Graphical abstract
Keywords: Assembly processes, Microbial communities, Single cell, Phyllosphere, Spatially explicit, Plant immunity
Highlights
-
•
The physicochemistry of leaves is unique and is a major driver of leaf colonisation.
-
•
Competition and cooperation may be major drivers of bacterial colonisation.
-
•
Leaves respond to bacterial colonisation locally and systemically.
-
•
How leaf responses shape bacterial colonisation patterns is unclear.
-
•
Plant-microbe interaction should be studied at the micrometer resolution.
Abstract
Bacteria establish complex, compositionally consistent communities on healthy leaves. Ecological processes such as dispersal, diversification, ecological drift, and selection as well as leaf surface physicochemistry and topology impact community assembly. Since the leaf surface is an oligotrophic environment, species interactions such as competition and cooperation may be major contributors to shape community structure. Furthermore, the plant immune system impacts on microbial community composition, as plant cells respond to bacterial molecules and shape their responses according to the mixture of molecules present. Such tunability of the plant immune network likely enables the plant host to differentiate between pathogenic and non-pathogenic colonisers, avoiding costly immune responses to non-pathogenic colonisers. Plant immune responses are either systemically distributed or locally confined, which in turn affects the colonisation pattern of the associated microbiota. However, how each of these factors impacts the bacterial community is unclear. To better understand this impact, bacterial communities need to be studied at a micrometre resolution, which is the scale that is relevant to the members of the community. Here, current insights into the driving factors influencing the assembly of leaf surface-colonising bacterial communities are discussed, with a special focus on plant host immunity as an emerging factor contributing to bacterial leaf colonisation.
Introduction
All the aboveground surfaces of a plant that represent microbial habitats are referred to as the phyllosphere [1]. In particular, leaf surfaces host a dense population of bacteria (i.e., epiphytes) estimated to reach 107 bacteria per cm2 of leaf surface [2]. Despite the high cell density, leaf surfaces are a challenging ecosystem to colonise and grow on. Epiphytes must cope with constant ultraviolet (UV) radiation exposure, low water and nutrient availability and large temperature fluctuations throughout the day, making leaves an extreme environment [3].
Recent culture-independent sequencing methods have shown that leaves host bacterial communities that are compositionally consistent within a plant species [4], [5], [6]. However, little is known about the factors that shape these communities. Although there is increasing evidence that non-pathogenic leaf-colonising bacteria may stimulate plant growth and provide protection against different stresses [7], [8], [9], [10], [11], [12], [13], [14], the functions of most of these bacteria, their dynamics at the community level, and their interactions with the plant host remain largely unknown.
The leaf surface
The leaf is a highly structured and multi-layered plant organ (Fig. 1). Its microtopography is determined by the first cell layer, namely, the epidermis, which consists of different cell types that regulate many aspects of leaf physiology, such as gas exchange, temperature regulation, and water and secondary metabolite secretion [15].
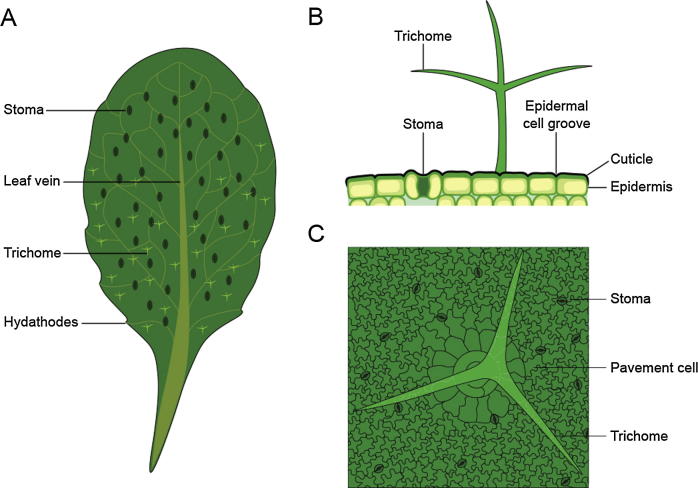
Fig. 1.
(A) Representation of an Arabidopsis thaliana leaf and its main features. (B) Cross-section and (C) top-view representations of the leaf surface, including different epidermal cell types that make up the leaf's surface relief.
The most common cell type in the epidermis is the pavement cell, which contributes to leaf shape. Within the layer of pavement cells, more specialised epidermal cell types are embedded [15]. Stomata, which are pores formed by two guard cells that act as turgor-driven valves to regulate gas exchange and transpiration, are an important feature of the epidermis [16]. Some plants develop modified stomata called hydathodes, which are pores found at the end of the vasculature on leaf margins [16]. Because these structures cannot regulate their pore aperture, hydathodes maintain a continuous pathway for water and solute secretion, a process known as guttation [17]. Another type of specialised epidermal cell are outgrowths called trichomes, which are either glandular or non-glandular [15]. Glandular trichomes are secretion organs that release a wide spectrum of exudates, such as polysaccharides, salts, lipids, volatile compounds, and proteins, the functions of which are associated with plant-plant, plant-insect and plant-microbe interactions [18], [19]. The functions of non-glandular trichomes may include water retention and absorption, light reflection to reduce the impact of UV radiation and heat, and increased freezing as well as drought tolerance [18].
The epidermis is covered by a cuticle, i.e., a waxy layer that provides a physical barrier against abiotic and biotic stresses and determines the physicochemical properties of the leaf surface. The cuticle is formed by an extracellular polymer membrane composed of a matrix of cross-linked polyhydroxy fatty acids and glycerol called cutin. This matrix is interspersed with polysaccharides and a complex mixture of long-chain aliphatic compounds, which are overlaid on and/or impregnated in the matrix (cuticular waxes) [20]. Aliphatic compounds render the cuticle hydrophobic and determine the physicochemical properties of the leaf surface, such as its permeability and wettability, which limits water and solute diffusion from inner cell layers to the leaf surface and the adherence of particles to the surface [21], [22], [23].
Impact of leaf topology and physicochemistry on microbial life on leaves
The organisation of leaf epidermal cell types defines leaf physiology and shapes an intricate microtopology that influences the distribution and abundance of microorganisms on the leaf surface [24], [25], [26]. The establishment of these microhabitats depends on the physicochemical properties of the leaf surface and the ability of microorganisms to adapt and modify this environment [27]. Epiphytes are often found in aggregates or biofilms, likely because these microenvironments protect bacteria from harsh environmental conditions [24], [28]. Large bacterial aggregates have been predominantly found at the bases of trichomes, above veins, and in epidermal cell grooves [29], [30], where water and nutrients are more prevalent.
The permeability and wettability of the leaf cuticle is likely to be one of the most important properties of the leaf surface that influences the ability of microbes to colonise this habitat [31]. Cuticular permeability determines the diffusion rate of compounds from the apoplast onto the leaf surface, while wettability influences the retention of water droplets on the leaf surface [22], [32].
Permeation plays an important role in the growth and survival of epiphytes by allowing the leaching of water and compounds to the phyllosphere, making nutrients accessible for microorganisms. An aqueous pathway contributes to compound permeation across the cuticle with facilitation from aqueous pores preferentially found on cuticular ledges of guard cells, at the base of trichomes, and over the cuticle of anticlinal cell walls [33]. Sites on the leaf surface that are characterised by higher permeation rates are also more densely colonised by bacterial communities [34]. Bacteria can modulate cuticular permeability and wettability through the production of biosurfactants such as syringafactin, which is released by Pseudomonas syringae [31], [35], [36]. Increased cuticular permeability not only affects water diffusion but also alters sugar availability for sustained epiphytic growth [37]. In situ fructose availability to the leaf-colonising bacterium Pantoea eucalypti 299R (formerly known as Erwinia herbicola and Pantoea agglomerans [38], [39]) in the bean phyllosphere was found in sites containing aqueous pores [32], [40], [41]. The patchy distribution of carbon sources on bean leaves promotes differentiation of the P. eucalypti population into subpopulations differing in access to fructose [40]. Thus, permeation of photosynthates across the cuticle is exploited by epiphytic microorganisms, allowing them to survive and thrive. Besides modulation of leaf physicochemistry by phyllosphere-associated microbes, changes in leaf chemistry can also be attributed to abiotic and biotic soil conditions [42], [43], [44]. However, the extent to which plant-soil feedbacks influence the assembly of phyllosphere microbial communities are yet to be determined.
The topography and physicochemical properties of leaves render the phyllosphere an oligotrophic and heterogeneous habitat for epiphytes. It is also possible for microorganisms to construct niches on the leaf surface in response to interaction with the plant [45]. Although discrete hotspots on the leaf form microhabitats in which bacterial populations can be sustained [46], the impact of these microhabitats on the assembly and establishment of bacterial communities is not yet understood.
Composition of phyllosphere-associated microbial communities
Advances in cultivation-independent methods and next-generation sequencing techniques have led to a better understanding of the composition and diversity of plant microbiota. Although plants host a wide variety of microbes, bacteria are far more abundant than eukaryotes and archaea [47], [48]. Bacterial communities on plants are dominated by only four phyla: the Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes [1], [49], [50]. Although bacterial community composition and abundance are host specific, members of the Alphaproteobacteria are predominant and ubiquitous in phyllosphere microbiotas, and within this class, the genera Methylobacterium and Sphingomonas are consistently detected among different hosts [47]. Plant-colonising bacteria are thought to share mechanisms of adaptation to leaf surfaces, considering the high overlap between the proteome of phyllosphere microbiotas and the identification of a core set of genes potentially involved in adaptation to plant colonisation in over 3000 plant-associated bacterial genomes [47], [51], [52].
Spatially explicit ecology of bacteria on leaves
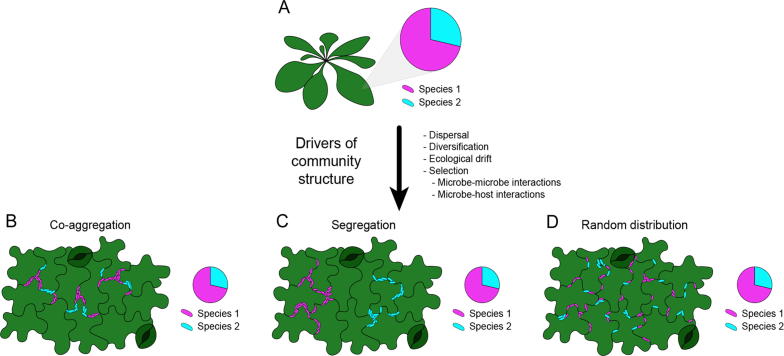
Due to the leaf's heterogeneous nature, the composition and abundance of bacterial communities at the whole-leaf scale are not sufficient to understand the drivers of community assembly [46]. Therefore, the importance of spatial information becomes increasingly apparent for understanding community structure (Fig. 2). In the example given in Fig. 2, the same community composition on a whole-leaf scale can be explained by different interspecies correlations such as different levels of co-aggregation, segregation, or random distributions (Fig. 2C, D, or E, respectively). Fluorescence in situ hybridisation (FISH), a method commonly used to visualise and identify microorganisms in their environment, has been used to describe the distribution patterns of taxonomically different bacterial groups within the phyllosphere [2]. This study, the first investigation of the spatial distribution of phylogenetically different taxa colonising leaf surfaces under different environmental conditions, estimated the likelihood of bacterial taxa co-aggregation in the Arabidopsis thaliana phyllosphere. Bacterial aggregates can be either monoclonal or polyclonal. Monoclonal aggregates represent the offspring of single cells, while polyclonal aggregates are formed by the aggregation of multiple cells in one location [53]. Distantly related taxa can form mixed aggregates, even though members of the same phylogenetic group have the highest probability of co-aggregation. However, due to the technical limitations of the FISH probes used in that study, it is unclear which individual species contribute to the observed aggregation patterns and whether co-aggregation of the same phylogenetic groups is a result of local monoclonal aggregate formation of an individual species or mixed populations. In general, aggregation between different taxa is observed at a distance of less than 5 µm. A similar approach taken to study the spatial distribution and colonisation patterns of two fluorescently tagged bacterial strains (P. eucalypti 299R and P. syringae B728a) on bean leaves provided similar results [26]. In an investigation of the spatial aggregation between the bacterial strains and topological features of the leaf, the strongest correlations were found between bacterial colonies and epidermal cell grooves within distances of up to 12 µm, and adjacent to glandular and non-glandular trichomes within 60 and 120 µm, respectively. Closer examinations of spatial relationships between bacterial species and their surroundings could shed light on the functional and metabolic diversity within aggregates and communities in the phyllosphere.
Fig. 2.
Relevance of spatial patterns in bacterial community structure on the leaf surface. (A) Considering a plant host with a defined microbial composition and relative abundance of species 1 and 2, factors driving community structure, i.e., dispersal, diversification, ecological drift, and selection, can lead to different aggregation patterns of these species while maintaining the same bacterial diversity and abundance. (B) Bacterial community of species 1 and 2 with a strong spatial co-aggregation pattern influenced by, for example, cooperation, resource partitioning, stochastic processes, and/or priority effects (niche modification). (C) Bacterial community of species 1 and 2 with a strong spatial segregation pattern influenced by resource overlap-driven exploitation competition, antibiosis (interference competition), stochastic processes, and/or priority effects (niche preemption). (D) Bacterial community of species 1 and 2 showing a random distribution, which might indicate benign interactions between the species.
Individual bacterial cells can significantly change the concentration of solutes, such as nutrients in their environments, within a distance of approximately ten times their cell's diameter. This distance can be effectively interpreted as an “interaction distance”, where fluxes of compounds and metabolites can diffuse from cell to cell [54]. This interaction distance of cells without direct physical contact is in good agreement with the observed scales of bacterial aggregation in the phyllosphere. Recently, cell-to-cell interactions on porous surfaces were found to be higher when the aqueous phase was fragmented, which increased the probability of direct physical interactions between cells [55]. These findings can be extrapolated to the leaf surface, as its segregated nature should result in a high prevalence of cell-to-cell interactions [56]. Evidently, community assembly on leaves is strongly determined by factors acting at a micrometre resolution [46].
Ecological drivers of bacterial community assembly
Recently, a framework of community assembly was proposed for microbial communities in the phyllosphere [57]. The framework proposes that community structure is driven by four main processes: (1) dispersal, (2) diversification, (3) ecological drift, and (4) selection (Fig. 2A). First, dispersal is the immigration of microorganisms onto the leaf surface, which can occur via seed inoculation, rainfall, animal transmission vectors, bud burst colonisation, and bioaerosols [57]. Second, the emergence of new genetic variation through evolutionary diversification may affect community diversity. As UV radiation and/or reactive oxygen species can lead to increased mutation rates, low-abundance taxa might be a genetic reservoir for horizontal gene transfer; furthermore, bacteria can exhibit dormancy [58]. Third, ecological drift relates to changes in the abundance of taxa due to stochastic events. This process is assumed to have greater effects on low-abundance taxa, which may become extinct at local scales [59]. Lastly, selection is the deterministic fitness differences between species within a community, which can be due to internal and external determinants, such as species interactions and environmental factors, respectively.
Microbe-microbe interactions
In general, the effect of microbial interactions in shaping community structure can be divided into cooperation, parasitism and competition. Cooperation describes interactions that are beneficial to at least one species and do not cause harm to the other, while the latter type refers to interactions that are detrimental to at least one species. Mutualism is a type of cooperative interaction in which both species benefit from each other, while commensalism is an interaction between two organisms in which one partner benefits while the other is not impacted. Microorganisms would be commensals when, for example, one microbe produces non-metabolisable substrates and/or growth factors that positively affect the commensal [60], [61]. However, to be considered a true commensal, the organism should not influence their interaction partner at all, which although theoretically possible, is highly unlikely in practice. The alternative term “tritagonist” has been proposed for these organisms instead [62]. The effect of cooperative interactions in shaping the structure of phyllosphere bacterial communities has not yet been investigated. However, an example of cooperation has been observed using synthetic communities from the maize rhizosphere, in which the removal of a keystone species led to the collapse of other bacterial populations [63]. Another kind of mutualism may occur between fast growing bacterial species and fungal pathogens infecting host plants, as the latter seemed to increase bacterial richness and diversity [64]. However, this depends on specific microbe-microbe interactions, as the fungal and oomycete species Dioszegia sp. and Albugo sp., respectively, have been shown to decrease the bacterial species diversity of the A. thaliana leaf microbiota [45].
Parasitism in the phyllosphere is mostly driven by virus-bacterium interactions. Bacterium-infecting viruses (i.e., bacteriophages or phages) are found in most (if not all) ecosystems and can alter community dynamics by influencing bacterial diversity, nutrient cycling, and species interactions [65]. Phages impose strong selection on bacterial members of the leaf microbiota at a local scale and in short time periods, affecting the microbiota composition [66], [67].
Competitive relationships involve detrimental effects for at least one species, which may be a result of interference or exploitation competition. When competition is the result of active mechanisms of species exclusion, this interaction is known as interference. The most common example of interference is antibiosis, in which a species secretes compounds that are toxic to the other. This effect is also the case for epiphytes; for example, P. agglomerans E325 has been shown to suppress the growth of the phytopathogen Erwinia amylovora on apple flowers through antibiotic activity [13]. Exploitation competition is the dispute for shared resources, such as nutrients or space. In this relationship, one of the species has compromised population growth, resulting in either complete spatial exclusion or coexistence [68]. Fructose and sucrose, for example, are limiting resources during phyllosphere colonisation [40]. In bean leaves, the amount of available sugars on the leaf surface decreases rapidly to a tenth of the initial concentration upon colonisation of Pseudomonas fluorescens A506 [69]. The availability of sugars is also limited by its permeation from the apoplast to the leaf surface, and sugars may not be replenished at rates supporting the survival of large bacterial populations [40].
Effect of resource overlap on species interactions
Cooperative and competitive interactions greatly depend on the metabolic capabilities of the members of a microbial community, that is, the potential of the microbes to uptake, metabolise, and secrete an array of nutrients and other compounds (e.g., siderophores or antimicrobials). The degree of shared nutritional requirements between species, i.e., nutrient or resource overlap, may shape microbial communities, as shown by the fact that nutrient overlaps in naturally occurring bacterial communities are higher than expected based on null models [70]. When estimating nutrient overlaps between over 6000 pair of bacterial species, Freilich et al. (2011) found a positive correlation with the competition potential between species in a community and negatively with the cooperation potential, suggesting that the higher the nutrient overlap between the species in a community is, the more likely these species are to compete for shared nutrients and, consequently, the less likely they are to cooperate with each other [71]. However, nutrient overlaps cannot solely predict the nature of species interactions e.g. it has recently been shown that close relatedness and similarity in gene expression between pairs of algal species, and thus similarity in their metabolisms, was correlated to facilitation, stabilising coexistence [72]. In addition, other factors such as priority effects can influence species interactions through, for example, niche preemptions or niche modifications [73]. Resource utilisation accounts for only a fraction of a species’ niche; thus, the spatial context, environmental conditions, and biotic relationships should be considered when positioning species in their ecosystems.
An operational approach for comparing species’ nutrient utilisation profiles is to determine a resource overlap index between pairs of species. In phyllosphere studies, Wilson and Lindow introduced the niche overlap index in 1994 [74], which relates to the number of carbon sources utilised by two species as a proportion of the total carbon sources used by one of the species. Two issues arise from this formulation: (1) the index is asymmetrical, such that one species can have total overlap while the other only partial overlap, as is the case for specialist and generalist species, respectively; and (2) by measuring only the ability of a strain to consume a carbon source independently, information is lost about the species’ affinity for that resource, which may lead to false interpretation of the potential for two species to coexist based on their nutrient preferences. Instead, symmetrical indices that account for the proportions of utilised nutrients are more informative and less biased [75].
The major drivers of community assembly in the phyllosphere are currently unclear. As the phyllosphere is generally oligotrophic, cooperation and competition may have a major impact on community assemblies in the phyllosphere.
The host as a driver of bacterial community structure
Plants provide habitats that support different bacterial communities [4], [76]. Although bacteria share a core set of adaptation mechanisms to colonise and thrive on plants [4], [47], [51], community composition is, to some degree, host specific [6], [76], [77], [78], [79]; thus, changes in bacterial diversity, abundance, and community structure can be attributed to host factors and specific bacterial adaptation mechanisms. For example, bacterial community composition is influenced by host species, genotype, plant traits (e.g., cuticle composition), leaf age and host developmental stage; the latter two factors are often indistinguishable from seasonal effects [25], [79], [80], [81], [82], [83], [84].
Plant responses to microbial colonisation
In addition to providing a habitat for microorganisms, plants interact with their associated microbiota. Recent findings suggest that the plant immune system plays a role in shaping the bacterial community [80], [85], thereby indicating an active role of the host plant in modulating the composition of its associated microbiota.
The plant immune system consists of two layers. The first layer of immunity, pattern-triggered immunity (PTI), is elicited by conserved molecular structures such as microbe/pathogen-associated molecular patterns (MAMPs/PAMPs) or damage-associated molecular patterns, which are perceived by plasma membrane-localised pattern recognition receptors [86], [87]. A prominent example of pattern recognition receptors is the receptor flagellin sensing 2 (FLS2), which recognises a 22-amino-acid peptide of bacterial flagellin (flg22) [88]. The significance of MAMP/PAMP recognition in limiting pathogen growth was shown in a fls2 mutant exhibiting enhanced susceptibility to the bacterial pathogen P. syringae pathovar (pv.) tomato DC3000 (Pst DC3000) [89]. Recently, comparison of plant responses to the growth-promoting rhizobacterium Pseudomonas simiae WCS417 and its cognate flg22 peptide showed that the whole organism elicited only about half of the plant transcriptional responses compared to the purified flg22 peptide alone. Genes that were only upregulated in the flg22-treated plants were enriched for defence-related transcriptional responses, suggesting that P. simiae WCS417 suppressed a significant number of defence-associated genes [90]. Whether plants recognise non-pathogenic bacteria and modulate their response to limit unnecessary costly defence are still unknown.
To subvert PTI, microbial pathogens release so-called effector proteins [91]. Effector proteins from different microbes target overlapping sets of plant proteins. Most of the targeted plant proteins are considered hubs of highly interconnected protein-protein interaction networks. Hence, targeting these proteins will likely result in strong perturbations of the host's immune response [92], [93]. As a countermeasure, plants have evolved additional intracellular receptors, which are often nucleotide-binding leucine-rich repeat proteins (NLRs), that directly or indirectly recognise effector proteins, thus forming the second layer of plant immunity, designated effector-triggered immunity (ETI) [94], [95]. Recently, a study on microbial genes important for adaptation to the plant environment identified 64 plant-resembling plant-associated and root-associated domains (PREPARADOs). Some PREPARADOs resemble NLR domains. This finding suggests that bacterial proteins containing PREPARADOs might compete with NLRs for effector binding, thereby restricting bacterial detection by the plant [51].
Plant responses are finely tuned
Although the same signaling networks seem to be employed during PTI and ETI, they are used in a different manner, generating specific outputs of immunity level. While the relationships between different signaling pathways in PTI are partly synergistic and partly compensatory, they are exclusively compensatory during ETI [96], [97]. As MAMPs can be found on pathogenic as well as non-pathogenic bacteria, it seems reasonable that PTI is less robust or more tunable than ETI, allowing the host to avoid recurrent fitness costs [98]. Synergism between signaling pathways enables the plant to elevate the output of its immune response when multiple MAMPs, providing more information than a single MAMP, signal a pathogen.
Different MAMPs have been shown to activate different immune signaling pathways with varying strengths, leading to diverse immune outputs (Fig. 3) [99]. Such differential use of the immune signaling network likely allows plants to induce appropriate defence mechanisms against pathogens with different lifestyles during PTI. For example, salicylic acid signaling is known to be effective against biotrophs, pathogens that feed on living host tissue, and hemibiotrophs, pathogens that first feed on living host tissue and later feed on dead host tissue. In contrast, jasmonic acid and ethylene signaling is effective against necrotrophs, pathogens that feed on dead host tissue [100]. Furthermore, tunability of the immune network presumably enables plants to limit costly immune responses to non-pathogenic colonisers.
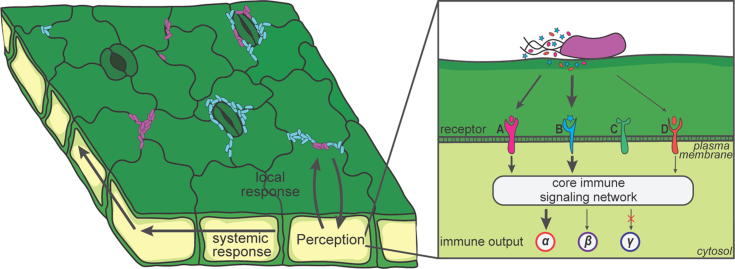
Fig. 3.
Plant responses to bacterial colonisation. Leaf-colonising bacteria elicit local and systemic responses. As shown on the left-hand side, a cell type-specific response to prevent bacteria from entering the apoplast is stomatal closure. As shown on the right-hand side, plants perceive bacteria via receptors localised in the plasma membrane (A, B, C, D), which recognise microbe-associated molecular patterns (depicted by hexagons, ovals and stars around the bacterium). Downstream signaling of these receptors is integrated in a highly interconnected immune signaling network. Integration of varying receptor inputs leads to specific immune outputs (α, β, γ).
Recently, two non-pathogenic bacteria were shown to elicit unique transcriptional and metabolic responses that differed from those of a pathogenic bacterium, indicating that plants differentiate bacteria [7], [101]. Since the availability of carbohydrates clearly affects bacterial community composition [102], [103] and plants actively deprive the apoplast of monosaccharides upon pathogen encounter to limit pathogen growth [104], plants likely also supply certain carbohydrates by sugar exporters [105] to support beneficial bacterial populations. Such responses are likely to be spatially explicit, as the bacterial composition on the leaf is heterogeneous and the same carbohydrate might promote the growth of spatially separated populations of beneficial and non-beneficial bacteria. However, plant responses at a high spatial resolution are currently underinvestigated in the context of plant-microbe interactions. Local plant responses may have important implications for leaf surface-colonising bacteria and bacterial community composition.
Local and systemic plant host responses to microorganisms
Plant immune responses to pathogens can be divided into local and systemic responses (Fig. 3). Local immune responses at the infection site comprise early calcium ion (Ca2+) influx and MAP kinase and calcium-dependent protein kinase activation within minutes, followed by reactive oxygen species production, defence gene activation and after several hours, callose deposition and hypersensitivity response resulting in programmed cell death, which is regarded as a hallmark of ETI [106]. A cell type-specific local response is stomatal closure, which occurs within an hour after bacterial recognition to restrict the bacteria from entering the apoplast (Fig. 3). However, some bacterial pathogens, such as Pst DC3000, are able to modulate stomatal aperture [107]. In addition to local responses, pathogens also elicit systemic responses, such as systemic acquired resistance, conferring broad spectrum resistance against biotrophic pathogens to plant parts that have not been in contact with the pathogen [108]. Recently, the impressively fast propagation of systemic Ca2+ signaling throughout the plant in A. thaliana (approximately 1 mm/s) was shown to be mediated by glutamate, which is released upon wounding [109].
Studying plant epigenetic, transcriptional, proteomic, and metabolomic changes in microorganisms at the whole-tissue level does not allow us to study local and systemic responses separately. Moreover, tissues are mixtures of different cell types that react differently to microorganisms. In a recent study, local responses to the biotrophic pathogen Hyaloperonospora arabidopsis were shown to differ markedly from systemic responses [110]. This finding highlights the importance of performing -omic studies at a high spatial (potentially single-host cell) resolution to identify the nature of plant responses. To perform cell type-specific and single-cell studies, the cell type of interest must be isolated from the surrounding tissue. Root hairs are likely the simplest cell type to isolate as they can be separated from frozen root tips by stirring with a glass rod [97]. Other cell types demand more sophisticated techniques for isolation; these types can be divided into two groups. Microscopy-assisted techniques such as capillary extraction [111], atomic force microscopy-based extraction [112] and laser capture microdissection [113] allow the isolation of cells or cell material with direct spatial context. With regard to plant-microorganism interactions, capillary extraction was used to study transcriptional changes of barley cells infected by powdery mildew [114]. Cell/nuclei sorting or affinity purification methods such as fluorescence-activated cell sorting of protoplasts [115], fluorescence-activated nuclei sorting [116], isolation of nuclei tagged in specific cell types [117], and “translating ribosome affinity purification” (TRAP) [118] allow the isolation of larger numbers of cells or more cell material. However, the spatial information is only indirect, and the techniques rely on cell type-specific markers.
Plants shape their associated microbiotas. Consequently, plants need to be considered as a driving factor of community composition in bacterial ecology studies. To study the role of the plant host in shaping microbiota, new controls must be found that distinguish between fast active plant signaling and comparably slow changes in physicochemical properties of leaves. To that end, studies can be performed on artificial plants, ranging from plastic tomato plants in a field context [119] to artificial leaves in the laboratory [120], [121], [122], [123]. Such studies should progress to a more refined spatial scale, as bacterial colonisation is heterogeneous, plant responses are likely also heterogenous.
Conclusions and future perspectives
The relatively static physicochemistry of the leaf surface as well as dynamic microbe-microbe, microbe-plant and plant-mediated microbe-microbe interactions are drivers of plant microbiota community composition. These factors do not homogeneously influence the bacterial communities on leaves; instead, there might be markedly different communities on the same leaf. Such spatial heterogeneity of leaf-colonising microbial communities is not only due to the heterogeneity of the leaf surface and heterogeneous microbial colonisation but also likely due to plant responses that may differ between individual cells of the same leaf. Since the plant immune network is highly tunable, it presumably enables plants to limit costly immune responses to non-pathogenic colonisers, and on a more refined spatial scale to respond differently to distinct microbial colonisers. Heterogeneous colonisation patterns are thus likely to translate into heterogeneous plant response patterns.
In the future, more emphasis should be placed on spatial heterogeneous differences in colonisation and plant responses to further our understanding of the underlying mechanisms that drive bacterial community composition and assembly. By resolving (1) the effect of microbe-microbe interactions, (2) the function of non-pathogenic bacteria on the plant and (3) the role of the host in shaping community structure, will give bottom-up insights into the intimate interplay of plant hosts and their bacterial communities.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
This work was funded by the Royal Society of New Zealand Marsden Fast Start grant (UOC1704) to MR-E. RS was supported by a New Zealand International Doctoral Research Scholarship, and MM was supported by a University of Canterbury Doctoral Scholarship.
Biographies

Rudolf O. Schlechter is a PhD student at the University of Canterbury, Christchurch, New Zealand under the supervision of Mitja Remus-Emsermann. He received a degree in biochemistry at the Pontificia Universidad Católica de Chile, Chile. In his thesis project, Rudolf characterised the immune response conferred by the loci RUN1 and REN1 in Vitis vinifera against Erysiphe necator. Currently, he is investigating the microbial ecology of plant leaf surfaces colonising bacteria at the single-cell resolution.

Moritz Miebach is a PhD student at the University of Canterbury, Christchurch, New Zealand under the supervision of Mitja Remus-Emsermann. He received a MSc. degree in biology at the Albert-Ludwigs-Universität, Freiburg, Germany. In his thesis project, Moritz investigated how chromatin modifications shape the spatio-temporal transcriptional control of the Arabidopsis root stem cell niche. Currently, he is investigating transcriptional host responses to microbial colonisation.

Mitja Remus-Emsermann is a Senior Lecturer in Microbiology at the University of Canterbury, Christchurch, New Zealand. Mitja received his Diploma in biology at the Rheinische Friedrich-Wilhelms-Universität Bonn, Germany before he joined the group of Prof. Johan Leveau at the Dutch Institute of Ecology (NIOO-KNAW), and received his PhD at the Free University Amsterdam. After 4.5 years of postdoctoral tenure in the laboratory of Julia Vorholt at ETH Zurich and Agroscope, Wädenswil, Switzerland, Mitja joined the School of Biological Sciences at the University of Canterbury as a tenured faculty member in 2016.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Vorholt J.A. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 2.Remus-Emsermann M.N.P., Lücker S., Müller D.B., Potthoff E., Daims H., Vorholt J.A. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol. 2014;16:2329–2340. doi: 10.1111/1462-2920.12482. [DOI] [PubMed] [Google Scholar]
- 3.Leveau J. Microbial Communities in the Phyllosphere. In: Riederer M, Müller C, editors. Biology of the Plant Cuticle, vol. 23, Oxford, UK: Blackwell Publishing Ltd; 2006, p. 334–67.
- 4.Bai Y., Müller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 5.Knief C., Ramette A., Frances L., Alonso-Blanco C., Vorholt J.A. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J. 2010;4:719–728. doi: 10.1038/ismej.2010.9. [DOI] [PubMed] [Google Scholar]
- 6.Redford A.J., Bowers R.M., Knight R., Linhart Y., Fierer N. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol. 2010;12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel C., Bodenhausen N., Gruissem W., Vorholt J.A. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 8.Vogel C., Innerebner G., Zingg J., Guder J., Vorholt J.A. Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp. strain Fr1 against Pseudomonas syringae DC3000. Appl Environ Microbiol. 2012;78:5529–5535. doi: 10.1128/AEM.00639-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Innerebner G., Knief C., Vorholt J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77:3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendes R., Garbeva P., Raaijmakers J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 11.Berg G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84:11–18. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 12.Stockwell V.O., Johnson K.B., Sugar D., Loper J.E. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9–1 applied as single strains and mixed inocula. Phytopathology. 2010;100:1330–1339. doi: 10.1094/PHYTO-03-10-0097. [DOI] [PubMed] [Google Scholar]
- 13.Pusey P.L., Stockwell V.O., Reardon C.L., Smits T.H.M., Duffy B. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology. 2011;101:1234–1241. doi: 10.1094/PHYTO-09-10-0253. [DOI] [PubMed] [Google Scholar]
- 14.Ritpitakphong U., Falquet L., Vimoltust A., Berger A., Métraux J.-P., L’Haridon F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–1043. doi: 10.1111/nph.13808. [DOI] [PubMed] [Google Scholar]
- 15.Glover B.J. Differentiation in plant epidermal cells. J Exp Bot. 2000;51:497–505. doi: 10.1093/jexbot/51.344.497. [DOI] [PubMed] [Google Scholar]
- 16.Nadeau J.A., Sack F.D. Stomatal development in Arabidopsis. Arabidopsis Book. 2002;1 doi: 10.1199/tab.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillitteri L.J., Dong J. Stomatal development in Arabidopsis. Arabidopsis Book. 2013;11 doi: 10.1199/tab.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werker E. Trichome diversity and development. Adv Bot Res. 2000:1–35. [Google Scholar]
- 19.Zhao Q., Development Chen X-Y. A new function of plant trichomes. Nat Plants. 2016;2 doi: 10.1038/nplants.2016.96. [DOI] [PubMed] [Google Scholar]
- 20.Riederer M., Schreiber L. Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot. 2001;52:2023–2032. doi: 10.1093/jexbot/52.363.2023. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber L, Schönherr J. Water and solute permeability of plant cuticles: measurement and data analysis. Springer Science & Business Media; 2009.
- 22.Barthlott W., Neinhuis C. Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta. 1997;202:1–8. [Google Scholar]
- 23.Sadler C., Schroll B., Zeisler V., Waßmann F., Franke R., Schreiber L. Wax and cutin mutants of Arabidopsis: quantitative characterization of the cuticular transport barrier in relation to chemical composition. Biochim Biophys Acta. 2016;1861:1336–1344. doi: 10.1016/j.bbalip.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Monier J.-M., Lindow S.E. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl Environ Microbiol. 2004;70:346–355. doi: 10.1128/AEM.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraishi K., Oku M., Kawaguchi K., Uchida D., Yurimoto H., Sakai Y. Yeast nitrogen utilization in the phyllosphere during plant lifespan under regulation of autophagy. Sci Rep. 2015;5:srep09719. doi: 10.1038/srep09719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esser D.S., Leveau J.H.J., Meyer K.M., Wiegand K. Spatial scales of interactions among bacteria and between bacteria and the leaf surface. FEMS Microbiol Ecol. 2015;91. doi: 10.1093/femsec/fiu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remus-Emsermann M.N.P., Tecon R., Kowalchuk G.A., Leveau J.H.J. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 2012;6:756–765. doi: 10.1038/ismej.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monier J.-M., Lindow S.E. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc Natl Acad Sci USA. 2003;100:15977–15982. doi: 10.1073/pnas.2436560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris C.E., Monier J., Jacques M. Methods for observing microbial biofilms directly on leaf surfaces and recovering them for isolation of culturable microorganisms. Appl Environ Microbiol. 1997;63:1570–1576. doi: 10.1128/aem.63.4.1570-1576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer C.A., Smith W.K., Vogelmann T.C. Functional interaction between leaf trichomes, leaf wettability and the optical properties of water droplets. Plant Cell Environ. 1991;14:955–962. [Google Scholar]
- 31.Burch A.Y., Zeisler V., Yokota K., Schreiber L., Lindow S.E. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ Microbiol. 2014;16:2086–2098. doi: 10.1111/1462-2920.12437. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber L. Polar paths of diffusion across plant cuticles: new evidence for an old hypothesis. Ann Bot. 2005;95:1069–1073. doi: 10.1093/aob/mci122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schönherr J. Characterization of aqueous pores in plant cuticles and permeation of ionic solutes. J Exp Bot. 2006;57:2471–2491. doi: 10.1093/jxb/erj217. [DOI] [PubMed] [Google Scholar]
- 34.Beattie G.A. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol. 2011;49:533–555. doi: 10.1146/annurev-phyto-073009-114436. [DOI] [PubMed] [Google Scholar]
- 35.Schreiber L., Krimm U., Knoll D., Sayed M., Auling G., Kroppenstedt R.M. Plant-microbe interactions: identification of epiphytic bacteria and their ability to alter leaf surface permeability. New Phytol. 2005;166:589–594. doi: 10.1111/j.1469-8137.2005.01343.x. [DOI] [PubMed] [Google Scholar]
- 36.Krimm U., Abanda-Nkpwatt D., Schwab W., Schreiber L. Epiphytic microorganisms on strawberry plants (Fragaria ananassa cv. Elsanta): identification of bacterial isolates and analysis of their interaction with leaf surfaces. FEMS Microbiol Ecol. 2005;53:483–492. doi: 10.1016/j.femsec.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 37.van der Wal A., Leveau J.H.J. Modelling sugar diffusion across plant leaf cuticles: the effect of free water on substrate availability to phyllosphere bacteria. Environ Microbiol. 2011;13:792–797. doi: 10.1111/j.1462-2920.2010.02382.x. [DOI] [PubMed] [Google Scholar]
- 38.Remus-Emsermann M.N.P., Kim E.B., Marco M.L., Tecon R., Draft Leveau JHJ. Genome Sequence of the Phyllosphere Model Bacterium Pantoea agglomerans 299R. Genome Announc. 2013;1 doi: 10.1128/genomeA.00036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tecon R., Leveau J.H.J. Symplasmata are a clonal, conditional, and reversible type of bacterial multicellularity. Sci Rep. 2016;6:31914. doi: 10.1038/srep31914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leveau J.H., Lindow S.E. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc Natl Acad Sci USA. 2001;98:3446–3453. doi: 10.1073/pnas.061629598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remus-Emsermann M.N.P., de Oliveira S., Schreiber L., Leveau J.H.J. Quantification of lateral heterogeneity in carbohydrate permeability of isolated plant leaf cuticles. Front Microbiol. 2011;2:197. doi: 10.3389/fmicb.2011.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohshiro M., Hossain M.A., Nakamura I., Akamine H., Tamaki M., Bhowmik P.C. Effects of soil types and fertilizers on growth, yield, and quality of edible Amaranthus tricolor lines in Okinawa, Japan. Plant Prod Sci. 2016;19:61–72. [Google Scholar]
- 43.Quan M, Liang J. The influences of four types of soil on the growth, physiological and biochemical characteristics of Lycoris aurea (L’ Her.) Herb. Scientific Reports 2017;7. doi: 10.1038/srep43284. [DOI] [PMC free article] [PubMed]
- 44.Meiners S.J., Phipps K.K., Pendergast T.H., 4th, Canam T., Carson W.P. Soil microbial communities alter leaf chemistry and influence allelopathic potential among coexisting plant species. Oecologia. 2017;183:1155–1165. doi: 10.1007/s00442-017-3833-4. [DOI] [PubMed] [Google Scholar]
- 45.Agler M.T., Ruhe J., Kroll S., Morhenn C., Kim S.-T., Weigel D. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remus-Emsermann M.N.P., Schlechter R.O. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018;218:1327–1333. doi: 10.1111/nph.15054. [DOI] [PubMed] [Google Scholar]
- 47.Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bodenhausen N., Horton M.W., Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand A., Maillard F., Alvarez-Lopez V., Guinchard S., Bertheau C., Valot B. Bacterial diversity associated with poplar trees grown on a Hg-contaminated site: community characterization and isolation of Hg-resistant plant growth-promoting bacteria. Sci Total Environ. 2018;622–623:1165–1177. doi: 10.1016/j.scitotenv.2017.12.069. [DOI] [PubMed] [Google Scholar]
- 51.Levy A., Salas Gonzalez I., Mittelviefhaus M., Clingenpeel S., Herrera Paredes S., Miao J. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50:138–150. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambais M.R., Barrera S.E., Santos E.C., Crowley D.E., Jumpponen A. Phyllosphere metaproteomes of trees from the Brazilian atlantic forest show high levels of functional redundancy. Microb Ecol. 2017;73:123–134. doi: 10.1007/s00248-016-0878-6. [DOI] [PubMed] [Google Scholar]
- 53.Tecon R., Leveau J.H.J. The mechanics of bacterial cluster formation on plant leaf surfaces as revealed by bioreporter technology. Environ Microbiol. 2012;14:1325–1332. doi: 10.1111/j.1462-2920.2012.02715.x. [DOI] [PubMed] [Google Scholar]
- 54.Franklin R.B., Mills A.L. Statistical analysis of spatial structure in microbial communities. The Spatial Distribution of Microbes in the Environment. 2007:31–60. [Google Scholar]
- 55.Tecon R., Ebrahimi A., Kleyer H., Erev Levi S., Or D. Cell-to-cell bacterial interactions promoted by drier conditions on soil surfaces. Proc Natl Acad Sci USA. 2018;115:9791–9796. doi: 10.1073/pnas.1808274115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monier J.-M., Lindow S.E. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl Environ Microbiol. 2005;71:5484–5493. doi: 10.1128/AEM.71.9.5484-5493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vacher C., Hampe A., Porté A.J., Sauer U., Compant S., Morris C.E. The phyllosphere: microbial jungle at the plant-climate interface. Annu Rev Ecol Evol Syst. 2016;47:1–24. [Google Scholar]
- 58.Nemergut D.R., Schmidt S.K., Fukami T., O’Neill S.P., Bilinski T.M., Stanish L.F. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Ning D. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 2017;81. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed]
- 60.Faust K., Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 61.Großkopf T., Soyer O.S. Synthetic microbial communities. Curr Opin Microbiol. 2014;18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freimoser F.M., Pelludat C., Remus-Emsermann M.N.P. Tritagonist as a new term for uncharacterised microorganisms in environmental systems. ISME J. 2016;10:1–3. doi: 10.1038/ismej.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu B., Paulson J.N., Zheng X., Kolter R. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA. 2017;114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suda W., Nagasaki A., Shishido M. Powdery mildew-infection changes bacterial community composition in the phyllosphere. Microbes Environ. 2009;24:217–223. doi: 10.1264/jsme2.me09114. [DOI] [PubMed] [Google Scholar]
- 65.Koskella B., Brockhurst M.A. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koskella B. Phage-mediated selection on microbiota of a long-lived host. Curr Biol. 2013;23:1256–1260. doi: 10.1016/j.cub.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 67.Morella N.M., Gomez A.L., Wang G., Leung M.S., Koskella B. The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol Ecol. 2018;27:2025–2038. doi: 10.1111/mec.14542. [DOI] [PubMed] [Google Scholar]
- 68.Begon M. John Wiley & Sons; 2010. Ecology: From Individuals to Ecosystems. [Google Scholar]
- 69.Mercier J., Lindow S.E. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol. 2000;66:369–374. doi: 10.1128/aem.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zelezniak A., Andrejev S., Ponomarova O., Mende D.R., Bork P., Patil K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freilich S., Zarecki R., Eilam O., Segal E.S., Henry C.S., Kupiec M. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 72.Narwani A., Bentlage B., Alexandrou M.A., Fritschie K.J., Delwiche C., Oakley T.H. Ecological interactions and coexistence are predicted by gene expression similarity in freshwater green algae. J Ecol. 2017;105:580–591. [Google Scholar]
- 73.Fukami T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. [Google Scholar]
- 74.Wilson M., Lindow S.E. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) pseudomonas syringae strains and a non-ice-nucleating (Ice-) biological control agent. Appl Environ Microbiol. 1994;60:3128–3137. doi: 10.1128/aem.60.9.3128-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith E.P., Zaret T.M. Bias in estimating niche overlap. Ecology. 1982;63:1248–1253. [Google Scholar]
- 76.Laforest-Lapointe I., Messier C., Kembel S.W. Tree phyllosphere bacterial communities: exploring the magnitude of intra- and inter-individual variation among host species. PeerJ. 2016;4 doi: 10.7717/peerj.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA 2014;111:585–92. [DOI] [PMC free article] [PubMed]
- 78.Coleman-Derr D., Desgarennes D., Fonseca-Garcia C., Gross S., Clingenpeel S., Woyke T. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016;209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner M.R., Lundberg D.S., Del Rio T.G., Tringe S.G., Dangl J.L., Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun. 2016;7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horton M.W., Bodenhausen N., Beilsmith K., Meng D., Muegge B.D., Subramanian S. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bodenhausen N., Bortfeld-Miller M., Ackermann M., Vorholt J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kembel S.W., O’Connor T.K., Arnold H.K., Hubbell S.P., Wright S.J., Green J.L. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rastogi G., Sbodio A., Tech J.J., Suslow T.V., Coaker G.L., Leveau J.H.J. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Copeland J.K., Yuan L., Layeghifard M., Wang P.W., Guttman D.S. Seasonal community succession of the phyllosphere microbiome. Mol Plant Microbe Interact. 2015;28:274–285. doi: 10.1094/MPMI-10-14-0331-FI. [DOI] [PubMed] [Google Scholar]
- 85.Lebeis S.L., Paredes S.H., Lundberg D.S., Breakfield N., Gehring J., McDonald M. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 86.Boller T., Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 87.Monaghan J., Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15:349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 88.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 90.Stringlis I.A., Proietti S., Hickman R., Van Verk M.C., Zamioudis C., Pieterse C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018;93:166–180. doi: 10.1111/tpj.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 92.Mukhtar M.S., Carvunis A.-R., Dreze M., Epple P., Steinbrenner J., Moore J. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weßling R., Epple P., Altmann S., He Y., Yang L., Henz S.R. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bonardi V., Cherkis K., Nishimura M.T., Dangl J.L. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jacob F., Vernaldi S., Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dong X., Jiang Z., Peng Y.-L., Zhang Z. Revealing shared and distinct gene network organization in Arabidopsis immune responses by integrative analysis. Plant Physiol. 2015;167:1186–1203. doi: 10.1104/pp.114.254292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsuda K., Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol. 2010;13:459–465. doi: 10.1016/j.pbi.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 99.Kim Y., Tsuda K., Igarashi D., Hillmer R.A., Sakakibara H., Myers C.L. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe. 2014;15:84–94. doi: 10.1016/j.chom.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 101.Ryffel F., Helfrich E.J.N., Kiefer P., Peyriga L., Portais J.-C., Piel J. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 2016;10:632–643. doi: 10.1038/ismej.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Badri D.V., Quintana N., El Kassis E.G., Kim H.K., Choi Y.H., Sugiyama A. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009;151:2006–2017. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reisberg E.E., Hildebrandt U., Riederer M., Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamada K., Saijo Y., Nakagami H., Takano Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science. 2016;354:1427–1430. doi: 10.1126/science.aah5692. [DOI] [PubMed] [Google Scholar]
- 105.Chen L.-Q., Hou B.-H., Lalonde S., Takanaga H., Hartung M.L., Qu X.-Q. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henry E., Yadeta K.A., Coaker G. Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol. 2013;199:908–915. doi: 10.1111/nph.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 108.Fu Z.Q., Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 109.Toyota M., Spencer D., Sawai-Toyota S., Jiaqi W., Zhang T., Koo A.J. Glutamate triggers long-distance, calcium-based plant defense signaling. Science. 2018;361:1112–1115. doi: 10.1126/science.aat7744. [DOI] [PubMed] [Google Scholar]
- 110.Coker T.L.R., Cevik V., Beynon J.L., Gifford M.L. Spatial dissection of the Arabidopsis thaliana transcriptional response to downy mildew using Fluorescence Activated Cell Sorting. Front Plant Sci. 2015;6:527. doi: 10.3389/fpls.2015.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karrer E.E., Lincoln J.E., Hogenhout S., Bennett A.B., Bostock R.M., Martineau B. In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA. 1995;92:3814–3818. doi: 10.1073/pnas.92.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guillaume-Gentil O., Grindberg R.V., Kooger R., Dorwling-Carter L., Martinez V., Ossola D. Tunable single-cell extraction for molecular analyses. Cell. 2016;166:506–516. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 113.Kerk N.M., Ceserani T., Tausta S.L., Sussex I.M., Nelson T.M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gjetting T., Hagedorn P.H., Schweizer P., Thordal-Christensen H., Carver T.L.W., Lyngkjaer M.F. Single-cell transcript profiling of barley attacked by the powdery mildew fungus. Mol Plant Microbe Interact. 2007;20:235–246. doi: 10.1094/MPMI-20-3-0235. [DOI] [PubMed] [Google Scholar]
- 115.Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 116.Zhang C., Barthelson R.A., Lambert G.M. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant. 2008 doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deal R.B., Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ottesen A.R., Gorham S., Reed E., Newell M.J., Ramachandran P., Canida T. Using a control to better understand phyllosphere microbiota. PLoS one. 2016;11 doi: 10.1371/journal.pone.0163482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Doan H.K., Leveau J.H.J. Artificial surfaces in phyllosphere microbiology. Phytopathology. 2015;105:1036–1042. doi: 10.1094/PHYTO-02-15-0050-RVW. [DOI] [PubMed] [Google Scholar]
- 121.Soffe R, Altenhuber N, Bernach M, Remus-Emsermann M, Nock V. Comparison of replica leaf surface materials for phyllosphere microbiology. engRxiv 2018. doi: 10.31224/osf.io/2pzrv. [DOI] [PMC free article] [PubMed]
- 122.Bernach M., Soffe R., Remus-Emsermann M.N.P., Nock V. Micropatterning of hybrid PDMS for replica leaves. Jpn J Appl Phys. 2019 [Google Scholar]
- 123.Soffe R, Bernach M, Remus-Emsermann MNP, Nock V. PDMS Replica leaf surfaces for phyllosphere microbiology. bioRxiv 2018. doi: https://doi.org/10.1101/523985.