Abstract
Benign prostatic hyperplasia (BPH) is the most common cause of lower urinary tract symptoms in men. Current treatments target prostate physiology rather than BPH pathophysiology and are only partially effective. Here, we applied next-generation sequencing to gain insight into BPH. By RNA-Seq, we uncovered transcriptional heterogeneity among BPH cases, where a 65-gene BPH stromal signature correlated with symptom severity. Stromal signaling molecules bone morphogenetic protein 5 (BMP5) and CXC chemokine ligand 13 (CXCL13) were enriched in BPH while estrogen-regulated pathways were depleted. Notably, BMP5’s addition to cultured prostatic myofibroblasts altered their expression profile toward a BPH profile that included the BPH stromal signature. RNA-Seq also suggested an altered cellular milieu in BPH, which we verified by immunohistochemistry and single-cell RNA-Seq. In particular, BPH tissues exhibited enrichment of myofibroblast subsets but also depletion of neuroendocrine cells and an estrogen receptor–positive fibroblast cell type residing near the epithelium. By whole-exome sequencing, we uncovered somatic single-nucleotide variants in BPH, of uncertain pathogenic significance but indicative of clonal cell expansions. Thus, genomic characterization of BPH has identified a clinically relevant stromal signature and new candidate disease pathways (including a likely role for BMP5 signaling) and reveals BPH to be not merely a hyperplasia, but rather a fundamental relandscaping of cell types.
Keywords: Cell Biology, Oncology
Keywords: Expression profiling, Molecular pathology, Prostate cancer
Next-generation sequencing technology applied to benign prostatic hyperplasia identifies a clinically-relevant stromal signature and a fundamental re-landscaping of cell types.
Introduction
Benign prostatic hyperplasia (BPH) is the most common cause of lower urinary tract symptoms (LUTS) in men due to bladder outlet obstruction (1). Symptoms range from increased urinary hesitancy, urgency, and frequency to acute urinary retention. BPH is also associated with serious consequences, including urinary bleeding, infections, bladder stones, and renal failure. Histologic BPH is found in approximately 50% of men aged 50 years, and its prevalence increases about 10% each subsequent decade (2). Likewise, significant LUTS are documented to occur in about 10%–20% of men aged 50–59 and increase to one-third of men by ages 70–79. In parallel, prostate volume increases between age 40 and age 79, with the greatest increases appearing in the sixth and seventh decades of life (2–4).
BPH develops in the prostate transition zone and periurethral glands and in advanced disease often appears grossly nodular (5). BPH represents a benign proliferative process and is not considered a precursor to prostate cancer. However, whether the prostatic hyperplasia reflects clonal cell expansions has never been examined. In BPH, increased cell numbers are found in both the epithelial and stromal compartments, though typically more so in the stroma (6, 7). Tissue architecture can vary, where in some cases glandular epithelial elements predominate, while in others fibromuscular-rich stroma comprises much of the BPH nodule, although most cases show a mixture of both histologies.
The pathophysiology of BPH is poorly understood, though androgen signaling, reactive stroma, and inflammation have been implicated (8). During prostate development, androgens appear to act primarily through the prostatic stroma, which in turn directs epithelial proliferation and differentiation (9). These stromal signals have been hypothesized to be reactivated during aging in BPH pathogenesis (5). Many stroma-derived growth factors appear to be controlled by androgen signaling in prostate tissue models (10–13), where they also act on prostatic stromal mesenchyme to drive proliferation and myofibroblastic differentiation (12, 14). Inflammation of the aging prostate has also been proposed to be an important driver of BPH (15–18). These inflammatory changes are thought to be linked to metabolic syndrome (19, 20) and may first exert their effects at the level of the prostatic stroma (17, 21–23).
Mirroring our incomplete understanding, BPH treatments have largely been developed empirically and likely do not target the underlying disease mechanisms but rather the physiology of the entire prostate. Androgen inhibition (most commonly by 5-α-reductase inhibition) reduces prostate volume but only partially relieves BPH symptoms, mainly in men with significant prostate enlargement (24). Likewise, α-adrenoceptor blockade relaxes prostatic smooth muscle tone. However, not all patients respond and α-blockade does not reduce the risk of needing surgery for BPH symptoms (25). Annual BPH treatment costs run in the billions of dollars (26). An improved understanding of BPH pathogenesis may suggest new strategies for precision therapy.
Genomics technologies have led to deeper understanding of human cancer, and more than 15 years ago researchers used DNA microarrays to begin to explore BPH (27, 28). Here, we revisit BPH using more sophisticated sequencing-based genomic approaches, including RNA-Seq to profile BPH transcriptomes and whole-exome sequencing to evaluate mutations and clonality. We have discovered potentially new signaling molecules elevated in BPH tissue, including bone morphogenetic protein 5 (BMP5) and CXC chemokine ligand 13 (CXCL13), and a stromal transcriptional signature correlated with BPH symptoms. We also uncovered significant changes in the cell types residing in both the BPH epithelium and BPH stroma, reframing BPH and the disease process from a simple hyperplasia to a more fundamental relandscaping of cells and tissue, with implications for disease prevention and treatment.
Results
Transcriptome profiling identifies a stromal signature correlated with BPH symptoms.
To investigate the transcriptional landscape of BPH, we performed RNA-Seq on BPH and matched normal prostate from FFPE tissue blocks, an optimal method for verifying histopathology. Cases were selected from radical prostatectomy specimens (performed for prostate cancer) with concurrent BPH, for which we had a detailed assessment of patients’ urinary symptoms from the preoperative International Prostate Symptom Score (IPSS) with Bother Score (29). We selected cases from patients not on 5-α-reductase inhibitors or α-adrenoceptor–blocking agents to eliminate confounding effects of BPH treatment. For about half the cases, we profiled matched histologically normal prostate tissue. For a smaller subset, we also profiled BPH stroma-rich nodules (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129749DS1), a recognized pathologic entity characterized as discrete nodules of stromal cells with an absence of epithelial cells, to understand this BPH tissue variant and to aid in the interpretation of BPH signatures. In all, 65 samples passed rigorous RNA-Seq quality control metrics (see Methods), including 37 BPH samples, 19 normal prostate tissues, and 9 BPH stromal nodules (Supplemental Table 1).
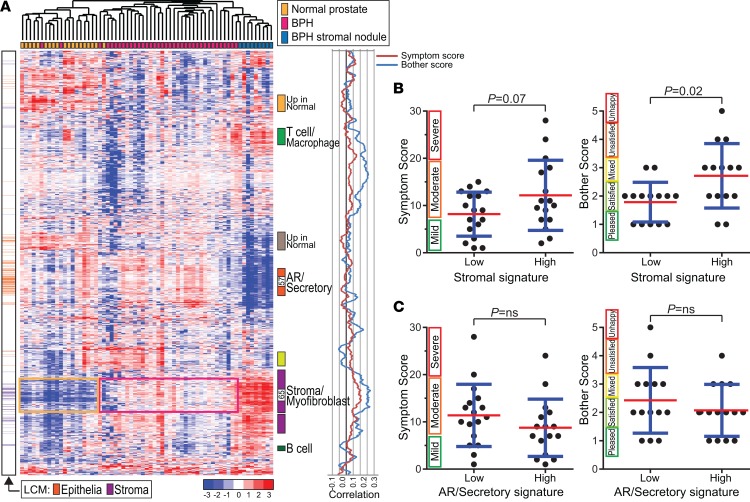
Unsupervised hierarchical clustering (of normalized log2 transcript levels) distinguished BPH from normal prostate, and the stromal nodules clustered distinctly from the BPH samples (Figure 1A and Supplemental Table 2). Gene expression features could be distinguished and interpreted based on characteristic marker genes, including features representing secretory epithelium/androgen receptor (AR) signaling (e.g., KLK3, TMPRSS2, KRT8), stroma (e.g., FBLN2, LUM, VCAN) (enriched in both BPH and stromal nodules), T cells/macrophages (e.g., CD2, CD5, LYZ) (enriched in BPH stromal nodules), and B cells (e.g., PAX5, CD79A, CXCR5) (also enriched in BPH stromal nodules). LCM was used to generate pure BPH epithelia and matched stroma from 5 independent cases followed by Smart-3SEQ (an RNA-Seq method developed for small cell numbers; ref. 30). The transcript profiles confirmed that genes assigned to epithelial or stromal compartments in the bulk transcriptome data were expressed in those compartments (Figure 1A).
Figure 1. BPH transcriptional landscape and clinically relevant stromal signature.
(A) Heatmap of unsupervised clustering of normal prostate, BPH, and BPH stromal nodules (samples color coded) across the 2548 most variably expressed genes. Transcript levels (median-centered log2 reads per kilobase million values) are indicated by color key. Select gene features (clusters) are annotated based on the expression of characteristic marker genes, including a stroma/myofibroblast feature (enframed on the heatmap) enriched in BPH over normal prostate and heterogeneous among BPH cases. Laser capture microdissection (LCM) panel indicates genes more highly expressed in laser microdissected BPH epithelium (orange) or BPH stroma (purple). The graph illustrates correlation of gene expression features (moving average 51 genes) with BPH IPSS and Bother Score. Note: peak correlations reside within the core stromal feature. The data set corresponding to the heatmap is available as Supplemental Table 2. (B) High expression of a 65-gene stromal signature is associated with elevated IPSS (strong trend) and Bother Score. Mean (red) and SD (blue) shown; P values generated from 2-tailed Student’s t test. The 65 genes correspond to the core of the stromal gene feature (indicated in A), with high expression defined as the top 50%. (C) A similarly derived 57-gene AR/Secretory signature showed no association with IPSS or Bother Score.
Across the BPH samples, unsupervised clustering revealed transcriptional heterogeneity, with most BPH cases partitioning into 2 main clusters (Figure 1A and Supplemental Figure 2). This subclass distinction appeared to be driven in part by a stromal gene feature, present with or without the stromal nodule samples (Supplemental Figure 2). Notably, when we compared patients with low versus high expression of a 65-gene stromal signature, comprising the core of the stromal gene feature (Figure 1A and Supplemental Table 3), patients with a more pronounced stromal signature reported higher IPSS (P = 0.07) and Bother Score (P = 0.02) (Figure 1B). In contrast, a similarly derived 57-gene AR/secretory signature did not associate with either IPSS or Bother Score (Figure 1C), nor did a comparable AR signature from the literature (ref. 31, Supplemental Figure 3A, and Supplemental Table 3). Indeed, upon plotting the correlation of genes with BPH IPSS and Bother Scores (Figure 1A), the peak correlations occurred within the core stromal feature. Though we lacked an independent BPH cohort to validate the association between stromal gene expression and patient-reported symptoms, we could build a predictor of Bother Score from the 65 stromal genes that by leave-one-out cross-validation reduced the null model mean squared error by about 70% (Supplemental Figure 3B).
Transcriptome profiling uncovers signaling molecules, including BMP5, upregulated in BPH.
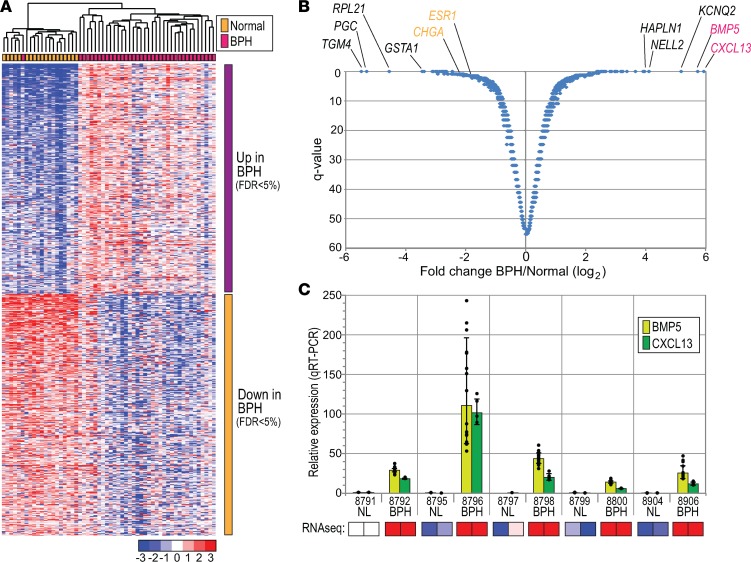
To more directly identify transcriptional differences between BPH and normal prostate, we performed a supervised analysis of the RNA-Seq data (significance analysis of microarrays [SAM]; ref. 32; Figure 2, A and B; and Supplemental Tables 4 and 5). In all, 301 genes were significantly more highly expressed in BPH, while 316 genes were more highly expressed in normal prostate (FDR < 0.05). The genes upregulated in BPH included growth factors previously linked to prostatic hyperplasia, including fibroblast growth factors (FGF7), insulin growth factors (IGF2), and TGF-β (TGFB3) (8). However, topping the list were 2 seldom-discussed signaling molecules, both markedly overexpressed in BPH: BMP5 (50-fold elevated) and CXCL13 (60-fold elevated). Elevated expression of both BMP5 and CXCL13 in BPH was confirmed by quantitative reverse transcription PCR (qRT-PCR) (Figure 2C). As yet, we have been unable to verify elevated BMP5 and CXCL13 at the protein level because of inadequate performance of antibodies in immunohistochemistry (IHC) and Western blot.
Figure 2. Genes differentially expressed in BPH versus normal prostate.
(A) Heatmap of genes with significant differential expression in BPH versus normal prostate. Samples are clustered (see color key) and genes ordered by SAM score (t statistic value). (B) Volcano plot (Q value vs. log2 fold change) annotated with topmost genes differentially expressed in BPH. (C) Technical validation of RNA-Seq results by qRT-PCR, confirming elevated expression of BMP5 and CXCL13 in BPH. Five BPH-normal pairs assayed once with technical quadruplicates. Graphed are mean (±1 SD) relative expression levels, normalized to GAPDH and compared against a reference sample (normal prostate sample 8791). Each data point represents 1 of up to 16 pairwise ratios (calculated from the quadruplicate test and reference values). Below, heatmap depiction of corresponding RNA-Seq (median-centered log2 reads per kilobase million) values.
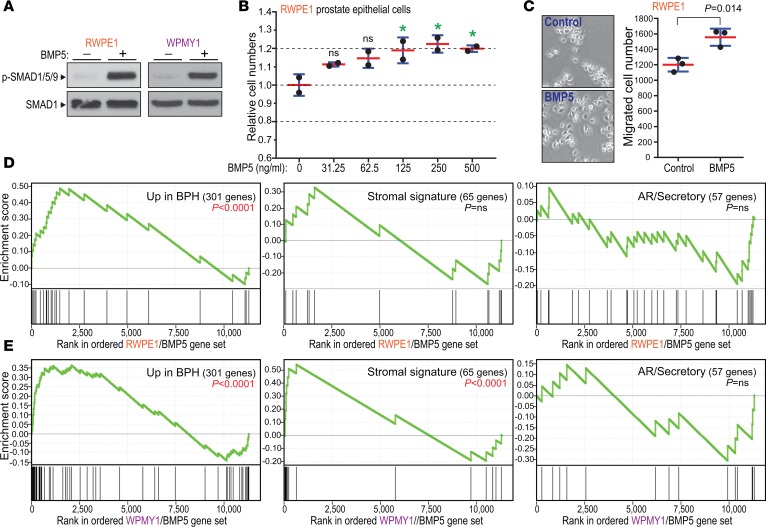
BMPs are known to regulate diverse tissue morphogenetic processes in mammals (33). To investigate a mechanistic connection between BMP5 and BPH, we exposed immortalized prostate epithelial cells (RWPE-1) (34) to a range of physiologic concentrations of BMP5 (35). Canonical BMP receptor/SMAD pathway activation was evidenced by increased phospho-SMAD1/5/9 (Figure 3A) and induction of canonical BMP transcriptional targets (e.g., ID3, SMAD6) (ref. 36 and Supplemental Figure 4A). BMP5 addition led to a significant and reproducible 20% increase in RWPE-1 cell proliferation (Figure 3B). In addition, BMP5 treatment led to a more dispersed cell growth pattern and to 30% increased cell migration by Boyden chamber assay (Figure 3C). In contrast, BMP5 addition to a different immortalized prostate epithelial cell line (BPH-1) (37) led to 15% decreased cell proliferation with no obvious cell dispersion (Supplemental Figure 4B). BMP5 also modestly reduced proliferation of WPMY-1 cells (Supplemental Figure 4B), an immortalized prostate myofibroblast line derived from the same prostate as RWPE-1 epithelial cells (38).
Figure 3. BMP5 addition to cultured prostate cells supports role in BPH disease process.
(A) Addition of BMP5 (250 ng/ml) to RWPE-1 prostate epithelial and WPMY-1 myofibroblast cells activates the canonical SMAD pathway, demonstrated by 15- to 20-fold increased phospho-SMAD1/5/9 on Western blot. (B) BMP5 addition (concentrations indicated) to RWPE-1 cells increases cell numbers. Mean (red) and SD (blue) shown. Multiplicity-adjusted P values generated from 1-way ANOVA with post hoc comparison to no BMP5 control (Dunnett’s test); *P < 0.05. Data are representative of 3 independent experiments, each done with 2 samples assayed per concentration. (C) Addition of BMP5 (250 ng/ml) to RWPE-1 cells leads to dispersed cell growth (with fewer cell clusters) and to increased Transwell migration. Mean and SD shown; P value generated from 2-tailed Student’s t test. Data are representative of 2 independent experiments, each done with 3 samples assayed per condition. (D and E) BMP5 addition to RWPE-1 and WPMY-1 cells generates a transcriptional response significantly enriched for BPH (vs. normal prostate) overexpressed genes (left), and with WPMY-1 cells for BPH stromal signature genes (center), but not AR/secretory signature genes (right). Gene set enrichment analysis (GSEA) enrichment score P values are indicated.
To further investigate the effects of BMP5 on cultured prostate cells, we profiled resultant changes in gene expression at 72 hours by RNA-Seq. Notably, BMP5 treatment of RWPE-1 cells drove gene expression changes that recapitulated the changes seen in BPH compared with normal prostate. Specifically, based on GSEA (39), the genes upregulated by BMP5 addition to RWPE-1 cells were significantly enriched for those genes more highly expressed in BPH (based on our SAM analysis; P < 0.0001) (Figure 3D and Supplemental Table 6). Likewise, BMP5 addition to WPMY-1 prostate myofibroblast cells led to the induction of genes more highly expressed in BPH (P < 0.0001) as well as genes of the BPH stromal signature (P < 0.0001) (Figure 3E and Supplemental Table 6). As a specificity control, we confirmed that neither the RWPE-1 nor WPMY-1 transcriptional response to BMP5 showed enrichment of AR/secretory genes (Figure 3, C and D). From the RNA-Seq data, RWPE-1 and WPMY-1 cells (as well as bulk BPH tissue) express presumptive BMP5 receptors (40), including type I receptors (BMPR1A, BMPR1B, ACVR1) and type II receptors (BMPR2, ACVR2A, ACVR2B) (Supplemental Table 7). RWPE-1 and WPMY-1 cells also express select other BMP ligands, though none are significantly upregulated in BPH (Supplemental Table 7). Our finding that BMP5 addition in 2 cell lines mimics the transcriptional changes of BPH suggests a role for BMP5 in driving BPH pathogenesis.
We also investigated CXCL13, the other ligand highly expressed in BPH tissue. From our RNA-Seq data, neither RWPE-1 nor WPMY-1 cells express the only known CXCL13 receptor, CXCR5, which we confirmed by qRT-PCR (Supplemental Table 8). Moreover, addition of recombinant CXCL13 to RWPE-1 and WPMY-1 cells did not affect cell proliferation or migration (Supplemental Figure 4, C and D). Rather, CXCL13 has been characterized primarily as a B cell chemoattractant (41). Indeed, from our BPH transcriptional heatmap (Figure 1A), we had identified a B cell gene feature that includes the CXCR5 gene, is prominent in BPH stromal nodules, and is correlated with stromal CXCL13 expression (Supplemental Figure 4E). Nonetheless, in contrast with CXCL13, the B cell feature did not show a significant increase in BPH versus normal prostate tissue.
Transcriptional changes uncover cell subsets altered in BPH.
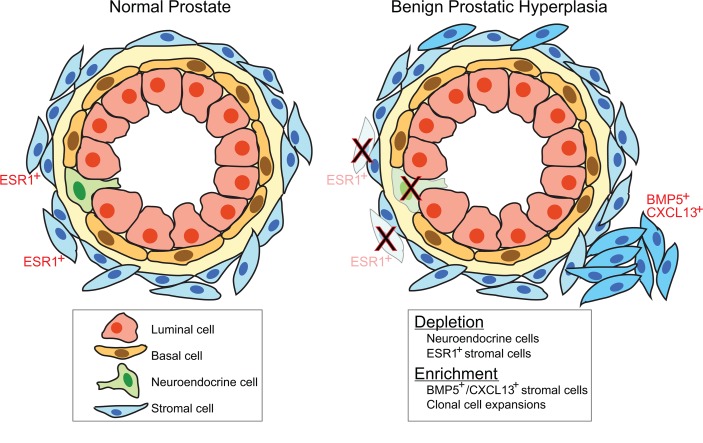
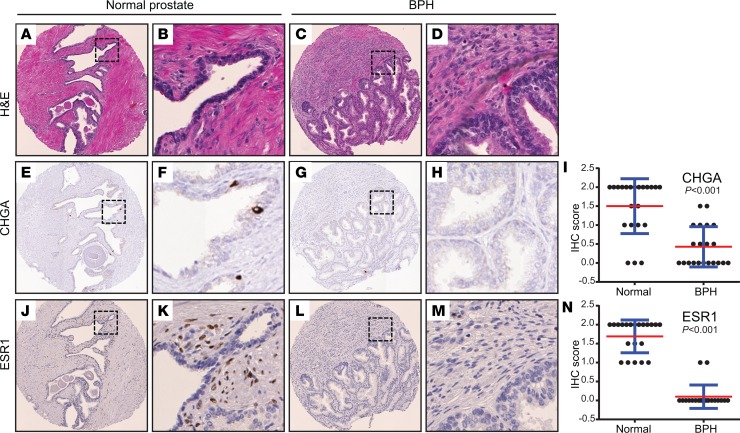
In analyzing the genes with significantly decreased expression in BPH (vs. normal prostate), several stood out as markers of specific cell types. For example, chromogranin A (CHGA), a marker of neuroendocrine cell lineage, was downregulated in BPH (Figure 2B). To further investigate this finding, we constructed a tissue microarray (TMA) containing over 100 normal prostate and BPH tissues, including most of the cases used for RNA-Seq. IHC demonstrated significant depletion of a subpopulation of CHGA-expressing neuroendocrine cells in the epithelium of BPH compared to matched normal (Figure 4, A–I).
Figure 4. IHC on prostate TMA identifies altered ESR1+ cell subset in BPH.
(A–D) Hematoxylin and eosin stains of representative normal prostate and BPH-matched pair. Original magnifications, ×200 and ×800 (insets). (E–H) CHGA immunostaining (marker of neuroendocrine cells) shown for same case; note depletion of neuroendocrine cells in BPH. (I) CHGA IHC scores across all TMA cases (n = 22 normal and n = 21 BPH). Mean (red) and SD (blue) shown; P values generated from 2-tailed Student’s t test. (J–M) ESR1 immunostaining shown for same case; note depletion of periglandular ESR1+ fibroblast-like cells in BPH. (N) ESR1 IHC scores across all TMA cases (n = 21 normal and n = 20 BPH).
Estrogen receptor (ESR1) transcript was also significantly decreased in the BPH samples (Figure 2B). Estradiol and estrogen receptor signaling have been implicated in rodent and canine studies as effectors of BPH (42, 43). By IHC, we observed that in normal prostate, ESR1 was expressed in a distinctive subset of periglandular fibroblast-like stroma cells, while in corresponding BPH stroma, ESR1 protein was not detected (Figure 4, J–N). Consistent with this finding, Ingenuity Pathway Analysis (IPA) of the transcriptome data identified estradiol as by far (P = 10–18) the top-ranked predicted regulator diminished in BPH (Supplemental Table 9). Based on these findings, BPH stroma appears to have lost a subset of ESR1+ fibroblast-like cells, with concomitant loss of downstream estrogen receptor signaling.
To investigate whether additional cell lineages might be affected in BPH, we carried out additional IHC using known lineage markers. We observed a trend toward fewer AR-positive nuclei in BPH epithelium, significantly reduced desmin staining (myofibroblasts/smooth muscle cells) in BPH stroma and BPH stromal nodules, a trend toward fewer CD3+ cells (T cells) in BPH, significantly increased CD20+ cells (B cells) in BPH stromal nodules, and significantly increased CD163+ cells (macrophages) in BPH and BPH stromal nodules (Supplemental Figure 5). Vimentin staining (fibroblasts, myofibroblasts) showed little variation among BPH samples by our semiquantitative IHC assay and so was not scored.
To independently evaluate altered cell subsets in BPH, we carried out single-cell RNA-Seq (scRNA-Seq) of matched BPH and normal tissue from a single prostate. Fresh tissue was enzymatically disaggregated, and then single-cell transcriptomes were sequenced using the 10× Genomics Chromium workflow. In total, we analyzed 9373 single cells with a median 1891 genes expressed per cell. Dimensional reduction and clustering revealed distinct cell clusters (identifiable by characteristic marker genes) representing epithelial cells (basal, intermediate, and secretory), myofibroblasts, immune cells, and endothelial cells (Figure 5, A–D, and Supplemental Figure 6, A–D). The BPH tissue showed depletion of neuroendocrine cells (CHGA; Figure 5, E, F, I, and J), as well as enrichment of a myofibroblast subset expressing BMP5 and CXCL13 (Figure 5, G, H, K, and L); both findings were consistent with the bulk RNA-Seq data. In contrast, ESR1 transcript was detected in a subset of myofibroblasts from both the normal prostate and BPH samples (Supplemental Figure 6, E–H). In the single prostate sampled, the BPH epithelial fraction also exhibited a shift toward more mature secretory epithelial cells (Figure 5, A, E, and I).
Figure 5. scRNA-Seq reveals altered cell subsets in BPH.
(A) Two-dimensional projection (t-SNE plot) of single-cell transcriptomes stratifies prostate tissue cells (dots) into distinct clusters, identifiable by characteristic expression of marker genes, including (B) KRT13 (basal epithelium) (red intensity scales to maximum log2 expression), (C) KLK3 (secretory epithelium), and (D) DCN (fibroblasts). Additional cell type markers are shown in Supplemental Figure 6. (E) Normal prostate tissue cell subset, illustrating expression of (F) CHGA (neuroendocrine cells), (G) BMP5, and (H) CXCL13. (I) BPH tissue cell subset, illustrating expression of (J) CHGA, (K) BMP5, and (L) CXCL13. Note relative depletion in BPH tissue of CHGA-expressing neuroendocrine cells and enrichment of BMP5- or CXCL13-expressing myofibroblasts. Insets magnify select cell clusters. Inset magnification ×1.6.
Exome sequencing reveals expansion of specific cell clones in BPH.
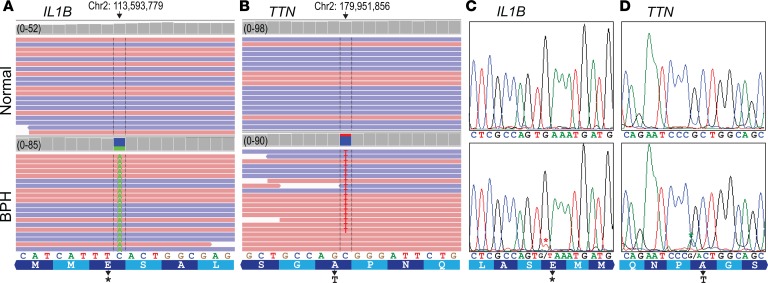
To address whether the underlying hyperplasia of BPH might represent a clonal process, we performed whole-exome sequencing (WES) on BPH tissues from 18 patients (fresh or FFPE tissue) along with matched normal DNA from either normal prostate or lymph node. Analysis of the WES data revealed 38 “high-evidence” (see Methods) single-nucleotide variants (SNVs) acquired among the BPH tissues (Figure 6, A and B, and Supplemental Table 10), with a median of 2 SNVs per patient (range, 0–6). We successfully validated 2 of 2 (100%) SNVs by PCR/Sanger sequencing and 15 of 16 (94%; randomly selected) by PCR amplicon deep resequencing (Figure 6, C and D, and Supplemental Table 11). The SNV allelic frequencies (many as high as 20%–30%) suggest that these presumed heterozygous mutations were present in up to 50% of the cells in the sample. The finding of an enriched population of cells with a somatically acquired mutation implies a clonal outgrowth of a population of cells from a single cell.
Figure 6. WES reveals somatic SNVs in BPH nodules.
(A) Integrated Genome Viewer (IGV) display of mapped reads from WES of BPH and matched normal DNA within the IL1B gene. Red reads correspond to the positive DNA strand, and blue reads correspond to the negative DNA strand; only a subset of mapped reads (totals indicated) shown. Note, the IL1B SNV in BPH leads to a frameshift with early translational termination. (B) Similar IGV display depicts a nonsynonymous SNV in the TTN gene in a different BPH case. (C) Sanger sequencing validation of IL1B SNV; red asterisk marks doublet peak indicative of SNV. (D) Sanger sequencing validation of TTN SNV; green asterisk marks doublet peak indicative of SNV. See Supplemental Table 10 for additional SNVs validated by deep amplicon resequencing.
The 38 high-evidence SNVs occurred across 33 genes, of which 2 genes were recurrently mutated; dentin sialophosphoprotein (DSPP) harbored 5 SNVs among 3 BPH samples (with 2 adjacent nucleotides identically mutated in 2 BPH samples), and titin (TTN) was represented by 2 SNVs over 2 BPH samples. The 38 SNVs included frameshift deletions, stop-gain mutations, and nonsynonymous substitutions, many predicted (by evolutionary conservation) to be deleterious. Interestingly, the 33 affected genes were significantly enriched for Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census (44) tier 1 genes (enriched genes: BARD1, CREBBP, NUP98, ZNF384) (P = 0.01; hypergeometric test). There was no enrichment for genes differentially expressed in BPH versus normal prostate (from our SAM analysis).
Recent studies have shown that tissues can acquire SNVs during cell expansion and turnover with normal tissue development and maintenance (45). To further evaluate the acquired SNVs in BPH, we performed a reverse analysis seeking to identify acquired SNVs in the matched normal tissue (using the BPH tissues as reference). Notably, the proportion of high-evidence SNVs identified among the BPH samples (forward analysis) was 7 times higher than among the normal tissues (reverse analysis) (P < 0.0001; χ2 test), suggesting the acquired SNVs in BPH are linked to the disease process.
Discussion
Our study describes the first sequencing-based genomic landscape of BPH to our knowledge. We observed transcriptional heterogeneity among BPH samples, including varied expression of a BPH stromal gene feature. Notably, elevated expression of a 65-gene BPH stromal signature (the core of the stromal gene feature) was associated with worse patient-reported LUTS and greater bother from those symptoms. This finding highlights the importance of prostatic stroma in driving BPH symptoms and agrees with previous observations that increased fibrosis and stiffness of the prostatic tissues correlate with worse LUTS (46). Our findings also echo a prior microarray study that reported an “extracellular matrix” gene expression feature that distinguished symptomatic from asymptomatic BPH (28). However, whether that feature corresponds to our stromal feature is unclear because those microarray data are not available for reanalysis. That our 65-gene stromal signature correlates with symptoms suggests that it is capturing an intrinsic tissue property robust to tissue sampling variables. Whether the signature reflects an increased stromal proportion or else increased stromal activation remains to be determined. In addition to validating the stromal signature in independent data sets, its association with treatment response to 5-α-reductase inhibitors and α-adrenoceptor–blocking agents should be tested. We also observed that an androgen-signaling signature (at least in prostatic epithelia) does not correlate with LUTS, underscoring that androgen signaling may play a permissive role for prostate growth but may not be an underlying driver of BPH.
The genes we found more highly expressed in BPH, which include many of the stromal signature genes, represent new candidate drivers and therapeutic targets. For example, BMP5 and CXCL13 transcript levels were highly overexpressed (at least 50-fold) and could be targetable because both are signaling molecules. BMP5 is a member of the bone morphogenetic protein family (and TGF-β superfamily) that has broad roles in tissue morphogenesis (33). In prior microarray studies, BMP5 was listed among genes found more highly expressed in the transition zone of the normal prostate (where BPH arises) (47) and in BPH (27), though it has never been investigated as a BPH driver. Buttressing our RNA-Seq results, we found that the addition of BMP5 protein to RWPE-1 prostate epithelial cells enhances cell proliferation. Although the proliferative effect is modest (20% increased cell numbers at 96 hours), the cumulative effect over years could more than explain the hyperplasia observed in BPH. BMP5 treatment of RWPE-1 cells also led to increased cell dispersion and Transwell migration, features of epithelial-mesenchymal transition (EMT). These findings are consonant with a prior study that reported features of EMT in BPH tissue (48). However, because the increased cell proliferation and dispersion were not replicated in a second immortalized prostate epithelial line (BPH-1), caution is warranted in not overinterpreting the findings. Likewise, that BMP5 modestly reduced WPMY-1 cell proliferation, while increased stromal cells were observed in BPH, suggests that the net impact of BMP5 on prostate tissue myofibroblasts is modulated by other signaling molecules.
Most remarkably, BMP5 treatment of both prostate epithelial (RWPE-1) and prostate myofibroblast (WPMY-1) cells resulted in gene expression programs that mirrored those upregulated in BPH tissue and (for WPMY-1 cells) in BPH stroma. Thus, BMP5 likely acts directly on prostatic epithelia and stroma to effect BPH changes, though possibly also indirectly by modulating stromal cells to secrete paracrine factors. Future work will need to confirm BMP5 overexpression at the protein level, define the most important receptors (in both prostatic epithelia and stroma) from candidates observed in our RNA-Seq data, and evaluate BMP5 signaling as a therapeutic target. Importantly, screening drugs/molecules for their ability to revert BPH cells/tissues to more closely mimic normal prostate transcriptomes might provide a valuable approach to evaluate new candidate BPH therapies.
CXCL13, the other highly overexpressed signaling molecule, has not been studied in the pathogenesis of BPH. CXCL13 is known as a B cell chemoattractant (41), and B cell numbers were largest in BPH stromal nodules, where CXCL13 was most highly expressed. However, apart from stromal nodules, B cells (or the B cell signature) were not more prevalent in BPH compared to normal tissue. Although a different chemokine (CXCL12; not found in our BPH transcript set) has been reported to enhance proliferation of prostatic epithelial cells in vitro (49), we did not observe CXCL13 effects on prostate cells. Therefore, the relevance of elevated CXCL13 to the BPH disease process (apart from BPH stromal nodules) remains uncertain but warrants further studies.
Beyond BPH genes and signatures, our bulk transcriptome analysis implied changes in the resident cell types, which we confirmed and extended by IHC and scRNA-Seq analysis. Our combined studies identified marked depletion of neuroendocrine cells in the BPH epithelial compartment. Neuroendocrine cells are a relatively uncommon cell type within the normal prostate epithelium and produce neurosecretory products (including serotonin) with paracrine effects on prostatic epithelium (50). A previous IHC study also reported reduced neuroendocrine cells in BPH tissue (51), though this finding has been contested (52), and serotonin has been implicated in negatively regulating prostate growth (53). Equally interesting was our discovery of an ESR1+ stromal cell type (with fibroblast morphology) residing adjacent to epithelium, present in normal prostate but absent from BPH. Moreover, pathway analysis identified estradiol as the top regulator decreased in BPH compared with normal tissue. Although speculative, these findings suggest that estrogen receptor signaling in prostatic stroma might constrain BPH, in marked contrast with rodent and canine models, in which estrogen administration produces prostatic enlargement (42, 43). Alternatively, the depletion of ESR1+ stromal cells might reflect the relative expansion of other stromal cell types or else loss of ESR1 expression rather than cell type depletion (though raising the question of what defines cell type vs. cell state; ref. 54). Concomitant with the observed cell type depletions, our bulk RNA-Seq and scRNA-Seq data suggested expansions of other cell types, including stromal (likely myofibroblast) subsets expressing BMP5 and CXCL13. Additional epithelial cell type shifts were also noted in the single scRNA-Seq sample, though not supported from our bulk RNA-Seq data; additional studies are warranted. Finally, immunostaining with cell lineage markers supported additional cell type shifts in BPH, including alterations in myofibroblast/smooth muscle proportions (decreased desmin staining) and increased tissue macrophages.
Whether BPH nodules represent clonal cell expansions has not been studied. Remarkably, by exome sequencing, we discovered somatically acquired mutations in BPH. Many of these mutations are predicted to affect protein function, though it remains unclear whether they are growth-promoting “drivers” or passenger genes that have no functional role in normal prostate biology. The genes did show enrichment for COSMIC Cancer Census Genes, suggesting the mutations might have a functional role in BPH development, though the gene numbers are small. We also found recurrently mutated genes across BPH samples, including TTN and DSPP, which have been reported to be mutated in human cancers (55, 56). TTN functions in striated muscle elasticity (57), while DSPP functions in dentin mineralization (58), neither with an obvious connection to BPH. Moreover, TTN is a large, late-replicating gene, a gene class that incurs increased mutations but is thought unlikely to represent cancer drivers (55), and DSPP contains a long stretch of triple codon repeats and therefore may be subject to increased mutation or sequence read-mapping errors. Although further studies are needed to determine whether the mutations observed in any of the 33 mutated genes are functionally relevant to BPH, the somatic SNVs nonetheless serve as lineage markers and are indicative of clonal cell expansions. We found significantly more SNVs in BPH compared with normal tissue (adjacent prostate or lymph node), suggesting that SNVs were acquired in recent cell expansions related to the BPH disease process, rather than during organ development. Moreover, the alternative allele frequencies suggest that the clonal population represents a substantial portion of the cell population in the BPH tissues. Future studies will determine which cell type(s) are clonally expanded and whether they are cellular drivers of the BPH disease process.
BPH has been thought of as prostate enlargement due to cellular hyperplasia. Taken together, our transcriptome and exome studies support a rather different picture, of a disease resulting from a fundamental relandscaping of cell types. It is possible that cell type depletions represent passive losses because of the expansion of others. However, the known intricacies of epithelial-stromal crosstalk and tissue homeostasis suggest that these cell type shifts are more likely to be intimately connected to disease pathogenesis and may fundamentally underlie the pathobiology and resulting symptoms. Based on our genomic characterization, we propose that BPH should be studied not only as altered molecules and signaling pathways but also as a fundamental relandscaping of cell types and tissue. Analysis of single cells in coculture models and from disaggregated prostate tissues and in situ in normal and diseased tissues will be illuminating.
Methods
Prostate tissue specimens.
Tissue specimens were acquired from radical prostatectomies for the diagnosis of prostate cancer that had concurrent BPH. For RNA-Seq, cases were selected from prostatectomies performed between 2011 and 2013, for which patients had completed the IPSS with Bother Score before surgery. Histopathologic samples of FFPE tissues were reviewed by a pathologist, and 1-mm–diameter core punch biopsies were taken from the blocks to ensure that the histologic entities were clearly identified and not contaminated by other histologic changes, such as prostate cancer. Cores were harvested from regions of BPH (mixed epithelia/stroma), BPH pure stromal nodules, and the normal peripheral zone of the prostate far away from any cancer and without other pathologic changes. For WES, cases were selected from freshly frozen tissue and FFPE blocks, archived in the Stanford Pathology Department from 2005 to 2013.
RNA-Seq and RNA-Seq data analysis.
RNA was isolated from FFPE cores using a Qiagen RNeasy FFPE kit and then quantified by NanoDrop spectrophotometer and Qubit fluorometer. Barcoded RNA-Seq libraries were constructed from 1 μg input RNA using Illumina TruSeq Stranded Total RNA LT kits with Ribo-Zero Gold depletion of rRNA. Barcoded libraries were then quantified by NanoDrop, qualified by Agilent BioAnalyzer, pooled (6 libraries per lane), and sequenced (unpaired, 100 bp) on an Illumina HiSeq2000 instrument (Stanford Genome Sequencing Service Center) to an average depth of 39,000,000 reads. Barcodes were deconvoluted using Illumina CASAVA software and reads were quality assessed using Babraham Institute FastQC software. Reads were then mapped to the RefSeq transcriptome using TopHat and Cufflinks (59) and transcripts quantified as reads per kilobase of transcript per million mapped reads (RPKM). Not all the FFPE RNAs yielded high-quality sequencing libraries; 65 samples (84%) passed rigorous quality control criteria based on RPKM histograms (>30% of genes with RPKM ≥1) and RPKM values for housekeeping genes (GAPDH > 20; ACTB > 100; B2M > 40). RNA-Seq data were then further processed by eliminating poorly measured genes, those absent from more than 50% of samples or having average expression less than 0.1 RPKM. Gene RPKM values were then bottom thresholded at 0.1 and transformed to log2 space. RNA-Seq libraries had been constructed by 2 individuals, necessitating batch normalization by separately median centering genes within each batch and then recentering the combined data set; equivalent results were obtained by batch normalization using ComBat (60). Subsequent analyses of variably expressed genes included 2548 genes with SD at least 1.2 across samples. Unsupervised hierarchical clustering (Pearson’s correlation; average linkage) was performed using Cluster 3.0 software (61) and visualized using Java Treeview (62). High versus low expression of a stromal (65 genes) and AR/secretory (57 genes) signature was defined using a median cutoff (across samples) of the average expression of all signature genes within each sample. In 2 patients with more than 1 BPH sample, signature scores were averaged before analysis. Gene-based models to predict BPH symptoms were built using lasso (63), implemented in R, and performance (added value) was evaluated by the reduction of the mean squared error compared to the null model (no genes). Supervised analysis of differentially expressed genes was done using SAM (2-class and unpaired, t statistic) (32), though DESeq2 (64) produced comparable results. Pathway analyses were done using IPA. qRT-PCR validation of BMP5 and CXCL13 was done using TaqMan Gene Expression Assays (Hs00234930_m1 and Hs00757930_m1) (Life Technologies) on an Applied Biosystems 7500 Fast Instrument.
Smart-3SEQ of LCM samples.
To confirm assignments of gene expression features to epithelial versus stromal compartments, LCM was performed on 5 independent BPH cases, using an Arcturus XT LCM system, and transcriptomes were profiled by Smart-3SEQ exactly as described (30). Genes significantly more highly expressed in BPH epithelia or stroma were identified by DESeq2 (64).
Prostate cell culture experiments.
RWPE-1, WPMY-1, and BPH-1 cell lines were obtained directly from the ATCC and DSMZ cell culture repositories, cultured as directed, and passaged for no more than 3 months. The 25,000–40,000 cells were plated in 6-well plates in duplicate, and then recombinant human BMP5 (R&D Systems), CXCL13 (R&D Systems), or vehicle control was added daily with media replacement. Relative cell counts were determined at 96 hours by WST-1 assay (MilliporeSigma). All cell growth experiments were repeated at least 3 times with comparable results. Western blots were used to quantify canonical SMAD pathway activation (30 minutes after BMP5 treatment), using qualified primary antibodies (Supplemental Table 12), chemiluminescence detection, and ImageJ (NIH) quantification. Boyden chamber (Transwell) migration assays were carried out in triplicate, using polyethylene terephthalate 8-μM pore inserts (Falcon) and a chemotactic gradient of complete media against a 1:20 dilution into base media. After 48 hours, migrated cells were stained with crystal violet (Sigma-Aldrich) and counted. For transcriptome sequencing studies, RWPE-1 and WPMY-1 cells (assayed in duplicate) were plated and 250 ng/ml BMP5 (or vehicle control) added with daily media replacement for 72 hours. RNA was isolated using a Qiagen RNeasy kit, RNA-Seq libraries were generated using TruSeq Stranded Total RNA LT kit (WPMY-1) and Illumina TruSeq RNA Library Prep Kit v2 (RWPE-1), and barcoded RNA-Seq libraries were sequenced and resultant data were processed as above. RPKM values were then averaged for the duplicate platings, and enrichment for gene sets was evaluated by GSEA (39).
IHC on TMA.
A TMA was constructed using a manual Beecher instrument and included duplicate 1-mm cores from over 100 BPH and matched normal prostate tissues. TMA 4-μM sections were processed for antigen retrieval (Ventana or Leica proprietary solution) and then used for IHC with primary antibodies (listed in Supplemental Table 12), followed by HRP-conjugated secondary antibody and DAB chromogenic detection (Stanford Pathology IPOX lab). IHC was interpreted by a trained pathologist (scoring criteria listed in Supplemental Table 12), and scores for duplicate cores were averaged.
scRNA-Seq.
Fresh prostate tissue was acquired from radical prostatectomy cases, where BPH was identifiable grossly by transition zone nodules, and normal prostate tissue was harvested from the peripheral zone opposite that having biopsy-confirmed cancer. Tissue was disaggregated using collagenase/hyaluronidase, trypsin, and dispase, following StemCell Technologies’ Prostate Tissue Dissociation protocol. Contaminating red blood cells were lysed using ammonium chloride solution (StemCell Technologies), and single cells were filtered through a 40-μM cell strainer. Half the remaining cells were further purified by flow cytometry, gating on viable (propidium iodide–negative) single cells. Single-cell RNA-Seq was then performed using the 10× Genomics Chromium workflow (by the Stanford Functional Genomics Facility), entailing microdroplet single-cell capture (with cell and molecular barcodes), cDNA synthesis, pooling, and library preparation, followed by sequencing on an Illumina HiSeq4000. Cells and transcripts were then enumerated using the 10× Genomics CellRanger pipeline and visualized using 10× Genomics Loupe software. Data from flow-sorted and unsorted cells were indistinguishable and combined for the illustrations shown. Note that the tissue processing procedure appears to have enriched for epithelial over stromal cell populations.
WES.
WES was carried out on individual BPH nodules, comprising mostly mixed epithelia and stroma. DNA was extracted using the Qiagen DNeasy kit (fresh tissue) or QIAamp DNA FFPE Tissue Kit and quantified by Nanodrop and Qubit. WES was done using the Agilent SureSelect Human All Exon V5 kit, following the modifications recommended for FFPE-derived DNA samples. Barcoded WES libraries (4 per lane) were sequenced on a HiSeq2000 to an average ×60 mean bp coverage. Raw reads were aligned to the hg19 human genome using Burrows-Wheeler Alignment (65), and Picard was used to mark duplicates. The GATK 3.x best practices were followed for Indel Realignment and Base Recalibration. Callable regions were calculated using bedtools followed by an ensemble approach of 4 variant callers: MuTect, VarScan2, VarDict, and Freebayes (66–69). Exonic and splicing SNVs called by at least 2 of the 4 callers were further processed by ANNOVAR (70) and custom python scripts for variant annotation, filtering, and ranking. High-evidence SNVs were defined as those with at least 10 alternative allele reads in the BPH sample and 0 alternative allele reads in the matched normal sample. Selected SNVs were validated by PCR/Sanger sequencing (PCR primers available in Supplemental Table 13). Additional SNVs were validated by amplicon deep resequencing (PCR primers available in Supplemental Table 13), with multiplexed barcoded amplicons sequenced on an Illumina NextSeq.
Data availability.
All RNA-Seq and WES data are available from the database of Genotypes and Phenotypes (phs001698.v1.p1). TMA IHC images are available from Stanford Tissue Microarray Database (https://tma.im/cgi-bin/home.pl), as array block TA-457.
Statistics.
Unless otherwise indicated, a 2-tailed Student’s t test was performed to assess statistical significance, using Prism version 7.0 (GraphPad), where P < 0.05 was considered significant. Two-class analysis of RNA-Seq data was done using SAM (32), which estimates FDRs against randomly permutated samples. Gene set enrichment analysis was done by GSEA (39), with significance determined by Kolmogorov-Smirnov statistic.
Study approval.
The study was approved by the Stanford University IRB. Written informed consent was received from participants before inclusion in the study.
Author contributions
LWM, ZS, JDB, RBW, and JRP conceived and planned the studies. LWM, ZS, ASP, S Varma, SZ, CZ, KT, JB, OE, and RN performed experiments. XG, JWF, S Vennam, RTS, RT, JDB, RBW, and JRP analyzed data. LWM, JDB, RBW, and JRP wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by funding from a National Institute of Diabetes and Digestive and Kidney Diseases P20 grant (P20 DK103093) to JDB, RBW, and JRP. We thank the Stanford Cancer Institute–supported core facilities providing services, including the Stanford Tissue Bank, the Stanford Functional Genomics Facility, and the Stanford Genome Sequencing Service Center.
Version 1. 05/16/2019
In-Press Preview
Version 2. 06/20/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e129749. https://doi.org/10.1172/jci.insight.129749.
Contributor Information
Lance W. Middleton, Email: lmiddl1@gmail.com.
Zhewei Shen, Email: zhewei.shen@gmail.com.
Sushama Varma, Email: svarma@stanford.edu.
Anna S. Pollack, Email: annapollack@stanford.edu.
Xue Gong, Email: gongxuesusan@gmail.com.
Shirley Zhu, Email: sxz@stanford.edu.
Chunfang Zhu, Email: cfzhu@stanford.edu.
Joseph W. Foley, Email: jwfoley@stanford.edu.
Sujay Vennam, Email: sxv@stanford.edu.
Robert T. Sweeney, Email: tripsweeney@gmail.com.
Karen Tu, Email: karen_tu@brown.edu.
Jewison Biscocho, Email: jlbiscocho@gmail.com.
Okyaz Eminaga, Email: okyaz.eminaga@stanford.edu.
Rosalie Nolley, Email: tinyn@stanford.edu.
Robert Tibshirani, Email: robert.tibshirani@stanford.edu.
James D. Brooks, Email: jdbrooks@stanford.edu.
Robert B. West, Email: rbwest@stanford.edu.
Jonathan R. Pollack, Email: pollack1@stanford.edu.
References
- 1.Chapple CR, et al. Lower urinary tract symptoms revisited: a broader clinical perspective. Eur Urol. 2008;54(3):563–569. doi: 10.1016/j.eururo.2008.03.109. [DOI] [PubMed] [Google Scholar]
- 2.Berry SJ, Coffey DS, Walsh PC, Ewing LL. The development of human benign prostatic hyperplasia with age. J Urol. 1984;132(3):474–479. doi: 10.1016/S0022-5347(17)49698-4. [DOI] [PubMed] [Google Scholar]
- 3.Roehrborn CG. Prostate size: does it matter? Rev Urol. 2000;2(2):95–98. [PMC free article] [PubMed] [Google Scholar]
- 4.Williams AM, et al. Prostatic growth rate determined from MRI data: age-related longitudinal changes. J Androl. 1999;20(4):474–480. [PubMed] [Google Scholar]
- 5.McNeal JE. Origin and evolution of benign prostatic enlargement. Invest Urol. 1978;15(4):340–345. [PubMed] [Google Scholar]
- 6.Bartsch G, Müller HR, Oberholzer M, Rohr HP. Light microscopic stereological analysis of the normal human prostate and of benign prostatic hyperplasia. J Urol. 1979;122(4):487–491. doi: 10.1016/S0022-5347(17)56476-9. [DOI] [PubMed] [Google Scholar]
- 7.Rohr HP, Bartsch G. Human benign prostatic hyperplasia: a stromal disease? New perspectives by quantitative morphology. Urology. 1980;16(6):625–633. doi: 10.1016/0090-4295(80)90577-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee KL, Peehl DM. Molecular and cellular pathogenesis of benign prostatic hyperplasia. J Urol. 2004;172(5 pt 1):1784–1791. doi: 10.1097/01.ju.0000133655.71782.14. [DOI] [PubMed] [Google Scholar]
- 9.Cunha GR, et al. The endocrinology and developmental biology of the prostate. Endocr Rev. 1987;8(3):338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- 10.Monti S, et al. Prevalent decrease of the EGF content in the periurethral zone of BPH tissue induced by treatment with finasteride or flutamide. J Androl. 1997;18(5):488–494. [PubMed] [Google Scholar]
- 11.Planz B, Wang Q, Kirley SD, Lin CW, McDougal WS. Androgen responsiveness of stromal cells of the human prostate: regulation of cell proliferation and keratinocyte growth factor by androgen. J Urol. 1998;160(5):1850–1855. doi: 10.1016/S0022-5347(01)62431-5. [DOI] [PubMed] [Google Scholar]
- 12.Niu Y, Xu Y, Zhang J, Bai J, Yang H, Ma T. Proliferation and differentiation of prostatic stromal cells. BJU Int. 2001;87(4):386–393. doi: 10.1046/j.1464-410x.2001.00103.x. [DOI] [PubMed] [Google Scholar]
- 13.Le H, Arnold JT, McFann KK, Blackman MR. DHT and testosterone, but not DHEA or E2, differentially modulate IGF-I, IGFBP-2, and IGFBP-3 in human prostatic stromal cells. Am J Physiol Endocrinol Metab. 2006;290(5):E952–E960. doi: 10.1152/ajpendo.00451.2005. [DOI] [PubMed] [Google Scholar]
- 14.Kassen A, Sutkowski DM, Ahn H, Sensibar JA, Kozlowski JM, Lee C. Stromal cells of the human prostate: initial isolation and characterization. Prostate. 1996;28(2):89–97. doi: 10.1002/(SICI)1097-0045(199602)28:2<89::AID-PROS3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Theyer G, et al. Phenotypic characterization of infiltrating leukocytes in benign prostatic hyperplasia. Lab Invest. 1992;66(1):96–107. [PubMed] [Google Scholar]
- 16.Roehrborn CG, Kaplan SA, Noble WD, Slawin KM, McVary KT, Kusek JW. The impact of acute or chronic inflammation in baseline biopsy on the risk of clinical progression of BPH. Results from the MTOPS study. J Urol. 2005;173(S4):346 [Google Scholar]
- 17.Begley LA, Kasina S, MacDonald J, Macoska JA. The inflammatory microenvironment of the aging prostate facilitates cellular proliferation and hypertrophy. Cytokine. 2008;43(2):194–199. doi: 10.1016/j.cyto.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickel JC, Roehrborn CG, O’Leary MP, Bostwick DG, Somerville MC, Rittmaster RS. The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial. Eur Urol. 2008;54(6):1379–1384. doi: 10.1016/j.eururo.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohrmann S, Smit E, Giovannucci E, Platz EA. Association between markers of the metabolic syndrome and lower urinary tract symptoms in the Third National Health and Nutrition Examination Survey (NHANES III) Int J Obes (Lond) 2005;29(3):310–316. doi: 10.1038/sj.ijo.0802881. [DOI] [PubMed] [Google Scholar]
- 20.Gacci M, et al. Metabolic syndrome and benign prostatic enlargement: a systematic review and meta-analysis. BJU Int. 2015;115(1):24–31. doi: 10.1111/bju.12728. [DOI] [PubMed] [Google Scholar]
- 21.Berger AP, et al. Increased growth factor production in a human prostatic stromal cell culture model caused by hypoxia. Prostate. 2003;57(1):57–65. doi: 10.1002/pros.10279. [DOI] [PubMed] [Google Scholar]
- 22.Gharaee-Kermani M, Kasina S, Moore BB, Thomas D, Mehra R, Macoska JA. CXC-type chemokines promote myofibroblast phenoconversion and prostatic fibrosis. PLoS One. 2012;7(11):e49278. doi: 10.1371/journal.pone.0049278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vignozzi L, et al. Fat boosts, while androgen receptor activation counteracts, BPH-associated prostate inflammation. Prostate. 2013;73(8):789–800. doi: 10.1002/pros.22623. [DOI] [PubMed] [Google Scholar]
- 24.Gormley GJ, et al. The effect of finasteride in men with benign prostatic hyperplasia. The Finasteride Study Group. N Engl J Med. 1992;327(17):1185–1191. doi: 10.1056/NEJM199210223271701. [DOI] [PubMed] [Google Scholar]
- 25.McConnell JD, et al. The long-term effect of doxazosin, finasteride, and combination therapy on the clinical progression of benign prostatic hyperplasia. N Engl J Med. 2003;349(25):2387–2398. doi: 10.1056/NEJMoa030656. [DOI] [PubMed] [Google Scholar]
- 26.Saigal CS, Joyce G. Economic costs of benign prostatic hyperplasia in the private sector. J Urol. 2005;173(4):1309–1313. doi: 10.1097/01.ju.0000152318.79184.6f. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, et al. Gene expression signature of benign prostatic hyperplasia revealed by cDNA microarray analysis. Prostate. 2002;51(3):189–200. doi: 10.1002/pros.10087. [DOI] [PubMed] [Google Scholar]
- 28.Prakash K, et al. Symptomatic and asymptomatic benign prostatic hyperplasia: molecular differentiation by using microarrays. Proc Natl Acad Sci U S A. 2002;99(11):7598–7603. doi: 10.1073/pnas.112191399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry MJ, et al. The American Urological Association Symptom Index for Benign Prostatic Hyperplasia. J Urol. 2017;197(2S):S189–S197. doi: 10.1016/j.juro.2016.10.071. [DOI] [PubMed] [Google Scholar]
- 30. Foley JW, et al. Gene-expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. bioRxiv website. https://www.biorxiv.org/content/10.1101/207340v3 Published January 9, 2019. Accessed May 23, 2019.
- 31.Nelson PS, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99(18):11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carreira AC, Alves GG, Zambuzzi WF, Sogayar MC, Granjeiro JM. Bone Morphogenetic Proteins: structure, biological function and therapeutic applications. Arch Biochem Biophys. 2014;561:64–73. doi: 10.1016/j.abb.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18(6):1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 35.Wutzl A, et al. Bone morphogenetic proteins 5 and 6 stimulate osteoclast generation. J Biomed Mater Res A. 2006;77(1):75–83. doi: 10.1002/jbm.a.30615. [DOI] [PubMed] [Google Scholar]
- 36.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16(3):251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Hayward SW, Dahiya R, Cunha GR, Bartek J, Deshpande N, Narayan P. Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev Biol Anim. 1995;31(1):14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 38.Webber MM, et al. A human prostatic stromal myofibroblast cell line WPMY-1: a model for stromal-epithelial interactions in prostatic neoplasia. Carcinogenesis. 1999;20(7):1185–1192. doi: 10.1093/carcin/20.7.1185. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadin D, Knaus P, Mueller TD. Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine Growth Factor Rev. 2016;27:13–34. doi: 10.1016/j.cytogfr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16(1):67–76. doi: 10.1016/S1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 42.Walsh PC, Wilson JD. The induction of prostatic hypertrophy in the dog with androstanediol. J Clin Invest. 1976;57(4):1093–1097. doi: 10.1172/JCI108353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson TM, et al. Testosterone and 17β-estradiol induce glandular prostatic growth, bladder outlet obstruction, and voiding dysfunction in male mice. Endocrinology. 2012;153(11):5556–5565. doi: 10.1210/en.2012-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang ML, et al. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc Natl Acad Sci U S A. 2016;113(35):9846–9851. doi: 10.1073/pnas.1607794113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Nieves JA, Macoska JA. Prostatic fibrosis, lower urinary tract symptoms, and BPH. Nat Rev Urol. 2013;10(9):546–550. doi: 10.1038/nrurol.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noel EE, et al. Differential gene expression in the peripheral zone compared to the transition zone of the human prostate gland. Prostate Cancer Prostatic Dis. 2008;11(2):173–180. doi: 10.1038/sj.pcan.4500997. [DOI] [PubMed] [Google Scholar]
- 48.Alonso-Magdalena P, et al. A role for epithelial-mesenchymal transition in the etiology of benign prostatic hyperplasia. Proc Natl Acad Sci U S A. 2009;106(8):2859–2863. doi: 10.1073/pnas.0812666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Begley L, Monteleon C, Shah RB, Macdonald JW, Macoska JA. CXCL12 overexpression and secretion by aging fibroblasts enhance human prostate epithelial proliferation in vitro. Aging Cell. 2005;4(6):291–298. doi: 10.1111/j.1474-9726.2005.00173.x. [DOI] [PubMed] [Google Scholar]
- 50.Abrahamsson PA, di Sant’Agnese PA. Neuroendocrine cells in the human prostate gland. J Androl. 1993;14(5):307–309. [PubMed] [Google Scholar]
- 51.Cockett AT, di Sant’Agnese PA, Gopinath P, Schoen SR, Abrahamsson PA. Relationship of neuroendocrine cells of prostate and serotonin to benign prostatic hyperplasia. Urology. 1993;42(5):512–519. doi: 10.1016/0090-4295(93)90260-H. [DOI] [PubMed] [Google Scholar]
- 52.Kyoda Y, Ichihara K, Hashimoto K, Kobayashi K, Fukuta F, Masumori N. Distribution of Neuroendocrine Cells in the Transition Zone of the Prostate. Adv Urol. 2017;2017:8541697. doi: 10.1155/2017/8541697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho-Dias E, et al. Serotonin regulates prostate growth through androgen receptor modulation. Sci Rep. 2017;7(1):15428. doi: 10.1038/s41598-017-15832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Linker SB, Bedrosian K, Conneally TA, Gage FH. Opinion: how to define cell type. The Scientist website. https://www.the-scientist.com/opinion/opinion-how-to-define-cell-type-30668 Published November 1, 2017. Accessed May 23, 2019.
- 55.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darcy DG, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget. 2015;6(2):755–770. doi: 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat Rev Mol Cell Biol. 2003;4(9):679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 58.Prasad M, Butler WT, Qin C. Dentin sialophosphoprotein in biomineralization. Connect Tissue Res. 2010;51(5):404–417. doi: 10.3109/03008200903329789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 62.Saldanha AJ. Java Treeview — extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 63.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Series B Stat Methodol. 1996;58(1):267–288. [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. Cornell University website. https://arxiv.org/abs/1207.3907 Published July 17, 2012. Accessed May 23, 2019.
- 70.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq and WES data are available from the database of Genotypes and Phenotypes (phs001698.v1.p1). TMA IHC images are available from Stanford Tissue Microarray Database (https://tma.im/cgi-bin/home.pl), as array block TA-457.