Abstract
During the assembly of dsDNA viruses such as the tailed bacteriophages and herpesviruses, the viral chromosome is compacted to near crystalline density inside a preformed head shell. DNA translocation is driven by powerful ring ATPase motors that couple ATP binding, hydrolysis, and release to force generation and movement. Studies of the motor of the bacteriophage phi29 have revealed a complex mechanochemistry behind this process that slows as the head fills. Recent studies of the physical behaviour of packaging DNA suggest that surprisingly long time scales of relaxation of DNA inside the head and jamming phenomena during packaging create the physical need for regulation of the rate of packaging. Studies of DNA packaging in viral systems have therefore revealed fundamental insight into the complex behaviour of DNA and the need for biological systems to accommodate these physical constraints.
Graphical abstract

Learning from living systems
Given that biological systems have adapted to the constraints of the physical world for a billion years, the study of biology serves to inform us as to how things work. The cross-section of the bird wing reveals the principles of aerodynamic lift. The surface of the Nelumbo leaf demonstrates the principles of superhydrophobicity. And the behavior of DNA in viruses reveals the complexities of how charged polymers interact.
For viruses, the key biological challenge is to deliver their parasitic genome from one host cell to the other. The study of virus assembly focuses on the events that yield a vehicle capable of accomplishing this task, with the double-stranded DNA viruses adopting a build-and-fill strategy whereby the protein capsid is constructed and then filled with genetic cargo[1]. The challenge this presents is formidable, given that most dsDNA viruses compact their DNA chromosome to near crystalline density inside the capsid[2].
The utility of DNA as an information bearing molecule is confounded by the complex physical behavior its structure yields. On the scale of viruses, it is stiff, having a persistence length longer than the radius of the virus capsid[3]. Its phosphate backbone resists compaction due to charge repulsion[4]. And confinement inside a capsid comes with an entropic penalty as it is driven into a space that is orders of magnitude smaller than it would otherwise occupy. Therefore, in order to assemble a DNA-filled capsid the dsDNA viruses have evolved a machine capable of overcoming these resistive forces in order to package their DNA.
At first blush the challenge is one that can simply be overcome by brute force. The DNA packaging machines that translocate DNA, powered by ATP, have been shown to be among the most powerful measured[5]. One of the best characterized is the DNA packaging motor of the Bacillus subtilis bacteriophage phi29[6], whose ring-motor gene product 16 (gp16) is of the ancient and diverse ASCE lineage that drive polymers across the biological continuum[7]. The gp16 pentamer forms the catalytic core of the packaging motor in phi29[8], and is anchored to a five-fold vertex of the icosahedral capsid by an RNA scaffold[9,10], forming a ribonucleoprotein complex that will be discarded after packaging the DNA through a portal assemblage embedded in the capsid (Fig. 1). The RNA scaffold, or pRNA, is unique to phi29, as most tailed dsDNA phages and the analogous herpesviruses employ only a portal and an ATPase[1] (referred to as terminase in these systems; see below). Single-molecule studies have revealed that the phi29 motor can pull DNA with a measured force of over 57pN[11], strong enough to begin to unwind the DNA helix if the DNA is tethered. But as with many things in biology, DNA packaging has revealed subtleties about the physical world that go beyond what were predicted or thought to be the limits of behavior.
Fig. 1.
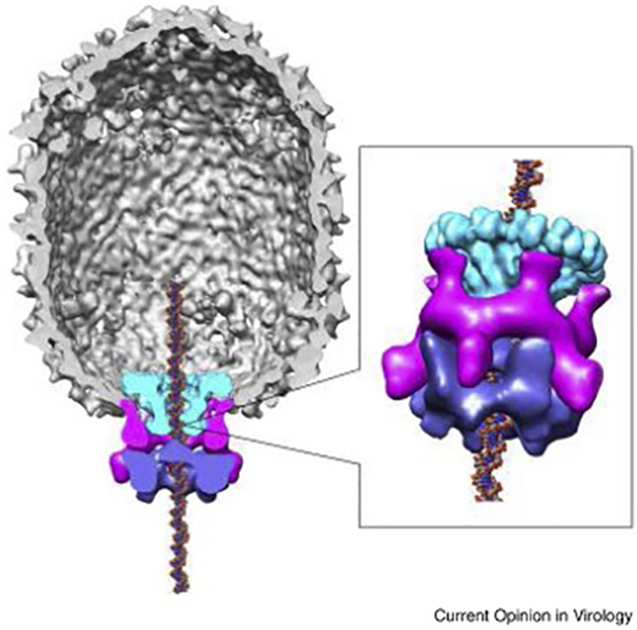
Architecture of the phi29 DNA packaging motor. The pentameric packaging ATPase, gp16, (blue) is anchored to the capsid through the pentameric prohead RNA (pRNA) scaffold (magenta) and pushes DNA through the dodecameric connector portal (green). Reprinted from Mao et al. 2016 with permission from Elsevier.
Virus DNA packaging
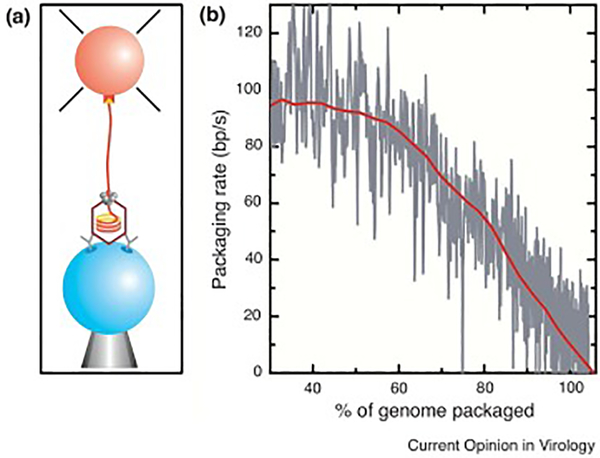
The first measurements of the physical parameters of DNA packaging for a viral system were achieved using single-molecule laser tweezers on the phi29 system by Smith et al. in 2000[11]. Adapted from a robust bulk in vitro assay, phi29 DNA translocation was observed to be highly processive and dynamic (Fig. 2). The first half of the full 19.6 kilobase complement of DNA moves at a rate of ~120bp/sec into the head shell. Thereafter the rate of packaging decreases noticeably, with the final segment moving at a snail’s pace of ~10bp/sec (Fig. 2b). The immediate inference was that this reduction in velocity was due to the motor encountering the resistive forces that are the result of DNA confinement. By emulating the response of the motor to increased load by pulling externally on the DNA in the laser tweezers, Smith, Tans, and colleagues calculated a resistive force curve relative to DNA filling, and estimated that a force of 80pN was required to drive the last of the DNA into the phi29 capsid. The magnitude of this force was unexpected, given that existing estimates of resistive force, derived from theoretical models that considered the stiff and self-repulsive nature of DNA, predicted a resistive force five-fold lower[12]. As a result of these early experiments, a flurry of theoretical models emerged to either validate or challenge these force calculations.
Fig. 2.
Early single-molecule measurements of phi29 DNA packaging using laser tweezers. (a) Packaging complexes were tethered between two microspheres, with the prohead/motor complex attached to one bead (blue) by antibodies against the capsid protein and the free end of the DNA attached to a second bead (orange) via a biotin-streptavidin linkage. (b) The change of tether length over time was used to calculate the change in packaging velocity over the course of head filling (grey trace is raw data; red trace is decimated and filtered). Reprinted from Chemla et al. 2005 with permission from Elsevier (a) and Smith et al. 2000 with permission from Springer Nature (b).
Nearly two decades later, a more complex picture of phi29 DNA packaging has emerged that sheds light on some of the assumptions made while interpreting the phenomena of reduced translocation velocity during head filling. Advanced, higher-resolution laser tweezers experiments in the Bustamante lab by Chemla, Moffit, Liu, and Chistol revealed the complex mechanochemistry of the phi29 packaging motor (Fig. 3). Much of the time the motor does not move the DNA, but rather holds the DNA static while the ATPase subunits exchange waste ADP for new ATP[13–15]. These protracted “dwells” take an average 125msec during early stages of filling when resistive forces are at their lowest, and end with a relatively rapid translocating “burst” where 10bp of DNA moves into the head in 10msec (hence the “real” rate of translocation is closer to 1000bp/sec). Given that much of the overall velocity of DNA packaging is dominated by the non-mechanical dwell, the change in the actual speed of DNA entry over the course of head filling was reassessed[16]. At the highest filling conditions assayed, it was found that the burst duration increased to ~80msec. Again, the internal resistive force was estimated by comparing the reduction in DNA movement velocity in response to external tensioning force, this time yielding a calculation of ~20pN. This value is much closer to that originally predicted by theory[12], thus reconciling the issue to this point. But whereas one discrepancy was addressed in the field, a new and more puzzling one emerged: what causes the composite speed of packaging to decrease so dramatically during head filling?
Fig. 3.
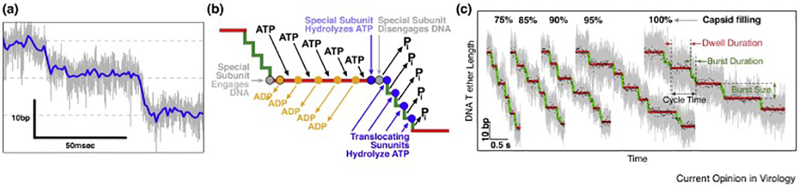
Mechanochemistry of the phi29 DNA packaging motor. (a) Plot of DNA tether length vs time showing the dwell-burst cycle of DNA packaging using high-resolution single-molecule laser tweezers (grey trace is raw data; blue trace is decimated and filtered). (b) The corresponding mechanochemical scheme showing the order and timing of the ATPase cycle within the motor ring and DNA movement, where ADP is exchanged for ATP during the static dwell (red line) and mechanical stepping (consisting of four 2.5 bp substeps) occurs during the burst (green line). (c) Changes in dwell and burst duration over the course of head filling. As the head fills, much of the reduction in DNA translocation rate is due to the lengthening of the static dwell (red lines) rather than the dynamic burst (green lines). Reprinted from Liu et al. 2014 with permission from Elsevier.
The answer, in part, resides in the enzymatic behaviour of the ATPase ring[15,16]. During low filling, when resistive forces are believed to negligible, the motor is able to cycle nucleotide substrate quickly, with a calculated Vmax of 120bp/sec and a Km of 30μM. As the head approaches becoming full, the Vmax drops to 10bp/sec and the Km to 10μM. Thus, the ratio of Vmax/Km goes from 4 to 1 bp/s/μM, in large part due to a reduction in ATP tight binding rate, which is the last step before entering the burst. It has been proposed that this is due to an allosteric signal from inside the head, possibly by or through the connector-portal complex, that reduces the nucleotide cycling efficiency of the ATPase. However, a more confounding question emerges: why does the motor slow down when it has ample force-generating capacity to continue moving DNA at a faster rate?
Insight into DNA behaviour
Over the past five years, evidence has emerged from parallel studies into the behaviour of DNA under confinement in the phi29 capsid that sheds light on this question. In studies conceived to determine the effects of charge screening on the resistive force opposing packaging and whether energy dissipates from the packaged DNA over time, Berndsen and Keller in the Smith lab began a series of experiments that reveal phenomena that provide insight into the slowing of the packaging motor.
The first was an experiment where DNA packaging was initiated in the laser tweezers and then halted by the introduction of a nucleotide analogue, gamma-S-ATP, into the motor causing it to stall[17]. After a range of times, the analogue was cleared and the velocity after the stall was measured and compared to the pre-stall measurement (Fig. 4). What was predicted was that, if the DNA relaxes inside the head after packaging, then the internal resistive force would decrease and velocity of translocation would increase relative to the speed just before the stall. This hypothesis was indeed supported, but the time-scale of relaxation observed was unexpectedly long. In most simulations, the rate of DNA relaxation is considered to be so fast that it is nearly irrelevant on the timescale of the viral DNA packaging process. The dissipation experiments from the Smith lab suggest that the time scale of relaxation for DNA under confinement on the order of minutes, over five orders of magnitude slower than used in simulations[12,18–20]. Arguably, DNA compacted to the high density seen in virus particles may in fact never completed reach an energy minimum, and is retained inside the head in a somewhat glassy state.
Fig. 4.
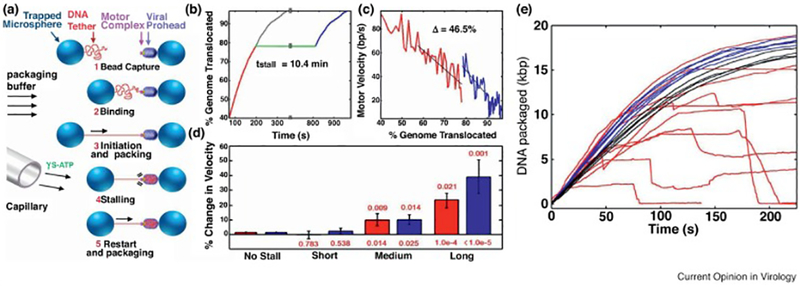
Long relaxation time-scale and jamming during DNA packaging. (a) Experimental design, where packaging is initiated in situ by bringing prohead motor complexes in close contact with tethered DNA (steps 1 and 2). After packaging is initiated and the rate of DNA translocation determined (step 3), the phi29 motor is reversibly stalled (step 4) and, after a prescribed time, restarted to determine if the rate of packaging has increased relative to the rate before the stall (step 5). (b) Measurement of DNA tether length over time taken before (red trace) and after (blue trace) the analogue induced stall (green trace). When the post-stall trace is transposed to the point of the stall (grey trace), the inflection in the trace reveals the increase in packaging velocity after the stall. (c) An example trace where the motor velocity before (red trace) and after (blue trace) the stall are plotted relative to the amount of head filling, showing that the motor increased in velocity by 46.5% due to the stall. (d) The length of the stall time corresponds to increase in packaging rate, for both packaging velocity (red) and motor velocity (blue; edited for motor pauses/slips), indicative of a time-dependent process of DNA relaxation during the stall. (e) Jamming appears with the addition of spermine as a DNA condensing agent can be seen when observing packaging over time. At low concentrations of spermine, packaging rate increases compared to controls (blue vs black traces). Higher concentrations of spermine causes abrupt decelerations and translocation failures (red traces). Reprinted from Berndsen et al. 2014 (a-d), and from Keller et al. 2014 with permission from Elsevier (e).
The second series of experiments from the Smith lab revealed another perspective on the issue. It is well understood that electrostatic screening of DNA charge reduces the repulsive force of charged phosphate backbones that are being forced together during packaging[21,22]. This was highlighted by a series of experiments where cations were varied in the phi29 packaging system and the force required for compaction showed a relative decrease when charge screening is high, i.e. in the presence of divalent magnesium, compared to less effective screening conditions, i.e. in the presence of monovalent sodium[23]. By extension, inclusion of more potent screening agents, such as cobalt hexamine or spermine, further reduced the force required to achieve high DNA density during filling[23,24]. In experiments designed to test the limits of this phenomenon (Fig. 4), higher levels of spermine were evaluated, but with an unexpected result: once spermine levels were increased to the point where the DNA could experience spontaneous condensation, rather than relieving the resistive force of charge repulsion in the virus head, stalling events appeared[24]. This “jamming” phenomena, interpreted as the result of a bolus of static DNA occluding the entrance of the head, is reminiscent of the broader jamming behaviour seen in macroscopic systems such as particle flow in piping systems. In hindsight, this observation is easy to rationalize, but revealed that some repulsion of the DNA inside the virus particle is required to prevent a terminal relaxation event, a final energy minimum, that occurs with condensation rather than compaction.
What emerges is a model whereby the complex physical behaviour of DNA places limits on packaging motor operation. It would seem preferable that, during virus assembly, the progression through the series of events required to produce virions be as swift as possible. However, the now apparent long time-scale of DNA relaxation during confinement requires that the DNA packaging event is throttled such that the packaged DNA is provided sufficient time to relax, thus preventing jamming.
Extending the model
These inferences are further supported by experiments on other dsDNA phage systems. The larger phages, rather than packaging a unit-length genome found in phi29, package DNA retrieved from long concatamers[1,25]: multiple genome lengths are linked as a result of a more complex DNA replication strategy employed by these phages. Therefore, during packaging, the amount of DNA packaged is determined, in part, by the capacity of the head. In these phages, packaging is “terminated” but the endonuclease action of the packaging ATPase in a sequence- dependent (ex. lambda) or sequence-independent (ex. T4) manner (hence the term “teminase” is applied to packaging machinery of these systems). It has been known for some time that such a head-full packaging mechanism relies on sensing of the amount of DNA in the capsid, ensuring that a chromosome of sufficient length to code for the entire genome is packaged. To this end, much effort has focused on the portal connector that is embedded in the head shell through which the DNA passes. Since the packaged DNA abuts the portal, and can therefore directly register the increasing pressure exerted by the DNA as the head fills, it may act as a pressure switch that allosterically signals the motor to slow[26–28]; it has been shown that mutations in the portal of phages P22 and SPP1 can alter the spatial timing of the termination event, since these mutants package shorter chromosomes than wild type.
Measurement of packaging velocity in the coliphage lambda[29] and T4[30] systems showed that these phages packaged DNA significantly faster than phi29. Phages lambda and T4, whose DNA are ~2.5X and 8X the length of phi29’s, respectively, package DNA faster than phi29 by roughly the same scale. The result is that packaging for all of these phages takes ~3 minutes in vitro, even though they range in chromosome size from less than 20kb to over 160kb. Hence, it is tempting to speculate that the time scale of packaging is determined not by the resistive force and work involved, but rather the time required for the DNA to relax upon and during confinement; this appears to be independent of the length of the viral chromosome since the length of the DNA scales proportionally with the size of the capsid receptacle. Although not yet measured experimentally, it is possible that DNA translocation during early filling in T4, which occurs at a velocity similar to the actual translocation rate measured in phi29, i.e. 1000bp/sec, can operate without the need for the throttling behaviour seen in phi29 that appears manifest in the relatively long and regulated dwell phase of the packaging mechanism; phage lambda would be predicted to fall between phi29 and T4, with a similar translocation velocity and intermediate dwell time. It also remains to be seen whether the phages with larger genomes such as lambda and T4 reduce their DNA translocation rate at high filling via an allosteric mechanism that reduces the rate of nucleotide cycling seen in phi29. As well as giving the DNA enough time to relax inside the head shell, this regulatory mechanism, a “head-filling sensor”, may play a role in the critical termination event not found in phi29. It remains to be determined whether and how these events are related, or whether the sensing function in phi29, with its unit length chromosome, has lost the signaling for filling that determines chromosome length and retained the throttling function or acquired a throttling function by other means.
Conclusion
Taken together, this series of experiments and observations reveal a complex physics at the submicroscopic level that DNA viruses experience, and have thus adapted to in order to produce infectious virus particles that can deliver DNA from cell to cell. Although speed would generally be considered an asset in terms of completing an assembly reaction to produce infectious virus during the short time scale of infection, the physical behaviour of the DNA demands patience. That such a fundamental observation of the time-scales of DNA relaxation be revealed by observing a biological system is compelling.
Funding
This work was supported by the National Institutes of Health (R01GM122979, R01GM071552 and R56AI127809) and the National Science Foundation (MCB 1715293).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rao VB, Feiss M: Mechanisms of DNA Packaging by Large Double-Stranded DNA Viruses. Annu Rev Virol 2015, 2:351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An excellent introduction to the field of dsDNA viral packaging.
- 2.Speir JA, Johnson JE: Nucleic acid packaging in viruses. Curr Opin Struct Biol 2012, 22:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia HG, Grayson P, Han L, Inamdar M, Kondev J, Nelson PC, Phillips R, Widom J, Wiggins PA: Biological consequences of tightly bent DNA: The other life of a macromolecular celebrity. Biopolymers 2007, 85:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knobler CM, Gelbart WM: Physical Chemistry of DNA Viruses. Annu Rev Phys Chem 2009, 60:367–383. [DOI] [PubMed] [Google Scholar]
- 5.Chemla YR, Smith DE: Single-molecule studies of viral DNA packaging. Adv Exp Med Biol 2012, 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimes S, Jardine PJ, Anderson D: Bacteriophage φ29 DNA packaging. Adv Virus Res 2002, 58:255–280. [DOI] [PubMed] [Google Scholar]
- 7.Iyer LM, Makarova KS, Koonin EV., Aravind L: Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 2004, 32:5260–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao H, Saha M, Reyes-Aldrete E, Sherman M, Woodson M, Atz R, Grimes S, Jardine P, Morais M: Structural and Molecular Basis for Coordination in a Viral DNA Packaging Motor. Cell Rep 2016, 14:2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG: Defining Molecular and Domain Boundaries in the Bacteriophage φ29 DNA Packaging Motor. Structure 2008, 16:1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A: Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci U S A 2011, 108:7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith DE, Bustamante C, Tans SJ, Smith SB, Grimes S, Anderson DL: The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 2001, 413:748–752. [DOI] [PubMed] [Google Scholar]; • The first single-molecule laser tweezers study of viral DNA packaging
- 12.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM: DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci 2001, 98:13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Early comprehensive modelling study of DNA confinement during packaging.
- 13.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C: Mechanism of force generation of a viral DNA packaging motor. Cell 2005, 122:683–692. [DOI] [PubMed] [Google Scholar]
- 14.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C: Intersubunit coordination in a homomeric ring ATPase. Nature 2009, 457:446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C: High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell 2012, 151:1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, Bustamante C: A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 2014, 157:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Extensive single-molecule studies of viral packaging motor coordination and DNA movement across the head-filling regime.
- 17.Berndsen ZT, Keller N, Grimes S, Jardine PJ, Smith DE: Nonequilibrium dynamics and ultraslow relaxation of confined DNA during viral packaging. Proc Natl Acad Sci U S A 2014, 111:8345–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Single-molecule laser tweezers study of relaxion of DNA inside the viral capsid revealing the time-scale of DNA relaxation is orders of magnitude slower than assumed.
- 18.Forrey C, Muthukumar M: Langevin dynamics simulations of genome packing in bacteriophage. Biophys J 2006, 91:25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrov AS, Harvey SC: Structural and Thermodynamic Principles of Viral Packaging. Structure 2007, 15:21–27. [DOI] [PubMed] [Google Scholar]
- 20.Ali I, Marenduzzo D, Yeomans JM: Polymer packaging and ejection in viral capsids: Shape matters. Phys Rev Lett 2006, 96:208102. [DOI] [PubMed] [Google Scholar]
- 21.Bloomfield VA: DNA condensation by multivalent cations. Biopolymers 1997, 44:269–282. [DOI] [PubMed] [Google Scholar]
- 22.Rau DC, Parsegian VA: Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys J 1992, 61:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE: Ionic effects on viral DNA packaging and portal motor function in bacteriophage φ29. Proc Natl Acad Sci U S A 2007, 104:11245–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller N, Grimes S, Jardine PJ, Smith DE: Single DNA molecule jamming and history-dependent dynamics during motor-driven viral packaging. Nat Phys 2016, 12:757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Single-molecule study of jamming behaviour during packaging in the presence of DNA-condensing ions that provide evidence that DNA charge repulsion is not simply a barrier but also permits DNA rearrangement during packaging.
- 25.Feiss M, Rao VB: The bacteriophage DNA packaging machine. Adv Exp Med Biol 2012, 726:489–509. [DOI] [PubMed] [Google Scholar]
- 26.Lokareddy RK, Sankhala RS, Roy A, Afonine PV., Motwani T, Teschke CM, Parent KN, Cingolani G: Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat Commun 2017, 8:14310. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Recent structural work examining the role of the dsDNA viral portal in packaging.
- 27.Tavares P, Zinn-Justin S, Orlova EV.: Genome gating in tailed bacteriophage capsids. Adv Exp Med Biol 2012, 726:585–600. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira L, Tavares P, Alonso JC: Headful DNA packaging: Bacteriophage SPP1 as a model system. Virus Res 2013, 173:247–259. [DOI] [PubMed] [Google Scholar]
- 29.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE: Measurements of Single DNA Molecule Packaging Dynamics in Bacteriophage λ Reveal High Forces, High Motor Processivity, and Capsid Transformations. J Mol Biol 2007, 373:1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE: Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci 2007, 104:16868–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]