Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the progressive loss of motor neurons, for which there is no effective treatment. Previously, we generated a Caenorhabditis elegans model of ALS, in which the expression of dnc-1, the homologous gene of human dynactin-1, is knocked down (KD) specifically in motor neurons. This dnc-1 KD model showed progressive motor defects together with axonal and neuronal degeneration, as observed in ALS patients. In the present study, we established a behavior-based, automated, and quantitative drug screening system using this dnc-1 KD model together with Multi-Worm Tracker (MWT), and tested whether 38 candidate neuroprotective compounds could improve the mobility of the dnc-1 KD animals. We found that 12 compounds, including riluzole, which is an approved medication for ALS patients, ameliorated the phenotype of the dnc-1 KD animals. Nifedipine, a calcium channel blocker, most robustly ameliorated the motor deficits as well as axonal degeneration of dnc-1 KD animals. Nifedipine also ameliorated the motor defects of other motor neuronal degeneration models of C. elegans, including dnc-1 mutants and human TAR DNA-binding protein of 43 kDa overexpressing worms. Our results indicate that dnc-1 KD in C. elegans is a useful model for the screening of drugs against motor neuron degeneration, and that MWT is a powerful tool for the behavior-based screening of drugs.
Subject terms: Cell death in the nervous system, Amyotrophic lateral sclerosis
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the progressive loss of motor neurons. Approximately 5–10% of ALS cases are familial, whereas approximately 90% are sporadic (SALS). To understand the pathological mechanisms of SALS and to develop effective drugs, we previously characterized the motor neuron-specific gene expression profile of SALS patients1,2 Among the dysregulated genes, dynactin-1was markedly downregulated in motor neurons from an early disease stage. Dynactin-1 is a crucial component of dynactin, which is a multiprotein complex associated with dynein3, the molecular motor for retrograde transport4. Interestingly, missense mutations in dynactin-1 are linked to familial lower motor neuron disease5, or Perry syndrome, a familial type of Parkinson disease involving TAR DNA-binding protein of 43 kDa (TDP-43) aggregation6. Recently, it has been demonstrated that Perry syndrome patients have diverse symptoms similar to those observed in Parkinson’s disease and a type of motor neuron disease that involves frontotemporal degeneration7. On the other hand, it was also reported that the loss of TDP-43 impaired the fusion of autophagosomes with lysosomes through the downregulation of dynactin-1, leading to the accumulation of immature autophagic vesicles8. These reports indicate the bidirectional regulation between TDP-43 pathology and dynactin-1 downregulation, which is a good candidate to explain the pathogenesis of SALS.
To analyze the effect of dynactin-1 downregulation in motor neuron degeneration, we generated a Caenorhabditis elegans (C. elegans) model of motor neuron disease, in which the expression of dnc-1, the homologous gene of human dynactin-1, is knocked down specifically in motor neurons9. This model shows progressive motor defects together with axonal and neuronal degeneration. Pathologically, we also observed axonal spheroids, degenerated mitochondria, ubiquitin-positive inclusions, and an increased number of autophagosomes in degenerated neurons, reflecting the pathology of SALS motor neurons.
In the present study, we used our new C. elegans model and established a behavioral screening assay using an automated tracking system, the Multi-Worm Tracker (MWT)10. Using this assay, we assessed 38 compounds from 5 categories: (1) approved by the FDA for ALS treatment, (2) approved by the FDA for other treatments and under or past clinical trials for ALS, (3) approved by the FDA for other treatments and found to have promising effects on ALS models, (4) autophagy activators based on our previous finding that rapamycin, an autophagy activator, improves the motor defects of a dnc-1 KD model9, and (5) histone deacetylase (HDAC) inhibitors based on our previous finding that trichostatin A, an HDAC inhibitor, improves the motor deficits of the dnc-1 KD model by activating tubulin acetylation and enhancing axonal transport9. Of the 38 compounds, we found that 12 compounds, including riluzole, the drug internationally approved for ALS, improved the motor deficits of dnc-1 KD animals. Among those 12 compounds, we found that nifedipine, a calcium-channel blocker, was the most effective drug for improving the dnc-1 KD motor defect. Follow-up experiments showed that nifedipine also ameliorated the neurodegeneration of motor neurons in dnc-1 KD animals.
Results
Adapting MWT for the screening of pharmacological effects on C. elegans locomotion
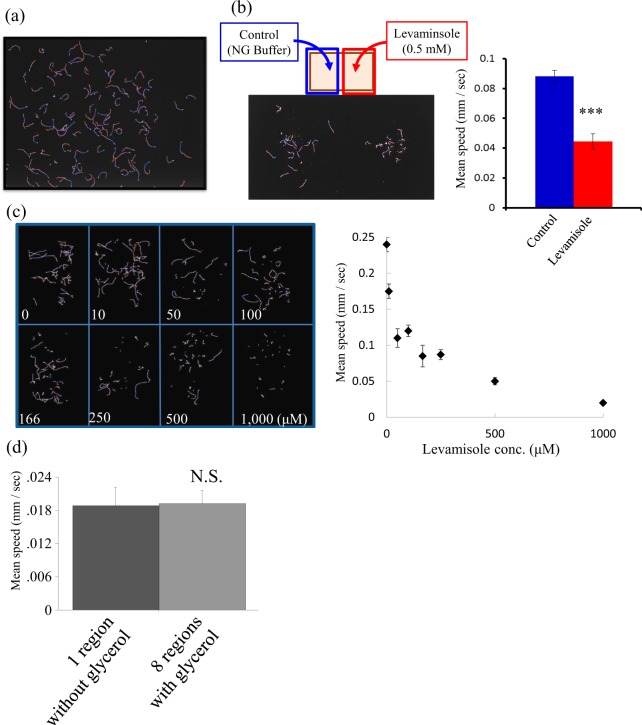
MWT was designed to quantify the behavior of many worms simultaneously on a 5-cm diameter Petri dish10. For the purpose of drug screening, we adapted the MWT to record a larger field of view using a higher resolution camera (12 Mpixels) and a larger rectangular assay plate (4 cm × 10 cm × 1.45 cm). This modified system enables us to analyze more animals at once (~500 animals) without increasing the population density (Fig. 1a). Thus, the assay plate can be divided into regions to test multiple treatments at once (i.e., Fig. 1b, 2 regions of ~250 worms each; Fig. 1c, 8 regions of ~40 worms each). Regions were separated by glycerol because worms avoid the high osmolarity of glycerol and will not cross between regions11. We confirmed that the separation into 8 compartments by glycerol did not change the moving speed of the animals compared to that without separation (Fig. 1d).
Figure 1.
Optimization of the drug-screening system using Multi-Worm Tracker (MWT) (a) A representative trajectory of an N2 worm for 30 sec. A 13 × 10 cm agar-filled plate was used for the assay, and images were captured using Toshiba-Teli Ultra-High-resolution 12 M pixel CMOS sensor camera-link camera. The locomotion of as much as about 500 worms can be detected and analyzed simultaneously. (b) Performance of the MWT assay for detection of levamisole treatment. Control worms treated with NG buffer only were placed on the left side of the assay plate (blue line, n = 18) and levamisole (0.5 mM) on the right side (red line n = 20). The average moving speed of each group was analyzed and quantified using Choreography. (c) The WMT assay is divided into 8 groups. To analyze multiple groups of worms simultaneously, assay plates were divided into 8 regions by glycerol. A representative trajectory of worms treated by different concentrations of levamisole is shown. The average moving speeds analyzed by Choreography are shown on the right side. (d) A comparison of the locomotion speed of the worms with and without separation into regions by glycerol The statistical analyses in b and d were performed using the Student t-test (***p < 0.0001).
We validated the effectiveness of the adapted system by analyzing whether this system could detect drug-induced motor defects. For this purpose, we used levamisole, a nicotinic acetylcholine receptor agonist, which causes muscle paralysis in C. elegans, and quantified the locomotor speed of levamisole-treated animals. We detected a dose-dependent, unequivocal motor defect caused by levamisole (Fig. 1b,c), showing that the system developed in this study was effective as a behavior-based drug-screening system.
MWT efficiently detected the motor defect in dnc-1 KD worms
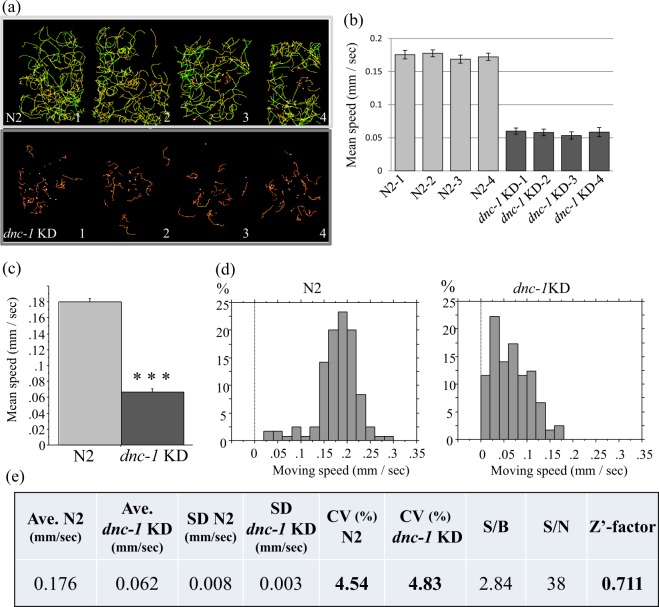
Previously, we analyzed the motor function of dnc-1 KD worms by the bending assay and thrashing assay, in which we manually scored the number of body bends of worms on an agar plate or in liquid, respectively. dnc-1 KD worms showed a 60–70% reduction in mobility compared with wild-type worms9.
We analyzed the effects of dnc-1 KD on locomotion using MWT (Fig. 2a–d). Consistent with the previous assays, the speed of dnc-1 KD worms was reduced by 70%. The variations in speed within a region and between regions were very small (Fig. 2c), suggesting that each region can be used to test a different treatment during a drug screen. Additionally, in contrast to the several hours it took for screening in previous methods, conducting the MWT assay and analyzing the results only took 15 min, thereby making it more feasible to conduct a screen to find drugs that improve the dnc-1 KD motor defect (Fig. 2e).
Figure 2.
Evaluation of dnc-1 KD worms as a model for drug screening by MWT. (a) Representative trajectories of N2 (upper four groups) and dnc-1 KD worms (lower four groups) analyzed in the same assay plate by WMT. Moving speeds are depicted as a linear rainbow-color gradient. (b) The average moving speeds of each region (N2-1 to 4: light gray bars; dnc-1 KD-1 to 4: dark gray bars). (c) The average moving speed of N2 and dnc-1 KD worms (N2, n = 120; dnc-1 KD worms, n = 121). (d) Representative histogram of moving speeds of N2 and dnc-1 KD worms. (e) Statistical analysis and evaluation of dnc-1 KD worms for the drug screening by MWT. The Z’-factor is defined as: Z’-factor = 1–3(SD_N2 + SD_dnc-1 KD)/(Ave._N2-Ave._dnc-1KD). The statistical analysis in C was performed using the Student t-test (***p < 0.0001).
Autophagy activation attenuated the motor defect of dnc-1 KD worms
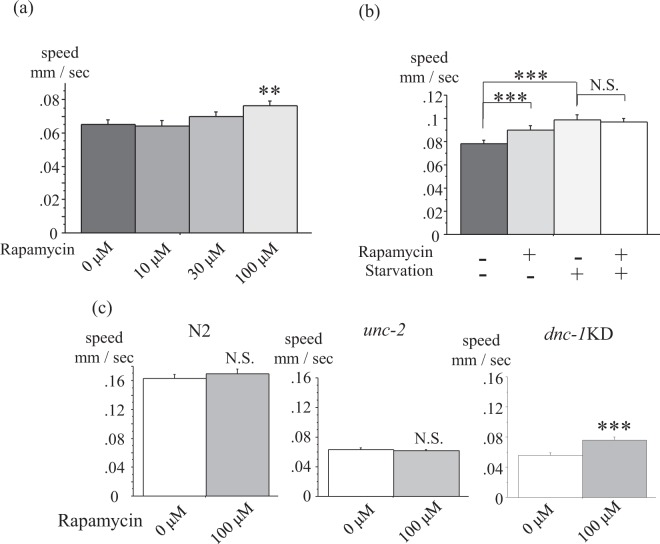
Autophagy activation by rapamycin or food restriction (starvation) ameliorates the motor defect of dnc-1 KD worms by attenuating axonal degeneration9. We hence tested whether MWT could detect the effect of rapamycin and starvation in dnc-1 KD worms. We confirmed the dose-dependent effect of rapamycin (Fig. 3a), and also showed that starvation had a similar effect to rapamycin without synergy (Fig. 3b).
Figure 3.
MWT detected the significant and specific effect of rapamycin on locomotion in dnc-1 KD worms. (a) Dose-dependent effects of rapamycin on locomotion in dnc-1 KD worms (n = 166, 120, 179, and 164 for rapamycin 0, 10, 30, and 100 μM, respectively). (b) Effects of single or combination therapy of rapamycin and starvation to dnc-1 KD worms (n = 248, 125, 67, and 70 for control, rapamycin only, starvation only, and a combination of both therapies, respectively). (c) The specificity of rapamycin to dnc-1 KD worms. No significant effect of rapamycin was observed in N2 and unc-2 mutant worms (n = 46 and 30 for control and rapamycin to N2, respectively; n = 20 and 35 for control and rapamycin to unc-2, respectively; and n = 124 and 93 for control and rapamycin to dnc-1 KD worms, respectively). Statistical analyses were performed by one-way ANOVA followed by the Bonferroni/Dunn post hoc test (a,b) and the Student t-test (c) (**p < 0.001 and ***p < 0.0001). Error bars indicate the S.E.M.
To verify the specificity of the effect of autophagy activation, we compared the effect of rapamycin between wild-type, unc-2 mutants, and dnc-1 KD worms (Fig. 3c). unc-2 encodes a C. elegans homologue of a voltage-gated calcium channel and its mutation causes a severe motor defect12. Although the dnc-1 KD worms were markedly affected by rapamycin, wild-type and unc-2 mutant worms did not show a significant change by rapamycin, indicating the specific effect of rapamycin on dnc-1 KD worms (Fig. 3c). Thus, we used rapamycin as a positive control for our drug screen to find drugs that improve the motor defects of dnc-1 KD worms.
Blinded screening of 38 compounds by MWT found that riluzole, an ALS drug, improved the motor defect of dnc-1 KD worms
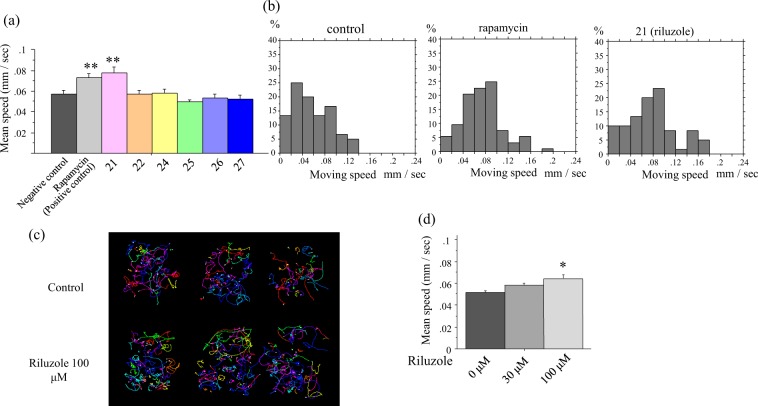
Using the automated behavior-based drug-screening system, we assessed in a blinded manner whether any of the 38 compounds, which have been reported or were expected to have a neuroprotective effect, could improve the motor defects of dnc-1 KD worms. Rapamycin and dimethyl sulfoxide (DMSO) were included as positive and negative controls, respectively. The results of the screening are listed in Table 1. We found that 12 compounds were as effective as or more effective than rapamycin. Among these 12 compounds, riluzole, the drug internationally approved for ALS, improved the locomotion speed of dnc-1 KD worms (Fig. 4). In Fig. 4a, we present a representative result of an assay, showing that among six compounds, only no. 21 (riluzole) ameliorated the speed of dnc-1 KD worms. The histograms of rapamycin-treated and riluzole-treated worms show a right shift compared with the negative control, demonstrating that these treatments improved the speed of dnc-1 KD worms (Fig. 4b). The tracks of control and riluzole-treated dnc-1 KD worms during the assay are also shown in Fig. 4c. We also found that the effect of riluzole is dose-dependent (Fig. 4d). Given that riluzole is an approved medication for ALS patients and was identified using the MWT automated behavior-based drug screen for dnc-1 KD worms, other compounds identified by the screen are potential candidates that may be useful for the treatment for ALS.
Table 1.
Result of drug screening by Multi Worm Tracker.
| Compound name | Blind number | Compound source | Mechanism of action | Highest phase for ALS treatment | MWT results (ratio to non treated dnc-1 KD worms) | |
|---|---|---|---|---|---|---|
| 1 | Nifedipine | 15 | FUJIFILM Wako Pure Chemical Corporation | Calcium channel blocker | Approved for other diseases | 1.69 |
| 2 | Masitinib | 14 | AK Scientific | Tyrosine kinase inhibitor | P-III | 1.34 |
| 3 | WN1316 | 2 | Synthetic sample in lab | NAIP enhancer | — | 1.32 |
| 4 | BYL-719 | 28 | MedChemExpress | PI3Kα inhibitor | P-III in other diseases | 1.32 |
| 5 | Riluzole | 21 | LKT Labs | Glutamate release inhibitor | Approved | 1.32 |
| 6 | Tubastatin A | 30 | MedChemExpress | HDAC6 inhibitor | — | 1.29 |
| 7 | GSK-2606414 | 5 | MedChemExpress | PERK inhibitor | Preclinical study | 1.29 |
| 8 | Retinoic acid | 16 | FUJIFILM Wako Pure Chemical Corporation | — | Preclinical study | 1.29 |
| 9 | VORINOSTAT | 29 | MedChemExpress | HDAC Inhibitor (HDACs 1, 2, 3, 6) | — | 1.29 |
| 10 | SA-4503 | 10 | MedChemExpress | σ1 receptor agonist | Clinical trial | 1.24 |
| 11 | CK-2017357 | 13 | ChemShuttle | Troponin activator | Preclinical study | 1.24 |
| 12 | Rasagiline | 7 | Sigma-Aldrich | MAO-B Inhibitor | Clinical trial | 1.23 |
| 13 | Rapamycin | Positive control | Tokyo Chemical Industry | mTOR inhibitor | Preclinical study | 1.22 |
| 14 | AICAR | 34 | Tokyo Chemical Industry | AMPK activator | — | 1.19 |
| 15 | Bromhexine | 17 | FUJIFILM Wako Pure Chemical Corporation | — | Preclinical study | 1.17 |
| 16 | — | 3 | Synthetic sample in lab | EPHA4 inhibitor | Preclinical study | 1.15 |
| 17 | ACY-1215 | 31 | MedChemExpress | HDAC6 inhibitor | — | 1.14 |
| 18 | METFORMIN | 33 | FUJIFILM Wako Pure Chemical Corporation | AMPK activator | — | 1.12 |
| 19 | AVex-73 | 20 | Synthetic sample in lab | Acts on M1 muscarinic sodium channels Receptor Agonist | Preclinical study | 1.12 |
| 20 | Tamoxifen | 33 | FUJIFILM Wako Pure Chemical Corporation | Estrogen receptor modulator | — | 1.09 |
| 21 | D-(+)-Trehalose | 36 | Tokyo Chemical Industry | Autophagy modulator | — | 1.09 |
| 22 | Methylcobalamin | 23 | FUJIFILM Wako Pure Chemical Corporation | — | Clinical trial | 1.07 |
| 23 | SB-431542 | 6 | MedChemExpress | ALK5 Inhibitor | Preclinical study | 1.07 |
| 24 | Dasatinib | 11 | MedChemExpress | Bcr-Abl inhibitor, tyrosine kinase inhibitor | Preclinical study | 1.06 |
| 25 | — | 4 | Synthetic sample in lab | EPHA4 inhibitor | Preclinical study | 1.05 |
| 26 | Kenpaullone | 1 | LKT Labs | GSK-3β inhibitor CDK inhibitor | Preclinical study | 1.03 |
| 27 | — | 19 | Synthetic sample in lab | mSOD1 aggregation inhibitor | Preclinical study | 1.01 |
| 28 | Fluoxetine | 38 | Tokyo Chemical Industry | Serotonin transporter inhibitor | — | 1.00 |
| 29 | SPAUTIN-1 | 35 | Sigma-Aldrich | Autophagy modulator | — | 0.98 |
| 30 | Edaravone | 22 | Tokyo Chemical Industry | Free radical scavenger | Approved | 0.98 |
| 31 | — | 18 | Synthetic sample in lab | P2X7 antagonist | Preclinical study | 0.97 |
| 32 | Pyrimethamine | 9 | Sigma-Aldrich | Dihydrofolate reductase inhibitor | Clinical trial | 0.95 |
| 33 | Arimoclomol | 12 | SEQUOIA | Heat-shock protein 70inducer | Clinical trial | 0.95 |
| 34 | Fingolimod | 37 | Tokyo Chemical Industry | S1P receptor agonist | Clinical trial | 0.94 |
| 35 | Deforolimus | 26 | MedChemExpress | mTOR inhibitor | — | 0.92 |
| 36 | Ibudilast | 8 | FUJIFILM Wako Pure Chemical Corporation | Phosphodiesterase PDE4 inhibitor | Clinical trial | 0.91 |
| 37 | AZD-8055 | 24 | MedChemExpress | mTOR inhibitor | — | 0.88 |
| 38 | PF-04691502 | 27 | MedChemExpress | mTOR inhibitor PI3K inhibitor | — | 0.84 |
| 39 | PP242 | 25 | Sigma-Aldrich | mTOR inhibitor | — | 0.83 |
Figure 4.
MWT identified riluzole, a drug widely used for ALS treatment, as a positive hit compound. (a) Results of the MWT assay from one screening plate. Rapamycin is a positive control and the other compounds were tested in a blind manner on dnc-1 KD worms and analyzed by MWT. Note: drug 21 is riluzole. (b) Histograms of the moving speed of control-, rapamycin-, and riluzole-treated dnc-1 KD worms, showing a right shift in rapamycin- and riluzole-treated groups. (c) Representative trajectories of control worms (upper three maps) and riluzole-treated worms (lower three maps). (d) Dose-dependent effect of riluzole on locomotion of dnc-1 KD worms. Statistical analyses were performed by one-way ANOVA followed by the Bonferroni/Dunn post hoc test (a,d) (*p < 0.05, and **p < 0.001). Error bars are S.E.M.
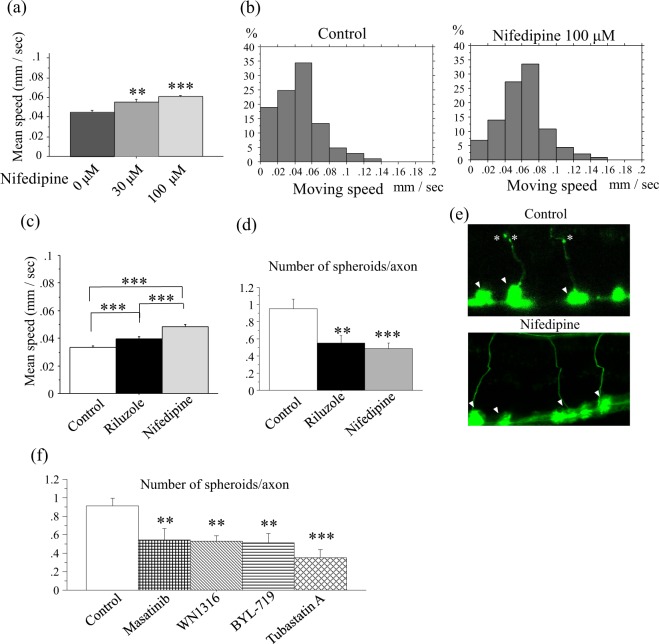
L-type calcium blocker nifedipine ameliorated the motor defects and axonal degeneration of dnc-1 KD worms
During the blinded screen, we found four compounds that were as effective as or more effective than riluzole (Table 1). Nifedipine, an L-type calcium channel blocker, showed the strongest effect on the locomotion of dnc-1 KD worms. Just as in the case of riluzole, the effect of nifedipine was dose-dependent (Fig. 5a). The distribution of the speed of nifedipine-treated dnc-1 KD worms was clearly shifted to the right compared with control-treated dnc-1 KD worms (median speed = 0.043 mm/sec and 0.061 mm/sec; DMSO-treated group and nifedipine-treated group, respectively), suggesting that nifedipine uniformly improves the motor function of dnc-1 KD worms (Fig. 5b). We also directly compared the effects of riluzole and nifedipine, and found that nifedipine had a stronger effect on the locomotion of dnc-1 KD worms than riluzole (Fig. 5c).
Figure 5.
Effects of nifedipine on the locomotion and axonal pathology of dnc-1 KD worms. (a) Dose-dependent effects of nifedipine on the locomotion of worms (n = 205, 170, and 230 for 0, 30, and 100 μM of nifedipine, respectively). (b) Histograms of the moving speed of control and nifedipine-treated dnc-1 KD worms. (c) Direct comparison of the effect of riluzole and nifedipine (n = 180, 210, and 170 for control, riluzole, and nifedipine). (d,e) The effects of riluzole and nifedipine on the pathology of dnc-1 KD worms. The number of axonal spheroids per transverse axon was counted after treatment by each compound (n = 20 worms for each group) (d), and representative images of a transverse axon with and without treatment by nifedipine are shown (e). Asterisks are axonal spheroids and arrowheads are the cell body of ventral motor neurons (e). (f) Number of axonal spheroids in a transverse axon treated by the hit compounds of the MWT assay (n = 20 worms for each group). Statistical analyses were performed by one-way ANOVA followed by the Bonferroni/Dunn post hoc test (a,c,d,f). (*p < 0.05, **p < 0.001, and ***p < 0.0001). Error bars indicate the S.E.M.
The motor defect in dnc-1 KD worms is caused by the degeneration of motor neurons9. Therefore, we tested whether nifedipine can improve the motor neuron degeneration in dnc-1 KD worms. In dnc-1 KD animals, axonal spheroids is a key feature of neurodegeneration, showing the accumulation of damaged mitochondria and autophagosomes9. We scored a number of axonal spheroids in a transverse section of ventral motor neuron axons (Fig. 5d), and found that riluzole and nifedipine significantly ameliorated the axonal degeneration in dnc-1 KD worms.
We also tested the effects of the remaining 4 of the top 6 compounds from MWT screening by scoring axonal spheroids. All 4 compounds significantly decreased the number of axonal spheroids in dnc-1 KD animals (Fig. 5f).
Given that in the dnc-1 KD model, dnc-1 shRNA is expressed under the ventral motor-neuron specific acr-2 promoter, other neurons and organs, including head neurons, and gut and pharyngeal muscles, which are essential for living, are intact in the dnc-1 KD model. Consistently, lifespan analysis showed no significant changes among DMSO-treated wild-type, DMSO-treated dnc-1 KD, and nifedipine-treated dnc-1 KD worms (Fig. 6).
Figure 6.
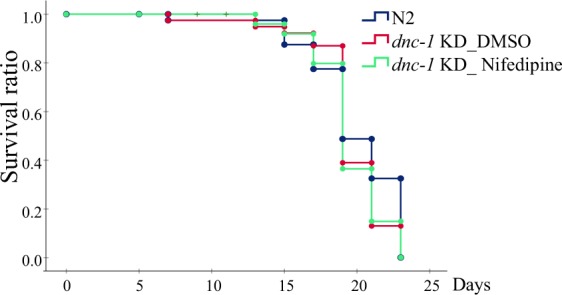
Lifespan analysis of nifedipine-treated animals. Kaplan meier curves of the wild type (N2) (n = 80) and dnc-1 KD animals treated with DMSO (n = 80) or 100 μM of nifedipine (n = 80). There were no significant differences between all groups according to the log-rank test.
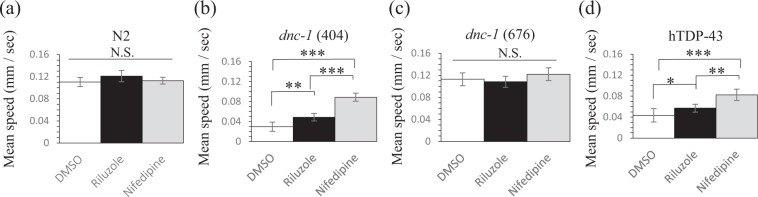
Nifedipine ameliorated the motor defects in other worm models with motor neuron degeneration
We further investigated the effects of riluzole and nifedipine on other worm models showing motor neuron degeneration and motor defects, to analyze the specificity and generality of these compounds. Both drugs did not show an increase in locomotion speed in wild-type worms (Fig. 7a). To evaluate the effect on dnc-1 mutants, we tested two alleles, dnc-1 (or404ts) and dnc-1 (or676ts), both of which show microtubule-associated defects in a temperature-dependent manner13,14. In our assay condition at 20 °C, only dnc-1 (or404ts) showed slow locomotion, and the drugs only affected the locomotion of dnc-1 (or404ts) (Fig. 7b,c). We also observed that both drugs ameliorated locomotor defects of human TDP-43-overexpressing strains (Fig. 7d). This strain expresses human wild-type TDP-43 under the pan-neuronal promotor, snb-1, and shows motor neuronal degeneration and progressive locomotor defects15. Importantly, the effect of nifedipine was significantly stronger than riluzole in dnc-1 KD animals, dnc-1 (or404ts) mutants, and hTDP-43 overexpressing animals, indicating the potential of nifedipine as a new candidate compound for the amelioration of motor neuron degeneration.
Figure 7.
Effects of riluzole and nifedipine on other models of motor neuron degeneration. Each worm line was treated with DMSO, riluzole, or nifedipine and then assayed by MWT. Statistical analyses were performed by one-way ANOVA followed by the Bonferroni/Dunn post hoc test. (*p < 0.05, **p < 0.001, and ***p < 0.0001). Error bars indicate the S.E.M.
Discussion
In this study, we developed a behavior-based drug screening system by combining a C. elegans model of ALS-like motor degeneration and dysfunction with an adapted version of the MWT. Our new screening system can evaluate eight compounds at once and only takes 15 min. We confirmed that the assay can detect the motor defects of dnc-1 KD worms, as well as the effects of rapamycin and food restriction on this dnc-1 KD model9.
We used this novel assay system to blindly test the effects of 38 compounds on dnc-1 KD worms. One of the most crucial results was that riluzole improved the motor defects of dnc-1 KD worms. As riluzole is an internationally approved drug for ALS, it is an indication that compounds identified by this approach will be worth pursuing as potential treatments for ALS. As we used compounds that have been previously reported or expected to have neuroprotective effects (see the criteria for selection in the Materials and Methods section), many compounds (25 out of 38) improved the motor phenotypes of dnc-1 KD worms. Among these compounds, we found that nifedipine, an L-type calcium-channel blocker, had the strongest effect on the locomotion of the worms.
Following the locomotor assay, we investigated the effects of the compounds on axonal degeneration. Importantly, all the top 6 compounds identified by the MWT assay had a significant effect on axonal degeneration. Moreover, we confirmed that nifedipine ameliorated the motor defects of other models of motor neuron disease, dnc-1 mutant animals, and human TDP-43 transgenic animals.
These results suggest that the adapted MWT assay can be a powerful system to perform behavior-based screening of drugs against motor neuron degeneration. The limitation of this assay is its throughputness. Several groups have developed worm-tracking systems to analyze many worms at once16–19. However, the application of these systems to drug screening in a high-throughput manner has been extremely challenging. Although our system, which enables the simultaneous analysis of 8 drugs using 500 worms, is the most powerful screening system to our knowledge to date, we still need faster and simpler screening systems for screening tens of thousands of drugs. Shunmoogum et al. performed a screening of 3,850 small molecules using mutant TDP-43-overexpressing C. elegans20. In their initial screening, they visually evaluated the motility of the worms and then confirmed the reproducibility and specificity of the results using a quantitative method, by manually scoring the percentage of paralyzed worms20. Considering that the big advantages of our system are its accuracy, reproducibility, and quantitativity, our newly established system will be a very strong tool as a second-throughput screening.
Materials and Methods
C. elegans culture
Standard methods were used to culture C. elegans on nematode growth medium (NGM) agar seeded with OP50 Escherichia coli (E. coli)21. The worms were maintained at 20 °C unless otherwise indicated.
C. elegans strains
The following strains were used in this study: N2 wild-type (Bristol), SBG8 Ex[Pacr-2::EmGFP::dnc-1miRNA#1; Pgcy-8::GFP]9, CB55 unc-2 (e55)22, EU1006 dnc-1 (or404)13, EU1257 dnc-1 (or676)14, and CL6049 dvIs62 [snb-1p::hTDP-43/3′ long UTR + mtl-2p::GFP], which were obtained from the C. elegans Genetics Center.
Compound selection
The compounds selected were as follows: (1) approved by the FDA for ALS treatment, (2) currently under/or finished clinical trials for ALS, and (3) show promising effects on ALS models. We also analyzed some of autophagy activators and HDAC inhibitors as positive controls because in a previous study we found that rapamycin, an autophagy activator, and trichostatin A, an HDAC inhibitor, were effective on this model9.
Synchronization of worms and compound treatment in liquid culture
Twenty gravid adult dnc-1 KD animals were selected by their uncoordinated phenotype and allowed to lay eggs on NGM plates with OP50 E. coli for 3 h to obtain approximately 200 synchronized eggs. Plates were cultivated for 3 days until the eggs had developed into adults. For the culture of temperature-sensitive dnc-1 mutant lines, they were incubated on NGM plates for 5 days at 15 °C until they had developed into the same age. Then worms were transferred into liquid medium (S basal medium with concentrated OP50 [80 mg/mL]) with compounds dissolved in DMSO at a final concentration of 100 μM (1% DMSO). We used drugs at 100 μM according to the most effective concentration of the positive control, rapamycin9. Two-hundred μL of the liquid culture (~50 worms) was transferred into each well of a 48-well plate with a flat bottom and without coating (Falcon/351178). The source of the compounds is listed in Table 1. The plate was incubated with shaking at 100 rpm for 16 h in 20 °C.
For levamisole treatment, approximately 300 synchronized adult worms (day 4) were collected from NGM plates and divided into 24-well plates with different concentrations of levamisole hydrochloride (Sigma-Aldrich, USA) (0, 10, 20, 50, 100, 200, 400, and 1,000 μM) dissolved in M9 buffer. After incubation for 10 minutes in 24-well plates, worms were transferred to the assay plate and analyzed by MWT.
Locomotion assay by MWT
After the incubation with a compound, worms were washed 3 times in NG buffer and gently transferred onto an assay plate. The assay plate was a 13 × 10-cm plate filled with agar, which was divided into 8 regions of equal area. Regions were surrounded with glycerol, an aversive stimulus for C. elegans, to keep animals from moving over to the other regions. Filter paper (Whatman/3MM paper) was used to remove excess NG buffer. Worms from a given compound-treated group were placed in 1 of the 8 regions with various compounds being tested simultaneously.
An adapted version of the MWT10 was used to record the locomotion of C. elegans on the agar plate. Adaptations included a Toshiba-Teli Ultra High Resolution 12 M pixel CMOS sensor camera-link camera (CSC12M25BMP19-01B), a lens (RICHO FL-YFL3528), and an adaptor (Toshiba-Teli FTAR-2). In addition, experiments were performed under dark-field lighting conditions using the ring LED light (CCS Inc. LDR-206SW2-LA1). C. elegans locomotion was recorded for 10 mins.
Analysis of the MWT data
Analysis of the recordings was performed using Choreography (part of the MWT software) and custom-written scripts to organize and summarize the data. Animal tracks were collected as a time series of the centroid position for each frame of the final 2 minutes of the recording. We used the final 2 minutes to enable animals to first adapt to the circumstances of the assay and to perform animal recognition by the tracker plateau. This was particularly important for slow-moving animals because the tracker only identifies animals that have moved from their initial position. The following Choreography filters were used to avoid image artifacts: ‒shadowless and -t 10. The speed of an individual animal was calculated as the sum of distances between sequential centroids divided by the duration of the track. Experimental groups were summarized using mean and standard errors of the mean, weighted by the duration of an animal’s track.
Microscopic analysis
Microscopic analysis of worms was performed as previously described9. Briefly, the worms were anesthetized by placing them in an 8-μL drop of levamisole (2 mM) on solidified pads of 2% agarose laid on slides with a coverslip. Worms were observed using a confocal microscope (Zeiss LSM 710).
For the evaluation of axonal degeneration, we scored the number of axonal spheroids per transverse axon.
Lifespan assay
A lifespan assay was performed as described previously13, with some modifications. Synchronized 3 day-old worms were collected and 20 worms were transferred to each NGM plate containing 100 μM 5-fluoro-2′-deoxyuridine (Sigma-Aldrich, USA), 100 μM drug (control or nifedipine), and 0.1% DMSO. For each group, 80 worms were transferred every 4 days to a freshly prepared plate. The animals were scored as dead if they did not move when prodded with a platinum pick and did not show pharyngeal pumping.
Statistical analysis
Statistical analyses were performed using StatView software version 5 (Hulinks, Tokyo, Japan). The Student t-test was used for the comparison of two independent groups and one-way analysis of the variance (ANOVA) with the Bonferroni/Dunn post-hoc test for more than three groups. We used the Kaplan-Meier and log-rank test to compare survivals of N2 and dnc-1 KD worms with our without treatment by riluzole or nifedipine. The application of these methods are indicated in each figure and legend.
Acknowledgements
This work was supported by a Grant-in-Aid for Challenging Exploratory Research JP14513047, the Japan Agency for Medical Research and Development (AMED) (no. JP18hm0102037), and a Grant-in-Aid for Scientific Research (no. JP16H02516). ACG was supported by a short-term (1-year) visiting postdoctoral fellowship from the Japanese Society for the Promotion of Science (JSPS). We thank Mr. Takumi Aoki (Toray, Industries, Inc.) for his contribution at the initial stage of this project.
Author Contributions
K.I., Y.T., A.C.G., T.A., S.N., M.K., G.S. and I.M. designed the experiments. K.I., Y.T., A.C.G. and S.N. accommodated the M.W.T. system. K.I., Y.N. and K.K. performed the compound screening. K.I. and T.A. selected the 38 compounds. K.I., Y.T., T.A., Y.N., S.N., H.M., M.K., G.S. and I.M. discussed the results and wrote the manuscript. All authors read and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jiang YM, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Annals of neurology. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 2.Jiang YM, et al. Gene expressions specifically detected in motor neurons (dynactin 1, early growth response 3, acetyl-CoA transporter, death receptor 5, and cyclin C) differentially correlate to pathologic markers in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2007;66:617–627. doi: 10.1097/nen.0b013e318093ece3. [DOI] [PubMed] [Google Scholar]
- 3.Gill SR, et al. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 5.Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 6.Farrer MJ, et al. DCTN1 mutations in Perry syndrome. Nature genetics. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araki K, et al. A small-angle X-ray scattering study of alpha-synuclein from human red blood cells. Scientific reports. 2016;6:30473. doi: 10.1038/srep30473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Q, et al. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. The EMBO journal. 2016;35:121–142. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikenaka K, et al. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PloS one. 2013;8:e54511. doi: 10.1371/journal.pone.0054511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nature methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer WR, Kenyon CJ. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- 13.Koushika SP, et al. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. Journal of Neuroscience. 2004;24:3907–3916. doi: 10.1523/Jneurosci.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Rourke SM, et al. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PloS one. 2011;6:e16644. doi: 10.1371/journal.pone.0016644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ash PE, et al. Neurotoxic effects of TDP-43 overexpression in C. elegans. Human molecular genetics. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronin CJ, Feng Z, Schafer WR. Automated imaging of C. elegans behavior. Methods Mol Biol. 2006;351:241–251. doi: 10.1385/1-59745-151-7:241. [DOI] [PubMed] [Google Scholar]
- 17.Ramot D, Johnson BE, Berry TL, Jr., Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PloS one. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husson, S. J., Costa, W. S., Schmitt, C. & Gottschalk, A. Keeping track of worm trackers. WormBook: the online review of C. elegans biology, 1–17, 10.1895/wormbook.1.156.1 (2013). [DOI] [PMC free article] [PubMed]
- 19.Itskovits E, Levine A, Cohen E, Zaslaver A. A multi-animal tracker for studying complex behaviors. BMC biology. 2017;15:29. doi: 10.1186/s12915-017-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patten, S. A. et al. Neuroleptics as therapeutic compounds stabilizing neuromuscular transmission in amyotrophic lateral sclerosis. JCI insight2, 10.1172/jci.insight.97152 (2017). [DOI] [PMC free article] [PubMed]
- 21.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews EA, et al. Critical residues of the Caenorhabditis elegans unc-2 voltage-gated calcium channel that affect behavioral and physiological properties. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:6537–6545. doi: 10.1523/JNEUROSCI.23-16-06537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]