ABSTRACT
Astrocytes display diverse morphologies in different regions of the central nervous system. Whether astrocyte diversity is attributable to developmental processes and bears functional consequences, especially in humans, is unknown. RNA-seq of human pluripotent stem cell-derived regional astrocytes revealed distinct transcript profiles, suggesting differential functional properties. This was confirmed by differential calcium signaling as well as effects on neurite growth and blood-brain barrier formation. Distinct transcriptional profiles and functional properties of human astrocytes generated from regionally specified neural progenitors under the same conditions strongly implicate the developmental impact on astrocyte diversity. These findings provide a rationale for renewed examination of regional astrocytes and their role in the pathogenesis of psychiatric and neurological disorders.
KEY WORDS: Human astrocyte development, Astrocyte heterogeneity, RNA-seq, Neuron-glia interaction, Blood-brain barrier
Summary: Astrocytes generated from hPSC-derived regional neural progenitors maintain their identity throughout their differentiation and exhibit differential gene expression and functions, suggesting developmental origins of astrocyte functional diversity.
INTRODUCTION
Astrocytes make up 20-40% of cells in the human brain (Oberheim et al., 2009; Zhang et al., 2016). Astrocytes not only provide structural support, but also play a crucial role in the formation (Kucukdereli et al., 2011) and elimination of synapses (Chung et al., 2013; Stevens et al., 2007; Tasdemir-Yilmaz and Freeman, 2014), as well as modulating the functioning of synapses through the release of gliotransmitters (Perea et al., 2009), formation and maintenance of the blood-brain barrier (BBB; Qosa et al., 2016; Venkatesan et al., 2015), regulation of coordinated firing of neurons (Chever et al., 2016) and metabolic support of neurons. In addition, astrocyte dysfunction has been implicated in many developmental and neurodegenerative diseases, including autism, epilepsy, Alzheimer's disease (AD), amyotrophic lateral sclerosis (ALS) and Alexander disease (Haidet-Phillips et al., 2011; Jo et al., 2014; Messing et al., 2012; Molofsky et al., 2012; Orre et al., 2014).
Astrocytes in different regions of the mouse central nervous system (CNS) exhibit distinct gene expression profiles and potentially also differential functions, including the expression of glutamate transporters, synaptic modulating genes and support of different neuronal subtypes (Bachoo et al., 2004; Morel et al., 2017; Regan et al., 2007). How these traits are acquired is not entirely clear. Knockout or forced expression of the basic helix-loop-helix (bHLH) transcription factor stem cell leukemia (SCL; also known as Tal1) regulates the generation of the S100β+ cells in the embryonic spinal cord (Muroyama et al., 2005). Similarly, mis-expression of neuronal transcription factors Pax6, Olig2 and Nkx2-2 disrupts the localization (migration) of reelin-expressing (dorso-lateral) and Slit1-expressing (ventro-medial) astrocytes (Hochstim et al., 2008). In addition, knockout of NKX6-1 in glial fibrillary acidic protein (GFAP)-expressing cells during development results in a lower number of astrocytes only in the ventral regions of the spinal cord (Zhao et al., 2014). These results suggest a role of transcriptional code for position-restricted astrocyte specification. Recent studies suggest that neighboring cells, including neurons, may instruct astrocytes to adopt a particular phenotype (Farmer et al., 2016). Thus, both developmental origins and interactions with neighboring cells contribute to the phenotypes of regional astrocytes. The functional properties of the regional astrocytes, however, remain largely unknown.
Studies of astrocyte heterogeneity have been conducted using mice. Although these studies are important, comparisons of mouse and human astrocytes have found meaningful differences. Human cortical astrocytes are nearly three times larger than their mouse counterparts and extend roughly ten times the number of processes (Oberheim et al., 2009). The ratio of astrocytes to neurons is also increased in humans to approximately 1.65 astrocytes for every neuron, making astrocytes the primary cell type of the human cortex (Nedergaard et al., 2003). In addition to their size, there are functional differences between mouse and human astrocytes. Chimerization of the mouse forebrain at postnatal day (P)0 with human astrocytes results in adult mice that have increased synaptic transmission in the hippocampus and are able to learn faster than their allografted littermates (Han et al., 2013). These studies give a rationale for the investigation of human astrocyte subtypes and their role in the development and pathogenesis of the human brain. Transcriptomic analysis of fetal and adult human astrocytes has been instrumental for increasing our understanding of human astrocyte development but has necessarily lacked analysis of regional subtypes (Zhang et al., 2016). Studies of regional astrocytes using human samples have focused on marker gene expression and have lacked functional analysis (Oberheim et al., 2012). In addition, these studies do not allow for the determination of the impact of developmental specification on regional astrocyte heterogeneity.
We have shown that astrocytes can be generated from human embryonic stem cells (ESCs) without contamination by neurons, oligodendrocytes or microglia (Krencik et al., 2011; Krencik and Zhang, 2011). This system allows for the specification of truly naïve astrocyte progenitors and, therefore, the study of developmental heterogeneity in isolation without the potential regionalization that other cell types, including neurons, may impart. In the present study, we show that naïve astrocytes with dorsal and ventral forebrain or dorsal and ventral spinal characteristics can be generated from human pluripotent stem cells (hPSCs). We show that regional astrocytes not only carry region-specific gene expression profiles, but also differential cell type-specific properties and functional attributes, suggesting that developmental origins instruct astrocyte phenotypes.
RESULTS
hPSC-differentiated astrocytes carry regional markers
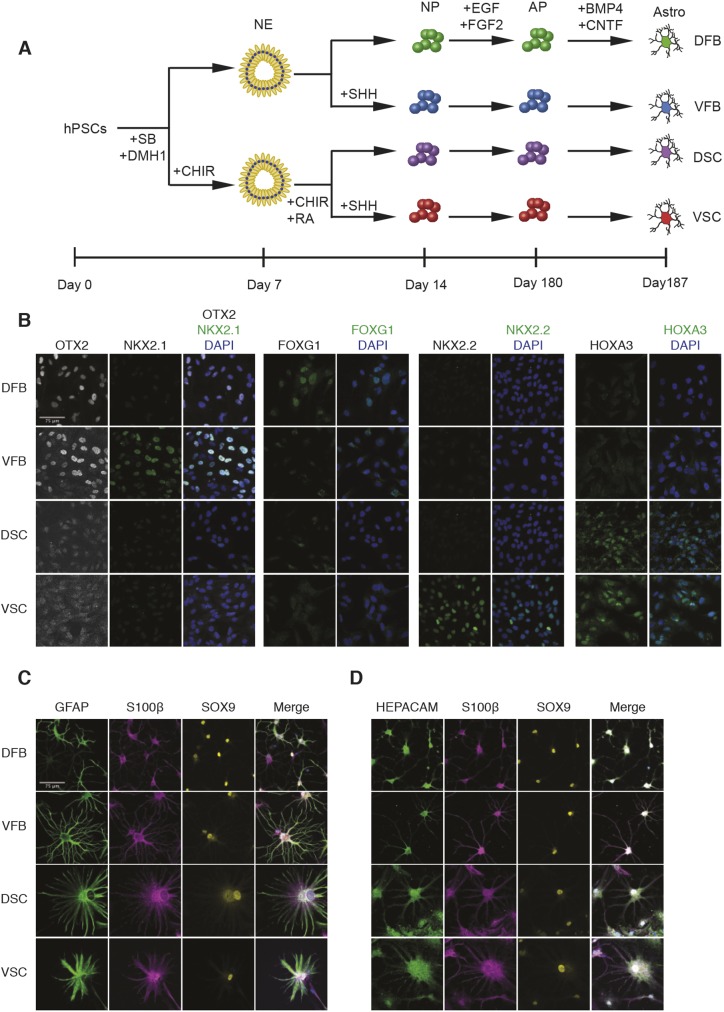
Neurons exhibit molecular and functional traits that are specific to their CNS region. As the same regionally specified neural progenitors give rise to glial cells after neurogenesis (Freeman, 2010; Molofsky and Deneen, 2015), we hypothesized that the regional identity of astrocytes is established during progenitor specification. Regionally defined astrocytes were generated from hPSCs according to our previous established protocols (Fig. 1A) (Krencik et al., 2011; Krencik and Zhang, 2011). hPSCs were first specified to dorsal and ventral progenitors in the absence (for dorsal) or presence (for ventral) of sonic hedgehog (SHH) signaling. Patterning to spinal cord fate was accomplished using the WNT agonist CHIR99021 and retinoic acid (RA) without (for dorsal spinal cord, DSC) or with an SHH agonist (for ventral spinal cord, VSC). These progenitors were expanded for 5.5 months under the same culture conditions before maturation to functional astrocytes for one week in the presence of BMP4 and CNTF (Fig. 1A). The regional identity of the neural progenitors at day 60 was confirmed via immunostaining for transcription factor markers of each region (Fig. 1B). Dorsal forebrain (DFB) and ventral forebrain (VFB) progenitors strongly stained for the forebrain marker OTX2. Only VFB progenitors expressed the medial ganglionic eminence marker NKX2-1. The DFB group was the only group to strongly express the cortical marker FOXG1, though weak staining can be observed in the VFB group as some progenitors express FOXG1 (Manuel et al., 2010; Roth et al., 2010). Both the DSC and VSC progenitors expressed the cervical spinal cord marker HOXA3, which is not found in the forebrain groups. Only VSC progenitors stained strongly for the ventral spinal cord marker NKX2-2 (Fig. 1B).
Fig. 1.
Differentiation and gene expression of region-specific astrocytes. (A) Schematic showing the process of subtype astrocyte differentiation. Human PSCs were induced to neuroepithelia (NE) in the presence of DMH1 and SB431542 (SB) in the first week, followed by patterning to region-specific progenitors in the second week in the presence or absence of morphogens. The neural progenitors (NP) were then expanded in suspension for 5.5 months before differentiating to astrocytes (Astro) in an adherent culture in the presence of BMP4 and CNTF for 1 week. AP, astrocyte progenitor. (B) ESC-derived regional neural progenitors (60 days) were stained for forebrain (OTX2), ventral forebrain (NKX2-1), cortical (FOXG1), ventral spinal cord (NKX2-2) and cervical spinal cord (HOXA3) markers. (C) Representative images of ESC-derived regional astrocytes that were stained for S100β, SOX9 and GFAP or HEPACAM. Cell nuclei were stained for Hoechst (blue). Scale bars: 75 μm.
Over 80% of cells in all regions expressed GFAP, with the exception of VSC astrocytes, in which 70-80% of cells express GFAP (Fig. S1). As not all astrocytes express GFAP (Sofroniew and Vinters, 2010; Walz and Lang, 1998), we stained our cultures with S100β and Sox9, markers of progenitors and mature astrocytes, as well as a recently identified mature astrocyte marker HEPACAM. These markers were expressed in nearly all cells (Fig. 1C,D, Fig. S1). We found that, although not all astrocytes expressed GFAP, the expression of S100β, Sox9 and their characteristic stellate morphology marked them as being definitive astrocytes (Fig. S2). This confirms observations of the heterogeneous expression of GFAP in astrocytes in mammalian brains (Kofler et al., 2002; Walz and Lang, 1998; Xu, 2018). Astrocytes differentiated from all regional progenitors exhibited a characteristic stellate morphology, though spinal cord astrocytes were larger in size and exhibited a slightly more rounded morphology (Fig. 1C, Fig. S3). In addition, we found virtually no expression of markers of neurons [neurofilament, heavy polypeptide (NEFH) and neuron-specific enolase (NSE; ENO2)], pluripotent cells (NANOG) or proliferating progenitors (KI-67; MKI67) in any regional astrocyte population (Fig. S4, Fig. S5).
These data show that the specification of the early progenitors is accurate to their anatomical region, and the final regional astrocyte populations are largely devoid of neurons and any proliferating progenitor cell types and display the characteristic morphology and gene expression.
RNA-seq reveals unique molecular profiles of regional astrocytes
Little is known about the molecular signature of regional human astrocyte subtypes. We performed an unbiased RNA-seq of each regional astrocyte population. Pearson correlation analysis revealed high correlation of gene expression among replicate samples within each region and differences between the regional populations (Fig. 2A). Both shared and uniquely expressed genes in different astrocyte types were identified (Fig. 2B). Gene expression in the two forebrain groups differed the most from the other regions, whereas the gene expression signatures of the two spinal cord regions had fewer differentially expressed genes (DEG; Fig. 2B). As expected, a large number (9283) of genes were expressed by all the subtypes of astrocytes (Fig. 2B). Astrocytes from all regions expressed genes associated with the astroglial lineage, including NFIA, NFIX, SOX9, CD44, vimentin, SLC1A3 (GLAST), SLC1A2 (GLT1), GJA1 (CX43), NOTCH1, GFAP and AQP4 (Table S1). None of the regional astrocyte groups expressed genes associated with neurons [NEUN (RBFOX3), NEUROD1, NEUROD2, NEFH] or oligodendrocytes [MAG, MBP, SOX10, GJC2 (CX47)] (Table S1). The astrocyte populations do express PDGFA and CSPG4 (Table S1) that are regarded as markers of oligodendroglia progenitor cells (OPCs). These genes are also expressed in human primary astrocytes and are not as specific for OPCs in humans as in mice (Zhang et al., 2016). Taken together, these results further confirm the highly enriched astrocyte identity of the cells.
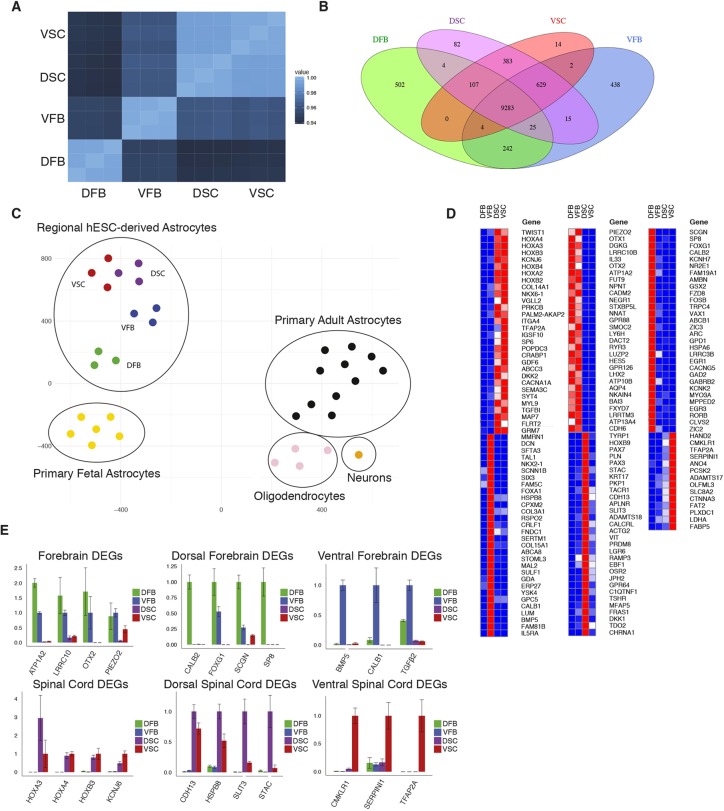
Fig. 2.
Differentially expressed genes in ESC-derived regional astrocytes. (A) Pearson correlation analysis showing the similarity of total RNA-seq data for all samples from each region (three biological replicates per region). (B) Venn diagram showing the number of genes that are differentially expressed between groups or combinations of groups. DEGs are defined as having expression >100 TPM and >2-fold over comparison groups. (C) t-SNE plot showing RNA-seq of primary human fetal and adult CNS cells with hESC-derived regional astrocytes. hESC-derived regional astrocytes are labeled by region. The top 1000 overdispersed genes were used for analysis. (D) Heatmap showing top 30 DEGs (14 DEGs in the VSC group) in each group of astrocytes (n=3 biological replicates). (E) qPCR of RNA-seq identified regional DEGs in iPSC-derived regional astrocytes (n=3 technical replicates). Error bars represent s.d.
Recently, Zhang et al. (2016) reported gene expression profiles of primary human glial cells and neurons from both fetal and adult brains. t-SNE analysis of the combined data from their primary cells with our human ESC (hESC)-derived astrocytes revealed that our ESC-derived astrocytes were most similar to primary fetal astrocytes and most distant to oligodendrocytes and neurons (Fig. 2C). Interestingly, the DFB astrocytes clustered the closest to the primary human fetal astrocytes, followed by the VFB group, with the spinal cord astrocytes clustering the furthest away. Cells collected for analysis by Zhang and colleagues were derived from the cortex. This analysis further confirms the astrocyte identity of our hESC-derived cells and similarity to fetal rather than adult astrocytes.
We further sought to corroborate the regional identity of our astrocytes by comparing our transcriptome data with those available from other sources. By comparing our data with the Allen Brain Atlas transcriptome database we found that the dorsal forebrain samples had a high correlation with the developing human telencephalon and cortex (Table S2). The ventral forebrain samples had a gene expression pattern similar to that of the lateral ganglionic eminence (LGE) and medial ganglionic eminence (MGE) as well as the developing diencephalon (Table S3). Therefore, the uniquely or differentially expressed genes in our regionally specified astrocytes in vitro coincide with the regions of the brain to which the astrocytes belong in vivo. Unfortunately, a comparable transcriptome database of the developing human spinal cord does not exist so similar analysis was not possible for these regions. To further analyze gene expression in regional astrocytes we compared DEGs between the mouse cortex and hypothalamus, regions that correspond most closely to the DFB and VFB groups, in two studies (Morel et al., 2017 and Boisvert et al., 2018). We found that many of the most differentially expressed genes between these regions were also differentially expressed in the DFB and VFB groups, confirming the in vivo relevance of these cells (Fig. S6).
Many of the DEGs that diverged in regional astrocytes were associated with regional CNS development; some of these were shown in the immunostaining pattern of early progenitors (Fig. 1B). The forebrain markers OTX1 and OTX2 were found in both forebrain regions, corroborating our immunofluorescence. Six of the top ten DEGs in the spinal cord were HOX family members that define locations along the anterior-posterior axis of the spinal cord. Between the dorsal and ventral forebrain cells, genes encoding cortical markers, especially FOXG1, were among the highest differentially expressed in the DFB astrocytes. In contrast, NKX2-1, FOXA1 and SIX3, which are associated with the development of ventral forebrain and the rostral portion of the ventral diencephalon, were enriched in VFB astrocytes (Fig. 2D). In the caudally specified astrocytes, HOX genes, including HOXA4, HOXB4 and HOXA3 were highly enriched, suggesting that the astrocytes are patterned to the cervical regions of the spinal cord and the hindbrain. RA and a high concentration of CHIR99021 are known to restrict cells to the hindbrain and anterior spinal cord fate (Sances et al., 2016; Xi et al., 2012). As previously noted, the differences within the spinal cord samples were much less pronounced than within the forebrain samples. Still, the dorsal spinal cord markers PAX3 and PAX7 were expressed only in the DSC astrocytes, whereas the ventral spinal cord marker NKX2-2 was expressed preferentially in VSC samples over DSC (Fig. 2D).
Analysis of the top 30 DEGs from the different regions using a pairwise Pearson correlation revealed that the dorsal and ventral forebrain samples had the largest degree of dissimilarity among the samples, with the dorsal forebrain (cortical) being the most unique (Fig. S7). The two spinal cord samples had a very high degree of correlation and the genes unique to the forebrain and the spinal cord had a very low level of correlation (Fig. S7). DEGs expressed highly in each region were confirmed in iPSC-derived regional astrocytes by qPCR (Fig. 2E). The differential gene expression suggests that the regional specification of neuroepithelial cells influences the gene expression of astrocytes derived from these cells.
The molecular profiles revealed expression of genes that have not been studied in astrocytes (Table S4) though their expression in astrocytes can be confirmed in other human astrocyte transcriptomic studies (Zhang et al., 2016). The reticulon 4 gene, which encodes the NOGO protein expressed by neurons during development and by oligodendrocytes in the adult brain, was expressed by all regional astrocytes, indicating a broad role for this protein in CNS development (Gil et al., 2006; Schwab, 2010). Upregulation of NOGO-A in hippocampal neurons is also associated with temporal lobe epilepsy (Bandtlow et al., 2004). Similarly, glycoprotein M6A (GPM6A), expressed in mouse neurons and known to be involved in anxiety, depression and claustrophobia (Boks et al., 2008; El-Kordi et al., 2013), was highly expressed in all astrocyte groups. HSPA8, a cognate heat-shock chaperone and ATPase involved in the disassembly of clathrin-coated vesicles, and heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), important for mRNA alternative splicing, metabolism and transport, were both expressed in all regional astrocytes. Both these molecules are expressed in neurons and are implicated in a number of neuropsychiatric and neurological disorders (Bozidis et al., 2014; Honda et al., 2015). The robust expression of genes that have previously been attributed to neurons suggests potentially novel roles of astrocytes in the neuropathology in these disorders.
Analysis of DEGs in regional astrocytes also yielded clues to their potential involvement in disease. Several genes enriched in forebrain (DFB and VFB) astrocytes have been linked to diseases and conditions that affect higher-order functions. ATP1A2, enriched in both DFB and VFB samples (Fig. 2D, Table S5), has been linked to epilepsy (Deprez et al., 2008). The DFB astrocytes had DEGs associated with behavioral disorders and drug addiction (FOSB and ARC) (Murphy et al., 2003), and schizophrenia (EGR1, EGR3 and ARC) (Duclot and Kabbaj, 2017; Huentelman et al., 2015). Ventral forebrain astrocytes had higher expression of genes that have been associated with the hippocampus and Alzheimer's disease, including UNC5C, A2M and CLU (APOJ) (Blacker et al., 1998; Lambert et al., 2009; Wetzel-Smith et al., 2014) (Table S5). Spinal astrocytes expressed a number of genes associated with the development and disorders of the spinal cord, including a potassium channel KCNJ6 (KIR3.6), a voltage-gated calcium channel CACNA1A (causing spinocerebellar ataxia type 6) (Tonelli et al., 2006), and GDF6 (causing Klippel-Feil syndrome) (Chacón-Camacho et al., 2012) (Fig. 2D, Table S5). The differences in expression of the functionally related genes suggest potential involvement of astrocyte subtypes in the pathogenesis of different diseases and the importance of including glial cell types in studies of neurodevelopmental and neurodegenerative diseases.
Pathway analysis suggests differential functional properties of astrocyte subtypes
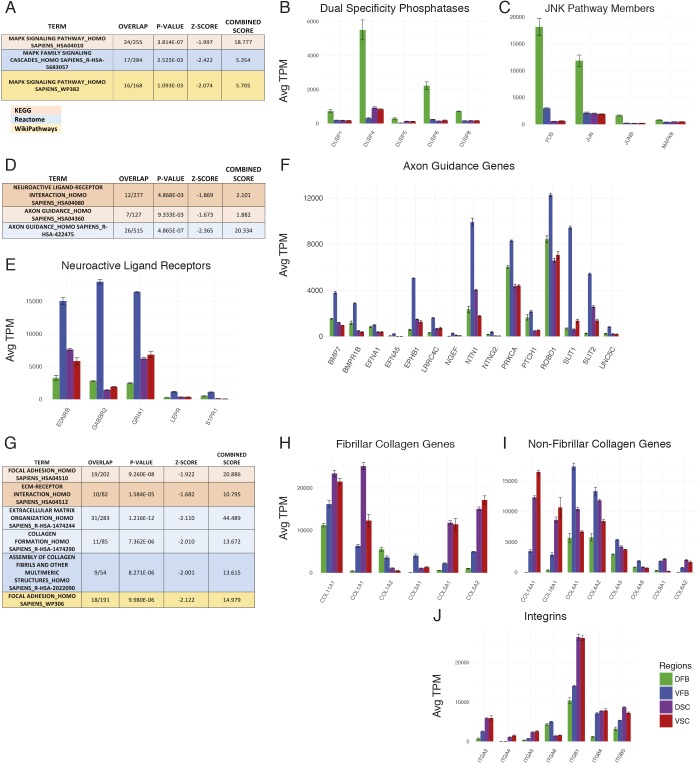
Three pathway databases were used to improve the accuracy of the analysis, and only pathways that were found differentially expressed in at least two of the databases were considered for further investigation. Pathway analysis of DFB astrocytes indicated that the MAPK pathway was the most differentially regulated (Fig. 3A, Fig. S8). Expression of genes within a signaling pathway alone is not sufficient to draw conclusions as to the activation status of the pathway. By analyzing the expression level of upstream regulators of the pathway, we found that several members of the dual specificity phosphatase (DUSP) gene family known to target different parts of the MAPK pathway, including DUSP1, DUSP4, DUSP5 and DUSP6 were highly expressed in the DFB group compared with other astrocyte types (Fig. 3B, Fig. S8). This expression pattern suggests that the ERK1/2 or p38 pathways may be differentially regulated. We also found the expression of JNK-Jun pathway target genes was highest in the DFB group (Fig. 3C, Fig. S8), suggesting that the MAPK pathway is potently active in the DFB group.
Fig. 3.
RNA-seq pathway analysis. (A) Analysis of the differentially utilized pathways in the DFB group; boxes show the MAPK pathway found in all analyses. (B) Expression of dual specificity phosphatase (DUSP) genes. (C) Expression of genes in the JNK/Jun pathway. (D) Analysis of the differentially utilized pathways in the VFB group; boxes show the axon guidance pathway and related neuroactive ligand receptor pathway found in KEGG and Reactome analyses. (E) Expression of selected genes in the neuroligand receptor pathway (KEGG HSA04080). (F) Expression of selected genes in the axon guidance pathway (KEGG HSA04360). (G) Analysis of the differentially utilized pathways in the spinal cord group; boxes show pathways involving extracellular matrix (ECM) function in all analyses. (H,I) Expression of fibrillary (H) and non-fibrillar (I) collagen genes. (J) Expression of integrin receptor genes. All gene expression values are average TPM (n=3 biological replicates of ESC-derived astrocytes for each group). Error bars represent s.d. of TPM.
In the VFB astrocytes, the most striking pathways are neuroligand receptors and axonal guidance/repellence (Fig. 3D, Fig. S7). The GABA receptor gene GABBR2 is highly expressed only in VFB astrocytes, along with genes encoding receptors for endothelin (EDNRB) and leptin (LEPR) (Fig. 3E, Fig. S8). Several genes in the axon guidance pathway were highly expressed only in VFB astrocytes, including members of the Ephrin/Eph pathway, which is crucial in neuron-astrocyte communication at the synapse (Murai and Pasquale, 2011) (Fig. 3F, Fig. S8). SLIT1 and SLIT2 were also highly expressed in the VFB astrocytes and are ligands for the ROBO family of axonal guidance receptors. These molecules are important in inducing neurite growth and mediating midline crossing of projection neurons during development (Bagri et al., 2002). VFB astrocytes also strongly expressed NTN1 (Fig. 3F, Fig. S8) that can increase neurite outgrowth of commissural axons while repelling other axons (Bloch-Gallego et al., 1999). The expression of genes associated with neurite outgrowth and guidance in enriched populations of astrocytes (without neurons) suggests that VFB astrocytes have an innate role in the migration and development of neurons in this region.
The most notable differentially expressed pathways identified in the spinal cord astrocytes involved extracellular matrix and focal adhesion (Fig. 3G). Spinal cord astrocytes had high expression of several collagen genes, including fibrillary and non-fibrillary collagens (Fig. 3H,I, Fig. S8). In addition, spinal cord astrocytes also expressed several integrin subunits that act as receptors for collagen, including α3 and α11 which, in concert with β1, act as collagen receptors (Fig. 3J, Fig. S8). The expression of several collagen genes was more prevalent in astrocytes of the forebrain including COL1A2 and COL3A1, which is expressed only in the VFB group. Additionally, the α6 integrin receptor was only expressed in the forebrain, highlighting the diversity of expression in collagens and integrins between regional astrocytes. The fact that spinal astrocytes express high levels of collagens and integrins suggests that astrocytes may be involved in the response to spinal cord injury, so they may be a crucial target for intervention as collagens are important in inducing repair after spinal cord injury (Hatami et al., 2009; Klapka and Müller, 2006).
Regional astrocytes display differential physiological properties
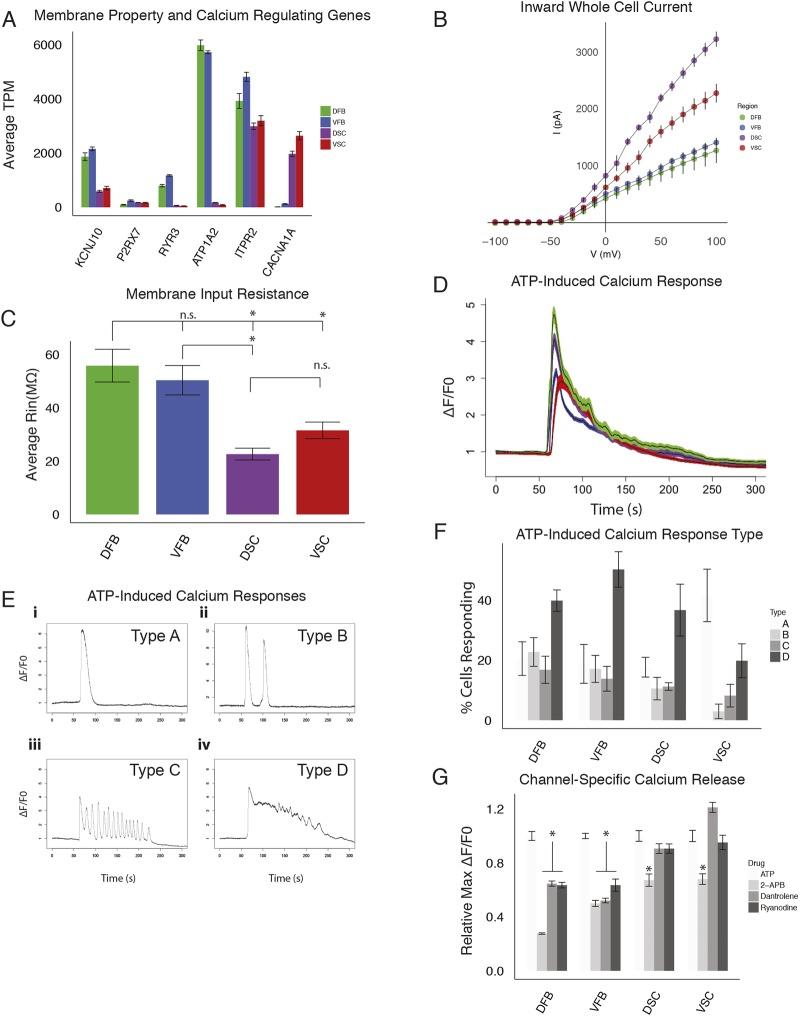
Astrocytes express inward-rectifying ion channels that endow astrocytes with a unique membrane property. Whole-cell patch clamping showed that spinal cord astrocytes had much larger whole-cell currents than those of the forebrain (Fig. 4B). DSC astrocytes had 3× the current of forebrain astrocytes and 1.5× that of the VSC group (Fig. 4B). The membrane resistance of forebrain astrocytes was 55.88±6.15 MΩ (DFB; data are mean±s.e.m.) and 50.42±5.52 MΩ (VFB), significantly higher than that in spinal cord astrocytes (22.66±2.21 MΩ for DSC and 31.58±3.11 MΩ for VSC) (Fig. 4C). The lower membrane resistance and higher inward current of the spinal cord astrocytes are likely due to the voltage-gated channel expression. We found a particular calcium channel (CACNA1A) to be highly expressed only in the spinal cord astrocytes (Fig. 4A). In addition, several membrane-bound Na+/K+ and H+/K+-ATPases of the ATP1 and ATP2 family of genes have much higher expression in forebrain over spinal cord astrocytes (Fig. 4A, Fig. S8). These data demonstrate a difference in the basic membrane properties of astrocytes between different regions.
Fig. 4.
Physiological properties of astrocyte subtypes. (A) ESC-derived astrocyte RNA-seq expression of calcium signaling- and membrane polarization-related genes in astrocyte subtypes (n=3 biological replicates, error bars represent s.d. of TPM). (B,C) I-V curves for subtype astrocytes measuring inward whole-cell current (B) and membrane resistance of astrocyte subtypes (C) (n=3 biological replicates, six cells measured per replicate, 18 total per region patched; *P<0.05, Student's t-test). (D) Average response (black line) and s.d. (colored bars) of ATP-induced calcium release for astrocyte subtypes visualized with Fluo-4AM (n=3 biological replicates totaling >50 analyzed cells per region). (E) Examples of characteristic responses of astrocytes to ATP visualized with Fluo-4AM. The four characteristic intracellular calcium release responses to ATP identified in all regional astrocyte populations are shown. (F) Percentage of cells exhibiting the calcium oscillations shown in panel E (n=3 biological replicates totaling >50 cells analyzed, error bars represent s.d.). (G) Maximum change in fluorescent intensity in the presence or absence of 2-APB (50 μM), dantrolene (10 μM) or ryanodine (50 nM) after the addition of ATP (n=3 biological replicates totaling >50 cells analyzed, error bars represent s.d.; *P<0.01, Student's t-test). n.s., not significant.
Intracellular calcium release in response to ATP stimulation in astrocytes regulates synaptic plasticity, gliotransmitter release and BBB function (Bazargani and Attwell, 2016; Khakh and McCarthy, 2015). Astrocytes from all regions exhibited a characteristic spike in intracellular calcium when ATP (10 μM) was applied, as indicated by intracellular calcium response using the calcium-sensitive dye Fluo-4AM (Fig. 4D). Interestingly, the astrocytes exhibited four distinct and consistent responses (which we have termed types A-D) based on the number and frequency of spontaneous calcium oscillations and the refractory period to baseline. Type A represents a single wave spike with a quick refractory period (Fig. 4Ei). Type B exhibits slow oscillations with multiple spikes as in Type A, with a complete return to baseline calcium levels before another calcium spike (Fig. 4Eii). Type C exhibits a fast oscillation response with multiple waves of calcium release of similar amplitude without a return to baseline levels (Fig. 4Eiii). Finally, Type D is similar to type A, with a long refractory period for each calcium release with overall downward trend in signal (Fig. 4Eiv). The regionally distinct astrocytes had broadly similar proportions of cells exhibiting each type of response, with the exception of the VSC group which had significantly more ‘A’ type waves (Fig. 4F). These results show that regional astrocytes exhibit characteristic intracellular calcium responses to stimulation with ATP.
Calcium release is regulated by the calcium channels in the endoplasmic reticulum (ER), including the ryanodine and inositol 1,4,5-triphosphate (IP3) receptors. The ryanodine receptors (RYR) regulate calcium release from the ER in several cell types, including skeletal and cardiac muscle, epithelial cells, neurons and astrocytes (Lanner et al., 2010). Of the three genes in the RYR family, skeletal and cardiac muscle express RYR1 and RYR2, respectively, whereas astrocytes express RYR3 (Lanner et al., 2010; Shigetomi et al., 2016). Our transcriptome analysis revealed that RYR3 is expressed in forebrain astrocytes but it is nearly absent in spinal cord astrocytes (Fig. 4A, Fig. S8). To test whether this expression has an effect on the calcium dynamics of regional astrocytes we inhibited the ryanodine receptor using the small molecule antagonists ryanodine (50 nM) and dantrolene (10 μM) in the presence of ATP. Inhibition of RYR3 in forebrain astrocytes results in a marked decrease in maximum calcium release, whereas there was no effect on spinal cord astrocytes (Fig. 4G). In contrast, the IP3 receptor ITPR2 was expressed in all of our astrocyte types (Fig. 4A). Expectedly, treatment of astrocytes with the IP3 receptor inhibitor 2-APB (50 μM) reduced the maximum calcium signal in all groups in the presence of ATP. These data demonstrate the functional consequence of the regionally specific expression of RYR3.
Regional astrocytes exert differential effects on neurons and endothelial cells
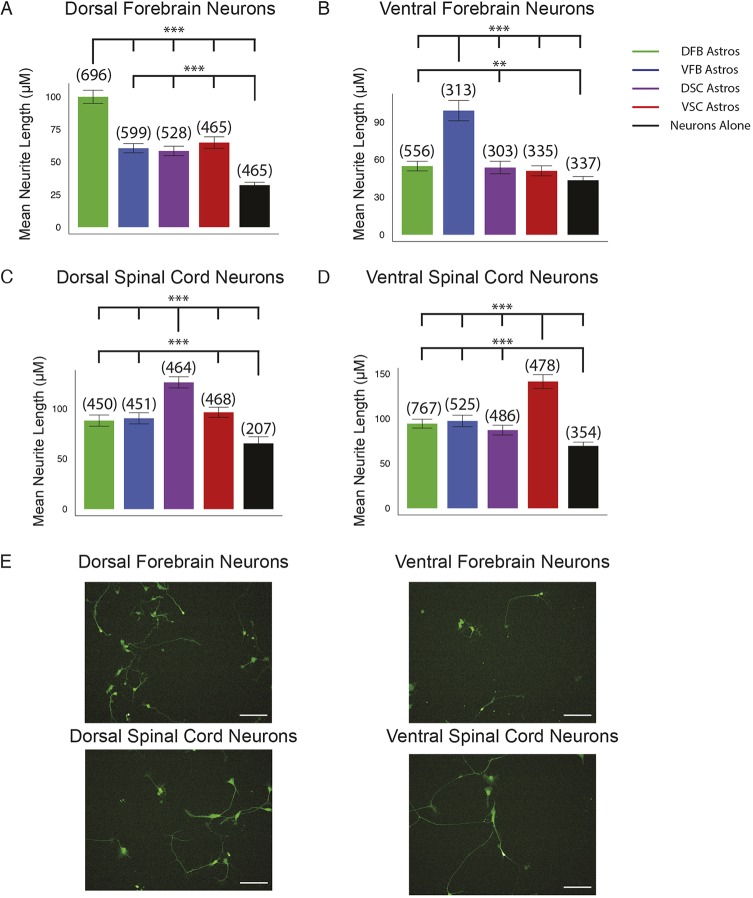
Astrocytes are important for neurite outgrowth. When regionally specified GFP-labeled human neurons were co-cultured with the four regional astrocyte groups individually for three days, neurite outgrowth in the co-cultures was always faster than in neurons cultured alone. However, regional neurons had faster neurite outgrowth on the matched regional astrocytes over non-matched regional astrocytes and neurons alone, although they exhibited normal neuronal morphology (Fig. 5). One exception is VFB neurons, which had no significant difference in neurite length on VSC astrocytes compared with neurons alone. It should be noted that VFB neurons are GABAergic interneurons as opposed to the projection neurons found in the other regions and this likely partially accounts for the smaller difference (Fig. 5B). The effect of the regional astrocytes on neurite length was not limited to neurites of certain length and increased the length of all neurites proportionally (Fig. S9). This result indicates that although there are likely broadly applicable mechanisms for astrocyte-induced neuronal maturation, each regional astrocyte promotes the maturation and growth of regionally matched neuronal subtypes.
Fig. 5.
Neurite outgrowth of regional neurons in co-culture. (A-D) The length of neurites from regionally specified ESC-derived neurons in co-culture with the indicated ESC-derived regional astrocytes measured after 3 days in co-culture or plated alone. The length of the longest neurite of each neuron was measured. Bar graph is of average neurite length. Error bars represent 95% confidence interval. Numbers in parenthesis represent total number of neurites counted for each group over three biological replicates. (**P<0.01, ***P<0.001, Kruskal–Wallis test). (E) Representative images of regional neurons plated on matched regional astrocytes. Scale bars: 100 μm.
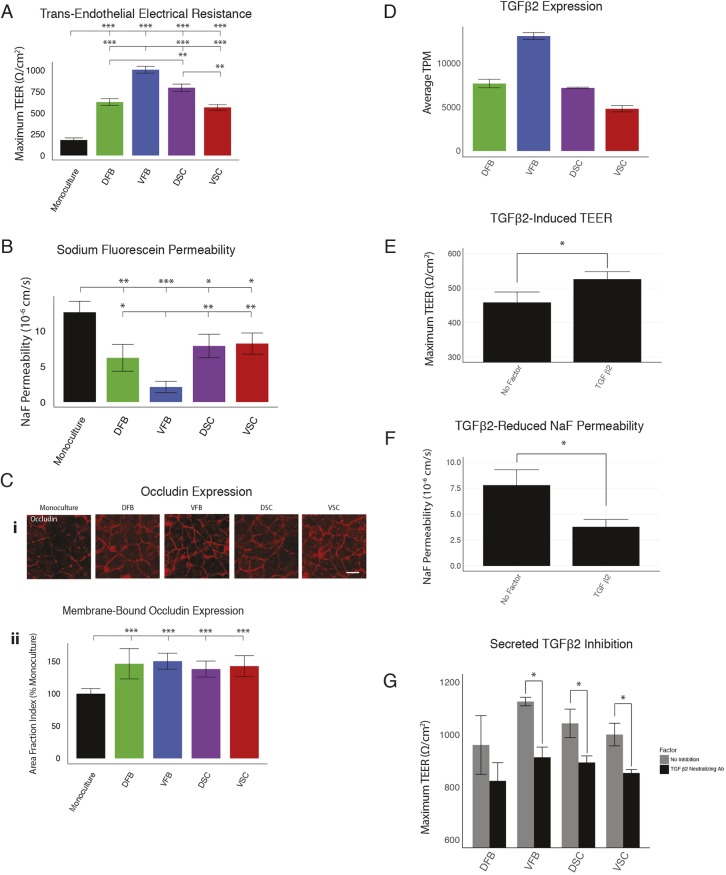
Formation and support of the BBB is one of the most important astrocyte functions. We first assessed barrier tightening by measuring trans-endothelial electrical resistance (TEER) across the human iPSC-derived brain endothelial cell monolayers (brain microvascular endothelial cells; BMECs) after co-culture with astrocytes (Canfield et al., 2016). All astrocyte sub-types increased the TEER significantly compared with the control group (without astrocytes), indicating that all astrocytes have the capability to induce barrier properties in BMECs. Among the astrocyte types, VFB astrocytes induced the highest TEER (1050 Ω/cm2), significantly higher than all of the other regional astrocyte groups (Fig. 6A). We assayed the BMEC permeability to a small hydrophilic solute, sodium fluorescein. Barrier induction should be correlated with more restricted diffusion of fluorescein across the BMEC monolayer. As with the TEER experiment, the fluorescein permeability was the lowest in the group co-culturing with VFB astrocytes (2.12×10−6 cm/s, Fig. 6B) and was significantly different than the permeability of each of the other groups. Correspondingly, the remaining astrocyte groups all showed higher and indistinguishable fluorescein permeability (DFB=6.22×10−6 cm/s, DSC=7.89×10−6 cm/s, VSC=8.22×10−6 cm/s) (Fig. 6B). Commensurate with the increased BMEC barrier function, quantitative analysis of the localization of the tight junction protein occludin indicated that astrocyte co-culture significantly increased occludin localization to the cell-cell junctions (Fig. 6C).
Fig. 6.
Effects of astrocyte subtypes on BBB. (A,B) Analysis of astrocyte-induced tight junction formation using TEER (A) and analysis of BBB permeability of sodium fluorescein across human iPSC-derived BMEC monolayers (B) with or without ESC-derived astrocyte co-culture (50,000 astrocytes per well, n=3 biological replicates; ***P<0.0001, **P<0.01, *P<0.05, Tukey's test). (C) Images of occludin immunostaining (i) and measurement of occludin membrane localization by quantification of area fraction index (ii) relative to monoculture (n=18 images analyzed over three biological replicates). (D) Expression of TGFβ2 as measured by RNA-seq (average TPM of ESC-derived astrocytes, three biological replicates, error bars represent s.d. of TPM). (E,F) TEER (E) and sodium fluorescein permeability (F) measurements of BMEC monoculture with the addition of recombinant TGFβ2 (n=3 biological replicates per group; *P<0.05, Student's t-test). (G) TEER measurements of BMEC co-cultured with regional astrocytes with or without the addition of a neutralizing antibody to TGFβ2 (100,000 astrocytes per well, n=3 biological replicates; *P<0.05, Student's t-test). Scale bars: 50 μm.
To identify potential factors responsible for the differential effects on BBB tightening, we searched our transcriptome data for secreted factors. We found a member of the TGFβ family, TGFβ2, as being expressed differentially among the astrocyte types and qualitatively mapping onto the TEER profiles (Fig. 6A,D, Fig. S8). When recombinant TGFβ2 was added to a monoculture of BMECs, TEER was increased and fluorescein permeability was reduced (Fig. 6E,F). When a neutralizing antibody for TGFβ2 was added to the co-cultures, TEER was significantly reduced in all the groups but DFB, which did not see a statistically significant reduction (Fig. 6G). In addition, the neutralization of TGFβ2 reduced the TEER levels to similar levels for all astrocyte types, suggesting that the difference in TGFβ2 expression is largely responsible for the differential barrier tightening observed for astrocytes subtypes. These results show that although all astrocyte types are able to induce BBB tightening, the extent of the tightening can vary between regions, and can be explained through the expression of secreted factors such as TGFβ2.
DISCUSSION
By generating astrocytes from regionally specified neural progenitors, we have shown that all astrocytes share common gene expression profiles that are characteristic of astrocytes, including typical astroglial genes and those that are less known to be associated with astrocytes. Astrocytes derived from regionally specified progenitors also exhibit distinct transcript profiles, including the predicted homeodomain transcription factors and transcripts that suggest differential functional properties. Functional analyses revealed differential membrane properties, intracellular calcium release and effects of the astrocyte subtypes on neurite growth and BBB formation. In particular, the intracellular calcium oscillation properties of and contribution to BBB tightening by regional astrocytes are associated with differential expression of calcium channels and use of cytokines, respectively. We propose that astrocyte regional identity and their functional properties are at least partly dependent on their developmental origins.
The in vitro astrocyte differentiation paradigm follows the developmental process (Krencik et al., 2011). It begins with neuroepithelial induction in the first week and regional patterning by morphogens in the second week, followed by expansion of progenitors and glial differentiation. Hence, the differentiation process is the same for all astrocyte subtypes, except when the neuroepithelia are treated with different morphogens to induce regional patterning. Yet, astrocytes differentiated from the regional progenitors carry gene expression profiles that correspond to the region of origin. The differential expression of regional markers, especially homeodomain transcription factors in astrocytes 6 months post-patterning suggests that specification of the regional progenitors in the second week of differentiation is sufficient to ‘lock in’ the regional identity of these progenitors throughout their life. This notion agrees with observations made in embryonic mouse forebrain and spinal cord, in which transcriptional codes in neural progenitors dictate the differentiation of certain groups of astrocytes (Hochstim et al., 2008; Muroyama et al., 2005). Loss of these homeodomain transcription factors can result in loss of astrocytes in only those specific regions (Zhao et al., 2014).
The differential expression of ion channels signals different properties of the astrocyte subtypes. Na+- and K+-ATPase that maintain the sodium and potassium gradients across the membrane, are highly expressed in forebrain astrocytes. Interestingly, ATP1A2 is highly expressed in the astrocytes isolated from the P1 and P2 neonatal mouse forebrain over more caudal regions (Yeh et al., 2009), similar to our stem cell-differentiated astrocytes. These ion channels are important in maintaining the membrane potentials and regulating the astrocytic response to stimuli. Indeed, electrophysiological analysis of the regional astrocytes revealed that the forebrain astrocytes exhibit higher resistance and lower inward currents than spinal astrocytes.
ATP-mediated intracellular calcium release is one of the crucial astrocyte functions to coordinate neuronal network activities in different brain regions. All regional astrocyte types are able to signal robustly with a characteristic calcium response when stimulated with ATP, with the exception of VSC astrocytes that exhibit much less spontaneous intracellular calcium oscillations. This pattern is strikingly similar to that observed in rat primary astrocytes, which show A, B and D types of waves, though there was no report of type C waves (Vaarmann et al., 2010). This phenomenon is consistent with the fact that all regional astrocytes had similar expression of the ATP receptor P2X7 and the ER-bound calcium uptake transporter SERCA2 (ATP2A2), though the VSC region did have increased expression of SLC8A2. SLC8A2 encodes the NCX2 protein, a cell membrane-bound sodium-calcium antiporter that is responsible for reducing intracellular calcium levels after transient increases (Khananshvili, 2013). On the other hand, our RNA-seq data revealed differential expression of RYR3 in regional astrocytes. The RYRs regulate calcium release from the ER in several cell types, including astrocytes. This suggests that regional astrocytes may respond to stimuli differently, thus regulating calcium signaling differentially. Indeed, inhibition of RYR3 using dantrolene and ryanodine results in a marked decrease in calcium release in forebrain but not spinal astrocytes when stimulated with ATP. Thus, regional astrocytes possess different molecular machineries to regulate calcium signaling.
Astrocytes promote neurite growth, at least in vitro. However, how regional astrocytes influence local neuronal growth is less well known (Bayraktar et al., 2015). Our systematic co-culture analysis revealed a somewhat surprising and intriguing phenomenon, i.e. although all astrocyte types support neurite growth, they tend to promote the growth from homotopic neurons better than heterotopic neurons. Similar results have been reported from primary mouse forebrain neurons and astrocytes, corroborating this finding (Morel et al., 2017). This phenomenon may be related to the differential expression of genes in regional astrocytes. Indeed, DFB astrocytes express higher levels of vascular endothelial growth factor (VEGF) and brain-derived neurotrophic factor (BDNF), which has been shown to increase neurite outgrowth and increase synaptic formation when co-cultured with cortical astrocytes (Buosi et al., 2017; Jin et al., 2006; Sang and Tan, 2003). VFB astrocytes strongly express a number of morphogens that influence neurite growth and growth cone pathfinding, including SLIT1 and SLIT2. VFB astrocytes also express high levels of fibroblast growth factor 1 (FGF1), also known as acidic FGF, which promotes the outgrowth of neurites (Tsai et al., 2015). Spinal cord astrocytes differentially express a large number of extracellular matrix proteins, including a wide variety of collagens and their associated integrin receptors, which are shown to facilitate motor neuron outgrowth in chick embryos (Varnum-Finney et al., 1995). It is likely that the homotopic neurons express corresponding receptors or ligands so that their neurite growth is enhanced. Future work is needed to systematically compare the gene expression profiles between neurons and astrocytes in a corresponding region. In addition, there are likely changes in gene expression in all regional astrocytes when co-cultured with neurons, which may further explain differences in their response to different regional neurons.
Astrocytes play a role in the formation and maintenance of the BBB (Abbott et al., 2006), partly through secreted factors (Engelhardt and Liebner, 2014). However, whether and how different types of regional astrocytes influence BBB development has not been directly studied in vitro. We found that, although all regional astrocytes are capable of inducing tightening of endothelial cells in a chamber co-culture model, VFB astrocytes exert the strongest effect on endothelial tightening. This differential effect may be associated with the varying levels of genes that are known to influence BBB formation, including TGFB2, which is highly expressed in VFB astrocytes. Inhibition of this secreted protein using a neutralizing antibody had the effect of reducing the TEER levels in all regions and eliminating the difference in TEER levels induced by the astrocyte subtypes. Changes in BBB permeability in specific brain regions under certain disease conditions have been reported. In AD and in aging brains, there is a substantial increase in the permeability of the BBB in the hippocampus that may contribute to or exacerbate the disease (Montagne et al., 2015). Whether regional astrocytes contribute to BBB alteration in pathogenesis remains to be seen.
Together, the substantial difference in gene expression profiles in astrocytes that are differentiated under the identical conditions from regional progenitors strongly suggests a developmental impact on astrocyte heterogeneity. Such molecular signatures likely underlie the various properties of astrocyte subtypes and their interaction with neurons. In the mature CNS, additional factors, especially the interaction of astrocytes with surrounding neurons, glia and blood vessels, will almost certainly further modify astrocyte diversity (Gomes et al., 2001; Sakimoto et al., 2012). Intrinsic molecular and functional astrocyte heterogeneity arising from their region of origin may contribute to the pathogenesis of neurological and psychiatric disorders in distinct brain regions as well as with neurodegenerative diseases (Molofsky et al., 2012; Phatnani and Maniatis, 2015). Indeed, our profiling analysis revealed genes that are known to be involved in various neurological and psychiatric conditions but not with astrocytes. Hence, different astrocyte types may be targets for therapeutic intervention.
MATERIALS AND METHODS
Astrocyte differentiation from hPSCs
hESCs (WA09, H9 line; RRID: CVCL_9773) and induced pluripotent stem cells [iPSCs; GM1-4, a control iPSC line reprogrammed from fibroblasts from an apparently healthy donor (GM00498) obtained from Coriell Cell Repository; RRID: CVCL_7290] were grown on Matrigel with Essential-8 medium (Stemcell Technologies) to 25% confluency. On day 0 of the differentiation, the hPSCs were cultured in the neural differentiation medium (NDM; DMEM/F12:Neurobasal 1:1+0.5× N2 supplement+0.5× B27+1 mM L-Glutamax) with the SMAD inhibitors SB431542 (Stemgent) and DMH1 (Tocris Bioscience) (both at 2 μM) with daily medium change for 7 days until formation of primitive neuroepithelia. Spinal cell differentiation also received the GSK3β antagonist CHIR99021 (Tocris Bioscience) (3 μM) from day 0. On day 8 the neuroepithelia were split 1:6 and patterned to their final regions. Forebrain progenitors were specified to dorsal forebrain progenitors with the addition of smoothened antagonist cyclopamine (Stemgent, 2 μM), or ventralized to ventral forebrain progenitors with the smoothened agonist purmorphamine (Tocris Bioscience, 500 nM) for 7 days with daily medium changes. Spinal progenitors were given RA (Tocris Bioscience) (100 nM) and the CHIR99021 concentration was reduced to 1 μM. Dorsal spinal progenitors were dorsalized with cyclopamine (2 µM) and ventral spinal progenitors ventralized with purmorphamine (500 nM) for 7 days with daily medium change. On day 14 neural progenitors were lifted from the Matrigel with 500 μM EDTA and resuspended in neurosphere media (NSM; DMEM/F12+1× N2 Supplement+1× non-essential amino acids) supplemented with fibroblast growth factor 2 (FGF2, 10 ng/ml). Neural progenitors were expanded until day 30 in NSM+FGF2, after which 10 ng/ml epidermal growth factor was supplemented to encourage the expansion of glial progenitors. After 180 days in culture, glial progenitors were plated on Matrigel and differentiated with NSM+BMP4 (Stemgent, 10 ng/ml) and CNTF (R&D Systems, 10 ng/ml) for 7 days.
Neuronal differentiation and maturation
Cortical neurons were derived from hESCs (WA09) following the above procedure for DFB astrocytes. At 21 days post differentiation, neuronal progenitors were plated in neuronal maturation media (NMM; NDM+10 ng/ml BDNF+10 ng/ml GDNF) for 10 days with 50% media changes every 3 days. Astrocytes which were analyzed for neuronal gene expression were cultured in astrocyte maturation media (NSM+10 ng/ml CNTF+10 ng/ml BMP4) for 10 days to allow for any potentially contaminating neuronal progenitors to mature.
Immunofluorescent staining and microscopy
Cells were fixed for 20 min with 4% paraformaldehyde in PBS at room temperature. Samples were blocked with 4% donkey serum and 0.2% Triton X-100 for 1 hour. Primary antibodies were diluted in 4% donkey serum and 0.1% Triton X-100 and applied to samples overnight at 4°C. Samples were washed with PBS, incubated with fluorescein-conjugated secondary antibodies for 1 hour at room temperature, and counterstained with Hoechst for 20 min. Samples were imaged on a Nikon A1s confocal microscope. See Table S6 for primary antibodies.
Automated imaging of astrocyte marker expression and morphology
Following fixation and staining, cells were imaged on a Perkin Elmer Operetta high content imager. Following imaging, cells were analyzed for marker expression as well as size and roundness using custom scripts on Perkin Elmer Columbus analysis software.
RNA-seq procedure
H9-derived astrocytes for RNA-seq analysis were cultured for 1 week with BMP4 and CNTF as described above. Samples were collected in biological triplicates. RNA was harvested using the RNeasy Plus Mini kit (Qiagen) following manufacturer's instructions. RNA quality was assessed using an Agilent RNA PicoChip with all samples passing quality control measurements. Sample libraries were prepared using poly-A selection using an Illumina TruSeq RNAv2 kit following the manufacturer's instructions.
Prepared libraries were sequenced for 101-bp single-read and performed on an Illumina HiSeq 2500 using 1×100 sequencing to a read depth of >25 million reads per sample by the University of Wisconsin-Madison DNA Sequencing Facility in the University of Wisconsin-Madison Biotechnology Center, USA. FastQC (RRID: SCR_014583) was performed on all samples with every sample passing all quality control measurements.
RNA-seq analysis
The empirical Bayes hierarchical modeling approach EBSeq (RRID: SCR_003526) was used to identify DEGs across two or more conditions. Median normalization technique of DESeq (RRID: SCR_000154) (Anders and Huber, 2010) was used to account for differences in sequencing depth. We evaluated the latent level of expression among the condition 1 (μ1; DFB; C1), condition 2 (μ2; VFB; C2), condition 3 (μ3; DSC; C3) and condition 4 (μ4; VSC; C4). Specifically, P1: μ1=μ2=μ3=μ4 (Equally expressed; EE); P2: μ1≠μ2=μ3=μ4 (DFB DEG); P3: μ2≠μ1=μ3=μ4 (VFB DEG); P4: μ4=μ1=μ2≠μ3 (DSC DEG); P5: μ1=μ2=μ3≠μ4 (VSC DEG).
EBSeq calculates the posterior probability (PP) of a gene being in each expression pattern. Genes were declared differentially expressed at a false discovery rate controlled at 100×(1−α) % if the PP of P1 (EE) is less than 1−α. Given this list of DEGs, the genes are further classified into each pattern and sorted by PP.
Over-dispersion and t-SNE analysis
All analyses were carried out using R (https://www.rproject.org) and RStudio (http://www.rstudio.com/) software. Over-dispersion and t-SNE analysis were conducted as previously described (Fan et al., 2016; van der Maaten and Hinton, 2008; Sloan et al., 2017). Briefly, over-dispersion was calculated as described in Fan et al. (2016) and in Sloan et al. (2017). The top 1000 over-dispersed genes between hESC-derived regional astrocytes and primary human cell data from Zhang et al. (2016) were selected. Dimensionality was reduced using t-SNE as described in van der Maaten and Hinton (2008) and in Sloan et al. (2017). For t-SNE, the R package tsne (van der Maaten and Hinton, 2008) was used. Clustering was performed using k-means and plotted using the ggplot2 package (Wickham, 2009).
Pathway analysis
DEGs from each group as well as combined forebrain and spinal cord groups were analyzed for differentially regulated pathways using ENRICHR (RRID: SCR_001575) (http://amp.pharm.mssm.edu/Enrichr/, accessed 5/30/2016) (Chen et al., 2013; Kuleshov et al., 2016) which uses several pathway databases for general pathway analysis. For our analysis, we used the KEGG, Reactome and Wikipathway databases. DEGs for a group were defined as above as well as expression >100 transcripts per million (TPM) and >2-fold change over each group in pairwise comparisons. Pathways that were statistically significant were highlighted as differentially regulated. Only pathways that were found significant in more than one of the three general analyses were considered for further evaluation.
qRT-PCR
RNA samples were obtained using the RNeasy Plus Mini kit (Qiagen) following the manufacturer's instructions. cDNA libraries were constructed using iScript cDNA Synthesis kit (Bio-Rad) using 500 ng of purified RNA from each sample as input following manufacturer's instructions. qRT-PCR was performed on a CFX Connect qPCR machine (Bio-Rad; RRID: SCR_003375) using iTaq SYBR green supermix (Bio-Rad) and equal amounts of cDNA samples. Results were normalized to GAPDH or 18s rRNA levels using the ΔΔCt method.
Calcium analysis
Samples were loaded with Fluo-4AM (Invitrogen) calcium sensitive dye at a final concentration of 4.1 ng/ml for 30 min before visualization. Cells were imaged on a Nikon A1s confocal microscope with live cell chamber incubation. We added 10 μM ATP to cells to a final concentration of 50 nM. Imaging was performed at 15 frames/second with a resonant sensor for at least 5 min. Inhibitors to calcium release from the ER were added along with Fluo-4AM for 30 min before visualization. We used 2-APB (Tocris Bioscience, 50 μM), Ryanodine (Tocris Bioscience, 50 nM) and Dantrolene (Tocris Bioscience, 10 μM) to inhibit calcium release.
Videos were analyzed by highlighting regions of interest in each cell soma and recording the average intensity for each frame in every region of interest using ImageJ (Schneider et al., 2012). This data was analyzed using R (RRID: SCR_001905) to graph the average change in fluorescence over the initial fluorescence (ΔF/F0) for each frame. Average intensity was defined as the average intensity for the thirty seconds immediately before ATP administration for each region of interest.
Electrophysiology
The cultured astrocytes were continuously perfused with artificial cerebrospinal fluid (ACSF) saturated with 95% O2/5% CO2. The composition of ACSF was (in mM) 124 NaCl, 3.5 KCl, 1.5 CaCl2, 1.3 MgSO4, 1.24 KH2PO4, 18 NaHCO3, 20 glucose (pH 7.4). The electrodes were filled with a solution consisting of (in mM) 140 K-gluconate, 0.1 CaCl2, 2 MgCl2, 1 EGTA, 2 ATP K2, 0.1 GTP Na3, and 10 HEPES (pH 7.25) (290 mOsm) and had a resistance of 4-6 MΩ. Astrocytes were visualized using an Olympus Optical BX51WI microscope with differential interference contrast optics at 40×. Voltage and current clamp recordings were obtained at 30°C using a MultiClamp 700B amplifier (Axon Instruments). Signals were filtered at 4 kHz using a Digidata 1322A analog-to-digital converter (Axon Instruments). Access resistance was monitored before and after recordings and cells with resistances >25 MΩ at either point were discarded from analyses.
Neurite outgrowth
Astrocytes from each region were cultured for 7 days on Matrigel in the presence of 10 ng/ml BMP4 (Stemgent) and CNTF (R&D Systems). hESC (H9 line)-derived neurons constitutively expressing eGFP were differentiated to the same regions as the astrocyte groups using the same patterning protocol and then plated on the astrocytes as single cells and allowed to grow for 3 days before imaging for neurite outgrowth. Neurons were imaged live on a Perkin Elmer Operetta automated confocal microscope. Neurite outgrowth was measured using FIJI (RRID: SCR_02285) using the NeuronJ package (RRID: SCR_002074) (Schindelin et al., 2012).
Measurement of permeability of endothelia co-cultured with astrocytes
BMECs were differentiated from IMR90-C4 (WiCell) iPSCs as previously reported (Canfield et al., 2016; Lippmann et al., 2012). BMECs were dissociated into single cells using Accutase (Life Technologies) and plated onto collagen IV (400 μg/ml; Sigma-Aldrich) and fibronectin (100 μg/ml; Sigma-Aldrich) at a density of 1×106 cells/cm2 on Transwell-Clear permeable inserts (0.4 µm pore size; Corning). BMEC-seeded transwells were placed onto plates with either 5×105 or 1×106 regionally specific astrocytes seeded onto the bottom of the plate in 200 ml human Endothelial Serum Free Media (hESFM; Life Technologies) supplemented with 20 ng/ml FGF2 and 1% platelet-derived bovine serum (Biomedical Technologies) for 24 h before removal of FGF2 for the following measurements.
TEER was measured every 24 h following the co-culture of BMECs with astrocytes. Resistance was recorded using an EVOM ohmmeter with STX2 electrodes (World Precision Instruments). TEER values were presented as Ω×cm2 following background subtraction on a non-seeded transwell. TEER data were measured three independent times from each sample and from at least three triplicate filters for each experimental group.
Sodium fluorescein (10 μM, 376 Daltons; Sigma-Aldrich) was used to determine the permeability of the BMEC barrier. Following 48 h of monoculture or co-culture conditions, sodium fluorescein was added to the top chamber. 150 μl aliquots were taken from the bottom chamber at 0, 15, 30, 45 and 60 min, and immediately replaced with pre-warmed medium. Permeability coefficients were calculated based on the cleared volume of fluorescein from the top chamber to the bottom chamber.
TGFβ2 (Peprotech, 100-35B) was all used at 10 ng/ml in a monoculture with BMECs. TEER and sodium fluorescein permeability was measured as described above. Neutralizing antibody to TGFβ1/2/3 (1 μg/ml; R&D Systems, MAB1835R, RRID: AB_357931) was used at concentrations above reported LD50 concentrations in co-cultures with regional astrocytes and BMECs for 3 days and maximum TEER was reported. TEER was measured as described above.
Immunocytochemistry was used to visualize occludin in iPSC-derived BMECs following 48 h of co-culture with region-specific astrocytes as previously described (Canfield et al., 2016). iPSC-BMECs were fixed in cold methanol (100%; Sigma-Aldrich) for 15 min followed by three washes in PBS. Cells were blocked in 10% goat serum (Sigma-Aldrich) for 1 hour at 20°C on a rotational platform. Primary antibody (Occludin, Sigma-Aldrich, OC-3F10, 1:50) was diluted in blocking solution and incubated overnight on iPSC-BMECs at 4°C on a rotational platform. Following three washes, a secondary antibody [AlexaFluor anti-mouse (H+L) 546; 1:200] was incubated on the cells at 20°C for 1 hour on a rotational platform. Images were taken using an Olympus epifluorescence microscope. Junctional occludin was quantified by the area fraction index; representing the area of each image that exhibited occludin immunoreactivity. Images were processed in ImageJ with a minimum of ten fields with approximately 30 cells/field quantified. All experimental groups remained blinded until completion of the study.
Supplementary Material
Acknowledgements
We thank Andy Pohl for assisting with analysis in R.
Footnotes
Competing interests
S.-C.Z. is the co-founder of BrainXell.
Author contributions
Conceptualization: R.A.B., S.-C.Z.; Methodology: R.A.B., S.G.C., C.K.; Software: R.A.B., J.C., C.K.; Formal analysis: R.A.B., J.C., S.G.C.; Investigation: R.A.B., J.S., C.M., S.G.C., Y.D., K.L., B.L., J.R.J., A.P.; Resources: J.R.J., A.P.; Writing - original draft: R.A.B., S.G.C., Y.D., A.B., E.V.S., S.-C.Z.; Writing - review & editing: R.A.B., S.G.C., Y.D., A.B., E.V.S., S.-C.Z.; Visualization: R.A.B.; Supervision: A.B., S.P.P., E.V.S., C.K., S.-C.Z.; Project administration: S.P.P., E.V.S., C.K., S.-C.Z.; Funding acquisition: S.P.P., E.V.S., C.K., S.-C.Z.
Funding
This study was supported in part by the National Institute of Mental Health (MH099587, MH100031), the National Institute of Neurological Disorders and Stroke (NS086604) and the National Institute of Child Health and Human Development (HD076892) and the National Institutes of Health (R01GM102756). This study was supported in part by a core grant to the Waisman Center from the National Institute of Child Health and Human Development (P30 HD03352 and U54 HD090256). S-C.Z. acknowledges the Steenbock Professorship in Behavioral and Neural Sciences. R.A.B. was supported by Molecular Biosciences Training Grant T32 (funded by the National Institute of General Medical Sciences, GM007215). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in GEO under accession number GSE133489.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.170910.supplemental
References
- Abbott N. J., Rönnbäck L. and Hansson E. (2006). Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 7, 41-53. 10.1038/nrn1824 [DOI] [PubMed] [Google Scholar]
- Anders S. and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, 1-12. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo R. M., Kim R. S., Ligon K. L., Maher E. A., Brennan C., Billings N., Chan S., Li C., Rowitch D. H., Wong W. H. et al. (2004). Molecular diversity of astrocytes with implications for neurological disorders. Proc. Natl. Acad. Sci. USA 101, 8384-8389. 10.1073/pnas.0402140101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagri A., Marín O., Plump A. S., Mak J., Pleasure S. J., Rubenstein J. L. R. and Tessier-Lavigne M. (2002). Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33, 233-248. 10.1016/S0896-6273(02)00561-5 [DOI] [PubMed] [Google Scholar]
- Bandtlow C. E., Dlaska M., Pirker S., Czech T., Baumgartner C. and Sperk G. (2004). Increased expression of Nogo-A in hippocampal neurons of patients with temporal lobe epilepsy. Eur. J. Neurosci. 20, 195-206. 10.1111/j.1460-9568.2004.03470.x [DOI] [PubMed] [Google Scholar]
- Bayraktar O. A., Fuentealba L. C., Alvarez-Buylla A. and Rowitch D. H. (2015). Astrocyte development and heterogeneity. Cold Spring Harbor Perspect. Biol. 7, a020362 10.1101/cshperspect.a020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani N. and Attwell D. (2016). Astrocyte calcium signaling: the third wave. Nat. Neurosci. 19, 182-189. 10.1038/nn.4201 [DOI] [PubMed] [Google Scholar]
- Blacker D., Wilcox M. A., Laird N. M., Rodes L., Horvath S. M., Go R. C. P., Perry R., Watson B., Bassett S. S., McInnis M. G. et al. (1998). Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat. Genet. 19, 357-360. 10.1038/1243 [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E., Ezan F., Tessier-Lavigne M. and Sotelo C. (1999). Floor plate and netrin-1 are involved in the migration and survival of inferior olivary neurons. J. Neurosci. 19, 4407-4420. 10.1523/JNEUROSCI.19-11-04407.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert M. M., Erikson G. A., Shokhirev M. N. and Allen N. J. (2018). The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269-285. 10.1016/j.celrep.2017.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks M. P. M., Hoogendoorn M., Jungerius B. J., Bakker S. C., Sommer I. E., Sinke R. J., Ophoff R. A. and Kahn R. S. (2008). Do mood symptoms subdivide the schizophrenia phenotype? association of the GMP6A gene with a depression subgroup. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 707-711. 10.1002/ajmg.b.30667 [DOI] [PubMed] [Google Scholar]
- Bozidis P., Hyphantis T., Mantas C., Sotiropoulou M., Antypa N., Andreoulakis E., Serretti A., Mavreas V. and Antoniou K. (2014). HSP70 polymorphisms in first psychotic episode drug-naïve schizophrenic patients. Life Sci. 100, 133-137. 10.1016/j.lfs.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Buosi A. S., Matias I., Araujo A. P. B., Batista C. and Gomes F. C. A. (2017). Heterogeneity in Synaptogenic profile of astrocytes from different brain regions. Mol. Neurobiol. 55, 751-762. 10.1007/s12035-016-0343-z [DOI] [PubMed] [Google Scholar]
- Canfield S. G., Stebbins M. J., Morales B. S., Asai S. W., Vatine G. D., Svendsen C. N., Palecek S. P. and Shusta E. V. (2016). An isogenic blood-brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J. Neurochem. 140, 874-888. 10.1111/jnc.13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-Camacho O., Camarillo-Blancarte L., Pelaez-González H., Mendiola J. and Zenteno J. C. (2012). Klippel–Feil syndrome associated with situs inversus: Description of a new case and exclusion of GDF1, GDF3 and GDF6 as causal genes. Eur. J. Med. Genet. 55, 414-417. 10.1016/j.ejmg.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R. and Ma'ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chever O., Dossi E., Pannasch U., Derangeon M. and Rouach N. (2016). Astroglial networks promote neuronal coordination. Sci. Signal. 9, ra6 10.1126/scisignal.aad3066 [DOI] [PubMed] [Google Scholar]
- Chung W.-S., Clarke L. E., Wang G. X., Stafford B. K., Sher A., Chakraborty C., Joung J., Foo L. C., Thompson A., Chen C. et al. (2013). Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394-400. 10.1038/nature12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez L., Weckhuysen S., Peeters K., Deconinck T., Claeys K. G., Claes L., Claes L. R. F., Suls A., Van Dyck T., Palmini A. et al. (2008). Epilepsy as part of the phenotype associated with ATP1A2 mutations. Epilepsia 49, 500-508. 10.1111/j.1528-1167.2007.01415.x [DOI] [PubMed] [Google Scholar]
- Duclot F. and Kabbaj M. (2017). The role of Early Growth Response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Frontier. Behav. Neurosci. 11, 35 10.3389/fnbeh.2017.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kordi A., Kästner A., Grube S., Klugmann M., Begemann M., Sperling S., Hammerschmidt K., Hammer C., Stepniak B., Patzig J. et al. (2013). A single gene defect causing claustrophobia. Transl. Psychiatr. 3, e254 10.1038/tp.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B. and Liebner S. (2014). Novel insights into the development and maintenance of the blood–brain barrier. Cell Tissue Res. 355, 687-699. 10.1007/s00441-014-1811-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Salathia N., Liu R., Kaeser G. E., Yung Y. C., Herman J. L., Kaper F., Fan J.-B., Zhang K., Chun J. et al. (2016). Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241-244. 10.1038/nmeth.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer W. T., Abrahamsson T., Chierzi S., Lui C., Zaelzer C., Jones E. V., Bally B. P., Chen G. G., Theroux J.-F., Peng J. et al. (2016). Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351, 849-854. 10.1126/science.aab3103 [DOI] [PubMed] [Google Scholar]
- Freeman M. R. (2010). Specification and morphogenesis of astrocytes. Science 330, 774-778. 10.1126/science.1190928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil V., Nicolas O., Mingorance A., Ureña J., Tang B., Hirata T., Sáez-Valero J., Ferrer I., Soriano E. and del Río J. A. (2006). Nogo-A expression in the human hippocampus in normal aging and in Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 433-444. 10.1097/01.jnen.0000222894.59293.98 [DOI] [PubMed] [Google Scholar]
- Gomes F. C. A., Spohr T. C. L. S., Martinez R. and Moura Neto V. (2001). Cross-talk between neurons and glia: highlights on soluble factors. Braz. J. Med. Biol. Res. 34, 611-620. 10.1590/S0100-879X2001000500008 [DOI] [PubMed] [Google Scholar]
- Haidet-Phillips A. M., Hester M. E., Miranda C. J., Meyer K., Braun L., Frakes A., Song S. W., Likhite S., Murtha M. J., Foust K. D. et al. (2011). Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 29, 824-828. 10.1038/nbt.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Chen M., Wang F., Windrem M., Wang S., Shanz S., Xu Q., Oberheim N. A., Bekar L., Betstadt S. et al. (2013). Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 12, 342-353. 10.1016/j.stem.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatami M., Mehrjardi N. Z., Kiani S., Hemmesi K., Azizi H., Shahverdi A. and Baharvand H. (2009). Human embryonic stem cell-derived neural precursor transplants in collagen scaffolds promote recovery in injured rat spinal cord. Cytotherapy 11, 618-630. 10.1080/14653240903005802 [DOI] [PubMed] [Google Scholar]
- Hochstim C., Deneen B., Lukaszewicz A., Zhou Q. and Anderson D. J. (2008). Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 133, 510-522. 10.1016/j.cell.2008.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda H., Hamasaki H., Wakamiya T., Koyama S., Suzuki S. O., Fujii N. and Iwaki T. (2015). Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions. Neuropathology 35, 37-43. 10.1111/neup.12153 [DOI] [PubMed] [Google Scholar]
- Huentelman M. J., Muppana L., Corneveaux J. J., Dinu V., Pruzin J. J., Reiman R., Borish C. N., De Both M., Ahmed A., Todorov A. et al. (2015). Association of SNPs in EGR3 and ARC with Schizophrenia supports a biological pathway for schizophrenia risk. PLoS ONE 10, e0135076 10.1371/journal.pone.0135076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K., Mao X. O. and Greenberg D. A. (2006). Vascular endothelial growth factor stimulates neurite outgrowth from cerebral cortical neurons via Rho kinase signaling. J. Neurobiol. 66, 236-242. 10.1002/neu.20215 [DOI] [PubMed] [Google Scholar]
- Jo S., Yarishkin O., Hwang Y. J., Chun Y. E., Park M., Woo D. H., Bae J. Y., Kim T., Lee J., Chun H. et al. (2014). GABA from reactive astrocytes impairs memory in mouse models of Alzheimer's disease. Nat. Med. 20, 886-896. 10.1038/nm.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B. S. and McCarthy K. D. (2015). Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb. Perspect. Biol. 7, a020404 10.1101/cshperspect.a020404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khananshvili D. (2013). The SLC8 gene family of sodium–calcium exchangers (NCX) – Structure, function, and regulation in health and disease. Mol. Asp. Med. 34, 220-235. 10.1016/j.mam.2012.07.003 [DOI] [PubMed] [Google Scholar]
- Klapka N. and Müller H. W. (2006). Collagen matrix in spinal cord injury. J. Neurotrauma 23, 422-436. 10.1089/neu.2006.23.422 [DOI] [PubMed] [Google Scholar]
- Kofler B., Bulleyment A., Humphries A. and Carter D. A. (2002). Id-1 expression defines a subset of vimentin/S-100beta-positive, GFAP-negative astrocytes in the adult rat pineal gland. Histochem. J. 34, 167-171. 10.1023/A:1020946631937 [DOI] [PubMed] [Google Scholar]
- Krencik R. and Zhang S.-C. (2011). Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 6, 1710-1717. 10.1038/nprot.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R., Weick J. P., Liu Y., Zhang Z.-J. and Zhang S.-C. (2011). Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 29, 528-534. 10.1038/nbt.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukdereli H., Allen N. J., Lee A. T., Feng A., Ozlu I. M., Conatser L. M., Chakraborty C., Workman G., Weaver M., Sage E. H. et al. (2011). Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc. Natl Acad. Sci. USA 108, E440-E449. 10.1073/pnas.1104977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A. et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90-W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M. J., Tavernier B. et al. (2009). Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 41, 1094-1099. 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- Lanner J. T., Georgiou D. K., Joshi A. D. and Hamilton S. L. (2010). Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harbor Perspect. Biol. 2, a003996 10.1101/cshperspect.a003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann E. S., Azarin S. M., Kay J. E., Nessler R. A., Wilson H. K., Al-Ahmad A., Palecek S. P. and Shusta E. V. (2012). Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat. Biotechnol. 30, 783-891. 10.1038/nbt.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M., Martynoga B., Yu T., West J. D., Mason J. O. and Price D. J. (2010). The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development 137, 487-497. 10.1242/dev.039800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing A., Brenner M., Feany M. B., Nedergaard M. and Goldman J. E. (2012). Alexander Disease. J. Neurosci. 32, 5017-5023. 10.1523/JNEUROSCI.5384-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A. V. and Deneen B. (2015). Astrocyte development: a guide for the perplexed. Glia 63, 1320-1329. 10.1002/glia.22836 [DOI] [PubMed] [Google Scholar]
- Molofsky A. V., Krenick R., Ullian E. M., Tsai H.-H., Deneen B., Richardson W. D., Barres B. A. and Rowitch D. H. (2012). Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 26, 891-907. 10.1101/gad.188326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne A., Barnes S. R., Sweeney M. D., Halliday M. R., Sagare A. P., Zhao Z., Toga A. W., Jacobs R. E., Liu C. Y., Amezcua L. et al. (2015). Blood-brain barrier breakdown in the aging human hippocampus. Neuron 85, 296-302. 10.1016/j.neuron.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L., Chiang M. R., Higashimori H., Shoneye T., Iyer L. K., Yelick J., Tai A. and Yang Y. (2017). Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 37, 8706-8717. 10.1523/jneurosci.3956-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K. K. and Pasquale E. B. (2011). Eph receptors and ephrins in neuron–astrocyte communication at synapses. Glia 59, 1567-1578. 10.1002/glia.21226 [DOI] [PubMed] [Google Scholar]
- Muroyama Y., Fujiwara Y., Orkin S. H. and Rowitch D. H. (2005). Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature 438, 360-363. 10.1038/nature04139 [DOI] [PubMed] [Google Scholar]
- Murphy C. A., Russig H., Pezze M.-A., Ferger B. and Feldon J. (2003). Amphetamine withdrawal modulates FosB expression in mesolimbic dopaminergic target nuclei: effects of different schedules of administration. Neuropharmacology 44, 926-939. 10.1016/S0028-3908(03)00074-1 [DOI] [PubMed] [Google Scholar]
- Nedergaard M., Ransom B. and Goldman S. A. (2003). New roles for astrocytes: Redefining the functional architecture of the brain. Trends Neurosci. 26, 523-530. 10.1016/j.tins.2003.08.008 [DOI] [PubMed] [Google Scholar]
- Oberheim N., Takano T., Han X., He W., Lin J. H. C., Wang F., Xu Q., Wyatt J. D., Pilcher W., Ojemann J. G. et al. (2009). Uniquely hominid features of adult human astrocytes. J. Neurosci. 29, 3276-3287. 10.1523/JNEUROSCI.4707-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim N. A., Goldman S. A. and Nedergaard M. (2012). Heterogeneity of astrocytic form and function. Methods Mol. Biol. 814, 23-45. 10.1007/978-1-61779-452-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M., Kamphuis W., Osborn L. M., Jansen A. H. P., Kooijman L., Bossers K. and Hol E. M. (2014). Isolation of glia from Alzheimer's mice reveals inflammation and dysfunction. Neurobiol. Aging 35, 2746-2760. 10.1016/j.neurobiolaging.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Perea G., Navarrete M. and Araque A. (2009). Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32, 421-431. 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Phatnani H. and Maniatis T. (2015). Astrocytes in Neurodegenerative Disease. Cold Spring Harbor Perspect. Biol. 7, a020628 10.1101/cshperspect.a020628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qosa H., Lichter J., Sarlo M., Markandaiah S. S., McAvoy K., Richard J.-P., Jablonski M. R., Maragakis N. J., Pasinelli P. and Trotti D. (2016). Astrocytes drive upregulation of the multidrug resistance transporter ABCB1 (P-Glycoprotein) in endothelial cells of the blood–brain barrier in mutant superoxide dismutase 1-linked amyotrophic lateral sclerosis. Glia 64, 1298-1313. 10.1002/glia.23003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan M. R., Huang Y. H., Kim Y. S., Dykes-Hoberg M. I., Jin L., Watkins A. M., Bergles D. E. and Rothstein J. D. (2007). Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J. Neurosci. 27, 6607-6619. 10.1523/JNEUROSCI.0790-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M., Bonev B., Lindsay J., Lea R., Panagiotaki N., Houart C. and Papalopulu N. (2010). FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development 137, 1553-1562. 10.1242/dev.044909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimoto S., Kidoya H., Naito H., Kamei M., Sakaguchi H., Goda N., Fukamizu A., Nishida K. and Takakura N. (2012). A role for endothelial cells in promoting the maturation of astrocytes through the apelin/APJ system in mice. Development 139, 1327-1335. 10.1242/dev.072330 [DOI] [PubMed] [Google Scholar]
- Sances S., Bruijn L. I., Chandran S., Eggan K., Ho R., Klim J. R., Livesey M. R., Lowry E., Macklis J. D., Rushton D. et al. (2016). Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 19, 542-553. 10.1038/nn.4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q. and Tan S.-S. (2003). Contact-associated neurite outgrowth and branching of immature cortical interneurons. Cereb. Cortex 13, 677-683. 10.1093/cercor/13.6.677 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M. E. (2010). Functions of Nogo proteins and their receptors in the nervous system. Nat. Rev. Neurosci. 11, 799-811. 10.1038/nrn2936 [DOI] [PubMed] [Google Scholar]
- Shigetomi E., Patel S. and Khakh B. S. (2016). Probing the complexities of astrocyte calcium signaling. Trends Cell Biol. 26, 300-312. 10.1016/j.tcb.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S. A., Darmanis S., Huber N., Khan T. A., Birey F., Caneda C., Reimer R., Quake S. R., Barres B. A. and Pasca S. P. (2017). Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779-790.e776. 10.1016/j.neuron.2017.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. and Vinters H. V. (2010). Astrocytes: biology and pathology. Acta Neuropathol. 119, 7-35. 10.1007/s00401-009-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B., Allen N. J., Vazquez L. E., Howell G. R., Christopherson K. S., Nouri N., Micheva K. D., Mehalow A. K., Huberman A. D., Stafford B. et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164-1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz O. E. and Freeman M. R. (2014). Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28, 20-33. 10.1101/gad.229518.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli A., D'Angelo M. G., Salati R., Villa L., Germinasi C., Frattini T., Meola G., Turconi A. C., Bresolin N. and Bassi M. T. (2006). Early onset, non fluctuating spinocerebellar ataxia and a novel missense mutation in CACNA1A gene. J. Neurol. Sci. 241, 13-17. 10.1016/j.jns.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Tsai M. J., Tsai S. K., Huang M. C., Liou D. Y., Huang S. L., Hsieh W. H., Huang W. C., Huang S. S. and Cheng H. (2015). Acidic FGF promotes neurite outgrowth of cortical neurons and improves neuroprotective effect in a cerebral ischemic rat model. Neuroscience 305, 238-247. 10.1016/j.neuroscience.2015.07.074 [DOI] [PubMed] [Google Scholar]
- Vaarmann A., Gandhi S. and Abramov A. Y. (2010). Dopamine induces Ca2+ signaling in astrocytes through reactive oxygen species generated by monoamine oxidase. J. Biol. Chem. 285, 25018-25023. 10.1074/jbc.M110.111450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L. and Hinton G. (2008). Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579-2605. [Google Scholar]
- Varnum-Finney B., Venstrom K., Muller U., Kypta R., Backus C., Chiquet M. and Reichardt L. F. (1995). The integrin receptor α8β1 mediates interactions of embryonic chick motor and sensory neurons with tenascin-C. Neuron 14, 1213-1222. 10.1016/0896-6273(95)90268-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan C., Birch D., Peng C.-Y. Y. and Kessler J. A. (2015). Astrocytic β1-integrin affects cellular composition of murine blood brain barrier in the cerebral cortex. Int. J. Dev. Neurosci. 44, 48-54. 10.1016/j.ijdevneu.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Walz W. and Lang M. K. (1998). Immunocytochemical evidence for a distinct GFAP-negative subpopulation of astrocytes in the adult rat hippocampus. Neurosci. Lett. 257, 127-130. 10.1016/S0304-3940(98)00813-1 [DOI] [PubMed] [Google Scholar]
- Wetzel-Smith M. K., Hunkapiller J., Bhangale T. R., Srinivasan K., Maloney J. A., Atwal J. K., Sa S. M., B Yaylaoglu M., Foreman O., Ortmann W. et al. (2014). A rare mutation in UNC5C predisposes to late-onset Alzheimer's disease and increases neuronal cell death. Nat. Med. 20, 1452-1457. 10.1038/nm.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Xi J., Liu Y., Liu H., Chen H., Emborg M. E. and Zhang S.-C. (2012). Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 30, 1655-1663. 10.1002/stem.1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. (2018). New insights into GFAP negative astrocytes in calbindin D28k immunoreactive astrocytes. Brain Sci. 8, 143 10.3390/brainsci8080143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.-H., Lee D. Y., Gianino S. M. and Gutmann D. H. (2009). Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia 57, 1239-1249. 10.1002/glia.20845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S. A., Clarke L. E., Caneda C., Plaza C. A., Blumenthal P. D., Vogel H., Steinberg G. K., Edwards M. S. B., Li G. et al. (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37-53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen Y., Zhu Q., Huang H., Teng P., Zheng K., Hu X., Xie B., Zhang Z., Sander M. et al. (2014). Control of astrocyte progenitor specification, migration and maturation by Nkx6.1 homeodomain transcription factor. PLoS ONE 9, e109171 10.1371/journal.pone.0109171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.