Abstract
Proper regulation and maintenance of the epigenome is necessary to preserve genome function. However, in every cell division, the epigenetic state is disassembled and then reassembled in the wake of the DNA replication fork. Chromatin restoration on nascent DNA is a complex and regulated process that includes nucleosome assembly and remodeling, deposition of histone variants, and the re-establishment of transcription factor binding. To study the genome-wide dynamics of chromatin restoration behind the DNA replication fork, we developed nascent chromatin occupancy profiles (NCOPs) to comprehensively profile nascent and mature chromatin at nucleotide resolution. Although nascent chromatin is inherently less organized than mature chromatin, we identified locus-specific differences in the kinetics of chromatin maturation that were predicted by the epigenetic landscape, including the histone variant H2AZ, which marked loci with rapid maturation kinetics. The chromatin maturation at origins of DNA replication was dependent on whether the origin underwent initiation or was passively replicated from distal-originating replication forks, suggesting distinct chromatin assembly mechanisms surrounding activated and disassembled prereplicative complexes. Finally, we identified sites that were only occupied transiently by DNA-binding factors following passage of the replication fork, which may provide a mechanism for perturbations of the DNA replication program to shape the regulatory landscape of the genome.
The distribution and phasing of histone octamers on the DNA, as well as the location of DNA-binding proteins such as transcription factors, define the regulatory landscape of the genome and govern transcription (Jiang and Pugh 2009; The ENCODE Project Consortium 2012). In addition to regulating gene expression, the local chromatin environment is also critical for other DNA-templated processes such as DNA replication and repair (MacAlpine and Almouzni 2013; Dabin et al. 2016; Gutiérrez and MacAlpine 2016). Despite the central role of chromatin in genome function, every cell cycle the chromatin landscape must be disassembled ahead of the replication fork and then reassembled behind the fork to preserve epigenetic memory.
The assembly of nascent chromatin is tightly coupled to the replication fork. Early electron microscopy studies found a similar density of nucleosomes (“beads on a string”) on both the parental and nascent DNA strands (McKnight and Miller 1977), indicating that histone deposition and nucleosome formation must occur rapidly behind the replication fork. Elegant genetic and biochemical experiments have elucidated many of the factors and mechanisms involved in the assembly of chromatin behind the DNA replication fork (Smith and Stillman 1989; Chang et al. 1997; Li et al. 2008; Tyler et al. 1999; Schlesinger and Formosa 2000; Luk et al. 2007), and reconstitution of replication-coupled assembly revealed that nucleosome assembly occurred within ∼250 bp of the replication fork (Sogo et al. 1986; Cusick et al. 1989; Gasser et al. 1996). The ordered deposition of histone octamers behind the replication fork is critical for viability, genome stability, and maintenance of the epigenetic state (Exner et al. 2006; Jasencakova et al. 2010; Cheloufi et al. 2015; Ishiuchi et al. 2015).
The organization of mature chromatin is dictated by many factors, including primary DNA sequence (Segal et al. 2006; Mavrich et al. 2008), the presence of pioneer factors (Bai et al. 2010; Li et al. 2015; Yan et al. 2018), and active transcription (Weiner et al. 2010). Despite the rapid deposition of the histone octamer behind the fork, nascent chromatin is differentially sensitive to nuclease digestion compared with mature chromatin (>20 min post replication) (DePamphilis and Wassarman 1980; Klempnauer et al. 1980; Annunziato and Seale 1982; Stillman 1986), suggesting that nucleosomes are nonuniformly spaced in nascent chromatin.
The study of nascent chromatin has been facilitated by the use of nucleoside analogs that allow for the affinity capture and purification of newly synthesized DNA. The enrichment of labeled nascent chromatin has been used in proteomic studies to identify proteins and protein networks associated with normal, stalled, and collapsed replication forks (Sirbu et al. 2013; Alabert et al. 2014). Although these proteomic studies provided a wealth of data on the proteins that ensure the stability and progression of the DNA replication fork and the temporal order in which chromatin modifications occur, they fail to reveal information about locus-specific differences in chromatin maturation.
Recent work by multiple groups have combined the power of 5-ethynyl-2-deoxyuridine (EdU) labeling of nascent DNA with MNase nucleosome mapping to ascertain the positioning of nucleosomes genome-wide in nascent and mature chromatin (Fennessy and Owen-Hughes 2016; Vasseur et al. 2016). These studies focused on the chromatin maturation dynamics of nucleosomes within gene bodies and the rapid establishment of nucleosomal organization and phasing, highlighting the role of transcription and histone chaperones in shaping the chromatin landscape. Studies in Drosophila found that the re-establishment of chromatin architecture at gene regulatory elements (e.g., promoters and enhancers) was dependent on the reassociation of transcription factors (Ramachandran and Henikoff 2016).
We have combined MNase epigenome mapping (Henikoff et al. 2011; Belsky et al. 2015) with EdU labeling of recently replicated DNA to generate nascent chromatin occupancy profiles (NCOPs), allowing us to holistically explore chromatin maturation dynamics in Saccharomyces cerevisiae. Compared with prior work in S. cerevisiae (Fennessy and Owen-Hughes 2016; Vasseur et al. 2016), our NCOP methodology allows us to interrogate the DNA occupancy of both histone octamers and site-specific DNA-binding factors, thus providing a factor-agnostic view of chromatin maturation dynamics throughout the genome, including both genic and intergenic loci.
Results
Profiling nascent chromatin occupancy
We developed NCOPs to provide a factor-agnostic view of protein–DNA occupancy on newly synthesized DNA at nucleotide resolution. As a proof of principle to show the sensitivity and specificity of detecting EdU-labeled chromatin, we took advantage of the intra-S-phase checkpoint to specifically label recently synthesized DNA proximal to early origins of DNA replication. HU treatment results in replication fork stalling and activation of the intra-S-phase checkpoint (Santocanale and Diffley 1998; Shirahige et al. 1998). Only those sequences proximal to early activating efficient origins will incorporate EdU into the nascent daughter strands. Following EdU labeling, chromatin was isolated and digested by MNase. Then, the EdU-labeled DNA was biotin labeled by click chemistry before streptavidin affinity capture and paired-end sequencing (Henikoff et al. 2011; Belsky et al. 2015)
EdU incorporation was specific and restricted to sequences proximal to early activating origins of DNA replication. As expected from prior genomic experiments (Lengronne et al. 2001, Belsky et al. 2015), we detected strong peaks of EdU incorporation centered on early activating origins of DNA replication along Chromosome IV (Fig. 1A, middle). Sequences surrounding early origins of replication were enriched approximately 20-fold relative to late origins (Supplemental Fig. S1A).
Figure 1.
Enrichment of EdU-labeled chromatin at sequences proximal to early origins. (A) Sequencing coverage across Chromosome IV. NCOP at an early replicating origin shows a defined chromatin architecture (bottom) compared with a nonreplicated region (top). (Middle) Enrichment of EdU-labeled sequences along Chromosome IV. Shaded green background highlights early origins of replication (Belsky et al. 2015). (Bottom) The green box represents an origin of replication. In the case of the region depleted of nucleosomes at ∼437 kb, there is a transposable element (YDLWtau1 [Ty4 LTR]) that overlaps this position. (B–D) Chromatin profiles at genes proximal to an early origin that showed unaltered (B) and altered (C,D) chromatin structure. Nucleosome position and occupancy are depicted with red shaded ovals. Gray boxes represent genes on the positive (light gray) and negative (dark gray) strands. (D) RAD51 shows recruitment of a transcription factor and downstream nucleosome shift in the EdU pull-down experiment compared with total chromatin.
The innovative aspect of NCOPs is the limited MNase digestion of EdU-enriched chromatin followed by the recovery and sequencing of fragments ∼200 bp and smaller (Henikoff et al. 2011; Belsky et al. 2015). To visualize the NCOPs, we plot the length of the paired-end reads as a function of their chromosomal position; thus, nucleosomes are evident as well-phased clusters of fragments with lengths of ∼150 bp, and smaller DNA-binding factors are evident as discrete clusters of fragments with lengths <80 bp. We found that both nucleosomes and DNA-binding factors were readily distinguishable in the NCOPs from EdU-labeled chromatin (Fig. 1A, bottom). In contrast, no discernable EdU-enrichment of chromatin was detected in the origin distal regions (Fig. 1A, top).
We analyzed the aggregate nucleosome distribution surrounding 539 gene promoters that were within 3500 bp of early origins, and found that nucleosome phasing and occupancy were very similar between the NCOP and “bulk” chromatin (no EdU labeling/enrichment) (Supplemental Fig. S1B). We found that when examined at the level of individual genes, the recovered EdU-labeled NCOPs resemble those prepared from untreated bulk chromatin (Fig. 1B). We did detect a handful of locus-specific alterations in chromatin structure, but these differences were likely attributable to differential transcription of HU-responsive genes. For example, at the RAD23 locus, we observed displaced nucleosomes from the gene body (Fig. 1C). At the RAD51 locus, we detected an expansion of the nucleosome-free region at the promoter and the association of an additional DNA-binding factor upstream of the gene in the presence of HU (Fig. 1D). The recruitment of this factor, which we speculate to be the MBF complex (Leem et al. 1998; Mathiasen and Lisby 2014), was accompanied by a downstream shift in the phasing of the genic nucleosomes. To confirm that these alterations in chromatin structure were owing to the HU arrest and not a consequence of the EdU labeling and enrichment, we also performed an experiment in which cells were HU-arrested in the presence of EdU and then released from the arrest for 2 h. We found that HU-dependent chromatin changes detected by the NCOPs were restored following release from HU and re-entry into the cell cycle (Supplemental Fig. S1B, right panel, and S1C–S1E). Together, these results show the specificity and sensitivity of NCOPs in detecting EdU-labeled chromatin structure and dynamics.
Locus-specific differences in the re-establishment of chromatin architecture
We sought to survey the dynamics of chromatin maturation throughout the genome by pulse-chase labeling, replicating cells with EdU. To enrich for cells in S phase, we first arrested the cell population in G1 using α-factor followed by a synchronous release in S phase (Fig. 2A). When the majority of cells were in S phase with mid-S DNA content (Supplemental Fig. S2A), the cells were pulsed with EdU for 10 min followed by a thymidine chase for 30 min (Fig. 2A). This approach allowed us to capture replication forks emanating from both early and late firing origins. Consistent with this, we observed a relatively uniform distribution of EdU incorporation across the genome (Supplemental Fig. S2B). A potential concern for these pulse-chase experiments is the inappropriate incorporation of EdU during the chase period, which would confound our results. To address this, we looked for the presence of EdU labeling following the release from HU-arrested and EdU-labeled chromatin (Supplemental Fig. S1B–F). If EdU was inappropriately incorporated following the wash and chase, we would see a broadening of the EdU-labeled early origin peaks in the following cell cycle. We found that the patterns of EdU labeling were almost indistinguishable following the wash and additional cell cycle chase period (Supplemental Fig. S1F).
Figure 2.
Nascent chromatin occupancy profiles (NCOPs). (A) Schematic of experimental design for capturing EdU-labeled nascent and mature chromatin. (B) NCOPs reveal the maturation of chromatin behind the DNA replication fork at a representative locus, CSI2. Gray boxes represent genes on the positive (light gray) and negative (dark gray) strands. A representation of nucleosome positioning and occupancy (intensity) is highlighted with red ovals above the chromatin plots. (C) Nascent chromatin organization in gene bodies is less organized than mature chromatin. Distribution of correlation scores between nascent and bulk (teal) or mature and bulk chromatin (purple). (D) Differential chromatin maturation kinetics for genes with well-organized mature chromatin. The autocorrelation function (ACF) was used to identify the top 50% of genes with regularly phased arrays of nucleosomes from mature chromatin. These 2700 genes were then binned into quintiles based on the correlation between nascent and mature chromatin. The first and fifth quintiles represent genes with slow and fast chromatin maturation kinetics. The distribution of ACF values (as a proxy for gene organization) in nascent and mature chromatin is depicted for the genes with slow (light green) and fast (dark green) chromatin maturation kinetics. The difference in ACF values among fast and slow maturing chromatin was significant (P < 2.2 × 10−16). (E) Heatmap representing mean Z-score values for 25 histone post-translational modifications, the histone variant H2AZ (encoded by the HTZ1 gene), and NET-seq scores for the individual quintiles of the correlation between nascent and mature chromatin for the 2700 genes described above.
NCOPs at individual loci revealed that nascent chromatin was inherently more disorganized than mature chromatin. For example, at the CSI2 locus we observed a marked difference in chromatin organization both upstream and in the gene body between the nascent and mature chromatin samples (Fig. 2B). To systematically characterize the differences in nucleosome organization between nascent and mature chromatin, we first calculated a nucleosome occupancy profile for each gene in the nascent and mature chromatin state. The nascent and mature nucleosome occupancy profiles were then compared with profiles calculated for bulk chromatin isolated from an asynchronous sample that was not EdU enriched. We found that the structure of mature chromatin at genic regions was highly correlated with bulk chromatin (median R-value 0.901); in contrast, nascent chromatin was significantly less correlated (median R-value 0.799) and significantly different from the mature chromatin population (P < 2.2 × 10−16) (Fig. 2C). Similar patterns were observed with varying the EdU pulse length (5 or 10 min) (Supplemental Fig. S3). The differences in chromatin organization were not uniform across the genome but instead were specified on a gene by gene basis. In contrast to CSI2, the chromatin at the neighboring gene COQ10 was well organized in both the nascent and mature chromatin samples (Fig. 2B).
To explore the features associated with the maturation dynamics of individual genes, we focused on those genes that showed an organized chromatin structure in the mature chromatin state. We used the autocorrelation function (ACF) to determine the regularity of nucleosome phasing and organization within gene bodies and identified 2700 genes above the median (0.460) of the ACF values. The genes with well-organized mature chromatin were stratified into quintiles based on the correlation between nascent and mature chromatin. Because we only focused on those genes with well-organized mature chromatin, their organization in nascent chromatin likely reflects their maturation dynamics, with the low (first quintile) and high (fifth quintile) extremes of nascent organization representing slow and fast chromatin maturation dynamics, respectively.
The positioning of the nucleosomes was identical between nascent and mature chromatin for those genes in the fifth or fast chromatin assembly quintile. In contrast, the nascent chromatin structure for genes in the first or slow quintile was less organized and readily distinguishable from mature chromatin (Supplemental Fig. S2C). We then analyzed the distribution of ACF values to examine the regularity of nucleosome phasing and organization within the genes that showed fast or slow chromatin maturation dynamics. As expected, the mature state from both classes had a high ACF, indicating well-phased and organized nucleosomes. In contrast, the class of genes with slow chromatin maturation kinetics showed significantly poorer nucleosome organization and structure (low ACF) in the nascent compared with the mature state (P < 2.2 × 10−16) (Fig. 2D). Together, these data reveal locus-specific differences in chromatin maturation for a large subset of gene bodies.
To explore the relationship between chromatin maturation dynamics and the epigenetic landscape, we analyzed data describing the genome-wide distribution and enrichment of post-translational histone tail modifications and the histone variant H2AZ (Weiner et al. 2015). We calculated Z-scores of enrichment for each of the chromatin modifications and generated a heatmap of average Z-score values for each modification across the quintiles of nascent chromatin organization as described above (Fig. 2E). We found that genes with rapid chromatin maturation kinetics (fifth quintile) were enriched for H2AZ and acetylation marks, including those on lysine residues 5, 8, and 12 of histone H4. H3K4me3 was also enriched in this group of fast maturing genes, and this mark, along with H4K12ac, has been shown to colocalize with H2AZ (Chen et al. 2012). There was also a significant decrease in the chromatin marks associated with actively transcribed genes, including H3K36me3 and H3K79me3. Consistent with the relative depletion of marks associated with active transcription, we found that these genes with rapid chromatin maturation showed less active transcription (Churchman and Weissman 2011).
Nucleosomes show distinct patterns of positioning and occupancy genome-wide
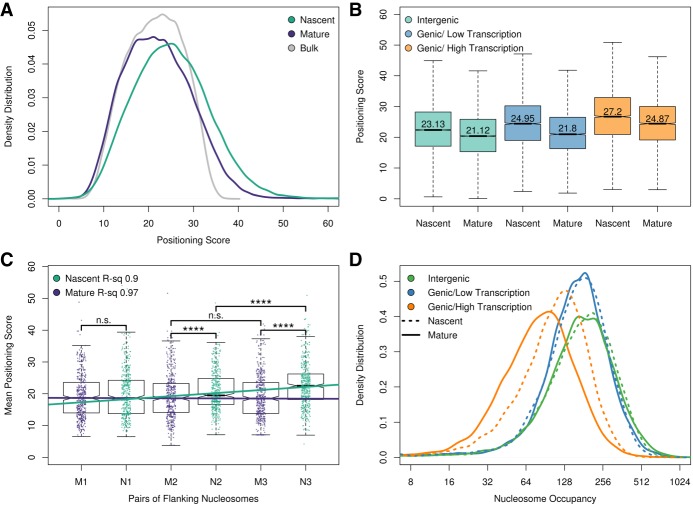
To study chromatin architecture at a genome-wide level and at intergenic regions, we focused on the positioning of individual nucleosomes, which eliminates the potential for confounding results owing to small DNA-binding proteins altering the nucleosome periodicity. We first identified the position of the nucleosome dyad for about 70,000 high-confidence nucleosomes from bulk chromatin (Supplemental Code). For every nascent or mature nucleosome-sized fragment, we calculated the distance of the fragment midpoint to the nearest nucleosome dyad in bulk chromatin. The average of these distances for each nucleosome represented a positioning score, with well-positioned nucleosomes having a low score and poorly positioned nucleosomes having a high score (Supplemental Code). We found that individual nucleosomes in nascent chromatin are more poorly positioned compared with mature or bulk chromatin (Fig. 3A).
Figure 3.
Genome-wide nucleosome positioning and occupancy in nascent and mature chromatin. (A) Genome-wide distribution of nascent, mature, and bulk nucleosome positioning. Approximately 70,000 high-confidence nucleosome dyads were identified in bulk chromatin. For each chromatin fraction (nascent, mature, and bulk), the distance from the midpoint of each sequencing read to the nearest nucleosome dyad was calculated as a nucleosome positioning score. (B) Nascent and mature chromatin organization (nucleosome positioning scores) at intergenic and intragenic nucleosomes. Intragenic nucleosomes were subdivided into high (top 10%) and low (bottom 10%) transcriptional activity. (C) Transcription factors influence chromatin maturation kinetics. Positioning scores of the first, second, and third pairs of nucleosomes flanking 436 transcription factors with strong occupancy in mature chromatin. t-test: (****) P ≤ 0.0001; (n.s.) P > 0.05. (D) Active transcription displaces nucleosomes from mature chromatin. Distribution of nucleosome occupancy scores (sequencing reads assigned to individual nucleosomes) for nascent and mature chromatin at intragenic and intergenic sequences. Actively transcribed genes have more nucleosome occupancy in nascent chromatin than in mature chromatin (P < 2.2 × 10−16).
Individual nucleosomes were broadly classified as either intergenic or genic. We found that nucleosomes within intergenic regions have decreased positioning scores (greater chromatin structure) than do nucleosomes within gene bodies. To understand the differences in chromatin maturation for intergenic and genic nucleosomes and their relationship to transcription, we identified the nucleosomes associated with the most (top 10%) and least (bottom 10%) expressed genes (Fig. 3B; Churchman and Weissman 2011). A similar number of nucleosomes were sampled from intergenic regions. In each category, we found that the positioning of the nucleosomes was decreased in nascent relative to mature chromatin. Nucleosomes in intergenic and poorly transcribed genes were better positioned than nucleosomes from active genes, consistent with transcription-dependent nucleosome eviction and remodeling (Lee et al. 2004; Boeger et al. 2004; Bernstein et al. 2004).
We reasoned that the greater nucleosome organization observed in intergenic regions was owing, in part, to the presence of transcription factors. To assess the chromatin maturation dynamics at intergenic regions, we examined nucleosome positioning for the first, second, and third pairs of nucleosomes surrounding 436 predicted transcription factor binding sites (MacIsaac et al. 2006) with defined occupancy footprints in mature chromatin (Fig. 3C). We found no significant difference in nucleosome positioning for the mature nucleosomes. In contrast, we identified a significant distance-dependent increase in nucleosome positioning scores for each successive nucleosome pair in the nascent chromatin. Together, these results underscore the role of transcription factors functioning as barrier elements in establishing nucleosome organization (Zhang et al. 2009) following DNA replication.
We also examined nucleosome occupancy for intergenic and genic regions of the genome. An occupancy score was calculated for individual nascent and mature nucleosomes as the number of fragment midpoints mapping within 70 bp of the high-confidence nucleosome dyads identified from bulk chromatin (see above). We found that nucleosomes in intergenic regions and genes that were not being actively transcribed showed similar nucleosome occupancies in both nascent and mature chromatin states (Fig. 3D). Thus, although nucleosomes are rapidly deposited behind the replication fork, they do not converge on their preferred position until maturation. We observed, in contrast to intergenic and nontranscribed regions, significantly more nucleosome occupancy in nascent relative to mature chromatin in actively transcribed genes, showing that the newly deposited nucleosomes behind the DNA replication fork are evicted by active transcription.
Actively and passively replicated origins have distinct maturation dynamics
Replication origins have their own inherent efficiency of activation during S phase (Aparicio 2013; Hawkins et al. 2013). A consequence of this is that highly efficient origins will initiate bidirectional DNA replication every cell cycle, whereas the least efficient origins will be passively replicated by forks from neighboring efficient origins. We reasoned that there might be distinct differences in chromatin maturation at active origins that initiate DNA replication versus those origins that are passively replicated. The distribution and strandedness of Okazaki fragments around origins were used as a proxy for origin efficiency (McGuffee et al. 2013). We first examined locus-specific NCOPs for ARS805 and ARS405, an efficient early origin and a passively replicating origin, respectively (Fig. 4A,B). As previously reported, we observed well-ordered and phased nucleosomes flanking either origin in mature chromatin (Belsky et al. 2015). We also detected smaller fragments (< 80 bp) in the mature chromatin that are indicative of origin recognition complex (ORC) binding in the nucleosome-free region at the autonomously replicating sequence (ARS) consensus sequence of both origins. In contrast, we observed a difference in nascent chromatin organization between ARS805 and ARS405. Specifically, we found that there was significantly more nascent chromatin organization at the active replication origin compared with the passively replicated origin.
Figure 4.
Chromatin maturation at actively and passively replicated origins. (A) Chromatin occupancy profile for nascent and mature chromatin at the active origin ARS805. (B) Chromatin occupancy profile for nascent and mature chromatin at the inactive origin ARS405. Green boxes in A,B represent origins of replication. Gray boxes represent genes on the positive (light gray) and negative (dark gray) strands. A representation of nucleosome positioning and occupancy (intensity) is highlighted with red ovals above the chromatin plots. (C) Passively replicated origins have slower chromatin maturation kinetics. Boxplots depicting the distribution of nucleosome positioning scores for nascent and mature chromatin at 100 active and 100 inactive origins. Nascent chromatin surrounding passively replicated origins is more disorganized than at active origins. t-test: (***) P ≤ 0.001; (n.s.) P > 0.05. (D) ORC occupancy is decreased at passively replicated origins. Density distribution of ORC occupancy footprints as determined from the NCOPs.
To comprehensively examine chromatin maturation dynamics at active and passive origins, we identified 269 origins with an ORC-dependent footprint and then stratified the origins by their efficiency score to identify the top 100 active and passive replication origins (Belsky et al. 2015). The mean efficiency for each class of origins was 0.65 (active) versus 0.017 (passive). For each actively or passively replicated origin, we examined the nucleosome positioning scores for the first three nucleosomes flanking each origin up- and downstream (Fig. 4C), where increasing chromatin organization is represented by lower nucleosome positioning scores. We found that the nascent chromatin surrounding passive origins was significantly more disorganized than at active origins (P ≤ 0.001). In contrast, there were no detectable differences in chromatin organization between active and passive origins in the chromatin that had matured behind the replication fork. Our NCOPs also revealed a decrease in small fragments (<80 bp) at the passively replicated origins, consistent with decreased ORC occupancy (Fig. 4D). We also examined the enrichment of histone variants and histone post-translational modifications associated with fast and slow chromatin maturation kinetics at each of the origins. We found that most of the examined histone marks were moderately depleted in the 1000 bp (500 up and 500 down) surrounding each origin of replication, and no meaningful epigenetic signature emerged (Supplemental Fig. S4). Together, these results suggest a feedback mechanism in place at active origins to promote ORC recruitment and immediately re-establish chromatin architecture for the next cell cycle.
Transient association of DNA-binding factors with nascent chromatin
DNA-binding proteins, such as transcription factors and ORC, associate with specific primary sequences; however, for any given factor, there are many more potential sequence motifs than occupied sites in the genome (Breier et al. 2004; Eaton et al. 2010; Wang et al. 2012; Slattery et al. 2014). Nucleosome occupancy is thought to limit access to many of these potential motif matches, thus defining the regulatory landscape. NCOPs provided an opportunity to comprehensively survey DNA occupancy in a factor-agnostic manner throughout the genome in both nascent and mature chromatin.
To identify all potential sites of DNA occupancy that were not protected by a nucleosome, we focused on the paired-end sequencing fragments that were < 80 bp and merged both the nascent and mature data sets to identify all peaks generated by the small size fragments. We identified 6272 loci that were significantly enriched (P < 0.05) for smaller fragments in either nascent or mature chromatin. To evaluate changes in DNA binding factor occupancy, we first determined the log2 ratio of normalized occupancy scores for nascent and mature chromatin at each of the 6272 sites and plotted these values as an ordered heatmap (Fig. 5A). The heatmap revealed three classes of DNA-binding profiles indicative of their chromatin maturation dynamics: (1) slow maturation, sites with greater occupancy in mature chromatin; (2) fast maturation, sites that were equally occupied in both mature and nascent chromatin; and (3) transient occupancy, sites that were enriched in nascent and not mature chromatin.
Figure 5.
Transcription factor association with nascent and mature chromatin. (A) Heatmap of small DNA-binding factor occupancy in mature and nascent chromatin (log2 [mature/nascent]) for 6272 high-confidence sites obtained from DNA fragment lengths between 20 bp and 80 bp. (B) Enrichment of small DNA fragments (DNA-binding factors) for the three classes of chromatin maturation: fast, equal occupancy in nascent and mature chromatin; slow, greater occupancy in mature than nascent chromatin; and transient, greater in nascent than mature. (C) Average plot of nucleosome organization at sites occupied by the factors described in B. (D) Sites of transient occupancy are promiscuously enriched at sites distal from the transcription start site (TSS) (P < 2.2 × 10−16 between fast and transiently associating factors).
To characterize the chromatin maturation of regulatory sites bound by transcription factors and other DNA-binding factors, we identified the extreme deciles of sites (approximately 625 sites for each decile) representing locations with slow or transient occupancy and an equal number of loci with fast maturing kinetics (Fig. 5B). As expected, the occupancy of small DNA-binding factors in the sites with slow maturation kinetics was less in nascent than mature chromatin (cf. dashed and solid red lines). Despite the different kinetics of maturation, similar occupancy levels were ultimately established in the mature chromatin (cf. solid red and gray lines).
We analyzed the nucleosome organization surrounding slow, fast, and transiently maturing sites of DNA-binding factor occupancy (Fig. 5C). We found that both the slow and fast maturing sites of DNA occupancy were surrounded by well-positioned nucleosomes in both nascent and mature chromatin. This suggests that for at least some sites, defined nucleosome positioning may be a requisite for factor occupancy. In contrast, we found that specific pioneer factors (e.g., Abf1p and Reb1p) showed occupancy footprints and chromatin organization profiles that were indistinguishable between nascent and mature chromatin, consistent with their immediate deposition behind the DNA replication fork (Supplemental Fig. S5).
The sites with transient occupancy only observed in the nascent chromatin may represent promiscuous binding to accessible motifs in regions of poor chromatin organization. The compact nature of the yeast genome dictates that most regulatory DNA-binding sites are proximal to gene promoters. Thus, we examined the location of each binding site for all three occupancy classes—fast, slow, and transient—relative to their gene start site. The DNA-binding sites with fast and slow maturation were found proximal to promoters (Fig. 5D). In contrast, the distribution of transient sites was not limited to promoters but rather occurred distally to promoters and likely in gene bodies. Together, these results suggest that potential binding motifs are exposed in poorly organized nascent chromatin and that they are ultimately removed by chromatin maturation and transcription.
Discussion
One of the consequences of semi-conservative DNA replication is that the chromatin landscape needs to be disrupted ahead of the replication fork and then re-established in its wake. Although genetic and biochemical experiments have elucidated many of the factors and mechanisms regulating the inheritance and assembly of chromatin behind the fork (Smith and Stillman 1989; Shibahara and Stillman 1999; Tyler et al. 1999; Li et al. 2008; Sirbu et al. 2013; Alabert et al. 2014), the spatiotemporal dynamics of DNA replication–dependent chromatin assembly across the genome are just starting to be revealed (Fennessy and Owen-Hughes 2016; Ramachandran and Henikoff 2016; Vasseur et al. 2016). We have used NCOPs to provide a factor-agnostic view of chromatin maturation at nucleotide resolution across the S. cerevisiae genome. The 30-fold increase in sequence coverage, afforded by the smaller S. cerevisiae genome, coupled with our unique visualization of the data provides a truly locus-specific view of chromatin occupancy that was not possible with the depth of sequencing performed for MINCE-Seq in Drosophila (Supplemental Fig. S6).
The establishment of chromatin organization is thought to be dependent on primary sequence (Segal et al. 2006; Kaplan et al. 2009), transcription (Weiner et al. 2010), and the positioning of nucleosomes relative to fixed barrier elements (Bai et al. 2010; Li et al. 2015; Yan et al. 2018). Our NCOPs revealed locus-specific differences in chromatin maturation kinetics at genes and origins of replication, suggesting that the maturation process is also influenced by the local chromatin state and origin activity (Fig. 6). We first focused on the chromatin maturation dynamics of genes with well-organized nucleosome architecture in mature chromatin. In contrast to prior reports describing transcription-dependent chromatin maturation dynamics in yeast (Vasseur et al. 2016), we found that maturation of nascent chromatin was fastest for genes with low transcriptional activity (Fig. 2E). The Vasseur study focused on the temporal maturation of nascent chromatin using the correlation with mature chromatin as a proxy for maturation within the gene body. As active transcription frequently results in nucleosome eviction and disorganized phasing in bulk chromatin (Lee et al. 2004; Boeger et al. 2004; Bernstein et al. 2004), they were looking at a de facto transcription-dependent effect. We also note in our studies that we detected the transcription-dependent eviction of nascent histone octamers from actively transcribed gene bodies (Fig. 3D), consistent with the role of transcription in shaping the mature chromatin landscape.
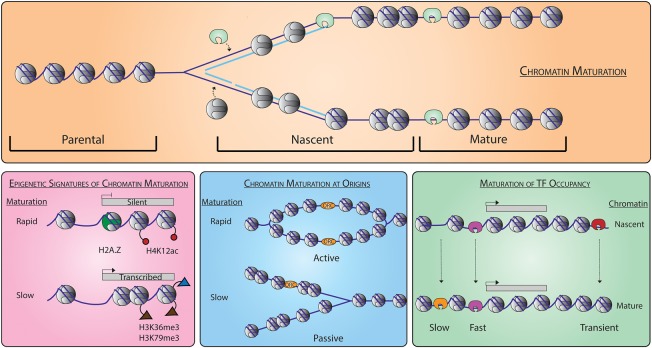
Figure 6.
Genome-wide chromatin maturation behind the DNA replication fork. Parental chromatin is re-established behind the replication fork (top). Locus-specific differences in chromatin maturation possess different epigenetic signatures (bottom left). Chromatin maturation at start sites of DNA replication is different for active and passive origins (bottom middle). Transcription factors (TFs) associate with different kinetics behind the DNA replication fork including sites of transient occupancy (bottom right).
Histone post-translational modifications and histone variants have long been postulated to form the basis of epigenetic memory (Probst et al. 2009; MacAlpine and Almouzni 2013). The local inheritance of parental H3-H4 histones at the replication fork, in part, contributes to the re-establishment of the parental chromatin state on the newly replicated DNA. We found that genes with rapid maturation kinetics were poorly expressed and depleted of modifications frequently associated with gene expression (e.g., H3K36me3 and H3K79me3) and instead were enriched for specific histone modifications, including H4K12ac, H3K4me3, and the histone variant H2AZ (Fig. 6, bottom left). In S. cerevisiae, H2AZ is located at the promoters of inactive or poorly transcribed genes and helps to stabilize their promoter architecture for recruitment of regulatory factors (Guillemette et al. 2005; Li et al. 2005). Deposition of parental H3-H4 tetramers containing H4K12ac and or H3K4me3 behind the fork may help facilitate the recruitment of H2AZ to inactive promoters, as single-molecule studies revealed that H2AZ variant–containing nucleosomes are enriched for H3K4me3 and H4K12ac (Chen et al. 2012). The recruitment of H2AZ may rapidly stabilize nascent nucleosome organization by inhibiting ATP-dependent chromatin remodeling activity (Li et al. 2005). It is important to note that we limited our analysis to only those genes that showed organized and well-positioned nucleosomes in mature chromatin. Thus, we argue that the observed epigenetic signature is associated with fast chromatin maturation kinetics behind the fork and is not just a property of genes with well-positioned nucleosomes. Future studies are needed to determine if this epigenetic signature represents an inherited state of parental chromatin or an inherent property of nascent chromatin maturation.
A long-standing model for how nucleosome positioning is achieved proposes that barrier elements such as transcription factors aid in establishing the fixed positions of their flanking nucleosomes (Fedor et al. 1988; Pazin et al. 1997; Mavrich et al. 2008; Li et al. 2015). As a result, this poses further positioning constraints on the subsequent +2 and +3 nucleosomes, thereby stabilizing their positioning. Pioneer factors like Abf1p and Reb1p are capable of positioning flanking nucleosomes without the need for chromatin remodeling (Yarragudi et al. 2004; Hartley and Madhani 2009; Ganapathi et al. 2011) and can position nascent nucleosomes following passage of the replication fork (Yadav and Whitehouse 2016). In agreement with this, we find that the first pair of nucleosomes immediately flanking factors bound to mature chromatin is tightly positioned in both nascent and mature chromatin (Fig. 3C).
Our work illustrates locus-specific differences in chromatin maturation following passage of the DNA replication fork. However, our current study does not discriminate between the leading and lagging strand of the replication fork. Recent work from the Groth and Zhang laboratories has begun to elucidate differential patterns of histone deposition on the leading and lagging strand and the mechanisms responsible for ensuring symmetric epigenetic inheritance on both daughter strands of DNA (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018). It will be interesting to determine if there are differential patterns of chromatin maturation for the leading and lagging strands, especially under conditions that perturb the normal symmetry of epigenetic inheritance.
We also investigated the chromatin maturation dynamics surrounding origins of DNA replication. The timing and efficiency by which individual origins of DNA replication are activated during S phase is determined, in part, by primary sequence, rate-limiting initiation factors (Mantiero et al. 2011; Tanaka et al. 2011), and the local chromatin structure (Berbenetz et al. 2010; Eaton et al. 2010; Kurat et al. 2017). We found that the maturation dynamics of the chromatin surrounding origins of DNA replication were dependent on whether the origin actively initiated DNA replication or was passively replicated by a replication fork from elsewhere in the genome (Fig. 6, bottom center). Efficient origins that initiate replication in the majority of cell cycles (McGuffee et al. 2013) rapidly reset their chromatin structure and show well-positioned nucleosomes flanking the origin immediately after initiation in nascent chromatin. The differential maturation of chromatin between efficient and passive origins likely reflects distinct molecular mechanisms in the reassembly of chromatin following either an initiation event or the disassembly of the prereplication complex at inefficient origins. The Fox laboratory has classified origins based on their ORC affinity as DNA dependent, chromatin dependent, or weak (Hoggard et al. 2013). As reported, we found that the weak class of origins showed poorly organized nucleosomes in both nascent and mature chromatin; however, the different classes did not discriminate between the fast and slow maturing origins (Supplemental Fig. S7). It is tempting to speculate that immediately following unwinding of the origin DNA and initiation of DNA replication, an active mechanism promotes the rapid reassociation of ORC and precise phasing of nucleosomes, which have become a hallmark for eukaryotic origins (Berbenetz et al. 2010; Eaton et al. 2010; Lubelsky et al. 2011; Cayrou et al. 2015; Miotto et al. 2016).
Chromatin maturation is governed by both replication-dependent and -independent processes. Whereas the deposition of nascent histone octamers behind the fork is replication dependent, the re-establishment of the regulatory landscape and binding of transcription factors is dependent on chromatin remodeling, local epigenetic signatures, and recruitment of other trans-acting factors (Allis and Jenuwein 2016). Competition between nucleosome occupancy and transcription factor binding has long been thought to be a determinant of which regulatory motifs are occupied (Wasson and Hartemink 2009; Li et al. 2015; Ramachandran and Henikoff 2016). The factor-agnostic NCOPs provided insight into how trans-acting factors and the regulatory landscape were established during chromatin maturation. We found a range of maturation kinetics for loci occupied by nonnucleosomal DNA-binding factors, including those that rapidly associate with nascent chromatin (fast) and those that associate later in mature chromatin (slow) (Fig. 6, bottom right). However, we also identified a significant number of sites that were only transiently occupied in nascent chromatin. Unlike the binding sites with slow or fast kinetics, which were promoter proximal, the transient sites were frequently located distal to the promoter and in gene bodies. This is consistent with a promiscuous or opportunistic mode of binding in the absence of organized chromatin immediately following DNA replication (Yan et al. 2018). During development in higher eukaryotes, the DNA replication program is characterized by changes in the number of firing origins, the length of S phase, and the timing of replication (Duronio 2012; Rhind and Gilbert 2013). The developmental plasticity in the DNA replication program may lead to promiscuous binding of regulatory factors through the chromatin changes that occur throughout this process and thus contribute to epigenetic regulation and cell-type–specific gene expression programs.
Methods
Yeast strains
The yeast strain DMM218 is in the W303 background and has the genotype MATa, leu2-3,112, BAR1::TRP, can1-100, URA3::BrdU-Inc, ade2-1, his3-11,15.
Chromatin occupancy profiling
To label cells arrested in early S phase, yeast was grown in rich medium at 30°C to an OD of ∼0.7 and arrested in G1 phase with α-factor (GenWay) at a final concentration of 50 ng/mL for 2 h. Cells were then washed twice in sterile water, resuspended in fresh medium containing 0.2 M hydroxyurea (Sigma-Aldrich) and 130 µM EdU (Berry & Associates), and grown for 2 h. Cells were washed twice with sterile water, and the pellets were quick-frozen and stored at −80°C.
To profile nascent and mature chromatin, yeast cells were grown at 25°C to an OD of ∼0.7 and arrested in G1 phase with α-factor at a final concentration of 50 ng/mL for 2 h. Cells were then washed twice in sterile water, resuspended in fresh medium, and allowed to grow for 45 min to enter S phase. EdU was then added to a final concentration of 130 µM and allowed to grow for 10 min (pulse), after which a sample was taken for nascent chromatin. Cells were washed, resuspended in fresh medium containing 1.3 mM thymidine (Sigma-Aldrich), and allowed to grow for 30 min (chase), when a sample was taken for mature chromatin. Cells were washed twice with sterile water, and the pellets were quick-frozen and stored at −80°C. All experiments were performed using independent biological replicates.
Chromatin preparation
MNase digestions were performed as previously described (Belsky et al. 2015).
Click reaction and streptavidin affinity capture
One hundred micrograms of MNase-digested DNA was concentrated in a speed vacuum. DNA was incubated in click chemistry reaction buffer as previously described (Kliszczak et al. 2011; Sirbu et al. 2012; Leung et al. 2013), with the exception that CuSO4 and ascorbic acid were replaced with CuBr and TBTA (Sigma-Aldrich). The click reaction proceeded for 1 h at room temperature with gentle shaking. DNA was recovered by ethanol precipitation and resuspended in 30 µL sterile water.
Biotin conjugated EdU-labeled DNA was enriched using 5 µL streptavidin magnetic beads (New England Biolabs). Beads were resuspended in blocking solution (2% I-Block [Thermo Fisher Scientific], 25 mM Tris, 150 mM NaCl, and 0.5% SDS) for 2 h with gentle shaking at room temperature. Beads were then washed twice with cold binding buffer (Leung et al. 2013). Recovered DNA was added to blocked beads and incubated in 200 µL cold binding buffer for 1 h at 4°C. Bead-bound DNA was washed once with binding buffer followed by three washes with EB buffer (Qiagen).
Flow cytometry
To analyze yeast cells by flow cytometry, cells were resuspended in 70% ethanol and fixed overnight at 4°C. Then, cells were washed, sonicated, and incubated in 50 mM sodium citrate (pH 7.4) with 0.3 mg/mL RNase A for 2 h at 50°C. Then, 0.6 mg/mL Proteinase K was added and incubated for an additional 2 h at 50°C. Finally, cell pellets were resuspended in 50 mM sodium citrate and 1:5000 SYTOX green (Invitrogen) and incubated for 1 h at room temperature. Flow cytometry was performed on a BD FACSCanto analyzer, and 30,000 cells were recorded for each sample.
Sequencing library preparation
Illumina sequencing libraries were prepared as previously described (Henikoff et al. 2011; Belsky et al. 2015), with the following modifications: All library preparations were performed on bead-bound DNA. After each step, clean up was accomplished by washing beads twice with binding buffer and three times with EB buffer. NEBNext multiplex oligos for Illumina kit (New England Biolabs) was used in adapter ligation and PCR steps. PCR reactions were cleaned using Agencourt AMPure XP beads (Beckman).
Data analysis
Data analysis was performed in R version 3.2.0 (R Core Team 2015). A detailed explanation of bioinformatic methods can be found in the Supplemental Material. Data processing scripts are available at https://gitlab.oit.duke.edu/dmm29/Gutierrez_2018 and as Supplemental Code.
Data access
The sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP158706.
Supplementary Material
Acknowledgments
We thank members of the MacAlpine laboratory for critical comments and suggestions. We also thank Jason Belsky for advice on data analysis. This work was supported by the National Institutes of Health (National Institute of General Medical Sciences) R35 GM127062 (D.M.M.) and F31-GM115158 (M.P.G.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.243386.118.
References
- Alabert C, Bukowski-Wills J-C, Lee S-B, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. 10.1038/ncb2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Annunziato AT, Seale RL. 1982. Maturation of nucleosomal and nonnucleosomal components of nascent chromatin: differential requirements for concurrent protein synthesis. Biochemistry 21: 5431–5438. 10.1021/bi00265a008 [DOI] [PubMed] [Google Scholar]
- Aparicio OM. 2013. Location, location, location: It's all in the timing for replication origins. Genes Dev 27: 117–128. 10.1101/gad.209999.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Charvin G, Siggia ED, Cross FR. 2010. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell 18: 544–555. 10.1016/j.devcel.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky JA, MacAlpine HK, Lubelsky Y, Hartemink AJ, MacAlpine DM. 2015. Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev 29: 212–224. 10.1101/gad.247924.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW. 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6: e1001092 10.1371/journal.pgen.1001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. 2004. Global nucleosome occupancy in yeast. Genome Biol 5: R62 10.1186/gb-2004-5-9-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14: 667–673. 10.1016/j.molcel.2004.05.013 [DOI] [PubMed] [Google Scholar]
- Breier AM, Chatterji S, Cozzarelli NR. 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22 10.1186/gb-2004-5-4-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau J-C, van Helden J, Méchali M. 2015. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25: 1873–1885. 10.1101/gr.192799.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36: 469–480. 10.1021/bi962069i [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. 2015. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528: 218–224. 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Miller A, Kirchmaier AL, Irudayaraj JMK. 2012. Single-molecule tools elucidate H2A.Z nucleosome composition. J Cell Sci 125: 2954–2964. 10.1242/jcs.101592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. 10.1038/nature09652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick ME, Wassarman PM, DePamphilis ML. 1989. Application of nucleases to visualizing chromatin organization at replication forks. In Methods in enzymology (ed. Wassarman PM, Kornberg RD), Vol. 170, pp. 290–316, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Dabin J, Fortuny A, Polo SE. 2016. Epigenome maintenance in response to DNA damage. Mol Cell 62: 712–727. 10.1016/j.molcel.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Wassarman PM. 1980. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem 49: 627–666. 10.1146/annurev.bi.49.070180.003211 [DOI] [PubMed] [Google Scholar]
- Duronio RJ. 2012. Developing S-phase control. Genes Dev 26: 746–750. 10.1101/gad.191171.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753. 10.1101/gad.1913210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Taranto P, Schönrock N, Gruissem W, Hennig L. 2006. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172. 10.1242/dev.02599 [DOI] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. 1988. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol 204: 109–127. 10.1016/0022-2836(88)90603-1 [DOI] [PubMed] [Google Scholar]
- Fennessy RT, Owen-Hughes T. 2016. Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable inter-nucleosome spacing. Nucleic Acids Res 44: 7189–7203. 10.1093/nar/gkw331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, Yu C, Zhang Z. 2018. The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol Cell 72: 140–151.e3. 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. 2011. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res 39: 2032–2044. 10.1093/nar/gkq1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R, Koller T, Sogo JM. 1996. The stability of nucleosomes at the replication fork. J Mol Biol 258: 224–239. 10.1006/jmbi.1996.0245 [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384 10.1371/journal.pbio.0030384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez MP, MacAlpine DM. 2016. Chromatin determinants of origin selection and activation. In The initiation of DNA replication in eukaryotes (ed. Kaplan DL), pp. 87–104, Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. 10.1016/j.cell.2009.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Retkute R, Müller CA, Saner N, Tanaka TU, de Moura APS, Nieduszynski CA. 2013. High-resolution replication profiles define the stochastic nature of genome replication initiation and termination. Cell Rep 5: 1132–1141. 10.1016/j.celrep.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323. 10.1073/pnas.1110731108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard T, Shor E, Müller CA, Nieduszynski CA, Fox CA. 2013. A link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet 9: e1003798 10.1371/journal.pgen.1003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AND, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. 2010. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743. 10.1016/j.molcel.2010.01.033 [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10: 161–172. 10.1038/nrg2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. 10.1038/nature07667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer KH, Fanning E, Otto B, Knippers R. 1980. Maturation of newly replicated chromatin of simian virus 40 and its host cell. J Mol Biol 136: 359–374. 10.1016/0022-2836(80)90395-2 [DOI] [PubMed] [Google Scholar]
- Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C. 2011. DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep 1: 95 10.1038/srep00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Yeeles JTP, Patel H, Early A, Diffley JFX. 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell 65: 117–130. 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-K, Shibata Y, Rao B, Strahl BD, Lieb JD. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905. 10.1038/ng1400 [DOI] [PubMed] [Google Scholar]
- Leem SH, Chung CN, Sunwoo Y, Araki H. 1998. Meiotic role of SWI6 in Saccharomyces cerevisiae. Nucleic Acids Res 26: 3154–3158. 10.1093/nar/26.13.3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. 2001. Monitoring S phase progression globally and locally using BrdU incorporation in TK+ yeast strains. Nucleic Acids Res 29: 1433–1442. 10.1093/nar/29.7.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KHT, Abou El Hassan M, Bremner R. 2013. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques 55: 204–206. 10.2144/000114089 [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutiérrez J, Chen J, Seidel C, Gerton J, Workman JL. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci 102: 18385–18390. 10.1073/pnas.0507975102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. 10.1016/j.cell.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, et al. 2015. Dynamic regulation of transcription factors by nucleosome remodeling. eLife 4: e06249 10.7554/eLife.06249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Sasaki T, Kuipers MA, Lucas I, Le Beau MM, Carignon S, Debatisse M, Prinz JA, Dennis JH, Gilbert DM. 2011. Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res 39: 3141–3155. 10.1093/nar/gkq1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Vu N-D, Patteson K, Mizuguchi G, Wu W-H, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, et al. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell 25: 357–368. 10.1016/j.molcel.2006.12.015 [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Almouzni G. 2013. Chromatin and DNA replication. Cold Spring Harb Perspect Biol 5: a010207 10.1101/cshperspect.a010207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814. 10.1038/emboj.2011.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiasen DP, Lisby M. 2014. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol Rev 38: 172–184. 10.1111/1574-6976.12066 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. 10.1101/gr.078261.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee SR, Smith DJ, Whitehouse I. 2013. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol Cell 50: 123–135. 10.1016/j.molcel.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, Miller OL Jr. 1977. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12: 795–804. 10.1016/0092-8674(77)90278-1 [DOI] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K. 2016. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci 113: E4810–E4819. 10.1073/pnas.1609060113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Bhargava P, Geiduschek EP, Kadonaga JT. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276: 809–812. 10.1126/science.276.5313.809 [DOI] [PubMed] [Google Scholar]
- Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A. 2018. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361: 1389–1392. 10.1126/science.aau0294 [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206. 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ramachandran S, Henikoff S. 2016. Transcriptional regulators compete with nucleosomes post-replication. Cell 165: 580–592. 10.1016/j.cell.2016.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. 2013. DNA replication timing. Cold Spring Harb Perspect Biol 5: a010132 10.1101/cshperspect.a010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618. 10.1038/27001 [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155: 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang J-PZ, Widom J. 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. 10.1038/nature04979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585. 10.1016/S0092-8674(00)80661-3 [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621. 10.1038/27007 [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D. 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7: 594–605. 10.1038/nprot.2012.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, Kavanaugh GM, Chen Y, Tabb DL, Cortez D. 2013. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem 288: 31458–31467. 10.1074/jbc.M113.511337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. 2014. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci 39: 381–399. 10.1016/j.tibs.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25. 10.1016/0092-8674(89)90398-X [DOI] [PubMed] [Google Scholar]
- Sogo JM, Stahl H, Koller T, Knippers R. 1986. Structure of replicating simian virus 40 minichromosomes: the replication fork, core histone segregation and terminal structures. J Mol Biol 189: 189–204. 10.1016/0022-2836(86)90390-6 [DOI] [PubMed] [Google Scholar]
- Stillman B. 1986. Chromatin assembly during SV40 DNA replication in vitro. Cell 45: 555–565. 10.1016/0092-8674(86)90287-4 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21: 2055–2063. 10.1016/j.cub.2011.11.038 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560. 10.1038/990147 [DOI] [PubMed] [Google Scholar]
- Vasseur P, Tonazzini S, Ziane R, Camasses A, Rando OJ, Radman-Livaja M. 2016. Dynamics of nucleosome positioning maturation following genomic replication. Cell Rep 16: 2651–2665. 10.1016/j.celrep.2016.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson T, Hartemink AJ. 2009. An ensemble model of competitive multi-factor binding of the genome. Genome Res 19: 2101–2112. 10.1101/gr.093450.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100. 10.1101/gr.098509.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hsieh T-HS, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N. 2015. High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58: 371–386. 10.1016/j.molcel.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T, Whitehouse I. 2016. Replication-coupled nucleosome assembly and positioning by ATP-dependent chromatin-remodeling enzymes. Cell Rep 15: 715–723. 10.1016/j.celrep.2016.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Chen H, Bai L. 2018. Systematic study of nucleosome-displacing factors in budding yeast. Mol Cell 71: 294–305.e4. 10.1016/j.molcel.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A, Miyake T, Li R, Morse RH. 2004. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol 24: 9152–9164. 10.1128/MCB.24.20.9152-9164.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu R-M, Zhang Z. 2018. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361: 1386–1389. 10.1126/science.aat8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852. 10.1038/nsmb.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.