Abstract
Peripheral nerve myelin facilitates rapid impulse conduction and normal motor and sensory functions. Many aspects of myelin biogenesis, glia–axonal interactions, and nerve homeostasis are poorly understood at the molecular level. We therefore hypothesized that only a fraction of all relevant myelin proteins has been identified so far. Combining gel-based and gel-free proteomic approaches, we identified 545 proteins in purified mouse sciatic nerve myelin, including 36 previously known myelin constituents. By mass spectrometric quantification, the predominant P0, periaxin, and myelin basic protein constitute 21, 16, and 8% of the total myelin protein, respectively, suggesting that their relative abundance was previously misestimated due to technical limitations regarding protein separation and visualization. Focusing on tetraspan-transmembrane proteins, we validated novel myelin constituents using immuno-based methods. Bioinformatic comparison with mRNA-abundance profiles allowed the categorization in functional groups coregulated during myelin biogenesis and maturation. By differential myelin proteome analysis, we found that the abundance of septin 9, the protein affected in hereditary neuralgic amyotrophy, is strongly increased in a novel mouse model of demyelinating neuropathy caused by the loss of prion protein. Finally, the systematic comparison of our compendium with the positions of human disease loci allowed us to identify several candidate genes for hereditary demyelinating neuropathies. These results illustrate how the integration of unbiased proteome, transcriptome, and genome data can contribute to a molecular dissection of the biogenesis, cell biology, metabolism, and pathology of myelin.
Introduction
Normal sensation and motor capabilities require fast nerve impulse transmission. In vertebrates, this is achieved by the ensheathment of axons with myelin that restricts action potentials to the nodes of Ranvier (Salzer et al., 2008). In the peripheral nervous system (PNS), myelin is comprised of multiple compacted layers of the molecularly specialized plasma membrane extended from Schwann cells. Adaxonal and abaxonal myelin are connected by cytoplasmic incisures that contain cytoskeleton, vesicles, and outposts of endoplasmic reticulum and Golgi apparatus (Perkins et al., 2008), which likely are relevant for myelin maintenance and turnover. So far, only ∼45 distinct proteins have been associated with PNS myelin, suggesting that knowledge of the proteins required for normal myelin biogenesis, metabolism, and interactions with axons has likely remained incomplete. Moreover, the causative genes of several demyelinating neuropathies remain unknown (Reilly and Shy, 2009).
The variety of PNS myelin proteins has been considered low as gel-electrophoretic separation followed by Fast Green or Coomassie staining yielded only three predominant bands which, in the order of increasing mobility, were designated P0, P1, and P2 (Greenfield et al., 1973; Whitaker, 1981). Their relative abundance was estimated to be 45–70%, 2–26%, and 1–15% of the total myelin protein, respectively. P0 [myelin protein zero (MPZ)], an immunoglobulin-like cell-adhesion molecule (Ig-CAM), mediates adhesion of the extracellular myelin membrane surfaces (D'Urso et al., 1990; Filbin et al., 1990; Giese et al., 1992), while P1 [now termed myelin basic protein, (MBP)] mediates intracellular adhesion (for review, see Boggs, 2006). P2 (also termed PMP2 or fatty acid-binding protein-8) is a lipid-associated protein of unknown function (Trapp et al., 1984; Chmurzyńska, 2006).
It became evident that PNS myelin contains more proteins than assumed upon the immunoblot-based demonstration that a known CNS myelin component, the Ig-CAM myelin-associated glycoprotein (MAG), is also present in PNS myelin (Figlewicz et al., 1981). Later, additional myelin proteins were identified by using myelin-directed antibodies or amino acid sequence information gained from purified myelin, such as the tetraspan peripheral myelin protein of 22 kDa (PMP22) (Welcher et al., 1992) and the scaffolding protein periaxin (PRX) (Gillespie et al., 1994). More recently, the analysis of sciatic nerve mRNA abundance profiles during myelin biogenesis or experimental hypomyelination (Verheijen et al., 2003; Buchstaller et al., 2004; D'Antonio et al., 2006; Ryu et al., 2008) increased the number of myelin proteins to >40.
Considering the complexity of the CNS myelin proteome (Ishii et al., 2009; Jahn et al., 2009), we hypothesized that many more PNS myelin proteins await discovery. By subjecting purified myelin to various separation techniques and mass spectrometry (MS), we identified 36 established myelin constituents and >500 proteins not previously associated with PNS myelin, and readjusted their relative abundance. Integrating our compendium with other systematic approaches and focusing on tetraspan-transmembrane proteins, we illustrate how this resource can be exploited to gain comprehensive knowledge on groups of myelin proteins defined by structural similarity, functional relationship, or coregulated expression. This first PNS myelin proteome analysis thus provides a versatile framework for systematic analyses of myelin and myelin-related diseases.
Materials and Methods
Animals.
Male wild-type mice (C57BL/6N) at the age of postnatal day 28 (P28) were used, unless indicated otherwise. All experiments were in compliance with the animal policies of the Max-Planck-Institute of Experimental Medicine, approved by the German federal state of Niedersachsen.
Myelin purification.
A light-weight membrane fraction enriched for myelin was purified from mouse sciatic nerves homogenized in 0.27 m sucrose according to Larocca and Norton (2007). The protein concentration was determined using the 2-D Quant kit according to the manufacturer (GE Healthcare). Where indicated, purified myelin was subjected to further high-salt and high-pH washing/centrifugation cycles as described previously (Werner et al., 2007) to enrich transmembrane proteins before proteome analysis.
Gel electrophoresis and identification of gel-separated proteins.
2D-isoelectric focusing (IEF)/SDS-PAGE was performed as described previously (Werner et al., 2007) with the difference that myelin was delipidated and precipitated by methanol/chloroform treatment (Wessel and Flügge, 1984) before IEF. The pellet was resuspended in lysis buffer (20 mm Tris/HCl pH 9.0, 7 m urea, 2 m thiourea, 2% ASB-14), centrifuged (2 min, 16,000 × g), and the protein concentration of the supernatant was determined using the 2-D Quant kit (GE Healthcare). A volume corresponding to 250 μg of myelin was diluted 1:1 with 2× IEF sample buffer (7 m urea, 2 m thiourea, 2% ASB-14, 1% ampholytes, 0.6% DTT) and incubated for 30 min at room temperature. Finally, the sample was filled up with rehydration buffer (7 m urea, 2 m thiourea, 2% ASB-14, 0.5% ampholytes, 0.3% DTT) to a volume of 350 μl, centrifuged (2 min 16,000 × g), and the supernatant subjected to IEF in immobilized pH gradient (18 cm Immobiline nonlinear, pH 3–11, GE Healthcare) for ∼45 kVh. After completion of IEF, the proteins were reduced with DTT and alkylated with iodoacetamide as described previously (Werner et al., 2007). Second dimension SDS-PAGE was performed on 20 × 20 cm gradient gels (8–16% acrylamide). Proteins were visualized by colloidal Coomassie staining with Coomassie Brilliant Blue G-250 according to Neuhoff et al. (1988). Approximately 500 gel plugs from each of two independent gels were manually excised and subjected to an automated platform for the identification of gel-separated proteins (Jahn et al., 2006), as recently applied in large-scale proteome studies (Reumann et al., 2007; Werner et al., 2007). An Ultraflex matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer (Bruker Daltonics) was used to acquire both peptide mass fingerprint (PMF) and fragment ion spectra, resulting in confident protein identifications based on peptide mass and sequence information. Database searches in the Swiss-Prot primary sequence database restricted to Mus musculus were performed using the Mascot Software 2.2 (Matrix Science). Carboxamidomethylation of Cys was specified as fixed and oxidation of Met as variable modification. One missed trypsin cleavage was allowed. Mass tolerances were set to 100 ppm for PMF searches and to 100 ppm (precursor ions) and 0.7 Da (fragment ions) for MS/MS ion searches. The minimal requirement for accepting a protein as identified was at least one peptide sequence match above identity threshold in coincidence with at least 20% sequence coverage in the PMF.
One-dimensional (1D) SDS-PAGE was performed on precast NuPAGE Bis-Tris 4–12% gradient gels using 2-(N-morpholino)ethanesulfonic acid (MES) or 3-(N-morpholino)propanesulfonic acid (MOPS) buffer systems according to the manufacturer (Invitrogen). Proteins were visualized by Coomassie staining (see above) or silver staining according to Blum et al. (1987). For systematic protein identification, 25 μg of untreated myelin was separated and a total of four lanes from independent gels were sliced into 24 uniform pieces. 2D-16-benzyldimethyl-n-hexadecylammonium chloride (16-BAC)/SDS-PAGE was performed as described previously (Werner et al., 2007). Briefly, 300–400 μg of purified myelin was subjected to washing/centrifugation cycles (see above), the final pellet was resuspended in 16-BAC sample buffer, and proteins were separated on 7.5% acrylamide 16-BAC gels (20 × 20 cm) as described previously (Hartinger et al., 1996; Werner et al., 2007). Two-dimensional SDS-PAGE was performed on 20 × 20 cm gradient gels (8–16% acrylamide). Proteins were visualized by colloidal Coomassie staining (see above), and approximately 100 gel plugs each from two independent gels were excised. To account for the lower resolution of 1D-SDS-PAGE and 2D-16-BAC/SDS-PAGE (resulting in more complex peptide mixtures after in-gel digestion), these gel systems were interfaced to protein identification by nano-flow HPLC-MS/MS. Bands from 1D-SDS-PAGE gels or plugs from 2D-16-BAC/SDS-PAGE gels were subjected to tryptic in-gel digestion, and the released peptides were extracted in two consecutive steps with 0.5% trifluoroacetic acid (TFA)/10% acetonitrile and 0.5% TFA/50% acetonitrile, respectively. Pooled extracts were dried, redissolved in 0.1% TFA, and subjected to reversed phase separation on an Ultimate nano-HPLC system (Dionex) as described previously (Galli et al., 2009). Peptides were desalted and preconcentrated on a 2 cm × 75 μm column (packed in-house with Reprosil-Pur C18-AQ material of 5 μm particle and 100 Å pore size; Dr. Maisch) with 0.1% TFA at a flow rate of 8 μl/min. Separation on 15 cm × 75 μm columns (packed in-house with the same material as above) was performed at 40°C with a flow rate of 250 nl/min. Peptides were directly eluted into an ESI ion trap LCQ Deca XP Plus mass spectrometer (Thermo Fisher Scientific) with the acquisition duty cycle set to a full-scan mass spectrum (mass-to-charge ratio 400–1500) followed by three data-dependent MS/MS scans. Mascot database searches were performed as for the MALDI-TOF-MS/MS data with the difference that the mass tolerance was 1.5 Da for the precursor ions and 1.0 Da for the fragment ions. The minimal requirement for accepting a protein as identified was at least two peptide sequence matches above identity threshold. Protein identifications on the basis of a single peptide were considered when the score was at least two times higher than the identity threshold and were accepted only after manual inspection of the mass spectrum.
2D-differential fluorescence-intensity gel electrophoresis (2D-DIGE) was used for differential proteome analysis of sciatic nerve myelin purified from 4-week-old mice with a deletion of the prion protein gene (Prnp0/0) backcrossed to the BALB/c background (Bremer et al., 2010) and wild-type BALB/c controls. 2D-DIGE was essentially performed as described for CNS myelin (Werner et al., 2007). Briefly, wild-type and Prnp0/0 myelin proteins from two independent preparations each were labeled twice, thereby swapping the dyes, resulting in four samples that were subjected to 2D-IEF/SDS-PAGE in parallel. Per channel, 80 μg of protein was labeled with 400 pmol of the respective Cy-dye. Fluorescence images were processed with the Proteomweaver software (version 4.0, Bio-Rad), and spots were accepted as significantly different for p < 0.05 (Student's t test).
Gel-free proteome analysis and GeLC-MSE.
For gel-free analysis, 25 μg of total myelin protein was precipitated using the ProteoExtract Protein Precipitation Kit (Calbiochem), solubilized in 50 mm NH4HCO3 with 0.1% Rapigest (Waters) (10 min, 85°C), reduced with 5 mm DTT (45 min, 56°C), and alkylated with 15 mm iodoacetamide (45 min, room temperature) in the dark. Solubilized proteins were digested with 0.5 μg of sequencing grade trypsin (Promega) for 16 h (37°C). After digestion, Rapigest was hydrolyzed by adding 10 mm HCl, the resulting precipitate was removed by centrifugation (13,000 × g, 15 min, 4°C), and the supernatant was transferred into an autosampler vial for ultraperformance liquid chromatography (UPLC) coupled to MS with an alternating low and elevated (E) energy mode of acquisition-(MSE). Alternatively, for gel-enhanced liquid chromatography (GeLC)-MSE analysis, 25 μg of total myelin protein were separated by 8–16% SDS-PAGE, the resulting gel lanes were cut into 11 equally sized pieces, and in-gel digestion was performed as described above. For UPLC-MSE-analysis, UPLC separation of tryptic peptides and on-line analysis by quadrupole-TOF (QTOF) mass spectrometry were performed as described previously (Krämer-Albers et al., 2007).
The continuum LC-MSE data were processed and searched using the IDENTITYE-algorithm of ProteinLynx Global Server (PLGS) version 2.3 (Waters). Protein identifications were assigned by searching the Uni-ProtKB/Swiss-Prot Protein Knowledgebase Release 52.3 for mouse proteins (12,920 entries) supplemented with known possible contaminants (porcine trypsin, human keratins) using the precursor and fragmentation data afforded by LC-MS acquisition as described previously (Krämer-Albers et al., 2007). Mass tolerances for peptide and fragment ions were set at 15 and 30 ppm, respectively. Peptide identifications were restricted to tryptic peptides with no more than one missed cleavage. Carboxamidomethylation of Cys was set as fixed modification, and oxidation of Met, acetylation of protein N termini, and deamidation of Asn and Gln were searched as variable modifications. For a valid protein identification, the following criteria had to be met: at least two peptides detected together with at least seven fragments. The false-positive rate for protein identification was set to 1% based on search of a 5× randomized database, which was generated automatically using PLGS 2.3 by reversing the sequence of each entry. By using replication rate as a filter, the false-positive rate was further minimized, as false-positive identifications do not tend to replicate across injections due to their random nature. By requiring a protein identification to be made in at least three technical replicates, the effective false-positive rate was lowered to <0.2%.
In situ hybridization.
Probes were generated by PCR amplification of total brain cDNA using primers specific for Cd151 (forward 5′-TACCTGCTCT TTACCTACAA CTGCT, reverse 5′-GAAGGTCTCC AGCTTAGTGA TACAG); Cmtm5 (forward 5′-CTGTCTGCTT CTCTAACCAG ATTTCT, reverse 5′-GTGGAATATA GATGGTGGGT ACTTG); or Sept9 (forward 5′-CTGGAGGAGA GGGTCTACTT CAAAC, reverse 5′-ACACATAGCTC TTCTAACCCCT CAG); and subsequent cloning into pGemT (Promega). The probes were sequence validated. One-month-old NMRI wild-type mice were retrocardially perfused with alkaline phosphate buffer (aPB; 0.1 m Na2HPO4, pH 9) containing 10% sucrose, followed by 6% formalin in aPB containing 20% sucrose. Sciatic nerves were isolated, fixed (1 h, 6% formalin in aPB containing 20% sucrose), rinsed (H2O), dehydrated (ascending alcohol row, 5 min/50% ethanol, 10 min/70% ethanol, 20 min/90% ethanol, 30 min/100% ethanol, 45 min isopropanol, 2× 60 min xylene), and embedded in paraffin wax (2× 90 min, 60°C). Cross sections (10 μm) were cut using a sliding microtome and collected on adhesive glass slides (SuperFrost/Plus). Sections were manually dried using tissue paper, incubated (37°C for 2 h and 60°C for 1 h), deparaffinized, and rehydrated (descending alcohol row 2× 20 min xylene, 10 min isopropanol, 10 min 100% ethanol, 5 min 90% ethanol, 5 min 70% ethanol, and 2 min 50% ethanol); fixed [15 min in 4% paraformaldehyde (PFA) in PBS, pH 7.4], washed (in PBS), and incubated with 20 μg/ml Proteinase K (in 20 mm Tris, pH 7.5/1 mm EDTA, pH 8, 2.5 min). Proteinase K was inactivated (0.2% glycine in PBS); sections were washed (in PBS), fixed (4% PFA and 0.2% glutaraldehyde in PBS, 20 min), washed (in PBS), prehybridized [hybridization buffer: 50% formamide, 5× SSC, 1% blocking reagent (Roche), 5 mm EDTA, 0.1% Tween, 0.1% CHAPS, 0.1 mg/ml heparin, and 100 μg/ml yeast RNA] at 48°C for 2 h, hybridized overnight at 48°C in hybridization buffer containing digoxigenin-labeled (DIG-RNA Labeling-Mix, Roche), and denatured (95°C, 5 min) with cRNA probe; washed (SSC, pH 4.5); treated with 20 μg/ml RNase in 0.5 m NaCl/10 mm Tris (30 min, 37°C); washed (2× SSC, pH 4.5); washed 3× in 50% formamide in 2× SSC, pH 4.5 (3 min, 37°C); washed (in KTBT: 50 mm Tris pH 7.5, 150 mm NaCl, 10 mm KCl, 1% Triton-X); blocked (2 h) in 20% sheep serum in KTBT (Ab-block); incubated with anti-DIG alkaline phosphatase antibody (Roche) 1:1000 in Ab-block (overnight, 4°C); washed in KTBT; washed 4× in KTBT for 30 min, washed in 100 mm Tris, pH 9.5, 100 mm NaCl, 50 mm MgCl2, and 0.05% Tween20 (NTMT); developed in NBT/BCIP in NTMT for 4–18 h at 37°C; washed 2× for 6 h in KTBT; counterstained (1 μg/ml DAPI in PBS); dehydrated using a quick ascending alcohol row; cleared in benzyl alcohol/benzyl benzoate 1:2; and mounted using Eukitt. If not noted otherwise, incubations were at room temperature and washing was 2× 5 min in a slowly (0.25 Hz) shaking cuvette. All solutions used before antibody incubation were prepared using RNase free (DEPC treated) H2O. The sense probes and a probe for an unrelated, exclusively neuronal mRNA (Neurod6) were processed in parallel as controls and gave no signal.
mRNA abundance profiles.
Male wild-type mice (C57BL/6N) at the ages of 1, 5, 15, or 65 d were subjected to cervical dislocation. The sciatic nerves were isolated and homogenized, and total RNA was extracted with Trizol reagent. RNA integrity and concentration were determined using an Agilent 2100 Bioanalyzer. Biotinylated cRNAs for hybridization to Affymetrix 3′-arrays were prepared from 1 μg of total RNA using GeneChip One-Cycle Target Labeling and Control Reagents (catalog #900493, Affymetrix). Labeled cRNA was fragmented and hybridized to Mouse Genome 430 2.0 Arrays (3′-arrays, catalog #900496, Affymetrix) according to the manufacturer's instructions. Raw image files were processed using Affymetrix GCOS 1.3 software to calculate individual probe cell intensity data and generate CEL Data Files. Statistical analyses have been performed with R using packages Biobase, limma, marray, affy, and masigpro. Data were filtered by reading out expression values for those 528 of 545 proteins that were identified in myelin by MS and represented on the mRNA arrays. Sept9 was also included, though SEPT9 protein was detected by MS only in Prnp0/0 myelin while it was below the significance threshold in wild-type myelin. For comparison, eight known myelin genes were included of which the corresponding proteins were not identified by MS (Pmp22, Mal, Cdh1, Ctnnb1, Mtmr2, Mp11/A330049/M08Rik, Jam4/Igsf5, and Cx32/Gjb1). Data were normalized using the robust multiarray average method. To facilitate clustering, we transformed the data and set the mean value to zero and the SD to 1/−1. Transformed data were then subjected to K-means clustering. The mRNA abundance dataset is available from the authors upon request.
Quantitative RT-PCR.
Sciatic nerves of 5-week-old PrnpO/O mice (Bremer et al., 2010) backcrossed to the c57Bl/6 background, and age-matched C57BL/6 wild-type mice were homogenized in TRIzol (Invitrogen) by using Polytron PT 3100 (Kinematica). RNA was extracted and purified on RNeasy columns (Qiagen). cDNA was synthesized with QuantiTect Reverse Transcription kit (Qiagen), and analyzed by real-time PCR using QuantiFast SYBR Green PCR kit (Qiagen) and 7900HT (Fast Real-Time PCR systems, Applied Biosystems). Primers were specific for Sept9 (forward 5′-ACTGCTGCCT CTACTTCAT, reverse 5′-TCTGCGTCCT CATCAAACTC) or GAPDH (forward 5′-CCACCCCAGC AAGGAGACT, reverse 5′-GAAATTGTGA GGGAGATGCT).
Immunoblotting.
Immunoblotting was performed as described previously (Werner et al., 2007). Antibodies were specific for PLP/DM20 (Jung et al., 1996), plasmolipin (Bosse et al., 2003), myelin and lymphocyte protein (MAL) (Schaeren-Wiemers et al., 2004), PMP22 (ab15506, Abcam), CD9 (PharMingen), CD81 (PharMingen), CMTM5 (CKLF-like MARVEL-transmembrane domain-containing protein 5, also termed PETA-3; ab88964, Abcam; or M-13, Santa Cruz Biotechnology, yielding similar results), CD151 [455807, R&D Systems (Cowin et al., 2006)], septin 9 (SEPT9) [10769, Protein Tech Group and Nagata et al., (2004) (yielding similar results)], MAG (clone 513, Millipore Bioscience Research Reagents), 2′,3′-cyclic nucleotide phosphodiesterase (CNP; 11–5B, Sigma), COX4 (MS404, MitoSciences), and actin (Mab1501, Millipore Bioscience Research Reagents).
Immunoelectron microscopy.
Immunolabeling of sciatic nerve cryosections and electron microscopy were as described previously (Werner et al., 2007). Primary antibodies were specific for CMTM5 (ab88964, Abcam; or M-13, Santa Cruz Biotechnology; yielding similar results), CD151 [455807, R&D Systems (Cowin et al., 2006)], or septin 9 (10769, Protein Tech Group).
Results
Purification of PNS myelin
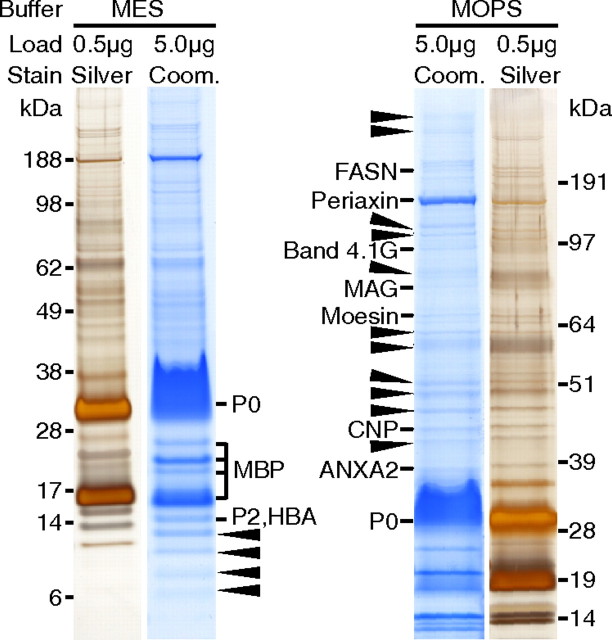
For the purification of myelin, we have subjected sciatic nerve homogenates of adult (P28) wild-type mice (C57BL6) to discontinuous sucrose-gradient centrifugation, obtaining the myelin-enriched fraction at the interface between 0.27 and 0.83 m sucrose (Larocca and Norton, 2007). When the myelin-enriched fraction was compared with nerve homogenate by 1D-SDS-PAGE and colloidal Coomassie stain, the number of bands was considerably reduced and the intensity of bands constituted by myelin proteins such as P0 was enhanced (data not shown), in accordance with numerous previous publications by several laboratories. As a pilot approach to systematically identify the proteins contained in the myelin-enriched fraction, 5 μg of myelin was separated by 1D-SDS-PAGE using two different buffer systems (MES and MOPS) for enhanced separation in the low- or high-molecular-weight range, respectively (Fig. 1). Bands visualized by Coomassie staining were excised and subjected to tryptic in-gel digestion, and proteins were identified based on peptide mass and sequence information obtained by MALDI-TOF-MS. While the coverage of known myelin proteins was high, it became apparent that several bands contained more than one protein. For example, P2 (calculated molecular mass 14.9 kDa) was not separated from hemoglobin A (HBA; 15.1 kDa), a common contamination from red blood cells. This suggests that insufficient protein separation has impaired previous 1D-gel-based estimations of the abundance of P2, and likely that of other myelin proteins. Importantly, the majority of bands were constituted by proteins not previously associated with PNS myelin (Fig. 1, arrows). Together, this pilot experiment supported our hypothesis that numerous myelin constituents remain to be identified. As an attempt to fill this gap, we have performed a systematic proteome analysis. For simplicity, we will use the term “myelin” for the myelin-enriched fraction isolated from sciatic nerves, and “myelin-associated” for all proteins identified in myelin, regardless of whether such localization has been validated with independent methods.
Figure 1.
One-dimensional separation of PNS myelin. Myelin purified from wild-type sciatic nerves was separated by SDS-PAGE in different buffer systems providing improved resolution either in the low-molecular-weight (MES) or high-molecular-weight (MOPS) range. Proteins were visualized by colloidal Coomassie staining (Coom.; 5 μg of protein load) or, for comparison, by silver staining (0.5 μg of protein load). Coomassie-stained bands were subjected to mass spectrometric protein identification. Bands constituted by known myelin proteins are denoted. In bands marked with arrowheads, only proteins not previously associated with myelin were identified, indicating that the complexity of the PNS myelin proteome is higher than previously assumed. Note that several bands contained more than one protein (e.g., P2 was not separated from HBA), indicating that one-dimensional separation does not allow reliable estimation of relative protein abundance. ANXA2, Annexin A2.
Proteome analysis of PNS myelin
With the goal to establish a comprehensive compendium of PNS myelin proteins, we have used a combination of various gel-based and gel-free methods. Similar to the procedure described above, 25 μg of myelin was prefractionated by 1D-SDS-PAGE (MES buffer system, data not shown). The complete lanes were sliced into 24 uniform pieces, and their analysis by HPLC coupled to MS (GeLC) led to the identification of 144 proteins (marked “S” in this article). The same technique was additionally applied to myelin that underwent several wash cycles for the enrichment of membrane proteins (wash-GeLC, 83 proteins, marked “wS”). When prefractionation of washed myelin by 1D-SDS-PAGE (25 μg of myelin, 11 gel slices) was combined with the LC-MSE technology (see below), the number of identified proteins increased by a factor of three (wash-GeLC-MSE 246 proteins, marked “wSE”).
To improve separation and proteome coverage, and to provide reference maps, also two-dimensional protein separation techniques were applied. First, 250 μg of delipidated and precipitated myelin was separated using IEF with immobilized pH gradients as the first dimension and SDS-PAGE as the second dimension, and proteins visualized by colloidal Coomassie staining (Fig. 2A) were identified by an automated platform based on MALDI-TOF-MS (2D-IEF/SDS-PAGE, 249 proteins, marked “I”). Next, we subjected 400 μg of myelin to the wash procedure and separated the remaining proteins by 2D-16BAC/SDS-PAGE, an alternative 2D gel system (Hartinger et al., 1996) suitable for displaying myelin proteins (Werner et al., 2007). Proteins visualized by colloidal Coomassie staining (Fig. 2B) were identified by HPLC-MS/MS (2D-16BAC/SDS-PAGE, 53 proteins, marked “B”). Collectively, gel-based techniques led to the identification of 436 nonredundant proteins in purified sciatic nerve myelin (Fig. 3A).
Figure 2.
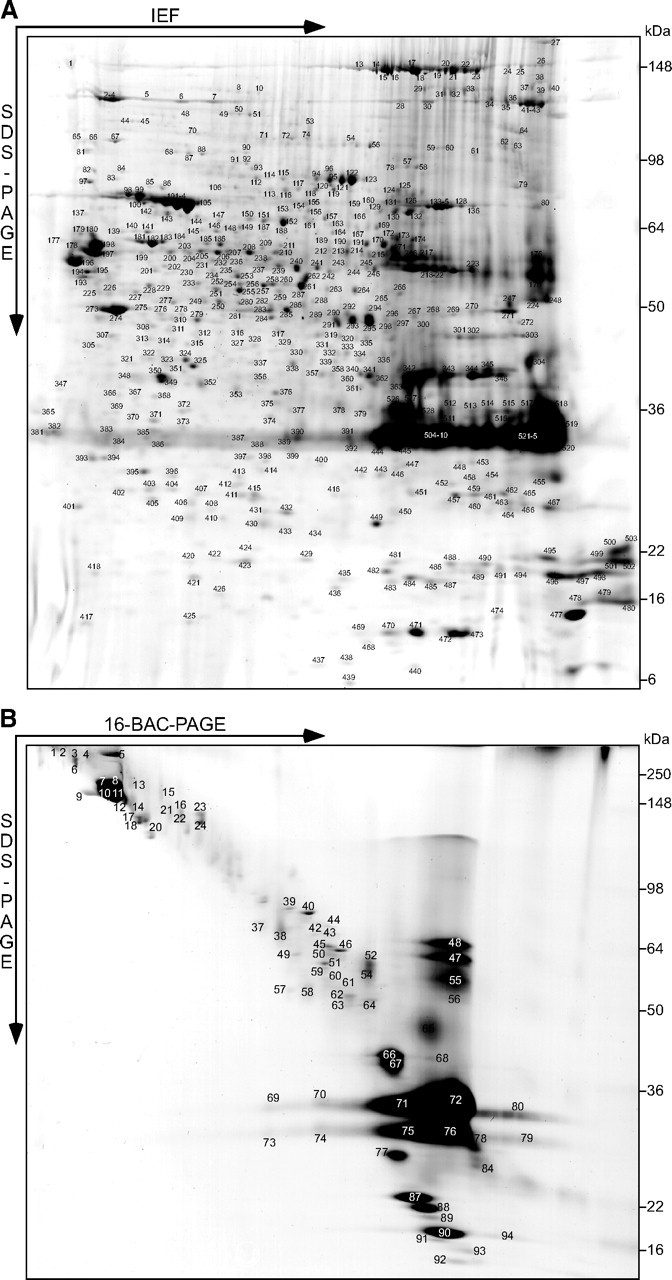
Two-dimensional myelin proteome maps. A, B, Myelin purified from wild-type sciatic nerves was separated in different gel systems, and proteins visualized by colloidal Coomassie staining were subjected to mass spectrometric protein identification. Representative gels are shown. The protein identities of all individual spots will be hosted on the author's web page (see Notes). This material has not been peer reviewed. A, 2D-IEF/SDS-PAGE with IEF in a nonlinear pH gradient (3–11) as the first and SDS-PAGE as the second dimension. Proteins were identified by an automated platform based on MALDI-TOF-MS (485 unequivocal identifications from 528 picked spots). B, 2D-16-BAC/SDS-PAGE with separation in a 16-BAC gel as the first and SDS-PAGE as the second dimension. Proteins were identified by HPLC-MS/MS to address the higher sample complexity resulting from the limited resolution of the gel (74 identifications from 94 picked spots).
Figure 3.
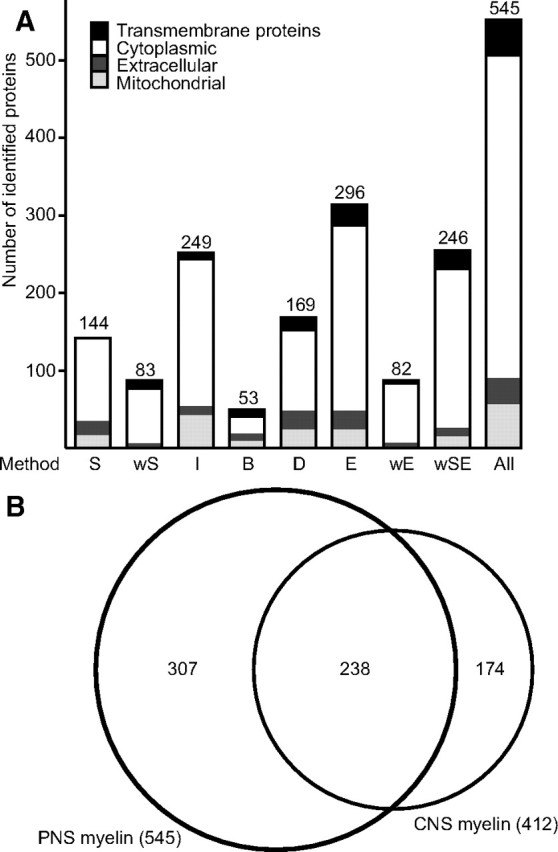
Comparison of approaches to the PNS myelin proteome, and comparison with the CNS myelin proteome. A, Proteins identified in purified wild-type PNS myelin by eight different technical approaches were categorized according to subcellular localization. Transmembrane domains were predicted with TMHMM2, PredHel/Phobius, and Scampi. Classification as mitochondrial was based on predictions using CELLO, TARGETP, WOLFPSORT, SUBLOC, and comparison with the brain mitochondrial proteome (MitoCarta). Extracellular proteins were classified based on CELLO, TARGETP, WOLFPSORT, SUBLOC, Swiss-Prot, NCBI-Gene, and according to the literature. Methods: S, GeLC; wS, wash-GelC; wSE, wash-GeLC-MSE; I, 2D-IEF/SDS-PAGE; B, 2D-16BAC/SDS-PAGE; D, LC-DDA; E, LC-MSE; wE, wash-LC-MSE (see text for details). B, Venn diagram comparing the number of proteins identified by MS in PNS myelin (this study) with those previously identified in rodent CNS myelin (Taylor et al., 2004; Vanrobaeys et al., 2005; Roth et al., 2006; Werner et al., 2007; Baer et al., 2009; Jahn et al., 2009). Note the high overlap of proteins identified in myelin from both sources.
As gel-based methods may exhibit a bias toward certain protein classes and a limited dynamic range, we also applied gel-free “shotgun” approaches to confirm and expand the myelin protein compendium. We subjected 20 μg of myelin to in-solution tryptic digestion and analyzed the resulting complex peptide mixture by UPLC coupled to a QTOF hybrid mass spectrometer. Data were acquired in the conventional data-directed automatic analysis mode (LC-DDA; 169 proteins, marked “D”) and with an alternating low/elevated collision energy acquisition mode (LC-MSE; 296 proteins, marked “E”). The latter circumvents the typical undersampling of LC-DDA, thereby improving proteome coverage (Fig. 3A) and allowing gel-free, label-free quantification of the relative abundance of proteins (see below) (Silva et al., 2005, 2006; Weinzierl et al., 2008). Finally, washed myelin was also subjected to LC-MSE (wash-LC-MSE; 82 proteins, marked “wE”). Together, gel-free methods led to the identification of 330 nonredundant myelin-associated proteins (Fig. 3A).
When the results of the gel-based and gel-free approaches were combined, a total of 545 nonredundant proteins were identified with a high overlap of 221 proteins (41%) detected by both methods (Fig. 3A). The numbers of proteins identified by the various methods are compared in Figure 3A. In particular, the proteins of higher abundance have been reproducibly identified with the majority of techniques. The identification of hundreds of proteins in PNS myelin supports our hypothesis that the complexity of its protein composition has been underappreciated so far because low-abundant components were overshadowed by dominant myelin components due to technical limitations in protein separation and staining.
Forty-four percent of the proteins identified in PNS myelin (this study) were also identified in rodent CNS myelin using similar techniques (Taylor et al., 2004; Vanrobaeys et al., 2005; Roth et al., 2006; Werner et al., 2007; Baer et al., 2009; Jahn et al., 2009) (Fig. 3B), which is surprisingly many when considering that Schwann cells and oligodendrocytes differ regarding their ontogenic origin (neural crest and subventricular zone, respectively), axonal segments myelinated per glial cell (1:1 and up to 60:1), and major myelin proteins (P0 and PLP) (for review, see Möbius et al., 2008).
Categorization of myelin proteins
We have categorized the proteins identified in sciatic nerve myelin as (1) membrane spanning, (2) cytosolic, and (3) extracellular myelin-associated proteins, and likely contaminants from (4) mitochondria, (5) nuclei, (6) blood, and (7) hair (Fig. 3A).
The category “membrane spanning” was based on calculations using TMHMM (v2.0) (smart.embl-heidelberg.de), Phobius/TMHMM (phobius.sbc.su.se), and Scampi (scampi.cbr.su.se). Sixty-three membrane-spanning proteins were identified (Table 1), including established myelin proteins such as P0 (D'Urso et al., 1990; Filbin et al., 1990; Giese et al., 1992); MAG (Figlewicz et al., 1981); sarcoglycan-δ (Cai et al., 2007), nectin-like protein-4 (NECL4) (Spiegel et al., 2007); and the tetraspan-transmembrane proteins proteolipid protein (PLP/DM20), plasmolipin (PLLP), CD81, and CD9 (Garbern et al., 1997; Bosse et al., 2003; Ishibashi et al., 2004). Tetraspans have been implicated in a variety of important myelin-related cellular events such as glial cell migration and growth regulation, organelle trafficking, myelin biogenesis and ultrastructure, and membrane adhesion (Bronstein, 2000; Gould et al., 2005). Moreover, several myelin tetraspans cause neuropathies when mutated. Therefore, we have chosen those tetraspans that have not previously been associated with myelin, CD151, and CMTM5, for antibody-based validation that myelin proteins were indeed newly identified in our compendium (see below).
Table 1.
Transmembrane proteins identified by MS in peripheral myelin
| Protein name | Gene | MS method | Phobius | Scampi | TMHMM |
|---|---|---|---|---|---|
| ADP/ATP translocase 1, Slc25a4 | Slc25a4 | wSE | 2 | 0 | 3 |
| ADP/ATP translocase 2, Slc25a5 | Slc25a5 | wSE | 1 | 0 | 2 |
| ADP/ATP translocase 4, Slc25a31 | Slc25a31 | wSE | 2 | 0 | 2 |
| ADP-ribosyltransferase 3 | Art3 | E | 1 | 0 | 0 |
| Amyloid-like protein 1 | Aplp1 | E | 1 | 2 | 1 |
| Basigin | Bsg | wSE | 1 | 2 | 1 |
| Ca2+ ATPase 1 | Atp2a1 | D, E, wSE | 8 | 8 | 7 |
| Ca2+ ATPase slow twitch 2 | Atp2a2 | wSE | 9 | 9 | 8 |
| Calnexin | Canx | S, D, wS, wSE | 1 | 2 | 1 |
| Caveolin 1 | Cav1 | S, E, wSE | 1 | 2 | 1 |
| CD151 | Cd151 | S, wS | 4 | 4 | 4 |
| CD81 | Cd81 | D, E, wE, wSE | 4 | 4 | 4 |
| CD9 | Cd9 | S, D, E, wS, wE, wSE | 4 | 4 | 4 |
| Cell adhesion molecule 3, NECL1 | Cadm3 | E, wSE | 1 | 2 | 1 |
| Cell adhesion molecule 4, NECL4 | Cadm4 | I, S, D, E, wS, wSE | 1 | 2 | 1 |
| Cell cycle exit and neuronal diff. | Cend1 | S, wSE | 1 | 1 | 1 |
| Choline transporter-like protein 1 | Slc44a1 | D, E | 9 | 12 | 9 |
| CKLF-like MARVEL TMD-containing 5 | Cmtm5 | wS, wSE | 4 | 4 | 4 |
| Claudin 19 | Cldn19 | wSE | 5 | 4 | 4 |
| Complement C3 | C3 | B, S, D, E, wSE | 0 | 3 | 1 |
| Cyclophilin B | Ppib | I, E | 0 | 1 | 1 |
| Cytochrome B5 | Cyb5a | wSE | 1 | 1 | 1 |
| Cytochrome c oxidase 2 | Mtco2 | wSE | 2 | 2 | 2 |
| Cytochrome c oxidase 4 isoform 1 | Cox4i1 | wS, wSE | 1 | 1 | 1 |
| Cytochrome c oxidase 6C | Cox6c | wSE | 1 | 1 | 1 |
| Cytoskeleton-associated protein 4 | Ckap4 | B, wSE | 1 | 1 | 1 |
| Dehydrocholesterol reductase | Dhcr7 | wSE | 9 | 8 | 6 |
| Dolichyl-oligosaccharide glycosyltransferase | Ddost | wSE | 1 | 2 | 2 |
| Ectonucleoside triphosphate diphosphatase 2 | Entpd2 | wSE | 1 | 3 | 2 |
| Endoplasmic reticulum protein ERp29 | Erp29 | I | 0 | 1 | 1 |
| Extended synaptotagmin-like 1 | Fam62a | wSE | 1 | 5 | 1 |
| Fucosyltransferase 8 | Fut8 | B | 1 | 1 | 1 |
| Hydroxysteroid (17-β) dehydrogenase 12 | Hsd17b12 | wSE | 1 | 1 | 1 |
| Integrin α6 | Itga6 | I, D, wSE | 1 | 3 | 1 |
| Integrin β1 | Itgb1 | wSE | 1 | 3 | 1 |
| K+ ATPase α1 | Atp4a | E | 8 | 8 | 8 |
| K+ ATPase α2 | Atp12a | E | 10 | 10 | 8 |
| Malectin | Mlec | I | 1 | 2 | 1 |
| Myelin-associated glycoprotein | Mag | S, D, E, wS, wSE | 1 | 2 | 1 |
| Myelin protein zero, P0 | Mpz | I, B, S, D, E, wS, wE, wSE | 1 | 2 | 2 |
| Myelin proteolipid protein | Plp1 | B, S, D, E, wS, wE, wSE | 4 | 4 | 4 |
| Na+/K + ATPase α1 | Atp1a1 | S, D, E, wS, wE, wSE | 8 | 10 | 10 |
| Na+/K + ATPase α2 | Atp1a2 | S, E, wS, wSE | 8 | 10 | 8 |
| Na+/K + ATPase α3 | Atp1a3 | S, D, E, wSE | 8 | 10 | 8 |
| Na+/K + ATPase α4 | Atp1a4 | E, wSE | 10 | 10 | 10 |
| Na+/K + ATPase β1 | Atp1b1 | D, E, wS, wE, wSE | 1 | 1 | 1 |
| Na+/K + ATPase β3 | Atp1b3 | E | 1 | 1 | 1 |
| NAD(P) dependent steroid dehydrogenase-like | Nsdhl | wSE | 1 | 1 | 1 |
| NADH-ubiquinone dehydrogenase 1a4 | Ndufa4 | wSE | 1 | 1 | 1 |
| Plasmolipin | Pllp | S, E, wS, wSE | 4 | 5 | 4 |
| Progesterone receptor membrane component 1 | Pgrmc1 | wSE | 1 | 1 | 1 |
| Receptor accessory protein 5 | Reep5 | wSE | 2 | 4 | 2 |
| Reticulon 3 | Rtn3 | wSE | 3 | 4 | 3 |
| Reticulon-4, NoGo | Rtn4 | B, S | 3 | 4 | 2 |
| Ribophorin 1 | Rpn1 | wSE | 1 | 2 | 1 |
| Ribophorin 2 | Rpn2 | B | 3 | 4 | 3 |
| S100-A10 | S100a10 | I, S | 1 | 0 | 1 |
| Sarcoglycan δ | Sgcd | wSE | 1 | 1 | 1 |
| Sideroflexin 3 | Sfxn3 | wSE | 2 | 3 | 2 |
| Solute carrier 25a3 | Slc25a3 | wS, wSE | 2 | 3 | 2 |
| Thy-1 membrane glycoprotein | Thy1 | wSE | 1 | 1 | 0 |
| Ubiquitin-specific peptidase 19 | Usp19 | wS | 1 | 3 | 1 |
| UDP galactosyltransferase 8 | Ugt8 | wSE | 1 | 4 | 2 |
For all proteins identified by MS in myelin, transmembrane domains (TMDs) were predicted using three algorithms (Phobius, Scampi, TMHMM). In this table, all proteins with at least one TMD were compiled, and tetraspan-TMD proteins (bold) were selected for experimental validation of their presence in myelin. Identification methods are as follows: S, GeLC; wS, wash-GelC; wSE, wash-GeLC-MSE; I, 2D-IEF/SDS-PAGE; B, 2D-16BAC/SDS-PAGE; D, LC-DDA; E, LC-MSE; wE, wash-LC-MSE. The complete compendium of MS-identified proteins, including nontransmembrane proteins, will be hosted on the author's web page (see Notes).
The category “cytoplasmic” contained the vast majority of identified proteins (Fig. 3A), including many known myelin proteins as MBP, fatty acid synthase (FASN), carbonic anhydrase (Cammer and Tansey, 1987), P2 (Trapp et al., 1984), CNP (Matthieu et al., 1980), the NAD+-dependent deacetylase Sirtuin-2 (SIRT2) (Werner et al., 2007), gelsolin (Gonçalves et al., 2010), cdc42 (Benninger et al., 2007), crystallin-α2 (D'Antonio et al., 2006), band 4.1 G-protein (Ohno et al., 2006), dystrophin-related protein-2 (DRP2) (Sherman et al., 2001), ERM proteins (ezrin, radixin, moesin) (Scherer et al., 2001), and small cytoskeleton-associated GTPases of the septin family (SEPT2, SEPT7, SEPT11) (Buser et al., 2009). The high coverage of known myelin proteins indicates that our proteomic approach is suitable for systematic analysis.
Since PNS myelin is tightly associated with the extracellular matrix, it was also expected to identify various collagen types and basal lamina proteins (Fig. 3A). Indeed, Laminin γ1 (Chen and Strickland, 2003) and various proteoglycans (decorin, lamican, mimecan/osteoglycin, perlecan) were identified that may modulate the function of recognition molecules for axo-glial interactions (Chernousov and Carey, 2000), such as integrin α6 (Nodari et al., 2008) and β1 (Feltri et al., 2002), which were also identified. Among the identified extracellular proteins, CNTF has been previously shown to be a Schwann cell-derived protein supporting axonal development and regeneration (Sendtner et al., 1994). This suggests that proteomic analysis of PNS myelin has the capacity to identify signaling proteins relevant for Schwann cell development.
Although we expect that a very high proportion of proteins identified for the first time in the myelin-enriched fraction are indeed true myelin constituents, some may only have copurified, such as proteins associated with mitochondria, which have flotation properties in sucrose gradients similar to myelin. Proteins were categorized as being of mitochondrial origin according to predictions using CELLO (cello.life.nctu.edu.tw), TARGETP (cbs.dtu.dk/services/TargetP), WOLFPSORT (wolfpsort.org), SUBLOC (bioinfo.tsinghua.edu.cn/SubLoc), and previous appearance in a mitochondrial proteome compendium (MitoCarta) (Pagliarini et al., 2008) (Fig. 3A). However, the fact that a considerable number of proteins display a dual cytoplasmic and mitochondrial localization (Pagliarini et al., 2008) prompted us to not exclude any protein from the compendium because of predicted subcellular localization. Only very few proteins were recognized as likely contaminations from nuclei, blood, and hair.
CMTM5 and CD151 are novel myelin tetraspan proteins
The proven relevance of tetraspan-transmembrane proteins for myelin (for review, see Bronstein, 2000) led us to select such proteins from our dataset for experimental validation that indeed myelin proteins were newly identified. Immunoblotting revealed that both CMTM5 and CD151 were abundant in the myelin-enriched fraction (Fig. 4A). For comparison, markers of compact myelin such as the tetraspans PMP22, PLP/DM20, and MAL (Schaeren-Wiemers et al., 2004) were enriched in myelin relative to lysates, markers of uncompact myelin (MAG, CNP) were detected with approximately equal abundance in myelin and lysates, while contaminating actin and the mitochondrial marker COX4 were barely detectable in the myelin-enriched fraction. In situ hybridization of sciatic nerves using cRNA probes specific for Cmtm5 and Cd151 led to signals consistent with strong expression in Schwann cells (Fig. 4B,C). To identify the exact localization of CMTM5 and CD151, we have applied cryo-immuno electron microscopy to cross-sectioned sciatic nerves. CMTM5 was prominently labeled in compact myelin (Fig. 4D) and at the endoplasmic reticulum of Schwann cells (Fig. 4D′), while CD151 was detected in compact myelin (Fig. 4E) and at the plasma membrane of myelinating Schwann cells (Fig. 4E′). Labeling was virtually absent from other structures (for overviews, see Fig. 4D″,4E″). In particular, Schmidt–Lanterman incisures and mitochondria were not labeled. Together, we have validated the presence of two novel tetraspans with distinct distributions in normal PNS myelin. Together with the finding that at least 36 proteins identified by MS in myelin are known myelin constituents, this raises confidence that our compendium has strong predictive value with respect to novel myelin constituents.
Figure 4.
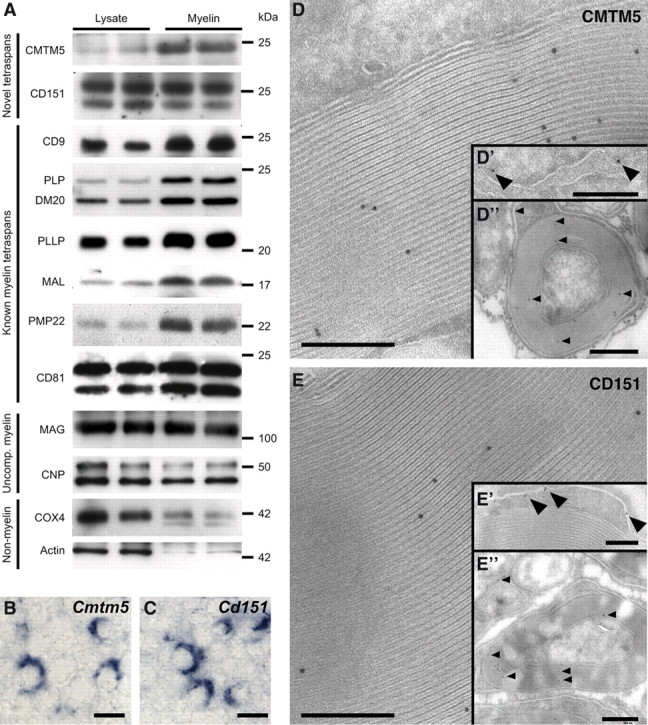
The tetraspan proteins CMTM5 and CD151 are novel constituents of PNS myelin. The myelin proteome compendium was systematically searched for tetraspan proteins, and those not previously associated with myelin were experimentally analyzed. A, Immunoblot analysis of CMTM5 and CD151 in sciatic nerve lysates and the myelin-enriched fraction, with equal amounts loaded. For comparison, previously known myelin tetraspans (CD9, PLP/DM20, PLLP, MAL, PMP22, CD81), markers of uncompact myelin (MAG, CNP), as well as a mitochondrial marker (COX4) and actin were tested. B, C, In situ hybridization of Cmtm5 (B) and Cd151 (C) in sciatic nerves using specific cRNA probes. Scale bar, 6 μm. Note that Schwann cells are strongly labeled. D, E, Immunodetection of CMTM5 (D) and CD151 (E) with 10 nm gold particles on cross sections of sciatic nerves. Labeling for CMTM5 was mostly confined to compact myelin (D, D″) and the endoplasmic reticulum of Schwann cells (D′), while CD151 was detected in compact myelin (E, E″) and the plasma membrane of Schwann cells (E′). Scale bars: D, D′, E, E′, 200 nm; D″, E″, 500 nm.
Relative abundance of myelin proteins
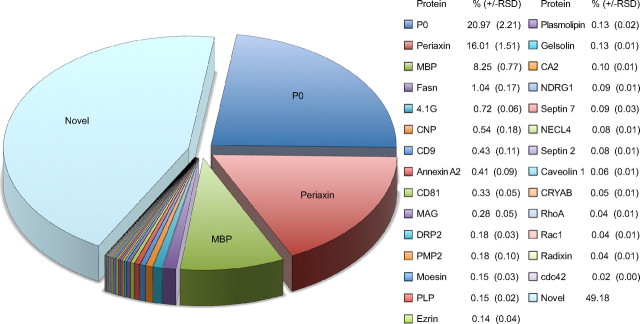
As a label-free MS-based quantification approach, LC-MSE can provide information regarding the relative abundance of peptides, and thereby the corresponding proteins, in a complex sample. We have calculated the relative abundance of myelin proteins as identified by LC-MSE based on the average intensity of the three most abundant unique peptides per protein. In the few cases for which only two peptides were identified, their average intensity was used. The 296 proteins identified by LC-MSE (see above) were identified with an average sequence coverage of 52%. Quantitative analysis of this dataset revealed that the four most abundant proteins of PNS myelin are P0, periaxin, MBP, and FASN, which constitute 21% [±2% relative SD (RSD)], 16 ± 2%, 8 ± 1%, and 1 ± 0.2% of the total myelin protein, respectively (Fig. 5). All previously known myelin proteins together constituted approximately half of the total myelin protein, while all newly identified proteins accounted for the other half. Our MS-based quantification data take into question previous estimates based on conventional techniques involving gel-electrophoretic separation and protein visualization with Sudan Black, Fast Green, or Coomassie Blue, in which P0, MBP, and P2 were suggested to be the most abundant myelin constituents with 45–70, 2–26, and 2–15% of the total myelin protein, respectively (Greenfield et al., 1973; Micko and Schlaepfer, 1978; Smith and Curtis, 1979; Whitaker, 1981). However, gel-based estimates of the relative abundance of myelin proteins are not very accurate. For example, the perceived high abundance of P2 in those estimates may mainly reflect the amount of contaminating blood, as indicated by our result that P2 and HBA are not well separated in 1D gels (see above). Indeed, by mass spectrometric quantification, P2 was of very low abundance (0.18 ± 0.04%) in PNS myelin.
Figure 5.
Quantification of PNS myelin proteins. Relative protein abundance was determined by LC-MSE and is given as a percentage (±RSD). Note that previously known myelin proteins constitute approximately half of the total myelin protein, while the other half are constituted by proteins not yet associated with myelin. Proteins derived from contaminations, such as mitochondria, blood, hair, and nuclei, which collectively constitute <5% of the total protein amount in the fraction, were not considered for this quantification. According to MS-based quantification, the most abundant myelin proteins are P0 (21%), Periaxin (16%), MBP (8%), and fatty acid synthase (1%). Note that MS-based quantification circumvents limitations of 1D gels such as limited resolving power and low dynamic range of protein staining, factors that have affected previous abundance estimates based on band intensity after 1D-PAGE.
Myelin proteome alteration in a pathological condition
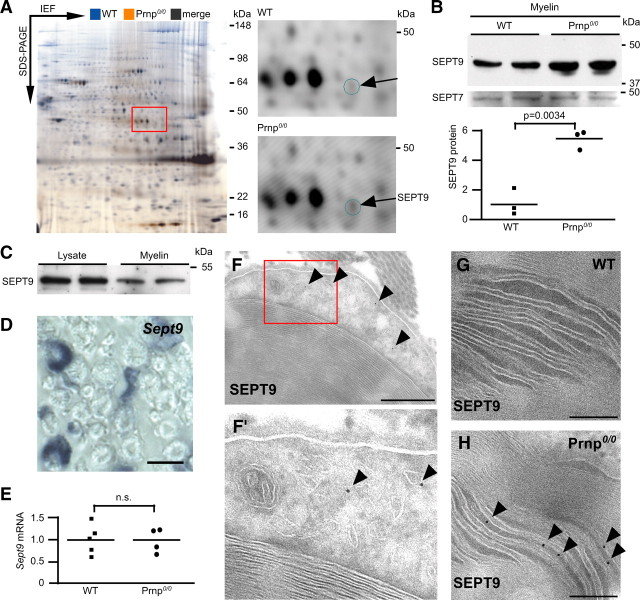
Mice harboring an inactivated prion protein gene (Prnp0/0) provide a model for a chronic polyneuropathy involving demyelination (Bremer et al., 2010). In this model, neuronally expressed prion protein (PrPC) acts as an axonal signal to prevent demyelination. To identify candidate proteins downstream of PrPC that could be implicated in the failure of Schwann cells to support peripheral myelin, we systematically screened Prnp0/0 mice for (secondary) alterations in the myelin proteome using 2D-DIGE, comprising the labeling of myelin proteins purified from mutant and wild-type mice with two different amine-reactive cyanine dyes (Fig. 6A). While the overall protein profiles of wild-type and Prnp0/0 myelin were very similar, we detected one single spot at ∼40 kDa that was indicative of increased abundance in mutant myelin (calculated regulation factor 1.4; p = 0.0053). The constituting protein was mass-spectrometrically identified as SEPT9, and its increased abundance was confirmed by immunoblotting (Fig. 6B). Septins are filament-forming proteins with GTPase activity (for review, see Barral and Kinoshita, 2008) and may contribute to the myelin cytoskeleton (Buser et al., 2009). SEPT9 is the causative gene in hereditary neuralgic amyotrophy (HNA), for which patients harboring point mutations (Kuhlenbäumer et al., 2005; Hannibal et al., 2009) or gene duplications (Collie et al., 2010) were described. While SEPT9 was detected by immunoblotting in myelin derived from both genotypes it was not mass-spectrometrically identified in wild-type myelin, indicating that its increased abundance in Prnp0/0 myelin is a prerequisite for identification with our stringent confidence level.
Figure 6.
Differential peripheral myelin proteome analysis in the Prnp0/0model of chronic demyelinating polyneuropathy. A, 2D-DIGE of wild-type (WT, false-colored in blue) and Prnp0/0 (false colored in orange) myelin. 2D-IEF/SDS-PAGE was performed with IEF in a nonlinear pH gradient (3–11) as the first and SDS-PAGE as the second dimension. Most protein signals appear as black spots, indicating that the overall protein profile is largely similar between the genotypes. One spot at ∼40 kDa was of increased abundance in Prnp0/0 myelin and identified as SEPT9. The corresponding region of a representative gel (red frame) is shown to the right with separated detection channels. Note that the abundance of SEPT9 in wild-type myelin was below the threshold of MS detection. Note also the high comparability with the reference map in Figure 2A. B, Comparison of SEPT9 abundance in myelin of WT and Prnp0/0 myelin by immunoblotting. Note that the abundance of the related SEPT7 was unchanged. Quantification of SEPT9 abundance was relative to that of GAPDH. C, Immunoblot analysis of SEPT9 in brain lysates and myelin-enriched fractions. Note that the abundance of SEPT9 was reduced with myelin purification, different from known constituents of the compact subcompartment of myelin. D, In situ hybridization of Sept9 in sciatic nerves. Note that Schwann cells are strongly labeled. Scale bar, 6 μm. E, Quantitative RT-PCR analysis of Sept9 in adult wild-type and Prnp0/0 sciatic nerves. Sept9-mRNA abundance was unaltered. F, Immunodetection of SEPT9 visualized with 10 nm gold particles in wild-type sciatic nerve cross sections. SEPT9 labeling was confined to the abaxonal noncompact subcompartment of myelin (bands of Cajal). The magnification shows that SEPT9 was associated with membranous structures, most likely intracellular transport vesicles (F′, arrowheads). G, No labeling was found in wild-type Schmidt–Lanterman incisures. H, SEPT9 was readily detected in Schmidt–Lanterman incisures of Prnp0/0 myelin (arrowheads). Scale bars: F, 500 nm; and G, H, 200 nm.
To independently confirm SEPT9 as a peripheral myelin protein, we analyzed normal sciatic nerve lysates and the myelin-enriched fraction by immunoblotting (Fig. 6C). One band was detected, corresponding to the smallest variant (334 aa, ENSEMBL splice isoform Sept9–004/005; calculated molecular weight 39 kDa). The amount of SEPT9 in myelin fractions was lower than in nerve lysates, indicating that SEPT9 is not exclusive to structures enriched with myelin purification such as compact myelin. On the basis of in situ hybridization of sciatic nerves, Sept9-mRNA is strongly expressed in Schwann cells (Fig. 6D). By qRT-PCR, its abundance was not different between wild-type and Prnp0/0 nerves, suggesting that the increased protein abundance is a post-transcriptional effect. To address the exact localization of SEPT9 in vivo, we applied cryo-immuno electron microscopy. In wild-type Schwann cells, SEPT9 labeling was restricted to the abaxonal, noncompacted myelin subcompartment (Fig. 6F), termed the bands of Cajal (Court et al., 2004), where it was associated with membranous structures (Fig. 6F′), most likely intracellular transport vesicles. Wild-type Schmidt–Lanterman incisures were devoid of SEPT9 labeling (Fig. 6G). Interestingly, SEPT9 was readily detectable in Schmidt–Lanterman incisures of Prnp0/0 nerves (Fig. 6H). This suggests that the adoption of a novel site of localization by SEPT9 underlies its increased abundance in myelin purified from Prnp0/0 nerves and may contribute to the observed chronic demyelinating polyneuropathy.
Abundance profiles of mRNAs encoding PNS myelin proteins
To systematically analyze the postnatal abundance profiles of the transcripts encoding the proteins in the compendium, we have performed microarray analysis of sciatic nerves at postnatal days 1, 5, 15, and 65. Unique gene identifiers were determined, and cluster analysis was performed for those 528 of 545 genes that were represented on our arrays (Fig. 7). Sept9 was also included, though detection of SEPT9 protein by MS in wild-type myelin was below the significance threshold. For comparison, eight known myelin proteins not identified by MS were included [E-cadherin (gene symbol Cdh1), β-catenin (Ctnnb1), myotubularin-related protein-2 (Mtmr2), MP11 (Mp11/A330049M08Rik), junctional adhesion molecule (JAM4/Igsf5), connexin-32 (Cx32/Gjb1), Pmp22, and Mal]. Among the resulting 537 mRNAs, 400 showed developmental abundance changes, including the majority of known myelin genes (Fig. 6). The majority of established myelin proteins displayed a postnatal mRNA abundance peak, corresponding either to an early phase of myelination [P5-UP: Mag, Ctnnb1, integrin-α6 (Itga6), septin 11 (Sept11)] or a later phase [P15-UP: Mpz, Mbp, Cnp, 4.1G (Epb4.1l2), Sirt2, Pmp2, periaxin (Prx), NECL4 (Cadm4), Mp11, and UDP-galactosyltransferase 8 (Ugt8) (Bosio et al., 1996)]. It has been noted previously that the abundance of a large number of mRNAs expressed in Schwann cells increases only late after myelination (e.g., genes involved in storage lipid metabolism) (Verheijen et al., 2003). The corresponding cluster (Late-UP) includes fatty acid synthase (Fasn) (Salles et al., 2002), Cdh1, Ndrg1 (Berger et al., 2004), dystrophin-related protein 2 (Drp2), caveolin-1 (Cav1) (Mikol et al., 2002), and the actin filament-severing gelsolin (Gsn) (Gonçalves et al., 2010). There is also a large cluster of mRNAs of which the abundance significantly decreases after the early stages (“descending”), though the protein product has been identified by MS in mature myelin. This cluster includes annexin A2 (Anxa2) (Hayashi et al., 2007), integrin β1 (Itgb1), septin 2 (Sept2), septin 7 (Sept7), Sept9, and ezrin (Ezr). The mRNA abundance profiles of only two genes (Rab5c, Ap2b1) did not fit into these clusters, indicating that the set mathematical parameters reflect the expression relationships well.
Figure 7.
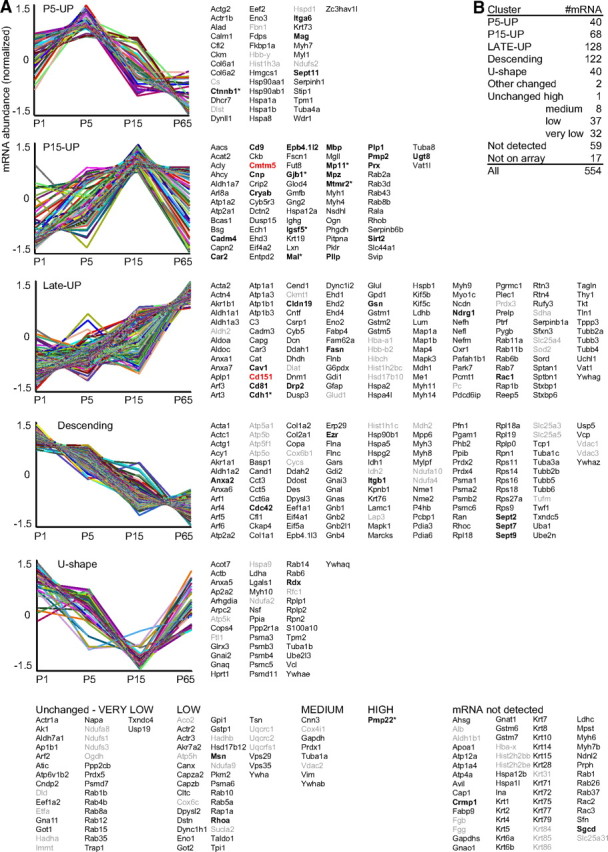
mRNA abundance profiles of myelin-associated genes in developing sciatic nerves. A, K-means clustering was performed for those 529 of 546 proteins in our compendium for which corresponding oligonucleotides were present on the arrays. Eight genes marked with an asterisk were additionally included for comparison, though the proteins were not identified by MS. The normalized mRNA abundance profiles are plotted with regard to the age (mouse postnatal days 1, 5, 15, and 65). Genes with significant mRNA abundance changes were categorized in five clusters, and genes with developmentally unchanged expression were grouped according to mRNA abundance. Known myelin genes are in bold, the novel myelin genes Cmtm5 and Cd151 are in red, and genes encoding mitochondrial, blood, hair, or nuclear proteins are in gray. In clusters P5-UP and P15-UP, the mRNA abundance is highest around the peak of myelination, while mRNA abundance in cluster Late-UP is highest after developmental myelination. mRNAs in cluster descending are significantly suppressed during development, though the corresponding proteins were identified in mature myelin. Note that a significant number of mRNAs is developmentally unchanged. B, The numbers of mRNAs per cluster are given. Note that only two mRNAs show developmental changes not fitting in any cluster.
Exploiting microarray data, mRNA abundance profiles in sciatic nerves can be evaluated systematically to determine coregulated expression, but also for individual genes of interest. Interestingly, the abundance profiles of the mRNAs encoding the newly identified CMTM5 and CD151 are similar to those of known myelin tetraspans (Fig. 7). Cmtm5-mRNA, like Cd9, Pllp, Mal, and Plp1, is most abundant at P15 with a subsequent decline (cluster “P15-UP”), while the abundance of Cd151 mRNA, like Cd81 and Claudin 19 (Cldn19) (Miyamoto et al., 2005), increases to a late peak after completion of the most active myelination (Late-UP). Interestingly, such reverse mRNA abundance changes affect a number of protein families, of which several members have been detected in myelin. For example, the myelin marker enzyme carbonic anhydrase 2 (Car2) mRNA abundance profile correlated with the peak of active myelination (P15-UP), while its later decrease in matured Schwann cells goes along with the increased abundance of carbonic anhydrase 3 (Car3, Late-UP). Similarly, there is a shift from the Rho-GTPase Cdc42 (descending) required for Schwann cell proliferation (Benninger et al., 2007) to the related Rho-GTPase Rac1 required for Schwann cell process extension and stabilization (Late-UP), and from RhoC (descending) to RhoB (P15-UP). We suggest that such reverse abundance changes within protein families may reflect different functional requirements during myelin biogenesis versus in mature myelin (e.g., regarding different enzymatic optima of proteins with principally similar functions).
Importantly, robust abundance though developmentally unchanged was observed for 46 mRNAs, including that for the glycolysis enzyme GAPDH (Gapdh), but also the established myelin genes moesin (Msn), RhoA (Brancolini et al., 1999), and Pmp22 (Fig. 7). Together, many mRNAs encoding myelin proteins exhibited the expected abundance profile with an early postnatal peak (at P5-P15). In contrast, the abundance of other myelin mRNAs decreased, increased, or did not change with maturation, indicating that myelin proteins are not necessarily signified by particular mRNA abundance profiles.
Ninety-one mRNAs were either not detected at all (59) or at very low abundance (32) (Fig. 7). The proteomic identification of the corresponding proteins thus may partly stem from contaminants, and indeed the group comprises all of the contaminating hair keratins and most of the nuclear and blood contaminants. However, some among them are false negatives. For example, sarcoglycan-δ (Sgcd) and collapsin-response mediator protein 1 (Crmp1; also termed dihydropyrimidinase-related protein 1) are established myelin proteins (D'Antonio et al., 2006; Cai et al., 2007), though their mRNAs were not detected in our microarray analysis. This suggests that at least some array probes were unsuitable to detect their target mRNA, justifying the categorization as “not detected” rather than “not expressed.”
Functional categorization
To examine whether certain functions are over-represented in the myelin proteome, we have systematically analyzed the associated gene ontology (GO) terms using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (david.abcc.ncifcrf.gov). As expected, many proteins were annotated as being involved in adhesion, but proteins involved in signaling, vesicle transport, protein translation, and the glucose metabolism also were over-represented (Fig. 8). For further subcategorization, we have analyzed the GO terms for the mRNA abundance clusters descending, Late-UP, and the combined P5/P15-UP. In accordance with the stages of Schwann cell development, proteins involved in protein translation were over-represented in the cluster with the highest mRNA-abundance at P1 (descending), while chaperones were over-represented in both the descending and combined P5/P15-UP cluster. Interestingly, proteins involved in the metabolism of lipids were over-represented during and after most active myelination (P5/P15-UP, Late-UP), and proteins involved in glucose metabolism were over-represented in the Late-UP cluster. Notwithstanding that functional classification according to previously annotated functions leads to a certain degree of ambiguity, we suggest that the developmental shift in mRNA abundance patterns reflects that the tasks of myelin shift from early functions mainly regarding protein synthesis and transport, and cellular adhesion, toward later functions requiring a more active lipid and glucose metabolism.
Figure 8.
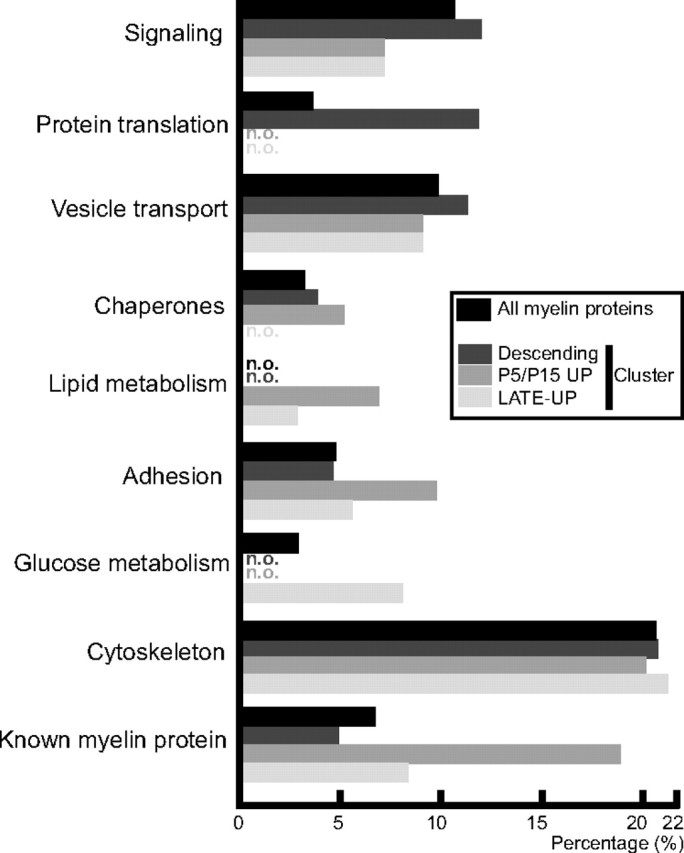
Functional categorization. The proteins identified in PNS myelin (in black) and the mRNA clusters descending (dark gray), P5/P15-UP (gray), and Late-UP (light gray) were analyzed for over-represented functional annotations using gene ontology terms. For comparison, myelin proteins known according to the literature were annotated. Mitochondrial, hair, blood, and nuclear proteins were omitted. The histogram indicates the percentage of proteins within each cluster that have been annotated with the particular function. n.o., Not over-represented.
Integrated proteomics and genetics data suggest novel positional neuropathy candidate genes
Hereditary neuropathies can be caused by mutations affecting a variety of genes, including many that encode myelin proteins (Barisic et al., 2008; Pareyson and Marchesi, 2009; Reilly and Shy, 2009). Indeed, our compendium comprises the causative genes MPZ, PRX, NDRG1, ras-related protein RAB7A, heat shock protein B1 (HSPB1), glycyl-tRNA-synthetase (GARS), and neurofilament light chain (NEFL) (Table 2). However, up to 50 causative genes remain unknown (Braathen et al., 2011). Therefore, we have explored whether novel candidate genes can be derived from our data. We have not restricted the analysis to demyelinating neurological subtypes (CMT1) but have included axonal subtypes (CMT2) because mutations affecting some genes such as MPZ or NEFL can cause either axonal or demyelinating pathology (Vance, 2000). We have also not excluded genes according to prior knowledge of spatiotemporal expression, as several disease-causing genes are expressed in both neurons and Schwann cells, such as GARS (Cuesta et al., 2002; Verheijen et al., 2003) and NEFL (Roberson et al., 1992; Previtali et al., 2003). For 543 (of 545) proteins in our compendium, we were able to determine the chromosomal localization of the corresponding human homologous gene using SNPper (snpper.chip.org), the University of California, Santa Cruz, Genome Bioinformatics Suite (genome.ucsc.edu), Entrez Gene (ncbi.nlm.nih.gov/gene), and blast search validation (when required). The comparison with 16 chromosomal segments previously associated with neuropathy suggested 59 candidate genes for 14 distinct loci (Table 2); for example, CMT3C maps to chromosomal segment 8q23–q24, comprising 204 genes. Three of them [oxidation resistance 1 (OXR1), N-myc downstream regulated gene-1 (NDRG1), and plectin (PLEC1)] encode proteins that were identified in our screen, making them attractive candidate neuropathy genes.
Table 2.
Comparison of neuropathy disease loci and proteins identified in PNS myelin
| Disease | OMIM no. | Locus | Genes in locus | Myelin associated genes |
|---|---|---|---|---|
| Neuropathy with known disease gene | Proven neuropathy gene | |||
| CMT1B/2I/2J/3B, DSS | 159440 | 1q23.3 | 28 | MPZ |
| CMT2B | 602298 | 3q21.3 | 42 | RAB7A |
| CMT2D, HMN5A | 600287 | 7p15 | 84 | GARS |
| CMT1F/2E | 162280 | 8p21 | 87 | NEFL |
| CMT2F, HMN2B | 602195 | 7q11.23 | 86 | HSPB1 (HSP27) |
| CMT3D/4F, DSS | 605725 | 19q13.13-q13.2 | 136 | PRX |
| CMT4D | 605262 | 8q24.22 | 108 | NDRG1* |
| HNA | 162100 | 17q25 | 94 | SEPT9 |
| Neuropathy lacking a known disease gene | Candidate neuropathy genes | |||
| CMT2C, DSMA | 606071 | 12q23-q24 | 304 | VPS29, RAN, CKAP4, DYNLL1, ERP29, ALDH2, HSP90B1, RAB35, SLC25A3 |
| CMT2G | 608591 | 12q12.2-q13.3 | 307 | ARF3, KRT, PRPH, ATP5B, CS, COL2A1, DCTN2, GPD1, FAM62A |
| DI-CMTA | 606483 | 10q24.1-q25.1 | 129 | PGAM1, INA, ARP1, GOT1, SFXN3 |
| CMT3C | NA | 8q23-q24 | 204 | OXR1, NDRG1, PLEC1 |
| CMT4G | 605285 | 10q23.2 | 18 | GLUD1 |
| CMT4C2 | NA | 8q21.2-q21.3 | 21 | CA2, CA3, GDAP, PMP2, FABP4 |
| CMTX2 | 302801 | Xp22.2 | 48 | NA |
| CMTX3 | 302802 | Xq26 | 54 | HPRT1 |
| CMTX4 | 310490 | Xq24-q26.1 | 77 | SLC25A5, PGRMC1 |
| HMN7A | 158580 | 2q14 | 53 | ACTR3, TSN |
| HMN3, HMN4, DSMA3 | 607088 | 11q13 | 268 | PC, STIP1, SERPINH1, CFL1, PRDX5, RAB6A, RAB1B, EHD1, GSTP1, DHCR7, RTN3 |
| HMN-J, DSMA2 | 605726 | 9p21.1-p12 | 123 | TLN1, VCP, TPM2, ALDH1B1 |
| HMN-X | NA | Xq13-q22 | 210 | DRP2, PLP1 |
| HSAN1B | 608088 | 3p22-p24 | 134 | PDCD6IP, RAB5A |
| HSAN with SPG | NA | 5q15.31-q14.1 | 82 | NA |
| HSAN with deafness | NA | Xq23-q27.3 | 174 | SLC25A5, HPRT1, PGRMC1 |
Proteins identified by MS in sciatic nerve myelin were analyzed with respect to the localization of the corresponding gene in the human genome for comparison with neuropathy disease loci. *Note that CMT4D has been previously associated with 8q24.3 while the causative gene (NDRG1) is located at 8q24.22. CMT, Charcot-Marie-Tooth disease; DSS, Déjerine-Sottas syndrome; HMN, hereditary motor neuropathy; HSAN, hereditary sensory and autonomic neuropathy; NA, not applicable; OMIM, Online Mendelian Inheritance in Man.
Discussion
We have established the first proteomic analysis of PNS myelin. Among 545 identified proteins, our compendium includes almost all known myelin proteins, indicating that the proteomic approach was appropriate for systematic analysis. Focusing on tetraspan-transmembrane proteins, we have proven that indeed myelin constituents were newly identified. We have also integrated the compendium with various systematic approaches, namely the following: (1) mRNA abundance profiles in peripheral nerves; (2) transmembrane domain prediction according to three algorithms; (3) subcellular localization prediction according to four algorithms and comparison with the brain mitochondria proteome; (4) over-represented functions; (5) association of the corresponding human genes with neuropathy disease loci; and (6) comparison with the CNS myelin proteome.
The identification of hundreds of myelin proteins necessitates reconsidering the long-standing view that PNS myelin contains only a few proteins, which was shaped in early studies in which only three bands (designated P0, P1, P2) were visible after myelin separation by 1D-SDS-PAGE followed by Fast-Green, Buffalo-Black, or Coomassie labeling (Greenfield et al., 1973; Micko and Schlaepfer, 1978). Later analyses revealed 15–25 bands (Gillespie et al., 1994), the majority of which were constituted by unidentified proteins. Since the 1970s, the number of myelin proteins increased to >40, often upon the genetic identification of neuropathy disease genes or mRNA microarray analyses filtering data for abundance changes during nerve development or in disease (Verheijen et al., 2003; Buchstaller et al., 2004; D'Antonio et al., 2006; Ryu et al., 2008). These studies clarified many developmental and pathological processes but were not suited to systematically analyze myelin protein composition because myelin proteins are not necessarily signified by particular mRNA abundance profiles and genes expressed in Schwann cells not necessarily encode myelin proteins. Unbiased proteomic approaches as pursued here circumvent these problems.
For the present study, myelin was purified using an essentially unchanged protocol. The key to comprehensiveness was the application of complementary proteomic approaches. In our hands, gel-based and gel-free methods identified rather similar numbers of proteins. However, gel-free analyses required less starting material by a factor of 10–100 compared with 2D gels. However, 2D-IEF/SDS-PAGE was important as it increased the coverage of myelin proteins (93 unique identifications), provided a two-dimensional reference map, and enabled differential myelin proteome analysis by 2D-DIGE. Gel-free LC-MSE was very powerful as it yielded quantitative data on thousands of tryptic peptides, allowing us to readjust the relative abundance of myelin proteins. The MS-based quantification of P0 (21%), periaxin (16%), and MBP (8%) supports that particular myelin proteins are of predominating abundance. However, the MS-based values are moderate compared with gel-based estimates, in which P0 and MBP were considered to constitute an extraordinarily high 50–70% and 15% of the total myelin protein, respectively (Garbay et al., 2000). Thus, the large-scale identification of low-abundant proteins goes along with a lower relative abundance of any individual myelin protein. Importantly, gel-based abundance estimates were hampered by insufficient 1D separation. For example, P2 was estimated to constitute 2–15%, depending on the species and the study (Whitaker, 1981). However, we show that in 1D gels P2 comigrates with hemoglobin-A and by MS quantification constitutes only <0.2% of the total myelin protein. Therefore, gel-based estimates of P2 abundance may actually represent a measure of contamination with blood.
Although we used state-of-the-art proteomic techniques, we failed to identify certain established PNS myelin proteins, reflecting technical limitations particularly affecting small membrane-tetraspans such as PMP22 (18.0 kDa) (Naef and Suter, 1998; Müller, 2000) and MAL (16.6 kDa) (Schaeren-Wiemers et al., 2004). Complete tryptic digestion of these proteins does not result in any peptide appropriate for MS sequencing because their tryptic cleavage sites are atypically distributed and membrane-spanning peptides are barely detected by MS. Thus, improved enrichment protocols for membrane-spanning proteins, usage of alternative proteases (e.g., elastase) (Rietschel et al., 2009), and technical advancements in MS may expand the myelin proteome even further.
Improved coverage of low-abundant proteins goes along with more identified contaminants. The myelin-enriched fraction from peripheral nerves doubtless contains contaminants, including mitochondria (with similar flotation properties) and axonal membranes (associated via adhesion proteins). As absolute purity may not be gained even with improved protocols, we attempted specifying likely contaminants. First, axonal contaminants were predicted by comparison with mRNA microarray data as the neuronal cell bodies are outside of the nerve segment used for proteome analysis. Neuronal mRNAs would be not detected in sciatic nerves (although some unsuitable oligonucleotides on microarrays lead to false negatives). Second, software-based prediction of subcellular localization and comparison with mitochondrial proteome datasets determined mitochondrial contaminants with reasonable probability (10.5% of proteins in our dataset). Interestingly, a considerable number of mitochondrial proteins have a dual localization (Pagliarini et al., 2008), suggesting that some may additionally be myelin constituents. Thus, software-based prediction of subcellular localization is valuable for proteins with sorting motifs but should be taken cautiously because some are ambiguous or irrelevant in vivo, and predictions by different algorithms can be conflicting. For example, MBP, DRP2, 4.1G, and periaxin are efficiently incorporated into myelin although they contain polybasic motifs consistently interpreted by software as nuclear targeting signals. Together, we estimate that contaminant proteins from hair, blood, and nuclei collectively comprise <5% of our compendium. Thus, we calculate that >400 proteins of the 545 in our dataset (representing >95% of the total protein amount in the myelin-enriched fraction) are true myelin constituents, though immuno-based validation will ultimately be required. All proteins identified by MS in PNS myelin that were also validated with independent methods are given in Table 3.
Table 3.
Mass-spectrometrically identified myelin proteins additionally validated with immuno-based methods
| Protein name | Gene | MS method | Reference | TMD | Cluster | CNS |
|---|---|---|---|---|---|---|
| Annexin A2 | Anxa2 | I,B,S,D,E,wS,wE,wSE | Hayashi et al., 2007 | Descend | N | |
| Band 4.1 protein G | Epb41l2 | B,S,D,E,wS,wE,wSE | Ohno et al., 2006 | P15-UP | N | |
| Carbonic anhydrase 2 | Ca2 | I,D,E | Cammer et al., 1987 | P15-UP | N | |
| Caveolin 1 | Cav1 | S,E,wSE | Mikol et al., 2002 | 1 | Late-UP | N |
| CD151 | Cd151 | S,wS | This article | 4 | Late-UP | N |
| CD81 | Cd81 | D,E,wE,wSE | Ishibashi et al., 2004 | 4 | Late-UP | Y |
| CD9 | Cd9 | S,D,E,wS,wE,wSE | Ishibashi et al., 2004 | 4 | P15-UP | N |
| Cell adhesion molecule 4, NECL4 | Cadm4 | I,S,D,E,wS,wSE | Spiegel et al., 2007 | 1 | P15-UP | Y |
| Cell division control protein 42 | Cdc42 | I,E,wSE | Benninger et al., 2007 | Descend | Y | |
| CKLF-like MARVEL TMD-containing 5 | Cmtm5 | wS,wSE | This article | 4 | P15-UP | N |
| Claudin 19 | Cldn19 | wSE | Miyamoto et al., 2005 | 4 | Late-UP | N |
| CNP | Cnp | I,B,S,D,E,wS,wE,wSE | Matthieu et al., 1980 | P15-UP | Y | |
| Crystallin α2 | Cryab | I,S,D,E,wS,wSE | d'Antonio et al., 2006 | P15-UP | Y | |
| Dihydropyrimidinase related 1 | Crmp1 | E | d'Antonio et al., 2006 | ND | Y | |
| Dystrophin-related protein 2 | Drp2 | E | Sherman et al., 2001 | Late-UP | N | |
| Ezrin | Ezr | I,E | Scherer et al., 2001 | Descend | Y | |
| Fatty acid synthase | Fasn | S,D,E,wS,wE,wSE | Salles et al., 2002 | Late-UP | Y | |
| Gelsolin | Gsn | I,S,D,E,wSE | Gonçalves et al., 2010 | Late-UP | Y | |
| Integrin α6 | Itga6 | I,D,wSE | Nodari et al., 2008 | 1 | P5-UP | N |
| Integrin β1 | Itgb1 | wSE | Feltri et al., 2002 | 1 | Descend | N |
| Moesin | Msn | I,B,S,D,E,wSE | Scherer et al., 2001 | Unchanged | Y | |
| Myelin associated glycoprotein | Mag | S,D,E,wS,wSE | Figlewicz et al., 1981 | 1 | P5-UP | Y |
| Myelin basic protein | Mbp | I,B,S,D,E,wS,wE,wSE | Boggs, 2006 | P15-UP | Y | |
| Myelin protein 2, PMP2 | Pmp2 | I,B,S,D,E,wS,wE,wSE | Trapp et al., 1984 | P15-UP | N | |
| Myelin protein zero, P0 | Mpz | I,B,S,D,E,wS,wE,wSE | Giese et al., 1992 | 1 | P15-UP | Y |
| Myelin proteolipid protein | Plp1 | B,S,D,E,wS,wE,wSE | Garbern et al., 1997 | 4 | P15-UP | Y |
| NDRG1, N-myc downstream regulated | Ndrg1 | I,D,E,wS,wE,wSE | Berger et al., 2004 | Late-UP | Y | |
| Periaxin | Prx | I,B,S,D,E,wS,wE,wSE | Gillespie et al., 1994 | P15-UP | N | |
| Plasmolipin | Pllp | S,E,wS,wSE | Bosse et al., 2003 | 4 | P15-UP | Y |
| Rac1 | Rac1 | I,S,D,E,wSE | Benninger et al., 2007 | Late-UP | Y | |
| Radixin | Rdx | I,E | Scherer et al., 2001 | U-shape | Y | |
| RhoA | Rhoa | I,D,E,wSE | Brancolini et al., 1999 | Unchanged | Y | |
| Sarcoglycan δ | Sgcd | wSE | Cai et al., 2007 | 1 | ND | N |
| Septin 11 | Sept11 | I | Buser et al., 2009 | P5-UP | Y | |
| Septin 2 | Sept2 | I,D,E | Buser et al., 2009 | Descend | Y | |
| Septin 7 | Sept7 | D,E,wSE | Buser et al., 2009 | Descend | Y | |
| Septin 9 | Sept9 | 2D-DIGE | This article | Descend | N | |
| Sirtuin 2, NAD-dependent deacetylase | Sirt2 | I,wSE | Werner et al., 2007 | P15-UP | Y | |
| UDP galactosyltransferase 8 | Ugt8 | wSE | Bosio et al., 1996 | 1 | P15-UP | N |
The proteins identified in peripheral myelin by MS have been compiled according to availability of validation with immuno-based methods. The methods of MS identification are as follows: S, GeLC; wS, wash-GelC; wSE, wash-GeLC-MSE; I, 2D-IEF/SDS-PAGE; B, 2D-16BAC/SDS-PAGE; D, LC-DDA; E, LC-MSE; wE, wash-LC-MSE. One selected reference is given in which presence in peripheral myelin was validated. Additionally given are the number of transmembrane domains (TMDs), the mRNA abundance profile cluster, and whether a protein was also identified by mass spectrometry in CNS myelin according to the compilation by Jahn et al. (2009) (CNS: Y, also identified in CNS myelin; N, not identified in CNS myelin). The complete compendium of MS-identified proteins will be hosted on the author's web page (see Notes). Tetraspan proteins are in bold. ND, mRNA not detected.
Systematic prediction of transmembrane domains suggested that CMTM5 and CD151 are novel myelin-tetraspans, a protein class with proven relevance for myelin (Bronstein, 2000). Indeed, we visualized their mRNAs in Schwann cells by in situ hybridization, revealed their mRNA abundance profiles in sciatic nerves, and detected both proteins in compact myelin by immuno-based methods. Like other myelin proteins, they are not exclusive to myelin but expressed in a range of tissues, though enriched in myelin. For example, expression of the tetraspanin CD151 was noted in the lung, kidney glomeruli, and platelet cells (Caplan et al., 2007; Goschnick and Jackson, 2007) where it regulates integrin signaling (Yunta and Lazo, 2003; Baldwin et al., 2008), which is also crucial for myelination (Berti et al., 2006). A characterization of native tissue expression of CMTM5 is yet unavailable, though it has been suggested as a potential tumor suppressor (Shao et al., 2007, 2009; Guo et al., 2009). CMTM5 is related to MAL and plasmolipin regarding the presence of a MARVEL domain implicated in membrane apposition (Sánchez-Pulido et al., 2002), and to chemokine-like proteins. Thus, CD151 and CMTM5 not only represent novel myelin markers, they are also likely relevant to the biogenesis and structural organization of PNS myelin, which will be specified in future gene-targeting experiments.
Differential myelin proteome analysis in the Prnp0/0 neuropathy model (Bremer et al., 2010) revealed the strongly increased abundance of SEPT9, related to its presence in Prnp0/0 mutant Schmidt-Lanterman incisures additional to its normal localization in the bands of Cajal. SEPT9 is expressed in many tissues (Scott et al., 2005) and more abundant in Schwann cells than in neurons (Sudo et al., 2007). Septins can assemble into filaments and have been implicated in regulating microtubules and vesicle trafficking (Peterson and Petty, 2010). Duplication of the whole SEPT9 gene causes the neuropathy HNA (Collie et al., 2010), and point mutations (Kuhlenbäumer et al., 2005) dramatically increase SEPT9–mRNA translation (McDade et al., 2007), suggesting that the SEPT9 dosage is pathophysiologically relevant (van Alfen, 2011). In comparison, the most frequent cause of CMT is a duplication of the PMP22 gene (Barisic et al., 2008). Together, the requirement of Prnp for the normal abundance and localization of SEPT9 in peripheral myelin implies that neuronal Prnp contributes to axon-to-Schwann cell signaling related to the myelin cytoskeleton and long-term myelin maintenance. The exact abundance of SEPT9 in Schwann cells may not only be related to the (yet speculative) pathomechanism of HNA (mainly affecting the brachial plexus) but also to other neuropathies, possibly by interfering with the normal formation or stability of SEPT9-containing filaments.
This first proteomic approach to peripheral myelin provides evidence that its molecular composition is much more complex and that the relative abundance of P0 and MBP is less extreme than thought. Two tetraspans were established as novel constituents of compact myelin. While we cannot ultimately validate the presence of all identified proteins, the integration with other systematic datasets allows the prediction of ∼400 true myelin proteins. We also provide categorization according to developmentally coregulated mRNA abundance. We noted that not only expected functional annotations such as adhesion and the lipid metabolism are over-represented in the myelin proteome but also the glucose metabolism. Finally, we have predicted novel candidate genes to cause (when mutated) hereditary neuropathies. Many proteins were identified in both PNS and CNS myelin, suggesting that Schwann cells and oligodendrocytes have recruited surprisingly similar proteins into myelin. However, differential protein distributions appear at least as relevant, as exemplified by the shift from P0 to PLP as the most abundant CNS myelin protein at the evolutionary transition from fish to tetrapods (Yoshida and Colman, 1996). Together, our compendium provides a framework to analyze myelination and glia–axonal interactions, the functions of myelin in insulating axons and preventing them from degeneration, and molecular changes in myelin-related disease.
Notes
The compendium of MS-identified proteins, including SwissProt/UniProt accession numbers, acronyms, subcellular localization according to three algorithms (Cello, Wolfpsort, Subloc), presence of signal peptides or mitochondrial targeting signals, spot numbers for proteins identified from 2D gels according to Figure 2, relative abundance of proteins identified by LC-MSE (in parts per million), and mRNA abundance profile, is available at http://www1.em.mpg.de/myelinproteome. This material has not been peer reviewed.
Footnotes
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF-DLR-Leukonet), the European Commission (FP7-LeukoTreat), and the Deutsche Forschungsgemeinschaft (DFG-SFB490, Z3). A.A. and K.-A.N. hold European Research Council (ERC) Advanced Investigator Grants. We dedicate this paper to the memory of our colleague C. Humml, who performed the mRNA array analysis for this study. We thank S. Papiol, M. J. Rossner, R. Stassart, L. Piepkorn, V. Tarabykin, and M. W. Sereda for discussions; and E. Leidmaa, M. Uecker, T. Liepold, and J. Forsch for technical assistance. We are grateful for antibody gifts by N. Schaeren-Wiemers, K.-I. Nagata, and F. Bosse.
The authors declare no competing financial interests.
References
- Baer AS, Syed YA, Kang SU, Mitteregger D, Vig R, Ffrench-Constant C, Franklin RJ, Altmann F, Lubec G, Kotter MR. Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain. 2009;132:465–481. doi: 10.1093/brain/awn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin G, Novitskaya V, Sadej R, Pochec E, Litynska A, Hartmann C, Williams J, Ashman L, Eble JA, Berditchevski F. Tetraspanin CD151 regulates glycosylation of (alpha)3(beta)1 integrin. J Biol Chem. 2008;283:35445–35454. doi: 10.1074/jbc.M806394200. [DOI] [PubMed] [Google Scholar]
- Barisic N, Claeys KG, Sirotković-Skerlev M, Löfgren A, Nelis E, De Jonghe P, Timmerman V. Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann Hum Genet. 2008;72:416–441. doi: 10.1111/j.1469-1809.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- Barral Y, Kinoshita M. Structural insights shed light onto septin assemblies and function. Curr Opin Cell Biol. 2008;20:12–18. doi: 10.1016/j.ceb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Benninger Y, Thurnherr T, Pereira JA, Krause S, Wu X, Chrostek-Grashoff A, Herzog D, Nave KA, Franklin RJ, Meijer D, Brakebusch C, Suter U, Relvas JB. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J Cell Biol. 2007;177:1051–1061. doi: 10.1083/jcb.200610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P, Sirkowski EE, Scherer SS, Suter U. Expression analysis of the N-Myc downstream-regulated gene 1 indicates that myelinating Schwann cells are the primary disease target in hereditary motor and sensory neuropathy-Lom. Neurobiol Dis. 2004;17:290–299. doi: 10.1016/j.nbd.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Berti C, Nodari A, Wrabetz L, Feltri ML. Role of integrins in peripheral nerves and hereditary neuropathies. Neuromolecular Med. 2006;8:191–204. doi: 10.1385/nmm:8:1-2:191. [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio A, Binczek E, Stoffel W. Functional breakdown of the lipid bilayer of the myelin membrane in central and peripheral nervous system by disrupted galactocerebroside synthesis. Proc Natl Acad Sci U S A. 1996;93:13280–13285. doi: 10.1073/pnas.93.23.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse F, Hasse B, Pippirs U, Greiner-Petter R, Müller HW. Proteolipid plasmolipin: localization in polarized cells, regulated expression and lipid raft association in CNS and PNS myelin. J Neurochem. 2003;86:508–518. doi: 10.1046/j.1471-4159.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- Braathen GJ, Sand JC, Lobato A, Høyer H, Russell MB. Genetic epidemiology of Charcot-Marie-Tooth in the general population. Eur J Neurol. 2011;18:39–48. doi: 10.1111/j.1468-1331.2010.03037.x. [DOI] [PubMed] [Google Scholar]
- Brancolini C, Marzinotto S, Edomi P, Agostoni E, Fiorentini C, Müller HW, Schneider C. Rho-dependent regulation of cell spreading by the tetraspan membrane protein Gas3/PMP22. Mol Biol Cell. 1999;10:2441–2459. doi: 10.1091/mbc.10.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J, Baumann F, Tiberi C, Wessig C, Fischer H, Schwarz P, Steele AD, Toyka KV, Nave KA, Weis J, Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- Bronstein JM. Function of tetraspan proteins in the myelin sheath. Curr Opin Neurobiol. 2000;10:552–557. doi: 10.1016/s0959-4388(00)00125-2. [DOI] [PubMed] [Google Scholar]
- Buchstaller J, Sommer L, Bodmer M, Hoffmann R, Suter U, Mantei N. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser AM, Erne B, Werner HB, Nave KA, Schaeren-Wiemers N. The septin cytoskeleton in myelinating glia. Mol Cell Neurosci. 2009;40:156–166. doi: 10.1016/j.mcn.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Cai H, Erdman RA, Zweier L, Chen J, Shaw JH, 4th, Baylor KA, Stecker MM, Carey DJ, Chan YM. The sarcoglycan complex in Schwann cells and its role in myelin stability. Exp Neurol. 2007;205:257–269. doi: 10.1016/j.expneurol.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Cammer W, Tansey FA. Immunocytochemical localization of carbonic anhydrase in myelinated fibers in peripheral nerves of rat and mouse. J Histochem Cytochem. 1987;35:865–870. doi: 10.1177/35.8.3110266. [DOI] [PubMed] [Google Scholar]
- Caplan MJ, Kamsteeg EJ, Duffield A. Tetraspan proteins: regulators of renal structure and function. Curr Opin Nephrol Hypertens. 2007;16:353–358. doi: 10.1097/MNH.0b013e328177b1fa. [DOI] [PubMed] [Google Scholar]
- Chen ZL, Strickland S. Laminin gamma1 is critical for Schwann cell differentiation, axon myelination, and regeneration in the peripheral nerve. J Cell Biol. 2003;163:889–899. doi: 10.1083/jcb.200307068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernousov MA, Carey DJ. Schwann cell extracellular matrix molecules and their receptors. Histol Histopathol. 2000;15:593–601. doi: 10.14670/HH-15.593. [DOI] [PubMed] [Google Scholar]
- Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Collie AM, Landsverk ML, Ruzzo E, Mefford HC, Buysse K, Adkins JR, Knutzen DM, Barnett K, Brown RH, Jr, Parry GJ, Yum SW, Simpson DA, Olney RK, Chinnery PF, Eichler EE, Chance PF, Hannibal MC. Non-recurrent SEPT9 duplications cause hereditary neuralgic amyotrophy. J Med Genet. 2010;47:601–607. doi: 10.1136/jmg.2009.072348. [DOI] [PubMed] [Google Scholar]
- Court FA, Sherman DL, Pratt T, Garry EM, Ribchester RR, Cottrell DF, Fleetwood-Walker SM, Brophy PJ. Restricted growth of Schwann cells lacking Cajal bands slows conduction in myelinated nerves. Nature. 2004;431:191–195. doi: 10.1038/nature02841. [DOI] [PubMed] [Google Scholar]
- Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. Wound healing is defective in mice lacking tetraspanin CD151. J Invest Dermatol. 2006;126:680–689. doi: 10.1038/sj.jid.5700142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta A, Pedrola L, Sevilla T, García-Planells J, Chumillas MJ, Mayordomo F, LeGuern E, Marín I, Vílchez JJ, Palau F. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet. 2002;30:22–25. doi: 10.1038/ng798. [DOI] [PubMed] [Google Scholar]
- D'Antonio M, Michalovich D, Paterson M, Droggiti A, Woodhoo A, Mirsky R, Jessen KR. Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia. 2006;53:501–515. doi: 10.1002/glia.20309. [DOI] [PubMed] [Google Scholar]
- D'Urso D, Brophy PJ, Staugaitis SM, Gillespie CS, Frey AB, Stempak JG, Colman DR. Protein zero of peripheral nerve myelin: biosynthesis, membrane insertion, and evidence for homotypic interaction. Neuron. 1990;4:449–460. doi: 10.1016/0896-6273(90)90057-m. [DOI] [PubMed] [Google Scholar]
- Feltri ML, Graus Porta D, Previtali SC, Nodari A, Migliavacca B, Cassetti A, Littlewood-Evans A, Reichardt LF, Messing A, Quattrini A, Mueller U, Wrabetz L. Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. J Cell Biol. 2002;156:199–209. doi: 10.1083/jcb.200109021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlewicz DA, Quarles RH, Johnson D, Barbarash GR, Sternberger NH. Biochemical demonstration of the myelin-associated glycoprotein in the peripheral nervous system. J Neurochem. 1981;37:749–758. doi: 10.1111/j.1471-4159.1982.tb12551.x. [DOI] [PubMed] [Google Scholar]
- Filbin MT, Walsh FS, Trapp BD, Pizzey JA, Tennekoon GI. Role of myelin P0 protein as a homophilic adhesion molecule. Nature. 1990;344:871–872. doi: 10.1038/344871a0. [DOI] [PubMed] [Google Scholar]
- Galli S, Jahn O, Hitt R, Hesse D, Opitz L, Plessmann U, Urlaub H, Poderoso JJ, Jares-Erijman EA, Jovin TM. A new paradigm for MAPK: structural interactions of hERK1 with mitochondria in HeLa cells. PLoS One. 2009;4:e7541. doi: 10.1371/journal.pone.0007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61:267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Garbern JY, Cambi F, Tang XM, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19:205–218. doi: 10.1016/s0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Giese KP, Martini R, Lemke G, Soriano P, Schachner M. Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell. 1992;71:565–576. doi: 10.1016/0092-8674(92)90591-y. [DOI] [PubMed] [Google Scholar]
- Gillespie CS, Sherman DL, Blair GE, Brophy PJ. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron. 1994;12:497–508. doi: 10.1016/0896-6273(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Gonçalves AF, Dias NG, Moransard M, Correia R, Pereira JA, Witke W, Suter U, Relvas JB. Gelsolin is required for macrophage recruitment during remyelination of the peripheral nervous system. Glia. 2010;58:706–715. doi: 10.1002/glia.20956. [DOI] [PubMed] [Google Scholar]
- Goschnick MW, Jackson DE. Tetraspanins-structural and signalling scaffolds that regulate platelet function. Mini Rev Med Chem. 2007;7:1248–1254. doi: 10.2174/138955707782795656. [DOI] [PubMed] [Google Scholar]
- Gould RM, Morrison HG, Gilland E, Campbell RK. Myelin tetraspan family proteins but no non-tetraspan family proteins are present in the ascidian (Ciona intestinalis) genome. Biol Bull. 2005;209:49–66. doi: 10.2307/3593141. [DOI] [PubMed] [Google Scholar]
- Greenfield S, Brostoff S, Eylar EH, Morell P. Protein composition of myelin of the peripheral nervous system. J Neurochem. 1973;20:1207–1216. doi: 10.1111/j.1471-4159.1973.tb00089.x. [DOI] [PubMed] [Google Scholar]
- Guo X, Li T, Wang Y, Shao L, Zhang Y, Ma D, Han W. CMTM5 induces apoptosis of pancreatic cancer cells and has synergistic effects with TNF-alpha. Biochem Biophys Res Commun. 2009;387:139–142. doi: 10.1016/j.bbrc.2009.06.148. [DOI] [PubMed] [Google Scholar]
- Hannibal MC, Ruzzo EK, Miller LR, Betz B, Buchan JG, Knutzen DM, Barnett K, Landsverk ML, Brice A, LeGuern E, Bedford HM, Worrall BB, Lovitt S, Appel SH, Andermann E, Bird TD, Chance PF. SEPT9 gene sequencing analysis reveals recurrent mutations in hereditary neuralgic amyotrophy. Neurology. 2009;72:1755–1759. doi: 10.1212/WNL.0b013e3181a609e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartinger J, Stenius K, Högemann D, Jahn R. 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal Biochem. 1996;240:126–133. doi: 10.1006/abio.1996.0339. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Nakashima K, Yamagishi K, Hoshi T, Suzuki A, Baba H. Localization of annexin II in the paranodal regions and Schmidt-Lanterman incisures in the peripheral nervous system. Glia. 2007;55:1044–1052. doi: 10.1002/glia.20529. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Ding L, Ikenaka K, Inoue Y, Miyado K, Mekada E, Baba H. Tetraspanin protein CD9 is a novel paranodal component regulating paranodal junctional formation. J Neurosci. 2004;24:96–102. doi: 10.1523/JNEUROSCI.1484-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Dutta R, Wark GM, Hwang SI, Han DK, Trapp BD, Pfeiffer SE, Bansal R. Human myelin proteome and comparative analysis with mouse myelin. Proc Natl Acad Sci U S A. 2009;106:14605–14610. doi: 10.1073/pnas.0905936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O, Hesse D, Reinelt M, Kratzin HD. Technical innovations for the automated identification of gel-separated proteins by MALDI-TOF mass spectrometry. Anal Bioanal Chem. 2006;386:92–103. doi: 10.1007/s00216-006-0592-1. [DOI] [PubMed] [Google Scholar]
- Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol. 2009;40:55–72. doi: 10.1007/s12035-009-8071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Sommer I, Schachner M, Nave KA. Monoclonal antibody O10 defines a conformationally sensitive cell-surface epitope of proteolipid protein (PLP): evidence that PLP misfolding underlies dysmyelination in mutant mice. J Neurosci. 1996;16:7920–7929. doi: 10.1523/JNEUROSCI.16-24-07920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer-Albers EM, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H, Nave KA, Schild H, Trotter J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clinical Applications. 2007;1:1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- Kuhlenbäumer G, Hannibal MC, Nelis E, Schirmacher A, Verpoorten N, Meuleman J, Watts GD, De Vriendt E, Young P, Stögbauer F, Halfter H, Irobi J, Goossens D, Del-Favero J, Betz BG, Hor H, Kurlemann G, Bird TD, Airaksinen E, Mononen T, Serradell AP, Prats JM, Van Broeckhoven C, De Jonghe P, Timmerman V, Ringelstein EB, Chance PF. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat Genet. 2005;37:1044–1046. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- Larocca JN, Norton WT. Isolation of myelin. Curr Protoc Cell Biol Chapter. 2007;3:Unit3.25. doi: 10.1002/0471143030.cb0325s33. [DOI] [PubMed] [Google Scholar]
- Matthieu JM, Costantino-Ceccarini E, Bény M, Reigner J. Evidence for the association of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase with myelin-related membranes in peripheral nervous system. J Neurochem. 1980;35:1345–1350. doi: 10.1111/j.1471-4159.1980.tb09008.x. [DOI] [PubMed] [Google Scholar]
- McDade SS, Hall PA, Russell SE. Translational control of SEPT9 isoforms is perturbed in disease. Hum Mol Genet. 2007;16:742–752. doi: 10.1093/hmg/ddm003. [DOI] [PubMed] [Google Scholar]
- Micko S, Schlaepfer WW. Protein composition of axons and myelin from rat and human peripheral nerves. J Neurochem. 1978;30:1041–1049. doi: 10.1111/j.1471-4159.1978.tb12397.x. [DOI] [PubMed] [Google Scholar]
- Mikol DD, Scherer SS, Duckett SJ, Hong HL, Feldman EL. Schwann cell caveolin-1 expression increases during myelination and decreases after axotomy. Glia. 2002;38:191–199. doi: 10.1002/glia.10063. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, Nakayama K, Okamura Y, Sasaki H, Miyachi Y, Furuse M, Tsukita S. Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol. 2005;169:527–538. doi: 10.1083/jcb.200501154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius W, Patzig J, Nave KA, Werner HB. Phylogeny of proteolipid proteins: divergence, constraints, and the evolution of novel functions in myelination and neuroprotection. Neuron Glia Biol. 2008;4:111–127. doi: 10.1017/S1740925X0900009X. [DOI] [PubMed] [Google Scholar]
- Müller HW. Tetraspan myelin protein PMP22 and demyelinating peripheral neuropathies: new facts and hypotheses. Glia. 2000;29:182–185. doi: 10.1002/(sici)1098-1136(20000115)29:2<182::aid-glia12>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- Naef R, Suter U. Many facets of the peripheral myelin protein PMP22 in myelination and disease. Microsc Res Tech. 1998;41:359–371. doi: 10.1002/(SICI)1097-0029(19980601)41:5<359::AID-JEMT3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nagata K, Asano T, Nozawa Y, Inagaki M. Biochemical and cell biological analyses of a mammalian septin complex, Sept7/9b/11. J Biol Chem. 2004;279:55895–55904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- Nodari A, Previtali SC, Dati G, Occhi S, Court FA, Colombelli C, Zambroni D, Dina G, Del Carro U, Campbell KP, Quattrini A, Wrabetz L, Feltri ML. Alpha6beta4 integrin and dystroglycan cooperate to stabilize the myelin sheath. J Neurosci. 2008;28:6714–6719. doi: 10.1523/JNEUROSCI.0326-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno N, Terada N, Yamakawa H, Komada M, Ohara O, Trapp BD, Ohno S. Expression of protein 4.1G in Schwann cells of the peripheral nervous system. J Neurosci Res. 2006;84:568–577. doi: 10.1002/jnr.20949. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009;8:654–667. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Sosinsky GE, Ghassemzadeh S, Perez A, Jones Y, Ellisman MH. Electron tomographic analysis of cytoskeletal cross-bridges in the paranodal region of the node of Ranvier in peripheral nerves. J Struct Biol. 2008;161:469–480. doi: 10.1016/j.jsb.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson EA, Petty EM. Conquering the complex world of human septins: implications for health and disease. Clin Genet. 2010;77:511–524. doi: 10.1111/j.1399-0004.2010.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previtali SC, Zerega B, Sherman DL, Brophy PJ, Dina G, King RH, Salih MM, Feltri L, Quattrini A, Ravazzolo R, Wrabetz L, Monaco AP, Bolino A. Myotubularin-related 2 protein phosphatase and neurofilament light chain protein, both mutated in CMT neuropathies, interact in peripheral nerve. Hum Mol Genet. 2003;12:1713–1723. doi: 10.1093/hmg/ddg179. [DOI] [PubMed] [Google Scholar]
- Reilly MM, Shy ME. Diagnosis and new treatments in genetic neuropathies. J Neurol Neurosurg Psychiatry. 2009;80:1304–1314. doi: 10.1136/jnnp.2008.158295. [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel B, Arrey TN, Meyer B, Bornemann S, Schuerken M, Karas M, Poetsch A. Elastase digests: new ammunition for shotgun membrane proteomics. Mol Cell Proteomics. 2009;8:1029–1043. doi: 10.1074/mcp.M800223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson MD, Toews AD, Goodrum JF, Morell P. Neurofilament and tubulin mRNA expression in Schwann cells. J Neurosci Res. 1992;33:156–162. doi: 10.1002/jnr.490330120. [DOI] [PubMed] [Google Scholar]
- Roth AD, Ivanova A, Colman DR. New observations on the compact myelin proteome. Neuron Glia Biol. 2006;2:15–21. doi: 10.1017/S1740925X06000068. [DOI] [PubMed] [Google Scholar]
- Ryu EJ, Yang M, Gustin JA, Chang LW, Freimuth RR, Nagarajan R, Milbrandt J. Analysis of peripheral nerve expression profiles identifies a novel myelin glycoprotein, MP11. J Neurosci. 2008;28:7563–7573. doi: 10.1523/JNEUROSCI.1659-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles J, Sargueil F, Knoll-Gellida A, Witters LA, Shy M, Jiang H, Cassagne C, Garbay B. Fatty acid synthase expression during peripheral nervous system myelination. Brain Res Mol Brain Res. 2002;101:52–58. doi: 10.1016/s0169-328x(02)00161-4. [DOI] [PubMed] [Google Scholar]
- Salzer JL, Brophy PJ, Peles E. Molecular domains of myelinated axons in the peripheral nervous system. Glia. 2008;56:1532–1540. doi: 10.1002/glia.20750. [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Bonnet A, Erb M, Erne B, Bartsch U, Kern F, Mantei N, Sherman D, Suter U. The raft-associated protein MAL is required for maintenance of proper axon—glia interactions in the central nervous system. J Cell Biol. 2004;166:731–742. doi: 10.1083/jcb.200406092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Xu T, Crino P, Arroyo EJ, Gutmann DH. Ezrin, radixin, and moesin are components of Schwann cell microvilli. J Neurosci Res. 2001;65:150–164. doi: 10.1002/jnr.1138. [DOI] [PubMed] [Google Scholar]
- Scott M, Hyland PL, McGregor G, Hillan KJ, Russell SE, Hall PA. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene. 2005;24:4688–4700. doi: 10.1038/sj.onc.1208574. [DOI] [PubMed] [Google Scholar]
- Sendtner M, Carroll P, Holtmann B, Hughes RA, Thoenen H. Ciliary neurotrophic factor. J Neurobiol. 1994;25:1436–1453. doi: 10.1002/neu.480251110. [DOI] [PubMed] [Google Scholar]
- Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang Y, Zhang Y, Ng KM, Han W, Ma D, Tao Q. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res. 2007;13:5756–5762. doi: 10.1158/1078-0432.CCR-06-3082. [DOI] [PubMed] [Google Scholar]
- Shao L, Guo X, Plate M, Li T, Wang Y, Ma D, Han W. CMTM5–v1 induces apoptosis in cervical carcinoma cells. Biochem Biophys Res Commun. 2009;379:866–871. doi: 10.1016/j.bbrc.2008.12.126. [DOI] [PubMed] [Google Scholar]
- Sherman DL, Fabrizi C, Gillespie CS, Brophy PJ. Specific disruption of a schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron. 2001;30:677–687. doi: 10.1016/s0896-6273(01)00327-0. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics. 2006;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- Smith ME, Curtis BM. Frog sciatic nerve myelin: a chemical characterization. J Neurochem. 1979;33:447–452. doi: 10.1111/j.1471-4159.1979.tb05174.x. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci. 2007;10:861–869. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum Mutat. 2007;28:1005–1013. doi: 10.1002/humu.20554. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE. Proteomic mapping provides powerful insights into functional myelin biology. Proc Natl Acad Sci U S A. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Dubois-Dalcq M, Quarles RH. Ultrastructural localization of P2 protein in actively myelinating rat Schwann cells. J Neurochem. 1984;43:944–948. doi: 10.1111/j.1471-4159.1984.tb12828.x. [DOI] [PubMed] [Google Scholar]
- van Alfen N. Clinical and pathophysiological concepts of neuralgic amyotrophy. Nat Rev Neurol. 2011;7:315–322. doi: 10.1038/nrneurol.2011.62. [DOI] [PubMed] [Google Scholar]
- Vance JM. The many faces of Charcot-Marie-Tooth disease. Arch Neurol. 2000;57:638–640. doi: 10.1001/archneur.57.5.638. [DOI] [PubMed] [Google Scholar]
- Vanrobaeys F, Van Coster R, Dhondt G, Devreese B, Van Beeumen J. Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J Proteome Res. 2005;4:2283–2293. doi: 10.1021/pr050205c. [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl AO, Rudolf D, Hillen N, Tenzer S, van Endert P, Schild H, Rammensee HG, Stevanović S. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur J Immunol. 2008;38:1503–1510. doi: 10.1002/eji.200838136. [DOI] [PubMed] [Google Scholar]
- Welcher AA, De Leon M, Suter U, Snipes GJ, Meakin SO, Shooter EM. Isolation of transcriptionally regulated sequences associated with neuronal and non-neuronal cell interactions. Prog Brain Res. 1992;94:163–176. doi: 10.1016/s0079-6123(08)61748-3. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Möbius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Whitaker JN. The protein antigens of peripheral nerve myelin. Ann Neurol. 1981;9(Suppl):56–64. doi: 10.1002/ana.410090710. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Yunta M, Lazo PA. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell Signal. 2003;15:559–564. doi: 10.1016/s0898-6568(02)00147-x. [DOI] [PubMed] [Google Scholar]