Abstract
Receptor-type protein tyrosine phosphatase, receptor type D (PTPRD) has likely roles as a neuronal cell adhesion molecule and synaptic specifier. Interest in its neurobiology and genomics has been stimulated by results from human genetics and mouse models for phenotypes related to addiction, restless leg syndrome, neurofibrillary pathology in Alzheimer’s disease, cognitive impairment/intellectual disability, mood lability, and obsessive-compulsive disorder. We review PTPRD’s discovery, gene family, candidate homomeric and heteromeric binding partners, phosphatase activities, brain distribution, human genetic associations with nervous system phenotypes, and mouse model data relevant to these phenotypes. We discuss the recently reported discovery of the first small molecule inhibitor of PTPRD phosphatase, the identification of its addiction-related effects, and the implications of these findings for the PTPRD-associated brain phenotypes. In assembling PTPRD neurobiology, human genetics, and mouse genetic and pharmacological datasets, we provide a compelling picture of the roles played by PTPRD, its variation, and its potential as a target for novel therapeutics.
Keywords: cell adhesion molecule, cocaine, addiction, neurofibrillary tangles, restless leg syndrome, obsessive compulsive disorder, neurotherapeutics
Graphical abstract
Receptor-type protein tyrosine phosphatase, receptor type D (PTPRD) is a neuronal cell adhesion molecule and synaptic specifier that has possible roles in addiction, Alzheimer’s disease, cognitive impairment, mood lability, and obsessive-compulsive disorder. This article reviews PTPRD’s discovery, gene family, candidate binding partners, phosphatase activities, brain distribution, human genetic associations with nervous system phenotypes, and mouse model data relevant to these phenotypes.
Context
Classical genetic studies support both genetic and environmental influences on vulnerability to virtually all common brain-based disorders and traits 1. Molecular genetic association studies now propose a modest number of oligogenic and large numbers of polygenic influences on these disorders. For each disorder or trait, the genetic influences thus typically arise from variations in a group of genes. There frequently are disease-associated variants in several distinct regions of each gene. There often is pleiotropy: effects of common functional variations that are sufficient to alter one brain-based phenotype often alter other phenotypes as well.
Here, we focus on receptor type protein tyrosine phosphatase, receptor type D (PTPRD) and the roles for its variants in brain, behavioral, and neurological disorders. We review this gene and some of the likely roles of PTPRD and its variants in brain. Data now support a working hypothesis that common and rarer PTPRD variants make pleiotropic contributions of differing strength to a number of brain phenotypes and disorders. Understanding the genomics and genetics of PTPRD is thus important for understanding the biology of interesting phenotypes for substance use disorders, restless leg syndrome, mood lability, cognitive abilities, and the neurofibrillary neuropathology of Alzheimer’s disease. Substance disorder phenotypes include dependence (polysubstance, opiate, alcohol), rewarding properties (stimulants), effects experienced during the first drinking sessions (alcohol), and ability to quit smoking (see references below). We thus review PTPRD, place its function and its variation in the context of these genetic findings, and describe the implications of these findings for improved understanding of pathogenesis and novel therapeutic opportunities. The recent elucidation of the actions of a novel small molecule inhibitor of PTPRD phosphatase 2 adds support for PTPRD roles in addiction-related phenotypes and solidifies PTPRD as an emerging target for novel therapeutics for addiction and possibly other PTPRD-associated disorders.
What is PTPRD?
Discovery
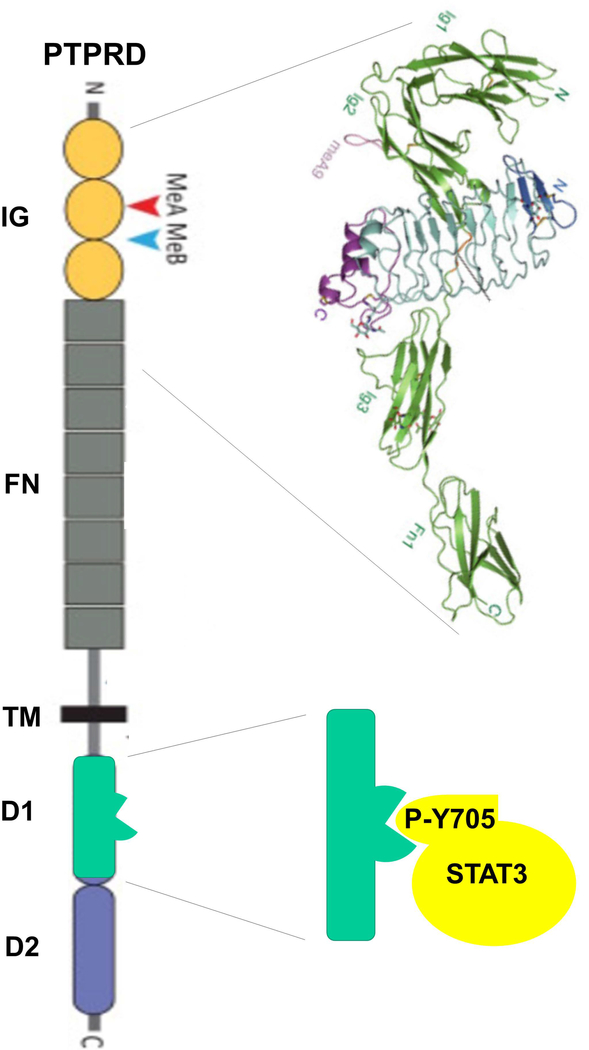
PTPRD was identified in 1990 by the low stringency screening of a human cDNA library with a Drosophila receptor-type protein tyrosine phosphatase (RPTP) hybridization probe 3. PTPRD contains extracellular immunoglobulin and fibronectin III domains, a transmembrane domain, and D1 and D2 intracellular phosphatase-like domains (Fig. 1). The D1 phosphatase domain is catalytically active (Fig. 1). PTPRD mRNAs and protein were identified in specific brain regions 4–6 and in the kidney. Human RNAseq data has now identified the highest expression in brain, followed by kidney, ovary, placenta, and intestine 7. Overall, the brain expresses moderate levels of PTPRD, with reads per kilobase per million (RPKM) values of about 18, compared to 90 for the mRNA encoding the very abundant brain protein THY1. The working hypothesis that PTPRD is one of the 400–500 genes that encode a cell adhesion molecule/synaptic specifier 8–9 initially arose from PTPRD’s homologies with other genes that serve these functions. This hypothesis has received substantial additional support, as we describe below.
Figure 1.
PTPRD domains. Immunoglobulin (IG, with miniexons (Me) A and B shown), fibronectin type III (FN), transmembrane (TM), active tyrosine phosphatase domain (D1), and enzymatically inactive regulatory domain (D2). Upper right: structure of the complex between the PTPRD IG and first FN domains with the leucine-rich repeat region of SLITRK2. Lower right: D1 domain dephosphorylation of phosphotyrosine 705 in STAT3, an example of PTPRD activity.
PTPRD gene and protein
The PTPRD gene is found on the short arm of human chromosome 9 (9p), at 8,314,246 to 10,612,841 basepairs (build 109, NC_000009.12). Its 5’ transcriptional start is centromeric to its telomeric 3’ last exon 10. There are many splicing and other variations; at least 50 PTPRD mRNA isoforms are described. As many as 49 exons that encode portions of mRNA-sense transcripts and two that encode natural antisense transcripts are annotated 11. The canonical PTPRD gene model contains 1912 amino acids, with a predicted mass of 215 kDa 12. Amino acids 24–114, 126–224, and 236–318 are immunoglobin-like (Ig) sequences; residues 325–415, 420–516, 518–607, 612–709, 714–822, 823–916, 921–1016, and 1020–1106 are fibronectin type III (FNIII) domains; and amino acids 1357–1612 and 1644–1903 represent the active D1 and inactive D2 protein tyrosine phosphatase–like sequences, respectively. The region of amino acids 180–189 is postulated to interact with IL1RAPL1 (see below). Functionally important splicing variants include those that include or exclude miniexons A and B and encode amino acids 181–189 and 227–230, respectively. Alternate splicing at the regions that encode fibronectin domains 4 to 7 yields long (Ig1–3, FNIII1–8) or short (Ig1–3, FNIII1–3,8) isoforms that are predominantly expressed postnatally in most brain regions 13.
PTPRD mRNAs include several that encode only extracellular sequences and might provide decoy soluble translation products. Such soluble isoforms could conceivably compete for binding interactions that might otherwise occur between PTPRD-containing membrane-bound homo- or hetero-oligomers 14.
Gene family
PTPRD is a member of a three-gene DSF mammalian subfamily of receptor type protein tyrosine phosphatases that also includes PTPRS (also known as PTPR sigma, PTPσ, or PTPRσ) and PTPRF (also known as leukocyte related antigen or LAR). Single Drosophila (Dlar) and C. elegans (ptp-3) genes 3, 15 encode homologs of these DSF family members, helping to explain the success of the homology cloning strategy that initially identified PTPRD.
Functions: Extracellular domains and binding partners
PTPRD’s immunoglobulin and fibronectin extracellular architecture (Fig. 1) suggested roles as a cell adhesion molecule and synaptic specifier. Wang and Bixby’s results from PTPRD expression in cultures support cell adhesion molecule roles for homophilic PTPRD–PTPRD interactions 16. Embryonic neuronal subpopulations from forebrain, retina, ciliary ganglion, and cerebellum bound to PTPRD-containing substrates and elaborated many more neuronal processes when plated on PTPRD-containing substrates, supporting positive neurotrophic and adhesive roles for PTPRD–PTPRD interactions. While this important data has not been substantially extended, the results provide significant evidence that homophilic PTPRD–PTPRD interactions can promote neuronal process growth.
In addition to the evidence for PTPRD homomeric binding, there is support for other candidate physiological PTPRD ligands (Table 1). Most evidence comes from studies of PTPRD interactions with the SLIT- and NTRK-like (SLITRK) proteins 17–18. Takahashi et al. screened candidate proteins for interactions with SLITRK3 and found that it bound to PTPRD, though not to neurexins, NCAM, CHL1, neurofascin, PTPRF, NTRK2 (also known as TRKB), or CNTNAP1 (also known as CASPR)). Each of the six SLITRKs displayed an affinity for PTPRD. Cells that express either SLITRK2 or −3 caused aggregation of PTPRD and of the vesicular GABA transporter, a marker for synaptic specialization, in contacting,expressing cells 18. Crystallographic structures of PTPRD/SLITRK1 and of PTPRD/SLITRK2 complexes are now available. These structures point to key roles for a PTPRD splice variant that includes amino acids encoded by miniexon B, lying between immunoglobulin domains 2 and 3, in facilitating strong PTPRD binding to SLITRK’s leucine-rich repeat domain 17, 19.
Table 1.
Some candidate homomeric and heteromeric PTPRD binding partners
| Homomeric |
| PTPRD |
| Heteromeric |
| SLITRK1 |
| SLITRK2 |
| SLITRK3 |
| IL-1RAcP |
| IL1RAPL1 |
| NGL3 |
| LRFN1 |
| LRFN5 |
Binding to PTPRD was also detected when investigators sought ligands for interleukin 1 receptor accessory protein (IL1RAP, also known as IL-1RAcP) and interleukin 1 receptor accessory protein like 1 (IL1RAPL1). These proteins contain extracellular immunoglobulin domains and single transmembrane regions, and display substantial expression in the brain. Testing for candidate ligands of IL-1RAcP and IL1RAPL1, including neurexins, SynCAMs, synaptic adhesion-like molecules (SALMs), and receptor-type protein tyrosine phosphatases, identified good interactions only with PTPRD 20. This binding again depends on PTPRD miniexon splicing. Crystallography confirms complexes between PTPRD immunoglobulin domains and the immunoglobulin domains of IL1RAPL1 and IL1RAP, again in a miniexon-dependent fashion 21.
Attention to PTPRD interactions with LRRC4B (also known as NGL-3) came from extensions of work that identified functional interactions between PTPRF and LRRC4B/NGL-3 22. Extracellular domains from PTPRD and expression constructs limited to PTPRD’s extracellular fibronectin domains I and II both interacted with LRRC4B in binding assays. However, these PTPRD sequences were weaker NGL-3 binders than analogous PTPRF or PTPRS sequences. These PTPRD sequences also failed to induce aggregation of the postsynaptic marker PSD-95, although PTPRF sequences triggered such aggregation in the same assay.
SALMs, especially those that are products of the LRFN1 and −5 genes, can also serve as binding partners of DSF family members, including PTPRD 23–26.
There are reported interactions between PTPRD and cytoplasmic proteins, including PPFIA1 5. However, follow-up work provides better evidence of interactions with other DSF family members than with PTPRD. Reported interactions of MTSS1 with cytoplasmic PTPRD sequences and actin filaments provide a plausible direct link between PTPRD and neuronal process formation 27. However, evidence for the effects of altered MTSS1 expression on neuronal process formation comes largely from studies of cerebellar neurons, which do not substantially express PTPRD 28.
Functions: Intracellular domains and phosphatase activity
PTPRD’s physiological functions are likely to derive from its dephosphorylation of phosphotyrosine residues in a group of cytoplasmic proteins. Its D1 phosphatase domain, proximal to the membrane, mediates this phosphatase activity. While the D2 domain displays sequence homology to phosphatases, it is largely inactive enzymatically but binds to and downregulates the activity of the D1 domains of PTPRD and several other related receptor type protein tyrosine phosphatases (especially in in vitro studies with PTPRA, PTPRE and PTPRS) 29–30.
Analysis of the selectivity of PTPRD’s D1 phosphatase domain reveals: (1) lack of a highly specific consensus sequence for the amino acids that surround the phosphotyrosine; (2) preference for large hydrophobic amino acids, especially adjacent (−1) to the phosphotyrosine; (3) preference for acidic residues in the vicinity of the phosphotyrosine; (4) aversion to basic residues at these locations; (5) relatively lower affinity for phosphotyrosine substrates when compared to the affinities of other protein tyrosine phosphatases; and (6) good catalytic activities (kcat values) that are close to those of other protein tyrosine phosphatases and thus the potential to act as a highly efficient enzyme with a high kcat/KM ratio 31.
The selectivity of the PTPRD phosphatase is likely reflected in the sequences of 45 C. elegans proteins whose phosphotyrosine residues are conserved in human homologs. Phosphorylation of these C. elegans proteins is altered with knockout of ptp-3 32 (A Pandey, personal communication, 2015).. PTPRD-mediated changes in the phosphorylation of STAT3 tyrosine 705 alter STAT3’s abilities to homodimerize, move into the nucleus, and change transcription of other genes 33. Altered tyrosine phosphorylation of FYN and SRC in homozygous Ptprd knockout mice has been related to differential elaboration of basal dendrites by layer V cortical pyramidal cells that normally express PTPRD 34.
As we discuss below, inhibition of the PTPRD D1 phosphatase domain provides an apparently successful route to modulating PTPRD activity. It is also theoretically possible to increase phosphatase activity of PTPRD by reducing the inhibitory effect of D2 domain binding to the active D1 phosphatase domain.
PTPRD distribution in the brain
Reports of the brain distribution of PTPRD mRNA or immunoreactivity (e.g. Ref. 35) fit well with Allen Brain Atlas images that describe the largely neuronal distribution of PTPRD mRNA in a number of brain regions whose function can often be related to human phenotypes associated with PTPRD variation 36. These data in turn mesh reasonably well with available single cell RNAseq datasets. We can thus describe pictures of the multifocal distribution of PTPRD mRNA and of the brain circuits that are likely to display altered activity when PTPRD function changes (Table 2).
Table 2.
Some of the neuronal types that express PTPRD mRNA2
| Neuronal type | PTPRD expression |
|---|---|
| Acetylcholine | Many, dense |
| Dopamine | Many, dense |
| Reticular thalamic | Many, dense |
| Cerebral cortical | Scattered, moderate |
| Olfactory bulb (inner plexiform) | Many, dense |
| Pyriform cortex (layer 2) | Many, dense |
We can relate some prominent neuronal PTPRD mRNA expression patterns to neurotransmitter phenotypes in the fore- and midbrain. Likely cholinergic neurons of the nucleus basalis, diagonal band, septum, striatum/accumbens, and reticular formation express high densities of PTPRD mRNA hybridization signal. Likely dopaminergic neurons of the substantia nigra pars compacta and the ventral tegmental area express PTPRD mRNA with moderately high hybridization signal densities. Human RNAseq data from pigmented, presumed dopaminergic neurons from ventral midbrain confirms this dopaminergic PTPRD mRNA localization 37.
Other areas of prominent PTPRD mRNA expression are easiest to characterize based on their relatively high levels of PTPRD mRNA hybridization in many or even most neurons. Most neurons of the reticular thalamic nucleus and specific olfactory bulb (inner plexiform) and pyriform cortex (layer 2) layers express relatively high densities of PTPRD mRNA.
Expressing neurons are also scattered in a number of other brain regions. There are scattered expressing neurons in several hypothalamic nuclei, in hippocampus, and in amygdaloid nuclei, especially in cortical and lateral amygdala. RNAseq data confirm higher hippocampal expression in CA2 neurons 38. This data supports levels of expression in hippocampal pyramidal neurons that lie in the highest decile of levels of expression for all cell adhesion molecules annotated by these authors 9.
Within neocortical regions, scattered neurons, especially in laminae II-III and V-VI, show moderate PTPRD mRNA hybridization densities. RNAseq data from the visual cortex 39 support PTPRD colocalization 40 with neurons containing vasoactive intestinal peptide, L4 and L5 (while somatostatin-expressing neurons express much less PTPRD). However, there is also significant PTPRD expression in opalin-expressing cells, suggesting some oligodendroglial expression that has also been demonstrated elsewhere 41. Such expression might modulate neuronal–oligodendroglial interactions in ways that have been supported by studies of other cell adhesion molecules 42.
Within the brainstem, there is moderately dense PTPRD mRNA hybridization in cholinergic neurons throughout, including parabigeminal and cranial nerve motoneurons. There is dense expression in olivary nuclei (especially inferior olive). There is modest expression in some dorsal raphe and locus coeruleus neurons and in scattered neurons in other brainstem regions.
In spinal cord, there is dense expression in motoneurons and significant expression in scattered dorsal horn neurons.
While it is not possible to comprehensively summarize these expression patterns in a simple fashion, patterns of expression do provide emphasis on (1) dopamine and acetylcholine systems that are implicated in reward, mnemonic, and motor functions in ways that involve substantial subcortical–cortical (and hippocampal) interactions; (2) thalamo-cortical interactions mediated through reciprocal reticular thalamic–cortical interactions; (3) likely roles in cortical interconnectivities; (4) roles in the strong cerebellar connections that emanate from the inferior olive; and (5) brainstem and spinal cord motoneuronal pools. PTPRD homomeric and heteromeric interactions could play differential roles in these circuits. We thus describe highlights of the distributions of expression of several candidate heteromeric PTPRD ligands below.
Brain distributions of other candidate PTPRD ligands
The mouse brain distribution of IL-1RAcP mRNA appears highest in the hippocampus, especially the dentate gyrus. There is moderate expression in many thalamic nuclei 43. There is also hybridization signal in pyriform cortex and in ventral midbrain areas, including the substantia nigra.
There is more hybridization signal for IL1RAPL1 mRNA, with prominent expression in many areas of the olfactory bulb and pyriform cortex, cerebral cortex (especially layers II and III), hippocampus (especially dentate gyrus), and mammillary body. There is moderate expression in cells of the striatum, many thalamic nuclei, hypothalamus, areas of the brainstem, including modest to moderate expression in ventral midbrain, and many spinal cord dorsal horn and motoneurons 44.
SLITRKs are also expressed in interesting brain patterns. We focus here on SLITRK3 since there is better documentation of its role as a PTPRD binding partner, as well as SLITRK1 and −2. SLITRK3 mRNA expression is abundant in many brain regions. SLITRK3 mRNA is expressed in the cerebral cortex (especially layer II), hippocampus (especially CA1/2 and dentate gyrus), olfactory bulb and tract, many thalamic regions, hypothalamic regions, midbrain regions (including ventral midbrain and red nucleus), and multifocal regions in the pons and medulla 45.
Much of the expression of SLITRK1 mRNA overlaps with the patterns of expression of PTPRD mRNA, including expression in likely cholinergic neurons of the basal forebrain, striatum, and brainstem motor nuclei, and in dopaminergic midbrain neurons 46. There is also abundant expression in the olfactory and pyriform cortex, cerebral cortex (especially layers II-III and V-VI), septum, many thalamic regions (including anterodorsal nucleus), hypothalamic regions (including supraoptic, paraventricular, and suprachiasmatic nuclei), hippocampal regions (especially dentate gyrus), and most amygdala regions.
SLITRK2 mRNA expression is also highest in the pyriform cortex, olfactory regions, and hippocampus (especially dentate gyrus) 47. There is again expression in presumed cholinergic neurons in the basal forebrain and brainstem, with moderate levels of expression in cortical layers I–VI, and thalamic, hypothalamic, and amygdaloid nuclei.
PTPRD genomic variation and PTPRD associations with human disorders and phenotypes
PTPRD genomic variation and association with levels of PTPRD brain mRNA expression and possible mechanism
A recent search of the dbSNP database identifies 700,229 PTPRD single nucleotide polymorphism (SNP) variants, with 56,540 involving short insertion or deletion variants. Of these, 1,346 are missense/nonsense or result in a premature stop codon. Though minor allele frequencies (MAFs) have not been determined for most, only a relatively few missense SNPs are known to display average MAFs above 2%. The SNP rs10977171 changes amino acid (aa) 444 in PTPRD’s second fibronectin type III domain from Q to E. The MAF of rs10977171 is above 0.11 in Asian samples but only 0.001 in African samples 48. The SNP rs35929428 changes aa 995 in the seventh fibronectin domain from R to either G or to C, with the MAF again lower in African samples (0.005) than in samples with European or Asian ancestries (MAF 0.08–0.14) 49. We are not aware of any experimental or in silico studies that elucidate the functional consequences of these relatively common missense variations. Nevertheless, their relative proximity (22,268 basepairs apart) and the similar distributions of their MAFs in different populations do suggest that they may mark an extended haplotype. The database of genomic variants lists 39 gold standard copy number variants (CNVs), of which all but two are deletions 50.
There is modest evidence for parent-of-origin effects at the locus, and thus possible imprinting. The associations between SNPs in PTPRD introns 7–9 with a metabolic trait, triglycerides in medium low density lipoproteins, display significantly less variation in homozygotes than in heterozygotes, one way in which parent-of-origin effects can manifest themselves 51.
We have sought evidence for cis effects of human PTPRD haplotypes on levels of PTPRD mRNA expression in postmortem human cerebral cortex samples. There are robust individual differences in levels of expression of PTPRD mRNA in these brain samples 52, with 70% differences in levels of PTPRD expression in brains from carriers of major versus minor alleles at an intron 10 SNP, rs2381970 (P = 0.002, remaining significant following Bonferroni correction; Fig. 2). There was also a nominal significance of P ≤ 0.05 for association with levels of PTPRD mRNA expression with SNPs rs4626664 (intron 9, P = 0.04) and rs1975197 (intron 12, P = 0.05; see discussion below of restless leg syndrome), as well as rs7470838 (intron 3, P = 0.04), rs2296094 (intron 36, P = 0.01), and rs10115782 (intron 39, P =0.003).
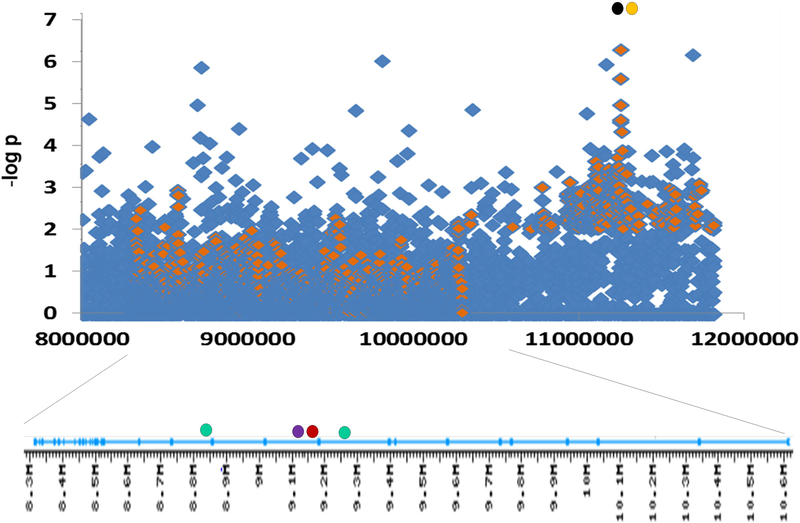
Figure 2.
Associations at PTPRD locus (top) and PTPRD introns (bottom) (Manhattan plot). Diamonds: –log P values for addiction-related phenotypes: smoking cessation success (blue; Uhl et al., unpublished observations) and positive responses to amphetamine (orange; data kindly provided by A. Hart). Filled circles: Intron positions of SNPs most strongly associated with RLS (green), levels of brain PTPRD mRNA expression (red), and neurofibrillary tangle density in Alzheimer’s disease brains (purple), and positions of SNPs most strongly associated with mood lability (orange) and obsessive-compulsive disorder (black). Left is telomeric and 3’, including the most 3’ exon encoding the 3’ untranslated region. Right: 5’ exons 1–12 encode the 5’ untranslated region of common adult brain transcripts.
These data support the possibility that cis-regulatory elements lie in several PTPRD genomic regions. The data do not indicate that any major toxicity is caused by the 70% differences in levels of human PTPRD expression noted above 52. The data also raise questions about genomic mechanisms, such as balancing selection, that might maintain such common, functional PTPRD gene variants in the population. Analyses of genomic variants do identify signatures of balancing selection 53. PTPRD variants, including a SNP in intron 9, display associations with human fecundity 54 that could plausibly contribute to balancing selection at this locus.
Association of PTPRD genomic markers with addiction phenotypes
Addiction phenotypes provided the first evidence for polygenic association with PTPRD variants 55 (Fig. 2). PTPRD has now been identified in human genome-wide SNP and CNV association studies 56–58 of several addiction-related phenotypes, usually by clusters of nearby SNPs or CNVs that each display association with a nominal significance of 10−8 < P < 10−2. Such associations were first identified with the 10K SNP array study of associations with polysubstance dependence (1 of 38 SNPs reported; now known to map to the 3’ flanking region of PTPRD) 55 and continued with studies that included 600K SNP array associations with polysubstance dependence (1 of 119 genes) 59, 1M SNP array associations for illegal substance abuse in African-American subjects from two independent samples (1 of 97 genes) 60, 1M SNP array studies of alcohol use disorder in an epidemiological sample (1 of 9 genes identified in both European- and African-American samples) 61, and genes identified by both SNP and CNV associations for opiate dependence (1 of 3 genes) 62.
The ability to quit smoking in clinical trial settings displayed PTPRD associations in several studies (1 of 33 genes in Ref. 63 and 1 of 116 genes in Ref. 64).
PTPRD was identified by one of the top two GWAS peaks in a study of acute responses to amphetamine administered in a research laboratory setting 65 (A Hart and A Palmer, personal communication, 2014). It was one of 156 genes identified by association with responses to drinking during the “first five” occasions for alcohol consumption 66.
PTPRD variants that reduce expression appear to be associated with reduced vulnerability to substance use disorder phenotypes. Rare CNVs likely to reduce PTPRD expression are more frequent in controls than in opiate-dependent individuals 62. Common haplotypes marked by the SNPs associated with lower PTPRD brain expression are associated with reduced addiction vulnerabilities 52. Some of the 10−8 < P < 10−2 SNP associations with addiction phenotypes do lie in regions of the PTPRD locus in which genomic markers have not been tested for associations with levels of PTPRD mRNA expression or do not display such associations.
Association of PTPRD genomic markers with restless leg syndrome
Variation at the PTPRD locus has provided robust, oligogenic associations with vulnerability to restless leg syndrome (RLS; also known as Willis-Ekbom disorder) 67–70. The magnitude of these association signals is consistent with observations that an RLS linkage peak was centered at the 9p location of the PTPRD gene in a family study 71. The two SNPs that displayed initial association and are the subject of most follow-up work, rs4626664 and rs1975197, lie in PTPRD introns 9 and 12 (Fig. 2). These SNPs display associations with both the RLS diagnoses and the periodic limb movements that are key features of this diagnosis 70. In different samples, the extent of the association signal differs from one of these SNPs to the other. In recent work corrected for covariates, rs1975197 alleles provided odds ratios of about 1.3 for either RLS diagnosis or periodic limb movements 70.
While PTPRD variants thus provide an oligogenic influence on the vulnerability to developing idiopathic RLS, effects on symptomatic RLS associated with other conditions are less clear. This may be due in part to smaller sample sizes in many of these studies. There is negative data from association efforts relevant to putative migraine and multiple sclerosis comorbidities with RLS 72–73. There was a nominally significant association of PTPRD rs4626664 alleles (odds ratio of 1.5) with symptomatic RLS in a sample of uremic patients, but no significant association of rs1975197 74. There was no association with either SNP in another report 75. Though reported as negative, rs1975197 associations with Parkinson’s disease, a RLS comorbidity, displayed odds ratios of 1.3 and 2 under recessive and dominant models, respectively. Suggestive odds ratios for PTPRD markers came from evaluation of association with age of Parkinson’s disease onset and dopamine-related symptoms (rigidity, bradykinesia) but not from association with tremor, a symptom less tightly tied to dopamine function 76.
While addiction phenotypes thus represent the first reported replicable associations with PTPRD, RLS provides the strongest PTPRD associations that have been repeatedly replicated. Furthermore, the two most RLS-associated SNPs display nominal/borderline-significant associations with levels of PTPRD expression and flank rs2381970, the PTPRD SNP that is the most strongly associated with these individual differences in levels of expression. Dopamine is involved in both addiction and RLS 77, and dopamine agonists are major RLS therapeutics 78. Virtually all addictive substances enhance dopamine release in the striatum 79. Though dopaminergic activities do not completely explain either disorder, the effects of PTPRD variation on dopaminergic brain systems could contribute to the noted associations with both addiction and RLS.
Association of PTPRD genomic markers with densities of neurofibrillary pathology in Alzheimer’s disease brain
A recent report examined genomic correlates of the densities of neurofibrillary tangles and of senile plaques in the brains of well-characterized patients who died with Alzheimer’s disease 80. There was a robust association of PTPRD SNPs with neurofibrillary tangle density. The strongest association was for the intron 10 SNP rs560380. There was specificity; there was no such association with the densities of senile plaques. The site of this association signal within the PTPRD gene provides a clear link with levels of PTPRD expression (Fig. 2). The PTPRD SNPs accounted for 3% of the individual differences in neurofibrillary pathology. The only other oligogenic influence was that of the well-known APOE4/TOMM40 variants. There was nominal association with Alzheimer’s pathological diagnosis in this report, though PTPRD has not previously displayed robust associations with Alzheimer’s disease diagnoses 81. MAPK5, a kinase associated with tangle-related tau hyperphosphorylation, is a likely PTPRD dephosphorylation target based on ptp-3 knockout data 32. This possible pathogenic link supports the hypothesis that PTPRD variants provide oligogenic influences on a key neurodegenerative pathogenic component. These data provide one robust link between PTPRD variation and neuropathology, since neither addicted nor RLS brains display reproducible neuropathological changes 82. The results present the exciting possibility that common PPTRD level-of-expression variants might influence neurofibrillary contributions to other neurodegenerative diseases.
Association of PTPRD genomic markers with self-reported mood lability
A recent report describes a robust genetic association between PTPRD SNPs and individual differences in response to the question “Does your mood often goes up and down?” in UK biobank participants 83. A >0.5 Mb zone in the 5’ flanking region of PTPRD is covered by SNPs with nominally significant association, with the minimal P value at rs10959826 (Fig. 2). The effect was the largest reported for mood lability in this sizable sample.
Association of PTPRD genomic markers with personality phenotypes
There are reports of associations between PTPRD genomic variants and personality phenotypes. SNPs in the PTPRD telomeric/3’ flanking region display genome-wide significant associations with openness in a female Korean sample 84 and with agreeableness (but not openness) in another Korean meta-analysis 85. A PTPRD intron 1 SNP displayed polygenic association with the N6 vulnerability neuroticism subscale in Korean samples 86. However, PTPRD is not identified by meta-analyses of personality data genetics in samples of largely U.S. origin 87. Impulsivity or venturesomeness, personality phenotypes that are related to addictions 88, have not been associated with PTPRD. However, such traits might be tested in future studies of the impulsiveness that is associated with substance use disorders.
Association of PTPRD genomic markers with obsessive compulsive disorder
SNPs in the 5’ flanking region of PTPRD were among those most highly associated with obsessive compulsive disorder (OCD) diagnosis in one report (minimal P value at rs2821204) 89; there was a more modest signal in a subsequent meta-analysis 90. These findings are reinforced by linkage of OCD to the chromosome 9p region that contains PTPRD 91–92, and by observations of a greater number of PTPRD copy number variants in pediatric OCD than in controls 93.
Association of PTPRD deletions with cognitive abilities
In each parent in a well-studied family, deletions of parts of one copy of PTPRD failed to cause any notable disability 94. However, their son displayed defects in both PTPRD copies, microcephaly, and failure to develop normal cognitive abilities. These data support human gene dose–dependent effects of PTPRD deletion on cognitive abilities. Failure of GWASs for cognitive abilities to identify PTPRD 95–96 is consistent with the lack of any large effect on cognitive abilities from the common variation that provides 70% individual differences in levels of PTPRD expression.
Mouse models: characterization and support for human PTPRD associations
Mouse model for Ptprd variation: initial characterization
Uetani and colleagues produced Ptprd knockout mice 97 in which the knockout cassette inserts into the tyrosine phosphatase domain of PTPRD. These workers focused largely on differences between wild-type and homozygous knockouts. Homozygotes were smaller than wild-type littermates. For survival, they required food placement on cage floors. Homozygotes were unable to learn the Morris water maze task. Performance on reinforced alternation and radial arm mazes was also reduced in homozygotes. Tests of hippocampal electrophysiological function identified enhanced long term potentiation in CA1 and CA3 hippocampal subfields in slices prepared from brains of homozygous Ptprd knockouts.
Initial histological analyses of brains of homozygous knockouts failed to identify gross or light microscopic differences from brains of wild-type mice 97, though more recent work identified less elaboration of basal dendrites in homozygous knockout cortical pyramidal neurons 34. Even Ptprd knockout combined with knockout of its subfamily member, Ptprs, produced no gross brain histological changes 98. However, spinal motoneurons were substantially lost between embryonic days 13.5 and 18.5 in mice with combined homozygous losses of both Ptprd and Ptprs. There was thus uniform lethality at C-section or birth that appeared to result from respiratory failure. Chick embryos treated with PTPRD antisense RNAs also displayed disorganization of certain peripheral nerves 99. While abnormalities increase in mouse models as knockout of Ptprd is added to knockout of Ptprs, abnormalities decrease in chick models as antisense RNA targets more PTPRD-related RPTPs. There may thus be different interactions between products of PTPRD-related genes in these two species.
Support for human PTPRD associations with mouse model data: addiction phenotypes
Consistent data now supports influences of the reduced Ptprd expression found in heterozygous Ptprd knockout mice on each of two models of stimulant reward 2, 100. Each of these reward models, conditioned place preference (CPP) 101 and self-administration 102, reliably separates virtually all of the substances that humans abuse from those with little human abuse potential, thus displaying substantial validity. Appropriate control experiments for the confounding factors that could muddy the interpretation of results are understood for each of these two tests.
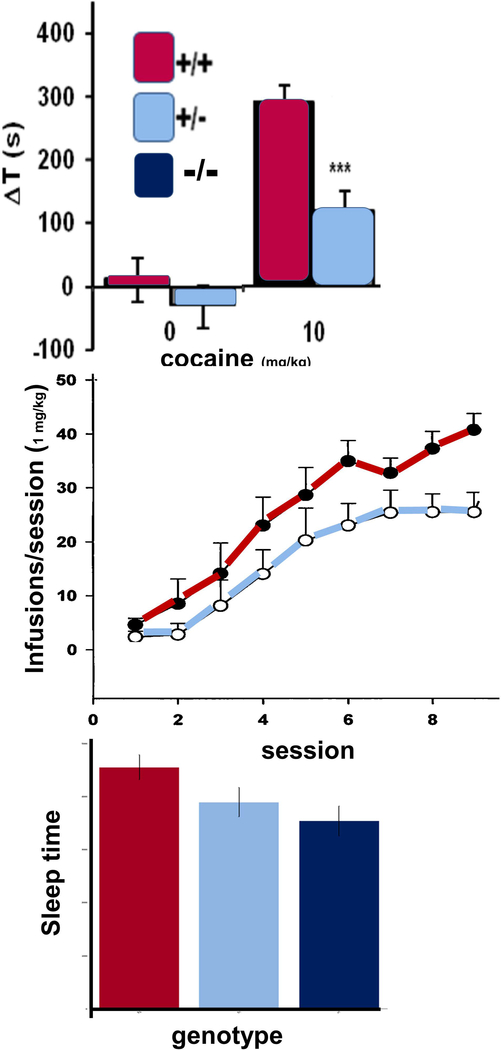
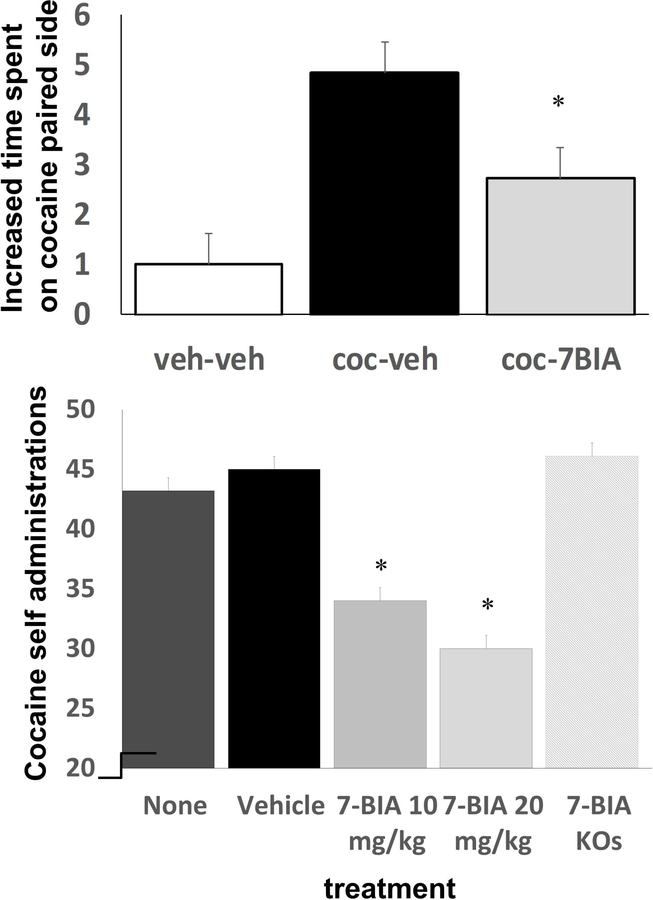
Heterozygous Ptprd knockout mice, which express half of the wild-type levels of Ptprd, display reduced preference for places paired with effects of normally highly rewarding 10 mg/kg doses of cocaine 52 (Fig. 3). Tests of strength, motor function, anxiety, and mnemonic function reveal no obvious confounding effects 52. These results provide additional support for human GWAS observations that variation at the PTPRD locus contributes to both vulnerability to develop a substance use disorder and to individual differences in the rewarding properties of amphetamine doses administered in a laboratory setting.
Figure 3.
Heterozygous Ptprd knockout mice display reduced cocaine-conditioned place preference (top), cocaine self-administration (middle, fixed ratio 1 schedule) and sleep (bottom, 2h around “lights on”). Homozygotes display larger sleep deficits than heterozygotes. All differences are statistically significant.
These CPP results have recently been reinforced by findings from self-administration assays 2. Ptprd heterozygous knockout mice took longer to establish high rates of 1 mg/kg/infusion cocaine self-administration than wild-type littermate controls. During the last three of eight successive three-hour sessions in which up to 50 cocaine infusions were available on fixed ratio 1 (FR1) schedules, heterozygotes self-administered about half of the maximal amounts available, while wild-type littermates self-administered about 70% of the maximal available drug. Heterozygotes continued to self-administer at lower rates than wild-type mice on days 6–8. There were no differences in control responding on inactive levers during FR1 responding.
There is additional support from the results of subsequent progressive ratio studies 2. After several days of experience with these progressive ratio schedules, Ptprd heterozygous knockouts displayed significantly lower break points. They thus self-administered about half the number of infusions that were self-administered by wild-type littermate control mice. Taken together, CPP and self-administration data provide strong support for the polygenic PTPRD associations identified in the human GWAS datasets.
Support for human PTPRD associations with mouse model data: RLS phenotypes
Mice with reduced Ptprd expression display behavioral evidence of sleep disruption during the beginning of the sleep period, the time period when sleep abnormalities are easiest to identify in humans with RLS 52. In assessments of sleep behaviors in the hour prior to and the hour following “lights on” (the start of the normal mouse sleep phase), mice with reduced Ptprd expression had a decreased number of sleep bouts, which decreased total sleep duration and increased the length of sleep bouts, providing evidence of sleep deprivation. While these observations provide face validity in relation to human RLS-associated sleep disturbances, there is a less clear relationship between RLS-associated periodic limb movements and either the twitch-like or the slower movements that can be quantified in mice. We found no effect of PTPRD genotype on either slower or faster twitch-like movements in the hour prior to or the hour after “lights on”. There were effects of Ptprd genotype on overall locomotor activity in several settings. Overall, there is thus good mouse model evidence that validates the effects of human PTPRD variation on sleep but no clear mouse model evidence to validate the effects of PTPRD variation on human periodic limb movements.
Support for human PTPRD associations with mouse model data: other phenotypes
There are striking parallels between the gene dose–dependent effects of reducing Ptprd expression and data relating to human cognitive abilities. Heterozygous knockout mice, humans with the common PTPRD variation that alters expression by perhaps 70%, and humans with a single PTPRD copy deletion all do not display abnormal cognitive test results. The case report of severe intellectual impairment when both copies of human PTPRD are deleted 94 accords well with the severe difficulties displayed by homozygous Ptprd knockout mice in the water maze task 52. Expression of at least 30–50% of wild-type levels of PTPRD appears sufficient for the development of apparently normal cognitive function.
Support for some other human PTPRD associations is not available, based in part on the lack of well-validated mouse models for mood instability, personality phenotypes, OCD, or neurofibrillary Alzheimer’s disease pathology.
Potential for future support of (as yet unelucidated) human motoneuron phenotypes by mouse model data
In mice, combined homozygous deletion of both Ptprd and Ptprs causes remarkable lethality 98. Even mice delivered by Caesarean section are paralyzed, never observed to breathe, and die with severe muscle dysgenesis and loss of spinal cord motoneurons. Phrenic nerves emerge normally from the cervical spinal cord, but stall on reaching the diaphragm. Preservation of a single functional copy of either the Ptprd or Ptprs genes allows significant survival in the background of a homozygous knockout of the other gene, though there were significantly reduced motoneuron densities in Ptprd+/– Ptprs–/– mice. These data predict that combined homozygous deletions of both the chromosome 9 sequence encoding PTPRD and the chromosome 16 sequence encoding PTPRS would cause severe motoneuron phenotypes, though there is no report of such humans. They also predict functional redundancy that should allow substantial differences in function of PTPRD or of PTPRS, individually, without any large consequence for motoneuron or neuromuscular physiologies.
Discovery and effects of the initial PTPRD ligand
Sites for targeting small molecule PTPRD ligands
As noted above, binding of PTPRD by its extracellular ligands is likely to alter the phosphatase activity of its key intracellular D1 phosphatase domain 103. Phosphatase activity of this D1 domain is likely to be reduced by binding interactions with the inactive D2 phosphatase-like domain 30. PTPRD could thus be targeted by compounds that could (1) bind extracellularly in ways that increase its phosphatase activity (e.g., agonist-like); (2) bind extracellularly in ways that decrease its phosphatase activity (e.g., antagonist-like): (3) bind to the active D1 phosphatase domain to decrease its phosphatase activity (e.g., inhibitor-like); or (4) disinhibit the active D1 phosphatase domain by reducing its inhibitory interactions with the D2 domain. Other interactions (e.g., allosteric effects, binding to other extracellular or intracellular modulators) are possible. Interest in the phosphatase portion of PTPRD as a drug target has been increased by the nonconservative amino acid differences between PTPRD and its other DSF subfamily members104.
Small molecule PTPRD phosphatase inhibitor
We have focused on inhibitors of PTPRD’s phosphatase domain as a novel approach to “drugging” PTPRD for several reasons 2 Natural product library screening identified ligands that inhibit PTPRF’s phosphatase activity. Illudalic acid displays micromolar Ki 105–106 in inhibiting PTPRF’s phosphatase. Illudalic acid analogs thus provided exciting clues to possible PTPRD ligands.
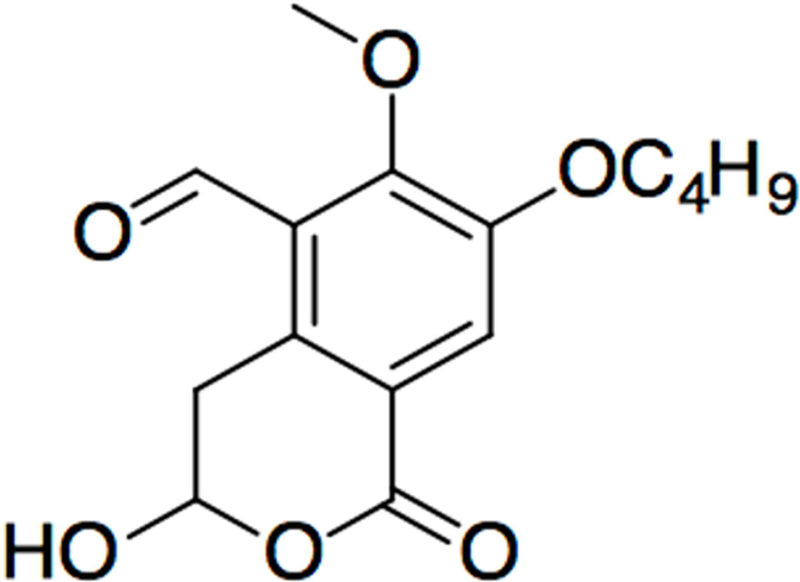
We were able 2 to (1) synthesize a 7-butoxy illudalic acid analog (7-BIA, Fig. 4); (2) produce enzymatically active recombinant human PTPRD and PTPRS D1 phosphatase domain fusion proteins; (3) develop in vitro assays for the activities of the PTPRD and PTPRS phosphatase domain proteins; and (4) use these assays to document significant 7-BIA potency in inhibiting PTPRD and PTPRS phosphatases. IC50 values for 7-BIA preincubated with the fusion protein are about 3 μM (PTPRD) and 40 μM (PTPRS)in assays in which preincubated protein is diluted 10-fold No tested precursor significantly inhibited PTPRD phosphatase fusion protein’s activity. 7-BIA failed to provide significant inhibition of any of the more than 85 transporter, receptor, enzymatic, or other sites assessed by Eurofins and Dr. A. Janowsky. Based on observation, behavioral tests, autopsy by an experienced veterinarian, and major organ histology evaluated by experienced veterinary pathologists, we found no dose-related behavioral or organ toxicity or peritoneal inflammation in mice sacrificed minutes or weeks following injections of doses limited by 7-BIA solubility in DMSO and maximal 5% DMSO volumes that mice could receive.
Figure 4.
Structure of the first PTPRD phosphatase inhibitor 7-BIA (7-butoxy illudalic acid analog).
Small molecule PTPRD phosphatase inhibitor reduces cocaine reward in two behavioral tests
Effects of 7-BIA on cocaine conditioned place preference.
We administered 7-BIA 90 min prior to each of the two 10 mg/kg cocaine conditioning sessions 2. Mice pretreated with DMSO vehicle displayed preferences for places paired with 10 mg/kg cocaine that were similar to those that we have previously reported in wild-type C57BL/6 mice 107. However, these preferences for cocaine-paired environments were significantly reduced in mice pretreated with 7-BIA (Fig. 5, top). There were no significant 7-BIA effects on locomotor speed or distance traveled. Apparent transient inhibition of PTPRD phosphatase activity influenced a measure of cocaine reward with behavioral specificity. This did not appear to be caused by preference or aversions induced by 7-BIA itself. Pairing with effects of 7-BIA provided no change in preference/aversion for environments in which it was administered.
Figure 5.
7-BIA pretreatment reduces cocaine-conditioned place preference (top) and self-administration (bottom) in wild-type but not Ptprd knockout (KO) mice.
Effects of 7-BIA on well-established cocaine self-administration.
Wild-type and Ptprd heterozygous mice were allowed to self-administer 0.5–1 mg/kg/infusion cocaine for at least 20 sessions so that they were self-administering >40 doses during 3-hour sessions with a 50-dose limit 2. 7-BIA–pretreated mice failed to obtain at least 40 cocaine doses during 52% of sessions, compared to the about 20% of sessions in which vehicle/non-pretreated mice failed to obtain at least 40 doses (Fig. 5, bottom). Mice also failed to self-administer at least 40 cocaine doses during the next session (1–2 days following single 7-BIA treatments) and returned to >40 doses/session in subsequent sessions. Most, if not all, of these 7-BIA effects appear to be attributable to the compound’s influences on PTPRD. Heterozygous Ptprd knockout mice treated with 7-BIA only failed to obtain >40 cocaine doses in 9% of sessions
PTPRD variation and brain phenotypes: current support
Substance reward and substance use disorders
There is compelling evidence for PTPRD roles in addiction that comes from the combination of data from: (1) human PTPRD associations with individual differences in (a) responses to stimulant and alcohol administration, (b) vulnerability to develop a substance use disorder, and (c) ability to quit smoking; as well as (2) mouse studies of effects of (a) lifelong alterations of the levels of PTPRD expression and (b) acute pharmacological inhibition of PTPRD’s phosphatase activity.
There remain caveats for the human findings by themself since no single PTPRD SNP provides consistent P < 10−8 association or any evidence for oligogenic influences on these addiction-related phenotypes. Taken together with the mouse model genetic and pharmacological data, however, there seems little a posteriori doubt that (1) PTPRD variation plays polygenic roles in human addiction-related phenotypes and that (2) changes in mouse PTPRD expression or activity can play larger roles in stimulant reward (when, of course, there is more control for genetic background and environmental influences than in human studies). PTPRD provides a novel and interesting target for anti-addiction medications development.
Restless leg syndrome
The combination of human and mouse model data also provides strong support for the roles of individual differences in PTPRD expression on the sleep disruption associated with RLS. Human data also supports roles for PTPRD variation in the RLS-associated periodic limb movements in sleep. The oligogenic human associations with RLS, indeed, are of at least as impressive magnitude as the significant, though moderate, differences in sleep parameters observed in mice with reduced Ptprd expression. There is no well-validated, well-established mouse model for periodic limb movements. Thus, it may not be surprising that our efforts to assess quicker and slower limb movements in mice around the “lights on” natural start of their sleep cycles might not reflect either bona fide RLS pathophysiology or PTPRD effects on this physiology. It is notable that studies of a mouse models of another RLS oligogene, Btbd9, also identified effects on sleep 108. Mice with altered expression of Btbd9, the RLS oligogene Meis1 109, and Ptprd 52 each exhibited increased locomotion (termed “restlessness” in one report), but none manifest clear-cut periodic limb movements.
Cognitive abilities
Human and mouse data provide mutual support for both effects of altered PTPRD expression on cognitive abilities and for gene dose-response relationships. Deletion of all PTPRD expression provides marked cognitive deficits in both mouse models and the very rare humans homozygous for such deletions. By contrast, the scope for substantial variation in levels of PTPRD function before deficits in cognitive function become notable is strongly supported by (1) the failure of human deletion heterozygotes or mouse Ptprd knockout heterozygotes to demonstrate significant cognitive impairments; (2) the failure to identify PTPRD in cognitive function GWAS; and (3) our findings that common haplotypes confer 70% individual differences in levels of PTPRD expression. Such data provide reassurance for the development of compounds that transiently reduce PTPRD function.
Neurofibrillary Alzheimer’s pathology, personality, and obsessive compulsive disorder
The careful recent report that identified PTPRD association with the extent of neurofibrillary pathology in Alzheimer’s disease brain specimens is impressive 110. However, this association comes from only a single publication to date. There is no supporting mouse model data of which we are aware. Personality associations are variable in human studies; there are only a few mouse models with substantial validation. There are a number of face-valid models for features of OCD, but relatively few of these models have been validated pharmacologically 111 and none have been applied to PTPRD, to our knowledge. These associations thus merit efforts for replication in humans and generalization to mouse models.
A note about cancer and other systemic phenotypes
There are reported associations between PTPRD variation and a number of cancers or their responses to treatment 112–128. While the details of these associations are beyond the scope of this paper, we note that most come from genotyping tumors or tumor-derived cell lines and that none of these associations focuses on the mid-gene regions that we have associated with a number of brain phenotypes and levels of PTPRD expression. Neither we nor others have identified any tumors in studies of thousands of mice with altered Ptprd expression.
There are also reported associations with susceptibilities to diabetes 129, treatment-resistant hypertension 130, weight gain associated with neuroleptic treatments 131, and craniofacial abnormalities with hearing deficits 94, though neither we nor others have identified these phenotypes in mice with altered Ptprd expression.
As we and others continue to develop drugs that target PTPRD, we must nevertheless continue to be alert to potential for unwanted carcinogenesis and other systemic phenotypes (as for all drugs that engage novel targets).
Neurobiology and genetics of PTPRD and brain phenotypes
The neurobiology of PTPRD, even incompletely understood, places this receptor type protein tyrosine phosphatase in a central position in regulating the development and modulation of interesting, PTPRD-expressing brain circuits. Clearer understanding of this neurobiology, and the consequences of acute and chronic modulation of PTPRD function, sharpens our pictures of the complex physiology and pathophysiology of PTPRD-expressing neurons and circuits.
PTPRD genetics, also partially understood, is consistent with both major/oligogenic and modest/polygenic contributions of common and rare PTPRD variation on common disorders: substance use disorders, RLS, Alzheimer’s disease and perhaps other neurodegenerative diseases. Mouse models link lifetime alterations in brain PTPRD expression to human phenotypes. Our lead compound work (gratifyingly) supports the idea that even acute changes in PTPRD function can have specific effects on important phenotypes, including in mouse models of stimulant reward.
Recent bioinformatic work has underscored the ways in which availability of pharmacological tools, publications, and grant funding focus on the products of a modest fraction of the genome, in ways that have not included PTPRD to date 132. PTPRD remains an interesting molecule, an apparently druggable drug target and a marker for and an important constituent of brain circuits of likely importance for major brain-based phenotypes. This interesting gene is thus a prime candidate target for funding, pharmacological work, and increasing therapeutic advances in addictions and other disorders.
Acknowledgments
We are grateful to support from the Biomedical Research Institute of New Mexico, Veterans’ Health Administration and NIH (NIDA and NIAAA).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Uhl GR; Grow RW, The burden of complex genetics in brain disorders. Arch Gen Psychiatry 2004, 61 (3), 223–9. [DOI] [PubMed] [Google Scholar]
- 2.Uhl GR; Martinez MJ; Paik P; Sulima A; Bi GH; Iyer MR; Gardner E; Rice KC; Xi ZX, Cocaine reward is reduced by decreased expression of receptor-type protein tyrosine phosphatase D (PTPRD) and by a novel PTPRD antagonist. Proc Natl Acad Sci U S A 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krueger NX; Streuli M; Saito H, Structural diversity and evolution of human receptor-like protein tyrosine phosphatases. EMBO J 1990, 9 (10), 3241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuno K; Hasegawa K; Katagiri T; Ogimoto M; Ichikawa T; Yakura H, MPTP delta, a putative murine homolog of HPTP delta, is expressed in specialized regions of the brain and in the B-cell lineage. Mol Cell Biol 1993, 13 (9), 5513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulido R; Serra-Pages C; Tang M; Streuli M, The LAR/PTP delta/PTP sigma subfamily of transmembrane protein-tyrosine-phosphatases: multiple human LAR, PTP delta, and PTP sigma isoforms are expressed in a tissue-specific manner and associate with the LAR-interacting protein LIP.1. Proc Natl Acad Sci U S A 1995, 92 (25), 11686–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulido R; Krueger NX; Serra-Pages C; Saito H; Streuli M, Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem 1995, 270 (12), 6722–8. [DOI] [PubMed] [Google Scholar]
- 7. https://www.ncbi.nlm.nih.gov/projects/mapview/maps.cgi?TAXID=9606&QUERY=PTPRD&chr=9&maps=genes-r&beg=8200000&end=10700000&links=off&verbose=on&compress=off&width=350&size=30.
- 8.Zhong X; Drgonova J; Li CY; Uhl GR, Human cell adhesion molecules: annotated functional subtypes and overrepresentation of addiction-associated genes. Ann N Y Acad Sci 2015, 1349, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foldy C; Darmanis S; Aoto J; Malenka RC; Quake SR; Sudhof TC, Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc Natl Acad Sci U S A 2016, 113 (35), E5222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. https://www.ncbi.nlm.nih.gov/gene/5789.
- 11. https://www.ncbi.nlm.nih.gov/genome/gdv/browser/?comtext=genome&acc=GCF_000001405.37A.
- 12. https://www.uniprot.org/uniprot/P23468.
- 13.Shishikura M; Nakamura F; Yamashita N; Uetani N; Iwakura Y; Goshima Y, Expression of receptor protein tyrosine phosphatase delta, PTPdelta, in mouse central nervous system. Brain Res 2016, 1642, 244–254. [DOI] [PubMed] [Google Scholar]
- 14.Forrest AR; Taylor DF; Crowe ML; Chalk AM; Waddell NJ; Kolle G; Faulkner GJ; Kodzius R; Katayama S; Wells C; Kai C; Kawai J; Carninci P; Hayashizaki Y; Grimmond SM, Genome-wide review of transcriptional complexity in mouse protein kinases and phosphatases. Genome Biol 2006, 7 (1), R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streuli M; Krueger NX; Tsai AY; Saito H, A family of receptor-linked protein tyrosine phosphatases in humans and Drosophila. Proc Natl Acad Sci U S A 1989, 86 (22), 8698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J; Bixby JL, Receptor tyrosine phosphatase-delta is a homophilic, neurite-promoting cell adhesion molecular for CNS neurons. Mol Cell Neurosci 1999, 14 (4–5), 370–84. [DOI] [PubMed] [Google Scholar]
- 17.Yamagata A; Sato Y; Goto-Ito S; Uemura T; Maeda A; Shiroshima T; Yoshida T; Fukai S, Structure of Slitrk2-PTPdelta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion. Sci Rep 2015, 5, 9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi H; Katayama K; Sohya K; Miyamoto H; Prasad T; Matsumoto Y; Ota M; Yasuda H; Tsumoto T; Aruga J; Craig AM, Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat Neurosci 2012, 15 (3), 389–98, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Um JW; Kim KH; Park BS; Choi Y; Kim D; Kim CY; Kim SJ; Kim M; Ko JS; Lee SG; Choii G; Nam J; Heo WD; Kim E; Lee JO; Ko J; Kim HM, Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion. Nat Commun 2014, 5, 5423. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida T; Shiroshima T; Lee SJ; Yasumura M; Uemura T; Chen X; Iwakura Y; Mishina M, Interleukin-1 receptor accessory protein organizes neuronal synaptogenesis as a cell adhesion molecule. J Neurosci 2012, 32 (8), 2588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamagata A; Yoshida T; Sato Y; Goto-Ito S; Uemura T; Maeda A; Shiroshima T; Iwasawa-Okamoto S; Mori H; Mishina M; Fukai S, Mechanisms of splicing-dependent trans-synaptic adhesion by PTPdelta-IL1RAPL1/IL-1RAcP for synaptic differentiation. Nat Commun 2015, 6, 6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo J; Kwon SK; Choi S; Kim S; Lee JR; Dunah AW; Sheng M; Kim E, Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci 2009, 12 (4), 428–37. [DOI] [PubMed] [Google Scholar]
- 23.Shen Y; Tenney AP; Busch SA; Horn KP; Cuascut FX; Liu K; He Z; Silver J; Flanagan JG, PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 2009, 326 (5952), 592–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coles CH; Shen Y; Tenney AP; Siebold C; Sutton GC; Lu W; Gallagher JT; Jones EY; Flanagan JG; Aricescu AR, Proteoglycan-specific molecular switch for RPTPsigma clustering and neuronal extension. Science 2011, 332 (6028), 484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lie E; Li Y; Kim R; Kim E, SALM/Lrfn Family Synaptic Adhesion Molecules. Front Mol Neurosci 2018, 11, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto-Ito S; Yamagata A; Sato Y; Uemura T; Shiroshima T; Maeda A; Imai A; Mori H; Yoshida T; Fukai S, Structural basis of trans-synaptic interactions between PTPdelta and SALMs for inducing synapse formation. Nat Commun 2018, 9 (1), 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodings JA; Sharp SJ; Machesky LM, MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem J 2003, 371 (Pt 2), 463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J; Lin S; Wang M; Liang L; Zou Z; Zhou X; Wang M; Chen P; Wang Y, Metastasis suppressor 1 regulates neurite outgrowth in primary neuron cultures. Neuroscience 2016, 333, 123–31. [DOI] [PubMed] [Google Scholar]
- 29.Wallace MJ; Fladd C; Batt J; Rotin D, The second catalytic domain of protein tyrosine phosphatase delta (PTP delta) binds to and inhibits the first catalytic domain of PTP sigma. Mol Cell Biol 1998, 18 (5), 2608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchetot C; Tertoolen LG; Overvoorde J; den Hertog J, Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J Biol Chem 2002, 277 (49), 47263–9. [DOI] [PubMed] [Google Scholar]
- 31.Selner NG; Luechapanichkul R; Chen X; Neel BG; Zhang ZY; Knapp S; Bell CE; Pei D, Diverse levels of sequence selectivity and catalytic efficiency of protein-tyrosine phosphatases. Biochemistry 2014, 53 (2), 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell CJ; Kim MS; Zhong J; Nirujogi RS; Bose AK; Pandey A, Unbiased identification of substrates of protein tyrosine phosphatase ptp-3 in C. elegans. Mol Oncol 2016, 10 (6), 910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julien SG; Dube N; Hardy S; Tremblay ML, Inside the human cancer tyrosine phosphatome. Nat Rev Cancer 2011, 11 (1), 35–49. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura F; Okada T; Shishikura M; Uetani N; Taniguchi M; Yagi T; Iwakura Y; Ohshima T; Goshima Y; Strittmatter SM, Protein Tyrosine Phosphatase delta Mediates the Sema3A-Induced Cortical Basal Dendritic Arborization through the Activation of Fyn Tyrosine Kinase. J Neurosci 2017, 37 (30), 7125–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer L; Rao M; Anderson DJ, RPTP delta and the novel protein tyrosine phosphatase RPTP psi are expressed in restricted regions of the developing central nervous system. Dev Dyn 1997, 208 (1), 48–61. [DOI] [PubMed] [Google Scholar]
- 36. http://mouse.brain-map.org/experiment/show?id=855.
- 37.Dong X; Liao Z; Gritsch D; Hadzhiev Y; Bai Y; Locascio JJ; Guennewig B; Liu G; Blauwendraat C; Wang T; Adler CH; Hedreen JC; Faull RLM; Frosch MP; Nelson PT; Rizzu P; Cooper AA; Heutink P; Beach TG; Mattick JS; Muller F; Scherzer CR, Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat Neurosci 2018, 21 (10), 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. http://hipposeq.janelia.org/full/t1/C4D0A824-369B-11E8-874A-A4F501797C37/
- 39.Tasic B; Menon V; Nguyen TN; Kim TK; Jarsky T; Yao Z; Levi B; Gray LT; Sorensen SA; Dolbeare T; Bertagnolli D; Goldy J; Shapovalova N; Parry S; Lee C; Smith K; Bernard A; Madisen L; Sunkin SM; Hawrylycz M; Koch C; Zeng H, Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 2016, 19 (2), 335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. http://casesstudies.brain-map.org/celltax#section_explorea.
- 41.Zhu Q; Tan Z; Zhao S; Huang H; Zhao X; Hu X; Zhang Y; Shields CB; Uetani N; Qiu M, Developmental expression and function analysis of protein tyrosine phosphatase receptor type D in oligodendrocyte myelination. Neuroscience 2015, 308, 106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feinberg K; Eshed-Eisenbach Y; Frechter S; Amor V; Salomon D; Sabanay H; Dupree JL; Grumet M; Brophy PJ; Shrager P; Peles E, A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron 2010, 65 (4), 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. http://mouse.brain-map.org/experiment/show?id=1462.
- 44. http://mouse.brain-map.org/experiments/show?id=70278045.
- 45. http://mouse.brain-map.org/experiment/show?id=69874150.
- 46. http://mouse.brain-map.org/experiment/show?id=74882837.
- 47. http://mouse.brain-map.org/experiment/show?id=69874144.
- 48. https://www.ncbi.nlm.gov/projects/SNP/snp_ref.cgi?rs=10977171.
- 49. https://www.ncbi.nlm.nih.gov/projects/SNP/snp-ref.cgi?rs=35929428.
- 50. http://dgv.tcag.ca/gb2/gbrowse.dgv2_hg19/?name=id:2888025;dbid=gene:database.
- 51.Pervjakova N; Kukushkina V; Haller T; Kasela S; Joensuu A; Kristiansson K; Annilo T; Perola M; Salomaa V; Jousilahti P; Metspalu A; Magi R, Genome-wide analysis of nuclear magnetic resonance metabolites revealed parent-of-origin effect on triglycerides in medium very low-density lipoprotein in PTPRD gene. Biomark Med 2018, 12 (5), 439–446. [DOI] [PubMed] [Google Scholar]
- 52.Drgonova J; Walther D; Wang KJ; Hartstein GL; Lochte B; Troncoso J; Uetani N; Iwakura Y; Uhl GR, Mouse Model for Protein Tyrosine Phosphatase D (PTPRD) Associations with Restless Leg Syndrome or Willis-Ekbom Disease and Addiction: Reduced Expression Alters Locomotion, Sleep Behaviors and Cocaine-Conditioned Place Preference. Mol Med 2015, 21 (1), 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitarello BD; de Filippo C; Teixeira JC; Schmidt JM; Kleinert P; Meyer D; Andres AM, Signatures of Long-Term Balancing Selection in Human Genomes. Genome Biol Evol 2018, 10 (3), 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barban N; Jansen R; de Vlaming R; Vaez A; Mandemakers JJ; Tropf FC; Shen X; Wilson JF; Chasman DI; Nolte IM; Tragante V; van der Laan SW; Perry JR; Kong A; Consortium B; Ahluwalia TS; Albrecht E; Yerges-Armstrong L; Atzmon G; Auro K; Ayers K; Bakshi A; Ben-Avraham D; Berger K; Bergman A; Bertram L; Bielak LF; Bjornsdottir G; Bonder MJ; Broer L; Bui M; Barbieri C; Cavadino A; Chavarro JE; Turman C; Concas MP; Cordell HJ; Davies G; Eibich P; Eriksson N; Esko T; Eriksson J; Falahi F; Felix JF; Fontana MA; Franke L; Gandin I; Gaskins AJ; Gieger C; Gunderson EP; Guo X; Hayward C; He C; Hofer E; Huang H; Joshi PK; Kanoni S; Karlsson R; Kiechl S; Kifley A; Kluttig A; Kraft P; Lagou V; Lecoeur C; Lahti J; Li-Gao R; Lind PA; Liu T; Makalic E; Mamasoula C; Matteson L; Mbarek H; McArdle PF; McMahon G; Meddens SF; Mihailov E; Miller M; Missmer SA; Monnereau C; van der Most PJ; Myhre R; Nalls MA; Nutile T; Kalafati IP; Porcu E; Prokopenko I; Rajan KB; Rich-Edwards J; Rietveld CA; Robino A; Rose LM; Rueedi R; Ryan KA; Saba Y; Schmidt D; Smith JA; Stolk L; Streeten E; Tonjes A; Thorleifsson G; Ulivi S; Wedenoja J; Wellmann J; Willeit P; Yao J; Yengo L; Zhao JH; Zhao W; Zhernakova DV; Amin N; Andrews H; Balkau B; Barzilai N; Bergmann S; Biino G; Bisgaard H; Bonnelykke K; Boomsma DI; Buring JE; Campbell H; Cappellani S; Ciullo M; Cox SR; Cucca F; Toniolo D; Davey-Smith G; Deary IJ; Dedoussis G; Deloukas P; van Duijn CM; de Geus EJ; Eriksson JG; Evans DA; Faul JD; Sala CF; Froguel P; Gasparini P; Girotto G; Grabe HJ; Greiser KH; Groenen PJ; de Haan HG; Haerting J; Harris TB; Heath AC; Heikkila K; Hofman A; Homuth G; Holliday EG; Hopper J; Hypponen E; Jacobsson B; Jaddoe VW; Johannesson M; Jugessur A; Kahonen M; Kajantie E; Kardia SL; Keavney B; Kolcic I; Koponen P; Kovacs P; Kronenberg F; Kutalik Z; La Bianca M; Lachance G; Iacono WG; Lai S; Lehtimaki T; Liewald DC; LifeLines Cohort S; Lindgren CM; Liu Y; Luben R; Lucht M; Luoto R; Magnus P; Magnusson PK; Martin NG; McGue M; McQuillan R; Medland SE; Meisinger C; Mellstrom D; Metspalu A; Traglia M; Milani L; Mitchell P; Montgomery GW; Mook-Kanamori D; de Mutsert R; Nohr EA; Ohlsson C; Olsen J; Ong KK; Paternoster L; Pattie A; Penninx BW; Perola M; Peyser PA; Pirastu M; Polasek O; Power C; Kaprio J; Raffel LJ; Raikkonen K; Raitakari O; Ridker PM; Ring SM; Roll K; Rudan I; Ruggiero D; Rujescu D; Salomaa V; Schlessinger D; Schmidt H; Schmidt R; Schupf N; Smit J; Sorice R; Spector TD; Starr JM; Stockl D; Strauch K; Stumvoll M; Swertz MA; Thorsteinsdottir U; Thurik AR; Timpson NJ; Tung JY; Uitterlinden AG; Vaccargiu S; Viikari J; Vitart V; Volzke H; Vollenweider P; Vuckovic D; Waage J; Wagner GG; Wang JJ; Wareham NJ; Weir DR; Willemsen G; Willeit J; Wright AF; Zondervan KT; Stefansson K; Krueger RF; Lee JJ; Benjamin DJ; Cesarini D; Koellinger PD; den Hoed M; Snieder H; Mills MC, Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet 2016, 48 (12), 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu QR; Drgon T; Walther D; Johnson C; Poleskaya O; Hess J; Uhl GR, Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proc Natl Acad Sci U S A 2005, 102 (33), 11864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishiguro H; Liu QR; Gong JP; Hall FS; Ujike H; Morales M; Sakurai T; Grumet M; Uhl GR, NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology 2006, 31 (3), 572–84. [DOI] [PubMed] [Google Scholar]
- 57.Fowler CD; Lu Q; Johnson PM; Marks MJ; Kenny PJ, Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature 2011, 471 (7340), 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson KJ; Marks MJ; Vann RE; Chen X; Gamage TF; Warner JA; Damaj MI, Role of alpha5 nicotinic acetylcholine receptors in pharmacological and behavioral effects of nicotine in mice. J Pharmacol Exp Ther 2010, 334 (1), 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu QR; Drgon T; Johnson C; Walther D; Hess J; Uhl GR, Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet 2006, 141B (8), 918–25. [DOI] [PubMed] [Google Scholar]
- 60.Drgon T; Johnson CA; Nino M; Drgonova J; Walther DM; Uhl GR, “Replicated” genome wide association for dependence on illegal substances: genomic regions identified by overlapping clusters of nominally positive SNPs. Am J Med Genet B Neuropsychiatr Genet 2011, 156 (2), 125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung J, Zhang H, Grant B, Chou P. Identification of novel genetic variants of DSM-5 Alcohol Use Disorder Am Soc Human Genetics presentation, 2017 [Google Scholar]
- 62.Li D; Zhao H; Kranzler HR; Li MD; Jensen KP; Zayats T; Farrer LA; Gelernter J, Genome-wide association study of copy number variations (CNVs) with opioid dependence. Neuropsychopharmacology 2015, 40 (4), 1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhl GR; Liu QR; Drgon T; Johnson C; Walther D; Rose JE, Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet 2007, 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhl GR; Liu QR; Drgon T; Johnson C; Walther D; Rose JE; David SP; Niaura R; Lerman C, Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry 2008, 65 (6), 683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hart AB; Engelhardt BE; Wardle MC; Sokoloff G; Stephens M; de Wit H; Palmer AA, Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CDH13). PLoS One 2012, 7 (8), e42646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joslyn G; Ravindranathan A; Brush G; Schuckit M; White RL, Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res 2010, 34 (5), 800–12. [DOI] [PubMed] [Google Scholar]
- 67.Schormair B; Kemlink D; Roeske D; Eckstein G; Xiong L; Lichtner P; Ripke S; Trenkwalder C; Zimprich A; Stiasny-Kolster K; Oertel W; Bachmann CG; Paulus W; Hogl B; Frauscher B; Gschliesser V; Poewe W; Peglau I; Vodicka P; Vavrova J; Sonka K; Nevsimalova S; Montplaisir J; Turecki G; Rouleau G; Gieger C; Illig T; Wichmann HE; Holsboer F; Muller-Myhsok B; Meitinger T; Winkelmann J, PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet 2008, 40 (8), 946–8. [DOI] [PubMed] [Google Scholar]
- 68.Yang Q; Li L; Yang R; Shen GQ; Chen Q; Foldvary-Schaefer N; Ondo WG; Wang QK, Family-based and population-based association studies validate PTPRD as a risk factor for restless legs syndrome. Mov Disord 2011, 26 (3), 516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim MK; Cho YW; Shin WC; Cho JW; Shon YM; Kim JH; Yang KI; Earley CJ; Allen RP, Association of restless legs syndrome variants in Korean patients with restless legs syndrome. Sleep 2013, 36 (12), 1787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore H. t.; Winkelmann J; Lin L; Finn L; Peppard P; Mignot E, Periodic leg movements during sleep are associated with polymorphisms in BTBD9, TOX3/BC034767, MEIS1, MAP2K5/SKOR1, and PTPRD. Sleep 2014, 37 (9), 1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen S; Ondo WG; Rao S; Li L; Chen Q; Wang Q, Genomewide linkage scan identifies a novel susceptibility locus for restless legs syndrome on chromosome 9p. Am J Hum Genet 2004, 74 (5), 876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuh JL; Chung MY; Yao SC; Chen PK; Liao YC; Hsu CL; Wang PJ; Wang YF; Chen SP; Fann CS; Kao LS; Wang SJ, Susceptible genes of restless legs syndrome in migraine. Cephalalgia 2016, 36 (11), 1028–1037. [DOI] [PubMed] [Google Scholar]
- 73.Vavrova J; Kemlink D; Sonka K; Havrdova E; Horakova D; Pardini B; Muller-Myhsok B; Winkelmann J, Restless legs syndrome in Czech patients with multiple sclerosis: an epidemiological and genetic study. Sleep Med 2012, 13 (7), 848–51. [DOI] [PubMed] [Google Scholar]
- 74.Lin CH; Chen ML; Wu VC; Li WY; Sy HN; Wu SL; Chang CC; Chiu PF; Liou HH; Lin CY; Chang HW; Lin SY; Wu KD; Chen YM; Wu RM, Association of candidate genetic variants with restless legs syndrome in end stage renal disease: a multicenter case-control study in Taiwan. Eur J Neurol 2014, 21 (3), 492–8. [DOI] [PubMed] [Google Scholar]
- 75.Schormair B; Plag J; Kaffe M; Gross N; Czamara D; Samtleben W; Lichtner P; Strohle A; Stefanidis I; Vainas A; Dardiotis E; Sakkas GK; Gieger C; Muller-Myhsok B; Meitinger T; Heemann U; Hadjigeorgiou GM; Oexle K; Winkelmann J, MEIS1 and BTBD9: genetic association with restless leg syndrome in end stage renal disease. J Med Genet 2011, 48 (7), 462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gan-Or Z; Alcalay RN; Bar-Shira A; Leblond CS; Postuma RB; Ben-Shachar S; Waters C; Johnson A; Levy O; Mirelman A; Gana-Weisz M; Dupre N; Montplaisir J; Giladi N; Fahn S; Xiong L; Dion PA; Orr-Urtreger A; Rouleau GA, Genetic markers of Restless Legs Syndrome in Parkinson disease. Parkinsonism Relat Disord 2015, 21 (6), 582–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Earley CJ; Uhl GR; Clemens S; Ferre S, Connectome and molecular pharmacological differences in the dopaminergic system in restless legs syndrome (RLS): plastic changes and neuroadaptations that may contribute to augmentation. Sleep Med 2017, 31, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anguelova GV; Vlak MHM; Kurvers AGY; Rijsman RM, Pharmacologic and Nonpharmacologic Treatment of Restless Legs Syndrome. Sleep Med Clin 2018, 13 (2), 219–230. [DOI] [PubMed] [Google Scholar]
- 79.Di Chiara G; Imperato A, Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A 1988, 85 (14), 5274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chibnik LB; White CC; Mukherjee S; Raj T; Yu L; Larson EB; Montine TJ; Keene CD; Sonnen J; Schneider JA; Crane PK; Shulman JM; Bennett DA; De Jager PL, Susceptibility to neurofibrillary tangles: role of the PTPRD locus and limited pleiotropy with other neuropathologies. Mol Psychiatry 2018, 23 (6), 1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raghavan N; Tosto G, Genetics of Alzheimer’s Disease: the Importance of Polygenic and Epistatic Components. Curr Neurol Neurosci Rep 2017, 17 (10), 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pittock SJ; Parrett T; Adler CH; Parisi JE; Dickson DW; Ahlskog JE, Neuropathology of primary restless leg syndrome: absence of specific tau- and alpha-synuclein pathology. Mov Disord 2004, 19 (6), 695–9. [DOI] [PubMed] [Google Scholar]
- 83.Ward J; Strawbridge RJ; Bailey MES; Graham N; Ferguson A; Lyall DM; Cullen B; Pidgeon LM; Cavanagh J; Mackay DF; Pell JP; O’Donovan M; Escott-Price V; Smith DJ, Genome-wide analysis in UK Biobank identifies four loci associated with mood instability and genetic correlation with major depressive disorder, anxiety disorder and schizophrenia. Transl Psychiatry 2017, 7 (11), 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim HN; Roh SJ; Sung YA; Chung HW; Lee JY; Cho J; Shin H; Kim HL, Genome-wide association study of the five-factor model of personality in young Korean women. J Hum Genet 2013, 58 (10), 667–74. [DOI] [PubMed] [Google Scholar]
- 85.Kim BH; Kim HN; Roh SJ; Lee MK; Yang S; Lee SK; Sung YA; Chung HW; Cho NH; Shin C; Sung J; Kim HL, GWA meta-analysis of personality in Korean cohorts. J Hum Genet 2015, 60 (8), 455–60. [DOI] [PubMed] [Google Scholar]
- 86.Kim SE; Kim HN; Yun YJ; Heo SG; Cho J; Kwon MJ; Chang Y; Ryu S; Shin H; Shin C; Cho NH; Sung YA; Kim HL, Meta-analysis of genome-wide SNP- and pathway-based associations for facets of neuroticism. J Hum Genet 2017, 62 (10), 903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Moor MH; Costa PT; Terracciano A; Krueger RF; de Geus EJ; Toshiko T; Penninx BW; Esko T; Madden PA; Derringer J; Amin N; Willemsen G; Hottenga JJ; Distel MA; Uda M; Sanna S; Spinhoven P; Hartman CA; Sullivan P; Realo A; Allik J; Heath AC; Pergadia ML; Agrawal A; Lin P; Grucza R; Nutile T; Ciullo M; Rujescu D; Giegling I; Konte B; Widen E; Cousminer DL; Eriksson JG; Palotie A; Peltonen L; Luciano M; Tenesa A; Davies G; Lopez LM; Hansell NK; Medland SE; Ferrucci L; Schlessinger D; Montgomery GW; Wright MJ; Aulchenko YS; Janssens AC; Oostra BA; Metspalu A; Abecasis GR; Deary IJ; Raikkonen K; Bierut LJ; Martin NG; van Duijn CM; Boomsma DI, Meta-analysis of genome-wide association studies for personality. Mol Psychiatry 2012, 17 (3), 337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kozak K; Lucatch AM; Lowe DJE; Balodis IM; MacKillop J; George TP, The neurobiology of impulsivity and substance use disorders: implications for treatment. Ann N Y Acad Sci 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mattheisen M; Samuels JF; Wang Y; Greenberg BD; Fyer AJ; McCracken JT; Geller DA; Murphy DL; Knowles JA; Grados MA; Riddle MA; Rasmussen SA; McLaughlin NC; Nurmi EL; Askland KD; Qin HD; Cullen BA; Piacentini J; Pauls DL; Bienvenu OJ; Stewart SE; Liang KY; Goes FS; Maher B; Pulver AE; Shugart YY; Valle D; Lange C; Nestadt G, Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol Psychiatry 2015, 20 (3), 337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.International Obsessive Compulsive Disorder Foundation Genetics C; Studies OCDCGA, Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry 2018, 23 (5), 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanna GL; Veenstra-VanderWeele J; Cox NJ; Boehnke M; Himle JA; Curtis GC; Leventhal BL; Cook EH Jr., Genome-wide linkage analysis of families with obsessive-compulsive disorder ascertained through pediatric probands. Am J Med Genet 2002, 114 (5), 541–52. [DOI] [PubMed] [Google Scholar]
- 92.Willour VL; Yao Shugart Y; Samuels J; Grados M; Cullen B; Bienvenu OJ 3rd; Wang Y; Liang KY; Valle D; Hoehn-Saric R; Riddle M; Nestadt G, Replication study supports evidence for linkage to 9p24 in obsessive-compulsive disorder. Am J Hum Genet 2004, 75 (3), 508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gazzellone MJ; Zarrei M; Burton CL; Walker S; Uddin M; Shaheen SM; Coste J; Rajendram R; Schachter RJ; Colasanto M; Hanna GL; Rosenberg DR; Soreni N; Fitzgerald KD; Marshall CR; Buchanan JA; Merico D; Arnold PD; Scherer SW, Uncovering obsessive-compulsive disorder risk genes in a pediatric cohort by high-resolution analysis of copy number variation. J Neurodev Disord 2016, 8, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choucair N; Mignon-Ravix C; Cacciagli P; Abou Ghoch J; Fawaz A; Megarbane A; Villard L; Chouery E, Evidence that homozygous PTPRD gene microdeletion causes trigonocephaly, hearing loss, and intellectual disability. Mol Cytogenet 2015, 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trampush JW; Yang ML; Yu J; Knowles E; Davies G; Liewald DC; Starr JM; Djurovic S; Melle I; Sundet K; Christoforou A; Reinvang I; DeRosse P; Lundervold AJ; Steen VM; Espeseth T; Raikkonen K; Widen E; Palotie A; Eriksson JG; Giegling I; Konte B; Roussos P; Giakoumaki S; Burdick KE; Payton A; Ollier W; Horan M; Chiba-Falek O; Attix DK; Need AC; Cirulli ET; Voineskos AN; Stefanis NC; Avramopoulos D; Hatzimanolis A; Arking DE; Smyrnis N; Bilder RM; Freimer NA; Cannon TD; London E; Poldrack RA; Sabb FW; Congdon E; Conley ED; Scult MA; Dickinson D; Straub RE; Donohoe G; Morris D; Corvin A; Gill M; Hariri AR; Weinberger DR; Pendleton N; Bitsios P; Rujescu D; Lahti J; Le Hellard S; Keller MC; Andreassen OA; Deary IJ; Glahn DC; Malhotra AK; Lencz T, GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry 2017, 22 (3), 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lam M; Trampush JW; Yu J; Knowles E; Davies G; Liewald DC; Starr JM; Djurovic S; Melle I; Sundet K; Christoforou A; Reinvang I; DeRosse P; Lundervold AJ; Steen VM; Espeseth T; Raikkonen K; Widen E; Palotie A; Eriksson JG; Giegling I; Konte B; Roussos P; Giakoumaki S; Burdick KE; Payton A; Ollier W; Chiba-Falek O; Attix DK; Need AC; Cirulli ET; Voineskos AN; Stefanis NC; Avramopoulos D; Hatzimanolis A; Arking DE; Smyrnis N; Bilder RM; Freimer NA; Cannon TD; London E; Poldrack RA; Sabb FW; Congdon E; Conley ED; Scult MA; Dickinson D; Straub RE; Donohoe G; Morris D; Corvin A; Gill M; Hariri AR; Weinberger DR; Pendleton N; Bitsios P; Rujescu D; Lahti J; Le Hellard S; Keller MC; Andreassen OA; Deary IJ; Glahn DC; Malhotra AK; Lencz T, Large-Scale Cognitive GWAS Meta-Analysis Reveals Tissue-Specific Neural Expression and Potential Nootropic Drug Targets. Cell Rep 2017, 21 (9), 2597–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uetani N; Kato K; Ogura H; Mizuno K; Kawano K; Mikoshiba K; Yakura H; Asano M; Iwakura Y, Impaired learning with enhanced hippocampal long-term potentiation in PTPdelta-deficient mice. EMBO J 2000, 19 (12), 2775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uetani N; Chagnon MJ; Kennedy TE; Iwakura Y; Tremblay ML, Mammalian motoneuron axon targeting requires receptor protein tyrosine phosphatases sigma and delta. J Neurosci 2006, 26 (22), 5872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stepanek L; Stoker AW; Stoeckli E; Bixby JL, Receptor tyrosine phosphatases guide vertebrate motor axons during development. J Neurosci 2005, 25 (15), 3813–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drgonova J; Walther D; Wang KJ; Hartstein GL; Lochte B; Troncoso J; Uetani N; Iwakura Y; Uhl GR, Mouse Model for Protein Tyrosine Phosphatase D (PTPRD) Associations with Restless Leg Syndrome or Willis-Ekbom Disease and Addiction: Reduced Expression Alters Locomotion, Sleep Behaviors and Cocaine-Conditioned Place Preference. Mol Med 2015, 21 (1), 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tzschentke TM, Measuring reward with the conditioned place preference (CPP) paradigm: update of the last decade. Addict Biol 2007, 12 (3–4), 227–462. [DOI] [PubMed] [Google Scholar]
- 102.Everitt BJ; Giuliano C; Belin D, Addictive behaviour in experimental animals: prospects for translation. Philos Trans R Soc Lond B Biol Sci 2018, 373 (1742). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi H; Craig AM, Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends Neurosci 2013, 36 (9), 522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hendriks WJ; Pulido R, Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim Biophys Acta 2013, 1832 (10), 1673–96. [DOI] [PubMed] [Google Scholar]
- 105.Ling Q; Huang Y; Zhou Y; Cai Z; Xiong B; Zhang Y; Ma L; Wang X; Li X; Li J; Shen J, Illudalic acid as a potential LAR inhibitor: synthesis, SAR, and preliminary studies on the mechanism of action. Bioorg Med Chem 2008, 16 (15), 7399–409. [DOI] [PubMed] [Google Scholar]
- 106.Ling Q; Zhou YY; Cai ZL; Zhang YH; Xiong B; Ma LP; Wang X; Li X; Li J; Shen JK, Synthesis and LAR inhibition of 7-alkoxy analogues of illudalic acid. Yao Xue Xue Bao 2010, 45 (11), 1385–97. [PubMed] [Google Scholar]
- 107.Drgonova J; Walther D; Hartstein GL; Bukhari MO; Baumann MH; Katz J; Hall FS; Arnold ER; Flax S; Riley A; Rivero-Martin O; Lesch KP; Troncoso J; Ranscht B; Uhl GR, Cadherin 13: human cis-regulation and selectively-altered addiction phenotypes and cerebral cortical dopamine in knockout mice. Mol Med 2016, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.DeAndrade MP; Johnson RL Jr.; Unger EL; Zhang L; van Groen T; Gamble KL; Li Y, Motor restlessness, sleep disturbances, thermal sensory alterations and elevated serum iron levels in Btbd9 mutant mice. Hum Mol Genet 2012, 21 (18), 3984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salminen AV; Garrett L; Schormair B; Rozman J; Giesert F; Niedermeier KM; Becker L; Rathkolb B; Racz I; German Mouse Clinic C; Klingenspor M; Klopstock T; Wolf E; Zimmer A; Gailus-Durner V; Torres M; Fuchs H; Hrabe de Angelis M; Wurst W; Holter SM; Winkelmann J, Meis1: effects on motor phenotypes and the sensorimotor system in mice. Dis Model Mech 2017, 10 (8), 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chibnik LB; White CC; Mukherjee S; Raj T; Yu L; Larson EB; Montine TJ; Keene CD; Sonnen J; Schneider JA; Crane PK; Shulman JM; Bennett DA; De Jager PL, Susceptibility to neurofibrillary tangles: role of the PTPRD locus and limited pleiotropy with other neuropathologies. Mol Psychiatry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoffman KL; Cano-Ramirez H, Lost in translation? A critical look at the role that animal models of obsessive compulsive disorder play in current drug discovery strategies. Expert Opin Drug Discov 2018, 13 (3), 211–220. [DOI] [PubMed] [Google Scholar]
- 112.Hsu HC; Lapke N; Chen SJ; Lu YJ; Jhou RS; Yeh CY; Tsai WS; Hung HY; Hsieh JC; Yang TS; Thiam TK; You JF, PTPRT and PTPRD Deleterious Mutations and Deletion Predict Bevacizumab Resistance in Metastatic Colorectal Cancer Patients. Cancers (Basel) 2018, 10 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lepikhova T; Karhemo PR; Louhimo R; Yadav B; Murumagi A; Kulesskiy E; Kivento M; Sihto H; Grenman R; Syrjanen SM; Kallioniemi O; Aittokallio T; Wennerberg K; Joensuu H; Monni O, Drug-Sensitivity Screening and Genomic Characterization of 45 HPV-Negative Head and Neck Carcinoma Cell Lines for Novel Biomarkers of Drug Efficacy. Mol Cancer Ther 2018, 17 (9), 2060–2071. [DOI] [PubMed] [Google Scholar]
- 114.Painter JN; O’Mara TA; Morris AP; Cheng THT; Gorman M; Martin L; Hodson S; Jones A; Martin NG; Gordon S; Henders AK; Attia J; McEvoy M; Holliday EG; Scott RJ; Webb PM; Fasching PA; Beckmann MW; Ekici AB; Hein A; Rubner M; Hall P; Czene K; Dork T; Durst M; Hillemanns P; Runnebaum I; Lambrechts D; Amant F; Annibali D; Depreeuw J; Vanderstichele A; Goode EL; Cunningham JM; Dowdy SC; Winham SJ; Trovik J; Hoivik E; Werner HMJ; Krakstad C; Ashton K; Otton G; Proietto T; Tham E; Mints M; Ahmed S; Healey CS; Shah M; Pharoah PDP; Dunning AM; Dennis J; Bolla MK; Michailidou K; Wang Q; Tyrer JP; Hopper JL; Peto J; Swerdlow AJ; Burwinkel B; Brenner H; Meindl A; Brauch H; Lindblom A; Chang-Claude J; Couch FJ; Giles GG; Kristensen VN; Cox A; Zondervan KT; Nyholt DR; MacGregor S; Montgomery GW; Tomlinson I; Easton DF; Thompson DJ; Spurdle AB, Genetic overlap between endometriosis and endometrial cancer: evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med 2018, 7 (5), 1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo Y; Li B; Zhang G; He Y; Bae JH; Hu F; Cui R; Liu R; Wang Z; Wang L, Integrated Oncogenomic Profiling of Copy Numbers and Gene Expression in Lung Adenocarcinomas without EGFR Mutations or ALK Fusion. J Cancer 2018, 9 (6), 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu X; Zhang F; Mao J; Lu Y; Li J; Ma W; Fan S; Zhang C; Li Q; Wang B; Song B; Li L, Protein tyrosine phosphatase receptor-type delta acts as a negative regulator suppressing breast cancer. Oncotarget 2017, 8 (58), 98798–98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Couto PP; Bastos-Rodrigues L; Schayek H; Melo FM; Lisboa RGC; Miranda DM; Vilhena A; Bale AE; Friedman E; De Marco L, Spectrum of germline mutations in smokers and non-smokers in Brazilian non-small-cell lung cancer (NSCLC) patients. Carcinogenesis 2017, 38 (11), 1112–1118. [DOI] [PubMed] [Google Scholar]
- 118.Schumacher SE; Shim BY; Corso G; Ryu MH; Kang YK; Roviello F; Saksena G; Peng S; Shivdasani RA; Bass AJ; Beroukhim R, Somatic copy number alterations in gastric adenocarcinomas among Asian and Western patients. PLoS One 2017, 12 (4), e0176045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Woodward EL; Biloglav A; Ravi N; Yang M; Ekblad L; Wennerberg J; Paulsson K, Genomic complexity and targeted genes in anaplastic thyroid cancer cell lines. Endocr Relat Cancer 2017, 24 (5), 209–220. [DOI] [PubMed] [Google Scholar]
- 120.Xu MD; Liu SL; Feng YZ; Liu Q; Shen M; Zhi Q; Liu Z; Gu DM; Yu J; Shou LM; Gong FR; Zhu Q; Duan W; Chen K; Zhang J; Wu MY; Tao M; Li W, Genomic characteristics of pancreatic squamous cell carcinoma, an investigation by using high throughput sequencing after in-solution hybrid capture. Oncotarget 2017, 8 (9), 14620–14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spina V; Khiabanian H; Messina M; Monti S; Cascione L; Bruscaggin A; Spaccarotella E; Holmes AB; Arcaini L; Lucioni M; Tabbo F; Zairis S; Diop F; Cerri M; Chiaretti S; Marasca R; Ponzoni M; Deaglio S; Ramponi A; Tiacci E; Pasqualucci L; Paulli M; Falini B; Inghirami G; Bertoni F; Foa R; Rabadan R; Gaidano G; Rossi D, The genetics of nodal marginal zone lymphoma. Blood 2016, 128 (10), 1362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Song L; Jiang W; Liu W; Ji JH; Shi TF; Zhang J; Xia CQ, Protein tyrosine phosphatases receptor type D is a potential tumour suppressor gene inactivated by deoxyribonucleic acid methylation in paediatric acute myeloid leukaemia. Acta Paediatr 2016, 105 (3), e132–41. [DOI] [PubMed] [Google Scholar]
- 123.Acun T; Demir K; Oztas E; Arango D; Yakicier MC, PTPRD is homozygously deleted and epigenetically downregulated in human hepatocellular carcinomas. OMICS 2015, 19 (4), 220–9. [DOI] [PubMed] [Google Scholar]
- 124.Ortiz B; Fabius AW; Wu WH; Pedraza A; Brennan CW; Schultz N; Pitter KL; Bromberg JF; Huse JT; Holland EC; Chan TA, Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci U S A 2014, 111 (22), 8149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Walia V; Prickett TD; Kim JS; Gartner JJ; Lin JC; Zhou M; Rosenberg SA; Elble RC; Solomon DA; Waldman T; Samuels Y, Mutational and functional analysis of the tumor-suppressor PTPRD in human melanoma. Hum Mutat 2014, 35 (11), 1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du Y; Su T; Tan X; Li X; Xie J; Wang G; Shen J; Hou J; Cao G, Polymorphism in protein tyrosine phosphatase receptor delta is associated with the risk of clear cell renal cell carcinoma. Gene 2013, 512 (1), 64–9. [DOI] [PubMed] [Google Scholar]
- 127.Clark O; Schmidt F; Coles CH; Tchetchelnitski V; Stoker AW, Functional analysis of the putative tumor suppressor PTPRD in neuroblastoma cells. Cancer Invest 2012, 30 (5), 422–32. [DOI] [PubMed] [Google Scholar]
- 128.Solomon DA; Kim JS; Cronin JC; Sibenaller Z; Ryken T; Rosenberg SA; Ressom H; Jean W; Bigner D; Yan H; Samuels Y; Waldman T, Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res 2008, 68 (24), 10300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Imamura M; Iwata M; Maegawa H; Watada H; Hirose H; Tanaka Y; Tobe K; Kaku K; Kashiwagi A; Kadowaki T; Kawamori R; Maeda S, Replication study for the association of rs391300 in SRR and rs17584499 in PTPRD with susceptibility to type 2 diabetes in a Japanese population. J Diabetes Investig 2013, 4 (2), 168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.El Rouby N; McDonough CW; Gong Y; McClure LA; Mitchell BD; Horenstein RB; Talbert RL; Crawford DC; e M. n.; Gitzendanner MA; Takahashi A; Tanaka T; Kubo M; Pepine CJ; Cooper-DeHoff RM; Benavente OR; Shuldiner AR; Johnson JA, Genome-wide association analysis of common genetic variants of resistant hypertension. Pharmacogenomics J 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maciukiewicz M; Gorbovskaya I; Tiwari AK; Zai CC; Freeman N; Meltzer HY; Kennedy JL; Muller DJ, Genetic validation study of protein tyrosine phosphatase receptor type D (PTPRD) gene variants and risk for antipsychotic-induced weight gain. J Neural Transm (Vienna) 2018. [DOI] [PubMed] [Google Scholar]
- 132.Oprea TI; Bologa CG; Brunak S; Campbell A; Gan GN; Gaulton A; Gomez SM; Guha R; Hersey A; Holmes J; Jadhav A; Jensen LJ; Johnson GL; Karlson A; Leach AR; Ma’ayan A; Malovannaya A; Mani S; Mathias SL; McManus MT; Meehan TF; von Mering C; Muthas D; Nguyen DT; Overington JP; Papadatos G; Qin J; Reich C; Roth BL; Schurer SC; Simeonov A; Sklar LA; Southall N; Tomita S; Tudose I; Ursu O; Vidovic D; Waller A; Westergaard D; Yang JJ; Zahoranszky-Kohalmi G, Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov 2018, 17 (5), 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]