Abstract
In river-lake systems, sediment and water column are two distinct habitats harboring different bacterial communities which play a crucial role in biogeochemical processes. In this study, we employed Phylogenetic Investigation of Communities by Reconstruction of Unobserved States to assess the potential functions and functional redundancy of the bacterial communities in sediment and water in a eutrophic river-lake ecosystem, Poyang Lake in China. Bacterial communities in sediment and water had distinct potential functions of carbon, nitrogen, and sulfur metabolisms as well as phosphorus cycle, while the differences between rivers and the lake were inconspicuous. Bacterial communities in sediment had a higher relative abundance of genes associated with carbohydrate metabolism, carbon fixation pathways in prokaryotes, methane metabolism, anammox, nitrogen fixation, and dissimilatory sulfate reduction than that of water column. Bacterial communities in water column were higher in lipid metabolism, assimilatory nitrate reduction, dissimilatory nitrate reduction, phosphonate degradation, and assimilatory sulfate reduction than that of sediment bacterial communities. Furthermore, the variations in functional composition were closely associated to the variations in taxonomic composition in both habitats. In general, the bacterial communities in water column had a lower functional redundancy than in sediment. Moreover, comparing to the overall functions, bacterial communities had a lower functional redundancy of nitrogen metabolism and phosphorus cycle in water column and lower functional redundancy of nitrogen metabolism in sediment. Distance-based redundancy analysis and mantel test revealed close correlations between nutrient factors and functional compositions. The results suggested that bacterial communities in this eutrophic river-lake system of Poyang Lake were vulnerable to nutrient perturbations, especially the bacterial communities in water column. The results enriched our understanding of the bacterial communities and major biogeochemical processes in the eutrophic river-lake ecosystems.
Keywords: Carbon metabolism, Nitrogen cycle, Tributaries, Bacterial community, Biogeochemistry, Eutrophication
Introduction
Lakes and their tributaries are highly linked ecosystems in multiple ways, especially through materials transported from the watershed to the lake through river systems (Cole et al., 2006; Marcarelli & Wurtsbaugh, 2009; Jones, 2010; Ylla et al., 2013). Microbial communities in lake and its tributaries have different taxonomic compositions (Ren et al., 2017a, 2019). In lake ecosystems, water and sediment are two distinct realms and interact closely through biogeochemical processes (Parker et al., 2016). These two habitats host tremendous diversity of microorganisms (Lozupone & Knight, 2007; Röeske et al., 2012; Huang et al., 2016), which constitute distinct microbial communities in sediment and water column (Briée, Moreira & López-García, 2007; Nishihama et al., 2008; Ren et al., 2019). However, the functional differences of bacterial communities in sediment and water column of lake-river systems were not well studied.
In aquatic ecosystems, bacterial communities play an extremely important role in transformation, accumulation, and migration of nutrients and other elements, as well as in energy conversion and material recycling (Cotner & Biddanda, 2002; Van Der Heijden, Bardgett & Van Straalen, 2008; Newton et al., 2011). Bacterial communities exhibit high compositional and functional variability (Newton et al., 2011). Functional traits are valuable ecological markers to understand the bacterial community assembly (Barberan et al., 2012). Moreover, microbial metabolic activities can influence water quality through the storage and release of nutrients (Nielsen et al., 2006; Hupfer & Lewandowski, 2008). Thus, it is crucial to understand the roles of bacterial communities in biogeochemical cycling and elucidate their responses to environmental changes by unraveling their functional potentials (Green, Bohannan & Whitaker, 2008; Fierer et al., 2012; Freedman & Zak, 2015; Ren et al., 2017b). In addition, previous studies suggested that distinct taxa can share specific functional attributes while closely related taxa may exhibit distinct functional features (Allison & Martiny, 2008; Philippot et al., 2010; Fierer et al., 2012; Dopheide et al., 2015). Thus, the relationships between taxonomic and functional differences can help to elucidate functional redundancy and stability of bacterial communities.
Changes in water quality and sediment properties drive the variation of bacterial communities which regulate the core biogeochemical processes such as carbon and nitrogen metabolisms in aquatic ecosystems (Liu et al., 2018; Wang et al., 2018; Yao et al., 2018). As the largest freshwater lake in China, Poyang Lake is fed by five tributaries and is experiencing aggravated nutrient loading from agriculture and urbanization of the catchment in recent decades (Wang & Liang, 2015; Liu, Fang & Sun, 2016). The increase in nutrient inputs caused by agriculture, urbanization, and industry has significantly degraded water quality and ecological integrity of Poyang Lake with serious eutrophication (Wang et al., 2015; Zhang et al., 2015; Liu, Fang & Sun, 2016). In the river-lake systems of Poyang Lake, our previous study has shown that the taxonomic composition of bacterial communities in lake sediment (SL), river sediment (SR), lake water (WL), and river water (WR) had distinct spatial distribution patterns and close relationships with nutrients (Ren et al., 2019). However, our understanding of the functions mediated by the bacterial communities in the sediment and water of this linked river-lake ecosystem is still limited. To reveal the functional potentials of bacteria, metagenomic sequencing has been used in a growing number of studies (Mackelprang et al., 2011; Fierer et al., 2012; Llorens-Marès et al., 2015). Alternatively, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) is cheaper, faster, and reliable (Wilkinson et al., 2018) and has been widely used to infer the functional profile of the bacterial communities using 16S rRNA genes and a reference genome database to predict the functional composition of a metagenome (Langille et al., 2013). In this study, we predicted metagenomes from 16S rRNA gene sequences and classified into Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs (KOs) using PICRUSt. The KOs associated with carbon, nitrogen, and sulfur metabolisms as well as phosphorus cycle were identified from KEGG database (Kanehisa & Goto, 2000). We aimed to reveal the functional properties of bacterial communities in SL, SR, WL, and WR in the river-lake system of Poyang Lake, including (1) metabolism pathways of major functions, (2) influences of nutrient variables on functional compositions, and (3) functional redundancy.
Materials and Methods
Study area and field sampling
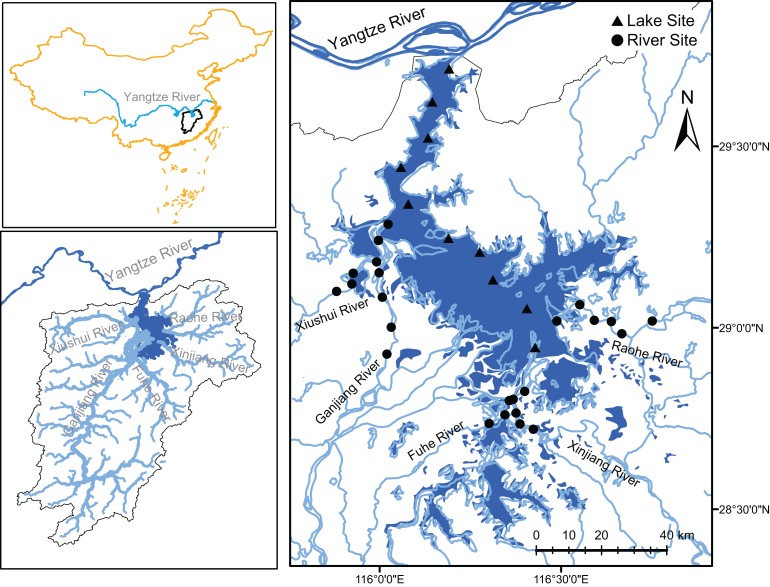
Poyang Lake is located in the lower reach of Yangtze River. With a surface area over 4,000 km2 (in summer), it is the largest freshwater lake in China. There are five rivers (Fuhe, Ganjiang, Xinjiang, Raohe, and Xiushui) feeding Poyang Lake and one outlet connecting to Yangtze River (Fig. 1). The annual runoff of Poyang Lake is 152.5 billion m3, accounting for 16.3% annual runoff of Yangtze River. Poyang Lake is a shallow seasonal lake and a typical water-carrying and throughput lake restricted by the water level of Yangtze River and the inflows of the five tributaries (Fang et al., 2011; Zhao et al., 2011). The high and low water levels of Poyang Lake are 20.69 and 9.82 m above the sea level, respectively (Liao, Yu & Guo, 2017). The average water depth is 8.4 m (Wang & Liang, 2015). Poyang Lake has been suffering persistent eutrophication (Liao, Yu & Guo, 2017). Previous research shown that cyanobacteria blooms have been observed in Poyang Lake since 2000 (Liu et al., 2016) but only occur periodically and regionally (Liu & Fang, 2017). We did not find cyanobacteria bloom during our sampling in early August 2017.
Figure 1. Study area and sampling sites.
Samples were collected from the surface water and sediment of Poyang lake and its fiver tributaries (Xiushui, Ganjiang, Fuhe, Xinjiang, and Raohe). This figure was modified from Ren et al. (2019).
We collected samples from Poyang Lake and its tributaries in 10 and 24 sample sites, respectively (Fig. 1). In each sample site, a handheld meter (YSI Professional Plus, Yellow Springs, OH, USA) was used to measure water temperature (Temp), dissolved oxygen (DO), pH, and conductivity (Cond) in situ. Secchi disk depth was measured as well. Water samples were collected at the depth of 0.5 m using a Van Dorn water sampler. A total of 200 mL water was filtered onto a 0.2-μm Polycarbonate Membrane Filter (Whatman, UK), which was immediately frozen in liquid nitrogen in the field and stored at −80 °C in the lab until DNA extraction. Another 500 mL water was acid fixed in the field and transported to the laboratory at 4 °C for chemical analyses. Sediment samples were collected using a Ponar Grab sampler at the depth of 5.5–6.5 m in Poyang Lake and of 3.9–5.8 m in the tributaries. The top five-cm sediment was homogenized by stirring with a spatula, collected in a sterile centrifuge tube, and immediately frozen in liquid nitrogen in the field for DNA extraction. The remaining sediment was collected in a clean Ziploc bag for chemical analyses.
For water samples, total nitrogen (TN), nitrate (NO3−), ammonium (NH4+), total phosphorus (TP), and soluble reactive phosphorus (SRP) were analyzed according to the Clean Water Act Analytical Methods (United States Environmental Protection Agency (EPA), 2017). DOC was analyzed using a TOC Analyzer (TOC-VCPH; Shimadzu Scientific Instruments, Kyoto, Japan). Detailed information of water sample analyses was provided in our previous study (Ren et al., 2019). For sediment samples, TN was analyzed using the modified Kjeldahl method (HJ717-2014). NO3− and NH4+ were analyzed using UV spectrophotometry method (HJ634-2012). TP was analyzed using alkali fusion-Mo-Sb Anti spectrophotometric method (HJ632-2011). Total organic carbon (OC) was analyzed using Potassium dichromate oxidation spectrophotometric method (HJ615-2011). Organic nitrogen was analyzed using acid hydrolysis method (Bremner, 1965). Organic phosphorus was analyzed using SMT method (Ruban et al., 1999).
DNA extraction, PCR, and sequencing
DNA was extracted from the filter and sediment (0.5 g) samples using the TIANGEN-DP336 soil DNA Kit (TIANGEN-Biotech, Beijing, China) following manufacturer protocols. Extracted DNA samples were quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). The V3 and V4 regions were amplified using the forward primer 347F 5′-CCTACGGRRBGCASCAGKVRVGAAT-3′ and the reverse primer 802R 5′-GGACTACNVGGGTWTCTAATCC-3′ (GENEWIZ, Inc., South Plainfield, NJ, USA) (Ren et al., 2019). PCR was performed using the following program: initial denaturation at 94 °C for 3 min, 24 cycles of denaturation at 94 °C for 30 s followed by annealing at 57 °C for 90 s and extension at 72 °C for 10 s, and final extension step at 72 °C for 10 min. Amplified DNA was verified by electrophoresis of PCR mixtures in 1.0% agarose in 1× TAE buffer and purified using the Gel Extraction Kit (Qiagen, Hilden, Germany). DNA libraries were validated by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and quantified by Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) according to manufacturer’s instructions.
Sequence analysis and functional gene prediction
Raw sequence data was processed using the software package QIIME 1.9.1 (Caporaso et al., 2010). The forward and reverse reads were joined and assigned to samples based on barcode and truncated by cutting off the barcode and primer sequence. Then the sequences were quality filtered, and the chimeric sequences were removed. Sequences which did not fulfill the following criteria were discarded: sequence length <200 bp, no ambiguous bases, mean quality score ≥20 (Ren et al., 2019). The effective sequences were grouped into operational taxonomic units (OTUs) at 97% sequence identity level against the Greengenes 13.8 database (McDonald et al., 2012). Then the functional potentials of the bacterial communities were predicted using PICRUSt 1.1.0 and the nearest sequence taxon index (NSTI) was calculated to indicate the accuracy of PICRUSt prediction (Langille et al., 2013). The average NSTI was 0.146, indicating high accuracy (Langille et al., 2013). Then, the predicted metagenomes were further classified into KEGG KOs. The KOs associated with carbon, nitrogen, and sulfur metabolism as well as phosphorus cycle were identified from KEGG database (Kanehisa & Goto, 2000; Bergkemper et al., 2016). The Raw sequence data are available at National Center for Biotechnology Information (PRJNA436872, SRP133903).
Statistical analysis
To reveal the functional differences (overall metagenomic functions and the major functions, including carbon metabolism, nitrogen metabolism, phosphorus cycle, and sulfur metabolism) between the bacterial communities in different habitats of the river-lake ecosystem of Poyang Lake, non-metric multidimensional scaling (NMDS) and analysis of variance using distance matrices (ADONIS) were applied using the Vegan package 2.4–6 (Oksanen et al., 2007) based on the relative abundance of KOs. Differences of the major pathways associated to carbon metabolism, nitrogen metabolism, phosphorus cycle, and sulfur metabolism between the bacterial communities in sediment and water column were tested using analysis of variance and the P-values were adjusted by FDR correction. Linear regression was used to assess the relationships between taxonomic and functional dissimilarities, revealing functional redundancy of the bacterial communities (stronger linear regression indicates lower functional redundancy) (Yang et al., 2017; Galand et al., 2018). Taxonomic and functional dissimilarities were calculated as Bray–Curtis distances based on the phylogenetic and metagenomic compositions (relative abundance of OTUs and KOs, respectively). The differences of linear regression slopes were compared using analysis of covariance (ANCOVA). Distance-based redundancy analysis (dbRDA) was conducted using Vegan package to reveal the relationships between environmental variables (normalized using “normalize” method) and overall functional compositions (relative abundance of KOs, Hellinger transferred) of bacterial communities in sediment and water column, and the significance of the nutrient variables was tested using Envfit function in R. Mantel tests were applied to assess the relationships between nutrient factors and major functions and the P-values were adjusted by FDR correction. All the analyses were conducted in R 3.4.4 (R Core Team, 2017).
Results
Functional differences
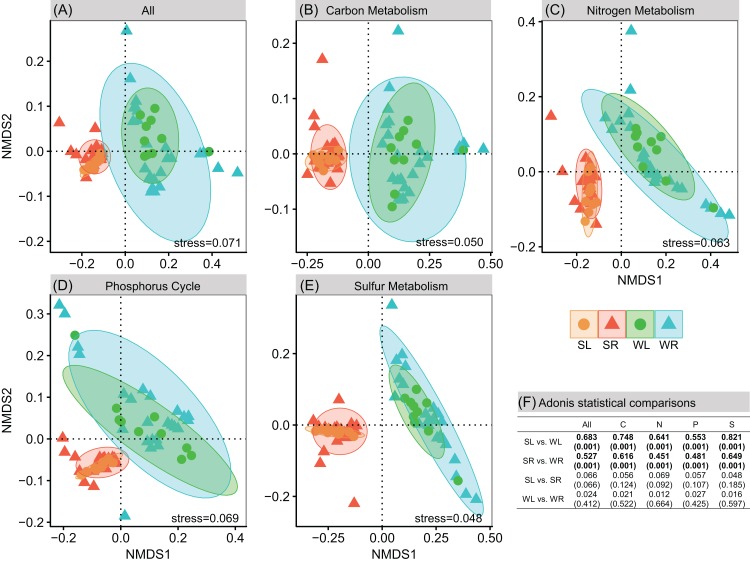
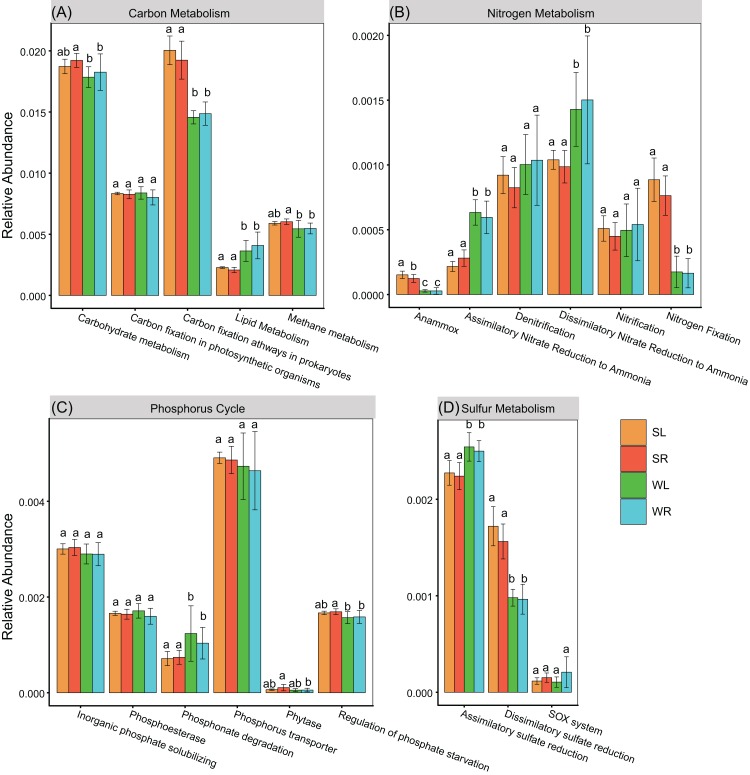
In total, 6,295 and 6,187 KOs were detected in bacterial communities in sediment and water column. Profound differences were detected in functional compositions between sediment and water. NMDS and ADONIS showed that bacterial communities in LS and RS were significantly (P < 0.05) different to LW and RW (LS vs. LW and RS vs. RW), respectively (Fig. 2). However, there was no difference between Poyang Lake and its tributaries (LS vs. RS and LW vs. RW, Fig. 2). For carbon metabolism, we detected 242 KOs associated to central carbon metabolism pathways (ko01200) based on the KEGG database. Carbohydrate metabolism, carbon fixation pathways in prokaryotes, and methane metabolism had a higher relative abundance of associated genes in the bacterial communities in sediment than in water (Fig. 3A). However, the lipid metabolism had a higher relative abundance in water than in sediment (Fig. 3A). For the nitrogen metabolism, we detected 41 KOs associated to nitrogen metabolism pathways (ko00910 in KEGG database). Bacterial communities had a higher relative abundance of genes associated to anammox and nitrogen fixation in sediment than in water (Fig. 3B). However, assimilatory nitrate reduction to ammonia (ANRA) and dissimilatory nitrate reduction to ammonia (DNRA) had a higher relative abundance in water than in sediment (Fig. 3B). For phosphorus, we detected 43 KOs associated to phosphorus cycle. Phosphonate degradation had a higher relative abundance in water than in sediment (Fig. 3C). For sulfur metabolism, we detected 45 KOs associated to the sulfur metabolism pathways (ko00920 in the KEGG database). Assimilatory sulfate reduction had a lower relative abundance while dissimilatory sulfate reduction had a higher relative abundance in sediment than in water (Fig. 3D).
Figure 2. Functional differences between habitats.
(A–E) Non-metric multidimensional scaling analysis of potential functions composition in terms of overall functions, carbon metabolism, nitrogen metabolism, phosphorus cycle, and sulfur metabolism. (F) Pairwise dissimilarity tests of functional composition between different habitats using ADONIS. The numbers outside the bracket are “R2.” P-values are in bracket.
Figure 3. Relative abundance of genes associated to major pathways in (A) Central carbon metabolism, (B) nitrogen metabolism, (C) phosphorus cycle, and (D) sulfur metabolism.
For each pathway, the same lowercase letter indicates a non-significant difference, whereas the different letter indicates a significant difference between habitats (ANOVA, P < 0.05). P-values were adjusted by FDR correction.
Environmental influences
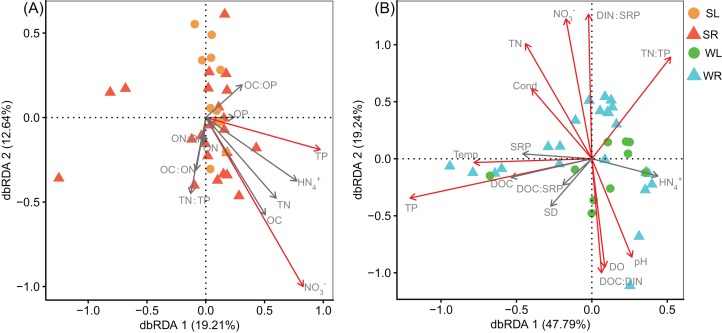
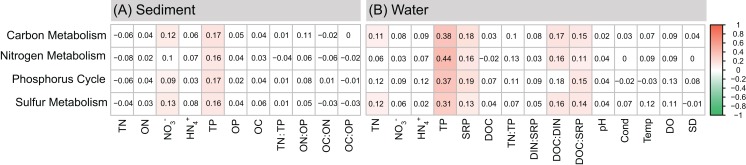
The results of dbRDA indicated that the overall functional compositions of bacterial communities in sediment were significantly correlated with TP and NO3− (Fig. 4A). The first two axes explained 31.85% of the functional variation (dbRDA 1: 19.21%; dbRDA 2: 12.64%). For the bacterial communities in water column, the overall functional compositions were significantly correlated with TN, NO3−, TP, TN:TP, DOC:DIN, DIN:SRP, as well as Cond, Temp, DO, and pH (Fig. 4B). The first two axes explained 67.03% of the functional variation (dbRDA 1: 47.79%; dbRDA 2: 19.24%). Mantel tests further demonstrated that the spatial variations of the major biogeochemical processes (C-metabolism, N-metabolism, P-cycle, and S-metabolism) were significantly influenced by TP and NO3− in sediment, and by TN, TP, SRP, DOC:DIN, and DOC:SRP in water column (Fig. 5).
Figure 4. Biplot of distance-based redundancy analyses (dbRDA) showing the relationship between functional composition and nutrient variables in (A) sediment and (B) water.
The red arrows represent the significant variables (envfit, P < 0.05).
Figure 5. Mantel tests between major functions and nutrient variables of (A) sediment and (B) water based on Spearman correlation.
Significant correlations (P < 0.05) were colored. P-values were adjusted by FDR correction.
Functional redundancy
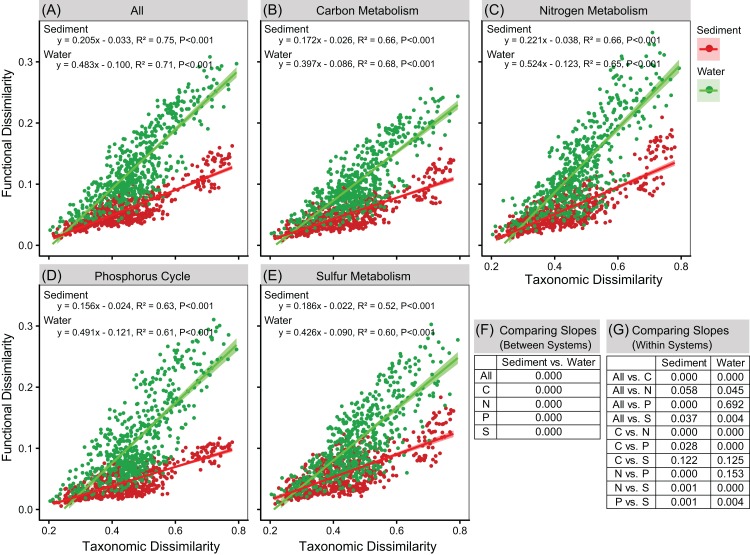
Linear regressions between taxonomic and functional dissimilarities showed that the variations in metagenomic functional composition (overall, C-metabolism, N-metabolism, P-cycle, and S-metabolism) were closely associated with the variations in phylogenetic composition (Fig. 6). However, sediment bacterial communities had significantly smaller slopes than bacterial communities in water column (ANCOVA, P < 0.05, Fig. 6F). For the major functions in sediment, nitrogen metabolism had a higher slope, followed by sulfur metabolism, carbon metabolism, and phosphorus cycle (Fig. 6). In the water column, however, nitrogen metabolism and phosphorus cycle had higher slopes than carbon and sulfur metabolisms (Fig. 6). The results suggested that bacterial communities in sediment had higher functional redundancy than in the water column. Moreover, bacterial communities had lowest functional redundancy for nitrogen metabolism but highest functional redundancy for phosphorus cycle in sediment, while had lowest redundancy for both nitrogen metabolism and phosphorus cycle in the water column.
Figure 6. Linear regressions between taxonomic and functional dissimilarities (A–E).
One point represents one sample pair. Shadow area denotes the 95% confidence interval. (F–G) Statistical test of the linear regression slopes between systems and within systems using ANCOVA.
Discussion
In this study, the functional composition of bacterial communities in the river-lake system of Poyang Lake were different between water and sediment (LS vs. LW and RS vs. RW), while no different between tributaries and the lake itself (LS vs. RS and LW vs. RW). In our previous study of the river-lake system of Poyang Lake (Ren et al., 2019), bacterial communities were taxonomically different between sediment and water. It has been well demonstrated that sediment and water had distinct bacterial communities (Jiang et al., 2006; Nishihama et al., 2008; Lu et al., 2016), which might determine significant functional differences (Fierer et al., 2012; Ren et al., 2017a). However, the taxonomical differences of bacterial communities between Poyang Lake and its tributaries were significant but smaller compared to the differences between sediment and water (Ren et al., 2019). In generally, bacterial communities were more taxonomically different than functional different (Louca et al., 2017; Ren et al., 2017a). Thus, the small differences in taxonomic composition of bacterial communities did not lead to their functional differences between Poyang Lake and its tributaries.
This study showed that carbohydrate metabolism, carbon fixation pathways in prokaryotes, and methane metabolism had a higher relative abundance in the bacterial communities in sediment than in water, and the lipid metabolism had a higher relative abundance in water than in sediment. The results suggested that bacterial communities in sediment and water had distinct carbon metabolism pathways. Organic matter (OM) transported by river provides fueling aquatic food webs as a major source of energy and is also a significant component of the global carbon cycle (Cole et al., 2007; Battin et al., 2008; Smith & Kaushal, 2015). In freshwater ecosystems, OM is a heterogeneous mixture including allochthonous materials contributed by soil and plant litter inputs from terrestrial ecosystems and autochthonous materials contributed by primary producers in freshwater ecosystems (Webster & Meyer, 1997). OM is consisted of carbohydrates, proteins, lipids, lignins, and other compounds in aquatic ecosystems (Thurman, 2012). Microorganisms are key biogeochemical agents in the generation, transformation, and mineralization of OM (Horvath, 1972).Variations of OM in its source and composition, as well as the bioavailability of its components determine the spatial patterns of bacterial composition and functional diversity (Hoostal & Bouzat, 2008; Wang et al., 2018). In aquatic ecosystems, sediment and water column have distinct redox environments (Röeske et al., 2012), and the OM derives from different sources with different compositions (Hedges, Clark & Come, 1988). These differences might lead to the distinct carbon metabolisms between water and sediment. For example, the reduction condition in sediment is benefit to methane production (Koyama, 1963; He et al., 2015; Liu & Xu, 2016). In sediment, methane-oxidizing and sulfate-reducing bacteria also play the roles in carbon fixation (Kellermann et al., 2012).
Our study also showed that sediment and water column were significantly different in nitrogen metabolism, suggesting different nitrogen use strategies. In the past century, the nitrogen entering freshwater ecosystem has been increased more than twofold by anthropogenic activities (Schlesinger, 2009; Meunier et al., 2016), contributing to eutrophication in lake and coastal ecosystems (Nixon, 1995; Smith, 2003). Poyang Lake has been facing serious threat of eutrophication (Wang et al., 2015; Zhang et al., 2015; Liu, Fang & Sun, 2016) because of the aggravated nutrient loading from agriculture and urbanization of the catchment in recent decades (Wang & Liang, 2015; Liu, Fang & Sun, 2016). Nitrogen has many different chemical forms from the oxidation state of nitrate (+5) to the reduction state of ammonia (−3) and is cycled by a suite of biogeochemical processes (Ollivier et al., 2011), including four reduction pathways (denitrification, nitrogen fixation, ANRA, and DNRA) and two oxidation pathway (anammox and nitrification) (Lamba et al., 2017). In aquatic ecosystems, denitrification is the main biological process turning nitrate to dinitrogen and nitrous oxide (Tiedje et al., 1983; Seitzinger, 1988) and anammox is another important pathway turning nitrite and ammonia to dinitrogen (Dalsgaard et al., 2003; Kuypers et al., 2003). Both denitrification and anammox play important roles in removing nitrogen from aquatic ecosystems. In our study, bacterial communities in both sediment and water had a high relative abundance of the genes associated to denitrification, suggesting strong potentials in nitrogen removal. Many previous studies have demonstrated that rivers and lakes are hot spots to remove N inputs to surface waters from terrestrial environments (Wollheim et al., 2008; Harrison et al., 2009; Beaulieu et al., 2011). Denitrification can be limited by the supply of NO3− and OC, as well as redox potential (Van Kessel, 1977; Seitzinger, 1988). Furthermore, nitrification is also an important process in the N cycle and couples with denitrification (Jenkins & Kemp, 1984; Nils, 2003), especially in the shallow lakes. In the eutrophic river-lake system of Poyang Lake, the high contents of OM and NO3− in water and sediment can facilitate denitrification. Moreover, the respiration in sediment can provide an anoxic environment and promote sediment denitrification. On the other hand, it has also been supported by many studies that aerobic denitrification can be performed by a broad range of bacterial organisms under an aerobic environment (Ji et al., 2015; Lv et al., 2017). In our study, the high potential denitrification (potential NO3− reductions) in water might be performed through aerobic denitrification with the facilitation of high supplement of OM and NO3−. In addition to denitrification, bacterial communities in sediment had a significantly higher relative abundance of the genes associated to anammox than in water. It was found that anammox can coupled to nitrate reduction to contribute substantially to produce dinitrogen in sediments (Thamdrup & Dalsgaard, 2002). These results suggested that the bacterial communities in water and sediment of this eutrophic river-lake system had strong functional potentials but different strategies in nitrogen removal. In contrast to nitrogen removal, bacterial communities in sediment also had a higher relative abundance of genes associated with nitrogen fixation. In fact, the genetic potential of nitrogen fixation is pervasive among the domains of Bacteria and Archaea (Zehr et al., 2003). Nitrogen fixation and denitrification can co-occur in sediments through heterotrophic nitrogen fixation (Newell et al., 2016). We have underestimated the importance of heterotrophic sediment nitrogen fixation in the past, which can be an important source of nitrogen even under higher inorganic nitrogen concentrations (Fulweiler & Heiss, 2014; Newell et al., 2016). Examining the expression of the genes encoding for nitrogenase (such as nifD, nifH, nifK, and anfG) in the bacterial communities can help us understand the nitrogen fixation potential in freshwater ecosystems. In our study, the high relative abundance of genes associated to nitrogen fixation suggested a significant nitrogen fixation potential in sediment in Poyang Lake and its tributaries. In nitrogen metabolism pathways, both ANRA and DNRA had a higher relative abundance in water than in sediment, suggesting strong potentials of nitrate reduction to ammonia for bacterial communities in water column. ANRA and DNRA serve distinct cellular functions (Lamba et al., 2017): ANRA consumes energy and provides ammonium for cell to synthesize amino acids and nucleotides, while NDRA generates ATP in absence of oxygen and retains the nitrogen in the form of NH4+ for further biological processes (Zumft, 1997).
Phosphorus is an essential element in all ecosystems used by all living organisms. Bacteria plays a pivotal role in natural phosphorus cycles on the earth (Ohtake et al., 1996; Kononova & Nesmeyanova, 2002). In this study, the results showed that phosphonate degradation had a higher relative abundance in water than in sediment. Phosphonates are characterized by direct carbon-to-phosphorus bonds, which are resistant to chemical hydrolysis and thermal degradation (Ohtake et al., 1996; Kononova & Nesmeyanova, 2002). In polluted freshwater ecosystems, large quantities of phosphonates are xenobiotics, such as pesticides, antibiotics, and detergent additives (Schowanek & Verstraete, 1990). It has been revealed that phosphonates are significantly removed from marine basin due to rapid release and remineralization (Benitez-Nelson et al., 2004). Our study suggested that bacterial communities in water column are important in phosphonates removal.
For sulfur metabolism in the eutrophic river-lake system of Poyang Lake, our study showed that the assimilatory reduction is more common than dissimilatory reduction and the bacterial communities in water had a higher assimilatory sulfur reduction potential while lower dissimilatory sulfate reduction than in sediment. Sulfur is an important element required for some cellular components related to proteins. In the sulfur metabolism of bacteria, assimilatory sulfate reduction commences with the incorporation of sulfide radical for the biosynthetic cycle. Thus, during assimilatory sulfate reduction, there is no sulfide produced. For some microorganisms, sulfur compounds are utilized in dissimilatory and energy-yielding metabolic processes, which takes place in anaerobic respiration. During dissimilatory sulfate reduction, sulfate ion is used as the terminal electron acceptor and is reduced to produce sulfide, in the meantime, OC is mineralized with producing of carbon dioxide. In SLs, dissimilatory sulfate reduction can account for a significant fraction of OC mineralization, especially in eutrophic lakes with high availabilities of OM and sulfate (Holmer & Storkholm, 2001). The differences of sulfur metabolism between water and sediment shed light on sulfur use strategies of bacterial communities in these two distinct habitats.
As discussed above, biogeochemical cycles of C, N, P, and S are important ecological functions in freshwater ecosystems. In our study, mantel tests showed significant correlations between taxonomic and functional dissimilarity matrixes (beta diversities), suggesting that the overall changes in potential functions, as well as the changes of potential metabolisms of C, N, P, and S were closely associated with changes in taxonomic compositions of the bacterial communities. Functional redundancy always exists in natural ecosystems (Cardinale, Nelson & Palmer, 2000; Rosenfeld, 2002; Allison & Martiny, 2008) and is measured by the correlation between taxonomic and functional gene diversities (Fierer et al., 2013; Yang et al., 2017). Functional redundancy occurs when different organisms execute a similar function, remaining functional stabilization of communities upon species loss (Rosenfeld, 2002; Nyström, 2006). Our results showed that the bacterial communities in water column had a lower redundancy of overall functions than in sediment. Moreover, compared to overall functions, sediment bacterial communities had lower functional redundancy of N metabolism, and bacterial communities in water column had lower functional redundancy of N metabolism and P cycle. In bacterial communities, functional redundancy is expected to allow bacterial communities to have a certain extent of resistance and resilience in facing environmental perturbations (Allison & Martiny, 2008; Bowen et al., 2011). The results suggested that the bacterial communities in water column were less stable than bacterial communities in sediment. Moreover, N metabolism and P cycle was more vulnerable to environmental perturbations than C and S metabolisms, influencing nutrient biogeochemical processes in the eutrophic river-lake system of Poyang Lake.
Conclusions
In this study, we assessed the functional properties of bacterial communities in SL, SR, WL, and WR in the river-lake system of Poyang Lake. In general, the results showed that bacterial communities in sediment and water had distinct potential functions in the biogeochemical processes of carbon, nitrogen, phosphorus, and sulfur. However, there was no difference between tributaries and the lake itself. Moreover, bacterial communities in water column had a lower functional redundancy than in sediment. Comparing to the overall functions within systems, bacterial communities had lower functional redundancy of nitrogen metabolism in sediment, and lower functional redundancy of nitrogen metabolism and phosphorus cycle in water column. In this eutrophic river-lake system, functional compositions of the bacterial communities were vulnerable to nutrient perturbations especially in water column. By revealing the metabolism pathways of major functions, the influences of nutrient variables on functional compositions, and functional redundancy, this study can provide insights into the microbial community structures and ecological processes in this river-lake system.
Supplemental Information
Acknowledgments
We are grateful to the anonymous reviewers for the comments, to Yuhang Zhang and Chenyu Yang for their assistances in the field and laboratory work.
Funding Statement
This study was supported by the Project of State Key Laboratory of Simulation and Regulation of Water Cycle in River Basin (SKL2018CG02), the National Natural Science Foundation of China (No. 51439007), and the IWHR Research and Development Support Program (WE0145B532017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Ze Ren conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Xiaodong Qu conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Wenqi Peng performed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Yang Yu performed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Min Zhang performed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw sequence data are available at National Center for Biotechnology Information (PRJNA436872, SRP133903).
References
- Allison & Martiny (2008).Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proceedings of the National Academy of Sciences of the United States of America. 2008;1051(Suppl 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan et al. (2012).Barberan A, Fernandez-Guerra A, Bohannan BJM, Casamayor EO. Exploration of community traits as ecological markers in microbial metagenomes. Molecular Ecology. 2012;21(8):1909–1917. doi: 10.1111/j.1365-294X.2011.05383.x. [DOI] [PubMed] [Google Scholar]
- Battin et al. (2008).Battin TJ, Kaplan LA, Findlay S, Hopkinson CS, Marti E, Packman AI, Newbold JD, Sabater F. Biophysical controls on organic carbon fluxes in fluvial networks. Nature Geoscience. 2008;1(2):95–100. doi: 10.1038/ngeo101. [DOI] [Google Scholar]
- Beaulieu et al. (2011).Beaulieu JJ, Tank JL, Hamilton SK, Wollheim WM, Hall RO, Mulholland PJ, Peterson BJ, Ashkenas LR, Cooper LW, Dahm CN, Dodds WK, Grimm NB, Johnson SL, McDowell WH, Poole GC, Valett HM, Arango CP, Bernot MJ, Burgin AJ, Crenshaw CL, Helton AM, Johnson LT, O’Brien JM, Potter JD, Sheibley RW, Sobota DJ, Thomas SM. Nitrous oxide emission from denitrification in stream and river networks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):214–219. doi: 10.1073/pnas.1011464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Nelson et al. (2004).Benitez-Nelson CR, O’Neill L, Kolowith LC, Pellechia P, Thunell R. Phosphonates and particulate organic phosphorus cycling in an anoxic marine basin. Limnology and Oceanography. 2004;49(5):1593–1604. doi: 10.4319/lo.2004.49.5.1593. [DOI] [Google Scholar]
- Bergkemper et al. (2016).Bergkemper F, Schöler A, Engel M, Lang F, Krüger J, Schloter M, Schulz S. Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environmental Microbiology. 2016;18(6):1988–2000. doi: 10.1111/1462-2920.13188. [DOI] [PubMed] [Google Scholar]
- Bowen et al. (2011).Bowen JL, Ward BB, Morrison HG, Hobbie JE, Valiela I, Deegan LA, Sogin ML. Microbial community composition in sediments resists perturbation by nutrient enrichment. ISME Journal. 2011;5(9):1540–1548. doi: 10.1038/ismej.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner (1965).Bremner JM. Organic forms of nitrogen. In: Black CA, editor. Method of Soil Analysis. Part II. Madison: American Society of Agronomy; 1965. [Google Scholar]
- Briée, Moreira & López-García (2007).Briée C, Moreira D, López-García P. Archaeal and bacterial community composition of sediment and plankton from a suboxic freshwater pond. Research in Microbiology. 2007;158(3):213–227. doi: 10.1016/j.resmic.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Caporaso et al. (2010).Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, Nelson & Palmer (2000).Cardinale BJ, Nelson K, Palmer MA. Linking species diversity to the functioning of ecosystems: on the importance of environmental context. Oikos. 2000;91(1):175–183. doi: 10.1034/j.1600-0706.2000.910117.x. [DOI] [Google Scholar]
- Cole et al. (2006).Cole JJ, Carpenter SR, Pace ML, Van De Bogert MC, Kitchell JL, Hodgson JR. Differential support of lake food webs by three types of terrestrial organic carbon. Ecology Letters. 2006;9(5):558–568. doi: 10.1111/j.1461-0248.2006.00898.x. [DOI] [PubMed] [Google Scholar]
- Cole et al. (2007).Cole JJ, Prairie YT, Caraco NF, McDowell WH, Tranvik LJ, Striegl RG, Duarte CM, Kortelainen P, Downing JA, Middelburg JJ, Melack J. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems. 2007;10(1):172–185. doi: 10.1007/s10021-006-9013-8. [DOI] [Google Scholar]
- Cotner & Biddanda (2002).Cotner JB, Biddanda BA. Small players, large role: Microbial influence on biogeochemical processes in pelagic aquatic Ecosystems. Ecosystems. 2002;5(2):105–121. doi: 10.1007/s10021-001-0059-3. [DOI] [Google Scholar]
- Dalsgaard et al. (2003).Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuña-González J. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature. 2003;422(6932):606–608. doi: 10.1038/nature01526. [DOI] [PubMed] [Google Scholar]
- Dopheide et al. (2015).Dopheide A, Lear G, He Z, Zhou J, Lewis GD. Functional gene composition, diversity and redundancy in microbial stream biofilm communities. PLOS ONE. 2015;10(4):e0123179. doi: 10.1371/journal.pone.0123179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang et al. (2011).Fang C, Lai Z, Yang J, Xu Y. Study on the nonuniform spatial distribution of water level in Poyang Lake based on ASAR images and DEM. Procedia Environmental Sciences. 2011;10:2540–2546. doi: 10.1016/j.proenv.2011.09.233. [DOI] [Google Scholar]
- Fierer et al. (2013).Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science. 2013;342(6158):621–624. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- Fierer et al. (2012).Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman & Zak (2015).Freedman ZB, Zak DR. Atmospheric N deposition alters connectance, but not functional potential among saprotrophic bacterial communities. Molecular Ecology. 2015;24(12):3170–3180. doi: 10.1111/mec.13224. [DOI] [PubMed] [Google Scholar]
- Fulweiler & Heiss (2014).Fulweiler RW, Heiss EM. (Nearly) a decade of directly measured sediment N-2 fluxes what can Narragansett Bay tell us about the global ocean nitrogen budget? Oceanography. 2014;27(1):184–195. doi: 10.5670/oceanog.2014.22. [DOI] [Google Scholar]
- Galand et al. (2018).Galand PE, Pereira O, Hochart C, Auguet JC, Debroas D. A strong link between marine microbial community composition and function challenges the idea of functional redundancy. ISME Journal. 2018;12(10):2470–2478. doi: 10.1038/s41396-018-0158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, Bohannan & Whitaker (2008).Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320(5879):1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Harrison et al. (2009).Harrison JA, Maranger RJ, Alexander RB, Giblin AE, Jacinthe P-A, Mayorga E, Seitzinger SP, Sobota DJ, Wollheim WM. The regional and global significance of nitrogen removal in lakes and reservoirs. Biogeochemistry. 2009;93(1–2):143–157. doi: 10.1007/s10533-008-9272-x. [DOI] [Google Scholar]
- He et al. (2015).He R, Wooller MJ, Pohlman JW, Tiedje JM, Leigh MB. Methane-derived carbon flow through microbial communities in arctic lake sediments. Environmental Microbiology. 2015;17(9):3233–3250. doi: 10.1111/1462-2920.12773. [DOI] [PubMed] [Google Scholar]
- Hedges, Clark & Come (1988).Hedges JI, Clark WA, Come GL. Organic matter sources to the water column and surficial sediments of a marine bay. Limnology and Oceanography. 1988;33(5):1116–1136. doi: 10.4319/lo.1988.33.5.1116. [DOI] [Google Scholar]
- Holmer & Storkholm (2001).Holmer M, Storkholm P. Sulphate reduction and sulphur cycling in lake sediments: a review. Freshwater Biology. 2001;46(4):431–451. doi: 10.1046/j.1365-2427.2001.00687.x. [DOI] [Google Scholar]
- Hoostal & Bouzat (2008).Hoostal MJ, Bouzat JL. The modulating role of dissolved organic matter on spatial patterns of microbial metabolism in Lake Erie sediments. Microbial Ecology. 2008;55(2):358–368. doi: 10.1007/s00248-007-9281-7. [DOI] [PubMed] [Google Scholar]
- Horvath (1972).Horvath RS. Microbial co-metabolism and the degradation of organic compounds in nature. Bacteriological Reviews. 1972;36(2):146–155. doi: 10.1128/br.36.2.146-155.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2016).Huang X, Hu B, Wang P, Chen X, Xu B. Microbial diversity in lake-river ecotone of Poyang Lake, China. Environmental Earth Sciences. 2016;75(1):1–7. doi: 10.1007/s12665-015-4873-x. [DOI] [Google Scholar]
- Hupfer & Lewandowski (2008).Hupfer M, Lewandowski J. Oxygen controls the phosphorus release from lake sediments - a long-lasting paradigm in limnology. International Review of Hydrobiology. 2008;93(4–5):415–432. doi: 10.1002/iroh.200711054. [DOI] [Google Scholar]
- Jenkins & Kemp (1984).Jenkins MC, Kemp WM. The coupling of nitrification and denitrification in two estuarine sediments. Limnology and Oceanography. 1984;29(3):609–619. doi: 10.4319/lo.1984.29.3.0609. [DOI] [Google Scholar]
- Ji et al. (2015).Ji B, Yang K, Zhu L, Jiang Y, Wang H, Zhou J, Zhang H. Aerobic denitrification: a review of important advances of the last 30 years. Biotechnology and Bioprocess Engineering. 2015;20(4):643–651. doi: 10.1007/s12257-015-0009-0. [DOI] [Google Scholar]
- Jiang et al. (2006).Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Applied and Environmental Microbiology. 2006;72(6):3832–3845. doi: 10.1128/AEM.02869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones (2010).Jones NE. Incorporating lakes within the river discontinuum: longitudinal changes in ecological characteristics in stream-lake networks. Canadian Journal of Fisheries and Aquatic Sciences. 2010;67(8):1350–1362. doi: 10.1139/F10-069. [DOI] [Google Scholar]
- Kanehisa & Goto (2000).Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann et al. (2012).Kellermann MY, Wegener G, Elvert M, Yoshinaga MY, Lin Y-S, Holler T, Mollar XP, Knittel K, Hinrichs KU. Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(47):19321–193216. doi: 10.1073/pnas.1208795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononova & Nesmeyanova (2002).Kononova SV, Nesmeyanova MA. Phosphonates and their degradation by microorganisms. Biochemistry (Moscow) 2002;67(2):184–195. doi: 10.1023/A:1014409929875. [DOI] [PubMed] [Google Scholar]
- Koyama (1963).Koyama T. Gaseous metabolism in lake sediments and paddy soils and the production of atmospheric methane and hydrogen. Journal of Geophysical Research. 1963;68(13):3971–3973. doi: 10.1029/JZ068i013p03971. [DOI] [Google Scholar]
- Kuypers et al. (2003).Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jørgensen BB, Kuenen JG, Sinninghe Damsté JS, Strous M, Jetten MSM. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422(6932):608–611. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- Lamba et al. (2017).Lamba S, Bera S, Rashid M, Medvinsky AB, Sun G-Q, Acquisti C, Chakraborty A, Li B-L. Organization of biogeochemical nitrogen pathways with switch-like adjustment in fluctuating soil redox conditions. Royal Society Open Science. 2017;4(1):160768. doi: 10.1098/rsos.160768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille et al. (2013).Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Yu & Guo (2017).Liao M, Yu G, Guo Y. Eutrophication in Poyang Lake (Eastern China) over the last 300 years in response to changes in climate and lake biomass. PLOS ONE. 2017;12:e01693191. doi: 10.1371/journal.pone.0169319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu & Fang (2017).Liu J, Fang S. Comprehensive evaluation of the potential risk from cyanobacteria blooms in Poyang Lake based on nutrient zoning. Environmental Earth Sciences. 2017;76(9):342. doi: 10.1007/s12665-017-6678-6. [DOI] [Google Scholar]
- Liu, Fang & Sun (2016).Liu J, Fang S, Sun J. Nutrient zoning of Poyang Lake based on aquatic eco-environment indices. Environmental Earth Sciences. 2016;75(1):1–12. doi: 10.1007/s12665-015-4873-x. [DOI] [Google Scholar]
- Liu et al. (2016).Liu X, Li Y-L, Liu B-G, Qian K-M, Chen Y-W, Gao J-F. Cyanobacteria in the complex river-connected Poyang Lake: horizontal distribution and transport. Hydrobiologia. 2016;768(1):95–110. doi: 10.1007/s10750-015-2536-2. [DOI] [Google Scholar]
- Liu & Xu (2016).Liu L, Xu M. Microbial biomass in sediments affects greenhouse gas effluxes in Poyang Lake in China. Journal of Freshwater Ecology. 2016;31(1):109–121. doi: 10.1080/02705060.2015.1046511. [DOI] [Google Scholar]
- Liu et al. (2018).Liu W, Yao L, Jiang X, Guo L, Cheng X, Liu G. Sediment denitrification in Yangtze lakes is mainly influenced by environmental conditions but not biological communities. Science of the Total Environment. 2018;616-617:978–987. doi: 10.1016/j.scitotenv.2017.10.221. [DOI] [PubMed] [Google Scholar]
- Llorens-Marès et al. (2015).Llorens-Marès T, Yooseph S, Goll J, Hoffman J, Vila-Costa M, Borrego CM, Dupont CL, Casamayor EO. Connecting biodiversity and potential functional role in modern euxinic environments by microbial metagenomics. ISME Journal. 2015;9(7):1648–1661. doi: 10.1038/ismej.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca et al. (2017).Louca S, Jacques SMS, Pires APF, Leal JS, Srivastava DS, Parfrey LW, Farjalla VF, Doebeli M. High taxonomic variability despite stable functional structure across microbial communities. Nature Ecology & Evolution. 2017;1(1):0015. doi: 10.1038/s41559-016-0015. [DOI] [PubMed] [Google Scholar]
- Lozupone & Knight (2007).Lozupone CA, Knight R. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(27):11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2016).Lu S, Sun Y, Zhao X, Wang L, Ding A, Zhao X. Sequencing insights into microbial communities in the water and sediments of Fenghe River, China. Archives of Environmental Contamination and Toxicology. 2016;71(1):122–132. doi: 10.1007/s00244-016-0277-5. [DOI] [PubMed] [Google Scholar]
- Lv et al. (2017).Lv P, Luo J, Zhuang X, Zhang D, Huang Z, Bai Z. Diversity of culturable aerobic denitrifying bacteria in the sediment, water and biofilms in Liangshui River of Beijing, China. Scientific Reports. 2017;7(1):10032. doi: 10.1038/s41598-017-09556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackelprang et al. (2011).Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480(7377):368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- Marcarelli & Wurtsbaugh (2009).Marcarelli AM, Wurtsbaugh WA. Nitrogen fixation varies spatially and seasonally in linked stream-lake ecosystems. Biogeochemistry. 2009;94(2):95–110. doi: 10.1007/s10533-009-9311-2. [DOI] [Google Scholar]
- McDonald et al. (2012).McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME Journal. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier et al. (2016).Meunier CL, Gundale MJ, Sánchez IS, Liess A. Impact of nitrogen deposition on forest and lake food webs in nitrogen-limited environments. Global Change Biology. 2016;22(1):164–179. doi: 10.1111/gcb.12967. [DOI] [PubMed] [Google Scholar]
- Newell et al. (2016).Newell SE, Pritchard KR, Foster SQ, Fulweiler RW. Molecular evidence for sediment nitrogen fixation in a temperate New England estuary. PeerJ. 2016;4(4):e1615. doi: 10.7717/peerj.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton et al. (2011).Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiology and Molecular Biology Reviews. 2011;75(1):14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen et al. (2006).Nielsen JL, Klausen C, Nielsen PH, Burford M, Jorgensen N. Detection of activity among uncultured Actinobacteria in a drinking water reservoir. FEMS Microbiology Ecology. 2006;55(3):432–438. doi: 10.1111/j.1574-6941.2005.00054.x. [DOI] [PubMed] [Google Scholar]
- Nils (2003).Nils R-P. Coupled nitrification-denitrification in autotrophic and heterotrophic estuarine sediments: on the influence of benthic microalgae. Limnology and Oceanography. 2003;48(1):93–105. doi: 10.4319/lo.2003.48.1.0093. [DOI] [Google Scholar]
- Nishihama et al. (2008).Nishihama S, Haraguchi A, Kawano T, Michiki K, Nakazawa K, Suzuki T, Uezu K, Yoshizuka K. Seasonal changes in the microbial population of the water column and sediments of the Ongagawa River, northern Kyushu, Japan. Limnology. 2008;9(1):35–45. doi: 10.1007/s10201-008-0236-6. [DOI] [Google Scholar]
- Nixon (1995).Nixon SW. Coastal marine eutrophication: a definition, social causes, and future concerns. Ophelia. 1995;41(1):199–219. doi: 10.1080/00785236.1995.10422044. [DOI] [Google Scholar]
- Nyström (2006).Nyström M. Redundancy and response diversity of functional groups: Implications for the resilience of coral reefs. Ambio. 2006;35(1):30–35. doi: 10.1579/0044-7447-35.1.30. [DOI] [PubMed] [Google Scholar]
- Ohtake et al. (1996).Ohtake H, Wu H, Imazu K, Anbe Y, Kato J, Kuroda A. Bacterial phosphonate degradation, phosphite oxidation and polyphosphate accumulation. Resources, Conservation and Recycling. 1996;18(1–4):125–134. doi: 10.1016/S0921-3449(96)01173-1. [DOI] [Google Scholar]
- Oksanen et al. (2007).Oksanen J, Kindt R, Legendre PO, Hara B, Stevens MHH, Oksanen MJ, Suggests MASS. The vegan package. Community Ecology Package. 2007;10:631–637. [Google Scholar]
- Ollivier et al. (2011).Ollivier J, Töwe S, Bannert A, Hai B, Kastl E-M, Meyer A, Su MX, Kleineidam K, Schloter M. Nitrogen turnover in soil and global change. FEMS Microbiology Ecology. 2011;78(1):3–16. doi: 10.1111/j.1574-6941.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- Parker et al. (2016).Parker SR, West RF, Boyd ES, Feyhl-Buska J, Gammons CH, Johnston TB, Williams GP, Poulson SR. Biogeochemical and microbial seasonal dynamics between water column and sediment processes in a productive mountain lake: Georgetown Lake, MT, USA. Journal of Geophysical Research: Biogeosciences. 2016;121(8):2064–2081. doi: 10.1002/2015JG003309. [DOI] [Google Scholar]
- Philippot et al. (2010).Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. The ecological coherence of high bacterial taxonomic ranks. Nature Reviews Microbiology. 2010;8(7):523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017).R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- Ren et al. (2017b).Ren Z, Gao HK, Elser JJ, Zhao QD. Microbial functional genes elucidate environmental drivers of biofilm metabolism in glacier-fed streams. Scientific Reports. 2017b;7(1):e12668. doi: 10.1038/s41598-017-13086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren et al. (2019).Ren Z, Qu XD, Peng WQ, Yu Y, Zhang M. Nutrients drive the structures of bacterial communities in sediments and surface waters in the river-lake system of Poyang Lake. Water. 2019;11(5):930. doi: 10.3390/w11050930. [DOI] [Google Scholar]
- Ren et al. (2017a).Ren Z, Wang F, Qu XD, Elser JJ, Liu Y, Chu LM. Taxonomic and functional differences between microbial communities in Qinghai Lake and its input streams. Frontiers in Microbiology. 2017a;8:2319. doi: 10.3389/fmicb.2017.02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld (2002).Rosenfeld JS. Functional redundancy in ecology and conservation. Oikos. 2002;98(1):156–162. doi: 10.1034/j.1600-0706.2002.980116.x. [DOI] [Google Scholar]
- Röeske et al. (2012).Röeske K, Sachse R, Scheerer C, Röeske I. Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany) Systematic and Applied Microbiology. 2012;35(1):35–44. doi: 10.1016/j.syapm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Ruban et al. (1999).Ruban V, López-Sánchez JF, Pardo P, Rauret G, Muntau H, Quevauviller P. Selection and evaluation of sequential extraction procedures for the determination of phosphorus forms in lake sediment. Journal of Environmental Monitoring. 1999;1(1):51–56. doi: 10.1039/a807778i. [DOI] [PubMed] [Google Scholar]
- Schlesinger (2009).Schlesinger WH. On the fate of anthropogenic nitrogen. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(1):203–208. doi: 10.1073/pnas.0810193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowanek & Verstraete (1990).Schowanek D, Verstraete W. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Applied and Environmental Microbiology. 1990;56:895–903. doi: 10.1128/aem.56.4.895-903.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzinger (1988).Seitzinger SP. Denitrification in freshwater and coastal marine ecosystems: ecological and geochemical significance. Limnology and Oceanography. 1988;33:702–724. [Google Scholar]
- Smith (2003).Smith VH. Eutrophication of freshwater and coastal marine ecosystems a global problem. Environmental Science and Pollution Research. 2003;10(2):126–139. doi: 10.1065/espr2002.12.142. [DOI] [PubMed] [Google Scholar]
- Smith & Kaushal (2015).Smith RM, Kaushal SS. Carbon cycle of an urban watershed: exports, sources, and metabolism. Biogeochemistry. 2015;126(1–2):173–195. doi: 10.1007/s10533-015-0151-y. [DOI] [Google Scholar]
- Thamdrup & Dalsgaard (2002).Thamdrup B, Dalsgaard T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Applied and Environmental Microbiology. 2002;68(3):1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman (2012).Thurman EM. Organic geochemistry of natural waters. Vol. 2. Dordrecht: Springer Science & Business Media; 2012. [Google Scholar]
- Tiedje et al. (1983).Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA. Denitrification: ecological niches, competition and survival. Antonie van Leeuwenhoek. 1983;48(6):569–583. doi: 10.1007/BF00399542. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency (EPA) (2017).United States Environmental Protection Agency (EPA) Clean water act analytical methods. 2017. https://www.epa.gov/cwa-methods https://www.epa.gov/cwa-methods
- Van Der Heijden, Bardgett & Van Straalen (2008).Van Der Heijden MGA, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecology Letters. 2008;11(3):296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Van Kessel (1977).Van Kessel JF. Factors affecting the denitrification rate in two water-sediment systems. Water Research. 1977;11(3):259–267. doi: 10.1016/0043-1354(77)90057-4. [DOI] [Google Scholar]
- Wang & Liang (2015).Wang L, Liang T. Distribution characteristics of phosphorus in the sediments and overlying water of Poyang Lake. PLOS ONE. 2015;10:e01258595. doi: 10.1371/journal.pone.0125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang J, Zhang Y, Yang F, Cao X, Bai Z, Zhu J, Chen E, Li Y, Ran Y. Spatial and temporal variations of chlorophyll-a concentration from 2009 to 2012 in Poyang Lake, China. Environmental Earth Sciences. 2015;73(8):4063–4075. doi: 10.1007/s12665-014-3691-x. [DOI] [Google Scholar]
- Wang et al. (2018).Wang K, Zou L, Lu X, Mou X. Organic carbon source and salinity shape sediment bacterial composition in two China marginal seas and their major tributaries. Science of the Total Environment. 2018;633:1510–1517. doi: 10.1016/j.scitotenv.2018.03.295. [DOI] [PubMed] [Google Scholar]
- Webster & Meyer (1997).Webster JR, Meyer JL. Organic matter budgets for streams: a synthesis. Journal of the North American Benthological Society. 1997;16(1):141–161. doi: 10.2307/1468247. [DOI] [Google Scholar]
- Wilkinson et al. (2018).Wilkinson TJ, Huws SA, Edwards JE, Kingston-Smith AH, Siu-Ting K, Hughes M, Rubino F, Friedersdorff M, Creevey CJ. CowPI: a rumen microbiome focussed version of the PICRUSt functional inference software. Frontiers in Microbiology. 2018;9:e1095. doi: 10.3389/fmicb.2018.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollheim et al. (2008).Wollheim WM, Vörösmarty CJ, Bouwman AF, Green P, Harrison J, Linder E, Peterson BJ, Seitzinger SP, Syvitski JPM. Global N removal by freshwater aquatic systems using a spatially distributed, within-basin approach. Global Biogeochemical Cycles. 2008;22(2):GB2026. doi: 10.1029/2007GB002963. [DOI] [Google Scholar]
- Yang et al. (2017).Yang S, Zhang Y, Cong J, Wang M, Zhao M, Lu H, Xie C, Yang C, Yuan T, Li D, Zhou J, Gu B, Yang Y. Variations of soil microbial community structures beneath broadleaved forest trees in temperate and subtropical climate zones. Frontiers in Microbiology. 2017;8:200. doi: 10.3389/fmicb.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao et al. (2018).Yao L, Chen C, Liu G, Liu W. Sediment nitrogen cycling rates and microbial abundance along a submerged vegetation gradient in a eutrophic lake. Science of the Total Environment. 2018;616-617:899–907. doi: 10.1016/j.scitotenv.2017.10.230. [DOI] [PubMed] [Google Scholar]
- Ylla et al. (2013).Ylla I, Peter H, Romani AM, Tranvik LJ. Different diversity-functioning relationship in lake and stream bacterial communities. FEMS Microbiology Ecology. 2013;85(1):95–103. doi: 10.1111/1574-6941.12101. [DOI] [PubMed] [Google Scholar]
- Zehr et al. (2003).Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environmental Microbiology. 2003;5(7):539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang D, Liao Q, Zhang L, Wang D, Luo L, Chen Y, Zhong J, Liu J. Occurrence and spatial distributions of microcystins in Poyang Lake, the largest freshwater lake in China. Ecotoxicology. 2015;24(1):19–28. doi: 10.1007/s10646-014-1349-9. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2011).Zhao J, Li J, Yan H, Zheng L, Dai Z. Analysis on the water exchange between the main stream of the Yangtze River and the Poyang Lake. Procedia Environmental Sciences. 2011;10:2256–2264. doi: 10.1016/j.proenv.2011.09.353. [DOI] [Google Scholar]
- Zumft (1997).Zumft WG. Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw sequence data are available at National Center for Biotechnology Information (PRJNA436872, SRP133903).