Abstract
Influenza-associated mortality continues to occur annually despite available antiviral therapies. New therapies that improve host immunity could reduce influenza virus disease burden. Targeting macrophage migration inhibitory factor (MIF) has improved the outcomes of certain inflammatory diseases, but its role in influenza viral infection is unclear. Here, we showed that, during influenza viral infection, Mif-deficient mice have less inflammation, viral load, and mortality compared with WT control mice; conversely, Tg mice, overexpressing Mif in alveolar epithelial cells, had higher inflammation, viral load, and mortality. Antibody-mediated blockade of MIF in WT mice during influenza viral infection improved their survival. Mif-deficient murine lungs showed reduced levels of parkin, a mitophagy protein that negatively regulates antiviral signaling, prior to infection and augmented antiviral type I/III IFN levels in the airspaces after infection as compared with WT lungs. Additionally, in vitro assays with human lung epithelial cells showed that treatment with recombinant human MIF increased the percentage of influenza virus–infected cells. In conclusion, our study reveals that MIF impairs antiviral host immunity and increases inflammation during influenza infection and suggests that targeting MIF could be therapeutically beneficial during influenza viral infection.
Keywords: Immunology, Infectious disease
Keywords: Cytokines, Influenza, Innate immunity
During influenza viral infection, macrophage migration inhibitory factor impairs antiviral host immunity to influenza infection.
Introduction
Influenza viral infections impose the heaviest disease burden of all infectious diseases due to acute respiratory inflammation, hospitalization, and mortality (1). Although vaccination against influenza viral infection provides the best defense, vaccine efficacy for the deadliest and most common strain influenza A virus (IAV) can be less than 40% (2–4). Once infected with influenza, there are 4 antiviral drugs available to attenuate the illness; however, only oseltamivir is recommended for patients with weakened immune systems, such as the elderly and infants (5). These limitations highlight the need to identify new therapeutic targets that would reduce influenza disease burden.
The lung’s mucosal innate immune system typically creates a strong defense against IAV infection (6). Activation of the innate immune response leads to the production of inflammatory cytokines and recruitment of leukocytes that can help quell IAV infection. However, exuberant innate immune responses can induce tissue injury (6). Antiviral protein production mitigates IAV replication and reduces the susceptibility to IAV-associated mortality (7, 8). Specifically, IFNs are antiviral proteins that limit IAV infection: IFN-λ provides antiviral protection without eliciting an inflammatory response, whereas IFN-α/β increase antiviral defense and initiate inflammation (7, 9). Once inflammation is induced after infection, it must be limited to reduce tissue damage. Inflammation is controlled by inhibiting several pathways, such as the stimulator of IFN genes (STING) pathway. The STING pathway can be activated by intracellular DNA, including that from mitochondrial sources (10). The critical mitophagy protein parkin was recently shown to diminish STING-mediated inflammation and, thus, the production of type I IFNs, such as IFN-α/β; however, its contribution to IAV infection is not known (11, 12).
During infection, IAV co-opts cellular machinery for transcription, translation, autophagy, and programmed cell death (apoptosis) to aid viral replication (13–15, 16). For example, IAV usurps autophagy by blocking autophagosome maturation and modulates apoptosis to increase viral replication (16, 17). Importantly, defects of either autophagy or apoptosis decrease IAV replication efficiency (16, 18, 19). However, IAV influences are not immunologically silent, and by 24 hours of infection, IAV causes cell death, specifically necrosis of bronchiolar cells, which results in enhanced inflammation (20).
The cytokine macrophage migration inhibitory factor (MIF) regulates innate immunity and has diverse influences on inflammation (21). Many cells express MIF, including epithelial, endothelial, and most immune cells. Upon an inflammatory stimulus, bioactive MIF is rapidly released from intracellular storage pools (21, 22). Elevated MIF levels occur in several human inflammatory conditions, such as West Nile virus infection, AIDS, Parkinson’s disease, and atherosclerosis (21, 23, 24). Upon release, MIF may enhance inflammation by counteracting glucocorticoids, inhibiting apoptosis, activating macrophages, enhancing autophagy, and stimulating the NLRP3 inflammasome (21, 22, 25–27). The 5′ promoter of the MIF gene contains a microsatellite repeat (CATT)5–8 that regulates transcription, with copy number directly related to enhanced MIF expression (28). High MIF levels are detrimental in some viral infections, such as dengue virus, Ross River virus, and West Nile virus (29–31). It was previously shown that IAV-infected human lung epithelial cells in vitro displayed increased MIF release without a concomitant increase in MIF transcription (20). Nevertheless, the role of MIF during IAV infection and its potential as a therapeutic target are unclear.
Results
IAV infection promotes MIF protein release into the airspace.
To evaluate MIF release during IAV infection in vivo, we infected mice with IAV strain PR8 and measured MIF levels in bronchoalveolar lavage (BAL) at 0, 3, and 6 days after infection (DPI). Throughout our study, we utilized mice lacking the Mif gene (Mif–/–), WT mice (Mif+/+), and Tg mice overexpressing Mif in alveolar epithelial cells (AECs) (Mif-lung-tg; ref. 32). The latter mice mimic the approximate 2-fold increase in Mif transcription observed in high MIF expressor genotypes (32). At all DPIs, Mif–/– samples contained no measurable MIF, as expected, and Mif-lung-tg BAL contained at least 2-fold more MIF relative to Mif+/+ BAL (Figure 1A). MIF protein levels in BAL increased from 0 DPI to 3 DPI, for both WT and Mif-lung-tg mice (Figure 1A), further supporting that IAV induces MIF release.
Figure 1. MIF increases in the airspace and enhances morbidity and mortality after infection with IAV.
MIF-knockout (Mif–/–, white circles), WT (Mif+/+, gray circles), and Mif-overexpressing Tg (Mif-lung-tg, pink circles) C57BL/6 background mice were challenged i.n. with 5 × 104 PFU H1N1 PR8. (A) MIF in the BAL measured by ELISA. The mice were monitored daily for (B) survival, (C) body weight, and (D) clinical score. (E) Survival curve of WT mice injected i.p. with anti-MIF antibody (black squares) or mouse IgG (black circles) on days labeled with blue arrows and infected with 5 × 104 PFU H1N1 PR8 at 0 DPI. Statistical differences were determined by (A) multiple t tests adjusted for multiple comparisons by Holm-Sidak test, (B and E) Gehan-Breslow-Wilcoxon test, or (C and D) 2-way ANOVA followed by Tukey multiple comparisons test. The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartile range, with each symbol representing a biological replicate (A). The data are displayed as mean ± SEM (C and D). There was no significant difference in survival rate between WT control groups in survival studies for Mif–/– or Mif-lung-tg mice; a representative group is shown in B–D for clarity. Data are representative of 2 independent experiments, with n = 4–5/group (A), 15–18 mice/group at time of infection (B–D), or (E) 8–10 mice/group at time of first injection. For only C and D, significance is denoted between Mif+/+ and Mif-lung-tg with a red asterisk; whereas, significant differences between Mif–/– and Mif+/+ were denoted with a black asterisk. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
MIF enhances morbidity and mortality after infection.
Next, we determined the influence of MIF on survival during IAV infection. Mif–/– mice infected with IAV exhibited the highest survival (93%), followed by Mif+/+ control mice (42%); finally Mif-lung-tg mice showed the lowest survival (18%; Figure 1B). Additionally, morbidity, as measured by body weight and clinical score, also showed an inverse correlation with Mif expression, with Mif–/– mice having the lowest morbidity and Mif-lung-tg mice having the highest (Figure 1, C and D). Finally, Mif+/+ mice treated with an anti-MIF-neutralizing antibody after IAV infection showed 63% survival, whereas Mif+/+ mice similarly treated with IgG control showed 41% survival (Figure 1E). Overall, these data indicate that MIF worsens morbidity and mortality during IAV infection and that MIF blockade during IAV infection improves survival.
MIF increases lung inflammation in airspaces and lung permeability during IAV infection.
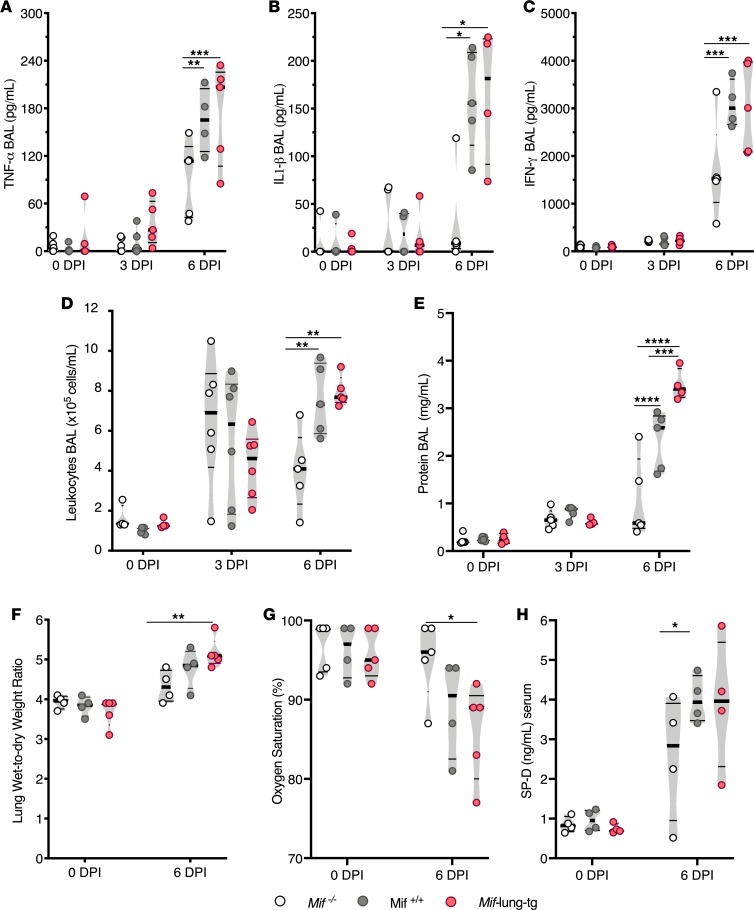
We examined the role of MIF on the innate immune response during IAV infection by measuring inflammatory cytokines, leukocyte infiltration in the airspace, and additional markers of lung permeability. At 6 DPI, Mif–/– BAL showed significantly lower TNF-α (1.3-fold), IL-1β (10-fold), and IFN-γ (2-fold) compared with Mif+/+ BAL; in addition, Mif-lung-tg mice had higher or similar levels of inflammatory cytokines compared with those of Mif+/+ mice (Figure 2, A–C). Similarly, leukocyte infiltration into the airspace was reduced approximately 2-fold in Mif–/– mice compared with MIF+/+ and Mif-lung-tg mice at 6 DPI (Figure 2D). At 6 DPI, there were significantly more macrophages and monocytes in the lungs of Mif-lung-tg and WT mice compared with Mif–/– mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128034DS1). We also assessed tissue injury by measuring lung permeability markers (i.e., total protein in BAL and lung wet-to-dry ratio) and circulating biomarkers (i.e., surfactant protein D and oxygen saturation). At 6 DPI, on average Mif–/– mice had at least 5-fold lower total protein levels in BAL compared with WT mice and up to 7-fold lower levels compared with Mif-lung-tg mice (Figure 2E) and 20% less lung edema compared with Mif-lung-tg mice (Figure 2F). Mif–/– mice also exhibited increased peripheral oxygen saturation (Figure 2G) and less circulating surfactant protein D (Figure 2H) than Mif-expressing mice at 6 DPI. Overall, these data indicate that elevated Mif expression correlates with enhanced IAV-induced lung inflammation and lung injury at 6 DPI.
Figure 2. Higher MIF promotes inflammation and lung permeability during IAV infection.
(A) TNF-α, (B) IL-1β, and (C) IFN-γ levels in BAL, as measure by ELISA. (D) Total leukocytes in BAL. (E) Total protein concentration in BAL. (F) Wet/dry weight ratios of lungs. (G) Arterial oxygen saturation determined by pulse oximetry. (H) Serum concentrations of surfactant protein D (SP-D). Statistical differences were determined by 2-way ANOVA followed by Tukey multiple comparisons test. The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartilerange, with each symbol representing a biological replicate. Data are representative of samples from 2 independent experiments, with n = 4–6 biological replicates/group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Mif–/– mice have lower viral load and higher antiviral protein levels in lung during IAV infection.
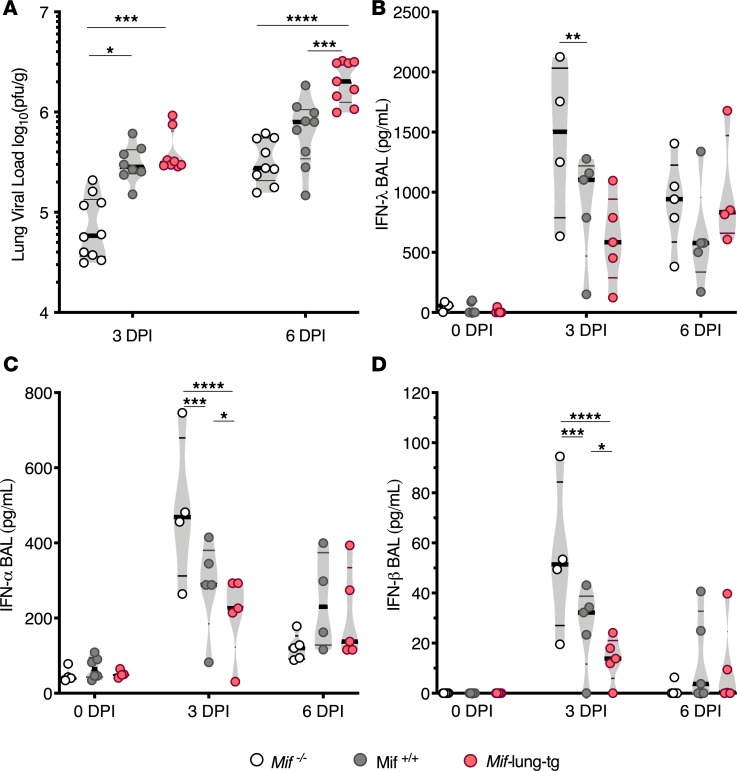
We quantified live virus in the lungs and the levels antiviral IFN proteins in the airspaces of Mif–/–, Mif+/+, and Mif-lung-tg mice after infection. At 3 DPI, Mif–/– lungs contained up to 5-fold less IAV PFU compared with Mif-expressing mice (Figure 3A). By 6 DPI, Mif-lung-tg lungs had the highest viral load compared with cohorts with lower Mif expression; however, the differences in viral load had narrowed between Mif–/– mice and Mif+/+ mice (Figure 3A). Interestingly, at 3 DPI, the BAL from Mif–/– mice had significantly higher levels of IFN-λ (up to 3-fold), IFN-α (up to 2-fold), and IFN-β (up to 5-fold) compared with BAL from Mif-lung-tg mice (Figure 3, B–D). IFN-α and IFN-β levels in BAL were significantly different between all 3 cohorts at 3 DPI (Figure 3, B–D). Thus, MIF levels inversely correlate with the levels of these antiviral proteins at 3 DPI. Together, these data indicate that increasing MIF levels attenuate antiviral cytokine levels and enhance viral titers during the early stages of IAV infection.
Figure 3. Mif expression correlates with higher viral load after infection and lower levels of IFNs.
(A) Quantification of viral load in lungs of mice at 3 and 6 DPI. IFN proteins in BAL by ELISA: (B) IFN-λ, (C) IFN-α, and (D) IFN-β. Statistical differences were determined by 2-way or 1-way ANOVA followed by Tukey multiple comparisons test. The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartilerange, with each symbol representing a biological replicate. Data are representative of pooled samples from 2 independent experiments, with total n = 7–8 biological replicates/group (A), or representative of 2 independent experiments, with n = 4–5 biological replicates/group (B–D). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Attenuated Mif expression results in less parkin in lungs prior to IAV infection.
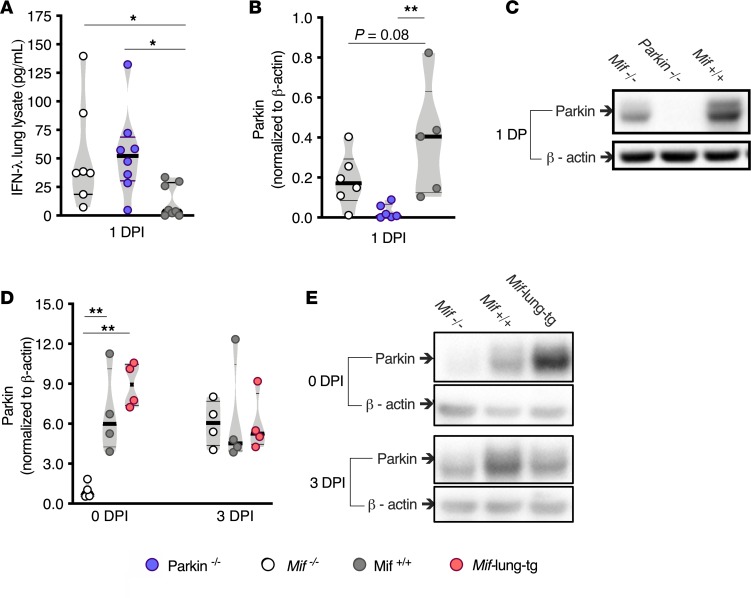
Previous work suggested that parkin is a negative regulator of the IFN pathway (10, 11). To examine the role of parkin in IFN production during IAV infection, we infected Mif+/+ and Parkin–/– mice with IAV and measured IFN-λ, which is released earlier and at higher concentrations than the other IFNs (8, 33). At 1 DPI, IFN-λ levels were more than 5-fold higher in lung lysates from Parkin–/– mice than in those from Mif+/+ mice (Figure 4A), consistent with negative regulation of antiviral signaling. Interestingly, lung levels of IFN-λ were similar between Parkin–/– mice and Mif–/– mice at 1 DPI (Figure 4A), suggesting that MIF might suppress IFN-λ via positive regulation of parkin. Indeed, we found that parkin levels directly correlated with MIF levels prior to infection: compared with Mif–/– mice, the parkin levels were 7.0-fold higher in Mif+/+ mice and 9-fold higher in Mif-lung-tg mice (Figure 4, D and E). However, by 1 DPI, MIF–/– mice exhibited increased parkin levels that were not significantly different compared with those of Mif+/+ mice (Figure 4, B and C). The correlation between MIF and parkin remained nonsignificant at 3 DPI (Figure 4, D and E). As parkin may negatively regulate antiviral cytokine production by degrading tumor necrosis factor receptor–associated factor 3 (TRAF3) in HEK293 cells in vitro, we measured TRAF3 in the lung lysates of the different experimental groups. However, we did not find differences between the experimental groups (Supplemental Figure 2 and ref. 11). Overall, these data suggest that the presence of MIF positively regulates parkin before infection, which in turn correlates with a diminished antiviral IFN response.
Figure 4. Mif expression correlates with higher parkin levels in lung lysate preinfection.
Lung lysate parkin/β-actin protein measured by Western blot at baseline and 3 DPI: (A) IFN-λ levels in 1 DPI lung lysate. Lung lysate protein measured by Western blot at 1 DPI: (B) parkin normalized to β-actin. (C) Representative image of blot. (D) Violin plot of parkin normalized to β-actin. (E) A representative image of blot. Statistical differences were determined by 1-way ANOVA followed by Tukey multiple comparisons test (D) or 1-way ANOVA comparing the mean of each group to Mif+/+ data followed by Holm-Sidak test for multiple comparisons (A and B). The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartilerange, with each symbol representing a biological replicate (A, B, and D). Data are representative of 2 independent experiments, with n = 4–5 biological replicates/group (D), or pooled samples from 5–8 biological replicates/group (A and B). *P < 0.05, **P < 0.01.
MIF release and viral load are similar to those of WT when Parkin is attenuated in AECs.
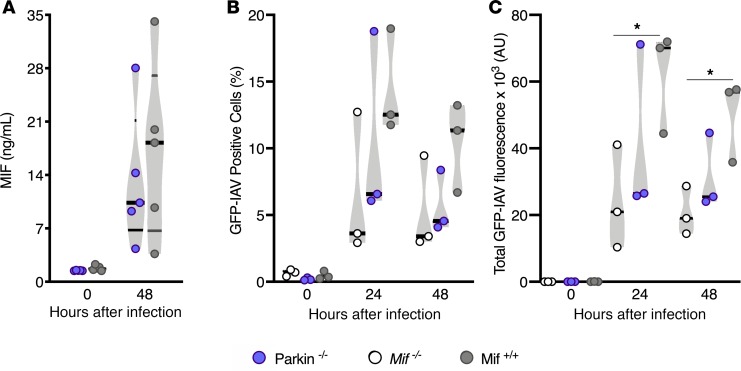
To examine the role of parkin in MIF release and influenza viral replication; we isolated AECs from Parkin–/–, WT, and Mif–/– mice. We chose AECs, because epithelial cells represent the first line of defense to influenza viral infection within the lung; additionally, Mif-lung-tg mice, which overexpress MIF in AECs had exhibited enhanced mortality (Figure 1) and viral load (Figure 3 and ref. 34). Hence, we cultured primary AECs from WT and Parkin–/– mice by either mock infection or IAV infection. We infected AECs and measured MIF in the cell culture supernatant at 48 hours. We found that release of MIF from infected cells was independent of parkin (Figure 5A). Consistent with parkin’s prior regulatory role in IFNs, we found that IFN-β production during infection with IAV PR8 was 4-fold higher in the Parkin–/– AECs compared with that in WT cells (Supplemental Figure 3). We next infected AECs from Parkin–/–, WT, and Mif–/– mice with a fluorescent reporter virus that expresses an mNeon green fluorescent reporter protein (GFP-IAV). We found no difference in the percentage of virally infected cells among the experimental groups during the course of infection (Figure 5B); however, the total fluorescence intensity was 3-fold lower in Mif–/– biological replicates compared with WT replicates at 24 hours and 48 hours (Figure 5C). This suggests that although the total proportion of cells infected was not different, the viral replication was lower in Mif–/– biological replicates. Parkin–/– AECs exhibited an intermediate total fluorescence compared with either WT or Mif–/– AECs, which was not significant between either group (Figure 5). This suggests that although parkin negatively regulates antiviral cytokines, it is not required for viral control in AECs, suggesting either that MIF may regulate viral control via parkin-independent mechanisms or that deficiency in parkin affects viral control by additional mechanisms independent of regulation of antiviral cytokines. To further elucidate mechanisms by which MIF may enhance viral replication, we continued our work in the human Calu-3 cell model.
Figure 5. MIF levels independent of parkin during IAV infection in alveolar epithelial cells.
Measurements from alveolar epithelial cells (AECs) at 24 or 48 hours after either PR8 IAV MOI 1 infection or mock infection: (A) MIF released from AECs, (B) green fluorescent protein IAV–positive (GFP-IAV–positive) cells, and (C) total GFP-IAV fluorescence intensity. Statistical differences were determined by unpaired 2-tailed Student’s t test (A–C). The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartilerange, with each symbol representing a biological replicate (A–C). Data are representative of pooled data, with n = 4 biological replicates/group (A) or 3 biological replicates/group (B and C). *P < 0.05.
MIF promotes greater spread of infection in human Calu-3 cells.
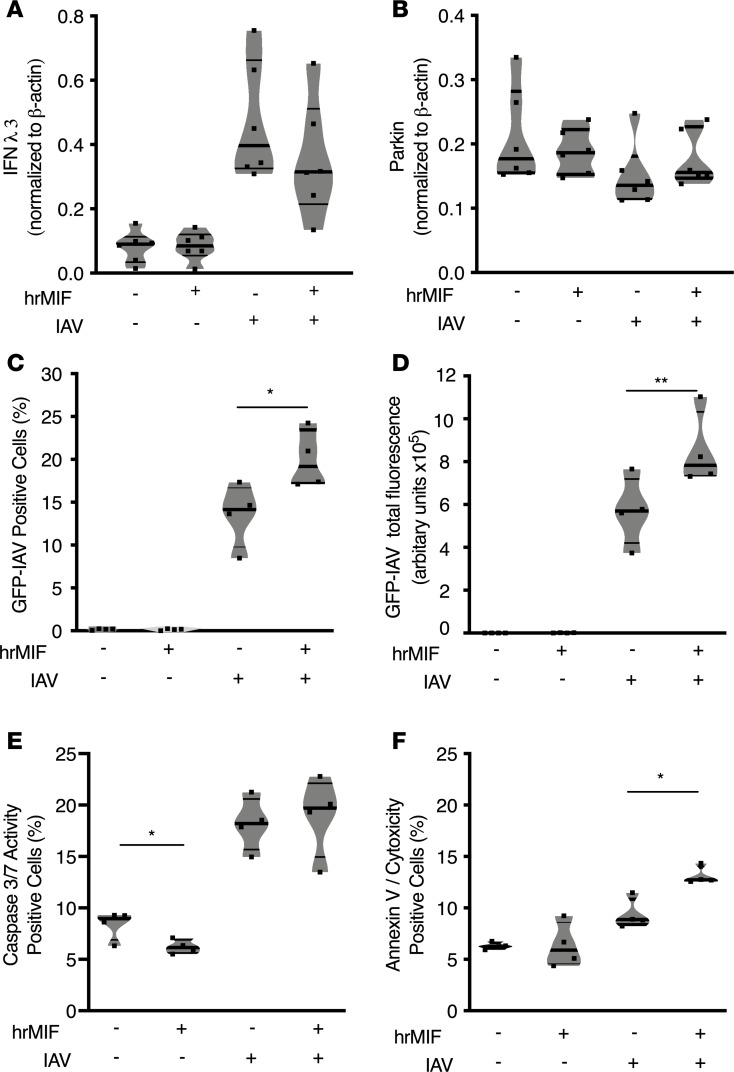
We employed Calu-3 human epithelial cells to examine the effect of MIF on viral replication and apoptosis at the cellular level during IAV infection in vitro. We found that infected Calu-3 cells released 3-fold more MIF compared with uninfected cells, in agreement with a previous study (Supplemental Figure 4 and ref. 20). We repeated the experiment with GFP-IAV or mock infection, including groups of Calu-3 cells treated with human recombinant MIF (hrMIF). We continued hrMIF treatment throughout the course of infection. We determined that hrMIF treatment did not have an influence on IFN-λ3 (closest subtype to mouse IFN-λ) release either before or after infection; however, IAV infection did increase IFN-λ3 levels 3- to 4-fold compared with mock-infected samples (Figure 6A). Parkin levels remained similar even after infection, despite treatment with hrMIF during the course of infection (Figure 6B).
Figure 6. MIF promotes spread of IAV infection in Calu-3 cells.
Measurements from Calu-3 cells at 24 hours after either PR8 IAV MOI 1 infection or mock infection: (A) IFN-λ3 in cell lysate by Western blot, (B) parkin in cell lysate by Western blot, (C) green fluorescent protein IAV–positive (GFP-IAV–positive) cells, (D) total GFP-IAV fluorescence, (E) cellular apoptosis initiation measured by caspase-3/7 activity, and (F) cells displaying necrotic/late apoptotic features, as measured by double positivity with annexin V and cytotoxicity dyes. Statistical differences were determined by unpaired 2-tailed Student’s t test (A–C). The violin plots are closed curves representing data distribution and encapsulate the median, range, and interquartilerange, with each symbol representing a biological replicate. Data are representative of at least 2 independent experiments with n = 6 biological replicates/group (A and B) or n = 4 biological replicates/group (C–F). *P < 0.05, **P < 0.01. Statistical differences were determined by unpaired 2-tailed Student’s t test.
We found that hrMIF treatment led to double the total of GFP-IAV–infected cells and total GFP-IAV per well by 24 hours (Figure 6, C and D), suggesting that MIF creates a permissive environment for both infection and viral replication. Next, we examined if MIF affected apoptosis in Calu-3 cells before and after IAV infection, as MIF is known to reduce apoptosis, which could affect viral replication (35–37). We used caspase-3/7 activity to determine apoptosis initiation in cells. Without infection, MIF reduced the frequency of caspase-3/7 active cells in agreement with prior studies showing an antiapoptotic effect of MIF (Figure 6E and refs. 35–37). IAV infection increased caspase-3/7 activity, which was not reduced by MIF, by 24 hours after infection (Figure 6E). Interestingly, we found that a higher frequency of MIF-treated cells exhibited necrotic or late apoptotic features at 24 hours after infection, based on annexin V and cytotoxic positivity (Figure 6F). This suggests that MIF could be enhancing inflammation during infection by necrosis, as it is known that necrosis or failed clearance of apoptotic cells enhances inflammation (20). Overall, MIF promotes the spread of influenza virus infection among human Calu-3 cells. Additionally, MIF may enhance necrosis to increase inflammation during IAV.
Discussion
Excessive IAV-associated inflammation contributes to morbidity and mortality (39). Our results indicate that MIF initially decreases antiviral IFN levels, which could contribute to the enhanced viral load and elevated IAV-associated mortality, morbidity, inflammation, and lung injury. We found that the preinfection levels of parkin, a mitophagy protein that negatively regulates antiviral signaling, correlated with Mif expression in the lung. However, by 3 DPI, parkin levels were similar in Mif-deficient and Mif-expressing mice (11, 12). Subsequently, Mif–/– mice and Parkin–/– mice had augmented antiviral type I/III IFN levels during influenza infection. In primary AEC cultures, Mif–/– cells had reduced viral load compared with WT cells, but Parkin–/– cells exhibited an intermediate response, which was not significantly different from that of WT cells. Our results suggest that MIF enhancement of viral replication could occur independently of parkin. An alternative explanation comes from our Calu-3 model, in which we found that MIF increases cellular necrosis or delayed clearance of apoptotic cells during late stages of IAV infection and enhances viral replication. This may explain MIF’s ability to enhance viral replication, as cells exhibiting increased late apoptotic and necrotic characteristics during late stages of infection have been a marker of enhanced IAV propagation (38, 40).
How MIF and parkin interact is not clear from our study and will require future investigation. Treatment of Calu-3 cells with recombinant MIF during the course of infection did not increase parkin levels, which indicates that exogenous MIF does not directly increase parkin. Although MIF deficiency led to increased parkin levels in the lung prior to infection, it is possible that MIF deficiency impacts parkin levels differently compared with exogenous treatment with recombinant MIF protein. Parkin is a mitophagy protein and can identify damaged mitochondria that require removal by the mitophagy machinery (41). Parkin’s role on mitophagy may be independent of its negative regulatory role on the type I and III IFN production. Hence, parkin likely has pleiotropic functions during IAV infection. Clearly future studies will be needed to investigate the role of parkin and other components of the mitophagy machinery, including PINK1, in antiviral immunity to IAV.
Our in vitro results indicate that during IAV infection MIF enhances inflammation. Necrosis or delayed clearance of apoptotic epithelial cells could promote inflammation either by direct release of intracellular cytokines or inflammatory mediators that trigger inflammation responses by surrounding cells (42). Such a process could explain the increase in the inflammatory cytokines, e.g., IL-1β and TNF-α noted in the WT mice of our study during IAV. The heightened inflammatory response likely increases tissue damage, noted by the elevated protein levels (Figure 2E), increased lung edema (Figure 2F), and decreased tissue oxygenation (Figure 2G). Future studies will be required to determine which inflammatory cytokines are critical for immune pathology induced by MIF during IAV.
Blockade of MIF in WT mice improved survival during IAV infection, implying that MIF neutralization could mitigate IAV-associated morbidity and mortality in humans. The present results also prompt an investigation into the role of the polymorphic MIF locus in human influenza infection outcomes. Indeed, variant MIF alleles have been shown to influence clinical outcomes in invasive pneumococcus and community-acquired pneumonia, which often develop as secondary infections after influenza virus (43, 44). Our study reveals that inhibiting MIF could represent a potential therapeutic target for reducing IAV-associated disease severity and mortality. In conclusion, our study has revealed what we believe to be a novel role for MIF in promoting immune pathology, viral replication, and mortality during IAV. Targeting MIF could improve outcomes to active influenza infection, particularly in susceptible populations.
Methods
Mice and cell cultures
In animal studies, the number of replicates and the time between repeated experiments were based on mouse availability and best standards of practice in order to minimize the number of total mice used during the study. All animals used for the study were male mice between 2 and 4 months of age and of pure C57BL/6 background. Mif–/– and Mif-lung-tg (32, 45) mice that were both on a pure C57BL/6 background were used for the study. The Mif-lung-tg mice overexpress Mif in alveolar type II epithelial cells; specifically, these mice had a 0.4-Kb DNA fragment with mouse Mif cDNA inserted into the expression plasmid under control of a CC10 promoter as previously reported (46). We noted that there was no statistical difference in survival nor morbidity between Mif-lung-tg littermate controls versus Mif–/– littermate controls. C57BL/6 Parkin–/– mice were ordered from The Jackson Laboratory (stock no. 006582). The Calu-3 HTB-55 cell line was purchased from ATCC. The cells were cultured in 5% CO2 at 37°C in Ham’s F12 Nutrient Mixture (Gibco), 100 U/mL penicillin/streptomycin, and 10% fetal calf serum (cell culture media).
Study design
The study objective was to determine the role and mechanistic implications of MIF protein during IAV infection in mouse models and the human cell line Calu-3. Mif expression in mice or recombinant MIF in vitro was varied to determine the influence of MIF during IAV infection. The study utilized mouse body measurement (clinical score/weight/oxygen saturation), whole lung lysate, BAL, and serum to conduct experiments. The study was conducted in a controlled laboratory setting using established immunological techniques. Initial sample size was determined through prior experience dependent on type of experiment and practical feasibility. Each experiment was repeated at minimum twice.
Based on prior experience, the survival experiments were conducted over a 15-day period. No biological samples were harvested from mice involved in the survival studies. Survival studies using Mif-lung-tg and littermate controls were conducted by a blinded investigator. There was no significant difference in survival rate between WT control groups in survival studies; thus, in the figures, only 1 set of Mif+/+ results were shown for clarity. Animals were assigned numerical identification by an individual uninvolved with genotyping, and genotypes were not revealed until completion of the survival experiment.
For tissue samples, each time point represents separate sets of animals, and time points were acquired a 1, 3, and 6 DPI. The animals sacrificed to harvest BAL fluid were probed using multiple cytokine ELISAs. A separate set of animals was sacrificed to harvest lung lysate and serum for use viral plaque assay, with Thermo Fisher Scientific IFN-λ ELISA (catalog BMS6028) at 1 DPI and Western blots.
Influenza infection of mice
Mice were anesthetized with isoflurane and instilled with 30 μL 5E4 PFU PR8 virus or 30 μL PBS control by i.n. route. H1N1 Influenza A/PR/8/34 virus was purchased from Advanced Biotechnologies Inc. Following infection, mice were monitored daily for weight change, clinical scores, and mortality. Clinical scores were determined by monitoring the mouse for 5 seconds and assigning a clinical score category according to the symptoms exhibited by the mouse as follows: 0, normal; 1, slight piloerection/orbital tightening/ear position forward; 2, moderate piloerection/orbital tightening/ear position back; 3, severe piloerection/orbital tightening/ear position back; 4, hunched back and all indicators in category 3; and 5, dead. Mice were euthanized after loss of 30% of their individual preinfection weight and then recorded as dead in survival data and censored from further weight loss and clinical scoring data. The mice utilized in the study were healthy and lacking any other co-morbidity. Anti-MIF mAb IIID.9 (IgG1), from the laboratory of Richard Bucala, was administered i.p. at 20 mg/kg at (time of dosing; ref. 47).
Tissue collection and preparation
Mice were euthanized with isofluorane immediately prior to tissue harvest.
Blood/serum.
Blood was collected by a terminal bleed via orbital sinus (48). The blood was collected in a 1.6-mL centrifuge tube and allowed to clot for 20 minutes on ice. The clotted blood was centrifuged at 5,000 g for 10 minutes, and the top serum layer was stored at –80°C.
BAL.
The mouse neck was wetted with 70% ethanol, and the trachea was exposed and cannulated using a 2.0-cm-long blunted 18-gauge catheter. The BAL was obtained by lavaging the lungs twice with 1.0 mL cold sterile 1× PBS. BAL was centrifuged at 400 g for 8 minutes and supernatant was stored at –80°C.
Lung dissection.
Lungs were excised, weighed, and then put on ice. For viral plaque assay and ELISA, lungs were placed in 1.0 mL Hank’s Balanced Salt Solution with 0.1% BSA with 2 stainless steel beads, homogenized in TissueLyser II (Qiagen) for 5.0 minutes set at 30 Hz, and centrifuged at 400 g for 10 minutes; supernatants were stored at –80°C.
Influenza viral load measurement
Lung viral titers were determined by plaque assay using Madin-Darby canine kidney (MDCK) cells (provided by Adam Lauring, University of Michigan). MDCK cells were cultured in DMEM medium at 37°C with 10% CO2 in 6-well plates (4 × 105 cells/well) for 20–24 hours until the cell monolayer reached 80%–90% confluency. Lung lysate supernatants were thawed and 10-fold dilutions (10–2 to 10–4) were made with 0.1% BSA in 1× PBS. MDCK cells were washed twice with 1× PBS followed by 1.0-hour incubation with 200 μL of the 10-fold dilutions of lung lysate supernatants at 37°C with gentle tapping every 15 minutes. Cells were washed twice with 1× PBS. The cells were overlaid with 2.0 ml of a medium containing a mixture of 1.0% agarose, DMEM, and 2.0 μg/mL acetylated trypsin. Plates were then incubated at 37°C for 72 hours. Next, the wells were filled with 70% ethanol for 15 minutes, the agarose mixture was removed from the plates, and the cells stained with 0.3% crystal violet. The plates were rinsed with tap water and viral plaques were counted.
Oxygen saturation
Mice were lightly anesthetized with isoflurane and placed prone on a warm pad. The Kent Scientific mouseSTAT Jr. pulse oximeter with connected mouse paw sensor was placed on the rear right paw and adjusted until the instrument displayed a stable heart rate and oxygen saturation reading for recording.
Wet-to-dry weight measurement and analysis
Lungs were weighed immediately after dissection and then dried in presence of desiccant for 14 days and weighed again. The wet-to-dry ratio is the lung weight immediately after dissection of the lung divided by the weight of the lung dried after dessication.
Flow cytometry
BAL were harvested from animals, and the cells were quantified in a hemacytometer. Cells were then centrifuged for 5 minutes at 400 relative centrifugal force and then the supernatant was poured off. The pellets were resuspended with 1.0 mL of ice cold 1× PBS containing 1:100 Zombie UV fixable viability dye (Biolegend, catalog 423108) and left to incubate for 30 minutes. The cells were again centrifuged, the supernatant was poured off, and the cell pellet was resuspended with 1:50 FCγR blocker (Biolegend) in 100 μL flow cytometry buffer (FACS buffer), 1× PBS containing 2% FCS. Cells were incubated for 15 minutes and then an additional 100 μL FACS buffer with 1:50 CD45 antibody conjugated to APC-EF780, from eBioscience, for another 25 minutes on ice. The cells were then centrifuged and washed with FACS buffer. For all samples, cells were fixed with 2.0% paraformaldehyde in 1× PBS before analysis. Flow cytometry was acquired with a Moflo Astrios, and resulting data were analyzed with FlowJo software. The following antibodies were used in this study: CD326 (PerCP-Vio700, Miltenyi Biotec, catalog 130-102-720), CD24 (BV421, Biolegend, catalog 101826), Ly6C (FITC, Biolegend, catalog 128006), CD11b (BV510, Biolegend, catalog 101263), MHC II (PerCP-EF710, eBioscience/Thermo Fisher Scientific, catalog 46-5321-82), Siglec F (PE, BD Biosciences, catalog 552126 ), CD64 (PE-Cy7, Biolegend, catalog 139314), CD11c (EF615, eBioscience/Thermo Fisher Scientific), Ly6G (APC, Biolegend, catalog 127614), and CD45 (APC-EF780, eBioscience/Thermo Fisher Scientific, catalog 47-0451-82). The comprehensive innate immune cell panel gating strategy was utilized as described in a prior publication (49).
IncuCyte
Data acquisition and analysis.
Cells were plated onto 96-well tissue culture-treated plates and grown to a confluence of 80%–90% before beginning the assay. Cells were infected with IAV MOI 1 or mock-infected with 1× Hank’s balanced salt solution for 60 minutes at 37°C. The cells were infected with GFP-IAV to assess viral replication. For viral replication experiments, we employed GFP-IAV, which was donated by Peter Palese (Icahn School of Medicine at Mount Sinai, New York, New York, USA), at 85.6% efficiency. Cells were washed once with cell culture media containing only 2% FCS with indicated treatments, such as 100 ng/mL hrMIF or 1× PBS (vehicle only). After infection, at least 4 representative images were collected per well, with an 20× objective at 3-hour intervals for at least 24 hours. After data acquisition, data were analyzed with IncuCyte Zoom system, and data were exported to Graphpad prism for statistical analysis and graphical display.
Reagents for the caspase-3/7 activity (catalog 4440), cytotoxicity (catalog 4633), and annexin V assays (catalog 4641) were purchased from IncuCyte, and experiments were conducted according to the manufacturer’s specifications.
Protein assays and immunoblotting
BAL samples were diluted 1:6 for measurement in all ELISAs or BCA assays.
ELISA.
The ELISA kits were purchased as follows: TNF-α (Invitrogen, catalog BMS607-3), IL-1β (R&D Systems, catalog MLB00C), SP-D (R&D Systems, catalog MsfPD0), IFN-γ (R&D Systems, catalog DY485), IFN-α (PBL assay science, catalog 42120-1), IFN-β (PBL assay science, catalog 42410-1), IL-28 (Invitrogen, catalog 50-246-674), and MIF (R&D Systems DY1978). Assays were then conducted according to each manufacturer’s specifications.
Immunoblotting.
For immunoblotting experiments, lungs were placed in 0.5 mL RIPA buffer containing 1% protease inhibitor and 1% phosphatase inhibitor, homogenized for 30 seconds, and centrifuged at 20,000 g for 15 minutes, and supernatant was stored at –80°C. Reduced tissue lysates were prepared using LDS reagent (Thermo Fisher Scientific) and reducing agent (Thermo Fisher Scientific) and were electrophoresed on 4%–12% gradient SDS-polyacrylamide gels and transferred to 0.20-μm polyvinylidene fluoride membranes (Thermo Fisher Scientific, catalog IB401001). Blots were blocked in PBS with 0.1% Tween-20 and 5% BSA at room temperature for 2 hours or 4oC overnight. Membranes were then incubated for 1 hour at room temperature with primary antibodies against parkin (Abcam, catalog ab77924; 1:500), IFN-λ3 (R&D Systems, catalog MAB5259), β-actin (Abcam, catalog ab8226; 1:1000). After washing in PBS plus 0.1% Tween-20, membranes were incubated in secondary antibody for 30 minutes (Abcam, catalog ab205719; 1:5000–1:10000) and then illuminated with chemiluminescent substrate (Thermo Fisher Scientific, catalog 34577) using a Bio-Rad ChemiDoc.
Pierce BCA protein assay kit.
The kit was utilized according to the manufacturer’s (Thermo Fisher Scientific) specifications.
AEC isolation and culture
Lungs of mice were harvested and processed as described previously (50). Single-cell suspensions of cells were processed using an EasySep Epithelial Cell Enrichment Kit (Stemcell Technologies, catalog 19758) in order to isolate AECs. We typically obtained approximately 80% purity of CD326+ cells as measured by flow cytometry when compared with single-cell lung suspensions. The total cells were counted and resuspended at a concentration of 1 million cells/ml in F12-Ham medium containing 10% FCS and allowed to adhere to tissue culture plates containing a Matrigel overlay for 48 hours followed by the infection, which follows the protocol detailed in the above IncuCyte section. In a separate isolation and infection, cell culture supernatants were collected 48 hours following infection following the protocol detailed in the above detailed IncuCyte section and were stored at −80 °C until analysis.
Statistics
Results are displayed as violin plots with curves encompassing the distribution of the data, with the following exceptions: survival studies are represented as Kaplan-Meier curves, weight and clinical scores are represented as line graphs displaying mean ± SEM for each phenotype per day post-infection. Kaplan-Meier curves were compared using Gehan-Breslow-Wilcoxon test. Assessment of normality was calculated with the Shapiro-Wilk test. Statistical comparisons were performed using 1- or 2-way ANOVA with Tukey’s post-hoc test or 2-tailed Student’s t test for unpaired samples with Holm-Sidak test for multiple comparisons where necessary. A value of P < 0.05 was considered significant. Data were graphed by Graphpad Prism 8.0. The number of mice tested are indicated in each figure.
Study approval
Animal work was conducted according to protocols approved by the University of Michigan’s Institutional Animal Care and Use Committee and adhered to the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
CAS, RB, RLZ, and DRG designed the research studies. CAS, DJT, UAK, and SW conducted experiments and acquired data. CAS, DJT, and UAK analyzed the data. CAS wrote the first draft and prepared the figures. DRG edited this version, and all authors were subsequently given the manuscript for input. LL developed Tg mice and the anti-MIF antibody. RB and DRG procured funding for the project.
Supplementary Material
Acknowledgments
This work was funded by NIH grants R01-HL130669 (to DG and RB), R01-AG028082 (to DG), and T32-HL007853 (to CS).
Version 1. 07/11/2019
Electronic publication
Footnotes
Conflict of interest: RB is a coinventor on patents describing the therapeutic utility of anti-MIF (US9139877B2) and MIF antagonists (US9221903B2) and has received royalty payments in the past.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(13):e128034. https://doi.org/10.1172/jci.insight.128034.
Contributor Information
Lin Leng, Email: lin.leng@yale.edu.
Rachel L. Zemans, Email: zemansr@med.umich.edu.
Richard Bucala, Email: richard.bucala@yale.edu.
Daniel R. Goldstein, Email: drgoldst@med.umich.edu.
References
- 1.Cassini A, et al. Impact of infectious diseases on population health using incidence-based disability-adjusted life years (DALYs): results from the Burden of Communicable Diseases in Europe study, European Union and European Economic Area countries, 2009 to 2013. Euro Surveill. 2018;23(16):1700454. doi: 10.2807/1560-7917.ES.2018.23.16.17-00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downey J, Pernet E, Coulombe F, Divangahi M. Dissecting host cell death programs in the pathogenesis of influenza. Microbes Infect. 2018;20(9-10):560–569. doi: 10.1016/j.micinf.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang PO, Mendes A, Socquet J, Assir N, Govind S, Aspinall R. Effectiveness of influenza vaccine in aging and older adults: comprehensive analysis of the evidence. Clin Interv Aging. 2012;7:55–64. doi: 10.2147/CIA.S25215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flannery B, Chung J, Ferdinands J. Preliminary Estimates of 2017–18 Seasonal Influenza Vaccine Effectiveness against Laboratory-Confirmed Influenza from the US Flu VE and HAIVEN Networks. Center for Disease Control and Prevention. https://www.cdc.gov/vaccines/acip/meetings/downloads/slides-2018-06/flu-02-Flannery-508.pdf Published June 20, 2018. Accessed June 25, 2019.
- 5. [No authors listed]. What You Should Know About Flu Antiviral Drugs. Center for Disease Control and Prevention. https://www.cdc.gov/flu/treatment/whatyoushould.htm Accessed June 25, 2019.
- 6.Banka TR, Ast MP, Parks ML. Early intraprosthetic dislocation in a revision dual-mobility hip prosthesis. Orthopedics. 2014;37(4):e395–e397. doi: 10.3928/01477447-20140401-63. [DOI] [PubMed] [Google Scholar]
- 7.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15(2):87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galani IE, et al. Interferon-λ Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 2017;46(5):875–890.e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 9.Stifter SA, et al. Functional Interplay between Type I and II Interferons Is Essential to Limit Influenza A Virus-Induced Tissue Inflammation. PLoS Pathog. 2016;12(1):e1005378. doi: 10.1371/journal.ppat.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paludan SR, Reinert LS, Hornung V. DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer. Nat Rev Immunol. 2019;19(3):141–153. doi: 10.1038/s41577-018-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin D, Gu H, Liu E, Sun Q. Parkin negatively regulates the antiviral signaling pathway by targeting TRAF3 for degradation. J Biol Chem. 2018;293(31):11996–12010. doi: 10.1074/jbc.RA117.001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sliter DA, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561(7722):258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ludwig S, Pleschka S, Planz O, Wolff T. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell Microbiol. 2006;8(3):375–386. doi: 10.1111/j.1462-5822.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 14.Oslund KL, Baumgarth N. Influenza-induced innate immunity: regulators of viral replication, respiratory tract pathology & adaptive immunity. Future Virol. 2011;6(8):951–962. doi: 10.2217/fvl.11.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gannagé M, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6(4):367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol. 2015;13(1):28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marjuki H, et al. Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion. Cell Microbiol. 2011;13(4):587–601. doi: 10.1111/j.1462-5822.2010.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeganeh B, Ghavami S, Rahim MN, Klonisch T, Halayko AJ, Coombs KM. Autophagy activation is required for influenza A virus-induced apoptosis and replication. Biochim Biophys Acta Mol Cell Res. 2018;1865(2):364–378. doi: 10.1016/j.bbamcr.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Dumit VI, Dengjel J. Autophagosomal protein dynamics and influenza virus infection. Front Immunol. 2012;3:43. doi: 10.3389/fimmu.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arndt U, et al. Release of macrophage migration inhibitory factor and CXCL8/interleukin-8 from lung epithelial cells rendered necrotic by influenza A virus infection. J Virol. 2002;76(18):9298–9306. doi: 10.1128/JVI.76.18.9298-9306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baugh JA, Bucala R. Macrophage migration inhibitory factor. Crit Care Med. 2002;30(1 Supp):S27–S35. [PubMed] [Google Scholar]
- 23.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 24.Savva A, et al. Functional polymorphisms of macrophage migration inhibitory factor as predictors of morbidity and mortality of pneumococcal meningitis. Proc Natl Acad Sci USA. 2016;113(13):3597–3602. doi: 10.1073/pnas.1520727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flaster H, Bernhagen J, Calandra T, Bucala R. The macrophage migration inhibitory factor-glucocorticoid dyad: regulation of inflammation and immunity. Mol Endocrinol. 2007;21(6):1267–1280. doi: 10.1210/me.2007-0065. [DOI] [PubMed] [Google Scholar]
- 26.Shin MS, et al. Macrophage Migration Inhibitory Factor Regulates U1 Small Nuclear RNP Immune Complex-Mediated Activation of the NLRP3 Inflammasome. Arthritis Rheumatol. 2018;0(0):1–12. doi: 10.1002/art.40672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Hua Y, Nair S, Bucala R, Ren J. Macrophage migration inhibitory factor deletion exacerbates pressure overload-induced cardiac hypertrophy through mitigating autophagy. Hypertension. 2014;63(3):490–499. doi: 10.1161/HYPERTENSIONAHA.113.02219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao J, et al. Transcription factor ICBP90 regulates the MIF promoter and immune susceptibility locus. J Clin Invest. 2016;126(2):732–744. doi: 10.1172/JCI81937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrero LJ, et al. Critical role for macrophage migration inhibitory factor (MIF) in Ross River virus-induced arthritis and myositis. Proc Natl Acad Sci USA. 2011;108(29):12048–12053. doi: 10.1073/pnas.1101089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arjona A, et al. Abrogation of macrophage migration inhibitory factor decreases West Nile virus lethality by limiting viral neuroinvasion. J Clin Invest. 2007;117(10):3059–3066. doi: 10.1172/JCI32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang YC, Chen HR, Yeh TM. Pathogenic roles of macrophage migration inhibitory factor during dengue virus infection. Mediators Inflamm. 2015;2015:547094. doi: 10.1155/2015/547094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreih AG, et al. Role of Macrophage Migration Inhibitory Factor in Granulomatosis With Polyangiitis. Arthritis Rheumatol. 2018;70(12):2077–2086. doi: 10.1002/art.40655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson S, Crotta S, McCabe TM, Wack A. Pathogenic potential of interferon αβ in acute influenza infection. Nat Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason RJ. Biology of alveolar type II cells. Respirology. 2006;11 Suppl:S12–S15. doi: 10.1111/j.1440-1843.2006.00800.x. [DOI] [PubMed] [Google Scholar]
- 35.Baumann R, Casaulta C, Simon D, Conus S, Yousefi S, Simon HU. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. FASEB J. 2003;17(15):2221–2230. doi: 10.1096/fj.03-0110com. [DOI] [PubMed] [Google Scholar]
- 36.Guo Y, Hou J, Luo Y, Wang D. Functional disruption of macrophage migration inhibitory factor (MIF) suppresses proliferation of human H460 lung cancer cells by caspase-dependent apoptosis. Cancer Cell Int. 2013;13(1):28. doi: 10.1186/1475-2867-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lue H, et al. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26(35):5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 38.Atkin-Smith GK, Duan M, Chen W, Poon IKH. The induction and consequences of Influenza A virus-induced cell death. Cell Death Dis. 2018;9(10):1002. doi: 10.1038/s41419-018-1035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thangavel RR, Bouvier NM. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhirnov OP, Klenk HD. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis. 2007;12(8):1419–1432. doi: 10.1007/s10495-007-0071-y. [DOI] [PubMed] [Google Scholar]
- 41.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393(7):547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herold S, Ludwig S, Pleschka S, Wolff T. Apoptosis signaling in influenza virus propagation, innate host defense, and lung injury. J Leukoc Biol. 2012;92(1):75–82. doi: 10.1189/jlb.1011530. [DOI] [PubMed] [Google Scholar]
- 43.Yende S, et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. 2009;23(8):2403–2411. doi: 10.1096/fj.09-129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savva A, et al. Functional polymorphisms of macrophage migration inhibitory factor as predictors of morbidity and mortality of pneumococcal meningitis. Proc Natl Acad Sci USA. 2016;113(13):3597–3602. doi: 10.1073/pnas.1520727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fingerle-Rowson G, et al. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci USA. 2003;100(16):9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun H, et al. A critical regulatory role for macrophage migration inhibitory factor in hyperoxia-induced injury in the developing murine lung. PLoS One. 2013;8(4):e60560. doi: 10.1371/journal.pone.0060560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly SC, et al. Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med. 1997;3(3):320–323. doi: 10.1038/nm0397-320. [DOI] [PubMed] [Google Scholar]
- 48.Hoff J. Methods of Blood Collection in the Mouse. Lab Anim (NY) 2000;29(10):47–53. [Google Scholar]
- 49.Yu YR, et al. A Protocol for the Comprehensive Flow Cytometric Analysis of Immune Cells in Normal and Inflamed Murine Non-Lymphoid Tissues. PLoS One. 2016;11(3):e0150606. doi: 10.1371/journal.pone.0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jansing NL, et al. Isolation of Rate and Mouse Alveolar Type II Epithelial Cells. In: Alper S, Jannsen W, eds. Lung Innate Immunity and Inflammation: Methods and Protocols. Berlin/Heidelberg, Germany: Springer Science + Business Media; 2018:69–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.