Abstract
Mycobacterium tuberculosis (Mtb) survives under oxidatively and nitosatively hostile niches inside host phagocytes. In other bacteria, adaptation to these stresses is dependent upon the redox sensitive two component systems (e.g., ArcAB) and transcription factors (e.g., FNR/SoxR). However, these factors are absent in Mtb. Therefore, it is not completely understood how Mtb maintains survival and redox balance in response to reactive oxygen species (ROS) and reactive nitrogen species (RNS). Here, we present evidences that a 4Fe-4S-cofactor containing redox-sensitive transcription factor (WhiB3) is exploited by Mtb to adapt under ROS and RNS stress. We show that MtbΔwhiB3 is acutely sensitive to oxidants and to nitrosative agents. Using a genetic biosensor of cytoplasmic redox state (Mrx1-roGFP2) of Mtb, we show that WhiB3 facilitates recovery from ROS (cumene hydroperoxide and hydrogen peroxide) and RNS (acidified nitrite and peroxynitrite). Also, MtbΔwhiB3 displayed reduced survival inside RAW 264.7 macrophages. Consistent with the role of WhiB3 in modulating host-pathogen interaction, we discovered that WhiB3 coordinates the formation of early human granulomas during interaction of Mtb with human peripheral blood mononuclear cells (PBMCs). Altogether, our study provides empirical proof that WhiB3 is required to mitigate redox stress induced by ROS and RNS, which may be important to activate host/bacterial pathways required for the granuloma development and maintenance.
1. Introduction
Approximately 30% of the world's population is infected with Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB). TB is the leading cause of human deaths globally from an infectious disease and is responsible for approximately 1.4 million deaths annually [9]. The capability of Mtb to acclimatize and counteract killing by the immune system facilitates its survival, replication, and long-term persistence. In this context, it has been shown that Mtb successfully senses and responds to host generated antimicrobial redox stresses such as reactive oxygen and nitrogen species (ROS and RNS). Impaired ability of mice lacking NADPH oxidase (NOX2) and nitric oxide synthase (iNOS) in controlling Mtb proliferation signifies the centrality of ROS and RNS in regulating TB infection [10,29,57]. Furthermore, children with defective NOX2 remain highly susceptible to TB infection [25].
Several studies have indicated that Mtb harbors sophisticated systems to continuously monitor and mount appropriate responses against host generated redox stresses. Mtb is known to mobilize several transcriptional regulators in response to ROS and RNS. For example, Mtb coordinates transcriptional response to oxido-reductive stress via activating sigma factors (SigH and SigE), two-component systems (SenX-RegX and DosR/S/T), serine-threonine kinase (PknG), and transcriptional regulators (MosR, Rv2745c, HpoR, and LtmA) [16,2,23,24,26,30,36,43]. Additionally, Mtb encodes seven members of WhiB family (WhiB1 to WhiB7) of redox-sensing transcription factors. These proteins are characterized by conserved Cys residues (Cys-Xn-Cys-X2-Cys-X5-Cys; where X is an amino acid and n is a variable number) that coordinate an Fe–S cluster [14]. Till date, WhiB proteins in mycobacteria have been reported to carry out diverse functions such as maintaining redox homeostasis (WhiB3, WhiB4 and WhiB7), regulating secretion systems (WhiB1, WhiB5 and WhiB6), virulence (WhiB3, WhiB4, WhiB5 and WhiB6), antibiotic resistance (WhiB7) and reactivation from dormancy (WhiB1 and WhiB5) [4,41,42,44,5,7,8]. Among all WhiB proteins, WhiB3 is the most widely studied. WhiB3 has been shown to influence Mtb pathogenesis by modulating host pathology, phagosomal-maturation, and cell-cycle [11,32,41,48]. Furthermore, WhiB3 has been recently shown to protect Mtb from acidic stress encountered inside macrophages by maintaining the redox potential of it's cytoplasmic redox buffer, mycothiol (MSH) [32].
Despite the great strides made in our understanding of the function of WhiB3 in Mtb, several gaps still exist. For example, studies have shown that WhiB3 iron-sulfur cluster is sensitive to oxidation and nitrosylation [42]. Atmospheric oxygen directly targets WhiB3 [4Fe-4S]2+ cluster to generate a [3Fe-4S]+ intermediate followed by a complete loss of Fe-S cluster. On the other hand, NO forms a protein-bound dinitrosyl–iron–dithiol complex (DINC) with the [4Fe-4S]2+ cluster of WhiB3 [42]. These results implicate WhiB3 in sensing and responding to oxidative and nitrosative conditions in Mtb. In other bacteria, Fe-S cluster transcription factors such as SoxR and FNR have been shown to be the primary sensors of NO and O2 coordinating metabolic adaptation under redox stress conditions [17]. However, a clear FNR-or SoxR -like protein is absent in Mtb. Therefore, it was proposed that WhiB3 functions as one of the crucial “redox sensors” involved in the adaptation of Mtb in response to ROS and RNS. In line with this, hydrogen peroxide (H2O2) and nitric oxide (NO) are known to up-regulate whiB3 expression in Mtb (55). Nevertheless, experimental evidences directly linking the role of WhiB3 in mediating mycobacterial adaptation to oxidative and/or nitrosative stress are lacking. Filling this knowledge gap will be crucial to firmly establish the role of WhiB3 as an intracellular redox sensor and a virulence regulator in Mtb.
In this study, we measured the dynamic changes in the cytoplasmic redox potential of wt Mtb and MtbΔwhiB3 in response to oxidative and nitrosative stresses in vitro. Furthermore, we performed survival experiments to assess the requirement of WhiB3 in mitigating stress induced by ROS and RNS. Lastly, we assessed the potential of wt Mtb and MtbΔwhiB3 to survive inside RAW264.7 macrophages and in inducing early granuloma development using the human PBMCs in vitro.
2. Results
2.1. Mtb WhiB3 is required to survive in response to ROS and RNS
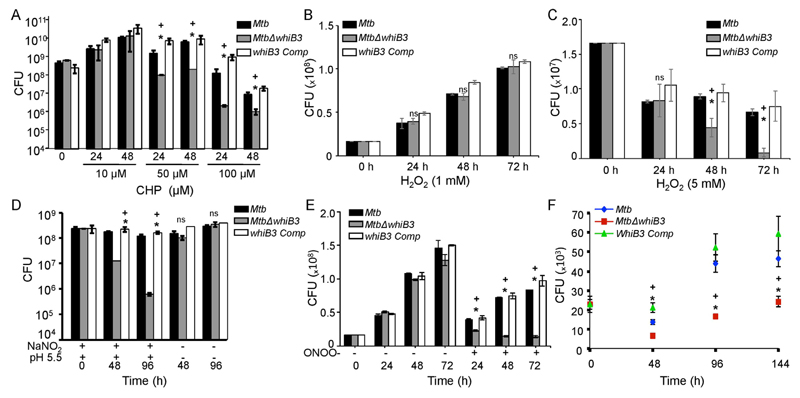
To examine if WhiB3 is required to tolerate oxidative stress, we first exposed wt Mtb, MtbΔwhiB3, and whiB3 Comp strains to a potent oxidant cumene hydroperoxide (CHP). The organic hydroperoxide such as CHP stimulates the production of the free radical intermediates peroxyl and alkoxyl radicals, which can permeate biological membranes and triggers the generation of hydroxyl radical [20]. Also, CHP is more hydrophobic than H2O2 and is known to induce lipid peroxidation as a redox-based mechanism to damage cell membrane [21]. The Mtb strains were exposed to non-lethal (10 μM), sub-lethal (50 μM), and lethal (100 μM) concentrations of CHP for 24 h and 48 h and survival was determined by enumerating viable bacteria. At 10 μM CHP, MtbΔwhiB3 survived equivalent to wt Mtb levels at 24 and 48 h, post-exposure (Fig. 1A). However, an ~10 to ~ 70 -fold reduction in MtbΔwhiB3 survival as compared to wt Mtb was detected upon treatment with 50 and 100 μM CHP at 24 and 48 h, post-exposure, respectively (Fig. 1A). Restoration of whiB3 expression in MtbΔwhiB3 fully complemented the oxidative stress survival defect (Fig. 1A). Additionally, we examined survival in response to 1 mM (non-lethal) and 5 mM H2O2 (sub-lethal) over time. While all there strains grew similarly at 1 mM H2O2 (Fig. 1B), MtbΔwhiB3 showed reduced survival upon exposure to 5 mM H2O2 as compared to wt Mtb and whiB3 Comp strains (Fig. 1C).
Fig. 1.
WhiB3 protects Mtb from Oxidative and Nitrosative stress. Exponentially grown cells of Mtb, MtbΔwhiB3 and whiB3 Comp strains expressing Mrx1-roGFP2 were exposed to (A) 10 μM, 50 μM and 100 μM CHP, (B) 1 mM H2O2, (C) 5 mM H2O2, (D) 0.5 mM acidified (pH 5.5) NaNO2 and (E) 50 μM sodium peroxynitrite (OONO-) and survival was monitored by enumerating CFUs at indicated time intervals. (F) Naïve RAW264.7 macrophages were infected with Mtb, MtbΔwhiB3 and whiB3 Comp and intramacrophage survival was monitored at indicated time points. Error bars represent SD from the mean. * p < 0.05 (as compared to Mtb), + (p < 0.05 as compared to whiB3 Comp), and ns (no significant difference). Data are representative of at least three independent experiments.
One of the major antimicrobial stresses induced by immune-activated macrophages include nitric oxide (NO) generated by iNOS. At acidic pH of 5.5 prevailing inside activated macrophages [39], nitrite (NO2−), which is a major oxidation product of NO protonates to form nitrous acid (HNO2). The HNO2 further dismutases to form other RNS such as NO2 [49]. These RNS inflict nitrosative stress by binding to metal, nitrosylating cysteines, and nitration of tyrosine residues in proteins [46,47]. Also, NO can react with low molecular weight (LMW) thiols such as mycothiol (MSH) to form S-nitrosomycothiols [54]. Studies have shown that RNS generated by acidified nitrite can affect Mtb survival by inactivating proteins involved in the metabolism and in antioxidant defense (lipoamide dehydrogenase and proteasome ATPase) via S-nitrosylation during infection [12,38]. Additionally, RNS oxidize cysteine thiols of some of the Mtb proteins to form mixed intermolecular disulfide [38]. The RNS generated by acidified nitrite not only induces nitrosative but also cause oxidative injury. For example, NO combines with superoxide from bacterial metabolism to generate peroxynitrite anion (OONO−), which is a potent oxidant of methionine (Met) residues in proteins, disrupts Fe-S clusters, nitrate tyrosine residues, and oxidize cysteines [35,45,46].
Given the fact that WhiB3 4Fe-4S cluster reacts with NO [42] and that OONO− oxidizes Fe-S clusters [45,51], we next investigated if WhiB3 protects Mtb from RNS. To mimic physiologically relevant conditions, we exposed wt Mtb, MtbΔwhiB3, and whiB3 Comp strains to 0.5 mM sodium nitrite at pH 5.5 (acidified nitrite), which is well known to induce nitrosative stress in Mtb without inducing significant killing [3]. As shown in Fig. 1B, acidified NaNO2 induces bacteriostasis in wt Mtb at 48 h post-exposure and killing at 96 h post-exposure as compared to unstressed bacteria. In contrast, ~15- and ~200- fold reduced survival of MtbΔwhiB3 as compared to wt Mtb was observed at 48 and 96 h, post-treatment, respectively (Fig. 1D). As expected, complementation of whiB3 restored wt Mtb growth phenotype in response to acidified NaNO2 (Fig. 1D). Because activated macrophages have the ability to generate ONOO− [33], we tested the antimicrobial activity of ONOO− on the survival of wt Mtb, MtbΔwhiB3, and whiB3 Comp strains. While wt Mtb and whiB4 Comp showed marginal sensitivity to 50 μM of ONOO− (Fig. 1E), MtbΔwhiB3 was relatively more sensitive to ONOO− (Fig. 1E). These findings are in line with an earlier report demonstrating remarkable resistance displayed by the virulent Mtb strains towards ONOO− [58]. Our results indicate that WhiB3 is likely to be one of the factors contributing towards ONOO− resistance in Mtb.
Since Mtb encounters ROS and RNS stresses inside macrophages, we assessed the growth of MtbΔwhiB3 inside murine macrophage cell line RAW 264.7. Wt Mtb, MtbΔwhiB3, and whiB3 Comp strains were used to infect RAW 264.7 cells (MOI = 2) and growth was monitored over time by enumerating CFUs. As shown in Fig. 1C, MtbΔwhiB3 showed growth comparable to wt Mtb at day 0 (i.e. 4 h, post-infection). However, while wt Mtb grew exponentially at later time points, MtbΔwhiB3 did not show any appreciable increase in growth overtime (Fig. 1F). Furthermore, whiB3 Comp strain displayed the growth phenotype comparable to wt Mtb (Fig. 1F).
2.2. Mtb WhiB3 maintains redox homeostasis in response to ROS and RNS
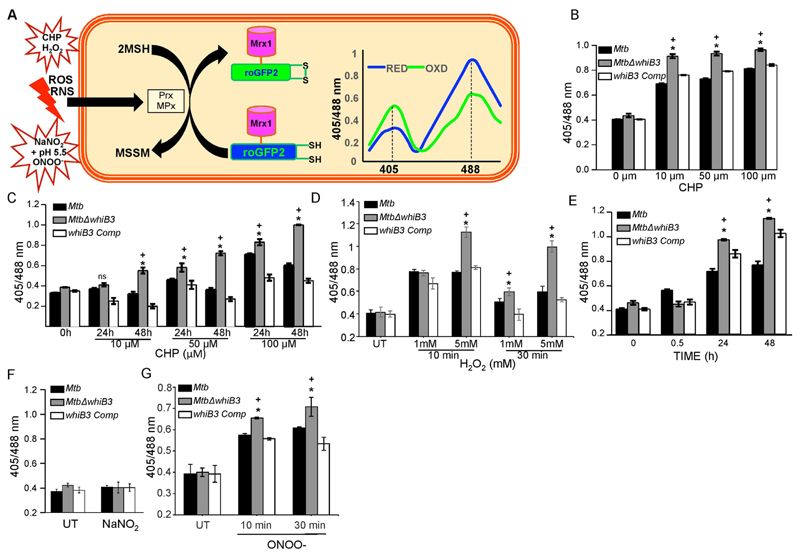
To examine if WhiB3 maintains redox homeostasis in response to ROS/RNS stress, we determined dynamic changes in mycobacterial redox state using a highly sensitive and specific biosensor of mycothiol (MSH) redox potential (EMSH; Mrx1-roGFP2) in wt Mtb [1]. In Mrx1-roGFP2, oxidation-reduction sensitive GFP (roGFP2) was genetically fused to MSH-specific oxidoreductase mycoredoxin-1 (Mrx-1), which reversibly transfers electrons between the mycothiol redox couple (MSH/MSSM) and thiol groups of roGFP2 [1]. Therefore, continuous formation and release of the roGFP2 disulfide bridge is coupled with the actual redox potential of the mycothiol buffer inside mycobacteria [1]. The redox state of Mrx1-roGFP2 thiols can be easily detected by measuring fluorescence intensity at 405 nm and 488 nm excitation wavelengths at a fixed emission of 510 nm [1]. An increase or decrease in 405/488 ratio indicates oxidation or reduction of Mrx1-roGFP2 thiols, respectively [1].
We have earlier reported that acidic pH induces reductive-EMSH in Mtb in a WhiB3-dependent manner [32]. Based on this, we envisaged that the influence of WhiB3 on mycobacterial reductive capacity will be more apparent against oxidative or nitrosative stress, wherein an efficient mobilization of mycothiol-reducing system is critical for the survival of Mtb [1,52]. To do this, wt Mtb, MtbΔwhiB3, and whiB3 Comp strains were exposed to various concentrations of CHP. We monitored short (10 min) – and long (24 and 48 h) term changes in intramycobacterial EMSH upon CHP treatment. The short-term measurements are required to capture rapid oxidation in EMSH, while long-term measurements will provide information about the efficiency by which the cellular antioxidant machinery dissipates oxidative stress to restore redox balance [1]. Addition of 10, 50, and 100 μM of CHP to wt Mtb cells resulted in a rapid (~10 min) increase in Mrx1-roGFP2 ratio (405/488 nm), indicating oxidative shift in EMSH (Fig. 2B). Longer exposure to 10 μM and 50 μM CHP led to a complete recovery from oxidative stress as evident by a substantial decrease in Mrx1-roGFP2 ratio at 48 h post-exposure, suggesting efficient recruitment of anti-oxidant response mechanisms in wt Mtb (Fig. 2C). As expected, treatment with higher concentration of CHP (100 μM) induced a greater oxidation and lesser recovery of biosensor at each time point examined (Fig. 2C). In contrast to wt Mtb, exposure to 10, 50, and 100 μM of CHP led to a significantly greater oxidative shift in EMSH of MtbΔwhiB3 at early time point (10 min) (Fig. 2B). Longer exposure of 10 μM (48 h), 50 μM (24 h and 48 h) and 100 μM CHP (24 h and 48 h) did not lead to recovery from oxidative stress in MtbΔwhiB3 as compared to wt Mtb (Fig. 2C). These findings implicate WhiB3 in orchestrating an efficient antioxidant response in Mtb. The whiB3 Comp strain displayed redox changes in EMSH similar to wt Mtb upon CHP treatment (Figs. 2B & 2C). Consistent with the CHP data, H2O2 treatment also induces greater oxidation of biosensor in MtbΔwhiB3 than wt Mtb and whiB3 Comp strains (Fig. 2D). Whereas wt Mtb and whiB3 Comp showed significant recovery from oxidative stress induced by 5 mM of H2O2 within 30 min, MtbΔwhiB3 remained oxidized (Fig. 2D).
Fig. 2.
WhiB3 maintains cytoplasmic redox potential of Mtb in response to oxidative and nitrosative stress. (A) Schematic representation of Mrx1-roGFP2 oxidation and reduction in response to redox stress. Prx/MPx denotes MSH-dependent peroxiredoxin (e.g., AhpE)/ MSH peroxidase. The graph represents the 405/488 nm ratios change upon oxidation or reduction of Mrx1-roGFP2 in response to oxidative or reductive stress. Exponentially grown cells of Mtb, MtbΔwhiB3 and whiB3 Comp strains expressing Mrx1-roGFP2 were exposed to 10 μM, 50 μM and 100 μM CHP and ratiometric sensor response was measured at (B) 10 min and (C) at 24 h and 48 h. Similarly, all the strains were exposed to (D) 1 mM and 5 mM H2O2 for 10 and 30 min, (E) 0.5 mM NaNO2 at pH 5.5 for 30 min, 24 h and 48 h, (F) 0.5 mM NaNO2 at pH 6.6 for 30 min, and (G) 50 μM sodium OONO- and biosensor response was measured using flow cytometry, as described in Materials and Methods. Error bars represent SD from the mean. * p < 0.05 (as compared to Mtb), + (p < 0.05 as compared to whiB3 Comp), and ns (no significant difference). Data are representative of at least three independent experiments.
Following oxidative stress, we examined dynamic changes in EMSH of Mtb in response to 0.5 mM of acidified NaNO2 for 0.5 h, 24 h and 48 h. In contrast to oxidative stress (Figs. 2B-2D), nitrosative stress did not induce any changes in EMSH of wt Mtb, MtbΔwhiB3, and whiB3 Comp strains at initial time point (30 min) (Fig. 2E). However, at 24 and 48 h, wt Mtb demonstrated a modest but significant increase in Mrx1-roGFP2 ratio, whereas a considerably greater increase was observed in case of MtbΔwhiB3 (Fig. 2E). Numerical assessment of intrabacterial EMSH at 48 h post-treatment with acidified nitrite revealed EMSH of − 261 ± 2 mV for wt Mtb and > −240 mV for MtbΔwhiB3, confirming a larger oxidative shift in the whiB3-deficient strain. As expected, complementation of whiB3 in MtbΔwhiB3 prevented excessive oxidation of EMSH caused by acidified NaNO2 (Fig. 2E). Addition of 0.5 mM of NaNO2 at a neutral pH (pH 6.6) did not influence the biosensor ratio (Fig. 2F), suggesting the role of RNS produced by acidified NaNO2 in perturbing mycobacterial redox balance. Interestingly, exposure to 50 μM ONOO− induces a rapid (within 10–30 min) increase in Mrx1-roGFP2 ratio with a slightly greater increase in MtbΔwhiB3 as compared to wt Mtb and whiB3 Comp (Fig. 2G).
It has been shown that a direct oxidation of mycothiol (MSH) by peroxides is kinetically slow [1]. Therefore, the relatively fast oxidative shift in EMSH upon exposure to CHP and ONOO− is likely to be mediated by MSH-dependent peroxiredoxins or peroxidases. In this context, recently a MSH-dependent peroxiredoxin (alkyl hydroperoxide reductase E; AhpE) has been reported to detoxify organic peroxides and ONOO− in Mtb [19]. Also, as shown in the case of Corynebacterium glutamicum, a yet-to-be-identified MSH-dependent peroxidase (MPx) might mediate degradation of H2O2 and CHP in Mtb [40]. Taken together, our findings confirm the role of WhiB3 in counteracting oxidative and nitrosative stresses commonly encountered by Mtb during infection.
2.3. Mtb WhiB3 is essential to induce granuloma formation in vitro
It has been previously shown that WhiB3 regulates the production of immunomodulatory complex polyketide lipids (e.g., SL-1, PAT/DAT) and secretory proteins (RD-1 antigens, espA) in Mtb [11,32,41]. Both secretory lipids and proteins play a critical role in host-pathogen interaction, cytokine response, and granuloma formation during TB infection [15,8]. Infection of mice with MtbΔwhiB3 resulted in the loss of granuloma formation and aberrant cytokine response, indicating a major role of WhiB3 in regulating immunopathology during infection [41,48]. However, to fully understand the potential of WhiB3 in the TB granuloma development, it is vital to examine the consequence of WhiB3 loss on the granuloma formation in human samples. To begin understanding this, we utilized the in vitro model for granuloma development using live mycobacteria and peripheral blood mononuclear cells (PBMCs) from humans. It has been shown that lymphocytes in human PBMCs aggregates around infected bacteria to form assembly of micro-granulomas [34]. Furthermore, micro-granulomas are formed specifically in response to Mtb, whereas other microbes such as E.coli and S. aureus did not form these structures when incubated with human PBMCs [37].
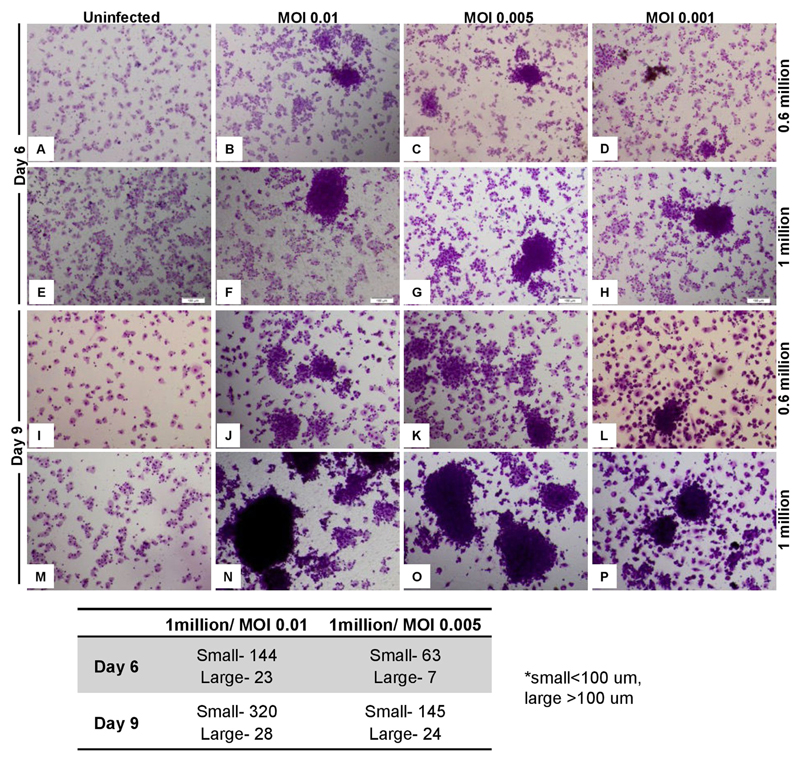
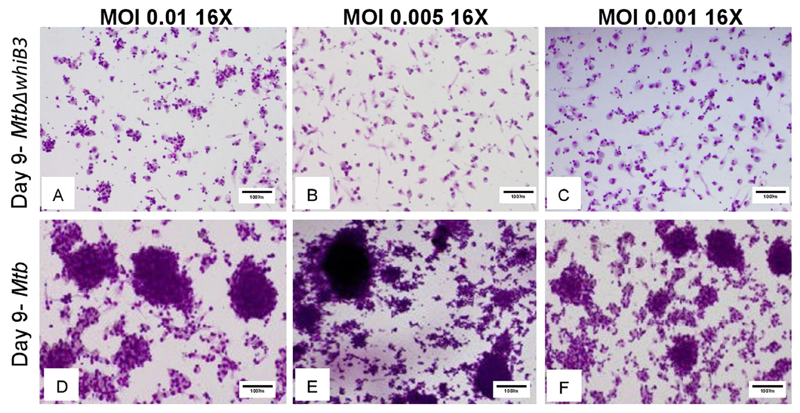
To optimize granuloma formation in an in vitro model, we infected 1 or 0.6 million human PBMCs with Mtb H37Rv at a MOI of 0.001, 0.005, and 0.01 and incubated for 6 or 9 days. As shown in the Fig. 3, human PBMCs tended to form cellular aggregates in the presence of wt Mtb at 6 and 9 days post-infection. Furthermore, aggregates were larger with higher number of PBMCs (1 million) infected with greater number of Mtb (MOI: 0.01) (Fig. 3). No granuloma development was observed in case of uninfected control samples (Fig. 3). In contrast to wt Mtb, infection of human PBMCs (1 million) with MtbΔwhiB3 showed no tendency of cells to aggregate and form granulomas (Fig. 4). Altogether, human data is in complete agreement with animal findings (mice and guinea pigs) [32,48] and suggest that WhiB3 is a major virulence factor that dictates the outcome of host-pathogen interaction during TB infection.
Fig. 3.
In vitro Granuloma Formation upon Mtb infection. 0.6 and 1 million PBMCs were infected by Mtb at MOI of 0.01, 0.005 and 0.001 and stained with May-grünwald giemsa stain at day 6 and day 9 p.i. Granulomas were classified as follows: < 100 μm clusters were classified as small and > 100 μm were classified as large granulomas. Images were taken at 16 S objective (10X with 1.6 zoom).
Fig. 4.
WhiB3 is required for the development of human Granuloma. Image shows granuloma formation upon incubation of 1 million human PBMCs with wt Mtb or MtbΔwhiB3 at MOI 0.01, 0.005 and 0.001. Note that all the aggregates after MtbΔwhiB3 infection were composed of less than 50 PBMCs (< 50 μm) and therefore not classified as granulomas. Images were taken 16× (10× with 1.6 zoom).
3. Discussion
Canonical regulators of oxidative and nitrosative stress such as OxyR, SoxR, and FNR are absent in Mtb. Despite this, Mtb is efficient in counteracting ROS and RNS in vitro, inside macrophages, and during infection in vivo. Furthermore, Mtb mounts effective transcriptional responses upon exposure to diverse ROS and RNS stress agents, indicating the presence of redox sensitive transcription factors in sensing and responding to redox stress. To this end, biochemical and genetic studies have clearly recognized the function of DosR/S/T three-component system in sensing NO, CO, and O2 through heme cofactor and inducing the expression of ~ 40 genes dormancy regulon [24]. Additionally, serine-threonine kinase, PknH, also phosphorylates response regulator DosR to mediate full induction of DosR regulation upon NO stress [6]. The ability of DosR/S/T system to sense O2 and NO was postulated to help Mtb in causing disease in the hypoxic environment of non-human primates and in the HIV patients [31,56]. It's been speculated that while the DosR/S/T system serves as an extracellular sensor of redox signals such as O2 and NO, WhiB3 via its 4Fe-4S cluster functions as an intracellular sensor of ROS and RNS. Overlapping function of both of the systems is required for the metabolic adaptation of Mtb in response to redox stress during infection [41]. Similar to this, in diverse bacteria, membrane localized ArcAB two-component system coordinates response to hypoxia, ROS, and RNS, whereas OxyR and SoxRS/FNR systems serve as intracellular redox sensors by exploiting thiols and Fe-S cluster redox chemistry, respectively [17,27,28]. Present study makes a clear case for at least one Fe-S cluster protein, WhiB3, in sensing and responding to ROS and RNS stress in Mtb. While biochemical studies showed that the 4Fe-4S cluster of WhiB3 interacts with ROS and RNS [42], lack of genetic evidences raised doubts on the role of WhiB3 in coordinating Mtb survival under oxidative or nitrosative stress. Since, whiB3 deficiency was already found to adversely affect Mtb survival under acidic stress [32], its role in ROS/RNS stress remain untested. We rest these speculations by providing unambiguous evidence linking the requirement of WhiB3 for survival and maintenance of redox balance in response to ROS and RNS. Agreeing to this, previous studies suggest that Mtb harbors acidic pH responsive factors that also provide cross-protection to other host defenses such as ROS, RNS and lysosomal hydrolases [53]. Our data indicate that WhiB3 is one such factor that coordinates survival in response to multitude of physiologically relevant stresses such as low pH, ROS and RNS. Since milder acidity (pH 6.2) is the earliest cue faced by Mtb inside naïve macrophages [50], we propose a model which indicates that WhiB3 mediated shift towards more reducing cytoplasm in response to acidic pH and induction of antioxidants machinery, secretory proteins, and virulence lipids [32], adequately prepares Mtb to counteract subsequent exposure to toxic ROS and RNS upon immune-activation (Fig. 5). Induction of immuno-modulatory lipids and secretory proteins by WhiB3 are likely to participate in the granuloma formation and blocking phagosomal maturation to facilitate survival of Mtb in vivo (Fig. 5). Agreeing to this, MtbΔwhiB3 showed survival defect inside naïve and activated macrophages [32]. Other WhiB family member, WhiB1 and WhiB6, have also been reported to respond to RNS via its 4Fe-4S cluster [44,8]. However, importance of these proteins in protecting Mtb and maintaining redox balance upon nitrosative stress remains to be determined.
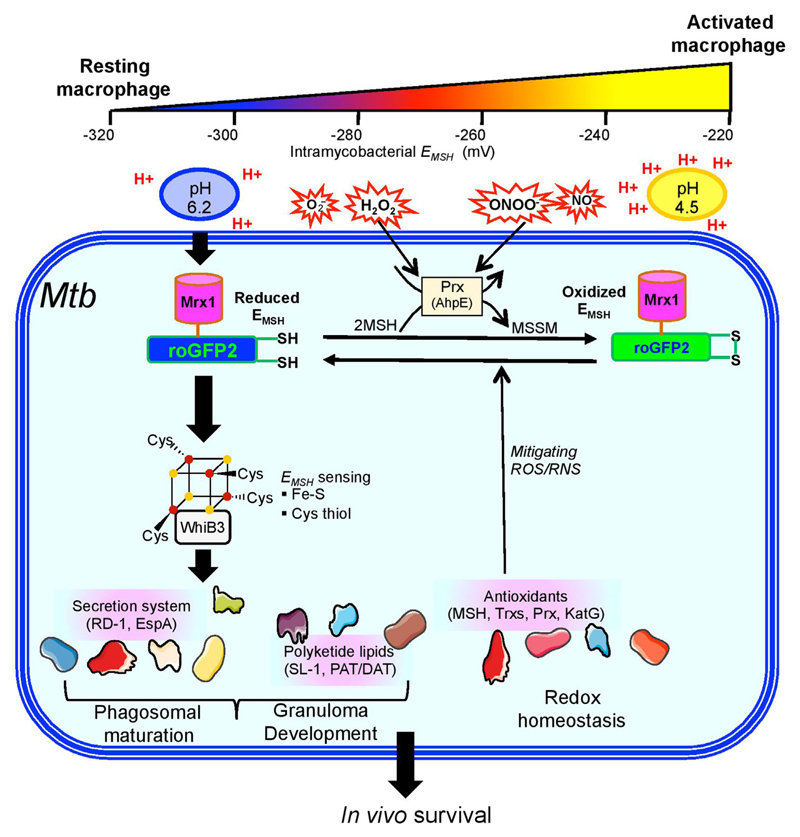
Fig. 5.
Model showing the role of WhiB3 in maintaining redox balance and survival in response to acidic pH, ROS, and RNS. In resting macrophages, Mtb is exposed to a milder form of acidic, ROS, and RNS stress. We have previously shown that a moderate acidity in the resting phagosomes (pH 6.2) induces a reductive shift in the intrabacterial EMSH in a whiB3-dependent manner. Under these conditions, WhiB3 regulates the expression of virulence genes involved in blocking phagosomal maturation (e.g., polyketides, RD-1 antigens), which allows Mtb to thrive in growth permissive environmental conditions. Moreover, secretory antigens and polyketides regulation via WhiB3 might be important for the development of granulomas during infection. Additionally, WhiB3 also induces the expression of several genes involved in maintaining redox homeostasis and protection from ROS and RNS. This facilitates Mtb persistence during activation of macrophages with IFN-γ, which induces phagosomal-lysosomal fusion to elevate the levels of proton (pH 4.5), ROI, and RNS in the microenvironment. Impaired ability of MtbΔwhiB3 to maintain cytoplasmic EMSH and survive in response to acidic pH, oxidative conditions and nitrosative stress, suggest a central role of WhiB3 in regulating redox balance, persistence, and granuloma formation in vivo.
Development and maintenance of granulomas in the lung is the hallmark of TB infection and is the major host defense against Mtb. Conventionally, TB granulomas have been considered host-protective structures to block the spread of infection and reduce bacterial growth. However, newer evidences suggest that nascent granulomas contribute to early mycobacterial growth. This early bacterial growth is dependent upon the ability of uninfected macrophages to efficiently locate and phagocyte-infected macrophages undergoing apoptosis, which lead to rapid, iterative expansion of infected macrophages, seeding of secondary granulomas and systemic dissemination [13]. More-importantly, mycobacterial secretion system RD-1 is crucial for granuloma-mediated expansion and dissemination of infection [13]. MtbΔwhiB3 showed reduced expression of genes encoding immuno-modulatory lipids and associated with RD-1 locus, and macrophages infected with the mutant showed reduced expression of genes associated with apoptosis, endosomal machinery, cell cycle and cytokine/chemokine [11,32]. Therefore, the reduced lung inflammation/pathology in MtbΔwhiB3 infected animals [32,48] strongly implicate the role of WhiB3 mediated granuloma formation in subsequent expansion/dissemination of TB infection. Since, the physiology and disease pathology differs considerably between animals and humans, we realized that it is imperative to examine if WhiB3 is required for granuloma development in humans. Our data clearly aligns with the animal data and showed the requirement of WhiB3 in granuloma formation using human PBMCs. The in vitro granuloma model is successfully leveraged to examine early changes in gene expression signatures (both host and bacteria) that take place during induction of granuloma formation in response to Mtb infection [18,37]. We extended this model to show that it can be exploited to characterize the function of Mtb genes associated with modulating host immune responses and pathogenesis. Future experiments will be aimed to determine specific profile of host gene expression and mechanisms influenced by WhiB3 in the human granuloma model.
4. Materials and methods
4.1. Culturing Mtb strains
Mtb H37Rv, MtbΔwhiB3, and whiB3 Comp strains were cultured aerobically in inkwell bottles with shaking at 150 rpm at 37 °C in liquid Middlebrook 7H9 media with 0.05% tween 80, 0.4% glycerol, 1X OADC, or solid 7H11 supplemented with 0.4% glycerol. Appropriate drugs kanamycin (25 μg/mL) or hygromycin (50 μg/mL) were added. For in vitro stress experiments, 0.02% tyloxapol (non-hydrozable detergent) was used instead of tween-80.
4.2. Culturing of cell lines and macrophage infections
The mice RAW 264.7 macrophages were maintained in an atmosphere containing 5% CO2 at 37 °C in the culture medium recommended by ATCC. 20,000 cells were seeded in 96 well cell culture plate and infected with all three strains at an MOI of 10 for 4 h as described [22], followed by amikacin treatment (200 μg/mL) for 2 h. After 2 h, cells were washed to remove any extracellular bacteria. At specific time points, cells lysed with 0.06% SDS in 7H9 (for growth curves) and N/10 and N/50 dilutions were plated on antibiotic-containing 7H11 plates. CFUs were enumerated by counting after 20–25 days. Statistical analysis was done using unpaired student's t-test using GraphPad Prism version 6 software.
4.3. Survival assays in presence of CHP and H2O2
Mtb H37RV, MtbΔwhiB3, whiB3 Comp cultures were grown at 37 °C in shaker incubator till log phase (OD 600 nm 0.5–0.8). 107/mL bacteria were seeded in 10 mL 7H9 with 0.05% Ty, 0.4% glycerol and 10% ADS. Appropriate concentration of hygromycin and kanamycin was added to the respective cultures. 7 M CHP stock (Sigma Aldrich) was diluted to 10 mM in 1X PBS and appropriate volumes were added so as to make it 10 μM, 50 μM and 100 μM, respectively. For H2O2, log phase (OD 600 nm 0.5–0.8) grown cultures of Mtb H37RV, MtbΔwhiB3, whiB3 Comp were treated with 1 and 5 mM H2O2 (Merck Life Sciences) for 72 h. Every 24 h, cells were suspended in fresh medium containing 1 or 5 mM H2O2. Day 0 CFUs were calculated by plating serially diluted strains just before CHP/ H2O2 addition. Experimental strains were kept at 37 °C in shaker incubator and plated on appropriate antibiotic-containing 7H11 plates at various time points after serial dilution. CFUs were enumerated after 20–25 days of plating. Statistical analysis was done using unpaired student's t-test using GraphPad Prism version 6 software.
4.4. Survival assays in presence of acidified nitrite and peroxynitrite
Mtb H37Rv, MtbΔwhiB3, whiB3 Comp cultures were grown at 37 °C in shaker incubator till log phase (OD 600 nm 0.5–0.8). 107 bacteria were seeded at pH 5.5 in 10 mL 7H9 with 0.05% tyloxapol, 0.4% glycerol and 10% ADS. pH of the media was adjusted using HCl. pH 5.5 adjusted medium was buffered using 100 mM MES. 1 M sodium nitrite solution was made in water. 0.5 mM sodium nitrite was added in +NO samples and same volume of water was added in -NO samples. For OONO- experiment, log phase (OD 600 nm 0.5–0.8) grown cultures of Mtb H37RV, MtbΔwhiB3, whiB3 Comp were treated with 50 μM of sodium peroxynitrite (Cayman Chemicals) for 72 h. Every 12 h, cells were suspended in fresh medium containing 50 μM of sodium peroxynitrite. Day 0 cells were serially diluted and plated on required antibiotic-containing 7H11-OADC plates. Bacteria were cultured in shaker incubator at 37 °C and plated again at various time points after serial dilution. CFUs were enumerated after 20–25 days of plating. Statistical analysis was done using unpaired student's t-test using GraphPad Prism version 6 software.
4.5. In vitro measurement of mycothiol redox potential (EMSH) using Mrx1-roGFP2
Mtb H37Rv, MtbΔwhiB3, whiB3 Comp cultures were grown in 37 °C shaker incubator (150 rpm) till log phase (OD 600 nm 0.5–0.8). 107 /mL bacteria were seeded at, 5.5 in 7H9 with 0.02% Ty, 0.4% glycerol and 10% ADS. pH of the media was adjusted using HCl and NaOH. pH 5.5 adjusted medium was buffered using 100 mM MES. For NO stress, bacterial cells at specified density were treated with 0.5 mM sodium nitrite at pH 5.5. For CHP stress, bacterial cells at specified density were treated with 10 μM, 50 μM and 100 μM CHP. For H2O2 stress, bacterial cells at specified density were treated with 1 mM and 5 mM of H2O2. For OONO- stress, bacterial cells at specified density were treated with 50 μM of sodium peroxynitrite. At indicated time points, samples were drawn, treated with 10 mM NEM followed by fixation 4% PFA and analyzed by Fluorescence Activated Cell Sorter (FACS) Verse Flow cytometer (BD Biosciences). The ratio of emission (510/10 nm) after excitation at 405 and 488 nm was calculated. Data was analyzed using the FACSuite software. Intramycobacterial EMSH was measured using Nernst Equation as described earlier [1].
4.6. Infection of human PBMCs with Mtb H37Rv and MtbΔwhiB3 strains
1 million or 0.6 million human PBMCs (Thermo Fisher Scientific) were seeded in 24-well plates and kept at 37 °C CO2 incubator. Single cell suspension of Mtb strains was made as described earlier [22]. Required MOI of Mtb and MtbΔwhiB3 strains was added, centrifuged at 700 rpm for 5 min, and incubated at 37 °C CO2 incubator for 4 h. After 4 h, 200 μg/mL amikacin was added to remove extracellular bacteria. After 2 h, cells were washed thrice and replaced with 10% GlutaMAX RPMI medium. At day 6 and 9 post-infection, cells were processed for may-grünwald giemsa staining.
4.7. May-grünwald giemsa staining protocol
The plate was centrifuged (at day 6 or day 9) at 700 rpm and fixed with acetomethanol fixative for 10 min at room temperature (RT). After incubation, the fixative was removed, and may-grünwald stain was added and kept for 30 min at RT. May-grünwald stain was removed after 30 min, and washed with giemsa buffer so as to remove excess of may-grünwald stain. 1X giemsa stain was added, incubated at RT for requisite time duration, and washed with 1X PBS. Images were taken at 16× (10× objective with 1.6 zoom), the number of granulomas were counted. Classification of granulomas was done as follows: < 100 μm-small and > 100 μm-large.
Acknowledgements
This work was supported by the following Wellcome Trust-DBT India Alliance grants, WT-DBT/500034-Z-09-Z (AS), IA/S/16/2/502700 (AS), and in part by Department of Biotechnology (DBT) Grant BT/PR11911/BRB/10/1327/2014, BT/PR5020/MED/29/1454/2012, BT/PR13522/COE/34/27/2015 (AS) and DBT-IISc Partnership Program (22-0905-0006-05-987436). AS is a senior fellow of Wellcome trust-DBT India Alliance and MM is a Department of Science and Technology (DST) National Post doctoral fellow.
References
- [1].Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, Singh A. Reengineering redox sensitive GFP to measure mycothiol redox potential of mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10:e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brugarolas P, Movahedzadeh F, Wang Y, Zhang N, Bartek IL, Gao YN, Voskuil MI, Franzblau SG, He C. The oxidation-sensing regulator (MosR) is a new redox-dependent transcription factor in mycobacterium tuberculosis. J Biol Chem. 2012;287:37703–37712. doi: 10.1074/jbc.M112.388611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bryk R, Gold B, Venugopal A, Singh J, Samy R, Pupek K, Cao H, Popescu C, Gurney M, Hotha S, Cherian J, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Burian J, Ramon-Garcia S, Sweet G, Gomez-Velasco A, Av-Gay Y, Thompson CJ. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J Biol Chem. 2012;287:299–310. doi: 10.1074/jbc.M111.302588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Casonato S, Cervantes Sanchez A, Haruki H, Rengifo Gonzalez M, Provvedi R, Dainese E, Jaouen T, Gola S, Bini E, Vicente M, Johnsson K, et al. WhiB5, a transcriptional regulator that contributes to mycobacterium tuberculosis virulence and reactivation. Infect Immun. 2012;80:3132–3144. doi: 10.1128/IAI.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chao JD, Papavinasasundaram KG, Zheng X, Chavez-Steenbock A, Wang X, Lee GQ, Av-Gay Y. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in mycobacterium tuberculosis. J Biol Chem. 2010;285:29239–29246. doi: 10.1074/jbc.M110.132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chawla M, Parikh P, Saxena A, Munshi M, Mehta M, Mai D, Srivastava AK, Narasimhulu KV, Redding KE, Vashi N, Kumar D, et al. Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol Microbiol. 2012;85:1148–1165. doi: 10.1111/j.1365-2958.2012.08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen Z, Hu Y, Cumming BM, Lu P, Feng L, Deng J, Steyn AJ, Chen S. Mycobacterial WhiB6 differentially regulates ESX-1 and the dos regulon to modulate granuloma formation and virulence in Zebrafish. Cell Rep. 2016;16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- [9].GBDT Collaborators. The global burden of tuberculosis: results from the global burden of disease study 2015. Lancet Infect Dis. 2018;18:261–284. doi: 10.1016/S1473-3099(17)30703-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. Transient loss of resistance to pulmonary tuberculosis in p47(phox-/-) mice. Infect Immun. 2000;68:1231–1234. doi: 10.1128/iai.68.3.1231-1234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cumming BM, Rahman MA, Lamprecht DA, Rohde KH, Saini V, Adamson JH, Russell DG, Steyn AJC. Mycobacterium tuberculosis arrests host cycle at the G1/S transition to establish long term infection. PLoS Pathog. 2017;13:e1006389. doi: 10.1371/journal.ppat.1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- [13].Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochim BiophysActa. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- [15].Dkhar HK, Nanduri R, Mahajan S, Dave S, Saini A, Somavarapu AK, Arora A, Parkesh R, Thakur KG, Mayilraj S, Gupta P. Mycobacterium tuberculosis ketomycolic acid and macrophage nuclear receptor TR4 modulate foamy biogenesis in granulomas: a case of a heterologous and noncanonical ligand-receptor pair. J Immunol. 2014;193:295–305. doi: 10.4049/jimmunol.1400092. [DOI] [PubMed] [Google Scholar]
- [16].Fernandes ND, Wu QL, Kong D, Puyang X, Garg S, Husson RN. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J Bacteriol. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Green J, Paget MS. Bacterial redox sensors. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- [18].Guirado E, Mbawuike U, Keiser J, Arcos J, Azad AK, Wang SH, Schlesinger LS. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio. 2015;6:e02537–02514. doi: 10.1128/mBio.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hugo M, Van Laer K, Reyes AM, Vertommen D, Messens J, Radi R, Trujillo M. Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from mycobacterium tuberculosis. J Biol Chem. 2014;289:5228–5239. doi: 10.1074/jbc.M113.510248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hwang YP, Yun HJ, Chun HK, Chung YC, Kim HK, Jeong MH, Yoon TR, Jeong HG. Protective mechanisms of 3-caffeoyl, 4-dihydrocaffeoyl quinic acid from Salicornia herbacea against tert-butyl hydroperoxide-induced oxidative damage. Chem Biol Interact. 2009;181:366–376. doi: 10.1016/j.cbi.2009.07.017. [DOI] [PubMed] [Google Scholar]
- [21].Jovanovic Z, Jovanovic S. Comparison of the effects of cumene hydroperoxide and hydrogen peroxide on Retzius nerve cells of the leech Haemopis sanguisuga. Exp Anim. 2013;62:9–17. doi: 10.1538/expanim.62.9. [DOI] [PubMed] [Google Scholar]
- [22].Karim AF, Chandra P, Chopra A, Siddiqui Z, Bhaskar A, Singh A, Kumar D. Express path analysis identifies a tyrosine kinase Src-centric network regulating divergent host responses to mycobacterium tuberculosis infection. J Biol Chem. 2011;286:40307–40319. doi: 10.1074/jbc.M111.266239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Khan MZ, Bhaskar A, Upadhyay S, Kumari P, Rajmani RS, Jain P, Singh A, Kumar D, Bhavesh NS, Nandicoori VK. Protein kinase G confers survival advantage to mycobacterium tuberculosis during latency-like conditions. J Biol Chem. 2017;292:16093–16108. doi: 10.1074/jbc.M117.797563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee PP, Chan KW, Jiang L, Chen T, Li C, Lee TL, Mak PH, Fok SF, Yang X, Lau YL. Susceptibility to mycobacterial infections in children with X-linked chronic granulomatous disease: a review of 17 patients living in a region endemic for tuberculosis. Pediatr Infect Dis J. 2008;27:224–230. doi: 10.1097/INF.0b013e31815b494c. [DOI] [PubMed] [Google Scholar]
- [26].Li W, Hu L, Xie Z, Xu H, Li M, Cui T, He ZG. Cyclic di-GMP integrates functionally divergent transcription factors into a regulation pathway for antioxidant defense. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Loui C, Chang AC, Lu S. Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol. 2009;9:183. doi: 10.1186/1471-2180-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lu S, Killoran PB, Fang FC, Riley LW. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002;70:451–461. doi: 10.1128/IAI.70.2.451-461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McGillivray A, Golden NA, Gautam US, Mehra S, Kaushal D. The mycobacterium tuberculosis Rv2745c plays an important role in responding to redox stress. PLoS One. 2014;9:e93604. doi: 10.1371/journal.pone.0093604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mehra S, Foreman TW, Didier PJ, Ahsan MH, Hudock TA, Kissee R, Golden NA, Gautam US, Johnson AM, Alvarez X, Russell-Lodrigue KE, et al. The DosR regulon modulates adaptive immunity and is essential for mycobacterium tuberculosis persistence. Am J Respir Crit Care Med. 2015;191:1185–1196. doi: 10.1164/rccm.201408-1502OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mehta M, Rajmani RS, Singh A. Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J Biol Chem. 2016;291:2888–2903. doi: 10.1074/jbc.M115.684597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Prolo C, Alvarez MN, Radi R. Peroxynitrite, a potent macrophage-derived oxidizing cytotoxin to combat invading pathogens. Biofactors. 2014;40:215–225. doi: 10.1002/biof.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Puissegur MP, Botanch C, Duteyrat JL, Delsol G, Caratero C, Altare F. An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell Microbiol. 2004;6:423–433. doi: 10.1111/j.1462-5822.2004.00371.x. [DOI] [PubMed] [Google Scholar]
- [35].Radi R. Peroxynitrite, a stealthy biological oxidant. J Biol Chem. 2013;288:26464–26472. doi: 10.1074/jbc.R113.472936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr, Husson RN. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in mycobacterium tuberculosis. J Bacteriol. 2001;83:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reyes N, Bettin A, Reyes I, Geliebter J. Microarray analysis of the in vitro granulomatous response to Mycobacterium tuberculosis H37Ra. Colomb Med. 2015;46:26–32. [PMC free article] [PubMed] [Google Scholar]
- [38].Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci USA. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- [40].Si M, Xu Y, Wang T, Long M, Ding W, Chen C, Guan X, Liu Y, Wang Y, Shen X, Liu SJ. Functional characterization of a mycothiol peroxidase in corynebacterium glutamicum that uses both mycoredoxin and thioredoxin reducing systems in the response to oxidative stress. Biochem J. 2015;469:45–57. doi: 10.1042/BJ20141080. [DOI] [PubMed] [Google Scholar]
- [41].Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Singh N, Kumar A. Virulence factor SenX3 is the oxygen-controlled replication switch of Mycobacterium tuberculosis. Antioxid Redox Signal. 2015;22:603–613. doi: 10.1089/ars.2014.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive ironsulfur cluster. Biochem J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Soum E, Brazzolotto X, Goussias C, Bouton C, Moulis JM, Mattioli TA, Drapier JC. Peroxynitrite and nitric oxide differently target the iron-sulfur cluster and amino acid residues of human iron regulatory protein 1. Biochemistry. 2003;42:7648–7654. doi: 10.1021/bi030041i. [DOI] [PubMed] [Google Scholar]
- [46].St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Peptide methionine sulfoxide reductase from Escherichia coli and mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stamler JS, Lamas S, Fang FC. Nitrosylation the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- [48].Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- [51].Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: mechanisms and relative contributions to aconitase inactivation. Free Radic Biol Med. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- [52].Ung KS, Av-Gay Y. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett. 2006;580:2712–2716. doi: 10.1016/j.febslet.2006.04.026. [DOI] [PubMed] [Google Scholar]
- [53].Vandal OH, Nathan CF, Ehrt S. Acid resistance in mycobacterium tuberculosis. J Bacteriol. 2009;191:4714–4721. doi: 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vogt RN, Steenkamp DJ, Zheng R, Blanchard JS. The metabolism of nitrosothiols in the mycobacteria: identification and characterization of S-nitrosomycothiol reductase. Biochem J. 2003;374:657–666. doi: 10.1042/BJ20030642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Walter ND, de Jong BC, Garcia BJ, Dolganov GM, Worodria W, Byanyima P, Musisi E, Huang L, Chan ED, Van TT, Antonio M, et al. Adaptation of mycobacterium tuberculosis to impaired host immunity in HIV-infected patients. J Infect Dis. 2016;214:1205–1211. doi: 10.1093/infdis/jiw364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe. 2012;12:301–312. doi: 10.1016/j.chom.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yu K, mitchell C, Xing Y, Magliozzo RS, Bloom BR, Chan J. Toxicity of nitrogen oxides and related oxidants on mycobacteria: M. tuberculosis is resistant to peroxynitrite anion. Tuber Lung Dis. 1999;79:191–198. doi: 10.1054/tuld.1998.0203. [DOI] [PubMed] [Google Scholar]