Abstract
BMP signaling is critical in embryogenesis and in the development of numerous tissues. Many genetically modified (knockout and transgenic) mice have been established to study BMP function in development and disease. Mice with altered BMP receptor genes (including global knockout, conditional knockout, and conditional constitutively active transgenic mouse lines) have been particularly informative. In this chapter, we describe how the genetically modified mice were generated and introduce genotyping methods. These methods include regular PCR and genomic real-time PCR using specific primers based on different constructs in different mice strains.
Keywords: Transgenic, BMP receptors, Regular PCR, Genomic real-time PCR, Primers
1. Introduction
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) superfamily. Similar to TGF-β, BMPs signal through type I and type II transmembrane serine/threonine kinase receptors. In response to the binding of BMP ligands, type I and type II BMP receptors form a membrane-bound heterotetrameric complex. Then, the constitutively active type II receptor transphosphorylates the type I receptor at a glycine-serine rich motif (GS domain). Consequently, the Smad signal transducers are phosphorylated, and the downstream signal is propagated [1–3]. There are three type I BMP receptors (BMPR-1A or ALK3, BMPR-1B or ALK6, ACVR-1 or ALK2) and three type II BMP receptors (BMPR-2, ACVR-2A, ACVR-2B). Both type I and type II receptors are required to form a heterotetrameric complex for BMP signal transduction [1–3]. However, the mechanism of the heterotetrameric signaling complex formation can vary. For example, BMP-2 and BMP4 interact with type I receptors and recruit type II receptors, whereas BMP6 and BMP7 preferentially bind to type II receptors and recruit type I receptors [4]. More importantly, the binding of a BMP ligand to preformed receptor complexes activate signaling pathways that differ from those activated by a receptor complex whose assembly was stimulated by BMP binding [5].
Genetic studies into the function of these complex receptors are essential for clarifying the role of BMP signaling in development and disease. As of now, several genetically modified BMP receptor mice have been established, including global knockout (KO), conditional knockout (cKO), and conditional constitutively active (ca) mice [6–17]. BMP receptor gene modifications were observed to result in embryonic lethality [18, 19] or abnormalities in many tissues, including the skeleton [20–22], craniofacial [23, 29], heart [24], vascular [25], lung [26], eye [27], and tooth [28], thus establishing that BMP signaling is critical for normal embryogenesis.
In this chapter, we first describe how the genetically modified mice were generated and then introduce genotyping methods. These methods include regular PCR and genomic real-time PCR using specific primers based on different constructs in different mice strains.
2. Materials
Unless otherwise noted, all solutions are prepared in water purified by double distillation or other methods.
2.1. Regular PCR
Lysis buffer: 100 mM NaCl, 1 mM Tris pH 7–8, 0.1 mM EDTA, 0.1% Triton X-100 in distilled water. Store at room temperature. Add 1/50 volume of 40 mg/mL proteinase K (0.8 mg/mL final concentration) immediately before use.
40 mg/mL proteinase K.
PCR oil.
Primers for genotyping of each receptor mutations are listed in Table 1. The positions of the primers are marked in the maps of targeted alleles shown in Figs. 1 and 2.
0.5 unit/μL Taq DNA polymerase.
1× Taq buffer: 10 mM Tris-HCl, 50 mM KCl, and 1.5 mM MgCl2, pH 8.3.
5 mM dNTP mix: 5 mM each of dATP, dCTP, dGTP, and dTTP.
25 mM MgCl2.
Thermo cycler.
10× Tris/Borate/EDTA (TBE) buffer: 1 M Tris, 0.9 M boric acid, and 0.01 M EDTA. Dilute 100 mL 10× TBE buffer in 900 mL water to make 1000 mL 1× TBE for agarose gel electrophoresis.
3% agarose gel prepared in 1× TBE.
10 mg/mL ethidium bromide stock.
Table 1.
Primers used for genotyping of different BMP receptors genetically modified mice
| Ref | Modified gene | Primers | Primer ratios | PCR products | Annealing | |
|---|---|---|---|---|---|---|
| Global knockout mutation | ||||||
| Fig. 1a | [6] | Alk2 null | A2–5 5′-ATGCTAGACCTGGGCAGCCATA-3′ PGK-Mot 5′-CGTGTGTAAGGTGTAGGTG GCC-3′ | A2–5:PGK-Mot: A2–3 = 2:1:1 | 195 bp (Alk2 null) | 65 °C, 40 cycles |
| A2–5 5′-ATGCTAGACCTGGGCAGCCATA-3′ A2–3 5′-CATGCTAGCAGCTCGGAGAAAC-3′ | 371 bp (wild type) | |||||
| Fig. 1b | [18] | Alk3 null | fx3 5′-AGACTGCCTTGGGAAAAGCGC-3′ fx5 3′-GGACTATGGACACACAATGGC-3′ | fx3:fx5:fx0 = 1:2:1 | 190 bp (Alk3 null) | 65 °C, 40 cycles |
| fx5 5′-GGACTATGGACACACAATGGC-3′ fx0 5′-CTCTGAATTTCTAGTCCACATCTGC-3′ | 280 bp (wild type) | |||||
| Fig. 1c | [7] | Alk6 null | Alk6-I 5′-TGGTGAGTGGTTACAACAAGATC AGCA-3′ | Neo-Al2: Alk6-I: Alk6-E = 1:2:1 | 300 bp (Alk6 null) | 65 °C, 40 cycles |
| Neo-Al2 5′-GAAAGAACCAGCTGGGGCTC GAG-3′ | ||||||
| Alk6-I 5′-TGGTGAGTGGTTACAACAAGATC AGCA-3′ | 350 bp (wild type) | |||||
| Alk6-E 5′-CTCGGCCCAAGATCCTACGTTG-3′ | ||||||
| Fig. 1d | [14] | Bmpr2 null | A 5′-GCTAAAGCGCATGCTCCAGACTGCC TT-3′ | Primer A:B:C = 2:1:1 | 260 bp (Bmpr2 null) | |
| C 5′-AGGTTGGCCTGGAACCTGAGGAAATC-3′ | ||||||
| A 5′-GCTAAAGCGCATGCTCCAGACTGCC TTG-3′ | 200 bp (wild type) | |||||
| B 5′-TCACAGCATGAACATGATGGAGGCGG-3′ | ||||||
| Fig. 1e | [16, 32] | Acvr2a null | A 5′-TGGGAAGACAATAGCAGGCATGC-3′ | 1:1:1:1 | 900 bp (Acvr2A null) | 70 °C, 30 cycles |
| B 5′-GCAGAGTGTGACCCGTACCCAC-3′ | ||||||
| C 5′-GTTGGTACCCGGAGGTATATGGC-3′ | 140 bp (wild type) | |||||
| D 5′-CCCTTACCATCTGCAGCAGTGCA-3′ | ||||||
| Acvr2b null | A 5′-ATGAACTGCAGGACGAGGCAGCG-3′ | 1:1:1:1 | 600 bp (Acvr2B null) | |||
| B 5′-GGCGATAGAAGGCGATGCGCTG-3′ | ||||||
| C 5′-CCGACAGCCCCCACCCTGCTCA-3′ | 241 bp (wild type) | |||||
| D 5′-GGCCCACCAGAGGGGATGGGGG-3′ | ||||||
| Conditional knockout mutation | ||||||
| Fig. 2a | [29] | Alk2 flox | Alk217F 5′-CCCCCATTGAAGGTTTAGAG AGAC-3′ | Alk217F: lk217R2 = 1:1 | 160 + 90 bp (flox) (see Note 4), 250 bp (wild type) | 65 °C, 40 cycles |
| Alk217R2 3′-CTAAGAGCCATGACAGA GGTTG-3′ | ||||||
| Alk2-recombined | Alk2RecF 5′-GAATTGCTAGAAGCCCATA GGC-3′ | Alk2RecF:Alk2WTR: Alk217F = 1:2:1 | 625 bp (recombined) | 60 °C, 40 cycles | ||
| Alk2WTR 5′-TGAGATTGTTCTAGCACTGC CC-3′ | ||||||
| Alk217F 5′-CCCCCATTGAAGGTTTAGAGA GAC-3′ | 530 bp (wild type) | |||||
| Alk2WTR 5′-TGAGATTGTTCTAGCACTGC CC-3′ | ||||||
| Fig. 2b | [8] | Alk3 flox | fx2 5′-GCAGCTGCTGCTGCAGCCTCC-3′ | fx2:fx4 = 1:1 | 230 bp (flox) | 65 °C, 40 cycles |
| fx4 3′-TGGCTACAATTTGTCTCATGC-3′ | 150 bp (wild type) | |||||
| Fig. 2b | Alk3-recombined (see Note 6) | fx1 5′-GGTTTGGATCTTAACCTTAGG-3′ | fx1:fx4:BMP2-A: BMP2-B = 1:1:1:1 | 180 bp (recombined) | 55 °C, 40 cycles | |
| fx4 5′-TGGCTACAATTTGTCTCATGC-3′ | ||||||
| BMP2-A 5′-AGCATGAACCCTCATGTGTT GG-3′ | 322 bp (wild type) | |||||
| BMP2-B 5′-GTGACATTAGGCTGCT GTAGCA-3′ | ||||||
| Fig. 2c | [15] | Bmpr2 flox | 5F 50-GGCAGACTCTGACTTTGACGCTAG-3′ | 315 bp (2loxP and 3loxP) | 60 °C, 40 cycles | |
| C 5′-TTATTGTAAGTACACTGTTGCTGTC-3′ | ||||||
| A 5′-CACACCAGCCTTATACTCTAG ATAC-3′ | 260 bp (wild type) | |||||
| 6R 5′-CACATATCTGTTATGAAACTTGAG-3′ | 350 bp (2loxP) | |||||
| A 5′-CACACCAGCCTTATACTCTAG ATAC-3′ | 270 bp (wild type) | |||||
| C 5′-TTATTGTAAGTACACTGTTGCTGTC-3′ | 500 bp (1loxP) | |||||
| Conditional constitutively active transgenic line | ||||||
| Before recombination | ||||||
| Fig. 2d | [12] | caAlk | TF41 5′-GTGCTGGTTATTGTGCTGTCTC-3′ | TF41:TF61:LbnFR1: LbnRev3 = 1:1:1:1 | 580 bp (caAlk transgene) | 65 °C, 40 cycles |
| TF61 5′-GACGACAGTATCGGCCTCAGGAA-3′ | ||||||
| LbnFR1 5′-GAGGACGCAGTCCAGTACCT-3′ | 334 bp (Internal control) | |||||
| LbnRev3 5′-TAGCCTCTGCCTCACGCCCT GC-3′ | ||||||
| After recombination | ||||||
| Fig. 2d | [11] | caAlk2 | TF41 (common) 5′-GTGCTGGTTATTGTG CTGTCTC-3′ | TF41:TF9 = 1:1 | 750 bp (caAlk2 transgene) | 65 °C, 40 cycles |
| TF9 (Alk2) 5′-CGAACACTACAGAGAGAAT AATG-3′ | ||||||
| [12] | caAlk3 | TF41 (common) 5′-GTGCTGGTTATTGTGC TGTCTC-3′ | TF41:TF17 = 1:1 | 300 bp (caAlk3 transgene) | ||
| TF17 (Alk3) 5′-CGGCGTAGCTGGGCTTTT GGA-3′ | ||||||
| caAlk6 | TF41 (common) 5′-GTGCTGGTTATTGTGC TGTCTC-3′ | TF41:TF25 = 1:1 | 300 bp (caAlk6 transgene) | |||
| TF25 (Alk6) 5′-GACATCCAGAGGTGACAA CAG-3′ | ||||||
Fig. 1.
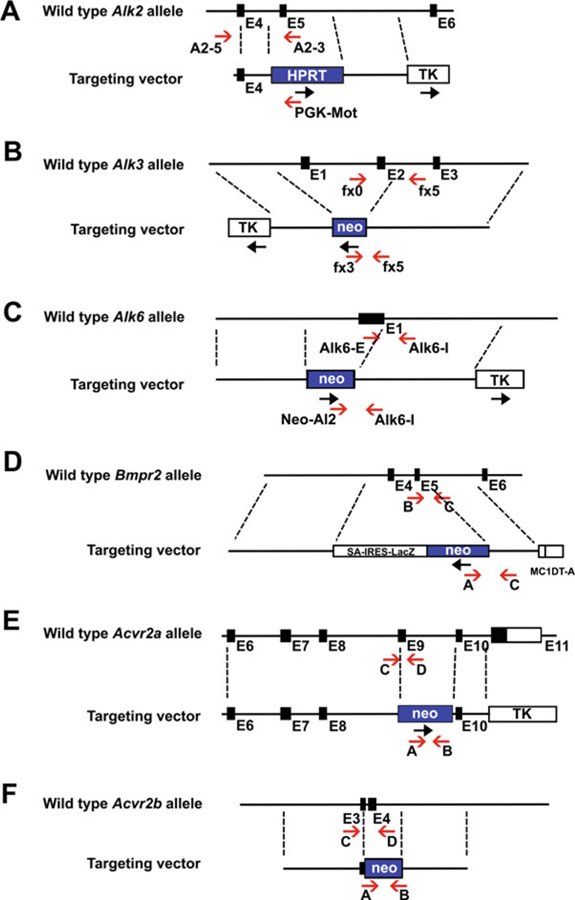
Structure representation of each BMP receptor KO mutation and PCR genotyping strategy. Schematic diagrams showing the wild-type locus of the Alk2 (a), Alk3 (b), Alk6 (c), Bmpr2 (d), Acvr2a (e), and Acvr2b (f) gene, and the targeting vector for each gene. The positions (red arrows) of PCR primers for genotyping are indicated below the locus
Fig. 2.
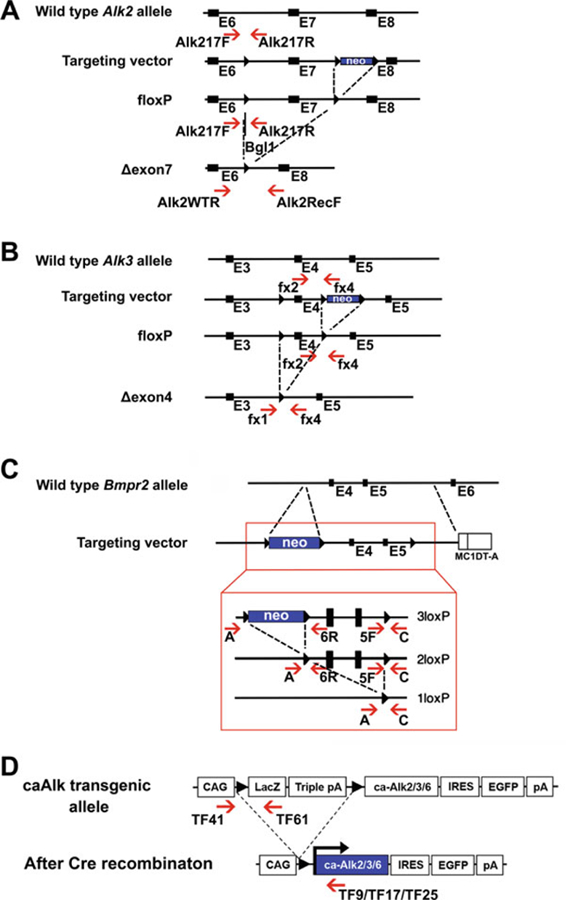
Structure representation of each BMP receptor cKO mutation and ca transgene and PCR genotyping strategy. (a–c) Schematic diagrams of the wild-type Alk2 (a), Alk3 (b), and Bmpr2 (c) locus, targeting vector, and mutant alleles after recombination. The positions of primers for genotyping by PCR are indicated by red arrows. (d) Schematic representation of the ca transgene of Alk2, Alk3, or Alk6. The primers used for genotyping are shown by red arrows below the locus
2.2. Genomic Real-Time PCR
TaqMan® Universal PCR Master Mix (Thermo Fisher, Cat: 4334437).
TaqMan primer sets of each receptor mutations are listed in Table 2.
Optical 96-well reaction plates compatible with your PCR machine.
Optical adhesive film.
Real-time PCR system.
Table 2.
TaqMan primers used for genomic real-time PCR of different BMP receptors genetically modified mice
| Ref | Modified gene | Primers |
|---|---|---|
| Conditional knockout mutation | ||
| [31] |
Alk2 (exon 7) (see Note 5) |
AIKAL5S_F 5′-CTCACTACTCTGGATACGGTTAGCT _3′, AIKAL5S_R 5′-GGGTCCCAAATATCTCTATGTGCAA-3′, AIKAL5S_M FAM 5′-CTATGGACAGTACAATCCG-3′ |
| [30, 31] |
Alk3 (exon 4) (see Note 5) |
AI89LJ8_F 5′-GACCAGAAGAAGCCAGAAAATGGA-3′, AI89LJ8_R 5′-TGTCCTGAGCAATAGCACTTTAAGAA-3′, AI89LJ8_M FAM 5′-CCTCTGGTGCTA AAGTC-3′ |
| Conditional constitutively active transgenic line | ||
|
Egfp (see Note 5) |
5′-GAGCGCACCATCTTCTTCAAG-3′, 5′-TGTCGCCCTCGAACTTCAC-3′, FAM 5′-ACGACGGCAACTACA-3′ |
|
3. Methods
3.1. Regular PCR
Collect small piece (less than 1 mm3) of tissues from the ear (ear notch), tail, or any organs. For embryos, the yolk sac or amniotic membrane may be used (do not use the placenta for genotyping). Place tissues into 96-format PCR tubes (do not cap).
Add 50 μL lysis buffer in each tube, overlaid with PCR oil.
Incubate at 55 °C for 6 h or more, then incubate at 85 °C for 30 min to inactivate proteinase K.
-
Take 4 μL DNA solution to mix with 76 μL of water to dilute samples, then use 4 μL of the diluted samples to set up 10 μL PCR reaction. Reaction mixture will be made as follows:
The conditions for thermal cycling are as follows (see Note 1):
Initial denaturation for 94 °C for 5 min followed by 30 to 40 cycles of denaturation at 94 °C for 30 s, annealing at 50–70 °C for 30 s, and extension at 72 °C for 1 min (30–40 cycles), then ending with 72 °C for 5–10 min followed by a cool down.
The number of cycles and annealing temperatures for different primer sets is shown in Table 1.
Run 3% agarose gel at 250–300 V, stain the gel with ethidium bromide, and photograph.
3.2. Genomic Real-Time PCR for In Vivo Deletion Efficiency
The in vivo deletion efficiency of conditional knockout by Cre-recombinase and other recombinases can vary. This protocol describes the quantification of Alk2 or Alk3 deletion in conditional knockout mice by genomic real-time PCR using custom-designed primer set [30, 31]. 5′ primers and 3′ primers are designed to amply the flox regions (exon 7 for Alk2, exon 4 for Alk3). FAM-labeled probes are designed within the PCR amplicons for detection (see Table 2).
Extract genomic DNA as in of Subheading 3.1, steps 1–3.
Mix the Gene Expression Master Mix thoroughly by swirling the bottle. Thaw Alk2 or Alk3 frozen primer set (Table 2) and templates DNA on ice. When thawed, vortex and then centrifuge the tubes briefly (see Note 2).
-
Prepare the PCR reaction mix (20 μL reactions):
TaqMan PCR Master Mix (2×) 10 μL Primer set (20×) 1 μL Template DNA (20x diluted) 5 μL Water 4 μL Perform three replicates of each reaction. Then vortex the tubes briefly to mix the solutions, centrifuge the tubes briefly to spin down the contents, and eliminate any air bubbles from the solutions.
Transfer 20 μL of each reaction mixture to each well of an optical plate.
Cover the plate with an optical adhesive film. Centrifuge the plate briefly to spin down the contents and eliminate air bubbles from the solutions.
Run the plate on a real-time PCR instrument using the following thermal cycling conditions: initial incubation at 50 °C for 2 min then denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 1 min, then ending with a cool down.
Determine the threshold cycles (CT) for the amplification curves. Use the comparative CT method to analyze target gene levels normalized to Gapdh level as described by the manufacturer of the instrument.
3.3. Genomic Real-Time PCR to Determine Homozygosity vs. Heterozygosity of Conditional Constitutively Active Transgenes
To generate conditional constitutively active transgenic mice, the EGFP cassette was inserted into the constructs (Fig. 2d) [10]. Therefore, the copy number of target genes can be quantified based on the copy number of Egfp via real-time PCR. This is a common strategy for the caALK2, caALK3, and caAlk6 mouse lines. The Egfp primer set is shown in Table 2.
Same as Subheading 3.2, steps 1 and 2.
-
Prepare the PCR reaction mix (20 μL reactions):
TaqMan PCR Master Mix (2×) 10 μL Egfp primer set (20×) 1 μL Template DNA (20× diluted) 5 μL Water 4 μL Perform three replicates of each reaction. Then vortex the tubes briefly to mix the solutions, centrifuge the tubes briefly to spin down the contents, and eliminate any air bubbles from the solutions. Wild-type, caAlk het, and caAlk homo samples, which genotypes are known, should be used as controls.
Same as Subheading 3.2, steps 4–7 (see Note 3).
Acknowledgments
We thank Drs. Ce Shi and Taocong Jin for designing probe sets for quantitative PCR. We are grateful to Dr. Kaitrin Kramer for critical reading of this manuscript. This work was supported by the National Institutes of Health (R01DE020843 to Y.M.), International FOP Association (Y.M.), the grant-in-aid from the National Natural Science Foundation of China (31500788 to J.Y.), and the Fundamental Research Fund for the Central Universities of China (410500114 to J.Y.).
4. Notes
Samples for which genotypes are known should be used as controls. For these primers and most others, these conditions work adequately. If not, try the following: (1) change the dilution of template DNA. The reaction will not work when the DNA concentration is too high; (2) optimize the annealing temperature; (3) change Taq DNA polymerase to Taq hot start DNA polymerase; or (4) purify DNA further.
Protect all reagents from light in the freezer until you are ready to use them. Excessive exposure to light may affect the fluorescent probes.
The calculated copy number of the Egfp in caAlk homozygous mice should be about twice as that of heterozygous mice. Homozygosity also can be genetically confirmed by crossing the mice to wild-type mice. All F1 pups should carry the transgene.
PCR amplification generates a 250 bp from both flox and wild-type alleles. Bgl1 digestion, which uniquely digests the flox gene product into 160 bp and 90 bp fragments, can be used to distinguish flox and wild-type PCR products.
All three primers are pre-mixed into one tube by the manufacturer.
The primers BMP2-A and BMP2-B are for an internal control at the Bmp2 locus. Please refer reference [33] for the specific positions of those primers.
References
- 1.Grafe I, Alexander S, Peterson JR, et al. (2017) TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb Perspect Biol 10 (5) 10.1101/cshperspect.a022202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadin D, Knaus P, Mueller TD (2016) Structural insights into BMP receptors: specificity, activation and inhibition. Cytokine Growth Factor Rev 27:13–34. 10.1016/j.cytogfr.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M (2016) Endocytosis and trafficking of BMP receptors: regulatory mechanisms for fine-tuning the signaling response in different cellular contexts. Cytokine Growth Factor Rev 27:35–42. 10.1016/j.cytogfr.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 4.De Caestecker M (2004) The transforming growth factor-beta superfamily of receptors. Cytokine Growth Factor Rev 15(1):1–11 [DOI] [PubMed] [Google Scholar]
- 5.Nohe A, Hassel S, Ehrlich M et al. (2002) The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem 277 (7):5330–5338 [DOI] [PubMed] [Google Scholar]
- 6.Mishina Y, Crombie R, Bradley A et al. (1999) Multiple roles for activin-like kinase-2 signaling during mouse embryogenesis. Dev Biol 213 (2):314–326 [DOI] [PubMed] [Google Scholar]
- 7.Yi SE, Daluiski A, Pederson R et al. (2000) The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 127(3):621–630 [DOI] [PubMed] [Google Scholar]
- 8.Mishina Y, Hanks MC, Miura S et al. (2002) Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32(2):69–72 [DOI] [PubMed] [Google Scholar]
- 9.Peterson JR, Eboda O, Agarwal S et al. (2014) Targeting of ALK2, a receptor for bone morphogenetic proteins, using the Cre/lox system to enhance osseous regeneration by adipose-derived stem cells. Stem Cells Transl Med 3 (11):1375–1380. 10.5966/sctm.2014-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon BS, Ovchinnikov DA, Yoshii I et al. (2005) Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A 102 (14):5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda T, Scott G, Komatsu Y et al. (2006) Generation of a mouse with conditionally activated signaling through the BMP receptor, ALK2. Genesis 44(4):159–167 [DOI] [PubMed] [Google Scholar]
- 12.Komatsu Y, Yu PB, Kamiya N et al. (2013) Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res 28 (6):1422–1433. 10.1002/jbmr.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Espinoza-Lewis RA, Sun C et al. (2010) Overexpression of constitutively active BMP-receptor-IB in mouse skin causes an ichthyosis-vulgaris-like disease. Cell Tissue Res 342 (3):401–410. 10.1007/s00441-010-1077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beppu H, Kawabata M, Hamamoto T et al. (2000) BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol 221(1):249–258 [DOI] [PubMed] [Google Scholar]
- 15.Beppu H, Lei H, Bloch KD et al. (2005) Generation of a floxed allele of the mouse BMP type II receptor gene. Genesis 41(3):133–137 [DOI] [PubMed] [Google Scholar]
- 16.Song J, Oh SP, Schrewe H et al. (1999) The type II activin receptors are essential for egg cylinder growth, gastrulation, and rostral head development in mice. Dev Biol 213 (1):157–169 [DOI] [PubMed] [Google Scholar]
- 17.Mayeur C, Leyton PA, Kolodziej SA et al. (2014) BMP type II receptors have redundant roles in the regulation of hepatic hepcidin gene expression and iron metabolism. Blood 124 (13):2116–2123. 10.1182/blood-2014-04-572644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishina Y, Suzuki A, Ueno N et al. (1995) Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev 9(24):3027–3037 [DOI] [PubMed] [Google Scholar]
- 19.Park C, Lavine K, Mishina Y et al. (2006) Bone morphogenetic protein receptor 1A signaling is dispensable for hematopoietic development but essential for vessel and atrioventricular endocardial cushion formation. Development 133(17):3473–3484 [DOI] [PubMed] [Google Scholar]
- 20.Salazar VS, Gamer LW, Rosen V (2016) BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol 12(4):203–221. 10.1038/nrendo.2016.12 [DOI] [PubMed] [Google Scholar]
- 21.Wu M, Chen G, Li YP (2016) TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 4:16009 10.1038/boneres.2016.9.eCollection [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing J, Hinton RJ, Feng JQ (2015) Bmpr1a signaling in cartilage development and endochondral bone formation. Vitam Horm 99:273–291. 10.1016/bs.vh.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 23.Graf D, Malik Z, Hayano S et al. (2016) Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev 27:129–139. 10.1016/j.cytogfr.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavrilov S, Lacy E (2013) Genetic dissection of ventral folding morphogenesis in mouse: embryonic visceral endoderm-supplied BMP2 positions head and heart. Curr Opin Genet Dev 23(4):461–469. 10.1016/j.gde.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García de Vinuesa A Abdelilah-Seyfried S, Knaus P et al. (2016) BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev 27:65–79. 10.1016/j.cytogfr.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 26.Eblaghie MC, Reedy M, Oliver T et al. (2006) Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol 291(1):67–82 [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Zhao JY, Wu D et al. (2012) Mutually inductive interactions between the lens and retina require ALK3 functions during mouse embryonic development. Int J Ophthalmol 5 (2):119–124. 10.3980/j.issn.2222-3959.2012.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Hai B, Qin L et al. (2013) Cessation of epithelial Bmp signaling switches the differentiation of crown epithelia to the root lineage in a beta-catenin-dependent manner. Mol Cell Biol 33(23):4732–4744. 10.1128/MCB.00456-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudas M, Sridurongrit S, Nagy A et al. (2004) Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev 121(2):173–182 [DOI] [PubMed] [Google Scholar]
- 30.Shi C, Zhang H, Louie K et al. (2017) BMP signaling mediated by BMPR1A in osteoclasts negatively regulates osteoblast mineralization through suppression of Cx43. J Cell Biochem 118(3):605–614. 10.1002/jcb.25746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan H, Zhang H, Abraham P et al. (2017) BmpR1A is a major type 1 BMP receptor for BMP-Smad signaling during skull development. Dev Biol 429(1):260–270. 10.1016/j.ydbio.2017.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh SP, Li E (1997) The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev 11(14):1812–1826 [DOI] [PubMed] [Google Scholar]
- 33.Singh AP, Castranio T, Scott G et al. (2008) Influences of reduced expression of maternal bone morphogenetic protein 2 on mouse embryonic development. Sex Dev 2 (3):134–141. 10.1159/000143431 [DOI] [PMC free article] [PubMed] [Google Scholar]


