Abstract
The social environment influences the circadian clock of diverse animals, but little is known about the functional significance, the specifics of the social signals, or the dynamics of socially mediated changes in the clock. Honey bees switch between activities with and without circadian rhythms according to their social task. Forager bees have strong circadian rhythms, whereas “nurse” bees typically care for the brood around-the-clock with no circadian rhythms in behavior or clock gene expression. Here we show that nurse-age bees that were restricted to a broodless comb inside or outside the hive showed robust behavioral and molecular circadian rhythms. By contrast, young nurses tended brood with no circadian rhythms in behavior or clock gene expression, even under a light-dark illumination regime or when placed with brood—but no queen—in a small cage outside the hive. This behavior is context-dependent because nurses showed circadian rhythms in locomotor activity shortly after removal from the hive, and in clock gene expression after ∼16 h. These findings suggest that direct interaction with the brood modulates the circadian system of honey bees. The dynamics of rhythm development best fit models positing that at least some pacemakers continue to oscillate and be entrained by the environment in nurses that are active around the clock. These cells set the phase to the clock network when the nurse is removed from the hive. These findings suggest that despite its robustness, the circadian system exhibits profound plasticity, enabling adjustment to rapid changes in the social environment.
Introduction
Social interactions influence circadian rhythms in diverse animals. In humans, it is thought that misalignment of the social environment and the clock can contribute to the expression of several mental and mood disorders (Frank et al., 2000; Lam and Levitan, 2000; De Leersnyder, 2006; Grandin et al., 2006). However, little is known about the function and mechanisms of this interplay between the social environment and the clock. There is no clear relationship between level of sociality (e.g., solitary vs living in groups) and sensitivity of the circadian system to social signals (Refinetti et al., 1992; Gattermann and Weinandy, 1997; Krupp et al., 2008; Knadler and Page, 2009). It is also not clear whether social influences are mediated by specific pathways connecting sensory systems to the clock or by general mechanisms such as arousal, food anticipation, or social gating of input pathways (Mistlberger and Skene, 2004).
Social insects such as honey bees provide a powerful model with which to study social influences on the clock because of their rich repertoire of social interactions, the feasibility of manipulating their social environment in an ecologically relevant context, and the apparent functional significance of temporal coordination of their behavior (Bloch, 2009). Honey bee workers show natural plasticity in circadian rhythms by switching between activities with or without circadian rhythms depending on their social role in the colony. “Nurses” tend the brood around-the-clock with no circadian rhythms whereas the typically older foragers have strong circadian rhythms that are necessary for timing their visits to flowers and for time-compensated sun-compass orientation (for review, see Moore et al., 1998; Bloch, 2009). Worker bees may switch between nursing and foraging activities according to age and colony needs (Robinson, 1992).
The animal clock is composed of a network of interacting clock cells (pacemakers). Each of these cells expresses a set of “clock genes” that are necessary for generating cell autonomous molecular oscillations (Dunlap, 1999; Dunlap et al., 2004; Bell-Pedersen et al., 2005). These defining oscillations in transcript and protein levels make it possible to assess the molecular mechanisms underlying overt behavioral rhythms. Analyses of brain clock gene expression have revealed strong oscillations in honey bee foragers, but not in nurses independent of the illumination regime (Toma et al., 2000; Bloch et al., 2001, 2004; Shemesh et al., 2007). These studies suggest that plasticity in activity rhythms is associated with plasticity in the molecular clockwork. The lack of molecular oscillations in nurses is not consistent with alternative hypotheses stating that in nurses internal circadian rhythms are masked by external social factors, or that their clock is uncoupled from locomotor activity controlling centers; both the “masking” and “uncoupling” hypotheses predict that the clock of nurses produce normal molecular oscillations (Shemesh et al., 2007).
In the current study we tested two hypotheses. The first is that plasticity in circadian rhythms is regulated by direct contact with the brood. The second is that activity around-the-clock in nurse bees is context dependent. Our findings lend credence to both hypotheses.
Materials and Methods
Observation hives.
The bees for most of the experiments were obtained from source colonies headed by a queen artificially inseminated with the semen of a single (different) drone (colonies S17, S20, S21, S26, S77, and S85). Because of the haplodiploid sex determination system in Hymenoptera, the workers in these colonies were full sisters with a relatedness of r = 0.75. Colony H7 was headed by a naturally mated queen (a queen typically mates with 10–20 drones).
For our experiments we set “triple-cohort” colonies which are commonly used in studies on the division of labor and sociobiology of bees (Giray and Robinson, 1994; Huang and Robinson, 1996). Triple-cohort colonies are typically smaller and more standardized than typical field colonies, but worker demography is set to mimics natural colonies and the development of workers is similar to that in typical colonies. Our triple-cohort colonies contained ∼1700 newly emerged bees (0–24 h of age), ∼1700 nurses, ∼1700 foragers, and their mother queen (the colony contained a total of ∼5000 worker bees). To obtain newly emerged bees, we transferred sealed brood combs from the source colonies to an incubator 24 h before the establishment of the colony. The bees that emerged in the incubator were marked (dot on their thorax) with a paint (Testors Enamel) and were introduced into the observation hive. To obtain nurses, we collected bees of unknown age from the source colony that were seen with their head inside brood-containing comb cells. To obtain foragers we blocked the hive entrance and collected bees returning to the hive with pollen loads on their hind legs (we did not collect foragers during the time of orientation flights). The observation hive was made of transparent glass and Plexiglas walls. The lower frame contained pollen and honey, and the upper frame had eggs and empty cells for the queen to lay in. We placed each observation hive in an environmental chamber [29 ± 1°C; relative humidity (RH) = 50 ± 5%] and connected it to the outside with a clear plastic tube (length = 60 cm, diameter = 3 cm). During the first 2 d, we kept the observation hives in constant darkness (DD, dim red light that bees cannot see) and prevented the bees from flying outside. Starting on day 3, we opened the hive entrance every morning at 8:00 A.M. and closed it at 8:00 P.M. On day 4, we changed the illumination regime to 12 h light/12 h dark with light on at 8:00 A.M., and light off at 8:00 P.M. (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). This experimental procedure set a similar illumination regime for the nest bees and foragers because foraging outside was restricted to the time during which the hive was illuminated. The illumination was arranged such that the light reached all parts of the observation hive, guaranteeing that the hive bees experienced a potent external cue that is known to entrain the circadian clock (Devlin and Kay, 2001). During days 4–6, we paint-marked (Testors Enamel) a few hundred foragers for identification during periods with no foraging activity (e.g., at night).
Experiment 1: The influence of the brood on behavioral and molecular rhythms in young bees in the hive.
In this experiment we prevented direct interactions with the brood for a subset of nurse-age bees by caging them on a broodless comb inside an observation hive (set up as described above). We cut approximately one third of a brood-containing honeycomb and replaced it with a three-chamber cage with empty pieces of comb. The vertical walls of the chambers were made of 8 hole per inch mesh, enabling contact with bees outside the cage and the diffusion of volatile compounds (supplemental Fig. S1B, available at www.jneurosci.org as supplemental material). The outside facing walls were made of transparent glass to allow observation of the behavior of the bees in the cage. In each chamber we placed a piece of empty honeycomb and pollen. The chambers were interconnected by plastic tubes, such that the bees could move freely between them. Each chamber was made of two parts that could be removed separately to facilitate the sampling of bees for RNA analysis at the end of the experiment. The cage contained 200 workers of unknown age. On day 3, we marked ∼600, 1-d-old bees with a paint dot on their thorax, and introduced 200 of these into the broodless cages, and the other 400 bees to the brood-containing comb. To enable individual identification of focal bees, we tagged an additional 100 caged and 100 freely moving workers with numbered plastic tags. Thus, the broodless cages contained ∼500 workers. The eventual number of bees was set such that the density inside and outside the cage was similar. On day 7 (the focal bees were 5–6 d of age), we observed the activity (walking continually for at least 3 s) of the number-tagged bees inside and outside the cages every 3 h (colonies H7 and S26) or every 2 h (colony S77) in LD. Observers used red goggles in observations conducted during the day (light phase) to minimize variation in visibility compared with the night observations, which were performed under dim red light. During each observation we scanned the two sides of the honeycomb six times, for 10 min each (as in Bloch and Robinson, 2001). The observers (n = 2) were trained before the experiment, to minimize interobserver variability. Following the last observation, we collected 15–20 foragers of unknown age (foragers are typically older than 21 d of age, (Robinson, 1992), 15–20 nurses (6–7 d of age), and 15–20 brood-deprived bees (same age as nurses) for analyses of locomotor activity (see below). Nurses and foragers were identified as in previous studies (Moore et al., 1998; Bloch and Robinson, 2001). On day 9 we collected bees for mRNA analysis from 3 different groups of bees: (1) 7-d-old nurses, (2) 7-d-old brood-deprived workers, and (3) foragers of unknown age.
Experiment 2: The influence of the brood on behavioral and molecular rhythms in young bees in small cages outside the hive.
We marked ∼1500 newly emerged bees (during 48 h) by placing a paint-dot (1300 bees) or a number-tag (200 bees) on their thorax, and reintroduced them to their source colony. After 4 d in the colony we transferred 600 marked bees (of which 100 were number-tagged) to a cage (27 × 49 × 5.5 cm) containing a honeycomb with brood (“+ brood”). We used 2- to 4-d-old larvae because preliminary experiments indicated that the survival of younger (1–2 d after egg hatching) larvae under these conditions is low. The honeycomb with the brood was obtained from the same colony from which we collected the workers. We transferred 600 additional marked bees (from which 100 were number-tagged) to a similar cage containing an empty honeycomb (“− brood”). In a third similar cage with a honeycomb with no brood we placed 600 foragers (identified as bees returning to the hive with pollen loads). Honey and pollen were provisioned ad libitum. We placed the three cages together in an environmental chamber (31 ± 1°C; RH = 50 ± 5%), which was subjected to a 12 h light/dark illumination regime. During the dark phase, the room was illuminated by dim red light. On the third and fourth days following the establishment of the cages (at 7:00 A.M. and 1:00 P.M., respectively), we introduced small pieces (11 × 10 cm) of honeycomb into each cage. A comb with 2- to 4-d-old larvae was introduced to the + brood cage, and similar honeycombs with no brood were introduced to the − brood and forager cages. Most of the introduced larvae successfully pupated; in a preliminary experiment in which we also examined the emerging adult bees, they appeared perfectly normal, suggesting that brood care in the small cages was effective.
On day 3 of the experiment (the young bees were 6–8 d of age), we recorded the activity and brood care behavior of the number-tagged bees. Each observation session lasted 1 h and was divided into two halves: during the first 30 min (six scans of 5 min each on both sides of the brood comb) we recorded brood care activity; namely, an event in which a bee is seen with her head in a cell containing larvae; during the second half hour (three scans of 5 min each, on the two cages containing nurse-age bees) we recorded overall activity: a bee was considered active if she moved continuously during one second). We repeated this observation protocol every 3 h (a total of 8 time points). To minimize variation in visibility between the light and dark phases, we used red goggles in observations conducted during the light phase. On the fourth day (the nurse bees were 7–9 d of age), we collected bees for mRNA analysis every 4 h at 7 different time points.
Experiment 3: Circadian rhythms in locomotor activity for bees transferred from the hive to individual cages in constant laboratory environment.
We removed nurses, foragers, and nurse-age brood-deprived bees from the colonies used for experiments 1 and 5 into a constant laboratory environment. We placed each focal bee in an individual monitoring cage made of a modified Petri dish (diameter = 90 mm, height = 15 mm), inside an environmental chamber (29 ± 1°C, RH = ∼50%). The chamber was illuminated with constant dim red light during the entire period of data acquisition. We provisioned each cage with sugar syrup (50%, w/w). We ensured that the bees were not exposed to light or other external factors while being transferred from the hive to the laboratory. We monitored locomotor activity with the ClockLab data acquisition system (Actimetrics) with 3 light-sensitive black and white Panasonic WV-BP334, 0.08 lux CCD cameras (each camera recorded activity from 30 cages), and a high-quality monochrome image acquisition board (IMAQ 1409, National Instruments) (Yerushalmi et al., 2006). Data were collected continuously at a frequency of 1 Hz. We determined circadian rhythmicity for the first 5 d in the laboratory with a χ2 periodogram analysis with 10 min bins (ClockLab circadian analyses software, Actimetrics).
Experiment 4: The molecular clockwork in nurses removed from the hive for ≥16 h.
This experiment was conducted in parallel to the in-hive caging experiment (three repetitions with bees from colonies S26, S77 and H7; see above). On day 6 of the experiment, we transferred 105 nurses into Libfield wooden cages (10 × 11 × 5 cm) (7 cages, each with 15 bees) that were placed next to the observation hive, in the same environmental chamber. After 16 h in the cages, we collected samples of these bees every 4 h over 1 d (7 time points over a total of 28 h) for RNA analysis. Thus, bees collected at the last time point had spent 40 h in the cages. We sampled bees from the cages at the same time as the collections of foragers, nurses, and brood-deprived bees from the same colonies (see above).
Experiment 5: The molecular clockwork in nurses removed from the hive for ≥8 h.
We established 3 additional observation hives with bees from colonies S17, S20 and S21. On day 6 we transferred nurses into Libfield cages as described above. After 8 h in the wooden cages in DD (on day 9), we collected samples of nurses, nurses removed to a cage, and foragers of unknown age for mRNA analyses. We sampled bees for mRNA analyses every 4 h, over one consecutive day (at 6 different time points). Thus, the bees at the last time point were collected after 28 h in the cage.
Measuring brain mRNA levels.
To minimize RNA degradation, we collected the bees directly into liquid nitrogen, and stored the frozen samples at −80°C until brain dissection. We measured mRNA levels with real-time reverse transcription (RT)-PCR as described previously by Rubin et al. (2006). Briefly, we removed and freeze-dried the bee head, and dissected out the brain on a frozen dissecting dish that was immersed in dry ice. We removed compound eyes, ocelli, hypopharyngeal glands, and any other glandular tissues during dissection; we discarded all brains in which pieces of tissue were lost and analyzed only intact brains. We stored each brain individually at −80°C until mRNA quantification. Total brain RNA was isolated (Invisorb Spin Tissue RNA Mini Kit, Invitek), treated with DNase (RQ1 RNase-Free DNase, Promega) and reverse-transcribed in 25 μl of 1× RT buffer + 2.5 U/μl Reverse Transcriptase (BioScript, BioLine), 4 mm deoxy NTPs mixture (Fermentas), 25 ng/μl random hexamers (Invitrogen), 1 U/μl RNase inhibitor (RiboLock Ribonuclease Inhibitor, Fermentas). RNA and random hexamers were incubated at 70°C for 5 min and immediately transferred to ice. Reverse transcription was performed at 25°C for 10 min, 42°C for 60 min, 70°C for 10 min and then incubated at 4°C.
We measured mRNA levels with real-time quantitative RT-PCR using an ABI Prism 7000 appliance (Winer et al., 1999). To measure Period (Per) mRNA levels in colonies S17, S20 and S21 we used a multiplex PCR protocol in which Per and EF-1α are amplified in the same reaction tube. Amplification reactions (25 μl) contained 1× TaqMan Universal PCR Master Mix (ABI Applied Biosystems), 0.1 μm concentration of each primer, 0.2 μm TaqMan probe, and 20–24 ng of cDNA (control samples had no reverse transcriptase). The amplification thermal profile was 50°C for 2 min, 95°C for 10 min (95°C for 15 s, 60°C for 1 min) × 40 cycles. We excluded outliers (SD among triplicates >0.3) from our analyses.
To measure Per mRNA levels in colonies H7, S26 and S77 and to measure Cryptochrome (Cry) mRNA levels in all six colonies, we used singleplex reactions with the SYBR Green dye protocol. The amplification reaction (20 μl) was similar to the above but contained SYBR green master mix (ABI Applied Biosystems) instead of TaqMan Universal PCR Master Mix and did not contain oligo probes. Each cDNA sample was analyzed in triplicate. PCRs for all focal genes and EF-1α were loaded on the same 96 well analysis plate. To prevent amplification of genomic DNA, we designed the PCR primers to span over an exon-exon boundary. Per, Cry and EF1α levels were measured from the same cDNA sample.
We compared clock gene mRNA levels with the 2−ΔΔCt method and EF-1α as a control gene for normalization (ABI User Bulletin #2) (see also Winer et al., 1999). For statistical analyses we used ΔΔCt values that are normally distributed. We assessed whether brain clock gene mRNA levels differed between bees collected at different time points with a one-way ANOVA (SPSS software) with time as a factor, and a Fisher LSD post hoc test. For more details see Rubin et al. (2006).
We also determined the correlation between the average brain mRNA levels for each time point and a cosine curve. To fit the best cosine model to the average RNA relative expression data we used the equation:
where f(x) is the average normalized RNA value at time x; a is the shift on the y-axis, b is the amplitude, and c is the period length. The regression analyses were performed with the MATLAB software package (MathWorks). We started with a value of 1 for parameters a–c, and looked for the parameters that produced the best fit (highest R 2). We then adjusted the R 2 value using the equation:
where n is the number of values (6–8 time points in our experiments), and m is the number of parameters (which was 3 in our analyses). R 2 adjusted was used because it is a more stringent parameter.
Results
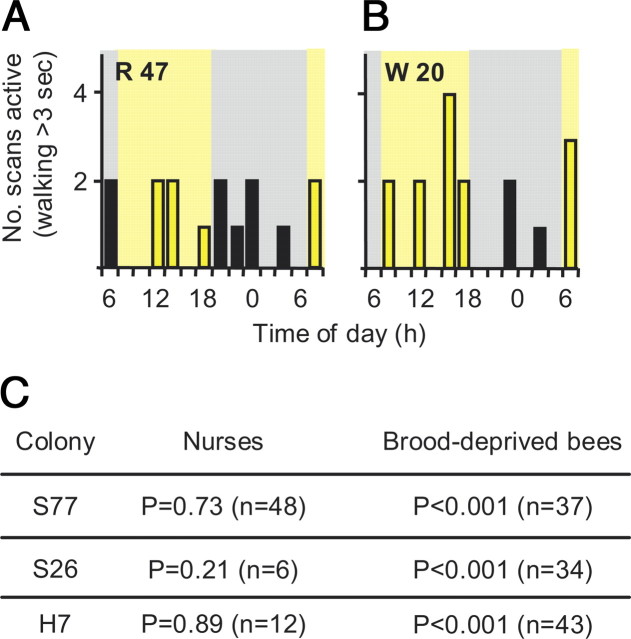
Table 1 summarizes the experiments detailed below. We first tested whether plasticity in circadian rhythms is modulated by specific social signals or by other environmental factors. In the first experiment we compared the behavior and clock gene expression of bees that were caged on broodless pieces of honeycomb inside the hive to those of bees with a similar age and genotype developing in the surrounding brood-containing comb (supplemental Fig. 1, available at www.jneurosci.org as supplemental material, see Materials and Methods). The caged bees experienced the physical environment of the hive and were exposed to its volatile odorants, but did not have direct contact with the brood. We found that at 6 d of age, which is typical of nurses in field colonies, the bees on the brood-containing comb frequently tended the brood cells and were active similarly during the light and dark phases of the illumination regime (Fig. 1 A). These observations are in line with previous studies of nurses (Crailsheim et al., 1996; Moore et al., 1998; Bloch and Robinson, 2001; Shemesh et al., 2007). By contrast, their same age full-sister bees that were caged on the broodless comb were significantly more active during the photophase (Fig. 1 B,C).
Table 1.
Summary of experiments
| Experiment | Experimental manipulation | Experimental conditions | Results |
|---|---|---|---|
| 1 | Nurse-age workers were caged on a broodless honey comb inside the hive | Hive: + | Behavior: caged bees showed activity rhythms similar to those of foragers, nurses were active around the clock. |
| Brood: − | |||
| Queen: +? | Clock gene expression: strong oscillations in caged bees and foragers but not in nurses. | ||
| Comb: + | |||
| IL: LD | |||
| 2 | Nurses were caged on a brood-containing or broodless comb outside the hive | Hive: − | Behavior: nurses with brood were active around the clock; nurses without brood were more active during the day. |
| Brood: +/− | |||
| Queen: − | Clock gene expression: strong oscillations in nurses caged without brood and in foragers, attenuated oscillations in nurses caged on a comb with brood. | ||
| Comb: + | |||
| IL: LD | |||
| 3 | Nurses were transferred to individual cages in constant lab environment | Hive: − | Nurses showed circadian rhythms in locomotor activity shortly after transfer to the lab. |
| Brood: − | |||
| Queen: − | |||
| Comb: − | |||
| IL: entrainment in LD, monitoring in DD | |||
| 4 | A group of nurses were removed from the hive to a cage for 16 h and then sampled every 4 h | Hive: − | Strong oscillations in foragers, weaker but significant oscillations in nurses that were transferred to a cage, attenuated oscillations in nurses. |
| Brood: − | |||
| Queen: − | |||
| Comb: + | |||
| IL: LD | |||
| 5 | A group of nurses were removed from the hive to a cage for 8 h and then sampled every 4 h | Hive: − | Strong oscillations in foragers, attenuated oscillations in nurses and in nurses that were transferred to a cage. |
| Brood: − | |||
| Queen: − | |||
| Comb: + | |||
| IL: LD |
IL, Illumination regime; LD, 12 h light/dark; DD, constant dim red light.
Figure 1.
Nurse-age bees on a broodless comb are more active during the photophase whereas nurses on an adjacent brood-containing comb are active similarly throughout the day. A, A representative nurse bee (R47, colony S77) was active similarly during the light (yellow columns and background) and dark (filled columns, gray background) phases of the illumination regime (two-tailed exact binomial test, p = 1). B, A representative nurse-age bee on a broodless comb (W20, colony S77, a full sister of the bee shown in A was more active during the light phase (p = 0.02). C, A summary of statistical analyses (Wilcoxon signed rank pair-comparison tests) for the overall level of activity during the light and dark phases of the illumination regime (for all tagged bees for which there were eight or more observations). n = number of bees analyzed. The results were similar in three repetitions, each with bees from a different source colony (S77, S26, H7).
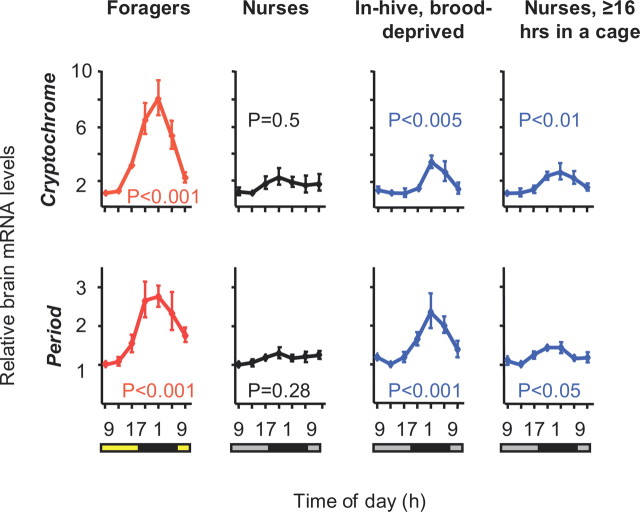
We further analyzed brain transcript levels for Cry and Per, which are the clock genes that show the most robust oscillations in honey bees (Rubin et al., 2006; Shemesh et al., 2007). We found that both Cry and Per transcript levels oscillated with higher levels during the night in foragers, but not in nurses, which is consistent with previous analyses of honey bees (Toma et al., 2000; Bloch et al., 2001, 2004; Shemesh et al., 2007) (Fig. 2; supplemental Fig. 2, available at www.jneurosci.org as supplemental material). The temporal expression profile for the in-hive caged bees was more similar to that of foragers than to nurses; both Cry and Per mRNA levels were significantly higher during the dark phase (one-way ANOVA, p < 0.05 in all three colonies) (Fig. 2; supplemental Fig. 2, available at www.jneurosci.org as supplemental material). These results indicate that the micro-environmental conditions (e.g., temp', humidity, CO2 concentration) and the volatile odor bouquet of the hive, which were similar for the bees inside and outside the mesh cages, are not likely to account for the around the clock activity and attenuated clock gene oscillations in nurse bees.
Figure 2.
The social context influences the temporal pattern of clock gene expression in young bees. The plots show brain Per and Cry mRNA levels (mean ± SE) for full sister bees from colony S77. “Foragers,” worker bees returning to the hive with pollen loads; “Nurses,” 7-d-old bees that were seen tending brood; “In hive, brood-deprived,” nurse-age bees that were caged on a broodless comb inside the hive; “Nurses, ≥16 h in a cage,” nurse bees that were transferred to a cage outside the hive for ≥16 h and sampled at the age of 7 d. Sample size = 5–6 bees/time point. The bar at the bottom of each plot depicts the illumination regime on the day of sample collection. Black bar, night or subjective night; gray bar, subjective day; yellow bar, day (light on). The p-values were obtained from one-way ANOVAs for the ΔΔCt values. Similar results were obtained in two additional trials with bees from different source colonies (S26 and H7; supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
The in-hive caging experiment is consistent with the hypothesis that plasticity in circadian rhythms is influenced by direct contact with the brood. However, caging bees on a broodless comb inside the hive could affect not only their interactions with the brood, but also with the queen and older bees. We therefore performed an additional experiment (experiment 2; see Table 1) in which we compared the behavior and clock gene expression of nurse bees that were caged outside the hive (with no queen or old workers) on honeycomb with or without brood. We found that the bees in small cages with a brood-containing comb tended the brood cells and were active similarly during the light and dark phases of the illumination regime (brood caring: Wilcoxon signed rank test, p > 0.4 in both colonies S85 and H7; activity: Fig. 3 A). By contrast, their same age full-sister bees that were placed in similar cages with no brood were more active during the photophase. Brain Cry and Per transcript levels oscillated, with higher levels during the night in foragers and in nurse-age bees caged with no brood (one-way ANOVA, p < 0.001, Fig. 3 B). Brain clock gene transcript levels also varied throughout the day in nurses in a cage with brood, but the oscillations were not as robust and the day–night differences were attenuated, specifically for Cry (Fig. 3 B). These results show that contact with brood outside the normal context of the hive is sufficient to induce nurse bees to be active around the clock with attenuated clock gene oscillation.
Figure 3.
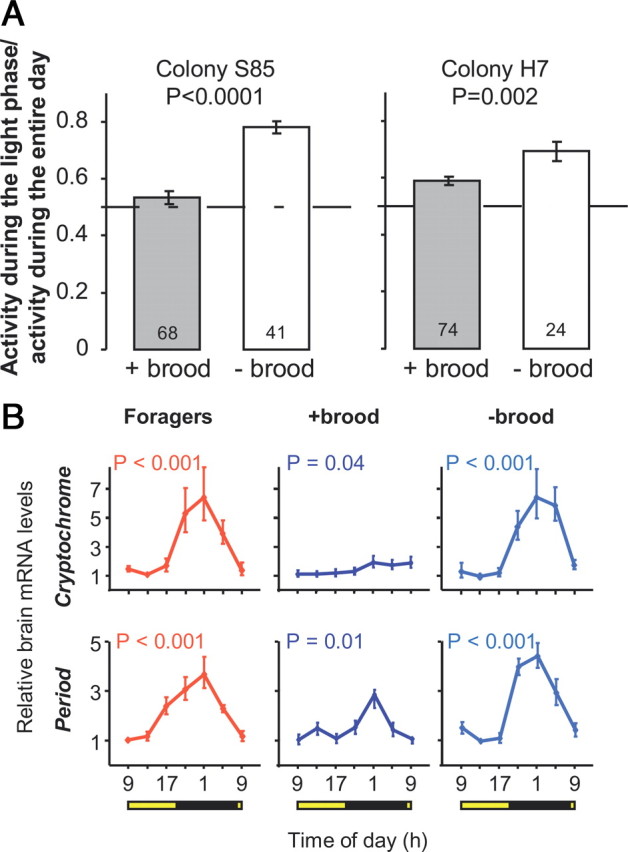
The brood influences behavioral and molecular rhythms for young bees in small cages outside the hive. A, Locomotor activity. The plot depicts the ratio between the levels of activity during the light and dark phases of the illumination regime (mean ± SE). Sample size within bars. The p-values were obtained from unpaired t tests. Left, A trial with bees from colony S85; right, a trial with bees from colony H7. B, Clock gene transcript levels of bees from colony S85. + brood, 8- to 9-d-old bees placed in a brood-containing comb outside the hive; − brood, 8- to 9-d-old bees placed on a similar broodless comb outside the hive. Other details of plot as in Figure 2.
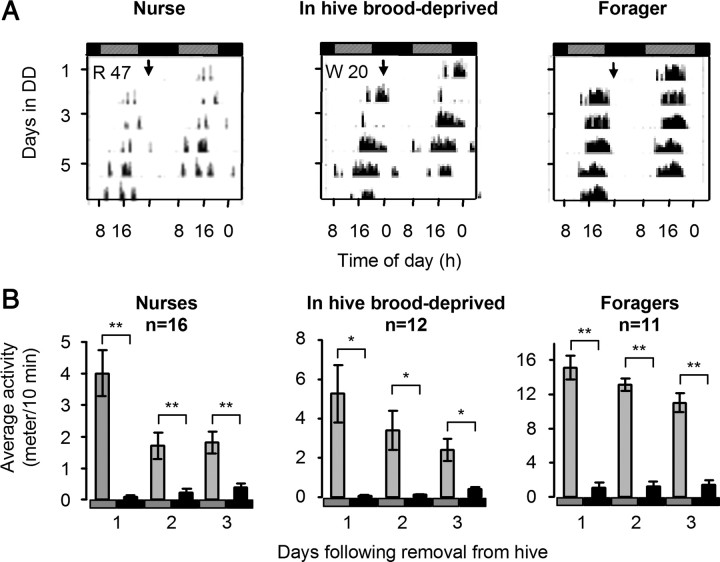
We next tested whether nurse bees manifest weak or no circadian rhythms because their circadian system cannot support strong rhythms (e.g., it is not fully developed), or because they are capable of generating strong rhythms but these are not expressed in the context of nursing behavior. In experiment 3 we monitored locomotor activity for nurses, foragers, and brood-deprived nurses age bees that we transferred from the lab to individual cages in a constant laboratory environment. We found that most of the nurses showed circadian rhythms in locomotor activity shortly after transfer to individual cages in constant darkness (Fig. 4 A; supplemental Fig. S3, available at www.jneurosci.org as supplemental material). The strength, phase, and period of circadian rhythms were similar to those expressed by the brood-deprived bees under similar conditions (supplemental Table S1, available at www.jneurosci.org as supplemental material). The bees showed a significant decrease in locomotor activity during their first subjective night outside the hive (occurring ∼5 h after transfer for the bees from colonies H7 and S26 that were transferred around noon, and immediately after transfer for bees from colony S77 that were transferred around 7:00 P.M.; Fig. 4 B). A significant increase in activity was only apparent the next subjective morning. These circadian rhythms in locomotor activity continued with a similar phase during the subsequent days and were similar to rhythms found in foragers from the same colonies (Fig. 4; supplemental Fig. S3, available at www.jneurosci.org as supplemental material). To further test the relationships between the phase of activity in individual cages in the laboratory, the illumination regime in the hive, and the time of removal from the hive, we examined six additional colonies (from this study and from Shemesh et al., 2007) in which we monitored locomotor activity for nurses removed from the hive. This analysis indicated that the daily onset of activity was correlated with the subjective morning (8:00 A.M. in all experiments), but not with the time the nurse bees were removed from the hive (supplemental Table 2, available at www.jneurosci.org as supplemental material). These behavioral analyses indicate that that the clock system of nurses is already developed and is synchronized to the light/dark regime which they experienced in the hive, but overt circadian rhythms are not expressed in the context of brood care activity. Thus, nurses that are active around-the-clock under a light/dark illumination regime with no apparent oscillations in brain clock gene expression are still capable of measuring time and are responsive to time givers in their environment.
Figure 4.
Nurse bees show circadian rhythms in locomotor activity shortly after transfer from a LD illuminated hive to a constant laboratory environment. A, Representative double-plot actograms for locomotor activity of a nurse (left, R47; data for the same bee are shown in Fig. 1 A), a nurse-age bee that was restricted to a broodless comb in the hive (middle, W20; data for the same bee are shown in Fig. 1 B), and a forager (right). Bees were monitored individually under constant conditions in the laboratory. The y-axis shows the days after removal from the hive. The height of the small bars for each day corresponds to locomotor activity in a 10 min bin. Horizontal bars at the top of the plot correspond to the illumination regime in the observation hive: striped bars, subjective day, filled bars, subjective night. The arrows point to the data acquisition start time. B, Summary of locomotor activity during the first 3 d in the laboratory for bees from colony H7 that were removed from the hive at noon. All the bees were significantly more active during the subjective day (gray bar) than during the subjective night (filled bar). This difference was already visible on the first day of isolation (Paired t tests, two-tailed,*p < 0.01; **p < 0.001). The subjective day and night were based on the Free Running Period (τ) of each bee. Similar results were obtained for bees from colonies S26 and S77 (see supplemental Fig. S3A, available at www.jneurosci.org as supplemental material).
In experiments 4 and 5 we further studied the molecular dynamics of this context-dependent plasticity in circadian rhythms, we transferred groups of young nurses from a LD illuminated hive to small wooden cages in a constant laboratory environment. In experiment 4, we collected these bees for RNA analysis every 4 h starting 16 h after removal from the hive. We found that 16 h in a cage were sufficient to produce a significant alteration in the temporal pattern of clock gene expression: the temporal pattern of expression was circadian with higher levels during the night for both Cry and Per (one-way ANOVA, p < 0.05; the differences did not reach statistical significance in the analysis of Per mRNA levels in colony S26) (Fig. 2; supplemental Fig. S2, available at www.jneurosci.org as supplemental material). This pattern is reminiscent of that in foragers, but the amplitude was lower than in foragers or in young bees developing on a broodless comb in the hive (Fig. 2; supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Because it is not clear whether these alternations in the pattern of clock gene expression stem from molecular changes within pacemaker cells, changes in the coupling of pacemaker cells or both, we use the term “molecular reorganization” in the clockwork to characterize the switch between strong and attenuated oscillations in whole brain clock gene expression. The results of this experiment suggest that molecular reorganization in the clockwork is already present but not yet complete after 16–40 h outside the hive.
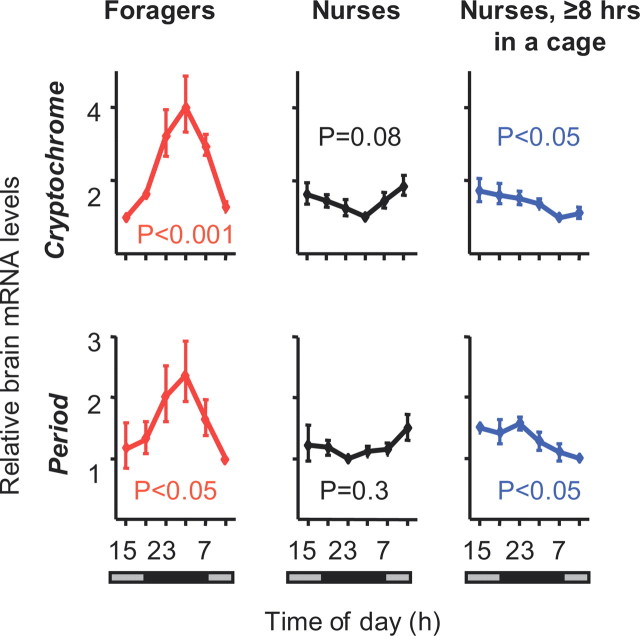
For a more fine-grained assessment of the dynamics of context-dependent plasticity in the molecular clockwork, we repeated the nurse removal experiment in three additional colonies, but this time (experiment 5, Table 1) sampled bees for RNA analysis starting as early as after 8 h in the cage. The analyses of foragers and nurses from these colonies were consistent with earlier studies: Per and Cry mRNA levels varied significantly during the day in foragers, but not in nurses (Fig. 5; supplemental Fig. S4, available at www.jneurosci.org as supplemental material). The temporal pattern of clock gene expression in nurses that were sampled after 8–28 h in cages was different from that of both nurses and foragers. Brain Cry and Per transcript levels appeared higher during the subjective night, which is more similar to foragers than to nurses, but the overall variation was low, which is reminiscent of the pattern in nurses. The phase was similar to the trend seen in nurses from these colonies at the earlier time points, and to that of foragers at the later time points (e.g., after midnight) (Fig. 5; supplemental Fig. S4, available at www.jneurosci.org as supplemental material). These observations suggest that molecular reorganization in the clockwork had already begun in bees removed from the hive for 8–28 h.
Figure 5.
Nurses removed from the hive for 8 h are at an initial stage of clockwork reorganization. “Nurses, ≥8 h in a cage,” nurse bees that were transferred to a cage outside the hive 8–28 h before collection for mRNA analysis. Other details of graph as in Figure 2. Similar results were obtained in two additional trials with bees from colonies S21 and S20 (see supplemental Fig. S4, available at www.jneurosci.org as supplemental material).
Finally, we were interested in comparing the pattern of clock gene expression for nurse-age bees (∼7 d of age) from the different experiments in which they experienced diverse social environments. To compare bees from different source colonies and different experiments we first fit a cosine model to the values of Per and Cry mRNA levels throughout the day (Fig. 6 A). Cosine models typically account for much of the variation in transcript levels during the day for genes that are regulated by the circadian clock (Rubin et al., 2006; Krupp et al., 2008). We normalized the obtained amplitude and R 2 for each group relative to those in nurse bees from the same colony and experiment (e.g., the normalized value is 1 if the amplitude for the nurse age bees is similar to that of nurse bees from the same colony). We found that the pattern of foragers, but not of nurses, fit very well with a cosine curve with high amplitude. The regression coefficient for bees developing on a broodless comb inside the hive was as good as for foragers, but the amplitude was lower. Nurses that were removed from the hive to small cages showed a gradual switch from a nurse-like to a forager-like pattern: after 8–28 h in the cages, the regression coefficient and the amplitude were low, as in nurses (Fig. 6 B). After 16–40 h outside the hive the regression coefficient was as good as for foragers, but the amplitude was notably lower (Figs. 2, 6). The amplitude was also lower than in their same-age sisters developing in cages inside the hive. These results suggest that molecular reorganization begins after 8–28 h, and is at a more advanced stage after 16–40 h outside the hive environment. However, 16–40 h outside the hive are apparently not sufficient to complete the molecular reorganization because the amplitude at this stage was still lower than in full sister bees of similar age that were caged on a broodless comb inside the hive (indicating that bees at this age can have stronger molecular oscillations).
Figure 6.
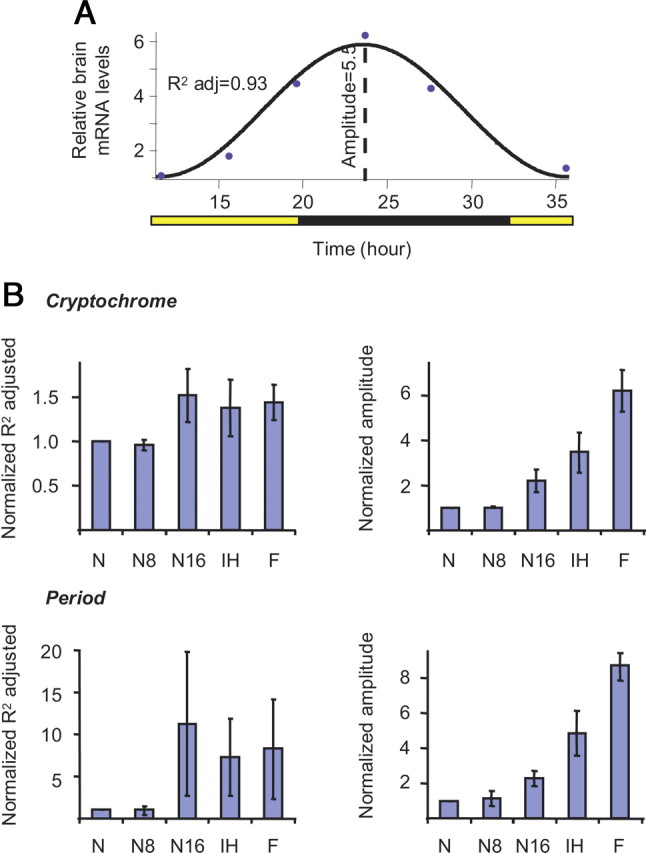
The social environment influences the temporal pattern of clock gene expression. A, Indices for the degree of oscillation in clock gene expression. The black line depicts the best fit cosine? model; the circles are the average measured brain clock gene mRNA levels for each time point. The regression coefficient (adjusted R 2) and amplitude were used for constructing the plots in B. B, Average regression coefficient (left column) and amplitude (right column) for Cry and Per mRNA expression (mean ± SE). N, nurses; N8, nurses assayed after ≥8 h outside the hive; N16, nurses assayed after ≥16 h outside the hive; IH, nurse-age workers that developed on a broodless cage inside the hive; F, foragers. The values were normalized relative to the value of nurses from the same colony (see the relative transcript levels for these bees in Figs. 2 and 5).
Discussion
The circadian behavior of bees is modulated by social interactions with the brood. The same bees that were active around-the-clock while nursing brood in the hive switched to activity with clear circadian rhythms shortly after release from the influence of the hive. This behavioral plasticity was associated with plasticity in clock gene expression. Nurses had no, or only weak, cycling in clock gene expression while in the hive, but built up a forager-like pattern, with higher Cry and Per mRNA levels during the night, shortly after transfer from the hive to small cages. Our results suggest that this capacity to rapidly reorganize the molecular clockwork is feasible because some components of the circadian system measure time even in nurses that are active around-the-clock. These characteristics of the circadian system of bees may enable nurses to care for the brood around-the-clock yet to be able to time their orientation flights to the appropriate time of day.
What in the hive environment induces nurses to be active around-the-clock with no circadian rhythms? The principal difference between the social environment inside and outside the cages in the in-hive caging experiment (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material) was the presence of brood, and brood care is the main activity of nurse bees. However, in-hive caging could affect not only interactions with brood, but also with the queen and old workers which are known to influence the physiology and behavior of young workers (Huang and Robinson, 1992, 1996; Robinson et al., 1998). Nevertheless, our observations and the storing of nectar inside the mesh cages provide direct and indirect evidence (respectively) for trophallactic interactions with bees outside the cage. Trophallaxis serves not only for mutual feeding, but also for the distribution of contact pheromones, including the queen pheromone (Naumann et al., 1991; Leoncini et al., 2004). In addition, in the complementary experiment, nurses that were removed from the hive to a cage with a brood comb but with no queen were similarly active during the day and night and had attenuated molecular oscillations, whereas their same age full-sister bees that were placed in a similar cage with a broodless comb were more active during the day and had strong cycling in clock gene expression (Fig. 3). Together, these experiments indicate that the stress of caging (which was similar for the bees in cages with or without brood) or interactions with the queen or with other workers cannot account for the observed plasticity in the circadian system. Rather, the results indicate that direct contact with the brood modulates the circadian system of bees. The identity of the brood signal(s) and the sensory modality by which it is detected have yet to be determined. Nevertheless, our experiments suggest that the signals are not volatile odorants or factors in the microenvironment of the hive because these were probably similar inside and outside the mesh-wall in the in-hive caging experiment (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Visual signals are also not very likely because the hive is typically dark. We hypothesize that the signals are probably contact pheromones, tactile, or short-distance sounds.
Circadian rhythmicity in bees is context dependent and the nurse removal experiments provide a first assessment of the dynamics of the molecular changes associated with this chronobiological plasticity. The daily pattern of clock gene expression in nurses began to resemble that of foragers 16–40 h after they had been removed from the hive influence; initial changes in clock gene oscillations could perhaps be detected even earlier, 8–28 h after removal from the hive. At 7–9 d of age, nurses that were removed from the hive and were placed for an additional 3–4 d on a broodless cage showed molecular cycling that was as strong as in foragers (Fig. 3 B). Given that Per and Cry are canonical clock genes in the honey bee and show robust cycling in the brain of foragers and other bees with strong circadian rhythms (Toma et al., 2000; Bloch et al., 2001, 2004; Rubin et al., 2006; Shemesh et al., 2007), we assume that the whole brain expression of these genes stems largely from clock cells that are part of the circadian network. It is noteworthy that the appearance of circadian rhythms in locomotor activity was faster than the molecular oscillations. Although the experimental design differed somewhat between the molecular (bees were caged in small groups) and behavioral experiments (bees were caged individually), the apparent temporal disparity between the appearance of molecular and behavioral rhythms is intriguing, as it may indicate that the bees manifested behavioral rhythms before the molecular reorganization of the clockwork was complete.
Although this study was not designed to characterize the mechanism underlying plasticity in the circadian system of the bee, our results may nevertheless contribute to a better understanding of this phenomenon. At least four hypotheses may account for this chronobiological plasticity. First, the circadian system of nurses in the hive is undeveloped or underdeveloped. This hypothesis predicts that nurses removed from the hive will show slow development of behavioral and molecular rhythms, with no specific phase (because individual bees differ in their rate of development). Although there is evidence for postembryonic development of the clock system in honey bees (Toma et al., 2000; Bloch et al., 2001, 2003, 2004), the rapid, almost instant, appearance of activity rhythms that are in phase with the hive environment in nurses removed from the hive (experiment 3) does not lend much credence to this hypothesis. The second hypothesis is that the molecular feedback loop in brain pacemaker cells of nurses is fixed at a certain state. The molecular cycling takes up again from this point when the nurse bee is released from the hive environment. This hypothesis predicts a rapid appearance of molecular cycling, with a phase that is determined by the time the bee is removed from the hive. Our findings that the onset of activity in the lab is correlated with the subjective morning in the hive and not with the time of removal from the hive are not consistent with this hypothesis (Table 2). The third hypothesis posits that at least some of the clock cells in the nurse brain continue to generate molecular rhythms and are entrained by the hive environment. The molecular oscillations in these cells are not detected in a whole brain analysis because they are masked by the constant levels in the majority of clock gene-expressing cells or because brain oscillators are desynchronized, or a combination of these two mechanisms. This hypothesis predicts that when the nurse bee is removed from the hive, the pacemakers that are entrained by the hive environment are coupled to other clock gene-expressing cells and set the phase for the entire clock network. The reorganization of molecular oscillations is predicted to be relatively slow, and with a phase entrained to the hive environment regardless of the time the nurse bee is removed from the hive. The fourth hypothesis is that in nurses, clock cells cycle, but at a low amplitude while in the hive. These cells switch to robust cycling when the nurse bee is removed from the hive. This hypothesis is consistent with studies in Drosophila suggesting that molecular amplitude below a critical threshold cannot support downstream rhythms in behavior and physiology. For example, mutations in Clock and Clockwork Orange (CWO) resulted in reduced amplitude in clock gene expression and severe attenuation in circadian rhythms in locomotor activity (Allada et al., 2003; Lim et al., 2007). Altered amplitude in clock gene oscillation in the brain and in pheromone producing peripheral clocks has been suggested to account for socially modulated changes in pheromone production (Krupp et al., 2008). Our current and previous (Shemesh et al., 2007) studies correspond best to the third and fourth hypotheses because the phase of the molecular and behavioral rhythms after removal from the hive was synchronized with the hive environment, the manifestation of activity rhythms was rapid, and the build up of molecular oscillations was gradual rather than abrupt.
Table 2.
The time of the daily onset of locomotor activity for nurse bees removed at different times from observation hives to individual cages in a constant laboratory environment
| Colony | Sample size | Time of removal | Onset of activity in the lab (mean hh:mm±SE) |
|---|---|---|---|
| S1 | 18 | 4:00 P.M. | 8:34 ± 0:16 |
| S4 | 19 | 3:30 P.M. | 9:22 ± 0:23 |
| S8 | 15 | 1:00 P.M. | 10:34 ± 0:45 |
| S17 | 19 | 10:00 A.M. | 11:08 ± 0:18 |
| S20 | 18 | 10:00 A.M. | 10:20 ± 0:47 |
| S21 | 21 | 12:30 P.M. | 9:18 ± 0:12 |
| S26 | 13 | 12:30 P.M. | 8:57 ± 0:15 |
| S77 | 14 | 8:00 P.M. | 10:47 ± 0:33 |
| H7 | 17 | 12:00 P.M. | 10:47 ± 0:35 |
All the observation hives experienced a 12 h light/dark illumination regime with the light on at 8:00 A.M.
Our findings show that plasticity in circadian rhythms of honey bees is modulated by signals from the brood that are transferred by close distance or direct contact. Circadian rhythmicity in bees is context-dependent; bees switch rapidly between activity with and without circadian rhythms. These findings for bees indicate that a conserved and robust system such as the circadian clock is capable of profound socially-regulated plasticity. The context dependence of the circadian behavior of bees has implications outside the field of chronobiology because it demonstrates the strong dependence of the physiology and behavior of animals on their social context. Nurses that were removed from the natural context of the hive not only showed altered circadian behaviors, but also differed in their clock gene expression. Thus, in some important facets of their physiology and behavior, they were not in a nurse state any more. This evidence of rapid changes in the molecular organization of a complex and conserved system such as the circadian clock illustrates the importance of preserving an ecologically relevant context in studies of natural complex behavior.
Footnotes
Financial support was provided by the Israeli Science Foundation (Grant 452/07 to G.B.), the Israel-US Binational Science Foundation (Grant 2003-151 to G.B.), and the German Israel Foundation (Grant I-822-73.1/2004 to G.B.). We thank Yair Halperin, Niv Bachanoff, and Noa Kahana for technical and experimental assistance, and Sebastian Kadener for valuable comments on an earlier version of this manuscript.
References
- Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–3375. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G. Socially mediated plasticity in the circadian clock of social insects. In: Gadau J, Fewell J, editors. Organization of insect societies—from genome to sociocomplexity. Boston: Harvard UP; 2009. pp. 402–431. [Google Scholar]
- Bloch G, Robinson GE. Chronobiology. Reversal of honeybee behavioural rhythms. Nature. 2001;410:1048. doi: 10.1038/35074183. [DOI] [PubMed] [Google Scholar]
- Bloch G, Toma DP, Robinson GE. Behavioral rhythmicity, age, division of labor and period expression in the honey bee brain. J Biol Rhythms. 2001;16:444–456. doi: 10.1177/074873001129002123. [DOI] [PubMed] [Google Scholar]
- Bloch G, Solomon SM, Robinson GE, Fahrbach SE. Patterns of PERIOD and pigment-dispersing hormone immunoreactivity in the brain of the European honeybee (Apis mellifera): age- and time-related plasticity. J Comp Neurol. 2003;464:269–284. doi: 10.1002/cne.10778. [DOI] [PubMed] [Google Scholar]
- Bloch G, Rubinstein CD, Robinson GE. Period expression in the honey bee brain is developmentally regulated and not affected by light, flight experience, or colony type. Insect Biochem Mol Biol. 2004;34:879–891. doi: 10.1016/j.ibmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Crailsheim K, Hrassnigg N, Stabentheiner A. Diurnal behavioural differences in forager and nurse honey bees (Apis mellifera carnica Pollm) Apidologie. 1996;27:235–244. [Google Scholar]
- De Leersnyder H. Inverted rhythm of melatonin secretion in Smith-Magenis syndrome: from symptoms to treatment. Trends Endocrinol Metab. 2006;17:291–298. doi: 10.1016/j.tem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Circadian photoperception. Annu Rev Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: biological timekeeping. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- Frank E, Swartz HA, Kupfer DJ. Interpersonal and social rhythm therapy: managing the chaos of bipolar disorder. Biol Psychol. 2000;48:593–604. doi: 10.1016/s0006-3223(00)00969-0. [DOI] [PubMed] [Google Scholar]
- Gattermann R, Weinandy R. Lack of social entrainment of circadian activity rhythms in the solitary golden hamster and in the highly social Mongolian gerbil. Biol Rhythm Res. 1997;28:85–93. [Google Scholar]
- Giray T, Robinson GE. Effects of intracolony variability in behavioral-development on plasticity of division-of-labor in honey-bee colonies. Behav Ecol Sociobiol. 1994;35:13–20. [Google Scholar]
- Grandin LD, Alloy LB, Abramson LY. The social zeitgeber theory, circadian rhythms, and mood disorders: review and evaluation. Clin Psychol Rev. 2006;26:679–694. doi: 10.1016/j.cpr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Huang ZY, Robinson GE. Honeybee colony integration: worker–worker interactions mediate hormonally regulated plasticity in division of labor. Proc Natl Acad Sci U S A. 1992;89:11726–11729. doi: 10.1073/pnas.89.24.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZY, Robinson GE. Regulation of honey bee division of labor by colony age demography. Behav Ecol Sociobiol. 1996;39:147–158. [Google Scholar]
- Knadler JJ, Page TL. Social interactions and the circadian rhythm in locomotor activity in the cockroach Leucophaea maderae . Chronobiol Int. 2009;26:415–429. doi: 10.1080/07420520902876634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Kent C, Billeter JC, Azanchi R, So AK, Schonfeld JA, Smith BP, Lucas C, Levine JD. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster . Curr Biol. 2008;18:1373–1383. doi: 10.1016/j.cub.2008.07.089. [DOI] [PubMed] [Google Scholar]
- Lam RW, Levitan RD. Pathophysiology of seasonal affective disorder: a review. J Psychiatry Neurosci. 2000;25:469–480. [PMC free article] [PubMed] [Google Scholar]
- Leoncini I, Le Conte Y, Costagliola G, Plettner E, Toth AL, Wang M, Huang Z, Bécard JM, Crauser D, Slessor KN, Robinson GE. Regulation of behavioral maturation by a primer pheromone produced by adult worker honey bees. Proc Natl Acad Sci U S A. 2004;101:17559–17564. doi: 10.1073/pnas.0407652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila . Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE, Skene DJ. Social influences on mammalian circadian rhythms: animal and human studies. Biol Rev. 2004;79:533–556. doi: 10.1017/s1464793103006353. [DOI] [PubMed] [Google Scholar]
- Moore D, Angel JE, Cheeseman IM, Fahrbach SE, Robinson GE. Timekeeping in the honey bee colony: integration of circadian rhythms and division of labor. Behav Ecol Sociobiol. 1998;43:147–160. [Google Scholar]
- Naumann K, Winston ML, Slessor KN, Prestwich GD, Webster FX. Production and transmission of honey-bee queen (Apis-mellifera L) mandibular gland pheromone. Behav Ecol Sociobiol. 1991;29:321–332. [Google Scholar]
- Refinetti R, Nelson DE, Menaker M. Social stimuli fail to act as entraining agents of circadian rhythms in the golden hamster. J Comp Physiol A. 1992;170:181–187. doi: 10.1007/BF00196900. [DOI] [PubMed] [Google Scholar]
- Robinson GE. Regulation of division-of-labor in insect societies. Annu Rev Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Winston ML, Huang Z, Pankiw T. Queen mandibular gland pheromone influences worker honey bee (Apis mellifera l.) foraging ontogeny and juvenile hormone titers. J Insect Physiol. 1998;44:685–692. doi: 10.1016/s0022-1910(98)00040-7. [DOI] [PubMed] [Google Scholar]
- Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, Bloch G. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006;16:1352–1365. doi: 10.1101/gr.5094806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh Y, Cohen M, Bloch G. Natural plasticity in circadian rhythms is mediated by reorganization in the molecular clockwork in honeybees. FASEB J. 2007;21:2304–2311. doi: 10.1096/fj.06-8032com. [DOI] [PubMed] [Google Scholar]
- Toma DP, Bloch G, Moore D, Robinson GE. Changes in period mRNA levels in the brain and division of labor in honey bee colonies. Proc Natl Acad Sci U S A. 2000;97:6914–6919. doi: 10.1073/pnas.97.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Yerushalmi S, Bodenhaimer S, Bloch G. Developmentally determined attenuation in circadian rhythms links chronobiology to social organization in bees. J Exp Biol. 2006;209:1044–1051. doi: 10.1242/jeb.02125. [DOI] [PubMed] [Google Scholar]