Abstract
Biofilm formation by Klebsiella pneumoniae on indwelling medical devices increases the risk of infection. Both type 1 and type 3 fimbriae are important factors in biofilm formation by K. pneumoniae. We found that a putative enzyme II (EII) complex of the phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS), etcA (EIIA)-etcB (EIIB)-etcC (EIIC), regulated biofilm and type 3 fimbriae formation by K. pneumoniae STU1. In this study, the regulatory mechanism of etcABC in K. pneumoniae type 3 fimbriae formation was investigated. We found via quantitative RT-PCR that overexpression of etcABC enhanced the transcription level of the mrk operon, which is involved in type 3 fimbriae synthesis, and reduced the transcription level of the fim operon, which is involved in type 1 fimbriae synthesis. To gain further insight into the role of etcABC in type 3 fimbriae synthesis, we analyzed the region upstream of the mrk operon and found the potential cyclic 3′5′-adenosine monophosphate (cAMP) receptor protein (CRP) binding site. After crp was deleted in K. pneumoniae STU1 and two clinical isolates, these three crp mutant strains could not express MrkA, the major subunit of the fimbrial shaft, indicating that CRP positively regulated type 3 fimbriae synthesis. Moreover, a crp mutant overexpressing etcABC could not express MrkA, indicating that the regulation of type 3 fimbriae by etcABC was dependent on CRP. In addition, deletion of cyaA, which encodes the adenylyl cyclase that synthesizes cAMP, and deletion of crr, which encodes the glucose-specific EIIA, led to a reduction in lac operon regulation and therefore bacterial lactose uptake in K. pneumoniae. Exogenous cAMP but not etcABC overexpression compensated for the role of cyaA in bacterial lactose uptake. However, either etcABC overexpression or exogenous cAMP compensated for the role of crr in bacterial lac operon regulation that would eventually restore lactose uptake. We also found via ELISA and the luxCDABE reporter system that overexpression of etcABC increased intracellular cAMP levels and the transcription level of crp, respectively, in K. pneumoniae. In conclusion, overexpression of etcABC positively regulated cAMP production and cAMP-CRP activity to activate the mrk operon, resulting in increased type 3 fimbriae synthesis in K. pneumoniae.
Keywords: Klebsiella pneumoniae, carbohydrate phosphotransferase system, biofilm, type 3 fimbriae, CRP-cAMP
Introduction
Patients using medical devices easily become vulnerable to microbial infection. Biofilm formation on medical device surfaces increases the risk of infection. A 10-year survey of healthcare-associated infections in intensive care units reported that 7% cases were related to Klebsiella pneumoniae (Kolpa et al., 2018). K. pneumoniae is a common pathogen associated with indwelling medical device infections, especially catheter-associated urinary tract infections, catheter-related blood stream infections, and ventilator-associated pneumonia (Singhai et al., 2012; Percival et al., 2015). K. pneumoniae can form biofilms on abiotic and human tissue surfaces (Reid et al., 1992; Clegg and Murphy, 2016). The capsular polysaccharide (CPS) and fimbriae of K. pneumoniae are reported to be the important factors contributing to biofilm formation (Vuotto et al., 2014; Chung, 2016). However, in different studies, results regarding the role of the CPS in biofilm formation by K. pneumoniae are conflicting (Vuotto et al., 2014; Wang et al., 2017). Genes encoding type 1 and type 3 fimbriae are common and well characterized in most K. pneumoniae strains. Type 1 fimbriae are found in the majority of Enterobacteriaceae family members. The type 1 fimbriae of K. pneumoniae are encoded by the genes in the fimAICDFGHK operon. The FimA protein constitutes the major fimbrial subunit. FimH is a mannose-binding adhesin (Struve et al., 2008). Type 3 fimbriae are produced by many members of the Enterobacteriaceae (Sebghati et al., 1998; Ong et al., 2010). The components of type 3 fimbriae are encoded by the genes in the mrkABCDF operon (Allen et al., 1991). The gene mrkA encodes the major fimbrial subunit that is polymerized to form the fimbrial shaft. MrkD is an adhesin located at the fimbrial tip (Chung, 2016). Type 1 fimbriae are essential for K. pneumoniae to cause urinary tract infections (Rosen et al., 2008; Struve et al., 2008). Type 3 fimbriae are reported to mediate the attachment of K. pneumoniae to the extracellular matrix, bind to human endothelial and bladder cells and promote biofilm formation on biotic and abiotic surfaces (Tarkkanen et al., 1997; Jagnow and Clegg, 2003; Schroll et al., 2010).
Cyclic 3′5′-adenosine monophosphate (cAMP) is a second messenger found in all cellular organisms and involved in global gene regulation (Lin and Green, 1989). cAMP is catalyzed from ATP by a group of enzymes known as adenylyl cyclases (ACs). These enzymes are divided into six classes based on their primary structures. Escherichia coli possesses a single class I AC (Cya). Mycobacterium tuberculosis H37Rv possesses at least 16 class III AC-like proteins, while Pseudomonas putida possesses a CyaA-type AC. cAMP binds to and then activates the transcriptional factor cAMP receptor protein (CRP), also called catabolite gene activator protein (CAP) (Green et al., 2014). The CRP-cAMP homodimer binds to CRP binding sites (TGTGA-N6-TCACA or TGCGA-N6-TCGCA) to enhance the ability of the RNA polymerase holoenzyme to initiate gene transcription (Gunasekera et al., 1992). The genome of E. coli contains approximately 200 CRP regulons. In addition to regulating genes responsible for carbon metabolism, CRP-cAMP regulates various stress-related genes, such as chaperone proteins and cold shock and heat shock proteins (Gosset et al., 2004; Zheng et al., 2004). In K. pneumoniae MGH78578, 198 operons including 378 genes are predicted to be regulated by CRP (Novichkov et al., 2010). Lin et al. (2016) reported that CRP indirectly repressed mrkA transcription via repression of mrkHI transcription in K. pneumoniae CG43S3. In K. pneumoniae AJ218 and K. pneumoniae IApc35, mrkH which is in mrkHI bicistronic operon and located immediately adjacent to the mrkABCDF operon encodes a transcriptional activator that contains PilZ domain which mediates c-di-GMP binding and DNA binding. MrkH directly activates transcription of mrkA and its own expression by binding to the region close to the promoter in the presence of c-di-GMP (Wilksch et al., 2011; Tan et al., 2015; Schumacher and Zeng, 2016). MrkI, a LuxR-type transcriptional regulator, is a co-activator for the expression of mrkA. However, overexpression of mrkH in a mrkI mutant background could restore fimbrial expression (Johnson et al., 2011).
Bacteria regulate diverse aspects of physiology in response to the carbohydrate availability via the phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS), which is a phosphorylation cascade that transfers phosphate sequentially from PEP to substrates (carbohydrates). Enzyme I (EI) and the histidine phosphocarrier protein (HPr) are the general cytoplasmic PTS proteins involved in the translocation of all PTS carbohydrates in most bacteria, whereas substrate specificity depends on the enzyme II (EII) complex. In most PTSs, the EII complex consists of membrane-bound EIIC component/domain and cytoplasmic EIIA and EIIB component/domain. Bacteria contain more than one EII complex; for example, E. coli contains at least 15 different EII complexes. In addition to regulating sugar (carbohydrates) transport, PTSs have been reported to regulate bacterial potassium uptake, nitrogen source utilization, and carbohydrate metabolic programs such as carbohydrate catabolite repression (CCR) and inducer exclusion (Deutscher et al., 2006, 2014). The glucose-specific EII complex of enteric bacteria consists of two distinct proteins: the cytoplasmic protein EIIAGlc, encoded by crr, and the membrane-associated protein EIICBGlc, encoded by ptsG, which contains the hydrophilic EIIB domain and the membrane-associated EIIC domain. EIIAGlc plays an important role in carbon metabolism in enteric bacteria not only by interacting with non-PTS permeases (such as LacY) to inhibit their activities but also by regulating AC to use glucose as a priority carbon source. These phenomena are called inducer exclusion and CCR, respectively. The phosphorylated or dephosphorylated state of EIIAGlc determines its regulatory role. In the absence of glucose and in the presence of PEP, EIIAGlc is mainly in a phosphorylated state and is required for the activation of AC. Unphosphorylated EIIAGlc can bind to and inhibit several non-PTS proteins, such as LacY (a lactose permease), MelB (a melibiose carrier protein), MalK (an ATP-hydrolyzing component of the maltose transport system), and GlpK (a glycerol kinase) (Deutscher et al., 2006).
We previously found the putative EII complex – KPN00353 (EIIA homolog), KPN00352 (EIIB homolog), and KPN00351 (EIIC homolog) – in the genome of K. pneumoniae MGH78578 (Jeng et al., 2017; Horng et al., 2018). We found the etcA, etcB, and etcC (etc for enzyme two complex), homologous to KPN00353, KPN00352, and KPN00351, respectively, in K. pneumoniae STU1 in this study. Overexpression of etcABC enhanced biofilm formation and type 3 fimbriae synthesis in K. pneumoniae STU1. We identified a putative CRP binding site located upstream of mrkA. In addition, we found that overexpression of etcABC compensated for the role of EIIAGlc in lactose uptake by K. pneumoniae. Furthermore, intracellular cAMP levels and the CRP transcriptional levels were observed in bacteria overexpressing etcABC. In summary, we provided a model to show the regulation of type 3 fimbriae by etcABC via the CRP-cAMP signaling pathway in K. pneumoniae.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Primers are listed in Supplementary Table S1. Unless otherwise stated, K. pneumoniae and E. coli were routinely cultured in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with appropriated antibiotics at the following concentrations: kanamycin (50 μg/mL), ampicillin (100 μg/mL), chloramphenicol (100 μg/mL), and gentamicin (20 μg/mL) on the rotatory shaker at 37°C and 200 rpm. For observation of lac operon activity, 2 μL of diluted overnight cultured bacterial solution was inoculated on MacConkey plate without/with 1 mM cAMP and LB agar plate containing 50 μg/mL of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-gal). Overnight cultured bacteria were diluted 1000-fold into the M9 minimal medium supplemented with 1% lactose and incubated for 24–48 h. The two clinical isolates of K. pneumoniae from bacterial storage bank in Tzu Chi Hospital to College of Medicine at Tzu Chi University was through official transfer.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain | Relevant genotype and phenotype | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | F-, φ80dlacZΔM15 (lacZYA-argF) U169, deoR, recA1, endA1, hsdR17(rk-, mk+), phoA, supE44, λ-, thi-1, gyrA96, relA1 | Invitrogen |
| S17-1 λpir | λ-pir lysogen of S17-1 [thi pro hsdR- hsdM+ recA RP4 2-Tc::Mu-Km::Tn7 (TpR. SmR.)]. Permissive host able to transfer suicide plasmids requiring the Pir protein by conjugation to recipient cells | Simon et al., 1983 |
| BL21(DE3)pLysS | F-, ompT, gal, dcm, lon, hsdSB(rB-mB-), λ(DE3), pLysS(cmR) | Novagen |
| K. pneumoniae | ||
| STU1 | Laboratory-maintained strain, Ampr | National Taiwan University |
| Δcrr | In frame deletion of crr gene in STU1 | This work |
| ΔcyaA | In frame deletion of cyaA gene in STU1 | This work |
| Δcrp | In frame deletion of crp gene in STU1 | This work |
| ΔcrrΔetcABC | In frame deletion of etcABC in Δcrr | |
| Clinical Kp-1 | Clinical strain labeled Kp 083535 which was isolated from blood specimen | Tzu Chi Hospital, Hualien, Taiwan |
| Clinical Kp-2 | Clinical strain labeled Kp 036749 which was isolated from urine specimen | Tzu Chi Hospital, Hualien, Taiwan |
| Plasmid | ||
| pET30b | Vector, Kmr | Novagen (Merck, Darmstadt, Germany) |
| pET30b::mrkA | pET30b derivative carrying structure gene of mrkA to express His-tagged MrkA, Kmr | This work |
| pET30b::fimA | pET30b derivative carrying structure gene of fimA to express His-tagged FimA, Kmr | This work |
| pET30b::crp | pET30b derivative carrying structure gene of crp to express His-tagged CRP, Kmr | This work |
| pUT::minTn5-km1 | Suicide plasmid requiring the Pir protein for replication and containing a mini-Tn5 cassette containing Kmr gene, Apr, Kmr | de Lorenzo et al., 1990 |
| pBlueScript SK+ (pBSK) | Vector containing lac promoter, pUC ori, Ampr | Stratagene (CA, United States) |
| pBSK-Gm | pBSK derivative carrying gentamicin resistance gene at the ScaI site, Gmr | Horng et al., 2018 |
| pBSK::Gm::etcABC | pBSK-Gm derivative carrying complete etcABC (promoter and structure), Gmr | This work |
| pBSK::Gm::cyaA | pBSK-Gm derivative carrying complete cyaA, Gmr | This work |
| pBSK::Gm::crr | pBSK-Gm derivative carrying complete crr, Gmr | This work |
| pBAD33 | Vector utilizing PBAD promoter, pACYC184 ori, Cmr | Guzman et al., 1995 |
| pBAD33::crp | pBAD33 derivative carrying complete crp, Cmr | This work |
| pK18mobsacB | Vector, pBR322 ori (ori V), RP4 mob, sacB, Kmr | Schafer et al., 1994 |
| pW18mobsacB | pK18mobsacB derivative that R6K ori with BamHI fragment was inserted into and pBR322 ori was removed from by using SacI restriction enzyme; a suicide plasmid in K. pneumoniae, Kmr | This work |
| pKO-Crp | yhfA’ (upstream of crp gene) and yhfK’ (downstream of crp gene) were inserted into pW18mobsacB with SmaI site, Kmr | This work |
| pACYC184 | Vector, p15A ori, Tcr, Cmr | Chang and Cohen, 1978 |
| pACYC184-Smr | pACYC184 derivative carrying streptomycin resistant gene in HindIII site, Smr | This work |
| pPless-lux | pACYC184-Smr derivative carrying promoterless luxCDABE in BamHI site, Smr | This work |
| pPcrp-lux | pPless-lux derivative carrying crp promoter in front of luxCDABE, Smr | This work |
Quantification of Biofilm Formation
The biofilm formation assay was conducted according to a previous published protocol with some modifications (O’Toole and Kolter, 1998). In brief, 2 mL of a bacteria suspension was inoculated into a Falcon polystyrene tube after dilution of the bacteria from an overnight culture with fresh LB to optical density at 600 nm (OD600) of 0.1. After incubation at 37°C for 24 h, the value of OD600 of the bacterial culture in the tube was measured. Thereafter, the bacterial suspension was discarded and the tube was rinsed twice with water. After incubation of 3 mL of 0.1% crystal violet at room temperature for 20 min, the tube was rinsed twice with water followed by air-drying. After addition of 95% ethanol, the absorbance of the ethanol solution was measured at 590 nm.
Construction of Gene Deletion Mutants
For unmarked mutagenesis in K. pneumoniae, we constructed a suicide vector, pW18mobsacB, based on the E. coli plasmid, pK18mobsacB (Schafer et al., 1994). In brief, R6Kori cut from the plasmid, pUT::minTn5-km1 (de Lorenzo et al., 1990), was inserted into BamHI site of pK18mobsacB and then pBR322 ori (oriV) was eliminated using SacI restriction enzyme to form the suicide vector, pW18mobsacB, which remained oriT_RP4 for RP4-mediated conjugation, sacB for negative selection and kan for kanamycin resistance. For specific gene deletion in K. pneumoniae, the approximately 700-bp upstream and downstream flanking DNA fragments of the specific gene were, respectively, amplified. These two fragments were inserted into pW18mobsacB. The plasmid, pW18mobsacB containing flanking DNA fragments, was transferred from E. coli S17-1 λ pir to K. pneumoniae by conjugation (Soo et al., 2007). The transconjugant was spread on LB plates containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL) for positive selection. Subsequently, the colonies from the positive selection were subcultured into LB broth containing 20% sucrose for negative selection. Then, the overnight bacterial culture was diluted and spread on LB plate containing 20% sucrose for getting the single colony of mutant candidate. The mutant strains were confirmed by PCR, followed by sequencing.
Cell Culture and Adhesion Assay
Cell culture and adhesion assay were performed as described previously with some modification (March et al., 2011). Human lung carcinoma cells A549 (ATCC CCL185) were grown to 90% confluence in F-12K medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin and 100 μg/mL streptomycin in 24-well cell culture dishes (4 × 104 cells per well) at 37°C under a humidified 5% CO2 atmosphere. For the adhesion assays, A549 cells were washed three times with phosphate-buffered saline (PBS) and then infected with a suspension of 2 × 107 bacterial cells in F12K medium. After 3-h infection, cells were washed three times with PBS. Subsequently, the adhered bacteria were released by 500 μL of 0.5% Triton X-100 for 5 min and serial dilutions were plated on LB agar plates for viable counts of bacteria.
Western Blotting
The bacterial concentration was determined by measuring the optical density (OD) at 600 nm. A fixed amount of bacteria was collected by centrifugation, re-suspended in SDS sample buffer and then lysed by heating for 10–15 min at 100°C. An aliquot of total bacterial proteins was analyzed by 12% SDS polyacrylamide gel (SDS-PAGE) and transferred to a nitrocellulose membrane, Amersham Hybond-C Extra (GE Healthcare, IL, United States), by Amersham Mini Trans-Blot semiphor transphor unit (GE Healthcare, IL, United States). The detection procedures were performed as described in the previous study using Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare, IL, United States) (Jeng et al., 2017). The intensities of the bands were detected using the gel catcher 2850 chemiluminescence camera system (CLUBIO, Taipei, Taiwan). For detection of MrkA, FimA, and mannose 6-phosphate isomerase (ManA), rabbit polyclonal antibody specific to the protein was the first antibody (LTK BioLaboratories, Taoyuan, Taiwan) and peroxidase-conjugated anti-rabbit IgG antibody was the second antibody (GE Healthcare, IL, United States). Rabbit anti-MrkA and anti-FimA polyclonal antibody were produced using His tagged proteins as immunogens in this study. Anti-ManA rabbit polyclonal antibody was produced in the previous study (Soo et al., 2014). For detection of CRP, mouse monoclonal antibody specific to CRP was the first antibody (BioLegend, CA, United States) and peroxidase-conjugated anti-mouse IgG antibody was the second antibody (Sigma-Aldrich, MO, United States).
Electrophoretic Mobility Shift Assay (EMSA)
DNA fragments for EMSA were amplified by PCR and using the specific primers (Supplementary Table S1). Purified His-tagged CRP was dialyzed using the dialysis buffer (400 mM NaCl, 25% glycerol, 10 mM DTT, 20 mM Tris–HCl, pH 7.5). For the serial dilution experiments, CRP protein was serially diluted in binding reaction buffer (20 mM Tris–HCl [pH 8], 0.1 mM MgCl2, 150 mM KCl, and 0.05 mM EDTA, 12.5% glycerol). The binding reaction comprised His-tagged CRP protein and DNA fragments was performed in binding reaction buffer supplemented with 30 μg/mL poly(dI-dC) and 1 μg/μL bovine serum albumin. The reaction mixtures were incubated for 30 min at room temperature before being loaded onto 7% non-denaturing polyacrylamide gels containing 0.5 × Tris-borate-EDTA buffer. After electrophoresis at 100 V for 1 h, the gel was stained with ethidium bromide solution.
Purification of His-Tagged Proteins
To purify His-tagged MrkA and His-tagged FimA for producing anti-MrkA and anti-FimA and to purify His-tagged CRP for EMSA, Novagen’s pET30b (Merck, Darmstadt, Germany) containing the structure gene of mrkA, fimA, or crp, respectively, was transformed to E. coli BL21(DE3) pLysS. After bacteria were cultured to mid-logarithmic phase, 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the culture, followed by further incubation for 3–4 h at 30°C. Bacterial cells were collected by centrifugation and then suspended in the LEW solution containing 30 mM imidazole. The preparation of LEW solution and protein purification using Protino Ni-TED 1000 Packed Columns followed the manufacturer’s instructions (Macherey-Nagel, Düren, Germany).
Quantitative Reverse Transcription PCR (RT-qPCR)
The bacterial RNA was extracted using TRI reagent (Sigma-Aldrich, MO, United States) and treated using RNase-free DNase I (New England Biolabs, MA, United States) for 30 min at 37°C to remove the DNA contamination. RNA was reversed transcribed by using QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). Quantitation of cDNA from the transcripts of fimA, fimH, mrkA, mrkD, and mrkH was performed by real time PCR in triplicate by using the specific primers (Supplementary Table S1) and QuantiNova SYBR® Green PCR Kit (Qiagen, Hilden, Germany) in the Rotor-Gene real-time genetic analyzer (Qiagen, Hilden, Germany). 16s rRNA was used as the internal reference gene for analysis.
Transmission Electron Microscopy (TEM)
After 1 mL of overnight bacterial culture was centrifuged, the bacterial pellet was fixed in a primary fixation solution (2.5% glutaraldehyde, 0.1 M cacodylate buffer, and 1% tannic acid) at room temperature for 1 h. Thereafter, 10 μl of bacterial suspension was absorbed onto 200-μm-pore-size mesh copper electron microscopy grids coated with carbon and Formvar. Subsequently, the grid was floated on a drop of 2% (w/v) uranyl acid for 15 s to negatively stain the bacterial sample. Bacterial cells were observed under a Hitachi H-7500 transmission electron microscope (Hitachi, Japan) operated under standard conditions with the cold trap in place. For immunogold electron microscopy, the bacteria were fixed with primary fixation solution and washed with PBS. After washed, the bacteria were blocked in blocking solution [5% w/v of bovine serum albumin (BSA) in PBS and 1% Tween 20] for 30 min. Subsequently, bacteria were incubated with rabbit anti-MrkA polyclonal antibody diluted in blocking solution (1:50) for 30 min at room temperature. After being washed three times with blocking solution, the bacteria were incubated with (goat) colloidal gold particle-conjugated anti-rabbit IgG (Sigma-Aldrich in Merck, Germany) for 30 min at room temperature. Before post-fixation (2% glutaraldehyde in PBS), the bacteria were washed twice with PBS. Ten microliters of bacterial suspension was absorbed onto carbon-coated grids for 2–3 min under a light bulb. After excess liquid was removed, the grids were rinsed with distilled water and negatively stained with 2% phosphotungstic acid (PTA) for 1 min. Samples were viewed on TEM.
Quantification of cAMP
After overnight culture, the bacterial OD was measured at 600 nm. A fixed amount of bacteria suspended in the lysis buffer (0.1 M HCl and 0.5% Triton X-100) was stored overnight at -70°C. After sonication and centrifugation, the amount of cAMP in the supernatant was measured by using the non-acetylated format provided in Direct cAMP ELISA kit (Enzo Life Sciences, NY, United States). All procedures followed the protocols supplied by the manufacturer.
Quantification of CRP Promoter Activity
To construct the luxCDABE reporter plasmid, the luxCDABE was cut from pBG (Soo et al., 2008) and subsequently inserted into pACYC184-Smr to form pPless-lux (Table 1). The 400-bp fragment containing crp promoter was amplified by PCR using primers pair, crp promoter FP/crp promoter RP (Supplementary Table S1), then digested with BamHI/EcoRV and inserted in front of luxCDABE in pPless-lux, forming pPcrp-lux (Table 1). Either pPless-lux or pPcrp-lux was transferred into ΔcrrΔetcABC, K. pneumoniae with double mutation of crr and etcABC, by electroporation. To observe the effect of EtcABC overexpression on CRP promoter activity, either pBSK::Gm::etcABC or pBSK-Gm was transferred into the strains, ΔcrrΔetcABC carrying pPless-lux or pPcrp-lux. After overnight culture in LB containing 0.5 mM IPTG, the bioluminescence was measured for 10 s by using Modulus single tube multimode luminescence value reader (Turner BioSystems, CA, United States). All procedures followed the protocols supplied by the manufacturer.
Nucleotide Sequence Accession Number
The nucleotide sequences of the etcABC genes have been deposited at GenBank nucleotide sequence database under the accession numbers MK675058.
Statistical Methods
Results of RT-qPCR, the concentration of intracellular cAMP and crp promoter activity were expressed as mean ± standard deviation from three independent experiments. Paired Student’s t-test was performed to determine statistically significant differences, and p-values of <0.05 were considered to indicate statistical significance.
Results
Overexpression of etcABC Enhanced Type 3 Fimbriae Production and Biofilm Formation
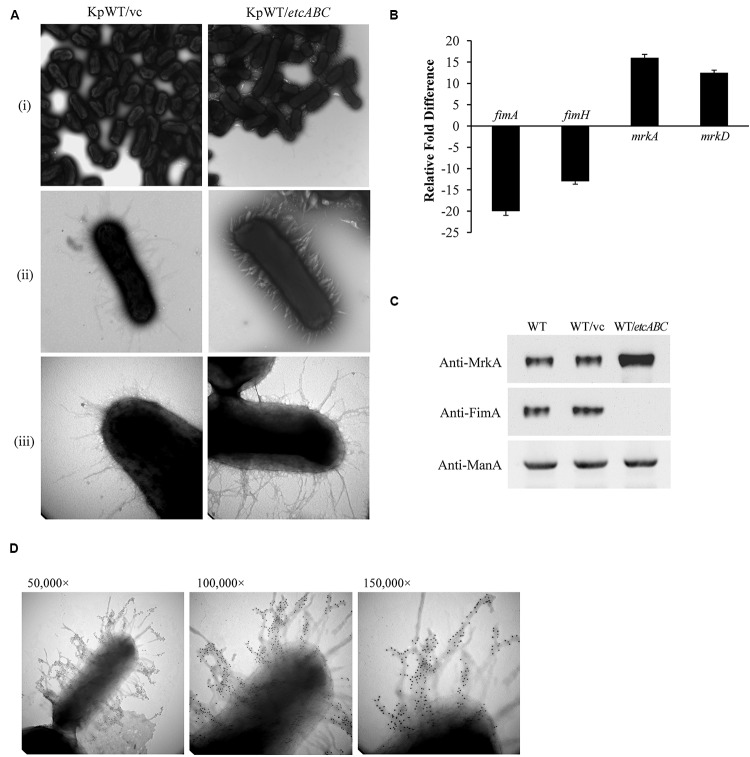
Our previous data showed that overexpression of KPN00353-KPN00352-KPN00351 increased biofilm formation by K. pneumoniae MGH78578 (Horng et al., 2018). In STU1, we found homologs having high sequence identity to KPN00353 (98%), KPN00352 (100%), and KPN00351 (99%) of MGH78578, respectively. We therefore refer to these genes as etcA, etcB, and etcC in this manuscript and the effect of EtcABC on biofilm formation was observed. Overexpression of etcABC in K. pneumoniae STU1 and two clinical K. pneumoniae isolates resulted in an increase in biofilm formation (Supplementary Figure S1A). The two clinical strains were isolated from blood and urine specimens, respectively (Table 1). Like STU1, these two clinical strains also have KPN00353-KPN00352-KPN00351 homologs, examined by PCR (data not shown). In addition, the effects of EtcABC overexpression on adhesion of K. pneumoniae STU1 to A549 epithelial alveolar cells were also examined. The amount of K. pneumoniae STU1 overexpressing etcABC adhered on A549 cells was more than that of K. pneumoniae STU1 carrying vector control (Supplementary Figure S1B). Fimbriae are reported to be an important factor contributing to bacterial biofilm formation and adhesion to cells (Hornick et al., 1992; Tarkkanen et al., 1997; Chung, 2016). Therefore, in this study, we examined the effects of etcABC on K. pneumoniae fimbriae production. Transmission electron microscopy (TEM) observation of bacteria grown in LB broth showed a greater presence of peritrichous pili on the surface of K. pneumoniae overexpressing etcABC than on the surface of the vector control (Figure 1A). However, the mutant strain lacking etcABC did not exhibit a difference from the wild-type strain (data not shown). To verify the type of pili expressed in K. pneumoniae overexpressing etcABC, we sought to quantify the transcriptional and translational levels of type 1 and type 3 fimbriae. Quantitative reverse transcription PCR (RT-qPCR) analysis showed that the mRNA levels of fimA and fimH were markedly decreased in K. pneumoniae overexpressing etcABC relative to their levels in the vector control. By contrast, the transcriptional levels of mrkA and mrkD in K. pneumoniae overexpressing etcABC were higher than those in the vector control (Figure 1B). Immunoblots of whole cell extracts of K. pneumoniae overexpressing etcABC probed with anti-MrkA polyclonal antibodies showed that MrkA, the component of the type 3 fimbrial shaft, was highly expressed relative to its expression in the vector control. However, K. pneumoniae overexpressing etcABC did not express FimA (Figure 1C). The amount of MrkA expressed by the mutant strain with the etcABC deletion was not different from that of the wild-type strain (data not shown). The expression of mannose 6-phosphate isomerase (ManA) in each strain confirmed the consistent sampling of bacteria (Soo et al., 2014). The fimbriae on the surface of K. pneumoniae overexpressing etcABC appeared to be in bundle or thicker than vector control (Figure 1A). To corroborate that fimbriae of K. pneumoniae overexpressing etcABC were type 3 fimbriae, the immunogold electron microscopy was performed. The immunogold electron microscopy micrograph of K. pneumoniae overexpressing etcABC using anti-MrkA showed gold particle localization on the fimbriae (Figure 1D). These results indicated that overexpression of etcABC positively regulated the mrk operon to enhance type 3 fimbriae production in K. pneumoniae.
FIGURE 1.
Overexpression of etcABC enhanced type 3 fimbriae production in K. pneumoniae. (A) The representative TEM images of bacteria incubated in LB broth in three independent experiments. KpWT/vc: K. pneumoniae STU1 carrying pBSK-Gm as the vector control. KpWT/etcABC: K. pneumoniae STU1 carrying pBSK::Gm::etcABC to overexpress etcABC. The magnifications of the images are (i) 15,000×, (ii) 50,000×, and (iii) 100,000×. (B) Transcriptional analysis of fimA, fimH, mrkA, and mrkD in KpWT/etcABC and KpWT/vc by RT-qPCR. The 16S rRNA gene was used as the reference. Relative gene expressions means amount of mrkA and mrkD mRNA in KpWT/etcABC compared to those in KpWT/vc and amount of fimA and fimH mRNA in KpWT/vc compared to those in KpWT/etcABC. The presented results are the means ± standard deviations of three replicates. The p-value of each group is less than 0.05 as compared with KpWT/vc. (C) Western blot analysis of MrkA, FimA, and ManA expression in each strain. WT: K. pneumoniae STU1, WT/vc: K. pneumoniae STU1 carrying pBSK-Gm as the vector control. WT/etcABC: K. pneumoniae STU1 carrying pBSK::Gm::etcABC to overexpress etcABC. ManA was used as the loading control. The blots are representatives of at least three independent experiments. (D) Immunogold electron microscopy using anti-MrkA was performed against K. pneumoniae STU1 carrying pBSK::Gm::etcABC. Bacteria displayed type 3 fimbriae. The magnification of the images is 50,000× (left), 100,000× (middle), and 150,000× (right), respectively.
CRP Positively Regulated Type 3 Fimbriae Production
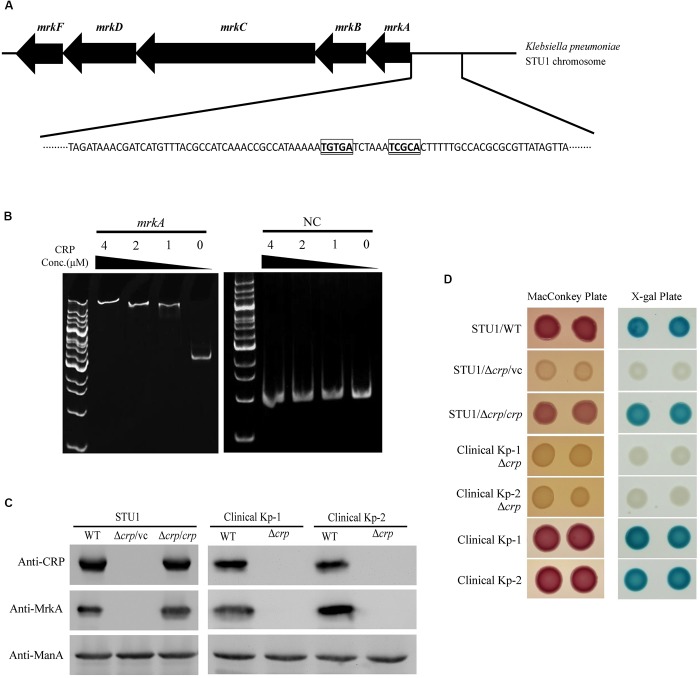
To identify possible factors regulating the transcription of the mrk operon, we performed sequence analysis and found a putative CRP binding site in the region upstream of the mrk operon (Figure 2A). An electrophoretic mobility shift assay (EMSA) was performed to confirm the binding of CRP to the region upstream of mrkA (Figure 2B). To confirm whether CRP regulated the expression of the mrk operon, the crp gene was deleted in K. pneumoniae STU1 and two clinical K. pneumoniae isolates to assess MrkA expression in crp mutant strains (Supplementary Figure S2). All three crp mutants and one complementation strain of the K. pneumoniae STU1 crp mutant strain were confirmed by Western blotting using anti-CRP monoclonal antibodies. None of the three crp mutants expressed MrkA, but all three of the wild-type and one complementation strain did (Figure 2C). In addition, the amount of mrkA transcript was reduced in all of three crp mutants containing vector, compared to their parent stains containing vector. The amount of mrkA transcript was restored in all of three crp complementation strains (Supplementary Figures S3A–C). These results indicated that CRP positively regulated type 3 fimbrial shaft synthesis in K. pneumoniae. Since fimbriae are important for bacterial biofilm formation and adhesion to cell, the effect of CRP on Klebsiella biofilm formation and adhesion to cell was observed. Even though the growth rate was slightly reduced, the K. pneumoniae STU1 crp mutant and crp mutant carrying vector showed the dramatical defects in both biofilm formation and adhesion to A549 cells, compared to wild type, K. pneumoniae STU1. The biofilm formation and adhesion ability were restored in crp complementation strain (Supplementary Figure S4). Therefore, these results suggested that CRP positively regulated type 3 fimbriae production in K. pneumoniae.
FIGURE 2.
The regulation of MrkA expression by CRP in K. pneumoniae. (A) The genetic organization of the mrk operon and putative CRP binding site located upstream of mrkA. The putative CRP core binding sites are indicated with bolded, underlined and boxed letters. (B) The EMSA demonstrating the interaction of the region upstream of mrkA and CRP. mrkA: a 394-bp DNA fragment containing the mrkA upstream region. NC: a 222-bp DNA fragment containing the EtcA structural region as the negative control. The concentration of purified His-tagged CRP ranged from 4 to 1 μM. The gel is representative of three independent experiments. (C) Western blot analysis of CRP, MrkA, and ManA expression in K. pneumoniae. ManA was used as the loading control. The blots are representatives of at least three independent experiments. (D) The representative colony phenotypes on MacConkey agar (MacConkey plate) and LB agar containing X-gal (X-gal plate) in three independent experiments. WT, wild-type K. pneumoniae. Δcrp: crp mutant. Δcrp/vc: crp mutant carrying pBAD33 as the vector control. Δcrp/crp: complementation strain of the crp mutant carrying pBAD33::crp. Clinical Kp-1 and Clinical Kp-2 are two clinical K. pneumoniae isolates. The phenotypes of the K. pneumoniae STU1 crp mutant (Δcrp) and its corresponding vector control (Δcrp/vc) were the same, so Δcrp/vc was used to represent of K. pneumoniae STU1 Δcrp in this figure.
CRP-cAMP was reported to be an activator of lactose fermentation in K. pneumoniae (formerly K. aerogenes) (Baldauf et al., 1988). To confirm the role of CRP in regulating the lac operon of the three K. pneumoniae strains in this study, the three crp mutants, one K. pneumoniae STU1 crp mutant carrying the vector (pBAD33; Δcrp/vc), three wild-type strains (K. pneumoniae STU1 and two clinical strains) and the complementation strain (K. pneumoniae STU1 Δcrp carrying pBAD33::crp; Δcrp/crp) were grown on MacConkey agar and LB agar containing X-gal. The three wild-type strains and the complementation strain formed pink colonies on MacConkey agar and blue colonies on LB agar containing X-gal. In contrast, the crp mutants formed colorless colonies on MacConkey agar and white colonies on LB agar containing X-gal (Figure 2D). These results indicated that CRP positively regulated the lac operon in these three K. pneumoniae strains.
Regulation of MrkA by etcABC Was Dependent on CRP
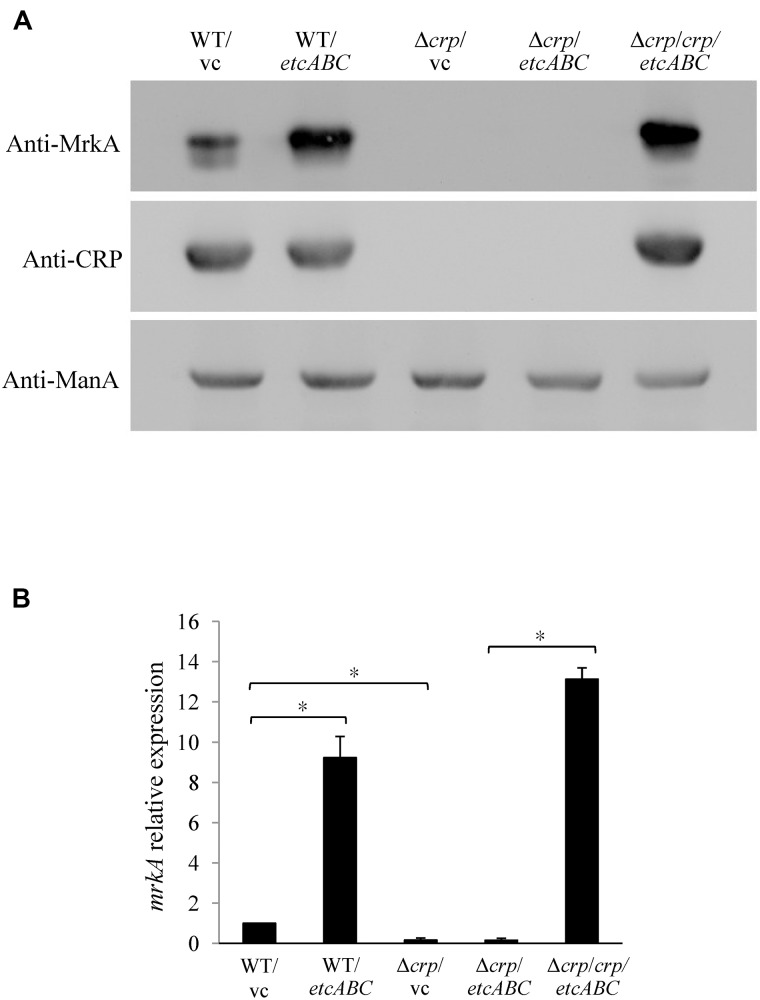
To investigate the mechanism by which overexpression of etcABC enhanced type 3 fimbriae expression (Figure 1B,C), we examined the effects of etcABC on MrkA expression in the crp mutant. The Western blotting results showed that overexpression of etcABC in the wild-type strain, K. pneumoniae STU1, increased MrkA expression compared to that in the wild-type strain carrying the vector (Figure 3A). The RT-qPCR results showed that the amount of mrkA transcript was increased in K. pneumoniae STU1 overexpressing etcABC, compared to the vector control (Figure 3B). However, MrkA was not expressed in either the crp mutant containing the vector or the crp mutant overexpressing etcABC (Figure 3A). The mrkA mRNA level also reduced in crp mutant containing the vector and the crp mutant overexpressing etcABC, compared to K. pneumoniae STU1 carrying the vector (Figure 3B). The phenotype of the crp mutant carrying the two plasmids pBAD33 and pBSK::Gm::etcABC was the same as that of the crp mutant carrying one plasmid, pBSK::Gm::etcABC (data not shown). Overexpression of etcABC in the crp complementation strain, the crp mutant carrying the two plasmids pBAD33::crp and pBSK::Gm::etcABC, restored both mrkA transcript and MrkA production (Figure 3). MrkA expression in the mutant strain lacking etcABC was not different from that of the wild-type strain (data not shown). Since MrkH was reported to positively regulate the mrkA transcription, the effect of etcABC and crp on mrkH transcription was examined by RT-qPCR. The results showed that overexpression of etcABC increased mrkH transcription in wild type, but not in crp mutant. Besides, the mRNA of mrkH was reduced in crp mutant containing vector, compared to wild type containing vector. Overexpression of etcABC in the crp complementation strain restored mrkH transcript (Supplementary Figure S5). These results suggested that overexpression of etcABC regulated type 3 fimbriae via CRP.
FIGURE 3.
The effects of EtcABC overexpression on MrkA was dependent on CRP. (A) Western blot analysis of MrkA, CRP, and ManA expression in each strain grown in LB. ManA was used as the loading control. The blots are representatives of at least three independent experiments. (B) RT-qPCR analysis of mrkA transcription in each strain grown in LB. The 16S rRNA gene was used as the reference. The transcriptional level of mrkA in each strain was compare to WT/vc. The presented results are the means ± standard deviations of three replicates. An asterisk (∗) represents p < 0.05 as compared with WT/vc or Δcrp/crp/etcABC compared with Δcrp/etcABC. WT/vc: K. pneumoniae STU1 carrying pBSK-Gm as the vector control. WT/etcABC: K. pneumoniae STU1 carrying pBSK::Gm::etcABC to overexpress etcABC. Δcrp/vc: K. pneumoniae STU1 crp mutant carrying pBSK-Gm as the vector control. Δcrp/etcABC: K. pneumoniae STU1 crp mutant carrying pBSK::Gm::etcABC to overexpress etcABC. Δcrp/crp/etcABC: K. pneumoniae STU1 crp mutant carrying pBAD33::crp and pBSK::Gm::etcABC. The phenotype of the crp mutant carrying the two plasmids pBAD33 and pBSK::Gm::etcABC was the same as that of the crp mutant carrying one plasmid, pBSK::Gm::etcABC.
Binding of cAMP to CRP has been reported to increase the affinity and specificity of CRP for the target DNA in E. coli (Takahashi et al., 1980, 1989; Harman, 2001). EIIAGlc can regulate the activity of adenylyl cyclase, also known as adenylate cyclase (AC or CyaA), to influence the cAMP concentration (Deutscher et al., 2006). Furthermore, EtcA was predicted to be an EIIA component. Therefore, we hypothesized that overexpression of etcABC can increase cAMP to enhance CRP activity in K. pneumoniae. Before testing this hypothesis, we examined whether the effects of cAMP on the lac operon in K. pneumoniae were the same as those in E. coli (Narang, 2009). The gene encoding CyaA was deleted, and growth of the cyaA mutant on MacConkey agar, on LB agar containing X-gal and in minimal medium containing lactose as the sole carbon source was observed. The cyaA mutant and its vector control formed colorless colonies on MacConkey agar and white colonies on LB agar containing X-gal and failed to grow in minimal medium containing lactose as the sole carbon source. In contrast, the wild-type and complementation strains formed pink colonies on MacConkey agar and blue colonies on LB agar containing X-gal and exhibited growth in minimal medium containing lactose as the sole carbon source (Figure 4A). Moreover, the phenotype of the cyaA mutant carrying the empty vector was restored by the addition of 1 mM cAMP to the MacConkey agar (Figure 5A). These results indicated that the effects of cyaA and cAMP on the lac operon were the same in both K. pneumoniae and E. coli.
FIGURE 4.

Colony morphology of K. pneumoniae STU1 and its derivative strains on MacConkey agar (MacConkey plate) and LB agar containing X-gal (X-gal plate) and bacterial growth in M9 minimal medium containing 1% lactose as the sole carbon source (M9 with 1% lactose) after overnight culture. KpWT: wild-type. (A) The phenotype of cyaA mutant and its derivative strains. ΔcyaA: cyaA mutant. ΔcyaA/cyaA: complementation strain of the cyaA mutant carrying pBSK::Gm::cyaA. ΔcyaA/vc: cyaA mutant carrying pBSK-Gm as the vector control. ΔcyaA/etcABC: cyaA mutant carrying pBSK::Gm::etcABC to overexpress etcABC. (B) The phenotype of crr mutant and its derivative strains. Δcrr: crr mutant. Δcrr/crr: complementation strain of the crr mutant carrying pBSK::Gm::crr. Δcrr/vc: crr mutant carrying pBSK::Gm as the vector control. Δcrr/etcABC: crr mutant carrying pBSK::Gm::etcABC to overexpress etcABC. The photos are representatives of three independent experiments.
FIGURE 5.
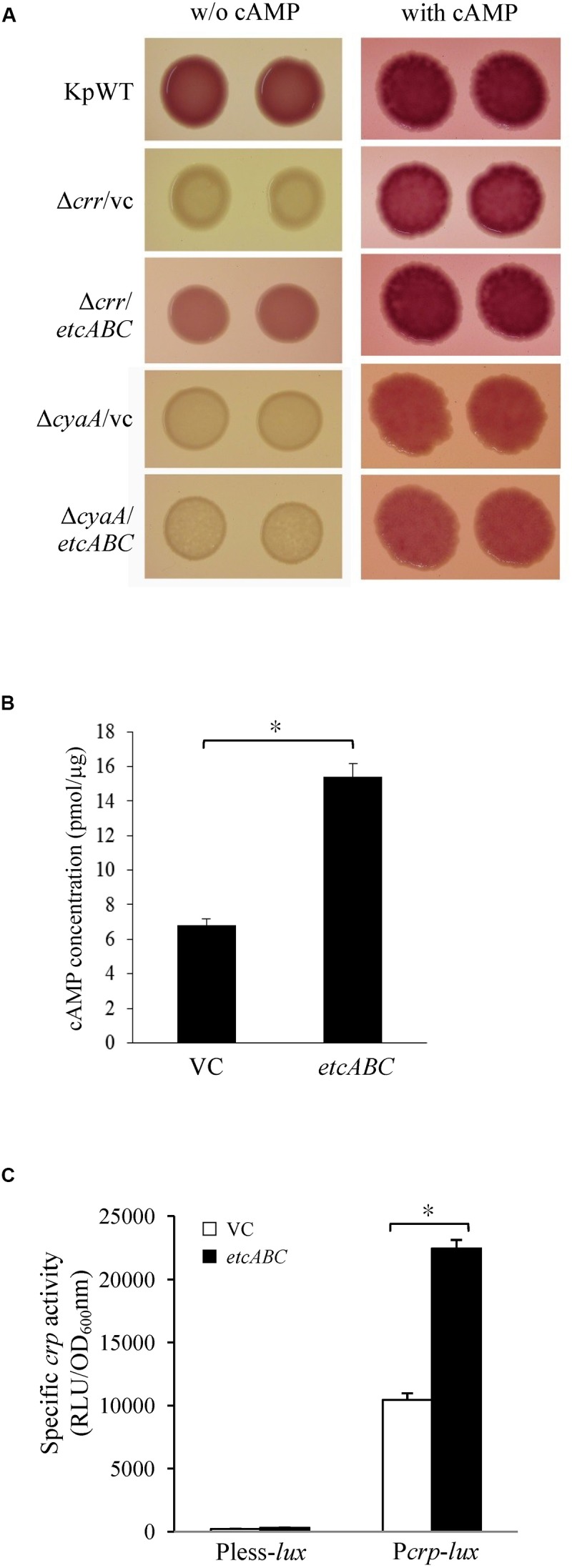
The effects of etcABC on cAMP production. (A) The representative colony morphology of K. pneumoniae STU1 and its derivative strains on MacConkey agar without (w/o) or with 1 mM cAMP in three independent experiments. KpWT: wild-type. Δcrr/vc: crr mutant carrying pBSK-Gm as the vector control. Δcrr/etcABC: crr mutant carrying pBSK::Gm::etcABC to overexpress etcABC. ΔcyaA/vc: cyaA mutant carrying pBSK-Gm as the vector control. ΔcyaA/etcABC: cyaA mutant carrying pBSK::Gm::etcABC to overexpress etcABC. (B) Quantification of intracellular cAMP levels by ELISA. (C) Quantification of luxCDABE luciferase activity in derivative strains of K. pneumoniae STU1. Pless-lux: Bacteria containing pPless-lux, a plasmid carrying a promoterless luxCDABE. Pcrp-lux: Bacteria containing pPcrp-lux, a plasmid carrying the crp promoter followed by the promoterless luxCDABE. VC in panels (B,C): K. pneumoniae ΔcrrΔetcABC carrying pBSK-Gm as the vector control. etcABC in panels (B,C): K. pneumoniae ΔcrrΔetcABC carrying pBSK::Gm::etcABC to overexpress etcABC. The data in panels (B,C) are presented as the averages ± standard deviations of at least three replicates. An asterisk (∗) represents p < 0.05 as compared with VC.
Overexpression of etcABC Compensated for the Role of crr in lac Operon Activation by CRP-cAMP Signaling
A Salmonella typhimurium mutant strain lacking crr, which encodes EIIAGlc, was reported to exhibit low levels of cAMP in medium containing galactose as the sole carbon source (Feucht and Saier, 1980). In this study, the K. pneumoniae crr mutant and its vector control formed very pale pink colonies on MacConkey agar and slightly blue colonies on LB agar containing X-gal and failed to grow in minimal medium containing lactose as the sole carbon source (Figure 4B). We assumed that deletion of crr resulted in a low level of cAMP in K. pneumoniae, leading to inactivation of the lac operon and, subsequently, pale colonies on MacConkey agar and LB agar containing X-gal, as well as death in medium containing lactose as the sole carbon source. To test this hypothesis, cAMP was added to MacConkey agar. The wild-type phenotype was restored in the crr mutant carrying the empty vector by the addition of cAMP (Figure 5A), indicating that Crr positively regulated the intracellular cAMP level in K. pneumoniae. However, overexpression of etcABC in the crr mutant restored the wild-type phenotype on MacConkey agar, LB agar containing X-gal and in minimal medium containing lactose as the sole carbon source but did not restore the wild-type phenotype in the cyaA mutant (Figure 4). This result strongly suggested that overexpression of etcABC compensated for the role of crr in CRP-cAMP-mediated activation of the lac operon.
Overexpression of etcABC Increased Intracellular cAMP Levels
Overexpression of etcABC in the cyaA mutant did not result in pink colonies on MacConkey agar (Figure 4A, 5A). However, the addition of cAMP restored the wild-type phenotype in not only the cyaA mutant strain but also the cyaA mutant strain overexpressing etcABC (Figure 5A). This result indicated that cAMP production was dependent on AC. In a previous study, the phosphorylated form of Crr, EIIAGlc, was reported to stimulate the activity of AC (CyaA) in E. coli by protein–protein interactions, leading to increased cAMP production in medium lacking glucose (Deutscher et al., 2006). Moreover, EtcA is predicted to be an EIIA component. Therefore, we hypothesized that overexpression of etcABC can increase the activity of CyaA to elevate the level of cAMP. To test this hypothesis, we quantified the intracellular cAMP levels by ELISA and showed that compared to the vector control, etcABC overexpression in K. pneumoniae ΔcrrΔetcABC could increase intracellular cAMP levels (Figure 5B). We also examined the effects of etcABC overexpression on the intracellular cAMP levels in K. pneumoniae STU1 wild type and Δcrr. The results showed that etcABC overexpression in wild type and crr mutant also increased intracellular cAMP levels, compared to their corresponding vector control. However, the difference of cAMP levels between etcABC overexpression and vector control in ΔcrrΔetcABC was larger than those in wild type and crr mutant (Supplementary Figure S6).
cAMP receptor protein was reported to be positively autoregulated at the transcriptional level by CRP-cAMP in E. coli (Ishizuka et al., 1994). Therefore, a crp promoter-controlled luxCDABE reporter plasmid was constructed and transformed into K. pneumoniae ΔcrrΔetcABC to test the effect of exogenous cAMP on crp expression. The results showed that 10 min after cAMP was added, crp promoter activity increased relative to that in medium with no exogenous cAMP (Supplementary Figure S7), indicating that the transcriptional level of crp was dependent on cAMP in K. pneumoniae, as in E. coli. Subsequently, we tested the effect of etcABC overexpression on crp expression. Analysis of the bioluminescence emission revealed that the activity of the crp promoter in the strain overexpressing etcABC was twofold higher than that in the strain containing the vector (Figure 5C), indicating that overexpression of etcABC increased the transcriptional level of the crp gene. Thus, these results allowed us to conclude that overexpression of etcABC increased the levels of cAMP and activity of CRP-cAMP.
Discussion
The knowledge about lac operon regulation by CRP-cAMP is mainly derived from studies in E. coli and S. typhimurium. The results of our study showed the same regulatory mechanisms in K. pneumoniae: (1) CRP and cyaA are essential for lac operon activation and lactose uptake (Figure 2D, 4A), and (2) cyaA is responsible for cAMP production (Figure 5). Furthermore, we observed that CRP positively regulated MrkA expression in K. pneumoniae STU1 grown in LB broth (Figure 2C). However, a conflicting result reported by Lin et al. (2016) showed that MrkA production in LB broth was markedly increased by deletion of cyaA or crp from the parent strain, K. pneumoniae CG43S3 and also reported that the mRNA levels of mrkABCDF genes and the activity of the mrkA promoter were increased in the crp mutant strain grown in LB broth. The authors suggested that CRP repressed type 3 fimbriae expression (Lin et al., 2016). After screening published reports, we found a report from Luo et al. (2017) showing that the transcriptional activity of the KP1-4563 gene was increased in the crp mutant compared to that in the parent strain, K. pneumoniae NTUH-K2044. In addition, Luo et al. (2017) observed that the KP1-4563 gene negatively regulated the function of type 3 fimbriae by indirect observation methods such as a mannose-resistant hemagglutination assay (MRHA) and bacterial adhesion assay. Although Luo et al. (2017) did not directly examine type 3 fimbriae synthesis in the crp mutant, they proposed that CRP positively regulated the function of type 3 fimbriae in K. pneumoniae NTUH-K2044. In addition, another research team found via yeast agglutination that CRP enhances fimbrial activity in K. pneumoniae NTUH-K2044 (Ou et al., 2017). To clarify the different findings about CRP and type 3 fimbriae, we randomly selected two clinical K. pneumoniae isolates for crp gene deletion. Neither of these two crp mutants grown in LB broth expressed MrkA, but their parent strains did (Figure 2C), suggesting that CRP positively regulated type 3 fimbriae synthesis in these two clinical K. pneumoniae isolates. Therefore, we believe that negative regulation of type 3 fimbriae by CRP is uncommon in K. pneumoniae.
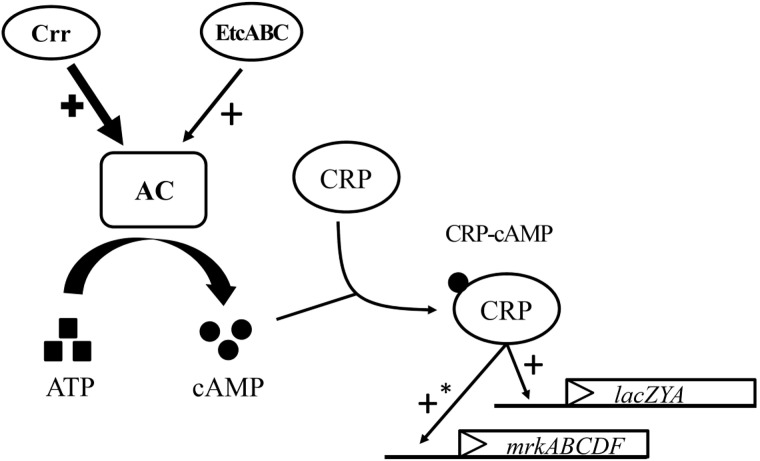
The K. pneumoniae crr mutant formed very pale pink colonies on MacConkey agar and slightly blue colonies on LB agar containing X-gal and failed to grow in minimal medium containing lactose as the sole carbon source (Figure 4B). The mutant strain lacking etcABC, like the wild-type strain, formed pink colonies on MacConkey agar and blue colonies on LB agar containing X-gal and exhibited growth in medium containing lactose as the sole carbon source (data not shown). The phenotype of the strain with the double mutation of etcABC and crr was similar to that of the crr mutant on MacConkey agar, on LB agar containing X-gal and in medium containing lactose as the sole carbon source (data not shown), indicating that compared to etcABC, crr played a more prominent role in the regulation of the lac operon by CRP-cAMP. However, overexpression of etcABC compensated for the role of crr in both the lac operon regulation phenotype and lactose uptake, which are mediated by CRP-cAMP. Moreover, overexpression of etcABC increased type 3 fimbriae synthesis, compared to the vector control (Figure 1, 3). However, type 3 fimbriae synthesis did not differ between the mutant strain lacking etcABC and the wild-type strain (data not shown). Therefore, we propose the following mechanism by which overexpression of etcABC enhances type 3 fimbriae synthesis in K. pneumoniae (Figure 6): in LB medium, the phosphorylation level of Crr, also known as EIIAGlc, is presumably high. Phosphorylated Crr enhances the activity of AC to produce cAMP (Deutscher et al., 2006). However, in unknown conditions in which the activity of Crr is likely reduced in K. pneumoniae, the activity of EtcABC (or EtcA, as discussed below) is increased to enhance the AC activity to produce cAMP. The increased level of intracellular cAMP leads to an increased level of CRP-cAMP and then activates the transcription of the lac and mrk operons. Activation of the latter operon results in the promotion of type 3 fimbriae synthesis, biofilm formation, and adhesion to cells in K. pneumoniae (Figure 6). However, how does CRP activate the transcription of mrk operon? Previous studies showed that MrkH positively regulates mrkHI bicistronic and mrkABCDF operon by direct binding on the upstream region of the target genes in the presence of c-di-GMP (Wilksch et al., 2011; Tan et al., 2015). In this study, the transcription of mrkH is reduced significantly in crp mutant, compared to wild type (Supplementary Figures S3D–F). We speculate that CRP positively regulates the transcription of mrkH. Then, MrkH activates the mrkABCDF operon to produce type 3 fimbriae. We are constructing the mrkH mutant overexpressing CRP and crp mutant overexpressing MrkH to confirm whether regulation of mrkABCDF by CRP can be independent on MrkH. However, the role of CRP binding to the upstream of mrkA in K. pneumoniae STU1 is unclear in the present study. Perhaps, binding of CRP to this site affects the neighbor region in the opposite direction of mrkA.
FIGURE 6.
Model for the role of etcABC in the regulation of type 3 fimbriae synthesis and lactose uptake in K. pneumoniae. In LB medium, Crr and/or EtcABC activate AC to produce cAMP. Then, cAMP binds to CRP to enhance the transcriptional activity of CRP. CRP-cAMP increases the transcription of lacZYA, resulting in bacterial lactose transportation and metabolism. CRP-cAMP increases the transcription of mrkABCDF, resulting in an increase in type 3 fimbriae production. The effect of Crr on AC is much greater than that of EtcABC in LB medium. An asterisk (∗) means indirect regulation. The possible mechanism was discussed in the Section “Discussion.”
Luo et al. (2017) reported that the KP1-4563 gene located upstream of the mrk cluster in K. pneumoniae NTUH-K2044 encodes a hypothetical protein with a putative conserved domain, DUF1471, with an unknown function. The authors reported that the CRP binding site located upstream of KP1-4563 and CRP negatively regulated KP1-4563. Compared to the wild-type strain, the KP1-4563-deleted mutant strain exhibited increased activity in the MRHA, mannan-binding assay and bacterial adhesion assay. Therefore, Luo et al. (2017) suggested that KP1-4563 negatively regulates the function of type 3 fimbriae. However, these three assays in the report by Luo et al. (2017) did not allow direct observation of type 3 fimbriae synthesis such as Western blotting using antibody specific to type 3 fimbriae. Some other factors in K. pneumoniae may affect the results of MRHA (Stahlhut et al., 2012), mannan-binding assay (Madison et al., 1994), and bacterial adhesion assay (Chung, 2016; Horng et al., 2018). Therefore, the regulation of MRHA, mannan-binding assay and bacterial adhesion assay by KP1-4563 depending on type 3 fimbriae or not is unclear. However, we found KPN03280, located upstream of mrkA in K. pneumoniae MGH78578, is homologous to KP1-4563 in K. pneumoniae NTUH-K2044. The putative CRP binding site was located upstream of KPN03280 in K. pneumoniae MGH78578. Although the upstream of mrkA in K. pneumoniae STU1 is not clarified, open reading frame homologous to KP1-4563 is possibly located upstream of mrkA in K. pneumoniae STU1. Thus, the effect of CRP on type 3 fimbriae should be further investigated to clarify whether the mechanism operates through indirect control via KP1-4563 homolog.
Overexpression of etcABC increased MrkA expression, compared to the vector control (Figure 3). We found that the strain overexpressing etcA showed greater MrkA production than the vector control but less MrkA expression than the strain overexpressing etcABC (data not shown). Therefore, here, we studied the effects of etcABC overexpression instead of etcA overexpression in K. pneumoniae. Previous studies showed that phosphorylation state of EIIA affects its activity (Deutscher et al., 2006; Deuschle et al., 2015). Therefore, we hypothesize that the different results from etcABC overexpression and etcA overexpression were due to the phosphorylation levels of EtcA in bacteria. However, we could not confirm the phosphorylation state of EtcA in this study. We previously found that KPN00353 is in the fructose-mannitol EIIA (EIIFru and EIIAMtl) family, a subfamily of the glucose-fructose-lactose PTS superfamily. There is 98% identity between EtcA and KPN00353 amino acid sequence. Crr is an EIIA in the glucose family, another subfamily of the glucose-fructose-lactose PTS superfamily. The identity between KPN00353 and Crr is not very high (Jeng et al., 2017). Therefore, the interaction of EtcA with AC and the phosphorylation state of EtcA need further study.
Because type 3 fimbriae play an important role in K. pneumoniae biofilm formation, several transcription factors have been reported to be involved in regulating type 3 fimbriae expression. These transcription factors include the histone-like nucleoid-structuring protein (H-NS); MrkHI, which depends on c-di-GMP; IscR, which also acts as a crucial transcriptional regulator to control iron–sulfur (Fe–S) cluster biosynthesis in bacteria; and OmpR, which is a response regulator in the OmpR/EnvZ two-component system, which senses osmotic conditions (Tan et al., 2015; Ares et al., 2016; Lin et al., 2017, 2018). We found that overexpression of EtcABC, EII components of a PTS, positively regulate type 3 fimbriae synthesis in K. pneumoniae via CRP. To our knowledge, EtcABC (homologous to KPN00353-KPN00352-KPN00351) is a unique EII complex of PTS in K. pneumoniae strains and is not found in other bacterial strains (Jeng et al., 2017). Therefore, EtcABC may play a unique role in K. pneumoniae pathogenesis. One limitation of this study is that the role of EtcABC in regulating type 3 fimbrial synthesis under various physiologic conditions is unclear. For example, we found that an etcABC mutant had similar expression of type 3 fimbriae when grown in LB media compared to wild type. There may, however, be environmental conditions in which EtcABC is more highly expressed and therefore does affect type 3 fimbriae expression as demonstrated by our experiments utilizing EtcABC overexpression in trans.
Conclusion
In conclusion, we showed that the CRP-cAMP signaling pathway positively regulated the lac operon and type 3 fimbriae synthesis by activating mrkABCDF in at least three K. pneumoniae isolates. Moreover, cAMP played a critical role in lac operon activation. The glucose-specific EIIA, Crr, positively regulated the lac operon via AC, which produces cAMP. Moreover, the putative EII complex EtcABC overexpression compensated for the role of Crr in lac operon activation and positively regulated type 3 fimbriae synthesis by increasing the transcription level of mrkABCDF in K. pneumoniae via CRP-cAMP. Thus, we concluded that K. pneumoniae modulated biofilm formation via the PTS and the CRP-cAMP signaling pathway.
Author Contributions
NP performed most of the experiments and wrote and revised the manuscript. Y-TH analyzed the data and wrote and revised the manuscript. S-WC constructed the suicide plasmid and crp mutant. W-TC purified the CRP and performed the EMSA. P-CS conceived and designed the study, performed and supervised the experiments, analyzed the data, and wrote and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Electron Microscopy Laboratory of the Tzu Chi University for technical assistance in TEM.
Footnotes
Funding. This work was supported by the grant MOST 107-2320-B-320-001-MY3 from the Ministry of Science and Technology and the grant TCRPP107007 from the Tzu Chi University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01558/full#supplementary-material
References
- Allen B. L., Gerlach G. F., Clegg S. (1991). Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 173 916–920. 10.1128/jb.173.2.916-920.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M. A., Fernandez-Vazquez J. L., Rosales-Reyes R., Jarillo-Quijada M. D., von Bargen K., Torres J., et al. (2016). H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 Pili and the Capsule Polysaccharide. Front. Cell. Infect. Microbiol. 6:13. 10.3389/fcimb.2016.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf S. L., Cardani M. A., Bender R. A. (1988). Regulation of the galactose-inducible lac operon and the histidine utilization operons in pts mutants of Klebsiella aerogenes. J. Bacteriol. 170 5588–5593. 10.1128/jb.170.12.5588-5593.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P. Y. (2016). The emerging problems of Klebsiella pneumoniae infections: carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 363:fnw219. 10.1093/femsle/fnw219 [DOI] [PubMed] [Google Scholar]
- Clegg S., Murphy C. N. (2016). Epidemiology and virulence of Klebsiella pneumoniae. Microbiol. Spectr. 4:UTI-0005-2012. 10.1128/microbiolspec.UTI-0005-2012 [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172 6568–6572. 10.1128/jb.172.11.6568-6572.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle M., Limbrunner S., Rother D., Wahler S., Chavarria M., de Lorenzo V., et al. (2015). Interplay of the PtsN (EIIA(Ntr)) protein of Pseudomonas putida with its target sensor kinase KdpD. Environ. Microbiol. Rep. 7 899–907. 10.1111/1758-2229.12323 [DOI] [PubMed] [Google Scholar]
- Deutscher J., Ake F. M., Derkaoui M., Zebre A. C., Cao T. N., Bouraoui H., et al. (2014). The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78 231–256. 10.1128/MMBR.00001-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Francke C., Postma P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70 939–1031. 10.1128/MMBR.00024-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht B. U., Saier M. H., Jr. (1980). Fine control of adenylate cyclase by the phosphoenolpyruvate:sugar phosphotransferase systems in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 141 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosset G., Zhang Z., Nayyar S., Cuevas W. A., Saier M. H., Jr. (2004). Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186 3516–3524. 10.1128/JB.186.11.3516-3524.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Stapleton M. R., Smith L. J., Artymiuk P. J., Kahramanoglou C., Hunt D. M., et al. (2014). Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr. Opin. Microbiol. 18 1–7. 10.1016/j.mib.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera A., Ebright Y. W., Ebright R. H. (1992). DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J. Biol. Chem. 267 14713–14720. [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman J. G. (2001). Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 1547 1–17. 10.1016/s0167-4838(01)00187-x [DOI] [PubMed] [Google Scholar]
- Horng Y. T., Wang C. J., Chung W. T., Chao H. J., Chen Y. Y., Soo P. C. (2018). Phosphoenolpyruvate phosphotransferase system components positively regulate Klebsiella biofilm formation. J. Microbiol. Immunol. Infect. 51 174–183. 10.1016/j.jmii.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Hornick D. B., Allen B. L., Horn M. A., Clegg S. (1992). Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect. Immun. 601577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka H., Hanamura A., Inada T., Aiba H. (1994). Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 13 3077–3082. 10.1002/j.1460-2075.1994.tb06606.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagnow J., Clegg S. (2003). Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149(Pt 9), 2397–2405. 10.1099/mic.0.26434-26430 [DOI] [PubMed] [Google Scholar]
- Jeng W. Y., Panjaitan N. S. D., Horng Y. T., Chung W. T., Chien C. C., Soo P. C. (2017). The Negative Effects of KPN00353 on Glycerol Kinase and Microaerobic 1,3-Propanediol Production in Klebsiella pneumoniae. Front. Microbiol. 8:2441. 10.3389/fmicb.2017.02441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. G., Murphy C. N., Sippy J., Johnson T. J., Clegg S. (2011). Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193 3453–3460. 10.1128/JB.00286-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpa M., Walaszek M., Gniadek A., Wolak Z., Dobros W. (2018). Incidence, microbiological profile and risk factors of healthcare-associated infections in intensive care units: a 10 year observation in a Provincial hospital in Southern Poland. Int. J. Environ. Res. Public Health 15:E112. 10.3390/ijerph15010112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. T., Lin T. H., Wu C. C., Wan L., Huang C. F., Peng H. L. (2016). CRP-Cyclic AMP Regulates the Expression of Type 3 Fimbriae via Cyclic di-GMP in Klebsiella pneumoniae. PLoS One 11:e0162884. 10.1371/journal.pone.0162884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. H., Chen Y., Kuo J. T., Lai Y. C., Wu C. C., Huang C. F., et al. (2018). Phosphorylated OmpR Is Required for Type 3 Fimbriae Expression in Klebsiella pneumoniae Under Hypertonic Conditions. Front. Microbiol. 9:2405. 10.3389/fmicb.2018.02405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. H., Tseng C. Y., Lai Y. C., Wu C. C., Huang C. F., Lin C. T. (2017). IscR Regulation of Type 3 Fimbriae Expression in Klebsiella pneumoniae CG43. Front. Microbiol. 8:1984. 10.3389/fmicb.2017.01984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. (1989). Similarities between prokaryotic and eukaryotic cyclic AMP-responsive promoter elements. Nature 340 656–659. 10.1038/340656a0 [DOI] [PubMed] [Google Scholar]
- Luo M., Yang S., Li X., Liu P., Xue J., Zhou X., et al. (2017). The KP1_4563 gene is regulated by the cAMP receptor protein and controls type 3 fimbrial function in Klebsiella pneumoniae NTUH-K2044. PLoS One 12:e0180666. 10.1371/journal.pone.0180666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison B., Ofek I., Clegg S., Abraham S. N. (1994). Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect. Immun. 62 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March C., Moranta D., Regueiro V., Llobet E., Tomas A., Garmendia J., et al. (2011). Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J. Biol. Chem. 286 9956–9967. 10.1074/jbc.M110.181008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang A. (2009). Quantitative effect and regulatory function of cyclic adenosine 5′-phosphate in Escherichia coli. J. Biosci. 34 445–463. 10.1007/s12038-009-0051-1 [DOI] [PubMed] [Google Scholar]
- Novichkov P. S., Laikova O. N., Novichkova E. S., Gelfand M. S., Arkin A. P., Dubchak I., et al. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38 D111–D118. 10.1093/nar/gkp894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C. L., Beatson S. A., Totsika M., Forestier C., McEwan A. G., Schembri M. A. (2010). Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC Microbiol. 10:183. 10.1186/1471-2180-10-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A., Kolter R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, pathways convergent signalling: a genetic analysis. Mol. Microbiol. 28 449–461. 10.1046/j.1365-2958.1998.00797.x [DOI] [PubMed] [Google Scholar]
- Ou Q., Fan J., Duan D., Xu L., Wang J., Zhou D., et al. (2017). Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J. Med. Microbiol. 66 1–7. 10.1099/jmm.0.000391 [DOI] [PubMed] [Google Scholar]
- Percival S. L., Suleman L., Vuotto C., Donelli G. (2015). Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64(Pt 4), 323–334. 10.1099/jmm.0.000032 [DOI] [PubMed] [Google Scholar]
- Reid G., Charbonneau-Smith R., Lam D., Kang Y. S., Lacerte M., Hayes K. C. (1992). Bacterial biofilm formation in the urinary bladder of spinal cord injured patients. Paraplegia 30 711–717. 10.1038/sc.1992.138 [DOI] [PubMed] [Google Scholar]
- Rosen D. A., Pinkner J. S., Walker J. N., Elam J. S., Jones J. M., Hultgren S. J. (2008). Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 76 3346–3356. 10.1128/IAI.00340-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Tauch A., Jager W., Kalinowski J., Thierbach G., Puhler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Schroll C., Barken K. B., Krogfelt K. A., Struve C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. 10.1186/1471-2180-10-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M. A., Zeng W. (2016). Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc Natl Acad Sci U.S.A. 113 10067–10072. 10.1073/pnas.1607503113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebghati T. A., Korhonen T. K., Hornick D. B., Clegg S. (1998). Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 66 2887–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Singhai M., Malik A., Shahid M., Malik M. A., Goyal R. (2012). A study on device-related infections with special reference to biofilm production and antibiotic resistance. J. Glob. Infect. Dis. 4 193–198. 10.4103/0974-777X.103896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo P. C., Horng Y. T., Chang Y. L., Tsai W. W., Jeng W. Y., Lu C. C., et al. (2014). ManA is regulated by RssAB signaling and promotes motility in Serratia marcescens. Res. Microbiol. 165 21–29. 10.1016/j.resmic.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Soo P. C., Horng Y. T., Lai M. J., Wei J. R., Hsieh S. C., Chang Y. L., et al. (2007). Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J. Bacteriol. 189 109–118. 10.1128/JB.00710-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo P. C., Horng Y. T., Wei J. R., Shu J. C., Lu C. C., Lai H. C. (2008). Regulation of swarming motility and flhDC(Sm) expression by RssAB signaling in Serratia marcescens. J. Bacteriol. 190 2496–2504. 10.1128/JB.01670-1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut S. G., Struve C., Krogfelt K. A. (2012). Klebsiella pneumoniae type 3 fimbriae agglutinate yeast in a mannose-resistant manner. J. Med. Microbiol. 61(Pt 3), 317–322. 10.1099/jmm.0.036350-36350 [DOI] [PubMed] [Google Scholar]
- Struve C., Bojer M., Krogfelt K. A. (2008). Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76 4055–4065. 10.1128/IAI.00494-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A. (1980). An equilibrium study of the cooperative binding of adenosine cyclic 3′,5′-monophosphate and guanosine cyclic 3′,5′-monophosphate to the adenosine cyclic 3′,5′-monophosphate receptor protein from Escherichia coli. Biochemistry 19 5124–5130. 10.1021/bi00563a029 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A., Hillen W. (1989). Ligand-modulated binding of a gene regulatory protein to DNA. Quantitative analysis of cyclic-AMP induced binding of CRP from Escherichia coli to non-specific and specific DNA targets. J. Mol. Biol. 207 783–796. [DOI] [PubMed] [Google Scholar]
- Tan J. W., Wilksch J. J., Hocking D. M., Wang N., Srikhanta Y. N., Tauschek M., et al. (2015). Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J. Bacteriol. 197 1659–1667. 10.1128/JB.02615-2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkkanen A. M., Virkola R., Clegg S., Korhonen T. K. (1997). Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect. Immun. 65 1546–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuotto C., Longo F., Balice M. P., Donelli G., Varaldo P. E. (2014). Antibiotic Resistance Related to Biofilm Formation in Klebsiella pneumoniae. Pathogens 3 743–758. 10.3390/pathogens3030743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wilksch J. J., Chen L., Tan J. W., Strugnell R. A., Gee M. L. (2017). Influence of fimbriae on bacterial adhesion and viscoelasticity and correlations of the two properties with biofilm formation. Langmuir 33 100–106. 10.1021/acs.langmuir.6b03764 [DOI] [PubMed] [Google Scholar]
- Wilksch J. J., Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. 10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Constantinidou C., Hobman J. L., Minchin S. D. (2004). Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic. Acids Res. 32 5874–5893. 10.1093/nar/gkh908 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.