1] One such tissue is bone marrow, which produces the peripheral white blood cells that comprise a common source of genomic DNA used in next-generation sequencing (NGS) hereditary pan-cancer panel testing.
NGS methods enable precise assessment of variant allele proportions and thus can reveal low-level mosaicism in an individual’s test sample.[1, 2] Many likely-mosaic variants identified in this manner reflect amplification of mutations acquired somatically in blood via clonal hematopoiesis of indeterminate potential (CHIP), which involves the expansion of clonal blood cell subpopulations without other hematologic disease.[3] Variants derived through CHIP are not thought to represent the germline genetic profile, hematologic malignancy, post-zygotic mosaicism, circulating tumor burden, or sequencing error. Previous studies have shown that CHIP rates increase with age and with exposure to cancer treatments such as chemotherapy and radiation (cytotoxic) therapy.[4–6] Several studies have associated CHIP with increased cardiovascular disease, all-cause mortality, and susceptibility to primary and secondary leukemias (i.e., therapy related leukemia).[3, 4, 7–9] Therefore, putative CHIP-derived somatic variants hold potential as independent clinical biomarkers, following clinical validation of methods for CHIP confirmation and variant classification.
Recent work demonstrated the identification of likely-somatic variants through NGS hereditary pan-cancer panel testing in a large clinical testing population.[10] The findings were consistent with previous CHIP studies, which showed likely-somatic variants occurring commonly in TP53 (MIM: 191170) and ATM (MIM: 607585) and having a higher prevalence among older individuals.[4, 7]
Having a large NGS hereditary pan-cancer testing population presented the unique opportunity to investigate the prevalence and characteristics of likely-somatic variants in cancer susceptibility genes. A better understanding of the etiology of putative CHIP-derived somatic variants is essential to improve test reporting, patient counseling, preventive screening and medical management. We hypothesized that cancer status (as a marker for likely cancer treatment) and/or germline mutation carrier status would contribute to the presence of likely-somatic variants. This is the first report to evaluate these factors from a hereditary cancer predisposition cohort. The analysis also examined the molecular characteristics of likely-somatic variants across the full testing population to identify trends that might provide further insight into their origins.
METHODS
Testing population
The full analysis set included 348,543 individuals who had NGS hereditary pan-cancer panel testing and results reported sequentially between September 2013 and July 2017. Testing was performed by Myriad Genetics Laboratories, Inc. (Salt Lake City, UT), a national Clinical Laboratory Improvement Amendments and College of American Pathology-certified facility. All individuals provided consent for clinical testing, and testing data were de-identified for analysis. The analysis set did not include individuals who met the following criteria: cancer history listed as unspecified or colorectal polyps only on the test request form; missing age at testing; and individuals from states with laws that prevent the use of de-identified genetic data. Individuals with known leukemias are not accepted for hereditary pan-cancer panel testing by the laboratory and therefore would not have been included in the analysis set.
Sequencing and variant classification
The NGS pan-cancer panel included 28 genes commonly associated with hereditary cancer predisposition: APC (MIM: 611731); ATM; BARD1 (MIM: 601593); BMPR1A (MIM: 602199); BRCA1 (MIM: 113705); BRCA2 (MIM: 600185); BRIP1 (MIM: 605882); CDH1 (MIM: 192090); CDK4 (MIM: 123829); CDKN2A (p16INK4a)(MIM: 600160); CDKN2A (p14ARF)(MIM: 600160); CHEK2 (MIM: 604373); EPCAM (MIM: 185535); GREM1 (MIM: 603054); MLH1 (MIM: 120436); MSH2 (MIM: 609309); MSH6 (MIM: 600678); MUTYH (MIM: 604933); NBN (MIM: 602667); PALB2 (MIM: 610355); PMS2 (MIM: 600259); POLD1 (MIM: 174761); POLE (MIM: 267800); PTEN (MIM: 601728); RAD51C (MIM: 602774); RAD51D (MIM: 602954); SMAD4 (MIM: 600993); STK11 (MIM: 602216); and TP53. All genes were available for the full time period except GREM1, POLD1, and POLE, which were added in July 2016. Testing was performed using blood or saliva samples. DNA extraction, testing methods, and variant identification have been described previously.[11, 12] The minimum depth of coverage for sequencing was 50X. For all tested genes except TP53, variants present with NGS allele frequencies of 30–70% were considered to be germline. Variants with allele frequencies between 10–30% were considered to be likely somatic. The laboratory does not maintain data on variants with allele frequencies lower than 10%. All somatic variants were confirmed, either by Sanger sequencing or by repeat NGS analysis, when the variant allele fraction was too low to be identified by Sanger sequencing (below approximately 20%).
Variant classification was consistent with guidelines from the American College of Medical Genetics and Genomics[13] and performed as described previously.[14] For the purposes of the analyses performed here, variants with a laboratory classification of Deleterious or Suspected Deleterious were considered to be Pathogenic Variants (PV). Variants with a laboratory classification of Polymorphism or Favor Polymorphism were considered to be Benign (clinically insignificant). Variants for which the clinical significance could not be determined were classified as Variants of Uncertain Significance (VUS).
The analysis included individuals who were found to carry a likely-somatic PV or VUS in a tested cancer susceptibility gene. Individuals with an apparent germline PV in TP53 (allele frequencies between 30–70%) were excluded, as it is difficult to determine whether TP53 variants are somatic or germline in origin without follow-up testing.[15] The analysis included somatic PV and VUS in order to examine both pathogenic and potentially-pathogenic variants across the testing population. This approach provided the most complete analysis set to support the characterization of likely-somatic variants.
Statistical Analyses
Multivariable logistic regression analysis was used to determine the risk of carrying a likely-somatic variant after adjusting for personal cancer history (affected or unaffected), age at testing (<50 versus ≥50 years), germline PV status (carrier or non-carrier), and clinical indication for testing. To determine the risk of carrying a likely-somatic variant, logistic regression analyses were performed for each tested gene separately in the subset of individuals who carried germline PVs, adjusting for personal cancer history, age at testing, and clinical indication for testing.
Chi-square tests were used to determine associations between affected status (breast/ovarian cancer vs unaffected) and type of likely-somatic variant (insertion/deletion, transversion, or transition). Odds ratios (OR), 95% confidence intervals (CI), and p-values are reported. Bonferroni multiple comparison p-value adjustments were made for the subset PV enrichment logistic regression analysis and for the breast or ovarian cancer versus variant type chi-square tests. All p-values of < 0.05 were considered statistically significant.
RESULTS
Prevalence of Likely-somatic Variants
Demographic characteristics of the testing population, including age at testing, personal cancer history and ancestry, are shown in Table 1. From NGS analysis of the hereditary pan-cancer gene panel, 0.22% of all tested individuals (753/348,853) were found to carry one or more likely-somatic PV or VUS. Most (97.2%; 732/753) carried only one. Among individuals with likely-somatic variants, 9.0% (68/753) also carried an apparent germline PV. A total of 775 likely-somatic variants (PV and VUS) were detected in these 753 individuals. Variants were seen most commonly in TP53 (28.9%; 224/775), CHEK2 (28.8%; 223/775), and ATM (20.9%; 162/775), with a steep drop-off in frequency of variants seen in all remaining panel genes (Table 2).
Table 1.
Demographic characteristics of the hereditary cancer genetic testing population.
| Likely-Somatic Variant Carriers (N = 753) |
Non-Likely- Somatic Variant Carriers (N = 347,790) |
TOTAL (N = 348,543) |
|
|---|---|---|---|
| Age at Testing | |||
| Mean (SD) | 62.6 (14.26) | 48.0 (13.64) | 48.0 (13.66) |
| Median (IQR) | 64 (54, 73) | 47 (38, 57) | 48 (38, 58) |
| Min, Max | 19, ≥90 | 11, ≥90 | 11, ≥90 |
| Age at Testing Category | |||
| <50 yrs. | 141 (18.7%) | 192604 (55.4%) | 192745 (55.3%) |
| ≥50 yrs. | 612 (81.3%) | 155186 (44.6%) | 155798 (44.7%) |
| Affected Status | |||
| Affected | 609 (80.9%) | 146269 (42.1%) | 146878 (42.1%) |
| Unaffected | 144 (19.1%) | 201521 (57.9%) | 201665 (57.9%) |
| Ancestry | |||
| White/Non-Hispanic | 445 (59.1%) | 169827 (48.8%) | 170272 (48.9%) |
| Black/African | 31 (4.1%) | 23695 (6.8%) | 23726 (6.8%) |
| Hispanic/Latino | 26 (3.5%) | 23336 (6.7%) | 23362 (6.7%) |
| Asian | 8 (1.1%) | 8349 (2.4%) | 8357 (2.4%) |
| Ashkenazi Jewish | 12 (1.6%) | 7256 (2.1%) | 7268 (2.1%) |
| Native American | 10 (1.3%) | 4571 (1.3%) | 4581 (1.3%) |
| Middle Eastern | 2 (0.3%) | 2378 (0.7%) | 2380 (0.7%) |
| Pacific Islander | 0 | 15 (<0.1%) | 15 (<0.1%) |
| Other | 8 (1.1%) | 2334 (0.7%) | 2342 (0.7%) |
| Multiple Ancestries | 47 (6.2%) | 25044 (7.2%) | 25091 (7.2%) |
| None Specified | 164 (21.8%) | 80985 (23.3%) | 81149 (23.3%) |
SD, standard deviation; IQR, interquartile range.
Table 2.
Gene distribution of likely-somatic variants (N=775).
| Gene | Frequency | Percent |
|---|---|---|
| TP53 | 224 | 28.9 |
| CHEK2 | 223 | 28.8 |
| ATM | 162 | 20.9 |
| BRCA2 | 23 | 3.0 |
| APC | 21 | 2.7 |
| BRCA1 | 12 | 1.5 |
| NBN | 11 | 1.4 |
| MLH1 | 10 | 1.3 |
| PMS2 | 10 | 1.3 |
| CDH1 | 9 | 1.2 |
| BRIP1 | 8 | 1.0 |
| PALB2 | 8 | 1.0 |
| MSH6 | 6 | 0.8 |
| BARD1 | 5 | 0.6 |
| PTEN | 5 | 0.6 |
| SMAD4 | 5 | 0.6 |
| CDK4 | 4 | 0.5 |
| RAD51D | 4 | 0.5 |
| STK11 | 4 | 0.5 |
| BMPR1A | 3 | 0.4 |
| CDKN2A (p14ARF) | 3 | 0.4 |
| MUTYH | 3 | 0.4 |
| POLD1 | 3 | 0.4 |
| RAD51C | 3 | 0.4 |
| CDKN2A (p16INK4a) | 2 | 0.3 |
| MSH2 | 2 | 0.3 |
| POLE | 2 | 0.3 |
| TOTAL | 775 | - |
Among individuals who reported no personal cancer history, 54.2% (78/144) carried a likely-somatic variant in one of these three genes. The cumulative frequency of likely-somatic variants increased to 81.1% (494/609) in individuals with a personal cancer history, most notably for CHEK2 and TP53. Likely-somatic variants in CHEK2 were seen in 16.7% (24/144) of individuals who had no cancer history and increased to 32.0% (195/609) among individuals with a cancer history. Similarly, the frequency of likely-somatic variants in TP53 was 18.8% (27/144) in individuals who had no cancer history, increasing to 30.2% (184/609) in individuals with a cancer history. For ATM, the frequency remained static at 18.8% (27/144) in individuals with no cancer history compared with 18.9% (115/609) in those who reported a personal cancer history.
Characteristics of Likely-somatic Variants and Association with Cancer
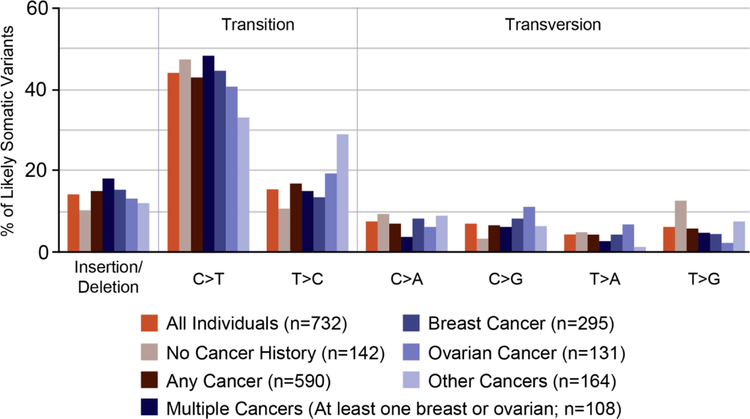
A total of 593 unique likely-somatic variants were identified. Substitutions comprised 85.2% (505/593) of unique variants, and insertions or deletions comprised 14.8% (88/593). C-to-T transition variants (C>T; reverse strand G>A) were the most common, accounting for 43.5% (337/775) of all variants identified (Figure 1). All likely-somatic variants identified in more than one individual (recurrent variants) are listed in Supplemental Table 1, with aggregate cancer type and age-at-diagnosis information for individuals with the variant who reported a personal history of cancer.
Figure 1.
Types of variants identified in individuals with or without personal history of cancer and a single likely-somatic variant.
Nucleotide substitution profiles for likely-somatic variants were examined to identify potential associations with clinical factors. Similar profiles were observed for all likely-somatic variants regardless of personal cancer history (Figure 1). Across all individuals, C>T/G>A transition variants were increased compared with all other variant types. There was no significant difference in the proportions of transition and transversion mutations based on reported breast and ovarian cancer histories. The numbers of individuals who had other types of cancers and carried likely-somatic variants were too small to analyze.
Of all unique variants identified, 11.5% (68/593) were observed in more than one individual. These variants were identified in TP53 (31), CHEK2 (24), ATM (11), CDKN2A (p14ARF) (1) and MLH1 (1) (Supplemental Table 1) and included five of the eight most common recurrent TP53 variants present in the International Agency for Research on Cancer (IARC) TP53 tumor database (c.524G>A [p.Arg175His]; c.742C>T [p.Arg248Trp]; c.743G>A [p.Arg248Gln]; c.818G>A [p.Arg273His]; and c.844C>T [p.Arg282Trp])(http://p53.iarc.fr/). These “hotspot” recurrent variants were detected most commonly in CpG dinucleotides, where deamination of 5-methylcytosine is known to lead to C>T transitions.[16] Recurrent variants accounted for 32.9% (241/732) of individuals carrying a single likely-somatic variant.
Age and Personal History of Cancer
Multivariable logistic regression analysis was performed to examine the relationships between carrying a likely-somatic variant and age or personal history of cancer (Table 3). Individuals who were 50 years of age or older at testing were found to be 3.1 times more likely to carry a likely-somatic variant compared with individuals under age 50 years (OR=3.1, 95% CI 2.5, 3.7; p<0.001).
Table 3.
Logistic regression analysis of relationships between clinical factors and risk of carrying a likely-somatic variant, adjusting for personal cancer history, age at testing, germline pathogenic variant carrier status, and clinical indication for testing. (All tested individuals; N=348,543.)
| Variable | Odds ratio | 95% confidence limits | p-value |
|---|---|---|---|
| Personal Cancer History (affected versus unaffected) | 3.3 | (2.7, 4.0) | <0.001 |
| Age at Testing (≥50 years versus <50 years) | 3.1 | (2.5, 3.7) A | <0.001 |
| Germline PV Carrier Status (positive versus negative/VUS) | 1.2 | (0.9, 1.5) | 0.203 |
PV, Pathogenic variant; VUS, Variant of uncertain significance.
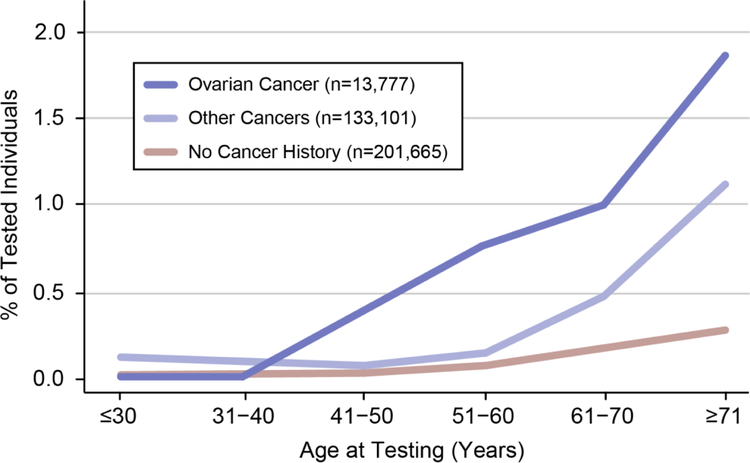
A personal history of cancer was reported for 42.1% (146,878/348,543) of individuals across the entire testing population. Among individuals with one or more likely-somatic variants, a personal cancer history was reported for 80.9% (609/753). Individuals who had a personal cancer history were 3.3 times more likely to have a likely-somatic variant compared to those with no personal cancer history (OR=3.3, 95% CI 2.7, 4.0; p<0.001) (Table 3). This trend was most prominent in individuals with a reported personal history of ovarian cancer (1.0%; 135 with likely-somatic variants/13,777 total) as compared with individuals reporting no personal cancer history (0.1%; 144 with likely-somatic variants/201,665 total). These individuals also showed the greatest increase in the frequency of likely-somatic variants at older ages (Figure 2). The frequency of likely-somatic variants in individuals with a reported personal history of ovarian cancer increased from 1.0% overall to 1.8% (61/3330) among individuals who were older than 71 years when tested.
Figure 2.
Percentage of individuals with a likely-somatic variant as a function of age and personal cancer history.
Germline PV Carrier Status
The testing population included 22,962 individuals who carried a germline PV in a cancer susceptibility gene. Logistic regression analysis showed no significant relationship between germline PV carrier status and increased likelihood of carrying a concomitant likely-somatic variant (OR=1.2, 95% CI 0.9, 1.5; p=0.203) (Table 3).
To examine the relationship between germline PV carrier status and risk of carrying a likely-somatic variant in any one gene in the panel, subset analyses were performed for each gene separately. After adjusting for age at testing, personal cancer history, and clinical indication for testing, there were no significant findings associated with germline PVs in individual genes, with the exception of ATM. For individuals who had a single germline PV in ATM (211), analysis showed enrichment of likely-somatic variants compared with single non-ATM PV carriers (521) (OR=2.8, 95% CI 1.6, 4.9, p=0.005). Interestingly, in 12 of 16 individuals who carried an apparent germline PV in ATM, the identified somatic variant also occurred in ATM. The remaining four individuals carried a somatic variant in BRCA1 (1), BRCA2 (1), CHEK2 (1) and PALB2 (1). Most individuals (75.0%; 9/12) who had a germline PV and a somatic PV or VUS in ATM had a personal history of cancer.
DISCUSSION
These analyses detail the frequency and characteristics of likely-somatic variants identified within the largest hereditary pan-cancer panel testing population described to date. Collectively, likely-somatic variants were rare (0.22%). Their presence in this testing population is consistent with CHIP, given the significant associations with advanced age and with a personal history of cancer.[4, 7, 17] In addition, they were enriched in genes associated previously with CHIP (ATM, CHEK2, and TP53).[4, 7, 17] Furthermore, C>T/G>A transition variants were notably more frequent than were other variant types across the testing population, consistent with observations made in studies of somatic variants and age in human cancers.[6, 18] The ubiquitous nature of CHIP makes this a plausible mechanism for most likely-somatic variants.[4, 7] A circulating tumor burden with allele frequencies between 10–30% is a possible explanation; however, overt hematologic malignancies, when noted, were not accepted by the laboratory and therefore should not confound the analysis. As the likely-somatic variants increased with age, multi-germ layer mosaicism would be a less probable explanation for most observed variants.
The presence of likely-somatic variants was significantly associated with a personal history of cancer, possibly related to the effects of cytotoxic treatment.[5, 6] The most prominent trend was seen with ovarian cancer, possibly due to the duration, amount, and/or type of chemotherapy used to treat ovarian cancer. Most individuals who have ovarian cancer undergo chemotherapy treatment, which usually involves platinum-containing compounds.[19] Therefore, of all individuals who had a reported cancer history, those with ovarian cancer might be most representative of a chemotherapy-treated population.
In this testing population, the proportion of individuals with likely-somatic TP53 and CHEK2 variants, but not ATM variants, increased among individuals with a personal history of cancer. Hematopoietic cells with a single TP53 PV may selectively expand with exposure to cytotoxic therapy.[20] Recent analyses of somatic variants in individuals who had previous cancer treatment have produced varied results with regard to TP53. [5, 6] Coombs et al. (2017) observed significant association of CHIP-related somatic TP53 mutations with both chemotherapy and radiation treatment. While Swisher et al. (2016) identified somatic TP53 mutations in individuals who received chemotherapy treatment for ovarian cancer, the association was not significant. Comparison among the current work and these recent reports is limited by differences in analysis population, cohort size, and available clinical information. The observed increase in CHEK2 somatic variants among individuals with a personal history of cancer, similar to the increase observed for TP53, suggests that hematopoietic cell sub-clones harboring CHEK2 PVs also may expand preferentially following exposure to cytotoxic therapy. The lack of a similar increase in likely-somatic ATM variants among individuals with a personal history of cancer indicates that hematopoietic cell sub-clones with ATM variants do not expand after cytotoxic therapy. These observations are consistent with the idea that haploinsufficiency of CHEK2 and TP53, but not of ATM, drives amplification of hematopoietic cell sub-clones in individuals who have had cancer.
Unlike other studies to date, this analysis specifically examined germline hereditary cancer gene mutation status relative to the appearance of likely-somatic variants. While germline PV carrier status overall showed no significantly increased risk of carrying a likely-somatic variant, subset analysis revealed a significant association for ATM heterozygous germline PV carriers, independent of personal cancer history. It has been hypothesized that ATM germline PV carriers are more susceptible to developing clonal hematopoiesis.[21] ATM regulates the DNA damage response to double-strand DNA breaks through its kinase activity, in a pathway that also involves TP53 and CHEK2.[22] Heterozygous germline ATM variants are enriched in women with breast cancer (1–2%)[12, 23–25] as well as in pancreatic cancer cohorts (1.2–4.1%).[26–29] As ATM germline PV carriers appear common in the population (~0.4% of non-Finnish Europeans) [23] a better understanding of the link between clonal hematopoiesis and ATM germline PV carrier status is warranted. Notably, ATM germline PV carriers frequently had a likely-somatic variant in ATM. A recent report showed similar association of inherited ATM variants with acquired loss of heterozygosity at the same locus in individuals found to have mosaic chromosomal alterations presumed to arise from clonal hematopoiesis.[30] Disruption of both ATM alleles in some portion of an individual’s blood might underpin development of chronic lymphocytic leukemia (CLL), as bi-allelic ATM inactivation is frequent in CLL.[28]
The strength of this study is the large, clinical-grade data set, which provides a rare opportunity to power statistical analyses of factors associated with likely-somatic variants. At the same time, analysis of de-identified NGS clinical testing population comes with inherent limitations. First, NGS allele frequencies suggest, but cannot confirm, CHIP as the mechanistic origin of most likely-somatic variants observed in this population. Ruling out constitutional or multi-germ layer mosaicism would require follow-up testing with additional tissues and/or family members [15], which was not possible. Second, clinical treatment details for individuals with a reported personal cancer history were unavailable, limiting the evaluation of relationships between cytotoxic therapy and the appearance of likely-somatic variants. Third, as the test was optimized to detect germline PVs, variants present at allele frequencies below 10% were not evaluated and thus were absent from reported totals. While germline testing is not optimized for somatic variant analysis, this testing pipeline provided a large and well-powered cohort in which to analyze prevalence and characteristics of likely-somatic variants, though with a variant allele frequency threshold greater than that used in other studies. Examining variants present at allele frequencies below 10% will bring needed dimension to future analyses. Finally, the analysis did not include common CHIP-associated genes, such as DNMT3A, TET2, and ASXL1, as they were not part of the clinical testing panel used here.[4, 7, 17] Even with these limitations, this analysis depicts a uniquely well-powered, real-world hereditary cancer risk testing scenario, further underscoring the value of laboratory precision and accurate result interpretation and reporting. Follow-up studies that include detailed cancer treatment history, variants with lower allele frequencies, and additional CHIP-associated genes will add needed dimension to the exploration of somatic variant origins.
In summary, NGS-based panel testing for hereditary cancer predisposition can identify and characterize somatic variants more readily than could previous testing methods. This work helps to highlight how frequently somatic variants can arise, likely from CHIP, with normal aging and with cancer. Several identified likely-somatic hotspot variants, particularly in ATM, CHEK2, and TP53, represent excellent candidates for future CHIP study. Somatic variant findings can meaningfully impact test result interpretation, as they may put patients at risk for CHIP-related morbidities such as primary and secondary leukemias, cardiovascular disease, and decreased overall survival.[4, 7, 17] Large, long-term follow-up studies are needed to better understand how somatic variants affect this spectrum of outcomes. Further, differentiating between germline and somatic acquisition of PVs in cancer-risk genes can have profound medical management implications for patients and their family members. Assay and data analysis customization, combined with follow-up confirmation using additional tissues and family member testing, is required to accurately interpret hereditary cancer predisposition test results. These findings highlight the investment that diagnostic laboratories must make to deliver a clinically accurate result.
Supplementary Material
Highlights.
NGS hereditary cancer genetic testing also detects somatic variants, which can arise via clonal hematopoiesis, and which impact cancer risk differently than do inherited variants.
This report describes the prevalence of somatic variants in a large clinical hereditary cancer risk testing population and spotlights genes that were most prone to harboring somatic variants in individuals with or without a personal history of cancer.
These findings highlight the importance of delivering clinically accurate NGS hereditary genetic testing results to healthcare providers and patients, along with the precision that is required to ensure accuracy.
ACKNOWLEDGMENTS
The authors thank Krystal Brown, PhD, an employee of Myriad Genetic Laboratories, Inc., for editorial assistance with the manuscript.
Funding: This work was supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) (grant numbers P30CA33572 and K08CA234394 [TJS]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Other sources of support include the Morris and Horowitz Families Endowed Professorship (SLN) and the Dr. Norman & Melinda Payson Professorship in Medical Oncology (JW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
BC, RB, JL, HCC, and DMD are employees of Myriad Genetic Laboratories, Inc., and receive salary and stock options as compensation. TPS, GM, JW, and SLN have no competing interests to disclose.
REFERENCES
- [1].Acuna-Hidalgo R, Sengul H, Steehouwer M, van de Vorst M, Vermeulen SH, Kiemeney LALM, Veltman JA, Gilissen C, Hoischen A. Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life. The American Journal of Human Genetics 2017;101:50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Slavin TP, Niell-Swiller M, Solomon I, Nehoray B, Rybak C, Blazer KR, Weitzel JN. Clinical Application of Multigene Panels: Challenges of Next-Generation Counseling and Cancer Risk Management. Front Oncol 2015;5:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Swisher EM, Harrell MI, Norquist BM, Walsh T, Brady M, Lee M, Hershberg R, Kalli KR, Lankes H, Konnick EQ, Pritchard CC, Monk BJ, Chan JK, Burger R, Kaufmann SH, Birrer MJ. Somatic Mosaic Mutations in PPM1D and TP53 in the Blood of Women With Ovarian Carcinoma. JAMA Oncol 2016;2:370–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, Arcila ME, Ladanyi M, Tallman MS, Levine RL, Berger MF. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell stem cell 2017;21:374–82.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landen M, Hoglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Gronberg H, Hultman CM, McCarroll SA. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, Balasis ME, Mesa TE, Sallman DA, Lancet JE, Komrokji RS, List AF, McLeod HL, Alsina M, Baz R, Shain KH, Rollison DE, Padron E. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. The lancet oncology 2017;18:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Takahashi K, Wang F, Kantarjian H, Doss D, Khanna K, Thompson E, Zhao L, Patel K, Neelapu S, Gumbs C, Bueso-Ramos C, DiNardo CD, Colla S, Ravandi F, Zhang J, Huang X, Wu X, Samaniego F, Garcia-Manero G, Futreal PA. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. The lancet oncology 2017;18:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Coffee B, Cox HC, Kidd J, Sizemore S, Brown K, Manley S, Mancini-DiNardo D. Detection of somatic variants in peripheral blood lymphocytes using a next generation sequencing multigene pan cancer panel. Cancer Genet 2017;211:5–8. [DOI] [PubMed] [Google Scholar]
- [11].Judkins T, Leclair B, Bowles K, Gutin N, Trost J, McCulloch J, Bhatnagar S, Murray A, Craft J, Wardell B. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC cancer 2015;15:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, Hartman AR, Winer EP, Garber JE. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2016;34:1460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine: official journal of the American College of Medical Genetics 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Eggington JM, Bowles KR, Moyes K, Manley S, Esterling L, Sizemore S, Rosenthal E, Theisen A, Saam J, Arnell C, Pruss D, Bennett J, Burbidge LA, Roa B, Wenstrup RJ. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clinical genetics 2014;86:229–37. [DOI] [PubMed] [Google Scholar]
- [15].Weitzel JN, Chao EC, Nehoray B, Van Tongeren LR, LaDuca H, Blazer KR, Slavin T, Facmg D, Pesaran T, Rybak C, Solomon I, Niell-Swiller M, Dolinsky JS, Castillo D, Elliott A, Gau CL, Speare V, Jasperson K. Somatic TP53 variants frequently confound germ-line testing results. Genetics in medicine: official journal of the American College of Medical Genetics 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Green PM, Montandon AJ, Bentley DR, Ljung R, Nilsson IM, Giannelli F. The incidence and distribution of CpG-TpG transitions in the coagulation factor IX gene. A fresh look at CpG mutational hotspots. Nucleic Acids Res 1990;18:3227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Della Pepa C, Tonini G, Pisano C, Di Napoli M, Cecere SC, Tambaro R, Facchini G, Pignata S. Ovarian cancer standard of care: are there real alternatives? Chin J Cancer 2015;34:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, Heath S, Baty JD, Klco JM, Ding L, Mardis ER, Westervelt P, DiPersio JF, Walter MJ, Graubert TA, Ley TJ, Druley T, Link DC, Wilson RK. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hinds DA, Barnholt KE, Mesa RA, Kiefer AK, Do CB, Eriksson N, Mountain JL, Francke U, Tung JY, Nguyen H, Zhang H, Gojenola L, Zehnder JL, Gotlib J. Germline variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shiloh Y ATM and related protein kinases: safeguarding genome integrity. Nature reviews Cancer 2003;3:155–68. [DOI] [PubMed] [Google Scholar]
- [23].Slavin TP, Maxwell KN, Lilyquist J, Vijai J, Neuhausen SL, Hart SN, Ravichandran V, Thomas T, Maria A, Villano D, Schrader KA, Moore R, Hu C, Wubbenhorst B, Wenz BM, D’Andrea K, Robson ME, Peterlongo P, Bonanni B, Ford JM, Garber JE, Domchek SM, Szabo C, Offit K, Nathanson KL, Weitzel JN, Couch FJ. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 2017;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cybulski C, Lubinski J, Wokolorczyk D, Kuzniak W, Kashyap A, Sopik V, Huzarski T, Gronwald J, Byrski T, Szwiec M, Jakubowska A, Gorski B, Debniak T, Narod SA, Akbari MR. Mutations predisposing to breast cancer in 12 candidate genes in breast cancer patients from Poland. Clinical genetics 2015;88:366–70. [DOI] [PubMed] [Google Scholar]
- [25].Li J, Meeks H, Feng BJ, Healey S, Thorne H, Makunin I, Ellis J, Campbell I, Southey M, Mitchell G, Clouston D, Kirk J, Goldgar D, Chenevix-Trench G. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. Journal of medical genetics 2016;53:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, Siddiqui A, Witmer PD, Tamura K, Song TJ, Almario JAN, Brant A, Borges M, Ford M, Barkley T, He J, Weiss MJ, Wolfgang CL, Roberts NJ, Hruban RH, Klein AP, Goggins M. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol;0:JCO.201772.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hu C, Hart SN, Bamlet WR, Moore RM, Nandakumar K, Eckloff BW, Lee YK, Petersen GM, McWilliams RR, Couch FJ. Prevalence of Pathogenic Mutations in Cancer Predisposition Genes among Pancreatic Cancer Patients. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2016;25:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tiao G, Improgo MR, Kasar S, Poh W, Kamburov A, Landau DA, Tausch E, Taylor-Weiner A, Cibulskis C, Bahl S, Fernandes SM, Hoang K, Rheinbay E, Kim HT, Bahlo J, Robrecht S, Fischer K, Hallek M, Gabriel S, Lander ES, Stilgenbauer S, Wu CJ, Kiezun A, Getz G, Brown JR. Rare germline variants in ATM are associated with chronic lymphocytic leukemia. Leukemia 2017;31:2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shieh A, Mohamed AA. A Case of Therapy-Related Acute Myeloid Leukemia in a Patient With Heterozygous Mutations in the Ataxia Telangiectasia Mutated Gene. Journal of Hematology; Vol 6, No 4, October 2017. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, Birmann BM, Talkowski ME, Bakhoum SF, McCarroll SA, Price AL. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018;559:350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.