Abstract
Recent advances in sequencing technologies have revealed the diversity of microbes that reside on the skin surface which has enhanced our understanding on skin as an ecosystem, wherein the epidermis, immune cells and the microbiota engage in active dialogues that maintain barrier integrity and functional immunity. This mutual dialogue is altered in atopic dermatitis (AD), in which an impaired epidermal barrier, the skin microbial flora and aberrant immunity can form a vicious cycle that leads to clinical manifestations as eczematous dermatitis. Microbiome studies have revealed an altered microbial landscape in AD and genetic studies have identified genes that underlie barrier impairment and immune dysregulation. Shifting from the long-standing notion that AD was mediated by conventional allergic responses, emerging data suggest that it is a disorder of an altered host–microbial relationship with sophisticated pathophysiology. In this review, we will discuss recent advancements that suggest the roles of the skin microbiota in AD pathophysiology, genetic factors that mediate barrier impairment, dysbiosis and inflammation. Studies in mice, classic AD and monogenic disorders that manifest as AD collectively facilitate our understanding of AD pathophysiology and provide a foundation for novel therapeutic strategies.
Keywords: epidermal barrier, dysbiosis, microbiome, skin
Introduction
Our understanding on the complexity of the microbial landscape that inhabits the barrier surfaces of our bodies has significantly deepened in parallel with the advancement of sequencing technologies. 16S ribosomal RNA as well as shotgun metagenomic sequencing have demonstrated that the skin is inhabited by a plethora of micro-organisms including bacteria, fungi and viruses, collectively referred to as microbiota, which has fueled research on barrier immunity (1, 2). The host barrier and immune systems have evolved hand-in-hand with the microbiota, forming mutually beneficial relationships. While epithelia and immune cells form barriers that protect the body from microbial invasion, they also create a surface environment that allows for the stable colonization by commensal microbes. Recent emerging evidence has revealed crucial roles of skin commensals in priming and harnessing local immunity (3, 4). Altered host–microbe cross-talk leads to immune dysregulation and is believed to be a driving force in inflammatory skin diseases.
This review will discuss the pathophysiology that underlies atopic dermatitis (AD), with particular focus on dialogues that take place between the host and the microbiota. While the majority of AD patients in the European population harbor loss-of-function mutations in the gene encoding Filaggrin (FLG) (5), a structural protein crucial for proper epidermal barrier formation, the association is variable across different ethnic backgrounds (6), wherein the majority of non-European AD patients do not harbor FLG mutations. In this regard, monogenic diseases that manifest as AD deliver insight into the pathogenesis of AD and mechanisms that underlie dysbiosis, both from the epithelial and immunological perspectives. Lastly, this review discusses the possibility of future interventions using microbiome transplants.
Skin as an ecosystem of the body surface
The skin is a physical barrier that insulates our body from the environment. While protecting the body from pathogen invasions, research in the recent years has established that the skin surface is far from an uninhabitable, harsh terrain. Indeed, its dry, yet lipid-rich nature and the invagination of hair follicles provide aerobic and anaerobic niches for a variety of microbial communities, rendering skin as a large ecosystem supporting symbiosis between the host and microbes. Analyses utilizing next-generation sequencing of bacterial 16S rRNA genes have demonstrated taxonomic diversity, and metagenomic sequences have revealed yeast, viruses and bacteriophages as natural inhabitants of skin (7).
Commensalism of microbes is mutually beneficial for the host. During development, the colonization of Staphylococcus epidermidis, a gram-positive cocci ubiquitous on human skin, leads to the generation of regulatory T cells that enable stable commensalism of the bacteria without eliciting immune responses (8). During adulthood, S. epidermidis is recognized by the skin immune system through intact barriers, priming skin-resident CD8+ T cells that confer heterologous protection against the yeast pathogen, Candida albicans (9, 10). While these studies were performed in mice using microbes that were derived from humans, they represent an example of how host and commensal bacteria communicate, and the broader picture of the interplay is an exciting field of research that is under active investigation.
The roles of skin barriers and immune cells have been well established in the setting of host-protective immune responses against pathogens. Recent studies have further unveiled homeostatic interactions between the immune cells and skin parenchyma that maintain structural, immunological and microbial homeostasis. Our previous studies highlight the hair follicles as control towers of skin immunity by producing chemokines to recruit and position skin-resident immune cells and to provide cytokines that enable their long-term persistence in skin (11, 12). We recently demonstrated that innate lymphoid cells (ILCs) exist in the upper parts of the hair follicles guided by a hair follicle-derived chemokine, CCL20, and that their residency was supported by Interleukin (IL)-7 and thymic stromal lymphopoietin (TSLP). There, the ILCs produced tumor necrosis factor receptor (TNFR) ligands that negatively controlled sebaceous gland function by suppressing the production of anti-bacterial lipids that restricted the commensalism of gram-positive cocci. This was in contrast to the actions of lymphocytes, whose absence led to the overgrowth of gram-positive cocci. Thus, the dynamic equilibrium of the skin microbiota is tuned by intricate dialogues between immune cells, epithelium and microbes (13). These fundamental mechanisms that regulate the skin microbiota provide insight into host–microbiota relationships, and further studies on how the dialogues go awry may provide better understanding of pathophysiology in inflammatory skin diseases such as AD.
Multifaceted pathophysiology of AD
AD is a chronic inflammatory skin disease that manifests as dry skin and eczematous dermatitis with relentless itch. Onset typically occurs during childhood and uncontrolled skin inflammation may lead to sequential onset of asthma, allergic rhinitis and food allergies, referred to as the atopic march (14). Other complications include cataracts, susceptibility to virus infections and mental health issues. Thus, significant comorbidities occur in AD, emphasizing the importance of understanding the pathophysiology involved. Note that while utilization of the term ‘eczema’ is not recommended from the clinical practice perspective, we herein utilize the term ‘eczematous dermatitis’ to reflect the spectrum of skin inflammation that may be observed in classic and monogenic AD, as well as in mouse models.
AD is a multifactorial, associated with impaired barrier formation, dysregulated type 2 immune response and increased susceptibility to Staphylococcus aureus colonization. Discoveries on genetic factors that underlie impaired epidermal barrier have provided momentum to the field, opening the door for detailed research also on immunological and microbiological aspects of the disease. Type 2 immune signatures and high levels of circulating IgE against inhaled allergens such as house dust and mite antigens have supported the notion that AD was an IgE-mediated allergic dermatitis elicited by chronic exposure to allergens. However, this long-held view is beginning to change with the observation that AD is closely associated with imbalanced balance of the skin microbiota, termed dysbiosis, which is predominated by S. aureus during the active phase of the disease (15).
These findings provide an opportunity for developing novel therapeutic strategies. A clearer picture on dysregulated immune networks has led to the emergence of new molecular targeting drugs such as IL-4R monoclonal antibody (16) and JAK inhibitors (17). Targeting the dysbiotic flora is also gaining considerable attention. However, there is still much to be learned to enable efficient and sustainable control of AD. Major unanswered questions include those regarding mechanisms that lead to exacerbated type 2 immune responses, cellular sources and targets of immune mediators, host-inherent factors that allow dysbiosis and characteristics of the dysbiotic flora themselves.
The skin microbiome in AD
It was first observed over five decades ago that AD skin was heavily colonized with S. aureus (18). More recently, 16S rRNA sequencing has revealed that the microbiome in AD skin is shifted toward an increased relative abundance of Staphylococcus species, particularly S. aureus. Staphylococcal colonization correlates with disease severity, suggesting the active involvement of Staphylococcus species during flares (15, 19). A study comparing S. aureus carrier and non-carrier AD patients has revealed that S. aureus carriers display higher disease severity and increased levels of type 2 immune biomarkers (20). Importantly, shifts in microbial compositions occur before the onset of AD (21, 22), suggesting microbial changes that are formed early in life may contribute to onset of AD.
It has been long debated whether the dysbiotic changes in AD were a consequence of chronic skin inflammation or whether they were actively involved in driving skin inflammation. This was, in part, due to the lack of mouse models that exhibited dysbiosis. Studies in mouse models have begun to reveal causal relationships. Mice deficient in a disintegrin and metalloproteinase 17 (ADAM17) spontaneously develop eczematous dermatitis with dysbiosis that was predominated by S. aureus and Corynebacterium species. Targeting the dysbiotic flora with an antibiotic cocktail in mice with pre-established eczematous dermatitis reversed dysbiosis and extinguished eczematous inflammation. Furthermore, the inoculation of S. aureus enhanced eczematous dermatitis and Corynebacterium bovis drove T helper 2 (Th2) responses, which presumably leads to IgE responses, revealing crucial and distinct roles of dysbiotic flora during atopic inflammation (23).
The mechanisms by which S. aureus initiate or exacerbate atopic inflammation remain to be further explored. Interestingly, S. aureus express molecules that mediate cytotoxic and immunological effects on host cells. α-Toxin is a pore-forming toxin that causes direct cellular damage to keratinocytes (24). Phenol soluble modulins (PSMs) are a family of Staphylococcal virulence factors that exert pro-inflammatory properties (25). In epicutaneous infection models of S. aureus in mice, PSM-α induces IL-17-dependent skin inflammation via IL-1R and IL-36R signaling (26, 27). PSM-δ (δ-toxin) induces skin inflammation by stimulating mast cell degranulation (28). Metagenomic analysis has revealed that particular strains of S. aureus dominate in individual patients during flares, and that the ability of S. aureus to induce immune responses is strain-dependent (19), warranting further investigations on whether strain-level differences in toxin production among S. aureus are associated with clinical phenotype and disease severity in AD.
Genetic factors in AD
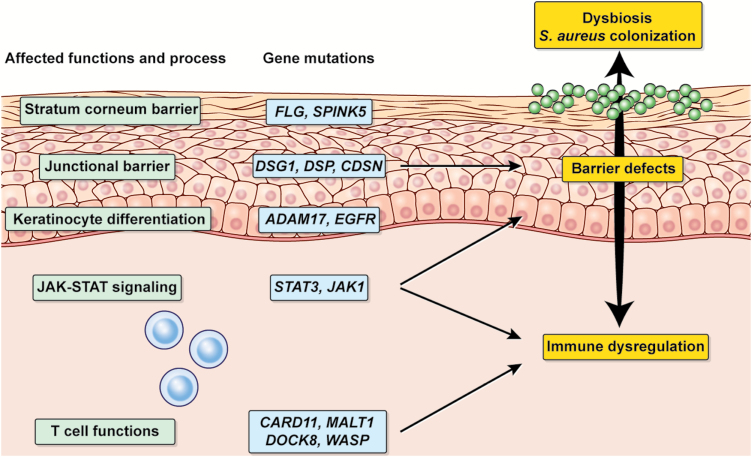
Studies on host genetic factors may reveal candidate genes that underlie barrier impairment, exacerbated immune responses and the inability to regulate the skin microbiota. Genome-wide association studies of AD have demonstrated the association of single-nucleotide polymorphisms in a number of genes and loci that are related to type 2 cytokines, T-cell proliferation and survival, innate immune response and epidermal barrier functions, such as IL4, IL13, IL18R1, IL6R, IL15RA, IL2RA, IL7R, CARD11, STAT3 and FLG (29–31). These results suggest that AD may either be impaired barrier-driven, immunity-driven or both. Multiple factors may synergistically trigger the development and exacerbation of AD, collectively contributing to the sophisticated pathophysiology. Importantly, single-nucleotide polymorphisms do not indicate functional alterations of the genes. In this regard, studies on monogenic diseases that manifest as AD with defined mutations in immune and non-immune genes are an attractive approach to elucidate the role(s) of single genes in the development of AD (Table 1; Fig. 1). The following sections will focus on dissecting molecular-level links between AD and dysbiosis from the genetic perspective.
Table 1.
Monogenic conditions that manifest as AD in humans.
| Affected cells or process | Gene mutations | Disease name |
|---|---|---|
| JAK-STAT signaling | Dominant negative mutations in STAT3 (OMIM: # 147060) | Autosomal dominant hyper IgE syndrome |
| Gain-of-function mutations in STAT3 (OMIM: # 615952) | Infantile-onset multisystem autoimmune disease-1 | |
| Gain-of-function mutations in JAK1 | Not applicable | |
| Dominant negative mutations in CARD11 (OMIM: # 617638) | Immunodeficiency-11B with atopic dermatitis | |
| T cell functions | Loss-of-function mutations in MALT1 (OMIM: # 615468) | Immunodeficiency-12 |
| Loss-of-function mutations in DOCK8 (OMIM: # 243700) | Autosomal recessive hyper IgE syndrome | |
| Loss-of-function mutations in WASP (OMIM: # 301000) | Wiskott-Aldrich syndrome | |
| Stratum corneum barrier | Loss-of-function mutations in FLG (OMIM: # 146700) | Icthyosis vulgaris |
| Loss-of-function mutations in SPINK5 (OMIM: # 256500) | Netherton syndrome | |
| Loss-of-function mutations in CDSN (OMIM: # 270300) | Peeling skin syndrome-1 | |
| Junctional barrier | Loss-of-function mutations in DSG1 (OMIM: # 615508) | Severe dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome |
| Dominant heterozygous mutations in DSP | ||
| Keratinocyte differentiation | Loss-of-function mutations in ADAM17 (OMIM: # 614328) | Neonatal inflammatory skin and bowel disease-1 |
| Loss-of-function mutations in EGFR (OMIM: # 616069) | Neonatal inflammatory skin and bowel disease-2 |
Fig. 1.
Genetic factors that underlie the pathophysiology of AD. Monogenic diseases with defined mutations in genes associated with epidermal barrier, JAK–STAT signaling and lymphocyte functions manifest as eczematous dermatitis and Staphylococcal dysbiosis.
Monogenic disorders manifesting as AD
The autosomal dominant hyper-IgE syndrome (STAT3-HIES; a.k.a. Job’s syndrome), caused by dominant-negative mutations in signal transducer and activator of transcription 3 (STAT3), is a monogenic disorder that has been long associated with AD. STAT3-HIES patients present elevated serum IgE levels, eczematous dermatitis and recurrent Staphylococcal infections and mucocutaneous candidiasis (32, 33). 16S rRNA sequencing has revealed altered microbial compositions represented by the emergence of Serratia marcescens, an environmental microbe that has not been previously identified as a component of the normal skin flora (34). Staphylococcus and Corynebacterium are highly over-represented, and the disease severity is positively correlated with the prevalence of these bacteria. In addition, analysis of oral mucosa in STAT3-HIES reveals fungal dysbiosis with dominance of C. albicans, which is consistent with recurrent mucosal fungal infections in the patients (35). Importantly, eczematous dermatitis in STAT3-HIES is attenuated with Staphylococcus clearance measures, reinforcing the role of S. aureus in driving eczematous dermatitis (36).
STAT3 is a key transcription factor for Th17 differentiation that is also involved in downstream signaling of IL-6, IL-21, IL-10 and IL-23 (37, 38). STAT3-HIES T cells show decreased expression of retinoid-related orphan receptor (ROR)-γt (a crucial transcription factor for Th17 cell differentiation) and are unable to differentiate into Th17 cells (39). Because IL-17 is an essential cytokine that promotes epithelial anti-microbial functions against extracellular bacterial and fungal infections, impaired Th17 differentiation is considered to be a major mechanism underlying the susceptibility to dysbiosis and recurrent infections. Notably, STAT3 is also expressed by keratinocytes and may be involved in barrier functions. Keratinocyte-specific depletion of Stat3 results in impaired hair cycling and wound healing in mice, which is associated with altered adhesion and migration of keratinocytes (40, 41). Intra-dermal lipopolysaccharide or epicutaneous ovalbumin challenge in mice that lack Stat3 in keratinocytes leads to high IgE and type 2 immune responses (42). The finding that neither T cell- nor keratinocyte-specific depletion of Stat3 leads to spontaneous onset of eczematous dermatitis suggests that impaired STAT3 signaling in immune cells and epithelial cells synergistically leads to eczematous dermatitis and dysbiosis (37, 43). Mice carrying the dominant-negative form of STAT3 as seen in STAT3-HIES exhibit elevated IgE and are susceptible to Citrobacter rodentium infection but do not develop eczematous inflammation, at least in SPF conditions (44), further suggesting that the involvement of additional environmental factors is required, such as signals from commensal microbes.
Interestingly, patients with gain-of-function mutations in STAT3 also present eczematous dermatitis (45), indicating that the JAK–STAT signaling pathway must be tightly regulated to maintain skin homeostasis. A JAK1 gain-of-function mutation also leads to systemic immune dysregulation and eczematous dermatitis in both humans (46) and mice (47), in which JAK inhibitors are efficacious. The use of JAK inhibitors in AD is under active investigation and preliminary results are promising (17). Further studies are required to understand how the JAK–STAT pathway operates in hematopoietic and non-hematopoietic compartments and contributes to the maintenance of skin barrier integrity and microbiome balance.
Mutations in genes involved in T cell receptor (TCR) signaling and T-cell responses also lead to atopic manifestations. Cytoskeletal remodeling proteins, Wiskott–Aldrich syndrome (WAS) protein and dedicator of cytokinesis 8 (DOCK8), mediate signaling during TCR-driven actin assembly in T cells (48). Both WAS- and DOCK8-deficient patients display elevated IgE, manifest eczematous dermatitis and have recurrent infections (49, 50). Similar to STAT3-HIES, skin microbiome analysis of WAS- and DOCK8-deficient patients has demonstrated significant shifts in the microbiome, represented by the predominance of Staphylococcus, Propionibacterium and Corynebacterium species (34). Strikingly, dysbiosis is not limited to bacteria and fungi. Deep metagenomic sequencing analysis of DOCK8-deficient skin revealed the predominance of DNA viruses with increased viral diversity and hundreds of novel human papillomavirus genomes (51). While DOCK8 is known to regulate T-cell function in anti-viral immunity, the shifts in the microbiome and virome of DOCK8-deficient skin underscore the importance of lymphocyte-mediated immune surveillance for micro-organisms. Why an atopic phenotype emerges as a result of impaired regulation of the actin cytoskeleton in T cells is not yet clear. Bias toward Th2 in CD4+ T cells from DOCK8-deficient patients might be related to enhanced IgE production and eczematous dermatitis (52). It is attractive to hypothesize that immune responses against the altered virome drive eczematous inflammation.
Mutations in genes encoding a signal complex that connects TCR signals to NF-κB activation result in immunodeficiency and atopic symptoms. Patients with defects in the caspase recruitment domain family member 11 (CARD11) (53, 54) and mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1) (55) present eczematous dermatitis and recurrent bacterial and viral skin infections. Taken together, these monogenic disorders due to dysregulated T-cell functions provide new insights into AD pathophysiology from an immunological perspective.
Epidermal barrier impairment underlying eczematous inflammation and dysbiosis
Mutations in a number of genes that are essential for epidermal barrier integrity have also been identified in both classic and monogenic AD (Table 1; Fig. 1). In particular, loss-of-function mutations in FLG, which were initially identified in patients with ichthyosis vulgaris, are a major predisposing factor for classic AD, particularly in the European population (5). FLG-derived peptides maintain structural integrity and hydration in the stratum corneum, in part through degradation into natural moisturizing factors (NMFs). NMFs influence skin surface pH, affecting the keratinization process as well as microbial colonization. Acidic conditions in the presence of NMFs disfavor S. aureus growth (56). Atopic individuals tend to have alkaline skin conditions, suggesting that S. aureus colonization may be facilitated in part by pH shifts in FLG-deficient skin. The increased pH in epidermis also influences the activity of serine proteases that cleave IL-1 family cytokines. Indeed, increased IL-1 cytokines are correlated with FLG deficiency and reduced NMFs both in human patients and in FLG-deficient mice (57). Incisional damage to skin in Flg-mutant (ft/ft) mice induces the release of intracellularly stored IL-1α in keratinocytes, resulting in IL-1α-mediated skin inflammation and dysbiotic conditions. Topical antibiotic treatment reverses the skin inflammation, supporting the idea that both barrier deficiency and dysbiosis are crucial components of skin inflammation (58).
Cytokines that are abundant in AD skin, IL-4 and IL-13, act negatively on FLG expression (59). IL-4 and IL-13 also down-regulate the expression of anti-microbial peptides, suggesting that dysregulated immunity also confers susceptibility to bacterial colonization in AD skin (37, 38, 60, 61). A mutation of Flg in the BALB/c background (BALB/c ft/ft) facilitates S. aureus penetration into viable skin layers, which correlates with increased expression of IL-4 and IL-13. These observations suggest a pathogenic loop formed by increased type 2 immunity, FLG deficiency and S. aureus colonization (62). Interestingly, S. aureus inoculation onto C57BL/6 mouse skin with genomic ablation of Flg does not induce type 2 responses, but rather, type 17 responses, and does not lead to overt eczematous inflammation (23). Both genetic backgrounds and the nature of FLG deficiency (mutation versus deletion) could contribute to these differences.
In addition to bacterial dysbiosis, patients with AD are known to be susceptible to viral infections such as herpes simplex virus (HSV) infections and vaccinia virus infections. Disseminated cutaneous HSV infections, called eczema herpeticum, are a common complication in AD. FLG mutations associated with AD confer a greater risk for eczema herpeticum (63), suggesting that impaired barrier and/or altered immune responses in the absence of FLG predisposes to viral infections. Smallpox vaccination is contraindicated in AD and may result in extensive rash and systemic illness, called eczema vaccinatum. Although the mechanism that allows for the spread of vaccinia virus remains largely unclear, it is reported in human skin explants that IL-4 and IL-13 suppress the expression of an anti-microbial peptide cathelicidin/LL-37, thereby enhancing vaccinia virus replication, and that neutralizing IL-4 and IL-13 rescues LL-37 expression and inhibits viral replication (64). Consistently, BALB/c ft/ft mice display increased susceptibility to cutaneous vaccinia virus inoculation wherein virus disseminates to internal organs (65). Future research that further elucidates barrier and immunological alterations in the absence of FLG might enable identification of potential therapeutic targets that prevent susceptibility to microbes.
Rare genetic disorders with mutations in barrier-associated genes also manifest symptoms that resemble AD (Table 1; Fig. 1). Autosomal-recessive loss-of-function mutations in Kazal type 5 (SPINK5), encoding the serine peptidase inhibitor LEKTI, are responsible for causing Netherton syndrome, which is characterized by congenital icthyosiform erythroderma, atopic manifestations and S. aureus colonization (66). LEKTI inhibits kallikrein family proteases which regulate desquamation of epidermis. Loss of LEKTI thereby results in enhanced proteolytic activity in the epidermis, leading to impaired stratum corneum formation and S. aureus colonization. In addition, unregulated kallikrein activates proteinase-activated receptor 2 (PAR2) and induces NF-κB-mediated over-expression of TSLP, which can trigger type 2 immune responses that may contribute to eczematous dermatitis (67, 68).
Impaired keratinocyte cell–cell adhesion also leads to atopic manifestations. Severe eczematous dermatitis, multiple allergies and Staphylococcal skin infections occur in patients with mutations in desmosomal proteins, desmoglein 1 (DSG1) and desmoplakin (DSP) (69, 70). Loss of corneodesmosin (CDSN), an adhesion protein in corneodesmosomes, leads to development of eczematous dermatitis, allergies and S. aureus skin infections (71). In aggregate, these genetic disorders emphasize barrier disruption as a fundamental element that leads to eczematous dermatitis and dysbiosis in skin.
Epithelial barrier integrity relies on proper terminal differentiation of keratinocytes, which is controlled, in part, by signaling through epidermal growth factor receptor (EGFR) (72). EGFR signaling is regulated by an upstream proteinase via release of membrane-bound form of EGFR ligands. ADAM17 is a transmembrane proteinase that cleaves a variety of membrane-anchored molecules including EGFR ligands and plays a critical role in the regulation of tissue integrity (73). A loss-of-function mutation in ADAM17 was reported in a patient who exhibited eczematous dermatitis and S. aureus skin infections (74). Three individual studies have demonstrated that keratinocyte-specific depletion of Adam17 in mice results in chronic AD-like skin inflammation with barrier impairment (23, 75, 76).
We have demonstrated that S. aureus colonization precedes eczematous dermatitis formation in the Adam17-deficient skin (23), suggesting that dysbiosis is attributed to impaired ADAM17 deficiency in keratinocytes and that it is not secondary to chronic inflammation. Epidermal deletion of EGFR in mice also leads to dysbiosis and eczematous dermatitis, suggesting that impaired EGFR signaling is in part responsible for the phenotype observed in ADAM17-deficient mice (23, 75, 77, 78). Consistently, patients with loss-of-function mutations in EGFR share clinical features with ADAM17-deficient patients (79). Furthermore, patients treated with EGFR inhibitors during cancer therapy experience cutaneous adverse effects including dry and erythematous skin and bacterial skin infections (80), from which S. aureus is commonly isolated (78). Antibiotics have beneficial effects in ameliorating these skin conditions (81). Increased susceptibility to bacterial infections might be related to altered expression of anti-microbial peptides that are regulated downstream of EGFR (78). Collectively, these studies identify the ADAM17–EGFR axis as crucial for tuning epidermal barrier functions and in restricting dysbiosis. Further studies that elucidate detailed upstream and downstream events may have broad impact on the development of new therapeutic strategies in AD and EGFR antagonist-related skin toxicities.
Intervention with microbiome therapies
Despite accumulating evidence on the contribution of dysbiosis and S. aureus in AD, the clinical efficacy of anti-bacterial interventions in AD remains controversial (82). Given that antibiotics treatment can have systemic effects on the normal flora and lead to the emergence of antibiotics-resistant bacteria, skin-targeted approaches without the use of antibiotics would be better suited. A high-throughput anti-microbial screening of coagulase-negative Staphylococcus (CoNS) species against S. aureus has revealed new anti-microbial peptides that inhibit S. aureus growth. Importantly, these anti-microbial peptide-producing CoNS strains are less frequent in atopic individuals, and reintroduction of CoNS decreases S. aureus colonization (83). An open-label trial was recently conducted to evaluate the safety and efficacy of a strain of Gram-negative bacteria on AD (84). Topical application of Roseomonas mucosa reduced S. aureus colonization and decreased both disease severity and the use of topical steroids. While R. mucosa affects the innate immune response and barrier function (85), whether R. mucosa mediates its effects by restricting dysbiotic flora, or by directly modulating immunity, remains unclear. Further work will be needed to elucidate the mechanisms of action and to determine the efficacy and safety of biological agent-based therapies.
Conclusion
Extensive research over the past decade has highlighted dynamic interactions that take place between the immune system and the microbiota. These dialogues are essential for tissue development, homeostatic maintenance of epithelial barriers and functional immunity. The host has developed an array of sophisticated physical and immunological strategies to maintain the mutualistic relationship with the microbiota. Impaired dialogue leads to chronic inflammatory conditions as seen in AD. The mechanisms underlying dysbiosis, its roles in AD and novel therapeutic strategies that target upstream and downstream pathways are only beginning to be revealed. While more multifaceted research from clinical, immunological and microbiological approaches is needed, the recent development opens the door to an exciting field of research that should unveil links between the host and the microbiota in health and disease.
Funding
This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health.
Conflicts of Interest statement: the authors declared no conflicts of interest.
References
- 1. Grice E. A. and Segre J. A. 2011. The skin microbiome. Nat. Rev. Microbiol. 9:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrd A. L., Belkaid Y. and Segre J. A. 2018. The human skin microbiome. Nat. Rev. Microbiol. 16:143. [DOI] [PubMed] [Google Scholar]
- 3. Belkaid Y. and Tamoutounour S. 2016. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 16:353. [DOI] [PubMed] [Google Scholar]
- 4. Chen Y. E., Fischbach M. A. and Belkaid Y. 2018. Skin microbiota-host interactions. Nature 553:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Irvine A. D., McLean W. H. and Leung D. Y. 2011. Filaggrin mutations associated with skin and allergic diseases. N. Engl. J. Med. 365:1315. [DOI] [PubMed] [Google Scholar]
- 6. Akiyama M. 2010. FLG mutations in ichthyosis vulgaris and atopic eczema: spectrum of mutations and population genetics. Br. J. Dermatol. 162:472. [DOI] [PubMed] [Google Scholar]
- 7. Oh J., Byrd A. L., Deming C., Conlan S., Kong H. H. and Segre J. A.; NISC Comparative Sequencing Program 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scharschmidt T. C., Vasquez K. S., Truong H. A. et al. 2015. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43:1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naik S., Bouladoux N., Wilhelm C. et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naik S., Bouladoux N., Linehan J. L. et al. 2015. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagao K., Kobayashi T., Moro K. et al. 2012. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 13:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adachi T., Kobayashi T., Sugihara E. et al. 2015. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 21:1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kobayashi T., Voisin B., Kim D., et al. 2019. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell. 176:982.doi: 10.1016/j.cell.2018.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bieber T. 2008. Atopic dermatitis. N. Engl. J. Med. 358:1483. [DOI] [PubMed] [Google Scholar]
- 15. Kong H. H., Oh J., Deming C. et al. ; NISC Comparative Sequence Program. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22:850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck L. A., Thaçi D., Hamilton J. D. et al. 2014. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N. Engl. J. Med. 371:130. [DOI] [PubMed] [Google Scholar]
- 17. Damsky W. and King B. A. 2017. JAK inhibitors in dermatology: the promise of a new drug class. J. Am. Acad. Dermatol. 76:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leyden J. J., Marples R. R. and Kligman A. M. 1974. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 90:525. [DOI] [PubMed] [Google Scholar]
- 19. Byrd A. L., Deming C., Cassidy S. K. B., et al. 2017. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 9:eaal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simpson E. L., Villarreal M., Jepson B. et al. 2018. Patients with atopic dermatitis colonized with Staphylococcus aureus have a distinct phenotype and endotype. J. Invest. Dermatol. 138:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kennedy E. A., Connolly J., Hourihane J. O. et al. 2017. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 139:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meylan P., Lang C., Mermoud S. et al. 2017. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J. Invest. Dermatol. 137:2497. [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi T., Glatz M., Horiuchi K. et al. 2015. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H. and Gouaux J. E. 1996. Structure of Staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859. [DOI] [PubMed] [Google Scholar]
- 25. Cheung G. Y., Joo H. S., Chatterjee S. S. and Otto M. 2014. Phenol-soluble modulins–critical determinants of Staphylococcal virulence. FEMS Microbiol. Rev. 38:698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu H., Archer N. K., Dillen C. A. et al. 2017. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 22:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakagawa S., Matsumoto M., Katayama Y. et al. 2017. Staphylococcus aureus virulent PSMα peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe 22:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakamura Y., Oscherwitz J., Cease K. B. et al. 2013. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paternoster L., Standl M., Waage J. et al. ; Australian Asthma Genetics Consortium (AAGC) 2015. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 47:1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamari M. and Hirota T. 2014. Genome-wide association studies of atopic dermatitis. J. Dermatol. 41:213. [DOI] [PubMed] [Google Scholar]
- 31. Bin L. and Leung D. Y. 2016. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin. Immunol. 12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minegishi Y., Saito M., Tsuchiya S. et al. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448:1058. [DOI] [PubMed] [Google Scholar]
- 33. Holland S. M., DeLeo F. R., Elloumi H. Z. et al. 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357:1608. [DOI] [PubMed] [Google Scholar]
- 34. Oh J., Freeman A. F., Park M. et al. ; NISC Comparative Sequencing Program 2013. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 23:2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abusleme L., Diaz P. I., Freeman A. F., et al. 2018. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight 3:e122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yong P. F., Freeman A. F., Engelhardt K. R., Holland S., Puck J. M. and Grimbacher B. 2012. An update on the hyper-IgE syndromes. Arthritis Res. Ther. 14:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Durant L., Watford W. T., Ramos H. L. et al. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kane A., Deenick E. K., Ma C. S., Cook M. C., Uzel G. and Tangye S. G. 2014. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 28:49. [DOI] [PubMed] [Google Scholar]
- 39. Milner J. D., Brenchley J. M., Laurence A. et al. 2008. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sano S., Itami S., Takeda K. et al. 1999. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18:4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kira M., Sano S., Takagi S., Yoshikawa K., Takeda J. and Itami S. 2002. STAT3 deficiency in keratinocytes leads to compromised cell migration through hyperphosphorylation of p130(cas). J. Biol. Chem. 277:12931. [DOI] [PubMed] [Google Scholar]
- 42. Domínguez-Hüttinger E., Christodoulides P., Miyauchi K. et al. 2017. Mathematical modeling of atopic dermatitis reveals “double-switch” mechanisms underlying 4 common disease phenotypes. J. Allergy Clin. Immunol. 139:1861. [DOI] [PubMed] [Google Scholar]
- 43. Takeda K., Kaisho T., Yoshida N., Takeda J., Kishimoto T. and Akira S. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652. [PubMed] [Google Scholar]
- 44. Steward-Tharp S. M., Laurence A., Kanno Y. et al. 2014. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123:2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milner J. D., Vogel T. P., Forbes L. et al. 2015. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 125:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Del Bel K. L., Ragotte R. J., Saferali A. et al. 2017. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J. Allergy Clin. Immunol. 139:2016. [DOI] [PubMed] [Google Scholar]
- 47. Yasuda T., Fukada T., Nishida K. et al. 2016. Hyperactivation of JAK1 tyrosine kinase induces stepwise, progressive pruritic dermatitis. J. Clin. Invest. 126:2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Janssen E., Tohme M., Hedayat M. et al. 2016. A DOCK8-WIP-WASp complex links T cell receptors to the actin cytoskeleton. J. Clin. Invest. 126:3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Q., Davis J. C., Lamborn I. T. et al. 2009. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 361:2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Imai K., Nonoyama S. and Ochs H. D. 2003. WASP (Wiskott-Aldrich syndrome protein) gene mutations and phenotype. Curr. Opin. Allergy Clin. Immunol. 3:427. [DOI] [PubMed] [Google Scholar]
- 51. Tirosh O., Conlan S., Deming C. et al. ; NISC Comparative Sequencing Program. 2018. Expanded skin virome in DOCK8-deficient patients. Nat. Med. 24:1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tangye S. G., Pillay B., Randall K. L. et al. 2017. Dedicator of cytokinesis 8-deficient CD4+ T cells are biased to a TH2 effector fate at the expense of TH1 and TH17 cells. J. Allergy Clin. Immunol. 139:933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma C. A., Stinson J. R., Zhang Y. et al. 2017. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 49:1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dadi H., Jones T. A., Merico D. et al. 2018. Combined immunodeficiency and atopy caused by a dominant negative mutation in caspase activation and recruitment domain family member 11 (CARD11). J. Allergy Clin. Immunol. 141:1818. [DOI] [PubMed] [Google Scholar]
- 55. McKinnon M. L., Rozmus J., Fung S. Y. et al. 2014. Combined immunodeficiency associated with homozygous MALT1 mutations. J. Allergy Clin. Immunol. 133:1458. [DOI] [PubMed] [Google Scholar]
- 56. Miajlovic H., Fallon P. G., Irvine A. D. and Foster T. J. 2010. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J. Allergy Clin. Immunol. 126:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kezic S., O’Regan G. M., Lutter R. et al. 2012. Filaggrin loss-of-function mutations are associated with enhanced expression of IL-1 cytokines in the stratum corneum of patients with atopic dermatitis and in a murine model of filaggrin deficiency. J. Allergy Clin. Immunol. 129:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Archer N. K., Jo J. H., Lee S. K. et al. 2018. Injury, dysbiosis, and filaggrin deficiency drive skin inflammation through keratinocyte IL-1alpha release. J. Allergy Clin. Immunol., [Epub ahead of print]. doi: 10.1016/j.jaci.2018.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Howell M. D., Kim B. E., Gao P. et al. 2009. Cytokine modulation of atopic dermatitis filaggrin skin expression. J. Allergy Clin. Immunol. 124:R7. [DOI] [PubMed] [Google Scholar]
- 60. Ong P. Y., Ohtake T., Brandt C. et al. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 347:1151. [DOI] [PubMed] [Google Scholar]
- 61. Howell M. D., Boguniewicz M., Pastore S. et al. 2006. Mechanism of HBD-3 deficiency in atopic dermatitis. Clin. Immunol. 121:332. [DOI] [PubMed] [Google Scholar]
- 62. Nakatsuji T., Chen T. H., Two A. M. et al. 2016. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J. Invest. Dermatol. 136:2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gao P. S., Rafaels N. M., Hand T. et al. 2009. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J. Allergy Clin. Immunol. 124:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Howell M. D., Gallo R. L., Boguniewicz M. et al. 2006. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity 24:341. [DOI] [PubMed] [Google Scholar]
- 65. Oyoshi M. K., Beaupré J., Venturelli N., Lewis C. N., Iwakura Y. and Geha R. S. 2015. Filaggrin deficiency promotes the dissemination of cutaneously inoculated vaccinia virus. J. Allergy Clin. Immunol. 135:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chavanas S., Bodemer C., Rochat A. et al. 2000. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 25:141. [DOI] [PubMed] [Google Scholar]
- 67. Briot A., Deraison C., Lacroix M. et al. 2009. Kallikrein 5 induces atopic dermatitis-like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J. Exp. Med. 206:1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Furio L., de Veer S., Jaillet M. et al. 2014. Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J. Exp. Med. 211:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Samuelov L., Sarig O., Harmon R. M. et al. 2013. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat. Genet. 45:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McAleer M. A., Pohler E., Smith F. J. et al. 2015. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J. Allergy Clin. Immunol. 136:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oji V., Eckl K. M., Aufenvenne K. et al. 2010. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am. J. Hum. Genet. 87:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pastore S., Mascia F., Mariani V. and Girolomoni G. 2008. The epidermal growth factor receptor system in skin repair and inflammation. J. Invest. Dermatol. 128:1365. [DOI] [PubMed] [Google Scholar]
- 73. Blobel C. P. 2005. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 6:32. [DOI] [PubMed] [Google Scholar]
- 74. Blaydon D. C., Biancheri P., Di W. L. et al. 2011. Inflammatory skin and bowel disease linked to ADAM17 deletion. N. Engl. J. Med. 365:1502. [DOI] [PubMed] [Google Scholar]
- 75. Franzke C. W., Cobzaru C., Triantafyllopoulou A. et al. 2012. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J. Exp. Med. 209:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Murthy A., Shao Y. W., Narala S. R., Molyneux S. D., Zúñiga-Pflücker J. C. and Khokha R. 2012. Notch activation by the metalloproteinase ADAM17 regulates myeloproliferation and atopic barrier immunity by suppressing epithelial cytokine synthesis. Immunity 36:105. [DOI] [PubMed] [Google Scholar]
- 77. Mascia F., Lam G., Keith C. et al. 2013. Genetic ablation of epidermal EGFR reveals the dynamic origin of adverse effects of anti-EGFR therapy. Sci. Transl. Med. 5: 199ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lichtenberger B. M., Gerber P. A., Holcmann M. et al. 2013. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci. Transl. Med. 5:199ra111. [DOI] [PubMed] [Google Scholar]
- 79. Campbell P., Morton P. E., Takeichi T. et al. 2014. Epithelial inflammation resulting from an inherited loss-of-function mutation in EGFR. J. Invest. Dermatol. 134:2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lacouture M. E. 2006. Mechanisms of cutaneous toxicities to EGFR inhibitors. Nat. Rev. Cancer 6:803. [DOI] [PubMed] [Google Scholar]
- 81. Lacouture M. E., Mitchell E. P., Piperdi B. et al. 2010. Skin toxicity evaluation protocol with panitumumab (STEPP), a phase II, open-label, randomized trial evaluating the impact of a pre-emptive skin treatment regimen on skin toxicities and quality of life in patients with metastatic colorectal cancer. J. Clin. Oncol. 28:1351. [DOI] [PubMed] [Google Scholar]
- 82. Bath-Hextall F. J., Birnie A. J., Ravenscroft J. C. and Williams H. C. 2010. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: an updated Cochrane review. Br. J. Dermatol. 163:12. [DOI] [PubMed] [Google Scholar]
- 83. Nakatsuji T., Chen T. H., Narala S. et al. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 99:eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Myles I. A., Earland N. J., Anderson E. D. et al. 2018. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3:e120608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Myles I. A., Williams K. W., Reckhow J. D. et al. 2016. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight 1:e86955. [DOI] [PMC free article] [PubMed] [Google Scholar]