Abstract
Background
Allergic disease is suspected to play a role in the development of childhood acute lymphoblastic leukemia (ALL). Studies conducted over the last several decades have yielded mixed results.
Methods
We examined the association between allergy, a common immune-mediated disorder, and ALL in the California Childhood Leukemia Study (CCLS), a case-control study of 977 children diagnosed with ALL and 1037 matched controls (1995–2015). History of allergies in the first year of life was obtained from interviews, mainly reported by mothers. Logistic regression analyses were conducted to estimate odds ratios (ORs) and 95% confidence intervals (CIs), controlling for birth order, day care attendance, and mode of delivery. In addition, we conducted meta-analyses with data from the CCLS and 12 published studies and employed a new method to estimate between-study heterogeneity (R_b).
Results
Overall, no associations were observed between childhood ALL risk and specific allergy phenotypes or any allergy, as a group. However, having any allergy was associated with an increased risk of ALL among the youngest study participants. In the meta-analysis random-effect models, a reduced odds of ALL was associated with hay fever (metaOR=0.65, 95% CI: 0.47, 0.90); however, restricting the analysis to studies that used medical records for assessment of allergy or recently published studies, led to null or attenuated results.
Conclusions
Overall, our findings do not support a clear association between allergy and childhood ALL.
Impact
The degree to which epidemiological studies can inform the relationship between allergies and risk of childhood ALL is limited by between-study heterogeneity.
INTRODUCTION
Childhood acute lymphoblastic leukemia (ALL) is a cancer of lymphoblasts, which are precursor immune cells. ALL is thought to arise through immune dysregulation, beginning in utero and extending throughout early life (1). Because allergies are also believed to result from aberrant immune function (i.e. an immune response to innocuous antigen), they have long been suspected as potential causes of childhood ALL (2). In the early 1950s, a case report suggested allergy could cause acute bone marrow injury, resulting in childhood leukemia (3). This report precipitated decades of research on the subject, ultimately yielding inconsistent results.
Epidemiologic evidence of shared risk factors between allergic disease and ALL as well as coinciding upward trends in their incidence suggest a relationship between the two conditions (4, 5). Surrogates of early life exposure to normal antigenic challenge, including a higher birth order (6, 7), daycare attendance (8, 9), and vaginal birth (10, 11), are protective against both ALL and allergy. In fact, these observations form the basis of the leading etiologic hypothesis for both diseases, the so-called “hygiene hypothesis”.
The hygiene hypothesis postulates that a lack of early-life exposure to normal immune challenges leads to aberrant immune system responses later in life. This hypothesis was initially employed to suggest that improvements to sanitation in the developed world during the 20th century led to the emergence of allergic diseases (12). Subsequently, Greaves et al. adapted this hypothesis by proposing that “delayed infection” early in childhood, followed by a hyper-responsiveness to infectious challenge later in childhood, generates the genetic mutations necessary to develop childhood ALL (13). The hypothesis implies that the two diseases – allergy and ALL – have related biological mechanisms.
Two separate but overlapping meta-analyses by Dahl et al. in 2009 (14) and Linabery et al. in 2010 (15) have been conducted on allergy and the risk of childhood ALL. Though the quantitative results of the two meta-analyses were nearly identical, disparate interpretations of the results by the two author teams (suggestion of causation and bias, respectively) leave the existence of a relationship between allergy and ALL in question. Here, we examine the association between allergy in the first year of life and the risk of ALL in children enrolled in the California Childhood Leukemia Study (CCLS). In addition, we update previous meta-analyses with four new epidemiologic studies of allergy and childhood leukemia published since 2010 (16–19) including results from the present study, increasing the previous sample from 30,763 to 40,123 children and including the largest medical record-based study conducted to date on this topic. Further, we provide a statistical evaluation of factors contributing to between-study heterogeneity using meta-regression. To estimate the percent variance due to between-study heterogeneity, we implement a new method with an alternative estimator that does not violate the assumption of constant within study variance, which is required for the I2% measure used in previous meta-analyses and is almost never met (20). Our methods and the addition of important new studies allow us to comprehensively evaluate the association of this challenging exposure with ALL risk.
MATERIALS AND METHODS
CCLS analysis
Study population
The CCLS is a case-control study conducted in California between 1995 and 2015 (21). Case ascertainment was rapid, usually within 72 hours from diagnosis, and ~84% of incident childhood leukemia cases were consented from participating hospitals. Eligibility criteria for cases and controls were as follows: 1) living in the study area at the time of recruitment; 2) having an English or Spanish-speaking parent; 3) being less than 15 years old at diagnosis or reference date for controls; and 4) having no previous diagnosis of cancer. From 1995 to 2008, one or two controls were selected using birth certificate information from the Office of Vital Records, and individually matched to cases on date of birth, sex, child’s Hispanic ethnicity, and maternal race. No controls were enrolled from 2009 to 2015, and the overall participation rate was ~68%. A summary of participation rates in the CCLS by study phase is summarized in Supplemental Table S1. The University of California, Berkeley’s Institutional Review Board, California Department of Health Services, and all participating clinical institutions approved the study. Informed consent was obtained from all study participants.
Sample size
A total of 1477 leukemia cases and 1226 controls were eligible to participate and interviewed. Participants with non-ALL leukemia (n=40), missing exposure or covariate data (n=252) or Down syndrome (n=57) were excluded. To ensure that all allergies occurred prior to the leukemia prodrome, participants whose date of diagnosis or reference date was less than 18 months after birth were excluded (n=153), leaving a minimum time interval of at least 6 months between potential exposure (allergy in the first year of life) and disease diagnosis. The final sample size was 977 ALL cases and 1073 controls (of which 653 were matched ALL/control pairs.
Exposure classification
We examined allergic diseases occurring in the first year of life as reported through interview with the biological parents (~98% mothers), wherein the parent was asked if the child had allergies, what the child was allergic to, and whether or not a physician was consulted. See the Supplementary Methods and Materials for structure of allergy-related questions by recruitment phase from the CCLS questionnaire. Allergens were categorized as hay fever (defined here as allergy to plants/grass/dust/mold), food, drugs and medications, soap/cosmetics, animal/insect, and other. Variables were constructed for any allergy (binary, defined as at least one of the above allergies) and total count of reported allergens. A total of 41 allergic participants were excluded from specific allergy phenotype analyses because the specific allergen(s) was unknown. A variable combining allergy to food and drugs was constructed for use in the subsequent meta-analysis. Of the resulting allergy phenotype categories, those with less than 10 exposed controls were not tested for association with disease (including animal/insect allergy and soap/cosmetic allergy).
Statistical Analysis
Matched and unmatched data analyzed with logistic regression analyses were conducted to estimate the odds ratios (OR) and 95% confidence intervals (CI). Individually matched and unmatched results were similar, therefore only those from the unmatched analyses using the entire set of available cases and controls are presented for increased statistical power. All models were adjusted for child’s age (centered on median age=4.61), sex, Hispanic ethnicity, maternal race, and for possible confounders determined a priori as being associated with both ALL and allergic disease through a literature search [i.e. mode of delivery(11, 22), birth order(6, 23), daycare attendance in the first year of life(8, 9), and household income]. Independent models included an interaction term to test for potential effect modification by age at diagnosis, sex, child’s Hispanic ethnicity, mode of delivery, and day care attendance. Models including interaction terms where the p-value for interaction was < 0.2 were then compared to the base model (no interaction term) for goodness-of-fit using the likelihood ratio test. For models in which the likelihood ratio test produced a p-value < 0.05, the marginal effects of allergy phenotype on ALL risk, derived from the model including the interaction term, are presented. To address potential control-selection bias potentially introduced by not recruiting controls to the study from 2009–2015, analyses were repeated excluding cases recruited during that time period; the results did not change (unpublished observation).
Meta-analysis
Search criteria
Using PubMed, we identified all epidemiologic studies published between November 1, 2008, the date of the first published meta-analysis(14), and June 1, 2016 using the following search terms: MeSH terms = “leukemia” OR “leukemia, lymphoid” AND “allergy and immunology” OR “hypersensitivity”; All Fields = “atopy” OR “atopic” OR “hypersensitivity” OR “allergy” OR “allergy and immunology” OR “allergic” AND “leukemia”. Results were filtered by age to include only children (birth to 18 years). In addition, all eligible and ineligible studies reported in the 2009 and 2010 meta-analyses (including unpublished data) were retrieved for the present study.(14, 15)
Study eligibility
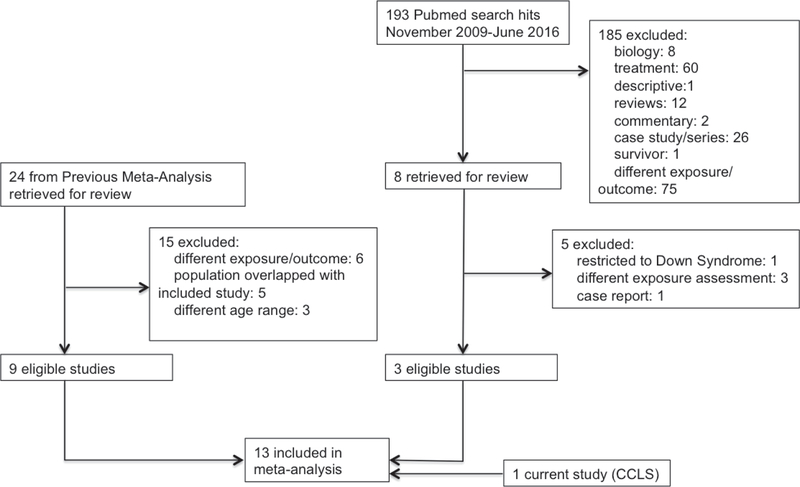
Studies eligible for the present meta-analysis included original reports, written in English, investigating the relationship of childhood ALL (age 18 and under) and allergy, asthma, or atopic diseases. Only studies presenting odds ratios and confidence intervals/standard error were included. If multiple studies utilized overlapping study populations, the study with the largest sample size was chosen for increased power. Reasons for exclusion included incompatible methods of exposure measurement, such as serum level IgE(24), restriction of studies to specific subgroups (e.g., Down syndrome(25), inclusion of individuals age ≥18(26–28)), and overlap with included studies (Figure 1).
Figure 1:
Flow chart - Meta-analysis study search and selection process. Application of selection criteria resulted in 13 studies to be included in the meta-analysis.
Data extraction
Information on authors; publication year; study design; assessment method for exposure, outcome, and covariates; and measures of association and standard error was extracted from eligible publications using a standardized form. We extracted six binary exposure variables including any allergy, eczema, asthma, hay fever, food/drug allergy, and hives in addition to the binary outcome of ALL. For studies presenting results using multiple latency periods between exposure and disease (29) or multiple methods of exposure assessment (30), reported results with the highest precision were included to improve statistical power.
Statistical analyses
Using the Metafor and hetmeta packages (20, 31) in R (32), summary associations between ALL risk and allergy phenotypes were estimated using data from 4 to 13 studies.
An inverse-variance-weighted (DerSimonian-Laird) random-effects model was used to estimate summary odds ratios for each allergy phenotype. A new estimator published in Crippa et al. (2016) was used to estimate the percent of variation attributable to between-study heterogeneity (R_b%), rather than the typical I2%, to reduce bias in the estimate. The p-value for heterogeneity was also estimated using Cochran’s Q test. Study-level characteristics, referred to as moderators, thought to potentially contribute to heterogeneity were selected and abstracted prior to analyses. These include medical record vs. self-reported exposure assessment, nested case-control vs. other study design, ≥80% vs. <80% control response rate, hospitalized vs. healthy controls, specified vs. not specified latency period, and publication year. Potential moderators were tested for correlation using an a priori determined Pearson correlation coefficient r2 threshold of 0.3 (33). Among correlated moderators, the one that could be used to characterize the most studies was selected for inclusion in the mixed-effects models. Mixed-effects models were constructed when significant heterogeneity was detected for models of ALL risk with any of the allergy phenotypes (R_b>0). Separate models were built for each potential moderator in turn and a full model including all uncorrelated moderators was constructed. Stratified measures of effect and associated p-values, PQm, were estimated from the omnibus test of model coefficients. Because the CCLS study only measured allergy exposure during the first year of life, analyses were also conducted excluding the CCLS study to assess potential sources of bias.
Publication bias was formally assessed for analysis of any allergy via a regression test of funnel plot asymmetry.
RESULTS
CCLS analysis
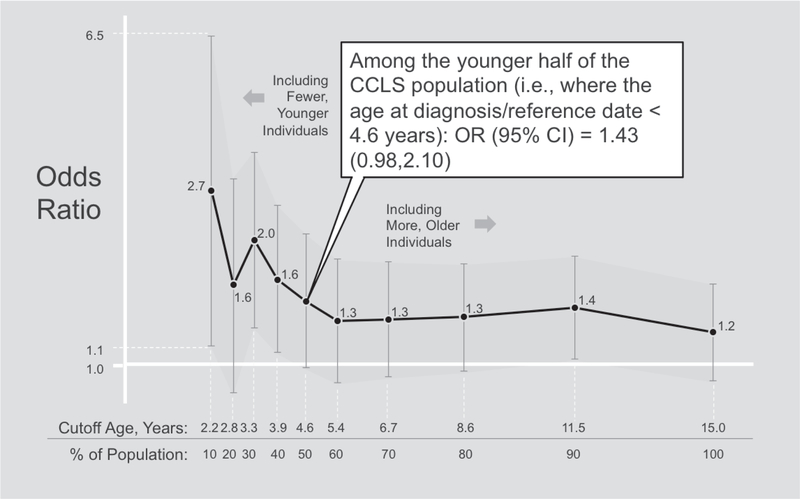
In general, controls were comparable to cases except for the fact that controls tended to be of higher income than cases (Supplemental Table S2). The odds ratios for any allergy and most allergy phenotypes ranged from 0.75 (hay fever) to 1.29 (any allergy), but there was no strong statistical evidence of association overall (Table 1). However, multiplicative interaction was detected between any allergy and age at diagnosis/reference date. Stratified analyses show that risk of ALL was associated with occurrence of any allergy during the first year of life only amongst the youngest study subjects (Figure 2). Food allergy and allergy count were also suggestive of interaction with age, though the goodness of fit test for including the interaction term did not meet the significance threshold P<0.05.
Table 1.
Odds ratios and 95% confidence intervals for ALL cases vs. controls who reported having allergy to specific allergens in the first year of life in the CCLS
| Allergen | Cases (n=977) | Controls (n=1073) | Odds Ratioa | 95% Confidence Interval | P-Value | |
|---|---|---|---|---|---|---|
| Any Allergy | No | 827 | 942 | |||
| Yes | 128 | 119 | 1.29b | 0.97,1.72 | 0.08 | |
| Hay Fever | No | 927 | 1020 | |||
| Yes | 22 | 30 | 0.75 | 0.42,1.32 | 0.32 | |
| Food | No | 906 | 995 | |||
| Yes | 40 | 45 | 1.09 | 0.69,1.71 | 0.71 | |
| Drugs and Medications | No | 909 | 1011 | |||
| Yes | 38 | 26 | 1.63 | 0.96,2.75 | 0.07 | |
| Food/Drug | No | 873 | 970 | |||
| Yes | 74 | 70 | 1.24 | 0.87,1.76 | 0.24 | |
| Allergy Count | No | 851 | 954 | |||
| 1 Allergen vs. None | Yes | 88 | 91 | 1.08 | 0.79,1.49 | |
| 2+ Allergens vs. None | Yes | 16 | 16 | 1.24 | 0.60,2.56 | 0.56c |
Adjusted for age, sex, maternal race, child’s Hispanic status, income, mode of delivery, daycare attendance, and birth order
Marginal effect where model additionally includes interaction term median-centered age*any allergy. See figure 2 for results from age strata
Test for linear trend
Figure 2:
Sensitivity analysis testing the association of any allergy and ALL risk among the youngest individuals in the CCLS by decile. Models were adjusted for sex, maternal race, child’s Hispanic status, income, mode of delivery, daycare attendance, and birth order. Text annotation box provides example interpretation for stratification at the 50th percentile.
Meta analysis
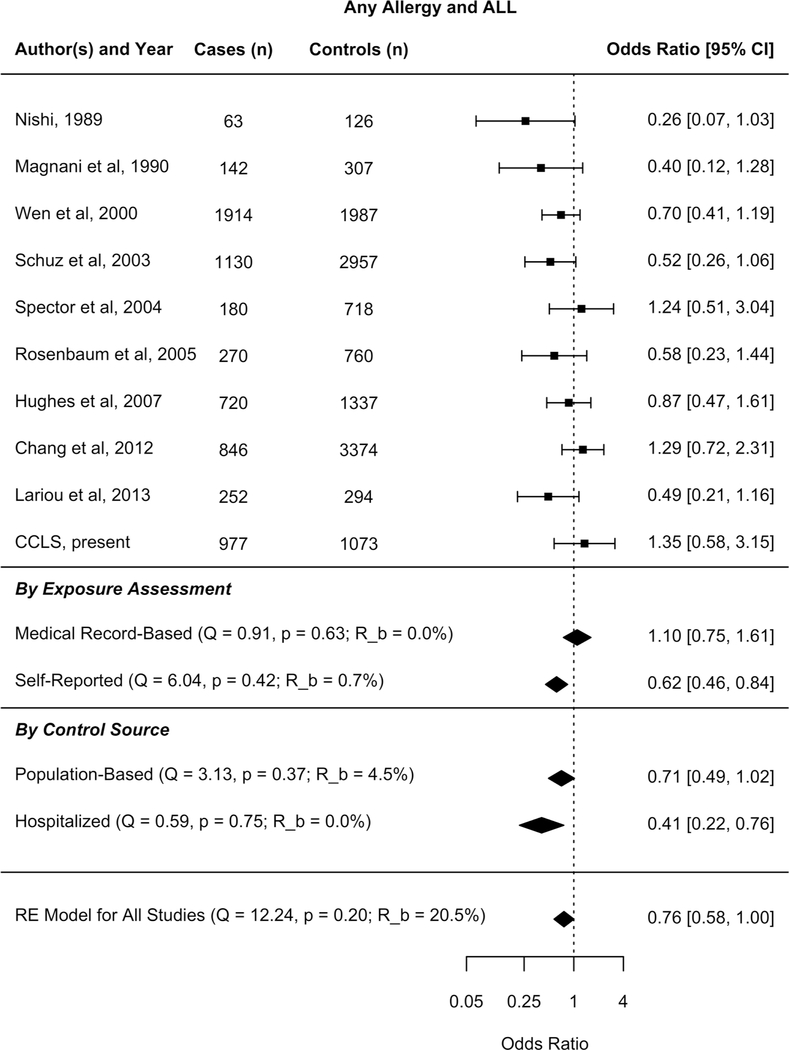
Characteristics of the 13 studies included in the meta-analysis are listed in Supplemental Table S3 (16, 18, 19, 23, 30, 34–40). Thirteen studies reported results specific to ALL and sample sizes ranged from less than 200 to several thousands (Figure 3).
Figure 3:
Forest plots - Any Allergy and Acute Lymphoblastic Leukemia random-effects model, meta-regression medical record vs. self report, and meta-regression hospitalized vs. population-based controls
Results from the random-effects models are shown in Figure 3 and Table 3 for any allergy and specific allergy phenotypes, respectively. Child’s history of hay fever was the sole allergy to be associated with a reduced risk of ALL (OR 0.65, 95% CI: 0.47, 0.90). No statistical evidence of association was seen for other types of allergies.
Table 3.
Results from 3 Random Effects Meta-Regression Models in the Association Between Any Allergy and ALL
| Model | Summary Beta | 95% CI | P value | R_b% | PQm | |
|---|---|---|---|---|---|---|
| Model 1 | ||||||
| Intercept | −0.46 | −0.76, −0.16 | 0.003 | 0 | 0.026 | |
| Medical Record | 0.55 | 0.07, 1.04 | 0.026 | |||
| Model 2 | ||||||
| Intercept | −0.98 | −1.63,−0.34 | 0.003 | 0 | 0.017 | |
| Publication year | 0.04 | 0.01,0.07 | 0.017 | |||
| Model 3 | ||||||
| Intercept | −0.98 | −1.63, −0.34 | 0.003 | 0 | 0.020 | |
| Medical Record | 0.41 | −0.10, 0.92 | 0.114 | |||
| Publication year | 0.03 | −0.003,0.07 | 0.073 |
CI, confidence interval; I2%, percent of total variability due to heterogeneity; PQm, P-value for Cochran’s test of Moderators
About 22% of the variability in effect estimates for any allergy is due to heterogeneity (R_b CI: 0,79%), while no heterogeneity was reported within allergy types (Table 2). To formally assess potential study-level characteristics contributing to heterogeneity of any allergy and ALL, a meta-regression was performed using a priori selected, uncorrelated potential moderators, including medical record vs. self-reported exposure assessment. Medical record-based studies (Figure 3) were associated with an attenuated relationship between any allergy and ALL risk in the mixed-effects model (PQm <0.05, Model 1, Table 3). Model 1 was then applied to four allergy phenotypes and the same trend was observed; i.e., the odds ratio for ALL risk associated with each of the allergy phenotypes was larger (and consistently null) in medical record-based studies compared to non-record based studies (Supplementary Figure 1). However, with fewer studies analyzed by subcategory, none of the formal tests of moderators (PQm) were statistically significant (unpublished observation).
Table 2.
Results of Random Effects Meta-Analyses for Various Allergic Exposures and ALL Risk from Current and Prior Meta-Analyses
| Any Allergy | Eczema | ||||||||
| Author | Year | N | Summary OR | 95% CI | I2% or R_b% | N | Summary OR | 95% CI | I2% or R_b% |
| Dahl et al | 2009 | 8 | 0.67 | 0.54,0.82 | NR | 5 | 0.68 | 0.56,0.83 | 29 |
| Linabery et al | 2010 | 6 | 0.69 | 0.54,0.89 | 80 | 5 | 0.74 | 0.58,0.96 | 62 |
| Present study | 10 | 0.76 | 0.58,1.01 | 22 | 7 | 0.86 | 0.64,1.14 | 0 | |
| Hay Fever | Food/Drug | ||||||||
| Author | Year | N | Summary OR | 95% CI | I2% or R_b% | N | Summary OR | 95% CI | I2% or R_b% |
| Dahl et al | 2009 | 5 | 0.53 | 0.43,0.65 | 28 | ||||
| Linabery et al | 2010 | 3 | 0.55 | 0.46,0.66 | 3 | ||||
| Present study | 8 | 0.65 | 0.47,0.90 | 0 | 6 | 0.73 | 0.52,1.03 | 0 | |
| Asthma | Hives | ||||||||
| Author | Year | N | Summary OR | 95% CI | I2% or R_b% | N | Summary OR | 95% CI | I2% or R_b% |
| Dahl et al | 2009 | 6 | 0.82 | 0.63,1.10 | 43 | ||||
| Linabery et al | 2010 | 7 | 0.79 | 0.61,1.02 | 44 | 2 | 0.93 | 0.73,1.19 | 0 |
| Present study | 9 | 0.85 | 0.63,1.16 | 0 | 4 | 1.23 | 0.80,1.87 | 0 | |
CI, 95% confidence interval; I^2%, percent of total variability due to heterogeneity; OR, odds ratio; N, number of included studies; R_b%, percent of total variability due to heterogeneity
We furthered explored whether control selection method added heterogeneity to the meta-analysis. Within studies where exposure assessment was self-reported, the odds ratio for ALL risk associated with any allergy was larger (null) in studies with population-based controls compared to studies with hospital-based controls; however the mixed-effects model comparing studies with hospital-based controls (3 studies) to those with population-based controls (6 studies) produced a test of moderators statistic that was not statistically significant (PQm =0.12)(Figure 3).
Finally, we hypothesized that publication year could contribute to between-study heterogeneity because the incidence rates of allergy and ALL are both changing over time, study quality is improving, and public perception of allergy is changing (41, 42). In a univariate mixed-effects model, publication year was associated with an increase in the odds ratio relating any allergy to ALL (PQm <0.05, Model 2, Table 3), indicating that early studies tended to report an inverse relationship but more recent studies have reported null or positive associations (Supplementary Figure 2). In the full model containing both the medical record indicator and publication year (Model 3, Table 3) the independent effects of medical-record exposure assessment and publication year remain compelling, though no longer reach statistical significance. All three models applied reduced the residual between-study heterogeneity estimate (R_b%) to 0.
The observed asymmetry in the funnel plot estimated using the random-effects model suggested evidence of publication bias for studies examining the relationship between any allergy and ALL, (Supplementary Figure 3, P=0.06). There were not enough studies that included specific allergy phenotypes to formally assess publication bias by subcategory.
DISCUSSION
Based on the CCLS study and the largest meta-analysis to date -- combining data for over 8000 ALL cases and 25,000 controls – our findings suggest there is no clear association between allergy and the risk of ALL. However, several observations have emerged from our analyses, raising methodological concerns and biological issues discussed below.
In the CCLS, we observed an increased risk of ALL given any allergy occurring in the first year of life. However, age-stratified analyses showed that the youngest study participants drove this association. It was more common for allergy to precede ALL when diagnoses occurred at 1.5 −3 years of age than if disease arose later in life. Allergy beyond the first year of life was not assessed in the CCLS and so it is unclear whether allergy occurs more frequently in cases proximal to diagnosis at any age, or if this phenomenon is limited to the youngest cases. Results from a large, record-based study in Taiwan support the hypothesis that any allergy is more common proximal to diagnosis across the life course, where compared to allergy in the first year of life and >1 year before diagnosis, allergy <1 year before diagnosis conferred the highest risk of childhood ALL (18). The association could also be explained by differential recall, where parents of younger children at the time of interview could more accurately recall allergy occurring in the first year of life than parents of older children.
Early-onset allergy to drugs or medications conferred a larger increase in the risk of ALL than any other specific allergy phenotype evaluated in the CCLS population, although the association was not statistically significant. While potentially a chance finding, there are several factors that could be contributing to this observation. First, true drug hypersensitivities have the potential to influence the allergy - ALL association. However, many adverse drug reactions are not immune-mediated and further, those that are are heterogeneous in type and can be IgE-, IgG-, or T cell-mediated (43) and may have differential effects on ALL etiology. If a causal relationship exists between allergies to drugs and ALL further functional studies are needed for full characterization. The relatively high odds ratio for ALL associated with an allergy to drugs or medications could also be due to confounding. We could not control for the number or type of medications taken in the first year of life, which was likely higher in cases than controls, yielding more opportunity for cases to develop a drug allergy that is causally-independent of ALL. This line of reasoning is supported by the observation that cases experience more frequent infection-related medical office visits prior to leukemia onset than controls, in both self-report (17) and medical record-based studies (29, 44) of ALL; and are therefore more likely to be exposed to a broader spectrum of medications. There is little epidemiologic evidence from previous studies to support an association between ALL and allergy to drugs per se, as previous studies have typically examined a combined food/drug allergy phenotype (36, 37, 40).
The CCLS population is large and unique. However, the study also presents some limitations. Selection bias is of concern in the CCLS, wherein participating controls are of higher socioeconomic status than cases and non-participating controls (45). Higher socioeconomic status has been previously associated with increased odds of allergic disease in children (46), potentially biasing our results in the direction of an inverse association. Parental interview assessed only allergy occurring in the first year of life. Allergic disease is relatively rare in infants (5) and while it may be a useful measure of early-life immune dysregulation, low exposure prevalence in our study limited the power to detect statistically significant associations with ALL. Further, certain allergies, like hay fever, can be difficult to diagnose in very young children. This restriction on exposure assessment may also limit the generalizability of our results to the childhood allergy – ALL relationship overall. However, Chang et al. (2012) also reported null associations between allergy phenotypes occurring before age one and ALL, with the exception of an increased risk for atopic dermatitis, an exposure that was not included in the CCLS questionnaire. A limitation of our study is that we do not have medical record data for comparison and the contribution of uncontrolled confounding to our result cannot be ruled out. However, we believe that several analytic advantages may have contributed to the discrepancy between previous studies and ours. First, we incorporated a latency period into our study, wherein we excluded cases and controls for which allergy occurred within 6 months of leukemia diagnosis or reference date, respectively. Thus, we reduced the potential that our observed associations were due to reverse causality. Second, our study controlled for shared risk factors for allergy and ALL, including birth order, day care attendance, and mode of delivery, which potentially confounded the results from previous studies.
In this updated meta-analysis, most associations with allergy were attenuated, compared to previous meta-analyses(14, 15), and only hay fever maintained a statistically significant inverse association with ALL. With the addition of recent studies including a large, medical-record based analysis, it is clear that between-study heterogeneity limits the interpretation of any summary measure of effect. Hay fever is the most common allergy under study (5) but is also the most commonly misdiagnosed (47), as its symptoms are similar to and easily mistaken for upper respiratory tract infections and vice versa. Using any allergy as an exposure variable allowed for inclusion of the greatest number of studies for meta-analysis and thus allowed us to statistically assess study-level sources of heterogeneity for the first time. Medical-record exposure assessment or correlated features (record-based or nested design, incorporation of a latency period, hospital vs. healthy controls, and control response rate) were important sources of between-study heterogeneity. Our meta-regression (Model 1) showed that, in medical record-based studies, no association was observed between allergy of any type and ALL; and within self-report-based studies, any allergy and hay fever resulted in statistically significant inverse relationships. A parallel pattern has emerged with the association of early-life infection and ALL, wherein questionnaire-based studies have found an inverse association between infection and ALL and medical-record based studies have found infection to be a risk factor (1). This difference in association is possibly the result of reporting bias, which may also be occurring in the allergy-ALL relationship. Linabery et al. argued that misclassification of the allergy phenotype is likely occurring in both the case of parental report and medical record; however, the record-based studies are more likely to be non-differential with respect to case status, and thus less subject to bias. Further, a recent systematic review showed a large and significant difference in prevalence of childhood atopic disorders comparing those diagnosed by a physician to parental-report (48). For example, the annual prevalence of allergic rhinitis according to physician report ranged from 0.4–4.1% whereas the self-reported annual prevalence ranged from 15.1–37.8% (41). The authors speculate that consistent under-reporting among general medical practitioners and over-reporting by parents create this difference. There is thus potential for self-report studies of allergy and childhood leukemia to produce study results that have both higher power and higher susceptibility to bias, potentially explaining the strong inverse associations between the two diseases in self-reported studies, whereas medical record-based studies present more attenuated relationships.
Within studies using self-reported exposure assessment, the test for heterogeneity between studies using population-based controls and hospital-based controls was not significant, supporting our decision to include both types of study. However, utilizing hospitalized controls does exaggerate the inverse relationship between allergy and ALL among self-reported studies, possibly due to control selection bias, as potential controls receiving treatment for allergy-related symptoms were included in some of these studies and not in others.
Another significant source of heterogeneity is publication year. Over time, the strong inverse association between allergy and ALL is shifting towards no association, with some of the most recent studies finding a positive association between the two diseases. Several factors could contribute to this observation, including improvements in study quality over time, changing prevalence of both allergy and ALL, and potentially changing opinions or knowledge regarding allergy and hygiene among the general public (49, 50).
Numerous sources of bias complicate interpretation of these results. Results from the CCLS and another recent study (18) suggest that the timing of allergy relative to ALL diagnosis may modify observed associations. Across the 13 studies included in the meta-analysis, timing of exposure assessment relative to diagnosis was largely consistent. Thus, tests of between-study heterogeneity related to timing of allergy could not be assessed, though it may be contributing to the underlying variation in effect estimates. Publication bias may be limiting the interpretability of our meta-analysis. Within questionnaire-based studies, questionnaire structure can result in differential parental response between studies. For example, a questionnaire may ask whether allergy was ever diagnosed by a physician, or have only asked about a specific subset of allergens, or as in the case of the CCLS, restricted to a particular time window. These factors may make the interpretation of the effect of combined allergy categories difficult. Medical records are likely more easily standardized across studies, but combining those studies with self-reported studies poses similar problems. Recall bias is also of concern in questionnaire-based studies, but not in record-based studies; wherein parents of children with cancer may not remember a comparably insignificant allergic event. On the other hand, medical-record based studies are likely capture only a subset of more severe allergy, an exposure that may not be representative of allergy overall. Similarly, selection bias is on concern particularly in studies where participant contact is required (i.e. to conduct a questionnaire). Our results show a clear exaggeration of associations when controls are selected in clinics, and in the CCLS study, like many modern case-control studies, controls are not exchangeable with cases in terms of socio-economic status, opening the door to residual confounding.
Concluding remarks
Due to the complexities outlined above, understanding the causal relationship between allergy and childhood leukemia cannot be achieved with additional status quo epidemiologic studies, rather it will require a combination of large, prospective, perhaps biomarker-based cohort studies and basic biological and mechanistic studies. Functional studies can demonstrate biologically meaningful relationships between the two diseases (51). For example, the tumor surveillance hypothesis has been well demonstrated in biological studies (51), wherein individuals who are prone to allergic response are considered to have a hyper-vigilant antigen recognition and response thus producing more successful cancer cell surveillance and elimination. The vast heterogeneity of the effect of allergy observed within and between cancer sites suggests that the mechanisms by which these two families of disease intersect are not mutually exclusive and speak to the potential pleiotropic effects of allergy across individuals and tissue sites (51).
In the absence of complex and expensive studies, germ-line genetic predictors of allergy could serve as an informative proxy for unbiased exposure assessment of allergy in the context of cancer risk. From the current analysis, the once apparently strong inverse relationship between allergy and ALL has waned to an absent or ambiguous association. As the tide changes on the story of allergy and ALL, so must our tactics for studying the intersection of these complex diseases.
Supplementary Material
ACKNOWLEGEMENTS
The authors thank the families that participated in the CCLS. Without their time and effort none of our studies would be possible. The authors also thank our clinical collaborators throughout California for their continued support of our research and commitment to their patients: University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. Mignon Loh and Katherine Matthay), Children’s Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children’s Hospital (Dr. Gary Dahl), Children’s Hospital Oakland (Dr. James Feusner), Kaiser Permanente Roseville (former Sacramento; Drs. Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo, Alan Wong, and Denah Taggar), Kaiser Permanente San Francisco (Dr. Kenneth Leung), and Kaiser Permanente Oakland (Drs. Daniel Kronish and Stacy Month). Finally, the authors thank the entire California Childhood Leukemia Study staff and the former UCB Survey Research Center for their effort and dedication. This work was supported by grants from the U.S. National Institute of Environmental Health Sciences (2R01ES009137–11A1, P50ES018172/EPA RD83615901, to C. Metayer and R01 CA175737, to J. Wiemels and X. Ma).
Footnotes
Conflict of Interest Statement: The authors declare no conflicts of interest
REFERENCES
- 1.Wiemels J Perspectives on the causes of childhood leukemia. Chem Biol Interact. 2012;196(3):59–67. doi: 10.1016/j.cbi.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang JS, Wiemels JL, Buffler PA. Allergies and childhood leukemia. Blood Cells Mol Dis. 2009;42(2):99–104. doi: 10.1016/j.bcmd.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Black AB, Meynell MJ. Aleukaemic myeloid leukaemia presenting as aplastic anaemia. Br Med J. 1951;1(4720):1430–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel DA, King J, Tai E, Buchanan N, Ajani UA, Li J. Cancer incidence rates and trends among children and adolescents in the United States, 2001–2009. Pediatrics. 2014;134(4):e945–55. doi: 10.1542/peds.2013-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson KD, Howie LD, Akinbami LJ. Trends in allergic conditions among children: United States, 1997–2011. NCHS Data Brief. 2013(121):1–8. [PubMed] [Google Scholar]
- 6.Bernsen RM, de Jongste JC, van der Wouden JC. Birth order and sibship size as independent risk factors for asthma, allergy, and eczema. Pediatr Allergy Immunol. 2003;14(6):464–9. [DOI] [PubMed] [Google Scholar]
- 7.Westergaard TA P; Pedersen J; Olsen J; Frisch M; Sorensen H; Wohlfahrt J; Melbye M;. Birth Characteristics, Sibling Patterns, and Acute Leukemia Risk in Childhood: a Population-Based Cohort Study. J Natl Cancer Inst. 1997;89(13):939–47. [DOI] [PubMed] [Google Scholar]
- 8.Hagerhed-Engman L, Bornehag CG, Sundell J, Aberg N. Day-care attendance and increased risk for respiratory and allergic symptoms in preschool age. Allergy. 2006;61(4):447–53. doi: 10.1111/j.1398-9995.2006.01031.x. [DOI] [PubMed] [Google Scholar]
- 9.Ma X, Buffler PA, Selvin S, Matthay KK, Wiencke JK, Wiemels JL, et al. Daycare attendance and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2002;86(9):1419–24. doi: 10.1038/sj.bjc.6600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thavagnanam SF J; Bromley A; Shields MD; Cardwell CR;. A meta-analysis of the association between Caesarian section and childhood asthma. Clin Exp Allergy. 2007(38):629–33. doi: 10.1111/j.1365-2222.2007.02780.xClinical. [DOI] [PubMed] [Google Scholar]
- 11.Francis SS, Selvin S, Metayer C, Wallace AD, Crouse V, Moore TB, et al. Mode of delivery and risk of childhood leukemia. Cancer Epidemiol Biomarkers Prev. 2014;23(5):876–81. doi: 10.1158/1055-9965.EPI-13-1098. [DOI] [PubMed] [Google Scholar]
- 12.Yazdanbakhsh M, Kremsner PG, van Ree R. Allergy, parasites, and the hygiene hypothesis. Science. 2002;296(5567):490–4. doi: 10.1126/science.296.5567.490. [DOI] [PubMed] [Google Scholar]
- 13.Greaves M Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 14.Dahl S, Schmidt LS, Vestergaard T, Schuz J, Schmiegelow K. Allergy and the risk of childhood leukemia: a meta-analysis. Leukemia. 2009;23(12):2300–4. doi: 10.1038/leu.2009.162. [DOI] [PubMed] [Google Scholar]
- 15.Linabery AM, Jurek AM, Duval S, Ross JA. The association between atopy and childhood/adolescent leukemia: a meta-analysis. Am J Epidemiol. 2010;171(7):749–64. doi: 10.1093/aje/kwq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudant J, Orsi L, Menegaux F, Petit A, Baruchel A, Bertrand Y, et al. Childhood acute leukemia, early common infections, and allergy: The ESCALE Study. Am J Epidemiol. 2010;172(9):1015–27. doi: 10.1093/aje/kwq233. [DOI] [PubMed] [Google Scholar]
- 17.Urayama KY, Ma X, Selvin S, Metayer C, Chokkalingam AP, Wiemels JL, et al. Early life exposure to infections and risk of childhood acute lymphoblastic leukemia. Int J Cancer. 2011;128(7):1632–43. doi: 10.1002/ijc.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang JS, Tsai YW, Tsai CR, Wiemels JL. Allergy and risk of childhood acute lymphoblastic leukemia: a population-based and record-based study. Am J Epidemiol. 2012;176(11):970–8. doi: 10.1093/aje/kws263. [DOI] [PubMed] [Google Scholar]
- 19.Lariou MS, Dikalioti SK, Dessypris N, Baka M, Polychronopoulou S, Athanasiadou-Piperopoulou F, et al. Allergy and risk of acute lymphoblastic leukemia among children: a nationwide case control study in Greece. Cancer Epidemiol. 2013;37(2):146–51. doi: 10.1016/j.canep.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Crippa A, Khudyakov P, Wang M, Orsini N, Spiegelman D. A new measure of between-studies heterogeneity in meta-analysis. Stat Med. 2016;35(21):3661–75. doi: 10.1002/sim.6980. [DOI] [PubMed] [Google Scholar]
- 21.Ma X, Buffler PA, Layefsky M, Does MB, Reynolds P. Control selection strategies in case-control studies of childhood diseases. Am J Epidemiol. 2004;159(10):915–21. [DOI] [PubMed] [Google Scholar]
- 22.Renz-Polster H, David MR, Buist AS, Vollmer WM, O’Connor EA, Frazier EA, et al. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy. 2005;35(11):1466–72. doi: 10.1111/j.1365-2222.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 23.Jourdan-Da Silva N, Perel Y, Mechinaud F, Plouvier E, Gandemer V, Lutz P, et al. Infectious diseases in the first year of life, perinatal characteristics and childhood acute leukaemia. Br J Cancer. 2004;90(1):139–45. doi: 10.1038/sj.bjc.6601384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JS, Buffler PA, Metayer C, Chokkalingam AP, Patoka J, Kronish D, et al. Maternal immunoglobulin E and childhood leukemia. Cancer Epidemiol Biomarkers Prev. 2009;18(8):2221–7. doi: 10.1158/1055-9965.EPI-09-0212. [DOI] [PubMed] [Google Scholar]
- 25.Nunez-Enriquez JC, Fajardo-Gutierrez A, Buchan-Duran EP, Bernaldez-Rios R, Medina-Sanson A, Jimenez-Hernandez E, et al. Allergy and acute leukaemia in children with Down syndrome: a population study. Report from the Mexican inter-institutional group for the identification of the causes of childhood leukaemia. Br J Cancer. 2013;108(11):2334–8. doi: 10.1038/bjc.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson R, Graham S, Lilienfeld A, Schuman L, Levin M, Swanson M. Epidemiology of diseases in adult males with leukemia. J Natl Cancer Inst. 1976;56(5):891–8. [DOI] [PubMed] [Google Scholar]
- 27.Viadana E, Bross ID. Use of the medical history to predict the future occurrence of leukemias in adults. Prev Med. 1974;3(1):165–70. [DOI] [PubMed] [Google Scholar]
- 28.Zheng W, Linet MS, Shu XO, Pan RP, Gao YT, Fraumeni JF Jr. Prior medical conditions and the risk of adult leukemia in Shanghai, People’s Republic of China. Cancer Causes Control. 1993;4(4):361–8. [DOI] [PubMed] [Google Scholar]
- 29.Chang JS, Tsai CR, Tsai YW, Wiemels JL. Medically diagnosed infections and risk of childhood leukaemia: a population-based case-control study. Int J Epidemiol. 2012;41(4):1050–9. doi: 10.1093/ije/dys113. [DOI] [PubMed] [Google Scholar]
- 30.Hughes AM, Lightfoot T, Simpson J, Ansell P, McKinney PA, Kinsey SE, et al. Allergy and risk of childhood leukaemia: results from the UKCCS. Int J Cancer. 2007;121(4):819–24. doi: 10.1002/ijc.22702. [DOI] [PubMed] [Google Scholar]
- 31.Viechtbauer W Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36(3):1–48. [Google Scholar]
- 32.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 33.Cohen J Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale NJ: L. Erlbaum Associates; 1988. xxi, 567 p. p. [Google Scholar]
- 34.Magnani C, Pastore G, Luzzatto L, Terracini B. Parental occupation and other environmental factors in the etiology of leukemias and non-Hodgkin’s lymphomas in childhood: a case-control study. Tumori. 1990;76(5):413–9. [DOI] [PubMed] [Google Scholar]
- 35.Nishi M, Miyake H. A case-control study of non-T cell acute lymphoblastic leukaemia of children in Hokkaido, Japan. J Epidemiol Community Health. 1989;43(4):352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbaum PF, Buck GM, Brecher ML. Allergy and infectious disease histories and the risk of childhood acute lymphoblastic leukaemia. Paediatr Perinat Epidemiol. 2005;19(2):152–64. doi: 10.1111/j.1365-3016.2005.00634.x. [DOI] [PubMed] [Google Scholar]
- 37.Schuz J, Morgan G, Bohler E, Kaatsch P, Michaelis J. Atopic disease and childhood acute lymphoblastic leukemia. Int J Cancer. 2003;105(2):255–60. doi: 10.1002/ijc.11054. [DOI] [PubMed] [Google Scholar]
- 38.Soderberg KC, Jonsson F, Winqvist O, Hagmar L, Feychting M. Autoimmune diseases, asthma and risk of haematological malignancies: a nationwide case-control study in Sweden. Eur J Cancer. 2006;42(17):3028–33. doi: 10.1016/j.ejca.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Spector L, Groves F, DeStefano F, Liff J, Klein M, Mullooly J, et al. Medically recorded allergies and the risk of childhood acute lymphoblastic leukaemia. Eur J Cancer. 2004;40(4):579–84. doi: 10.1016/j.ejca.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Wen W, Shu XO, Linet MS, Neglia JP, Potter JD, Trigg ME, et al. Allergic disorders and the risk of childhood acute lymphoblastic leukemia (United States). Cancer Causes Control. 2000;11(4):303–7. [DOI] [PubMed] [Google Scholar]
- 41.Howard LM, Wessely S. The psychology of multiple allergy. BMJ. 1993;307(6907):747–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altman DR, Chiaramonte LT. Public perception of food allergy. J Allergy Clin Immunol. 1996;97(6):1247–51. [DOI] [PubMed] [Google Scholar]
- 43.Roujeau JC. Clinical heterogeneity of drug hypersensitivity. Toxicology. 2005;209(2):123–9. doi: 10.1016/j.tox.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 44.Roman E, Simpson J, Ansell P, Kinsey S, Mitchell CD, McKinney PA, et al. Childhood acute lymphoblastic leukemia and infections in the first year of life: a report from the United Kingdom Childhood Cancer Study. Am J Epidemiol. 2007;165(5):496–504. doi: 10.1093/aje/kwk039. [DOI] [PubMed] [Google Scholar]
- 45.Slusky DA, Does M, Metayer C, Mezei G, Selvin S, Buffler PA. Potential role of selection bias in the association between childhood leukemia and residential magnetic fields exposure: A population-based assessment. Cancer Epidemiol. 2014;38(3):307–13. doi: 10.1016/j.canep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galobardes B, Granell R, Sterne J, Hughes R, Mejia-Lancheros C, Davey Smith G, et al. Childhood wheezing, asthma, allergy, atopy, and lung function: different socioeconomic patterns for different phenotypes. Am J Epidemiol. 2015;182(9):763–74. doi: 10.1093/aje/kwv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378(9809):2112–22. doi: 10.1016/S0140-6736(11)60130-X. [DOI] [PubMed] [Google Scholar]
- 48.Pols DH, Wartna JB, Moed H, van Alphen EI, Bohnen AM, Bindels PJ. Atopic dermatitis, asthma and allergic rhinitis in general practice and the open population: a systematic review. Scand J Prim Health Care. 2016;34(2):143–50. doi: 10.3109/02813432.2016.1160629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadley C Food allergies on the rise? Determining the prevalence of food allergies, and how quickly it is increasing, is the first step in tackling the problem. EMBO Rep. 2006;7(11):1080–3. doi: 10.1038/sj.embor.7400846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bloomfield SF, Stanwell-Smith R, Crevel RW, Pickup J. Too clean, or not too clean: the hygiene hypothesis and home hygiene. Clin Exp Allergy. 2006;36(4):402–25. doi: 10.1111/j.1365-2222.2006.02463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoste E, Cipolat S, Watt FM. Understanding allergy and cancer risk: what are the barriers? Nat Rev Cancer. 2015;15(3):131–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.