Abstract
Susceptibility to chronic beryllium (Be) disease (CBD) is linked to HLA-DP molecules possessing a glutamic acid at the 69th position of the β-chain (βGlu69), with the most prevalent βGlu69-containing molecule being HLA-DP2. We have previously shown that HLA-DP2–transgenic (HLA-DP2–Tg) mice exposed to Be oxide (BeO) develop mononuclear infiltrates in a peribronchovascular distribution and a beryllium-specific, HLA-DP2–restricted CD4+ T cell response. In addition to T cells, B cells constituted a major portion of infiltrated leukocytes in the lung of BeO-exposed HLA-DP2–Tg mice and sequester BeO particles within ectopic lymphoid aggregates and granulomas. B cell depletion was associated with a loss of lymphoid aggregates and granulomas as well as a significant increase in lung injury in BeO-exposed mice. The protective role of B cells was innate in origin, and BeO-induced B cell recruitment to the lung was dependent on MyD88 signaling. Similar to BeO-exposed HLA-DP2–Tg mice, B cells also accumulate in the lungs of CBD subjects, located at the periphery and surrounding the granuloma. Overall, our data suggest what we believe is a novel modulatory role for B cells in the protection of the lung against sterile particulate exposure, with B cell recruitment to the inflamed lung occurring in an antigen-independent and MyD88-dependent manner.
Keywords: Immunology, Pulmonology
Keywords: Adaptive immunity, B cells, Chemokines
Following BeO exposure in the lung, B cells accumulate in ectopic lymphoid aggregates and granulomas that sequester beryllium particles and protect the lung from injury.
Introduction
Exposure to high levels of beryllium (Be) in the workplace causes activation of the innate immune system with the recruitment of neutrophils to the lung and the development of intra-alveolar edema, consistent with acute lung injury (1). With ongoing exposure, a subset of these individuals develop chronic beryllium disease (CBD), a granulomatous lung disorder that is characterized by an influx of Be-specific CD4+ T cells into the lung and fibrosis (2–4). The development of Be sensitization and CBD has been strongly linked to HLA-DPB1 alleles expressing a glutamic acid at position 69 of the β-chain (βGlu69) (5–9), and the majority of Be-specific CD4+ T cells recognize antigen in an HLA-DP–restricted manner, suggesting that these molecules are crucial for disease development due to their ability to bind and present Be to antigen-specific CD4+ T cells (10–14). In mice transgenic (Tg) for HLA-DP2, the most prevalent βGlu69-expressing allele, Be oxide (BeO) exposure results in the accumulation of mononuclear cells in a peribronchovascular distribution in the lung and an HLA-DP2–restricted Be-specific adaptive immune response (15, 16). Importantly, a subset of these Be-specific CD4+ T cells recognize the same HLA-DP2 peptide/Be epitope as T cells derived from the lungs of CBD subjects (15). Thus, this murine model replicates many of the features of the human disease (15, 16). However, the mechanisms linking innate immune activation and the development of a Be-specific adaptive immune response remain unknown.
Intratracheal instillation of crystalline sterile particulates [e.g., Be hydroxide Be(OH)2 and aluminum hydroxide (Al(OH)3)] induces alveolar macrophage cell death and the subsequent release of damage-associated molecular pattern molecules (DAMPS), including IL-1α and DNA, that drive inflammation (17–19). Despite similarities in the mechanism by which Be(OH)2 and Al(OH)3 induce inflammation in the lung, significant differences exists. For example, the adjuvant properties of Be(OH)2 in the lung were dependent on MyD88 and independent of the IL-1 pathway (17), while IL-1α was shown to play a key role in the adjuvant effects of Al(OH)3 (18). These differing adjuvant effects translate into the generation of different CD4+ T cell responses, with Be(OH)2 driving the expansion of Th1-type T cells (20, 21) and Al(OH)3 generating Th2-type T cells and enhancing IgE production and eosinophilic inflammation (18).
Here, we noted that, in addition to CD3+ T cells, intratracheal BeO exposure resulted in the recruitment of large numbers of activated follicular B cells to the lung. These B cells were organized in ectopic lymphoid aggregates (ELAs) equipped with high endothelial venules (HEVs) and containing CXCL13-expressing stromal-like cells. Depletion of B cells had no effect of the quantity of Be-specific CD4+ T cells in the lung. However, B cell depletion eliminated the ELAs and enhanced lung injury, suggesting that B cells and these tertiary lymphoid structures play a protective role after BeO exposure. In MD4 BCR–Tg mice, B cell recruitment to the lung continued even in the absence of both the HLA-DP2 transgene and a Be-specific CD4+ T cell response, suggesting that the protective role of these B cells was innate in origin and independent of antigen. Finally, BeO-induced B cell recruitment to the lung was dependent on MyD88 and CXCL13 and independent of DNA signaling pathways such as TLR9 and STING. B cell accumulation was also noted in the lungs of CBD patients, surrounding the granulomas. Taken together, our findings suggest what we believe is a novel role of B cells in the sequestration of particulate antigen within ELAs to protect the lung from BeO-induced innate immune injury.
Results
B cell accumulation in the lungs of BeO-exposed HLA-DP2–Tg mice.
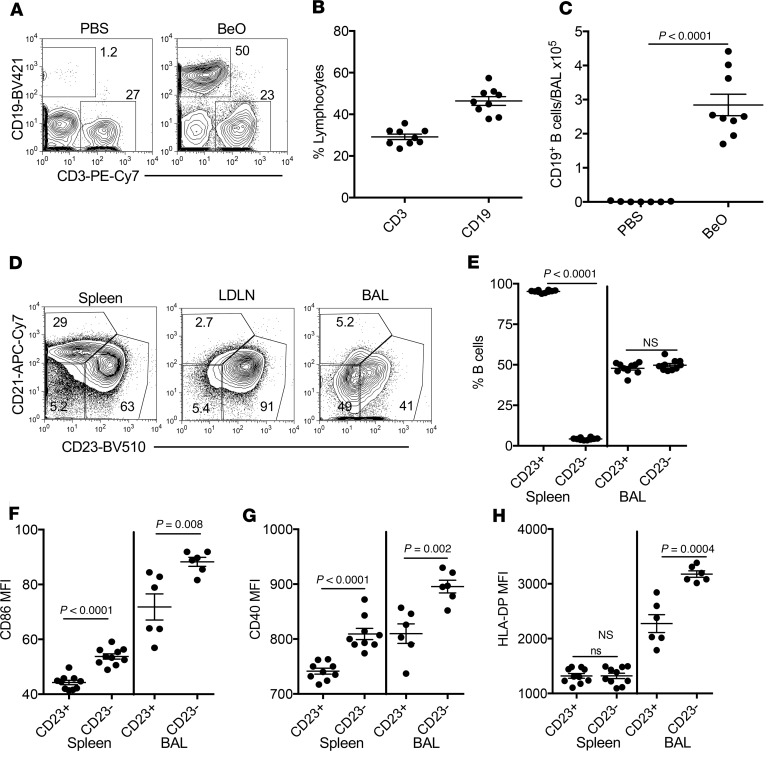
We have previously shown that HLA-DP2–Tg FVB/N mice exposed to 100 μg BeO via oropharyngeal aspiration on days 0, 1, and 2 and challenged with BeO (100 μg) on days 14, 15, 18, and 19 prior to sacrifice on day 21 develop a CD4+ T cell alveolitis (15). A subset of these CD4+ T cells are Be specific and recognize the same T cell epitopes as those T cells derived from the lungs of HLA-DP2–expressing CBD patients (15). In addition to CD3+ T cells, the lungs of BeO-exposed HLA-DP2–Tg mice contain neutrophils, recruited monocyte/macrophages (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125494DS1), and large populations of CD19+ B cells at day 21 (Figure 1). As shown in the representative density plots in Figure 1A and cumulative data in Figure 1B, 46% of bronchoalveolar lavage (BAL) cells in the lymphocyte gate expressed CD19 compared with 29% expressing CD3. Overall, BeO exposure resulted in the accumulation of a significantly increased number of CD19+ B cells in the BAL fluid (BALF) of BeO-exposed mice compared with PBS-treated control mice (Figure 1C). B cells recruited to the BAL, lung, and lung-draining lymph nodes (LDLNs) coexpressed CD19 and B220 (Supplemental Figure 2). BAL B cells were mature, expressing CD22 as well as surface IgM and IgD (data not shown). Using CD21 and CD23 to phenotype these cells, the predominant B cell subset in the spleen and LDLNs was follicular B cells (Figure 1D). As expected, marginal zone B cells were absent in BAL or LDLNs, and follicular B cells in BAL were equally divided between cells that expressed CD23 and those that had lost CD23 expression (Figure 1, D and E). Few CD23–CD21– B cells were present in the spleen and LDLNs (Figure 1D). We were unable to assess CD23 expression on lung B cells because collagenase digestion of lung tissue strips CD23 from the cell surface (22).
Figure 1. B cell accumulation and activation in the lungs after BeO exposure in FVB/N HLA-DP2–Tg mice.
HLA-DP2–Tg FVB/N mice were exposed to PBS or BeO as described in Methods, and BAL was harvested on day 21 and analyzed by flow cytometry. (A) Representative density plots show the percentage of CD19+ B cells and CD3+ T cells in the BAL of PBS- (left) and BeO-exposed (right) mice. The numbers in each density plot indicate the percentage of cells in each population. (B) Percentage of lymphocytes that are CD3+ T cells and CD19+ B cells in the BAL are cumulatively shown for PBS- and BeO-exposed mice. (C) Number of CD19+ B cells in the BAL of HLA-DP2–Tg mice exposed to either PBS or BeO is shown. (D) B220+CD19+ B cells in spleen were stained for CD21 and CD23 expression to establish gates for marginal zone B cells (CD21+CD23–) and follicular B cells (CD23+CD21–) in BAL and lung-draining lymph nodes (LDLNs). The numbers in each gate indicate the percentage of cells in each B cell subset. (E) Percentage of CD23+ and CD23– B cells in the spleen and BAL of HLA-DP2–Tg mice after BeO exposure is shown. The expression (mean fluorescence intensity [MFI]) of costimulatory molecules CD86 (F) and CD40 (G), and HLA-DP (H) on CD19+CD23+ and CD19+CD23– B cells derived from spleen (n = 10 mice/group) and BAL (n = 6 mice/group) of HLA-DP2–Tg mice after BeO exposure is shown. Data are representative of 3 independent experiments having 5 to 10 mice per group. Solid lines and error bars depict the mean ± SEM. Student’s t test (2 tailed) (B and C) and 2-way ANOVA were used to test for differences. A P value of <0.05 was considered statistically significant.
BAL CD23– B cells in BeO-exposed HLA-DP2–Tg mice were significantly more activated than their CD23+ counterparts based on increased expression of CD86 (Figure 1F), CD40 (Figure 1G), and HLA-DP (Figure 1H). Similar findings were seen in the spleen for CD86 and CD40, with no difference in HLA-DP2 expression seen in CD23– and CD23+ B cells. Compared with spleen B cells, BAL B cells expressed significantly greater CD86 (Figure 1F), CD40 (Figure 1G), and HLA-DP (Figure 1H), regardless of CD23 expression. Collectively, these data show an influx of activated, mature follicular B cells into the BAL of HLA-DP2–Tg mice exposed to BeO.
B cell localization in BeO-induced granulomatous inflammation.
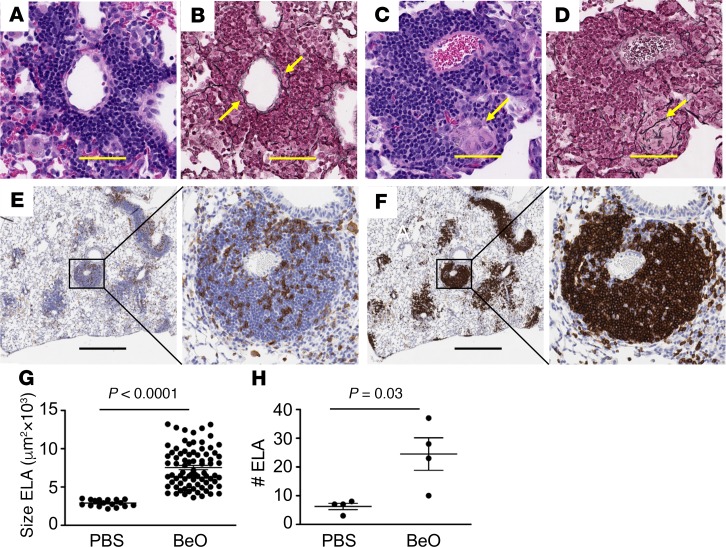
We have previously shown that WT FVB/N mice exposed to BeO develop few scattered cellular infiltrates (15). Conversely, HLA-DP2–Tg FVB/N mice exposed to BeO developed lymphocytic infiltrates in a peribronchovascular distribution (Figure 2A) with scattered granulomas (yellow arrow in Figure 2C and ref. 15). Reticulin fibers were intermixed within lymphocytic infiltrates in the lungs of mice after BeO exposure (Figure 2B, yellow arrows). In Figure 2D, a high-power view shows particulate-laden giant cells within a granuloma with reticulin fibers surrounding and likely supporting the structure of the granuloma. In order to localize the lymphocyte subsets within these infiltrates, we performed immunohistochemistry (Figure 2, E and F). CD4+ T cells were scattered throughout the infiltrates (Figure 2E), while B220+ B cells made up the majority of these cellular infiltrates and were concentrated into structures resembling primary B cell follicles in secondary lymphoid organs (Figure 2F). Consistent with our findings in the BAL, B cells were the predominant lymphocyte subset in these peribronchovascular lymphoid aggregates. Using morphometric analysis, BeO-induced ELAs were significantly larger in size (P < 0.0001; Figure 2G) and more numerous (P = 0.03; Figure 2H) compared with those in mice instilled with PBS.
Figure 2. BeO exposure resulted in the formation of ectopic lymphoid aggregates (ELAs) and granulomatous inflammation in HLA-DP2–Tg mice.
HLA-DP2–Tg mice were exposed to BeO as described in Methods, lungs were harvested on day 21, and serial sections were analyzed by H&E and reticulin staining (A–D) or were analyzed by immunohistochemical staining for CD4 and B220 (E and F). BeO exposure of HLA-DP2–Tg mice induced ELAs (A and B) and granuloma formation (C and D) in the lungs of HLA-DP2–Tg mice (H&E stained in A and B; reticulin stained in C and D; original magnification, ×40). Yellow arrows in B and D depict the distribution of type IV collagen within the aggregates and granulomas, respectively. The yellow arrow in C depicts a multinucleated giant cell. (E) Immunohistochemical staining of CD4+ cells and (F) B220+ cells infiltrating the lungs of BeO-exposed HLA-DP2–Tg mice. Original magnification (E and F), ×2 (left) and ×20 (right). Histology images are representative of 5 to 6 animals per group from 3 independent experiments. Scale bars: 20 μm. The average size (G) and number (H) of ELAs in the lungs of BeO-exposed HLA-DP2–Tg mice are shown. Statistical significance was determined using a Student’s t test (2 tailed), and the solid lines and error bars depict the mean ± SEM. A P value of <0.05 was considered statistically significant.
BeO exposure results in the formation of ELAs.
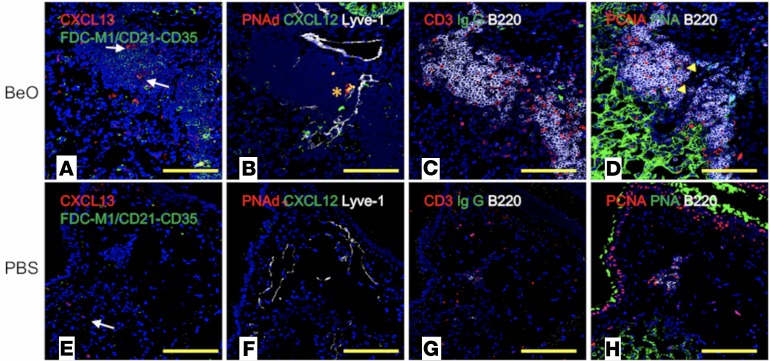
B cells are attracted to lymphoid follicles by homeostatic B cell chemokines, such as CXC chemokine ligand 13 (CXCL13), produced by follicular dendritic cells (FDCs) (23, 24) and stromal cells (25). We stained formalin-fixed, paraffin-embedded lung sections with mAbs specific for CXCL13 and FDCs (FDCM1, CD21/CD35) and identified CXCL13+ cells scattered throughout the ELAs of BeO-exposed HLA-DP2–Tg mice (Figure 3A). Although we did not detect FDC networks, B cells were numerous in the ELAs of mice treated with BeO (Figure 3A). In contrast, PBS-exposed mice had few CXCL13+ cells with round-shape morphology in the interstitial space, suggesting that these cells are CXCL13+ mononuclear phagocytes (Figure 3E). Recently, Fleige et al. (26) reported CXCL12-dependent induction of inducible bronchus-associated lymphoid tissue (iBALT) in the absence of FDCs in mice infected with Pseudomonas. Therefore, we stained lung sections with antibodies directed against CXCL12, peripheral node addressin (PNAd) expressed in HEVs, and lymphatic vessel endothelial hyaluronan 1 (Lyve-1). CXCL12+PNAd+ HEVs and lymphatics were detected at the border of B cell areas in lungs of HLA-DP2–Tg mice treated with BeO (Figure 3B). PNAd+ structures and lymphatics were evident in control mice; however, HEVs were not positive for CXCL12 and associated with no immune cell recruitment (Figure 3F).
Figure 3. BeO exposure of HLA-DP2–Tg mice induced the formation of ectopic lymphoid aggregates (ELAs).
Representative multicolor immunofluorescence lung images of serial sections from HLA-DP2–Tg mice exposed to either BeO (A–D) or PBS (E–H) are shown. Original magnification, ×200. (A and E) Lung sections were stained for CXCL13 (red), FDC-M1 (green), and nuclei were stained with DAPI (blue). (A) FDC-M1+ cells with branched stromal cell morphology were not present; however, CXCL13+ cells with stromal cell morphology (white arrows) were located inside lymphoid aggregates of BeO-exposed mice. (E) CXCL13+ cells with monocyte/macrophage–like morphology (white arrows) were present in PBS-treated HLA-DP2–Tg mice in the absence of ELAs. (B and F) Lung sections were stained for expression of CXCL12 (green), PNAd/high endothelial venule (HEV) cells (red), lymphatic vessels/Lyve-1 (white), and with DAPI (blue). (B) ELAs with CXCL12+ (green) PNAd+ HEVs (orange asterisk), and Lyve-1+ lymphatics were present in BeO-treated HLA-DP2–Tg mice. (F) A small PNAd+ HEV and Lyve-1+ lymphatics in PBS-treated HLA-DP2–Tg mice are shown. (C and G) Lung sections were stained for expression of IgG (green), CD3 (red), B220 (white), and with DAPI (blue). (C) CD3+ T cells outside and inside of B220+ B cell aggregates in the lungs of BeO-treated HLA-DP2–Tg–mice are shown. (G) Few dispersed B220+ B cells and CD3+ T cells were seen in the PBS-treated HLA-DP2–Tg mice. (D and H) Lung sections were stained for expression of PCNA (red), PNA (green), B220 (white), and with DAPI (blue). (D) Large B cell blasts (PCNA+PNA+B220lo, yellow arrowheads) were present in B cell aggregates of BeO-treated HLA-DP2–Tg mice. (H) Rare B220+ B cells were seen in the small ELAs observed in PBS-treated HLA-DP2–Tg mice and were not expressing PCNA or PNA. Yellow scale bars: 100 μm.
To analyze B and T cell compartmentalization as well as the presence of IgG+ antibody-secreting cells (ASCs), we stained lung sections with antibodies specific for CD3ε, CD45R (B220), and IgG. B cell aggregates containing T cells within as well as T cells in extrafollicular locations were seen in HLA-DP2–Tg mice receiving BeO (Figure 3C). In addition, cells staining for cytoplasmic IgG were located outside of B cell follicles, suggesting ASCs are likely derived from ELAs (Figure 3C). Conversely, B and T cells were scarce in the lungs of mice exposed to PBS (Figure 3G). Surprisingly, few large proliferating centroblasts positive for proliferating cell nuclear antigen and peanut agglutinin (PCNA+PNA+B220lo) were seen in the B cell aggregates of mice instilled with BeO (Figure 3D), while none were detected in the lungs of PBS-treated HLA-DP2–Tg mice (Figure 3H).
LDLNs served as internal controls for our multicolor fluorescent staining and confirmed that differentiation of pulmonary stromal cells into FDCs was not occurring after BeO instillation (Supplemental Figure 3A) or after PBS exposure (Supplemental Figure 3E). Immunofluorescence staining of LDLNs from BeO-exposed (Supplemental Figure 3B) or PBS-exposed (Supplemental Figure 3F) mice confirmed that PNAd+ HEVs were positive for CXCL12 and that there was no activation of B cells, at least based on the scarce numbers of IgG+ ASCs (Supplemental Figure 3, C and G) and the absence of proliferating B cell blasts (Supplemental Figure 3, D and H). Thus, our systematic characterization of these ELAs shows that BeO instillation induces the formation of lymphoid structures equipped with Lyve-1 lymphatics, PNAd+ HEVs decorated with CXCL12, and stromal cells that produce CXCL13, which attract CXCR5+ B and T cells to the lungs. Finally, the lack of germinal centers in the LDLNs suggest that BeO instillation is preferentially inducing a local innate immune response in the lung and that ASCs are potentially being produced in the absence of a germinal center.
Elevated CXCL13 in the BALF of BeO-exposed HLA-DP2–Tg mice.
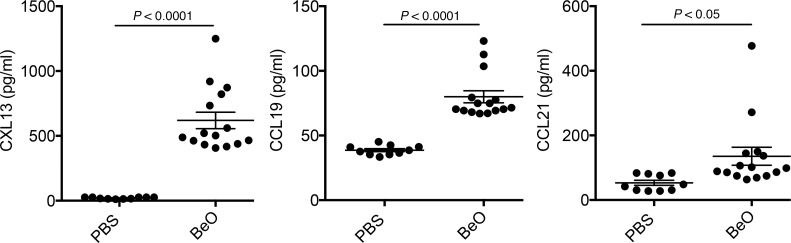
Homeostatic chemokines, such as CXCL13, CC chemokine ligand 19 (CCL19), and CCL21 are expressed in secondary lymphoid organs, directing the recruitment and organization of lymphocytes and dendritic cells for the induction of highly regulated immune responses (25, 27, 28). In addition, these chemokines are expressed in nonlymphoid tissues after inflammation or infection, recruiting lymphocytes to sites of inflammation (29, 30). To determine the stimulus for B cell recruitment to the lungs during BeO exposure, we performed ELISAs for CXCL13, CCL19, and CCL21. Compared with PBS-exposed HLA-DP2–Tg mice, BALF obtained from BeO-exposed HLA-DP2–Tg mice contained large quantities of CXCL13 (P < 0.0001; Figure 4). Similarly, significantly larger quantities of both CCL19 (P < 0.0001) and CCL21 (P < 0.05) were present in the BALF of HLA-DP2–Tg mice exposed to BeO compared with PBS-treated control mice (Figure 4).
Figure 4. BeO exposure induced secretion of homeostatic B cell chemokines.
HLA-DP2–Tg mice were exposed to BeO as described in Methods and sacrificed on day 21. Quantification of the homeostatic B cell chemokines CXCL13 (left), CCL19 (middle), and CCL21 (right), in the bronchoalveolar lavage fluid (BALF) obtained from HLA-DP2–Tg mice treated with either PBS or BeO is shown. Cumulative data from 3 separate experiments having 3 to 5 mice per group are shown, and the mean concentration (pg/ml) is shown as a solid line. Statistical significance was determined using a Student’s t test (2 tailed), and the solid line and error bars depict the mean ± SEM. A P value of <0.05 was considered statistically significant.
Next, we examined the expression of CXCR4 and CXCR5, receptors for CXCL12 and CXCL13, respectively, on B cells in BAL, lungs, LDLNs, and spleen and noted increased expression of CXCR4 on BAL B cells compared with the other organs (Supplemental Figure 4). CXCR5 was highly expressed on B cells in all of the compartments analyzed, with slightly decreased expression in lung tissue (Supplemental Figure 4), suggesting that the recruitment of B cells in the alveolar space is guided by coordinated expression of chemokines and their receptors. Collectively, these findings confirm a key role for CXCL12 and CXCL13 in the recruitment of CXCR4+CXCR5+ B cells to the BAL following BeO inhalation.
Effects of B cell depletion on lung inflammation.
In order to determine the role of B cells in BeO-induced lung inflammation, we depleted B cells with an anti–mouse CD20 mAb, 5D2, that was injected intraperitoneally (i.p.) 2 days prior to BeO exposure and weekly for the duration of the experiment. As shown in Supplemental Figure 5A, treatment with 5D2 depleted B cells from the BAL, lung, LDLNs, and spleen at day 21, with a greater than 97% reduction in B cell numbers. B cell depletion was also associated with a significantly decreased total serum IgG (Supplemental Figure 5B) as well as a decrease in serum IgG1, IgG2a, and IgG2b, with no change in IgG3 (Supplemental Figure 5C).
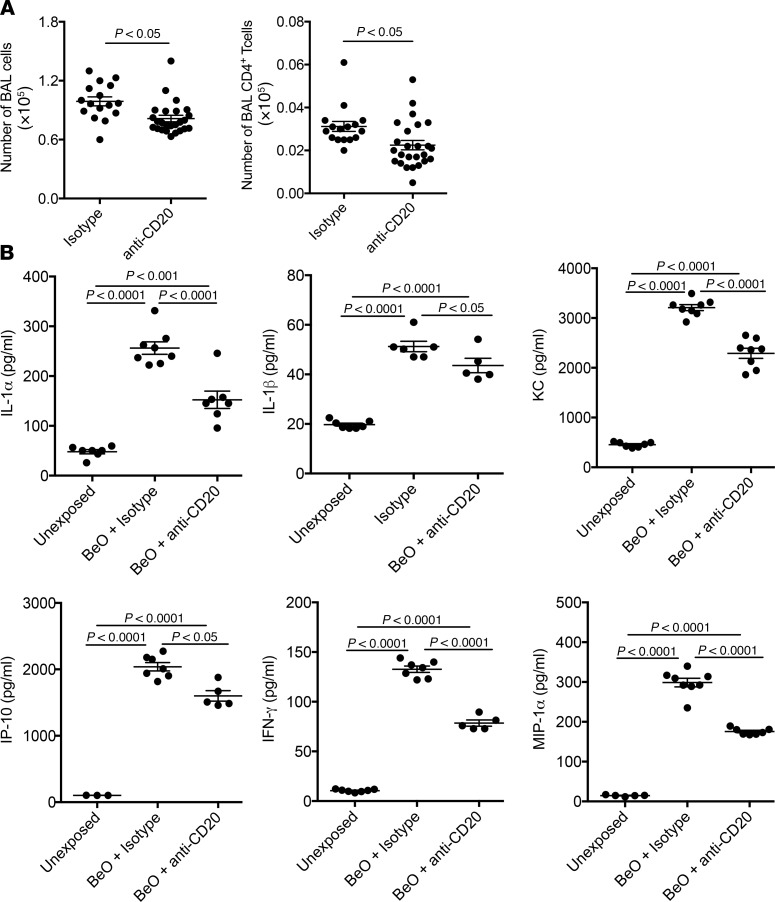
B cell depletion resulted in a modest, albeit significant, decrease in the number of WBCs in the BAL (P < 0.05; Figure 5A). In addition to the loss of B cells, a significant reduction in the number of BAL CD4+ T cells contributed to the overall reduction in the cellularity of the BAL (P < 0.05; Figure 6A). Compared with unexposed HLA-DP2–Tg mice, BeO exposure significantly increased secretion of IL-1α, IL-1β, KC, IFN-γ–induced protein 10 (IP-10), IFN-γ, and MIP-1α (P < 0.0001 for all cytokines) in lung homogenates (Figure 5B). Following B cell depletion, significant reductions in lung IL-1α (P < 0.0001), IL-1β (P < 0.05), KC (P < 0.0001), IP-10 (P < 0.05), IFN-γ (P < 0.0001), and MIP-1α (P < 0.0001) were seen, although the reductions in all of these cytokines remained significantly elevated compared with unexposed mice (P < 0.0001; Figure 5B). Collectively, these data suggest that B cells contribute to the BeO-induced inflammatory response.
Figure 5. B cell depletion reduced cellular recruitment to the lung and reduced inflammatory cytokine/chemokine secretion in BeO-exposed HLA-DP2–Tg mice.
HLA-DP2–Tg FVB/N mice were injected intraperitoneally with either anti–mouse CD20 mAb (5D2) or a murine IgG2a isotype control antibody 2 days prior to BeO exposure, and then weekly for the duration of the experiment. BALF was harvested on day 21 and analyzed by flow cytometry and cytokine bead array. (A) Total number of cells (left) and CD4+ T cells (right) in the BAL of BeO-exposed HLA-DP2–Tg mice and treated with either isotype or anti-CD20 mAb is shown. (B) Cytokines and chemokines measured by cytokine bead array in BALF collected from unexposed HLA-DP2–Tg mice or BeO-exposed HLA-DP2–Tg mice treated with either isotype control or anti-CD20 mAb is shown. Cumulative data from 2 separate experiments having 3 to 5 mice per group are shown, and the mean ± SEM concentration (pg/ml) is shown as a solid line with error bars. Statistical significance was determined using a 2-way ANOVA, and a P value of <0.05 was considered statistically significant.
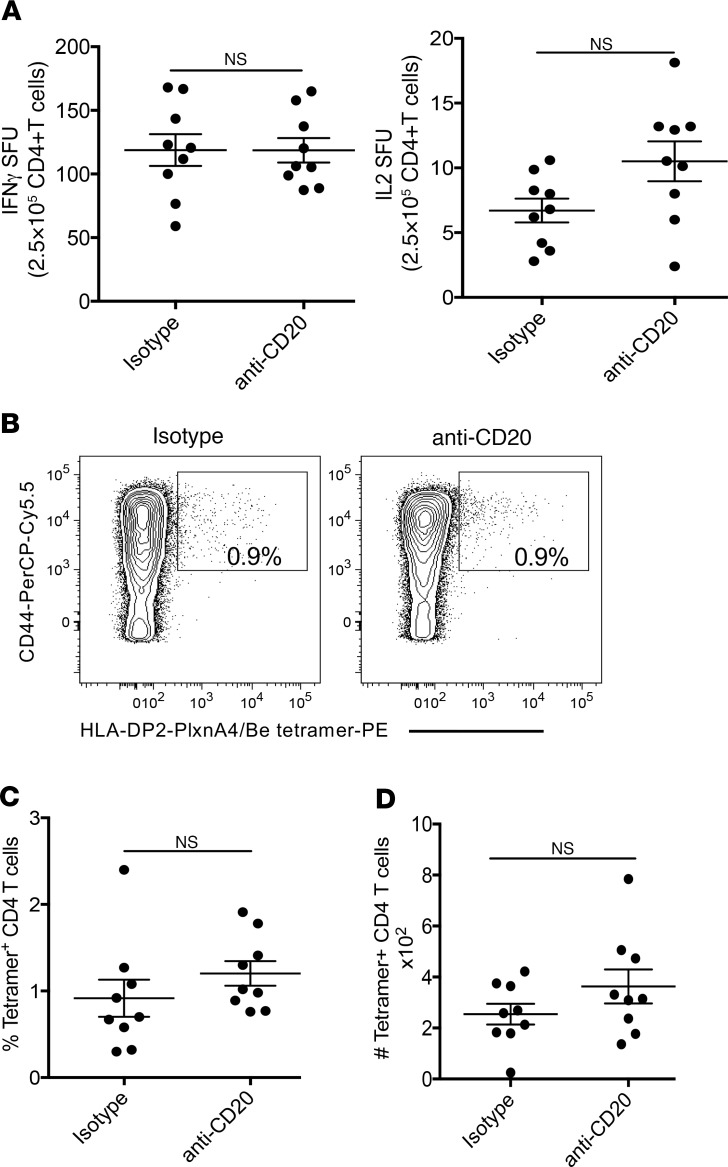
Figure 6. B cell depletion had no effect on the development of a Be-specific CD4+ T cell response in BeO-exposed HLA-DP2–Tg mice.
(A) ELISPOT assay shows beryllium-specific spot-forming units (SFU) for IFN-γ (left) and IL-2 (right) production by purified CD4+ T cells derived from lung cells of BeO-exposed HLA-DP2–Tg mice treated with either isotype control or anti-CD20 mAb (5D2). (B) Representative density plots showing HLA-DP2-PLXNA4/Be tetramer staining of gated CD4+ T cells from ex vivo BAL cells of HLA-DP2–Tg mice treated with either isotype control (left) or anti-CD20 mAb (right). The numbers in each density plot represent the percentage of tetramer-binding CD4+ T cells. Summary of the (C) frequency and (D) number of tetramer-positive ex vivo BAL CD4+ T cells in the BALF of BeO-exposed HLA-DP2–Tg mice treated with either isotype or anti-CD20 mAb. Dots on graphs indicate values from individual mice (n = 9/group), and bars indicate means. Data are representative of 3 independent experiments. A lack of statistical significance was determined using a Student’s t test (2 tailed), and the solid line and error bars depict the mean ± SEM. A P value of <0.05 was considered statistically significant.
B cell depletion had no effect on the generation of the Be-specific adaptive immune response.
Based on the importance of B cells as antigen-presenting cells in other immune-mediated disease, such as type 1 diabetes (31, 32), we initially hypothesized the lung B cells were serving as antigen-presenting cells for the induction of the Be-specific Th1-polarized adaptive immune response. Using ELISPOT to measure beryllium-specific T cell responses, we determined the number of IFN-γ– and IL-2– secreting CD4+ T cells in the lungs of BeO-exposed HLA-DP2–Tg mice that were treated with either anti-CD20 mAb or an isotype control mAb. To ensure that equal numbers of CD4+ T cells were added per well, CD4+ T cells were positively selected via magnetic beads, and 2.5 × 105 CD4+ T cells were cultured with equal numbers of purified CD19+ splenic B cells from an unexposed HLA-DP2–Tg mouse in the presence or absence of BeSO4. As shown in Figure 6A, the number of IFN-γ–secreting (left panel) and IL-2–secreting (right panel) CD4+ T cells in the lung was unchanged regardless of the presence of B cells. To determine whether the frequency of epitope-specific CD4+ T cells in the lung was altered by B cell depletion, we stained lung CD4+ T cells with Be-pulsed HLA-DP2-plexin A4 tetramers as previously described (14, 15). Total lung cells were analyzed by flow cytometry, with gating on live, B220+, F4/80+, CD8+, CD44hi, and CD4+ T cells, followed by examination of tetramer binding. No HLA-DP2 tetramer staining was detected in the lung of PBS-exposed mice or in the spleen of PBS, BeO-exposed mice (data not shown). Conversely, representative density plots show a distinct population of CD4+CD44hi T cells binding to the HLA-DP2-plexin A4/Be tetramer in the lungs of BeO-exposed mice in the presence or absence of B cells (0.9% and 0.9%, respectively; Figure 6B). Cumulatively, no difference was seen in the percentage or number of HLA-DP2-plexin A4/Be tetramer–binding CD4+ T cells in the lungs of BeO-exposed HLA-DP2–Tg mice in the presence or absence of B cells (Figure 6, C and D).
B cells modulate ELA and granuloma formation and protect against BeO-induced lung injury.
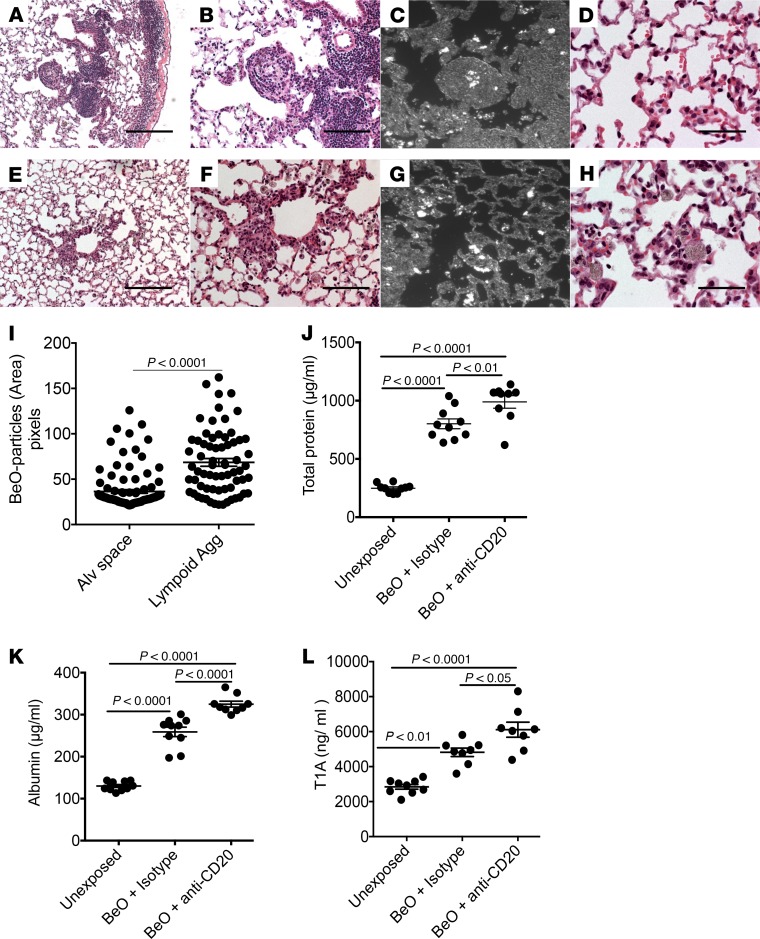
One of the earliest noted toxic effects of Be exposure in the workplace was the induction of tracheobronchitis and acute lung injury (1). As shown in Figure 2A and Figure 7, A and B, BeO exposure in HLA-DP2–Tg mice induces ELA and granuloma formation around bronchi and blood vessels. With B cell depletion, lymphocytic cell infiltration and granuloma formation in the lung were significantly diminished (Figure 7, E and F). To localize BeO distribution within the lungs of BeO-exposed mice, we performed dark-field microscopy. As shown in Figure 7C, BeO was sequestered within the ELAs in HLA-DP2–Tg mice with intact B cells, while BeO particles were randomly distributed within the interstitium and alveolar walls of B cell–depleted mice (Figure 7G). Quantitative analysis of BeO particles within the lungs showed their sequestration within ELAs as compared with the alveolar space (Figure 7I). As compared with the delicate alveolar structures in BeO-exposed mice (Figure 7D), B cell–depleted, BeO-exposed HLA-DP2–Tg mice showed epithelial cell injury and diffuse particulate-containing monocytes (Figure 7H). To confirm alveolar injury, we compared total protein, albumin, and podoplanin (T1A) in the BALF of HLA-DP2–Tg mice that were either unexposed or exposed to BeO in the presence or absence of B cells. As shown in Figure 7, J–L, BeO significantly increased BAL total protein, albumin, and T1A compared with unexposed mice. In addition, the absence of B cells further increased total protein (P < 0.01), albumin (P < 0.0001), and T1A (P < 0.05) in the BAL compared with isotype-treated BeO-exposed HLA-DP2–Tg mice (Figure 7, J–L). Taken together, these data suggest that B cells play a critical role in sequestering toxic particles within lymphocytic cell infiltrates and granulomas, thus protecting the lung from injury.
Figure 7. BeO exposure enhanced acute lung injury in HLA-DP2–Tg mice deficient in B cells.
(A and E) Low-magnification view of H&E-stained lung tissue collected from BeO-exposed HLA-DP2–Tg mice treated with isotype control (A) or anti-CD20 (E) mAb is shown. (B and F) Higher-magnification view of A and B is shown. Dark-field microscopic images show BeO particle distribution (white nodules) in the lungs of BeO-exposed mice treated with either isotype control (C) or anti-CD20 (G) mAb. High-magnification view of alveolar tissue in BeO-exposed HLA-DP2–Tg mice treated with isotype control (D) or anti-CD20 (H) mAb is shown. Scale bars: 20 μm. (I) Quantitative analysis of BeO particles within the granuloma and alveolar space. Concentrations of total protein (J), albumin (K), and podoplanin (TIA) (L) measured by ELISA in BALF from isotype control or anti-CD20 mAb–treated BeO-exposed HLA-DP2–Tg mice is shown. Cumulative data from 2 independent experiments having 3 to 5 mice per group are shown, and the mean ± SEM concentration (pg/ml) is shown as a solid line with error bars. Statistical significance was determined using a Student’s t test (2 tailed) (I) or 2-way ANOVA (J–L), and a P value of <0.05 was considered statistically significant.
BeO-induced B cell recruitment to the lungs is dependent on MyD88 signaling pathways.
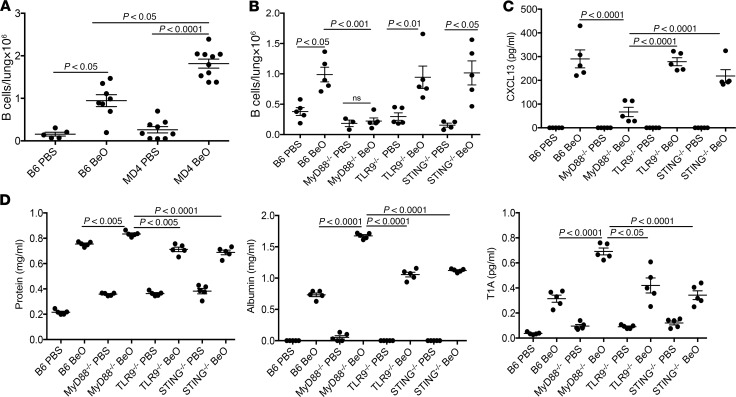
Although B cells are not required for the generation of a Be-specific CD4+ T cell response in the lung of HLA-DP2–Tg mice, we queried whether B cells would be recruited to the lung in the absence of the HLA-DP2 transgene. We exposed WT C57BL/6J (B6) and MD4 BCR–Tg C57BL/6J (MD4) mice to BeO using an identical exposure method to that used in the HLA-DP2–Tg mice (15). In heterozygous MD4 mice, greater than 95% of B cells are specific for hen egg lysozyme (33). We observed a significant recruitment of B cells to the lungs of BeO-exposed MD4 mice compared with PBS-treated MD4 mice (Figure 8A). In addition, the number of B cells recruited to the lung was significantly greater in MD4 mice compared with B6 mice (P < 0.05). These data show that B cells expressing a defined BCR that does not recognize Be are recruited to the lungs following exposure to BeO, suggesting an antigen-independent recruitment of B cells to the target organ in response to BeO exposure.
Figure 8. B cell recruitment to the lung after BeO exposure was independent of antigen and dependent on MyD88 signaling.
(A) Total numbers of B cells in the lungs of WT C57BL/6J (B6) and MD4 BCR–Tg (MD4) mice exposed to either PBS or BeO is shown. Cumulative data from 2 separate experiments having 5 mice per group are shown. (B) Total numbers of B cells in the lungs of WT C57BL/6J (B6), MyD88–/–, TLR9–/–, and STING–/– mice exposed to either PBS or BeO are shown. (C) CXCL13 (pg/ml) and (D) total protein (left), albumin (middle), and T1A (right) levels in the bronchoalveolar lavage fluid of WT C57BL/6J (B6), MyD88–/–, TLR9–/–, and STING–/– mice exposed to either PBS or BeO are shown. Data are representative of 3 independent experiments having 5 mice per group. Statistical significance was determined using a 2-way ANOVA, and the solid line and error bars depict the mean ± SEM. A P value of <0.05 was considered statistically significant.
To investigate the innate mechanism of B cell recruitment to the lungs of BeO-exposed mice, we queried whether the MyD88 pathway was involved in the recruitment of B cells to the lungs of BeO-exposed mice. Given that Be exposure induced cellular death and the release of DNA (17), we also examined the role of TLR9 and STING, innate immune sensors involved in the recognition of DNA. Thus, we exposed WT B6, MyD88-KO, TLR9-KO, and STING-KO mice to BeO and analyzed the number of B cells in lung tissue at day 21. We found similar numbers of B cells in the lungs of BeO-exposed WT, TLR9-KO, and STING-KO mice that were significantly increased compared with PBS-exposed mice (P < 0.05; Figure 8B). Conversely, mice lacking MyD88 showed an impaired recruitment of B cells to the lung in response to BeO exposure (Figure 8B). To delineate the underlying mechanism of MyD88 dependence of B cell recruitment, we measured CXCL13 in the BALF of WT B6, MyD88-KO, TLR9-KO, and STING-KO mice exposed to either PBS or BeO. As shown in Figure 8C, CXCL13 secretion was significantly diminished in BeO-exposed MyD88-KO compared with BeO-exposed WT, TLR9-KO, and STING-KO mice, and no differences were noted in CXCL13 levels between PBS- or BeO-exposed MyD88-KO. Because decreased B cells in the BALF and lung tissue were associated with the enhanced level of lung injury (Figure 7, J–L), we therefore examined lung injury in BeO-exposed WT B6, TLR9-KO, STING-KO, and MyD88-KO mice. BeO-exposed MyD88-KO mice showed elevated levels of total protein, albumin, and T1A in the BALF after BeO exposure compared with WT B6, TLR9-KO, and STING-KO mice exposed to BeO (Figure 8D) as well as to PBS-exposed control mice. Conversely, total protein, albumin, and T1A levels in BALF were similar between BeO-exposed WT B6, TLR9-KO, and STING-KO mice. Collectively, these data show that the BeO-induced damage response is sensed in a MyD88-dependent manner with the resultant secretion of CXCL13 in the lung, thus promoting an antigen-independent B cell recruitment to the lung.
B cells surround granulomas in the lungs of CBD patients.
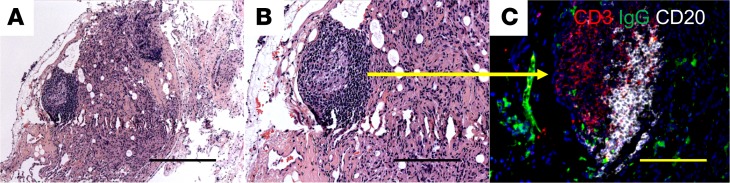
To determine the clinical relevance of our murine findings, we obtained transbronchial lung biopsies from CBD patients (n = 5) and performed multicolor immunofluorescence microscopy to assess the presence and location of B cells. Figure 9, A and B, shows H&E staining of a granuloma with its centrally located macrophages and surrounding lymphocytic infiltrates. Immunofluorescence staining of the granuloma depicted in Figure 9, A and B, show CD20+ B cells (white) surrounding the granuloma in a pattern that resembles B cell accumulation in BeO-exposed HLA-DP2–Tg mice (Figure 9C). Similar findings were seen in the lungs of the other CBD patients.
Figure 9. B cells accumulate at the periphery of granulomas in the lungs of a CBD patient.
H&E staining of a transbronchial biopsy specimen from a CBD patient. Original magnification, ×100 (A) and ×200 (B). (C) Immunofluorescence staining (×200) shows CD3+ cells and CD20+ B cells in the lung granuloma of a CBD patient. Data are representative of images from 5 CBD subjects. Scale bar: 100 μm.
Discussion
The study of the immunopathogenesis of rare diseases is facilitated by the generation of murine models that reproduce the major features of the human disease. In the case of CBD, we previously utilized an HLA-DP2–Tg FVB/N mouse and showed that these mice developed Be-specific CD4+ T cells in spleen and lung that were HLA-DP2 restricted and secreted IFN-γ and IL-2 after BeO exposure, replicating the major features of the disease (15). In addition to CD3+ T cells, in the present study we identified large populations of activated follicular B cells in the lung of these mice after BeO exposure that were organized into ELAs. CD20+ B cells were also present in the lungs of CBD patients, located at the periphery of the granuloma. Although B and T cells were compartmentalized in ELAs, the structures lacked germinal centers as well as FDC networks, histological features compatible with the early stages of organization in iBALT (29, 30, 34). Based on the ability of these lymphoid aggregates to serve as sites of immune-response generation in the lung (34), we hypothesized that the generation of the Be-specific CD4+ T cell response would be diminished in the absence of B cells, the predominant cell comprising these ELAs. Although B cell depletion abrogated the lymphoid aggregates and granuloma formation, the number of Be-responsive CD4+ T cells in the lung was unchanged, suggesting that the ELAs were dispensable for the generation of the T cell response and that B cells were not primarily functioning as APCs in the lung. However, in the absence of B cells and lymphoid aggregates, lung injury was significantly increased, suggesting that innate B cells were preventing lung injury induced by a sterile particulate through a MyD88- and CXCL13-dependent manner.
The induction of B cell aggregates in the lung has been documented in humans with rheumatoid arthritis (35), tuberculosis (36), chronic obstructive lung disease (37), and sarcoidosis (38), diseases that are characterized by chronicity and persistent antigen stimulation. CBD shares these features with the above disorders (39). At least for rheumatoid arthritis (35) and tuberculosis (36), the formation of iBALT and granulomas, respectively, was associated with increased expression of CXCL13, CCL19, and CCL21. These homeostatic chemokines are predominantly expressed in secondary lymphoid organs and facilitate the homing and localization of lymphocytes and dendritic cells in these organs (27, 28, 40). However, in the setting of lung infection and inflammation, CXCL13, CCL19, and CCL21 are induced in the lung and participate in the recruitment of immune cells (30, 41), as was the case in the current study where BeO exposure was associated with significantly increased expression of CXCL13, CCL19, and CCL21 and the recruitment of large populations of activated T and B cells to the lung.
Seminal studies by Randall and colleagues showed that protective primary immune responses could be generated in iBALT in the absence of secondary lymphoid organs and that early stages of iBALT formation were dependent on IL-17 (29, 30, 34). In the present study, BeO-induced ELAs were poorly organized without obvious B and T cell zones, FDC networks, or germinal centers. It is possible that these structures would further organize with continued BeO exposure beyond 21 days. It is also possible that the poorly organized nature of the BeO-induced lymphoid aggregates relates to the Th1-polarized Be-induced immune response and the absence of IL-17, which has been shown to be critical for iBALT formation (29). Of note, the BeO-induced lymphoid aggregates were equipped with HEVs and are likely a source of CCL19 and CCL21, suggesting their potential ability to support T cell responses in the lung. However, our data strongly support the hypothesis that the Be-specific CD4+ T cell response was initiated in LDLNs. For example, Be(OH)2 can function as an adjuvant, activating and inducing the trafficking of dendritic cells to LDLNs as well as enhancing the generation of antigen-specific memory CD4+ T cells (3, 17). In the present study, we show that the ELAs and granulomas were eliminated following depletion of B cells, while the quantity of Be-specific CD4+ T cells in the lung was unaltered. Thus, our findings are consistent with previously published studies suggesting that in mice with an intact immune system, the formation of ectopic lymphoid structures can support the recruitment of T and B cells that were antigen primed in the LDLNs to the target organ (30). In addition, the dark-field microscopy images show clusters of BeO within the lymphoid aggregates and granulomas that could function as an antigen depot, allowing for persistent antigen exposure, which is characteristic of the human disease (2, 4).
BeO-induced B cell recruitment to the lung was associated with enhanced lung inflammation, since B cell depletion resulted in significantly decreased IL-1α, IL-1β, KC, IP-10, IFN-γ, and MIP-1α in lung homogenates. However, despite diminished inflammatory cytokine/chemokine secretion, the loss of follicular B cells and the ELAs exacerbated BeO-induced acute lung injury. These findings suggest a novel innate role of B cells in protecting the lung from injury through sequestration of antigen within the ELAs and granulomas. In both influenza (30, 34) and Schistosoma mansoni (41) infections, these lymphoid aggregates can generate and maintain the local immune response in the lung. In the case of a toxic sterile particulate, like BeO, that is repeatedly inhaled and poorly cleared, sequestering BeO within granulomas and lymphoid aggregates limits lung exposure, thus constraining lung injury. This is not unprecedented, since Moyron-Quiroz et al. (34) showed that influenza-specific immune responses generated in iBALT were less pathogenic than those generated systemically. Our findings also suggest caution with the use of B cell–depleting therapies in granulomatous lung diseases, like sarcoidosis and CBD. Although B cell infiltrates have been identified in granulomas of sarcoidosis patients (38), this caution is reiterated with the results of a recent prospective, open-label, phase I/II trial of rituximab, a chimeric anti-CD20 mAb that depletes CD20+ B cells (42), in refractory sarcoidosis, which showed an inconsistent clinical response with 2 of 10 patients dying of respiratory failure during the study period.
The innate role of B cells in our model is further supported by BeO-induced recruitment of B cells to the lung in MD4 mice, which express a single BCR specific for an irrelevant antigen (hen egg lysozyme). In fact, significantly greater numbers of B cells were recruited to the lung of MD4 mice as compared with WT C57BL/6J mice. Importantly, BeO-induced B cell recruitment to the lungs was dependent on MyD88 signaling pathways, and the mechanism of MyD88 dependence was through the induction of CXCL13 secretion in the lung and the resultant promotion of an antigen-independent B cell recruitment to the lung. Although TLR9 and STING were strong candidates for the involved pathogen recognition receptor, B cell recruitment to the lung in response to BeO was not altered in these mice. Thus, the innate receptor upstream of MyD88 responsible for B cell recruitment to the lung remains unknown at the present time.
The expansion and maintenance of the Be-specific CD4+ T cells in the lungs of HLA-DP2–Tg mice in the absence of B cells demonstrates that B cells are not required antigen-presenting cells in CBD pathogenesis. This is in contrast to type 1 diabetes, where B cells serve as critical antigen-presenting cells for the initiation of T cell–mediated autoimmune diabetes in nonobese diabetic mice (32), and treatment with anti-CD20 mAb prevents and reverses autoimmune diabetes in mice (31). It remains possible that the recruited follicular B cells in the lung play an adaptive role in BeO-induced inflammation. This is supported by the increased serum IgG and its subtypes after BeO exposure compared with untreated mice that was subsequently diminished after anti-CD20 mAb treatment. In addition, the number of B cells in the lungs of HLA-DP2–Tg FVB/N was increased 6-fold over WT C57BL/6J mice, mice incapable of developing a Be-specific CD4+ T cell response in the absence of the HLA-DP2 transgene.
Collectively, these findings support a protective role for B cells and the ELAs in BeO-induced lung injury. The formation of these cellular aggregates sequesters this sterile particulate antigen and limits BeO-induced innate immune activation and injury. In CBD patients, B cells are also located at the periphery of the granuloma, providing clinical relevance to our murine findings. Our findings also suggest that the development of B cell clusters in the lung in response to inhaled antigen likely exists on a continuum from ELAs to iBALT. In the case of sterile particulates, we speculate that this protective innate mechanism teleologically evolved to enhance survival against repeated lung injury following exposure to antigen, with the wayward consequence being the development of an antigen-specific adaptive immune response, persistent T cell activation, and lung fibrosis.
Methods
Mice.
C57BL/6J, C57BL/6-Tg (IghelMD4)4 Ccg/J, B6.129P2 (SJL)-Myd88tm1.1Defr/J, C57BL/6J-Tlr9M7Btlr/Mm jax were purchased from Jackson Laboratory. STING-KO mice were donated by John Cambier. HLA-DP2–Tg FVB/N mice used in these studies were as previously described (43).
Exposure of mice to BeO.
Six- to 8-week-old FVB/N HLA-DP2–Tg mice or WT C57BL/6J, MyD88-KO, TLR9-KO, STING-KO, and MD4-Tg mice were exposed to 50 μl of either sterile PBS or PBS containing 100 μg BeO (NIST, standard reference material 1877; refs. 44, 45), via oropharyngeal aspiration as previously described (46) on days 0, 1, and 2. Mice were lightly anesthetized with isoflurane to allow inhalation of sterile PBS or BeO, and a limulus amebocyte assay (Sigma-Aldrich) was performed to confirm that preparations contained less than 20 μg of endotoxin. After sensitization, mice were boosted with either BeO or PBS (Hyclone) via oropharyngeal aspiration on days 14, 15, 18, and 19, and euthanized on day 21.
Anti-CD20 treatment.
The anti–mouse CD20 mAb, 5D2, and a murine IgG2a isotype control antibody were provided by Genentech. Pilot experiments were performed to determine optimal dosing for depletion of peripheral blood B cells, which was found to be 5 mg/kg i.p. once a week. Mice were injected i.p. with anti–mouse CD20 antibody or control antibody 2 days prior to BeO exposure and weekly for the duration of the experiment.
Preparation of single-cell suspensions of splenocytes and lung and LDLN cells.
CD45PE mAb (5 μg/mouse, Tonbo Biosciences; clone 30-F11) was injected intravenously via the retro-orbital route to separate tissue cells from circulating cells (47). Mice were euthanized under anesthesia, and the lungs were perfused with ice-cold PBS. Spleens were pressed through a 100-μm cell strainer, rinsing the strainer with PBS. Lungs were removed, minced, and digested in RPMI-C containing 1 mg/ml collagenase (Sigma-Aldrich). After 30 minutes, collagenase-digested lungs were sequentially disrupted with 16-G and 18-G needles. Collagenase was quenched with cold PBS, and lung cells were centrifuged at 250 g for 10 minutes. Erythrocytes were lysed, and lung cells were filtered through a 70-μm cell strainer and resuspended in RPMI-C. Spleens and LDLNs were processed through a 100-μm cell strainer, rinsing the strainer with PBS. Cells were pelleted by centrifugation at 250 g for 10 minutes. After ammonium chloride lysis of erythrocytes, splenocytes were filtered through a 70-μm cell strainer and resuspended in complete culture media (RPMI-C), consisting of RPMI 1640 (HyClone) supplemented with 10% heat-inactivated FBS (HyClone) and penicillin and streptomycin (Invitrogen).
Isolation of BAL cells and BALF.
Prior to the lavage, lungs were perfused with 5 ml of PBS, and lavage was accomplished by tracheal cannulation. For BALF acquisition, lavage consisted of 1 ml PBS instillation with a 0.75-ml return volume. BALF was centrifuged at 2,000 g for 2 minutes, and the acellular fraction was frozen at –80°C for analysis of lung injury. For BAL cell acquisition, lungs were sequentially lavaged 3 times with 1 ml of PBS, and return volumes were pooled and added to an equal volume of RPMI-C on ice. Cells were centrifuged at 250 g for 5 minutes.
Preparation of lung homogenates and cytokine analysis.
Lungs were snap-frozen in liquid nitrogen and stored at –80°C. Frozen lungs were thawed in 0.5 ml PBS containing 0.05% Tween 20 and protease inhibitors (Sigma-Aldrich) and homogenized at 35,000 rpm for 30 seconds on ice. Homogenates were centrifuged at 10,000 g for 10 minutes at 4°C. Supernatants were transferred to a 96-well microtiter plate and stored at –80°C. Supernatants were thawed and treated with nuclease (25 units Benzonase; Sigma-Aldrich) for 30 minutes at 37°C and centrifuged at 400 g for 5 minutes through a Multiscreen filter plate (Millipore). A Mouse Cytokine 20-Plex kit (Invitrogen) was used to evaluate mouse secretion of IL-1α, IL-1β IP-10, IFN-γ, and MIP-1α, according to the manufacturer’s guidelines.
Flow cytometry.
Fluorescently labeled antibodies against cell surface antigens were diluted in PBS containing 1% FCS, 0.05% sodium azide, and 0.5 μg/ml CD16/CD32 (Tonbo Biosciences; 2.4G2). Cells were incubated for 30 minutes at 4oC, and then washed and resuspended in PBS containing 1% formaldehyde. The following antibodies (vendor; clone) were used for multiparameter FACS analysis: CD3 (Tonbo Biosciences; 145-2C-11), CD4 (BioLegend; RM4-5), CD8 (BioLegend; 53-6.7), CD19 (BioLegend; 6D5), B220 (RA3-682), CD44 (Tonbo Biosciences; IM7), HLA-DP (purified from hybridoma, B7.21), CXCR4 (BD Pharmingen), and CXCR5 (BioLegend, L138D7). For staining of innate cells in BAL and LDLNs, antibodies directed against CD11c (N418), CD11b (M1/70), IA/IE (M5.114.15.2), and Ly6G (1A8) were purchased from eBioscience, and mAbs directed against CD64 (X54-5/7.1) and Siglec-F (E250-4440) were purchased from BD Biosciences. For detection of biotinylated antibodies, samples were incubated with a 1:250 dilution of streptavidin-PE (eBioscience; 12-4317-87) for 15 minutes on ice. For HLA-DP2-PLXNA4/Be tetramer staining, BAL cells were stained as previously described (14, 16). In brief, cells were incubated in 25 μl of culture medium containing tetramer (20 μg/ml) for 2 hours at 37°C in a humidified 10% CO2 incubator, with gentle mixing every 30 minutes, followed by surface staining as described above. Data were obtained using an LSR II Flow Cytometer (BD), and all postacquisition analyses were performed in FlowJo (v10.06). For absolute number per organ, the total cell number was multiplied by the percentage of cells in the live cell gate.
Analysis of IFN-γ and IL-2 production by ELISPOT assay.
ELISPOT plates (ImmunoSpot M200, BD Biosciences) were coated with IFN-γ or IL-2 capture mAb (eBioscience) overnight and blocked with RPMI-C for 2 hours at room temperature (48). Lung CD4+ T cells were negatively selected via magnetic beads, and 2.5 × 105 CD4+ T cells were combined with equal numbers of purified CD19+ splenic B cells in triplicate from an unexposed HLA-DP2–Tg mouse. Cells were incubated overnight at 37°C with medium or BeSO4 (100 μM). IFN-γ or IL-2 detection mAbs (eBioscience) were added, and spots visualized by avidin–horseradish peroxidase and 3-amino-9-ethylcarbazole substrate reagent (BD Biosciences). ELISPOT plates were analyzed using a CTL Immunospot Analyzer (Cellular Technology Ltd), and results reported as mean ± SD spot-forming units (SFUs) per well minus background SFUs.
IgG ELISAs.
Blood was collected by submandibular puncture, and serum obtained using Microtainer Serum Separator Tubes (BD). To assess immunoglobulin levels, serum was diluted 1:10,000. The ELISAs for total IgG and IgG subclasses were performed according to the manufacturer’s instructions (eBioscience). Absorbance readings at 450 nM were obtained using a VMax microplate reader (Molecular Devices), and concentrations were calculated using SoftMaxPro software (Molecular Devices).
Lung injury endpoints.
To quantitate protein, BALF was diluted 1:10. Ten microliters of diluted BALF was assayed in a Qubit protein fluorometric dye binding assay (ThermoFisher Scientific). Total protein was determined using a BSA standard curve according to the manufacturer’s instructions. To measure albumin, a bromocresol green (BCG) assay was performed as previously described (49) with modifications. Briefly, 100 μl of BALF was combined with 100 μl BCG reagent (BCG dissolved in 300 mM citric acid, pH 4.2 to a final concentration of 0.12 mM). Total albumin was determined using a BSA standard curve. Absorbance readings were measured at 620 nm using a VMax microplate reader. Concentrations were calculated using GraphPad Prism (v.5.0d). To assess relative T1A levels, BALF was diluted 1:100. A double-sandwich ELISA was performed with a goat anti–mouse podoplanin (R&D Systems) capture antibody (10 μg/ml) and biotinylated anti–mouse podoplanin (eBioscience; eBio8.1.1) (0.5 μg/ml) as the detection antibody. Coating buffer, assay diluent, SA-HRP, and TMB substrate were from eBioscience. The absorbance was measured at 450 nm and used to compare the levels of T1A. Absorbance readings were obtained using a VMax microplate reader.
Histology/scoring.
Lungs were inflated and stored in 10% neutral-buffered formalin for 24 hours and transferred to 70% ethanol for histopathological analysis of the lung injury. Immunohistochemistry on paraffin-embedded sections of lung slides was performed by Histowiz. Briefly, each section was stained with a 1:500 dilution of rabbit anti-CD4 (Abcam, EPR19533) or 1:10,000 dilution of rabbit anti-B220 (Novus Biologicals, RA3-6B2) using a Bond RX automated stainer (Leica Biosystems). Primary antibodies were detected using a rabbit-specific IHC polymer detection kit (Abcam) and the antibody-bound antigen sites were visualized by DAB. Sections were counterstained with hematoxylin and whole-slide scanning (×40) was performed on an Aperio AT2 (Leica Biosystems) and saved as single-file pyramidal tiled TIFF files. Enumeration of the positive-staining cells was performed using the digital pathology image analysis software Qupath (50). Hematoxylin and eosin (H&E) histological staining was done at the UCD Cancer Center–Research Histology Shared Resource core. Sequestration of BeO in the lungs was analyzed by ImageJ 1.52k (NIH) using the Renyi entropy method of thresholding.
Multicolor immunofluorescence microscopy of formalin-fixed, paraffin-embedded mouse and human lung sections.
Lung sections (5 μm) were placed on a plate at 60°C to melt paraffin and quickly transferred into xylene. Sections were sequentially transferred into ethanol, 95% ethanol, and finally placed into water. To unmask antigens, slides were boiled in DAKO antigen retrieval solution (DakoCytomation, catalog S-1699) for 30 minutes. Lung sections were blocked with 5% donkey serum (Jackson ImmunoResearch Laboratories, catalog 017-000-001) prior to staining with primary antibodies. A set of lung sections were first incubated with goat anti–mouse CXCL13 (R&D Systems, catalog AF470), biotinylated rat anti–mouse CD21/CD35 (Biolegend, clone 7E9), and rat anti–mouse FDCM1 (BD Pharmingen, catalog 551320). Alexa Fluor 568–donkey anti–goat IgG (ThermoFisher Scientific, catalog A-11057), Alexa Fluor 488–donkey anti–rat IgG (ThermoFisher Scientific, catalog A-21208), and Alexa Fluor 488–streptavidin (ThermoFisher Scientific, catalog S-11223) were added to lung sections to detect CXCL13-producing cells and CD21/CD35+/FDCM1+ FDCs. Another set of slides were probed with rat anti–mouse/human peripheral node addressin (PNAd, Biolegend, MECA-79), FITC–mouse anti–mouse/human C-X-C motif chemokine 12 (CXCL12, R&D Systems, 79018), and rabbit anti–mouse lymphatic vessel hyaluronan receptor 1 (Lyve-1, Acris, catalog DP3513). Cy-3–goat anti–rat IgM (Jackson ImmunoResearch Laboratories, catalog 112-166-075), and Alexa Fluor 647–donkey anti-rabbit (Jackson ImmunoResearch Laboratories, catalog 711-606-152) were added to visualize PNAd+ HEVs, CXCL12-producing cells, and Lyve-1+ lymphatics. Finally, a third group of lung sections were covered with a cocktail of goat anti-CD3ε (Santa Cruz Biotechnology, M-20), Alexa Fluor 488–donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, catalog 715-546-151), and APC–rat anti–mouse CD45R (B220, BD Pharmingen, RA3-6B2), followed by incubation with Alexa Fluor 568–donkey anti-goat (ThermoFisher Scientific, catalog A-11057) to label CD3+ T cells, IgG-producing cells, and CD45R/B220+ B cells.
Human lung sections were probed with goat anti-CD3ε (Santa Cruz Biotechnology, M-20), Alexa Fluor 488–donkey anti–human IgG (Jackson ImmunoResearch Laboratories, catalog 709-546-149), and mouse anti–human CD20 (GeneTex, L26). Bound primary antibodies were detected with Alexa Fluor 568–donkey anti–goat IgG (Life Technologies, catalog A-11057) and Alexa Fluor 647–donkey anti–mouse IgG (Jackson ImmunoResearch Laboratories, catalog 715-606-151). FDCs in human lungs were detected with mouse anti–human CD21 (ThermoFisher Scientific, 2G9). Biotinylated donkey anti-mouse (Jackson ImmunoResearch Laboratories, catalog 715-066-150) and Alexa Fluor 555–streptavidin (Thermo Fisher Scientific, catalog S-32355) were added to slides to reveal primary antibodies. Slides were briefly washed in PBS and mounted with Vectashield antifade mounting media with DAPI (Vector Laboratories, catalog H-1200). Pictures were taken with a Hamamatsu camera and recorded with a Zeiss Axioplan digital camera.
Statistics.
Statistical comparisons were analyzed with GraphPad Prism (v.5.0d) using unpaired Student’s t test (2-tailed) or 2-way ANOVA. P values < 0.05 were deemed statistically significant. No data sets were generated or analyzed during the current study.
Study approval.
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Colorado Anschutz Medical Campus (Aurora, CO), in accordance with the NIH guidelines for use of live animals. The University of Colorado Anschutz Medical Campus is accredited by the American Association for Accreditation of Laboratory Animal Care. Informed consent was obtained from each participant, and the study protocol was approved by the Colorado Multiple Institutional Review Board (Aurora, Colorado).
Author contributions
SMA, DGM, ASM, and APF designed the research. AG and JCC provided the MD4-Tg mice. SMA, DGM, ASM, JRM, AKM, LAM, RT, and APF performed the research. SMA, DGM, ASM, and APF analyzed the data. SMA, DGM, ASM, and APF wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants ES025534, HL062410, and HL102245 (to APF).
Version 1. 05/16/2019
In-Press Preview
Version 2. 06/20/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e125494. https://doi.org/10.1172/jci.insight.125494.
Contributor Information
Shaikh M. Atif, Email: SHAIKH.ATIF@UCDENVER.EDU.
Douglas G. Mack, Email: doug.mack@ucdenver.edu.
Amy S. McKee, Email: amy.mckee@ucdenver.edu.
Javier Rangel-Moreno, Email: Javier_Rangel-Moreno@URMC.Rochester.edu.
Allison K. Martin, Email: allison.martin@ucdenver.edu.
Andrew Getahun, Email: andrew.getahun@ucdenver.edu.
Lisa A. Maier, Email: maierl@njhealth.org.
John C. Cambier, Email: john.cambier@ucdenver.edu.
Rubin Tuder, Email: Rubin.Tuder@ucdenver.edu.
Andrew P. Fontenot, Email: andrew.fontenot@ucdenver.edu.
References
- 1.Hardy HL. Beryllium poisoning--lessons in control of man-made disease. N Engl J Med. 1965;273(22):1188–1199. doi: 10.1056/NEJM196511252732205. [DOI] [PubMed] [Google Scholar]
- 2.Fontenot AP, Falta MT, Kappler JW, Dai S, McKee AS. Beryllium-induced hypersensitivity: genetic susceptibility and neoantigen generation. J Immunol. 2016;196(1):22–27. doi: 10.4049/jimmunol.1502011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKee AS, Fontenot AP. Interplay of innate and adaptive immunity in metal-induced hypersensitivity. Curr Opin Immunol. 2016;42:25–30. doi: 10.1016/j.coi.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26(10):543–549. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 5.McCanlies EC, Kreiss K, Andrew M, Weston A. HLA-DPB1 and chronic beryllium disease: a HuGE review. Am J Epidemiol. 2003;157(5):388–398. doi: 10.1093/aje/kwg001. [DOI] [PubMed] [Google Scholar]
- 6.Maier LA, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171(12):6910–6918. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 7.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262(5131):242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 8.Rossman MD, Stubbs J, Lee CW, Argyris E, Magira E, Monos D. Human leukocyte antigen Class II amino acid epitopes: susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med. 2002;165(6):788–794. doi: 10.1164/ajrccm.165.6.2104002. [DOI] [PubMed] [Google Scholar]
- 9.Silveira LJ, et al. Chronic beryllium disease, HLA-DPB1, and the DP peptide binding groove. J Immunol. 2012;189(8):4014–4023. doi: 10.4049/jimmunol.1200798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+ T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci USA. 2000;97(23):12717–12722. doi: 10.1073/pnas.220430797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardi G, et al. HLA-DP allele-specific T cell responses to beryllium account for DP-associated susceptibility to chronic beryllium disease. J Immunol. 2001;166(5):3549–3555. doi: 10.4049/jimmunol.166.5.3549. [DOI] [PubMed] [Google Scholar]
- 12.Bowerman NA, et al. Identification of multiple public TCR repertoires in chronic beryllium disease. J Immunol. 2014;192(10):4571–4580. doi: 10.4049/jimmunol.1400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton GM, et al. Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity. Cell. 2014;158(1):132–142. doi: 10.1016/j.cell.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falta MT, et al. Identification of beryllium-dependent peptides recognized by CD4+ T cells in chronic beryllium disease. J Exp Med. 2013;210(7):1403–1418. doi: 10.1084/jem.20122426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack DG, et al. Regulatory T cells modulate granulomatous inflammation in an HLA-DP2 transgenic murine model of beryllium-induced disease. Proc Natl Acad Sci USA. 2014;111(23):8553–8558. doi: 10.1073/pnas.1408048111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falta MT, et al. Metal-specific CD4+ T-cell responses induced by beryllium exposure in HLA-DP2 transgenic mice. Mucosal Immunol. 2016;9(1):218–228. doi: 10.1038/mi.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKee AS, Mack DG, Crawford F, Fontenot AP. MyD88 dependence of beryllium-induced dendritic cell trafficking and CD4+ T-cell priming. Mucosal Immunol. 2015;8(6):1237–1247. doi: 10.1038/mi.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda E, et al. Inhaled fine particles induce alveolar macrophage death and interleukin-1α release to promote inducible bronchus-associated lymphoid tissue formation. Immunity. 2016;45(6):1299–1310. doi: 10.1016/j.immuni.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Wade MF, et al. TLR9 and IL-1R1 promote mobilization of pulmonary dendritic cells during beryllium sensitization. J Immunol. 2018;201(8):2232–2243. doi: 10.4049/jimmunol.1800303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontenot AP, Canavera SJ, Gharavi L, Newman LS, Kotzin BL. Target organ localization of memory CD4(+) T cells in patients with chronic beryllium disease. J Clin Invest. 2002;110(10):1473–1482. doi: 10.1172/JCI15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinkle SS, Kittle LA, Schumacher BA, Newman LS. Beryllium induces IL-2 and IFN-gamma in berylliosis. J Immunol. 1997;158(1):518–526. [PubMed] [Google Scholar]
- 22.Payet-Jamroz M, et al. Suppression of IgE responses in CD23-transgenic animals is due to expression of CD23 on nonlymphoid cells. J Immunol. 2001;166(8):4863–4869. doi: 10.4049/jimmunol.166.8.4863. [DOI] [PubMed] [Google Scholar]
- 23.Carragher DM, Rangel-Moreno J, Randall TD. Ectopic lymphoid tissues and local immunity. Semin Immunol. 2008;20(1):26–42. doi: 10.1016/j.smim.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randall TD, Carragher DM, Rangel-Moreno J. Development of secondary lymphoid organs. Annu Rev Immunol. 2008;26:627–650. doi: 10.1146/annurev.immunol.26.021607.090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16(1):67–76. doi: 10.1016/S1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 26.Fleige H, et al. IL-17-induced CXCL12 recruits B cells and induces follicle formation in BALT in the absence of differentiated FDCs. J Exp Med. 2014;211(4):643–651. doi: 10.1084/jem.20131737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cyster JG. Chemokines and the homing of dendritic cells to the T cell areas of lymphoid organs. J Exp Med. 1999;189(3):447–450. doi: 10.1084/jem.189.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cyster JG. Chemokines and cell migration in secondary lymphoid organs. Science. 1999;286(5447):2098–2102. doi: 10.1126/science.286.5447.2098. [DOI] [PubMed] [Google Scholar]
- 29.Rangel-Moreno J, et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol. 2011;12(7):639–646. doi: 10.1038/ni.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Kusser K, Randall TD. Pulmonary expression of CXC chemokine ligand 13, CC chemokine ligand 19, and CC chemokine ligand 21 is essential for local immunity to influenza. Proc Natl Acad Sci USA. 2007;104(25):10577–10582. doi: 10.1073/pnas.0700591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu CY, et al. Treatment with CD20-specific antibody prevents and reverses autoimmune diabetes in mice. J Clin Invest. 2007;117(12):3857–3867. doi: 10.1172/JCI32405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serreze DV, Fleming SA, Chapman HD, Richard SD, Leiter EH, Tisch RM. B lymphocytes are critical antigen-presenting cells for the initiation of T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Immunol. 1998;161(8):3912–3918. [PubMed] [Google Scholar]
- 33.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334(6184):676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 34.Moyron-Quiroz JE, et al. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med. 2004;10(9):927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 35.Rangel-Moreno J, Hartson L, Navarro C, Gaxiola M, Selman M, Randall TD. Inducible bronchus-associated lymphoid tissue (iBALT) in patients with pulmonary complications of rheumatoid arthritis. J Clin Invest. 2006;116(12):3183–3194. doi: 10.1172/JCI28756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slight SR, et al. CXCR5(+) T helper cells mediate protective immunity against tuberculosis. J Clin Invest. 2013;123(2):712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogg JC, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 38.Kamphuis LS, et al. Perigranuloma localization and abnormal maturation of B cells: emerging key players in sarcoidosis? Am J Respir Crit Care Med. 2013;187(4):406–416. doi: 10.1164/rccm.201206-1024OC. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer RT, Abraham JL, Daniloff E, Newman LS. Secondary ion mass spectroscopy demonstrates retention of beryllium in chronic beryllium disease granulomas. J Occup Environ Med. 2005;47(12):1218–1226. doi: 10.1097/01.jom.0000184884.85325.36. [DOI] [PubMed] [Google Scholar]
- 40.Khader SA, et al. In a murine tuberculosis model, the absence of homeostatic chemokines delays granuloma formation and protective immunity. J Immunol. 2009;183(12):8004–8014. doi: 10.4049/jimmunol.0901937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo JC, et al. Differential regulation of CCL21 in lymphoid/nonlymphoid tissues for effectively attracting T cells to peripheral tissues. J Clin Invest. 2003;112(10):1495–1505. doi: 10.1172/JCI19188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reff ME, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83(2):435–445. [PubMed] [Google Scholar]
- 43.Tarantino-Hutchison LM, et al. Genetic determinants of sensitivity to beryllium in mice. J Immunotoxicol. 2009;6(2):130–135. doi: 10.1080/15476910902977399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefaniak AB, et al. Surface area of respirable beryllium metal, oxide, and copper alloy aerosols and implications for assessment of exposure risk of chronic beryllium disease. AIHA J (Fairfax, Va) 2003;64(3):297–305. doi: 10.1080/15428110308984820. [DOI] [PubMed] [Google Scholar]
- 45.Winchester MR, et al. Certification of beryllium mass fraction in SRM 1877 Beryllium Oxide Powder using high-performance inductively coupled plasma optical emission spectrometry with exact matching. Anal Chem. 2009;81(6):2208–2217. doi: 10.1021/ac802251n. [DOI] [PubMed] [Google Scholar]
- 46.Rao GV, et al. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J Toxicol Environ Health Part A. 2003;66(15):1441–1452. doi: 10.1080/15287390306417. [DOI] [PubMed] [Google Scholar]
- 47.Anderson KG, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9(1):209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin AK, et al. Beryllium-specific CD4+ T cells in blood as a biomarker of disease progression. J Allergy Clin Immunol. 2011;128(5):1100–6.e1. doi: 10.1016/j.jaci.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doumas BT, Watson WA, Biggs HG. Albumin standards and the measurement of serum albumin with bromcresol green. Clin Chim Acta. 1971;31(1):87–96. doi: 10.1016/0009-8981(71)90365-2. [DOI] [PubMed] [Google Scholar]
- 50.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.