Abstract
Previously, we determined that genetic and environmental factors contributed equally towards rosacea in twins. To assess an environmental factor, we characterized the malar cheek bacterial microbiome from twins discordant for rosacea. We found no significant difference in facial microbiome alpha and beta diversity between related twins discordant for rosacea. However, the relative percentage abundance of Gordonia and Geobacillus, low-abundant genera, was positively and negatively associated with rosacea severity, respectively. Our data demonstrate a significant correlation between facial microbiome and severity of rosacea in genetically matched twins and importantly that overall microbiome composition is largely unchanged.
Keywords: 16S rRNA, alpha and beta diversity, microbiome, NRS score, relative percentage abundance
1 |. BACKGROUND
Rosacea is a chronic inflammatory disease of the facial skin and eyes that affects ~16 million individuals in the United States alone. Although its aetiology is unknown, we previously determined that both genetic and environmental factors contribute equally towards disease using identical (monozygotic) and fraternal (dizygotic) twins.[1] Previous studies have implicated microbial colonization and the cognate immune response as the critical driver of rosacea.[2,3] Increased levels of Demodex mites and associated bacteria (bacillus), antimicrobial peptides (eg cathelicidin/LL37), host immune variables (eg Toll-like receptor (TLR)-2) and nutrients (eg vitamin D3) are reported to alter the innate immune response and cause chronic inflammation and the facial skin sensitivity in rosacea.[2,3] In addition, the recent studies suggest significant associations between the severity of rosacea and systemic comorbidities (eg cardiovascular, allergic, respiratory, metabolic and gastrointestinal diseases) that are linked with chronic inflammation.[4,5] Rosacea and its reported comorbidities share involvement with barrier tissues that are colonized with a wide variety of micro-flora that constitute the microbiome.
2 |. QUESTION ADDRESSED
The beneficial use of therapeutic drugs for rosacea including oral and topical antibiotics, sulphur compounds and ivermectin that alter the facial microbiome,[6] suggests that altering the resident skin microbiome may play a role in the disease. However, no conclusive evidence has been established.
Microbial dysbiosis could be one of the factors associated with the pathogenesis of rosacea as well as its comorbidities. Harnessing the genetic and environmental control inherent in a twin study, we performed next-generation sequencing of the 16S rRNA gene to evaluate the hypothesis that facial bacterial microbiome dysbiosis will associate with the severity of rosacea in twins discordant for rosacea.
3 |. EXPERIMENTAL DESIGN
See Appendix S1.
4 |. RESULTS
4.1 |. Comparable overall facial skin microbiome composition across subjects
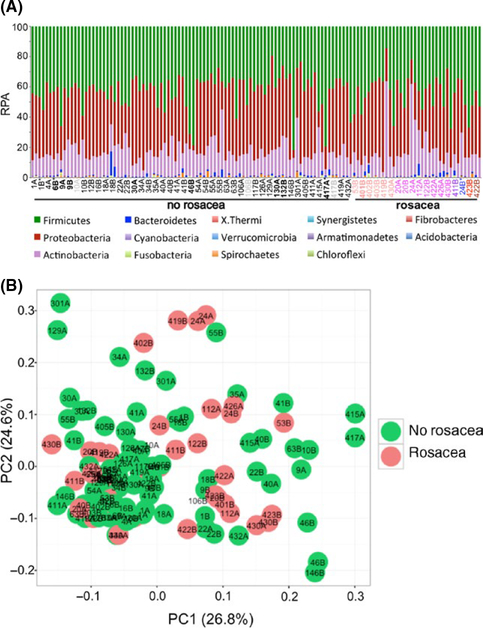
We estimated the relative percentage abundance (RPA) of facial bacterial microbiome at the phylum level in twins with and without rosacea. The top four abundant phyla were Firmicutes (mean ± SD; 42.98% ± 1.05%), Proteobacteria (39.29% ± 1.11%), Actinobacteria (15.88% ± 0.78%) and Bacteroidetes (1.04% ± 1.85%) in all subjects (Figure 1A and Table S1), and our data were generally consistent with the previous reports of other facial skin sites.[7,8] We found that Firmicutes were present in greater abundance than Actinobacteria. However, Proteobacteria were represented by a greater number of genera than Firmicutes and Actinobacteria (Table S1). There was no significant difference in mean abundance for all phyla when compared between different sites (left versus right cheek) or disease (no rosacea versus rosacea) groups. Thus, both inter- and intrapersonal variability in mean abundance of microbiome was observed. The principal coordinates analysis confirmed both inter- and intrapersonal variability and revealed no distinct difference between individuals with and without rosacea (Figure 1B).
FIGURE 1.
Comparable overall facial skin bacterial microbiomein twin subjects. (A) The relative percentage abundance (RPA) atthe phylum level displayed. Phylum ordered from greatest to least abundant from top to bottom in each bar. NRS score of individual twins with rosacea: NRS score (Twin ID): 1 (53B, 401B, 402B, 403B, 419B, 430A (orange)), 2 (20A, 20B, 24A, 112A, 122B, 422A, 426A, 430B (pink)), 3 (411B (violet)), 4 (24B (blue)), 6 (423B (red)) and 9 (422B (brown)). Twin numbers assigned in the order of recruitment with “A” being used per convention for the firstborn of the twin pair versus “B” for the second born twin. Two bars are displayed for each twin representing the composition of left and right cheeks, respectively. Twin IDs in bold font are fraternal (dizygotic) twins and in grey (10A, 106B, and 417B) did not have a validated NRS score (disease status) and thus were excluded from all further analysis. *Individuals used antibiotic within last 3 months. (B) Principal coordinates analysis (PCoA) based on weighted UniFrac distance of twin subjects with (n = 18) and without (n = 42) rosacea. Twin IDs (10A, 106B, and 417B) without a validated NRS score (disease status) shown without a filled circle
4.2 |. Comparable alpha and beta diversity of twin pairs discordant for rosacea
The assessment of facial bacterial microbiome alpha diversity (S1) by Shannon index showed comparable alpha diversity between monozygotic twin pairs with and without rosacea (Figure S1A).
There are earlier reports[9] of skin diseases that showed less bacterial diversity in disease states. Given the paired nature of data, we plotted the National Rosacea Society (NRS) scores differences versus alpha diversity differences between related twin pairs (Figure S1B). We found a negative association between NRS score differences and alpha diversity differences. These data suggest that rosacea negatively impacting bacterial diversity. The non-significance might be due to a low number of samples at high NRS score.
Next, we determined whether facial microbiome beta diversity (S1) is distinct between subjects with rosacea and healthy subjects. Principal component analysis for unweighted, weighted UniFrac (WUF) or Bray-Curtis beta diversity measures (S2) showed no distinct segregation between healthy versus rosacea subjects (data not shown). However, we found that WUF distance between siblings in which only one individual has rosacea was greater than WUF distance between siblings with rosacea and WUF distance between siblings without rosacea (Figure S2). These data indicate the trend that the facial microbiome is more diverse between siblings if rosacea is present, although it did not meet the statistical significance.
Consistent with a previous study,[8] we also found that the related monozygotic twins have more similar (lowest mean WUF distance) facial bacterial microbiome than related dizygotic twins and significantly dissimilar than unrelated twins (Figure S3).
4.3 |. Facial skin bacterial microbiome correlates with NRS score
We tested the hypothesis that the changes in the facial microbial community might be associated with rosacea severity. We found a significant (p < .05) correlation between NRS score and the RPA of 9 genera (Table S3).
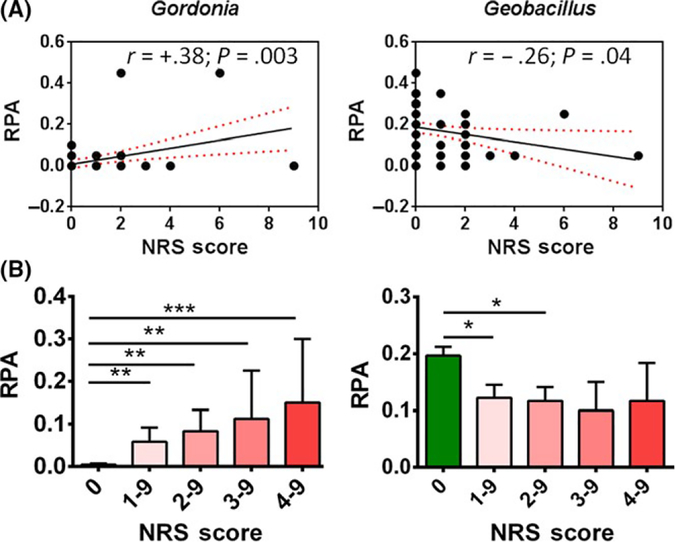
To evaluate the association between NRS score and RPA of bacterial genera and demographics, we performed random effect Poisson regression (REPR) analysis (see Section 3 Experimental Design for rationale and details). Univariate REPR analysis revealed a significant (P < .05) association between NRS scores and six genera: Gordonia, Blautia, Chryseobacterium, Wautersiella, Geobacillus and unknown genus (phylum Proteobacteria) but not with demographics listed in Table S1. Multivariate REPR analyses found that only two genera (Gordonia and Geobacillus), age group 30–60 years versus 0–30 years, and birth order B versus A remained significantly predictive of NRS score. REPR model suggests that a 10% increase in RPA of Gordonia or a 10% decrease in RPA of Geobacillus will increase a NRS score by a factor of 1.76 or 2.16, respectively. These data support that bacterial microbiome (Gordonia and Geobacillus) is the dominant predictor of NRS score in this cohort (Table S4). Concordantly, Gordonia and Geobacillus RPA correlated with NRS score (Figure 2).
FIGURE 2.
Facial skin bacterial microbiome correlates with NRS score. (A) The positive and negative correlation of relative percentage abundance (RPA; closed dots) of bacterial microbiome genera (Gordonia and Geobacillus) with NRS score displayed. The Pearson correlation coefficient (r), and p-values, and linear regression line fitting the data (black solid line) with 95% confidence interval (red dotted line) are shown on each graph. n = 60 (n = 42 (no rosacea); n = 18 (rosacea)). There are several closed dots that overlap with each other. Therefore, the number of total dots appears less than total number of sample. (B) Relative percentage abundance (RPA) of Gordonia and Geobacillus for subjects with “No Rosacea (subjects with NRS score = 0)” and “Rosacea (subjects with score NRS score = 1–9)” was plotted with increasing NRS score. The difference in mean of RPA between no rosacea and rosacea is greater with increasing NRS score. Data are shown as mean ± SEM of n = 42 (NRS score = 0), 18 (NRS score = 1–9), 12 (NRS score=2–9), 4 (NRS score=3–9) and 3 (NRS score = 4–9). p-values for no rosacea vs. rosacea are shown as *<.05; **<.001; ***<.0001. NRS, National Rosacea Society measured for severity of rosacea
5 |. CONCLUSIONS
This is the first study to characterize the facial bacterial microbiome of twins. We took advantage of the relatively controlled nature of our study cohort and found that changes in the facial microbiome are associated with the changes in the NRS score (severity of rosacea) more so than previously identified demographic correlates. Specifically, we uncovered a positive and a negative association for Gordonia and Geobacillus with rosacea, respectively. Importantly, this was in the background of a largely unchanged microbiome landscape. This twin study can serve as a reference for future studies to confirm and pursue specific microbial associations with rosacea.
Supplementary Material
APPENDIX S1 Experimental design
FIGURE S1 Comparable alpha diversity of complete twin pairs discordant for rosacea. (A), Alpha diversity (Shannon index) was plotted as box plot for complete twin pairs (n = 15 twin pairs). Box plot showing central rectangle spans the first quartile to the third quartile (the interquartile range). A segment inside the rectangle shows the median and “whiskers” above and below the box show the locations of the minimum and maximum values of the data. The alpha diversity was calculated for siblings in which neither has rosacea (n = 9 twin pairs), both siblings have rosacea (n = 4 twin pairs) or only one sibling has rosacea (n = 2 twin pairs). (B), A trend of negative association between alpha diversity and NRS score observed within twin pairs. Difference in alpha diversity (Shannon index) on Y-axis and NRS score on X-axis was plotted as values for twin B subtracted from related Twin A (n = 15 twin pairs). There were five twin pairs who have different NRS score including 2 twin pairs in which only one sibling has rosacea. All other twin pairs had the same NRS score resulting in a difference of zero. Pearson correlation coefficient r and p-values are displayed. Two data points have equal value hence 14 data points instead of 15 are visible on the plot
FIGURE S2 The facial microbiome is more diverse between siblings affected by rosacea than siblings not affected by rosacea. The Weighted UniFrac (WUF) distance was plotted between complete twin pairs (n = 15 pairs). The WUF distance was calculated between siblings in which neither has rosacea (no rosacea-no rosacea, n = 9 twin pairs), both siblings have rosacea (rosacea-rosacea, n = 4 twin pairs) or only one sibling has rosacea (no rosacea-rosacea: n = 2 twin pairs), from left to right. WUF distance was calculated between left versus left cheeks and right versus right cheeks
FIGURE S3 Monozygotic twin pairs have more similar facial microbial beta diversity to each other than dizygotic twin pairs and unrelated twin pairs. Facial skin microbiome beta diversity between a pair of related monozygotic (identical) twins, related dizygotic (fraternal) twins versus unrelated twins was calculated by weighted UniFrac (WUF) distance. WUF distance was calculated between left versus left cheeks and right versus right cheeks. Mean ± SEM between related twins versus unrelated subjects were plotted. n = 15 monozygotic pairs, 2 dizygotic pairs, and 60 unrelated pairs. p-values determined from t-test. One fraternal twin (417B) excluded from all other analysis was included in this analysis for statistical calculation. *p < .05
ACKNOWLEDGEMENTS
This study was supported in part by the Skin Diseases Research Center, NIH P30AR039750, start-up institutional funds to DLP and an award from the National Rosacea Society and Dermatology Foundation to DLP and MRG. We would like to acknowledge Mahmoud Ghannoum and Pranab Mukherjee for their critical and helpful suggestions.
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interests.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- [1].Aldrich N, Gerstenblith M, Fu P, Tuttle MS, Varma P, Gotow E, Cooper KD, Mann M, Popkin DL, JAMA Dermatol. 2015, 151, 1213. [DOI] [PubMed] [Google Scholar]
- [2].Holmes AD, Steinhoff M, Exp. Dermatol 2017, 26, 659. [DOI] [PubMed] [Google Scholar]
- [3].Two AM, Wu W, Gallo RL, Hata TR, J. Am. Acad. Dermatol 2015, 72, 749. [DOI] [PubMed] [Google Scholar]
- [4].Hua TC, Chung PI, Chen YJ, Wu LC, Chen YD, Hwang CY, Chu SY, Chen CC, Lee DD, Chang YT, Liu HN, J. Am. Acad. Dermatol 2015, 73, 249. [DOI] [PubMed] [Google Scholar]
- [5].Rainer BM, Fischer AH, Luz D da Silva Felipe, Kang S, Chien AL, J. Am. Acad. Dermatol 2015, 73, 604. [DOI] [PubMed] [Google Scholar]
- [6].van Zuuren EJ, Fedorowicz Z, Carter B, van der Linden MM, Charland L, Cochrane Database Syst. Rev 2015, CD003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA, Science 2009, 324, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Si J, Lee S, Park JM, Sung J, Ko G, BMC Genom. 2015, 16, 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Williams MR, Gallo RL, Curr. Allergy Asthma Rep 2015, 15, 65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1 Experimental design
FIGURE S1 Comparable alpha diversity of complete twin pairs discordant for rosacea. (A), Alpha diversity (Shannon index) was plotted as box plot for complete twin pairs (n = 15 twin pairs). Box plot showing central rectangle spans the first quartile to the third quartile (the interquartile range). A segment inside the rectangle shows the median and “whiskers” above and below the box show the locations of the minimum and maximum values of the data. The alpha diversity was calculated for siblings in which neither has rosacea (n = 9 twin pairs), both siblings have rosacea (n = 4 twin pairs) or only one sibling has rosacea (n = 2 twin pairs). (B), A trend of negative association between alpha diversity and NRS score observed within twin pairs. Difference in alpha diversity (Shannon index) on Y-axis and NRS score on X-axis was plotted as values for twin B subtracted from related Twin A (n = 15 twin pairs). There were five twin pairs who have different NRS score including 2 twin pairs in which only one sibling has rosacea. All other twin pairs had the same NRS score resulting in a difference of zero. Pearson correlation coefficient r and p-values are displayed. Two data points have equal value hence 14 data points instead of 15 are visible on the plot
FIGURE S2 The facial microbiome is more diverse between siblings affected by rosacea than siblings not affected by rosacea. The Weighted UniFrac (WUF) distance was plotted between complete twin pairs (n = 15 pairs). The WUF distance was calculated between siblings in which neither has rosacea (no rosacea-no rosacea, n = 9 twin pairs), both siblings have rosacea (rosacea-rosacea, n = 4 twin pairs) or only one sibling has rosacea (no rosacea-rosacea: n = 2 twin pairs), from left to right. WUF distance was calculated between left versus left cheeks and right versus right cheeks
FIGURE S3 Monozygotic twin pairs have more similar facial microbial beta diversity to each other than dizygotic twin pairs and unrelated twin pairs. Facial skin microbiome beta diversity between a pair of related monozygotic (identical) twins, related dizygotic (fraternal) twins versus unrelated twins was calculated by weighted UniFrac (WUF) distance. WUF distance was calculated between left versus left cheeks and right versus right cheeks. Mean ± SEM between related twins versus unrelated subjects were plotted. n = 15 monozygotic pairs, 2 dizygotic pairs, and 60 unrelated pairs. p-values determined from t-test. One fraternal twin (417B) excluded from all other analysis was included in this analysis for statistical calculation. *p < .05