Abstract
Cytochrome P450 (P450) enzymes are major catalysts involved in the oxidations of most drugs, steroids, carcinogens, fat-soluble vitamins, and natural products. The binding of substrates to some of the 57 human P450s and other mammalian P450s is more complex than a two-state system and has been proposed to involve mechanisms such as multiple ligand occupancy, induced-fit, and conformational-selection. Here, we used kinetic analysis of binding with multiple concentrations of substrates and computational modeling of these data to discern possible binding modes of several human P450s. We observed that P450 2D6 binds its ligand rolapitant in a mechanism involving conformational-selection. P450 4A11 bound the substrate lauric acid via conformational-selection, as did P450 2C8 with palmitic acid. Binding of the steroid progesterone to P450 21A2 was also best described by a conformational-selection model. Hexyl isonicotinate binding to P450 2E1 could be described by either a conformational-selection or an induced-fit model. Simulation of the binding of the ligands midazolam, bromocriptine, testosterone, and ketoconazole to P450 3A4 was consistent with an induced-fit or a conformational-selection model, but the concentration dependence of binding rates for varying both P450 3A4 and midazolam concentrations revealed discordance in the parameters, indicative of conformational-selection. Binding of the P450s 2C8, 2D6, 3A4, 4A11, and 21A2 was best described by conformational-selection, and P450 2E1 appeared to fit either mode. These findings highlight the complexity of human P450-substrate interactions and that conformational-selection is a dominant feature of many of these interactions.
Keywords: cytochrome P450, enzyme kinetics, pre-steady-state kinetics, steroid, enzyme mechanism, steroidogenesis, drug metabolism, conformational selection, CYP, induced fit, P450
Introduction
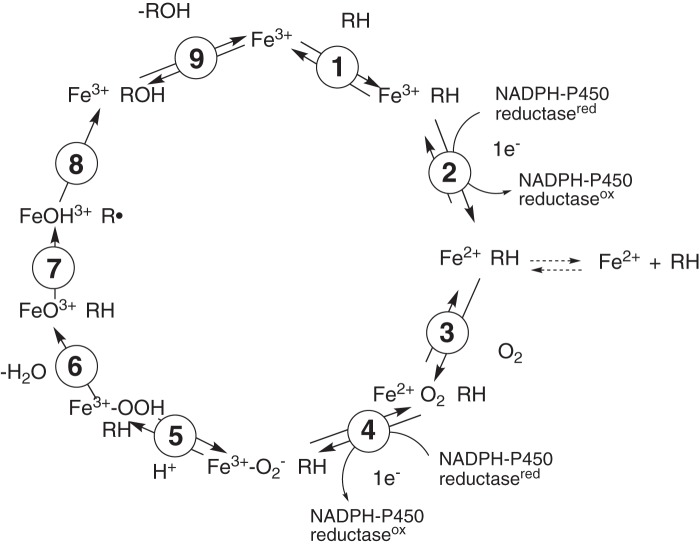
Cytochrome P450 (P450)3 (CYP) enzymes are the major catalysts involved in the metabolism of drugs, steroids, fat-soluble vitamins, chemical carcinogens, and numerous other chemicals of natural and industrial origin (1, 2). Collectively the P450s are involved in ∼95% of the reported oxidations and reductions of all chemicals (3). The oxidation of a chemical by a P450 is a complex process involving electron transfer, formation of a highly reactive iron-oxygen complex, and breaking of C-H and other bonds (Fig. 1) (1, 4, 5). The first step in the reaction cycle is generally agreed to be substrate binding, in that the presence of the substrate facilitates the introduction of an electron to the ferric iron in some but not all cases (6). Binding of substrates can also occur after initial iron reduction (7, 8).
Figure 1.
Although the binding of substrate to P450 enzymes might seem to be the simplest and most straightforward of the reaction steps (4), it can also be complex. An early observation was the change in heme Soret (and the α,β) spectra upon binding (9, 10), attributed to an iron low- to high-spin state shift (Type I difference spectra) associated with partial removal of the distal H2O ligand from the heme iron in the active site (1, 11–15). Some ligands, mainly inhibitors, bind directly to the heme iron via basic nitrogen atoms, yielding so-called Type II difference spectra, but a number of these ligands can also be substrates (16, 17). However, not all P450 substrates produce spectral changes (18), and several other spectroscopic and other methods have been utilized to study P450-substrate binding (19, 20), including X-ray crystallography (21). Several bacterial and human P450s have now been demonstrated to show multiple occupancy (22–25).
The binding of the substrate camphor by bacterial P450 101A1 (P450cam) is an apparently facile process that has been described in terms of a 2-state system with a kon rate of 4.6 × 106 m−1 s−1 and koff rate of 6 s−1 (Kd = 1.3 μm) (26). Rates of substrate binding have also been reported for a small number of mammalian P450s, including several human P450s (Table 1). Several mammalian P450s have been reported to show complex binding behavior, and some of these results may be attributable to multiple occupancy (31–33). However, multistep binding can be observed even for a substrate (e.g. bromocriptine) when only one molecule is present in the P450 enzyme (32, 34, 35).
Table 1.
Estimated rate constants for 2-state binding of substrates and human P450s from previous literature
| P450 | Substrate | Rate constants |
Reference | |
|---|---|---|---|---|
| kon | koff | |||
| 2A6 | Coumarin | 2.7 × 106 m−1 s−1 | 5.7 s−1 | 7 |
| 7-Hydroxycoumarin | 2.0 × 106 m−1 s−1 | 6.8 s−1 | ||
| 4A11 | Lauric acid | 2.0 × 106 m−1 s−1 | 4.0 s−1 | 27 |
| 19A1 | Androstenedione | 2.5 × 106 m−1 s−1 | 1.4 s−1 | 28 |
| 19-OH androstenedione | 2.0 × 107 m−1 s−1 | 240 s−1 | ||
| 19-Formyl androstenedione | 2.5 × 106 m−1 s−1 | 300 s−1 | ||
| 21A2 | Progesterone | 2.4 × 107 m−1 s−1 | 0.24 s−1 | 29 |
| 17α-OH progesterone | 2.2 × 106 m−1 s−1 | 0.66 s−1 | ||
| 27C1 | All-trans-retinol | 7.2 × 105 m−1 s−1 | 0.42 s−1 | 30 |
The multistep nature of ligand binding to some P450s (31–35) raises the issue of whether the basis of the phenomenon is mainly attributable to induced-fit or conformational-selection (Fig. 2), a general question in modern enzymology (36–41). The two pathways can be considered energetically equivalent in terms of a “thermodynamic box” diagram, and distinguishing between them is usually not trivial. Evidence for both models has been presented for P450s. Davydov et al. (42) reported high pressure spectroscopic evidence for conformational heterogeneity of P450 3A4 in the absence of ligand. Our laboratory presented kinetic evidence suggesting an induced-fit model for binding of testosterone to P450 3A4, based upon kinetic double-mixing experiments with testosterone and the (Type II) inhibitor indinavir (33). Studies with other P450s have provided evidence for both models, depending upon the case. For instance, an NMR study with an unnatural amino acid showed spectral heterogeneity of bacterial P450 119, which can be evidence for a conformational-selection model (43). NMR spectra of P450 17A1, in the presence of substrates and ligands, revealed peaks indicative of multiple conformations (44), and protein structures differed in the presence of the R- and S-enantiomers of orteronel (45). The existence of multiple structures of human P450 2E1 (46, 47), P450 1A1 (48, 49), and 3A4 (25, 34, 50, 51) can be interpreted as evidence for an induced-fit mechanism but cannot be excluded as support for an alternative conformational-selection model.
Figure 2.
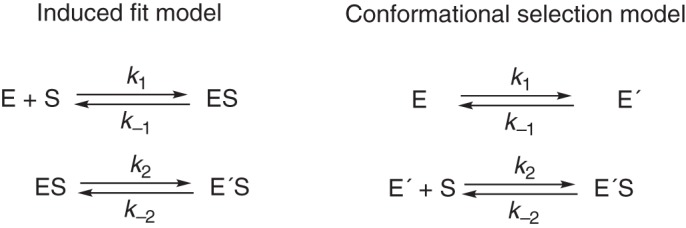
Induced-fit and conformational-selection kinetic schemes. E and E′ are conformationally distinct forms of the enzyme (S is substrate and P is product).
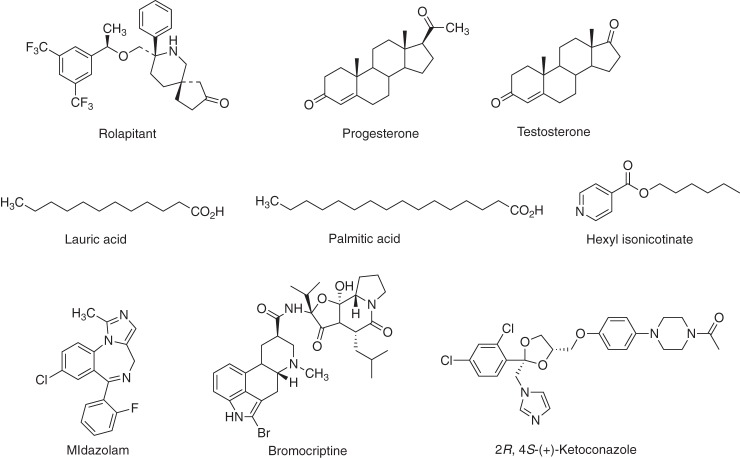
We investigated the binding of steroids to human P450 17A1 and concluded that the mechanism is dominated by conformational-selection, not induced-fit (52). The conclusion was based upon (i) the decreasing rates of ligand binding as a function of steroid concentration (37), (ii) the differing plots of concentration dependence seen with the ligand and the enzyme (53), and (iii) comparisons made by fitting into KinTek Explorer models (54). In this report we evaluated more human P450-substrate systems (Fig. 3) and also re-examined some previous conclusions, with a view to minimalization of models if possible. Accordingly, we investigated human P450 2C8, 2E1, 4A11, and 21A2 binding and also reinvestigated previous data obtained with P450s 3A4 and 2D6, as well as adding new experiments. We conclude that the P450s examined also primarily use conformational-selection mechanisms.
Figure 3.
Structures of P450 ligands used in this study.
Results
General
All experiments were done with ferric P450 enzymes. Although ferrous P450s can bind substrates (e.g. Ref. 7), in many (but not all) P450 systems the binding of substrate facilitates the kinetics of reduction (6, 55). Therefore most of the interest in substrate binding is with the ferric enzymes. The point should be made that even if binding is not the rate-limiting step, the absence of bound substrate may therefore change rates of other steps in the catalytic cycle (Fig. 1). As pointed out in the Introduction, we monitored the binding of substrates to P450s in most cases by observing the spectral changes associated with partial removal of the distal H2O ligand from the heme iron in the active site (Type I change), a relatively well-established principle (1, 11–15).
P450 2D6 and rolapitant
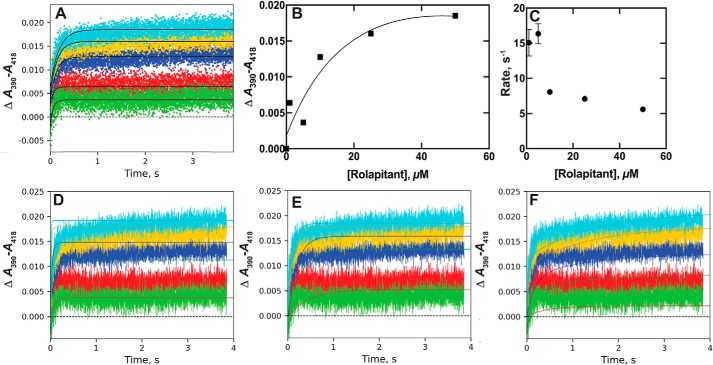
The binding of P450 2D6 and the inhibitory drug rolapitant have been described with a 2-state model using data we developed earlier (56). The single-exponential fits (Fig. 4A) were relatively good, and the amplitudes could be plotted versus the rolapitant concentration to yield a Kd,app of ∼10 μm (Fig. 4B) (cf. 1.2 μm in steady-state (56)). A plot of the binding rates versus rolapitant concentration yielded a negative slope (Fig. 4C), indicative of a conformational-selection process (37). A simple 2-state model did not provide an adequate fit to the experimental data (Fig. 4D). Although a reasonable fit could be achieved with an induced-fit model (Fig. 4E), the fit to a conformational model (Fig. 4F) was as good or better. We conclude, based largely on the relationship between rates of binding and rolapitant concentration (Fig. 4C), that the process involves conformational-selection.
Figure 4.
Binding of rolapitant to P450 2D6. P450 2D6 (2 μm) was mixed with rolapitant (2 (green), 10 (red), 20 (dark blue), 50 (gold), and 100 (light blue) μm) (raw data presented previously (56)). A, single exponential fits of ΔA390–A418 traces. B, plot of ΔA390–A418 amplitude versus final concentrations of rolapitant (Kd,app ∼33 μm). C, plot of single exponential rate fits versus final rolapitant concentration. D, fits to a simple 2-state kinetic model (E + S ⇄ ES, Fig. 2) with k1 = 1.0 × 106 m−1 s−1 and k−1 = 7.4 s−1 (ϵ390–418 5.3 mm−1 cm−1). E, fit to an induced-fit model (Fig. 2) with k1 = 1.1 × 106 m−1 s−1, k−1 = 26 s−1, k2 = 9.6 s−1, and k−2 = 1.8 s−1 (ϵ390–418 5.7 mm−1 cm−1). F, fit to a conformational-selection model (Fig. 2) with k1 = 0.65 s−1, k−1 = 0.46 s−1, k2 = 0.19 × 106 μm−1 s−1, and k−2 = 2.0 s−1 (ϵ390–418 4.2 mm−1 cm−1).
P450 4A11 and lauric acid
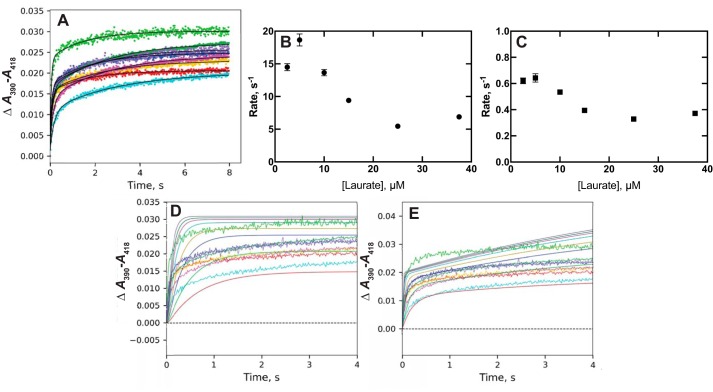
We previously described binding of lauric acid to P450 4A11 based on a simple second-order experiment with equal concentrations of enzyme and substrate (kon 2 × 106 m−1 s−1, koff 4 s−1) (Table 1) (27). Reinvestigation of the binding with multiple concentrations of lauric acid showed complex behavior, with a need to use biexponential fitting (Fig. 5A). Rates for both phases of binding showed inverse relationships with the concentration of lauric acid (Figs. 5, B and C), indicative of a conformational-selection model (37).
Figure 5.
Binding of lauric acid to P450 4A11. P450 4A11 (2 μm) was mixed with varying concentrations of lauric acid (5 (blue), 10 (red), 20 (magenta), 30 (gold), 50 (purple), 75 (dark green), and 150 (light green) μm). A, biexponential fits to traces of ΔA390–A418. B, plot of fast rate from A versus final concentration of lauric acid. C, plot of slow rate from A versus final concentration of lauric acid. D, fit of data (A) to an induced-fit model (Fig. 2) with k1 = 0.09 × 106 m−1 s−1, k−1 = 8.5 s−1, k2 = 24 s−1, and k−2 = 0.73 s−1 (ϵ390–418 8.0 mm−1 cm−1). E, fit of data (A) to a conformational-selection model with k1 = 0.15 s−1, and k−1 = 0.25 s−1, k2 = 0.55 × 106 m−1 s−1, and k−2 = 1.6 s−1 (ϵ390–418 9.5 mm−1 cm−1).
Fitting to an induced-fit model yielded a poor fit, particularly at the lower lauric acid concentrations (Fig. 5D). Fitting to a simple conformational-selection model showed good fits at the lower concentrations of lauric acid, although the fit at higher concentrations was less satisfactory (Fig. 5E).
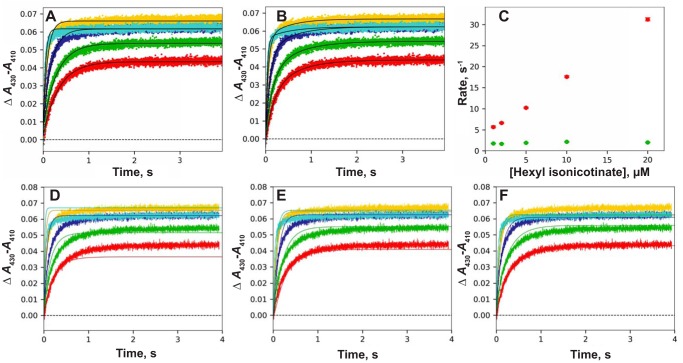
P450 2E1 and hexyl isonicotinate
Many of the classic substrates for P450 2E1 are small molecules (57) and do not give strong binding spectra (58). Alkyl isonicotinic acid esters have been shown to be substrates for ω-1 hydroxylation by P450 2E1 (at least in reactions supported by the oxygen surrogate cumene hydroperoxide), as well as generating Type II binding spectra (17). The binding of hexyl isonicotinate to P450 2E1 was rapid and could be fit with single exponential or biexponential equations (Fig. 6, A and B). The rate of the fast phase of binding increased with the ligand concentration (Fig. 6B) but the rate of the slower phase did not (Fig. 6C).
Figure 6.
Binding of hexyl isonicotinate to P450 2E1. P450 2E1 (2 μm) was mixed with hexyl isonicotinate concentrations of 1.0 (red), 2.0 (green), 5 (dark blue), 10 (light blue), and 20 (gold) μm. A, single exponential fits to traces of binding versus time. Linear regression analysis yielded kon = 0.98 × 106 m−1 s−1 and koff = 1.3 s−1 (results not shown). B, biexponential fit of data of A. C, fast (red points) and slow (green points) rates of binding as a function of substrate concentration. D, fits of binding data with a 2-state model (solid lines) for varying concentrations of hexyl isonicotinate, with kon = 1.5 × 106 m−1 s−1 and koff = 1.2 s−1. E, fits of data with an induced-fit model, with k1 = 1.9 × 106 m−1 s−1, k−1 = 5.5 s−1, k2 = 15 s−1, and k−2 = 2.8 s−1 (ϵ430–410 19.5 mm−1 cm−1). F, fits of data with a conformational-selection model, with k1 = 18 s−1, and k−1 = 110 s−1, k2 = 10 × 106 m−1 s−1, and k−2 0.41 s−1 (ϵ430–410 15.5 mm−1 cm−1).
A simple 2-state model was not adequate in fitting the data (Fig. 6D). Both an induced-fit model (Fig. 6E) and a conformational-selection model (Fig. 6F) yielded satisfactory fits, at least at the lower concentrations of the substrate, and a conclusion could not be reached as to which was superior.
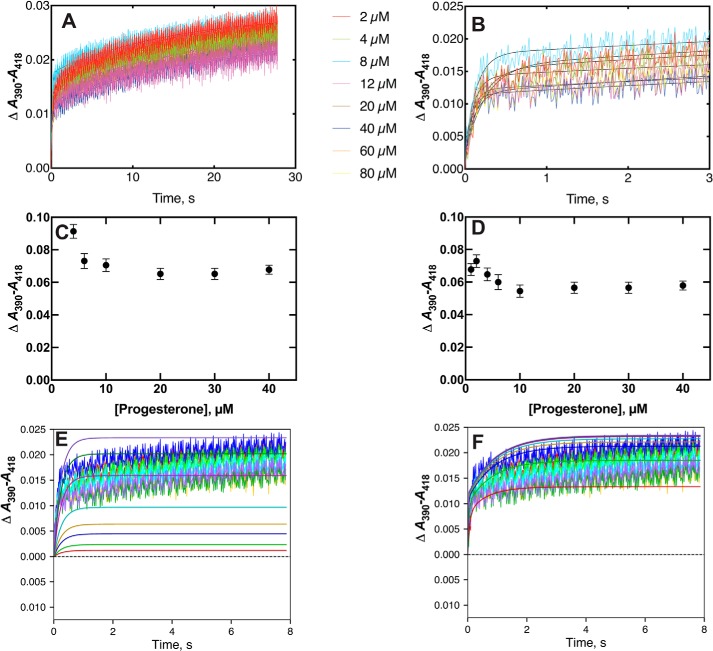
P450 21A2 and progesterone
P450 21A2 bound its substrate progesterone in a clearly biexponential mode (Fig. 7, A and B, showing separate time frames). Plotting of either the single-exponential rate (Fig. 7C) or the slow rate of the biexponential fit (Fig. 7D) yielded plots that showed decreasing rates with increasing substrate concentrations, suggesting a conformational-selection model (the faster of the biexponential rates were too fast to be useful). Fitting of the data yielded a generally better fit for a conformational-selection model than an induced-fit model (Fig. 7, E and F).
Figure 7.
Binding of progesterone to P450 21A2. P450 21A2 (2 μm) was mixed with varying concentrations of progesterone (2 (red), 4 (green), 8 (dark blue, lower trace), 12 (gold), 20 (light blue), 40 (magenta), 60 (red), and 80 (dark blue, upper trace) μm). A, traces of ΔA390-A418 measured with varying concentrations of progesterone. B, expansion of early phase (first 3 s) of A. C, plot of single exponential rates of binding versus progesterone concentration. D, plot of rates of the slow phase of biexponential fits (A) versus progesterone concentration. E, fits of data with an induced-fit model, with k1 = 1.2 × 106 m−1 s−1, k−1 = 100 s−1, k2 = 1.2 s−1, and k2 = 3.7 s−1 (ϵ390–418 46 mm−1 cm−1). F, fits of data with a conformational-selection model with k1 = 1.1 s−1, k−1 = 1.1 s−1, k2 = 6.6 × 106 m−1 s−1, and k−2 = 2.2 s−1 (ϵ390–418 12 mm−1 cm−1).
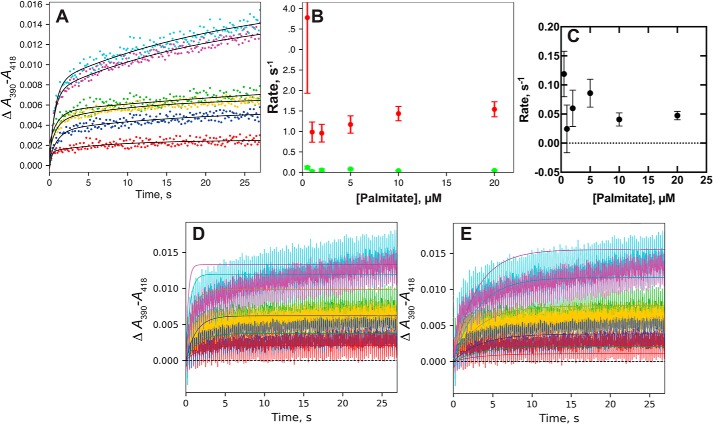
P450 2C8 and palmitic acid
The binding of the substrate palmitic acid to P450 2C8 yielded relatively weak spectral changes and the data were less robust (Fig. 8A). However, the traces could only be fit to biexponential plots (Fig. 8A). The rates of the faster phase increased slightly with the concentration of palmitic acid (Fig. 8B) but the rates of the slower phase decreased (Fig. 8C), indicative of a conformational-selection process. The fit to an induced-fit model (Fig. 8D) was generally not as good as that to a conformational-selection model (Fig. 8E).
Figure 8.
Binding of palmitic acid to P450 2C8. P450 2C8 (2 μm) was mixed with varying concentrations of palmitic acid (1.0 (red), 2.0 (dark blue), 4.0 (green), 10 (gold), 20 (magenta), and 40 (light blue) μm). A, biexponential fits to traces of ΔA390-A418. B, plots of fast (red points) and slow (green points) rates from A. C, plot of slow rate of binding from A, expanded from B. D, fit of data (A) to an induced-fit model (Fig. 2) with k1 = 0.11 × 106 m−1 s−1, k−1 = 6.8 s−1, k2 = 24 s−1, k−2 = 1.8 s−1 (ϵ390–418 4.0 mm−1 cm−1). E, fit of data (A) to a conformational-selection model (Fig. 2) with k1 = 0.4 s−1, k−1 = 110 s−1, k2 = 4.0 × 106 m−1 s−1, and k−2 = 0.26 s−1 (ϵ390–418 4.0 mm−1 cm−1).
P450 3A4 binding of midazolam and other ligands
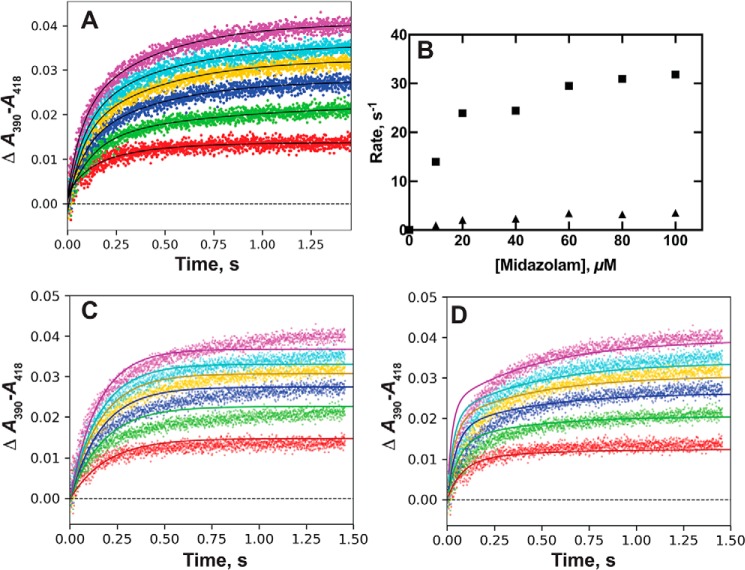
P450 3A4 interactions with several substrates and inhibitors were previously reported (32, 33), including the drug midazolam. We previously considered several possibilities for binding of midazolam to P450 3A4, including versions with multiple occupancy (32). The best of these fits, developed then using DynaFit software (59), involved a double induced-fit mechanism with two spectrally equivalent complexes (e.g. Scheme 1A of Ref. 32), and we tested some simpler models with the goal of finding less complex models that could adequately explain the data.
We re-evaluated some of the original data (32, 33) using our newer approaches, including the KinTek Explorer software (54). In our previous work, we reported rates of single-exponential fits for the rates of binding plotted versus midazolam concentration (32). The binding traces are clearly complex, as reported earlier (32) with biphasic absorbance changes (Fig. 9A). Neither plots of single-exponential fits nor either of the double-exponential rates yielded linear plots as a function of substrate concentration (Fig. 9B).
Figure 9.
Binding of midazolam to P450 3A4 (data from Ref. 32). P450 3A4 (2 μm) was mixed with midazolam (20 (red), 40 (green), 60 (dark blue), 80 (gold), 100 (light blue), and 150 (magenta) μm). A, double-exponential fits to data from (32) at varying midazolam concentrations. B, plots of biexponential rates (A) versus midazolam concentration: fast rate (■); slow rate (▴). C, fits to induced-fit model (Fig. 2) with k1 = 3.6 × 106 m−1 s−1, k−1 = 70 s−1, k2 = 3.4 s−1, and k−2 = 4.5 s−1 (ϵ390–418 28 mm−1 cm−1). D, fits to conformation selection model (Fig. 2) with k1 = 1.8 s−1, k−1 = 1.6 s−1, k2 = 0.2 × 106 m−1 s−1, and k−2 = 8.1 s−1 (ϵ390–418 10 mm−1 cm−1).
Reasonable fits were obtained with a simple induced-fit model (Fig. 9C). The kon rate (k1) was 4.4 × 106 m−1 s−1, which is realistic in light of other P450s (Table 1). The fit began to diverge at the higher midazolam concentrations. A basic conformational-selection model (which was not included earlier (32)) also fit well except at the higher midazolam concentrations (Fig. 9D). The kon rate of only 0.29 × 106 m−1 s−1 is low but probably not unrealistic. We also re-evaluated the binding of other ligands to P450 3A4, using the data files from our previous work (Figs. S1–S3).
With the substrate testosterone, a single-exponential fit was not unreasonable, and the rates increased with the substrate concentration (Fig. S1, A and B). A biexponential fit was better, and the rates for both reaction phases increased with testosterone concentration (Fig. S1, C and D). An induced-fit model (with kon 1.7 × 106 m−1 s−1, Fig. S1E) provided a credible fit, except for being somewhat too fast at the higher concentrations. Adjustment of the conformational-selection model (Fig. S1F) to fit the higher concentration data involved a kon rate of only 0.13 × 106 m−1 s−1, and the fit was inadequate at lower testosterone concentrations.
Bromocriptine binding was also re-examined (32), in that the size of this substrate and the crystal structure of the complex (34) rule out multiple ligand occupancy. Plotting the single-exponential fits of the data (Fig. S2A) versus the bromocriptine concentration showed increasing rates (Fig. S2B), as reported earlier (32). With biexponential fits (Fig. S2C), the faster rate also increased with bromocriptine concentration (Fig. S2D). Fitting to a simple induced-fit model was fair at low substrate concentrations (Fig. S2E) but attempts to fit to a conformational-selection model were much worse at multiplex bromocriptine concentrations (Fig. S2F).
Ketoconazole is an inhibitor of P450 3A4, producing a Type II difference spectrum with an azole nitrogen bonding to the heme iron (25). This inhibitory drug has also been reported to be a substrate and be oxidized by P450 3A4 (60). Fitting the previous data (33) to either single or biexponential plots of rate versus ketoconazole concentration (Fig. S3, A and C) gave plots in which the rates increased with the ketoconazole concentration (Fig. S3, B and D). The plots could be fit with an induced-fit or a conformational-selection model (Fig. S3, E and F), with deficiencies in each.
P450 3A4 and midazolam concentration dependence
The results with fitting of the previous P450 3A4 data (32, 33) were ambiguous, in that some could be fit with either an induced-fit or a conformational-selection model (Fig. 9, C and D, and Figs. S1, E and F, and S3, E and F). Furthermore, increased rates (hyperbolic) as a function of ligand concentration can be interpreted in terms of both induced-fit and conformational-selection models in the absence of more data (40, 53).
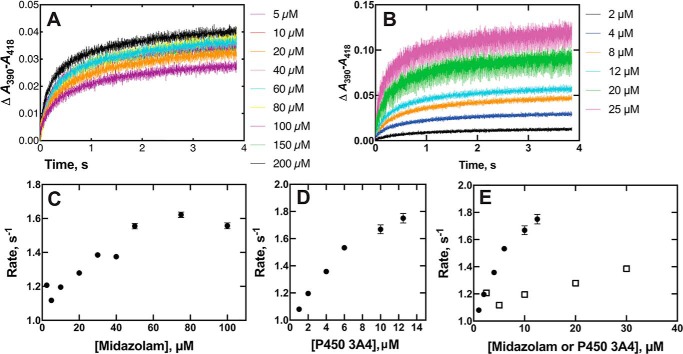
We repeated the rate measurements with a fixed concentration of P450 3A4 and varying concentrations of midazolam (Fig. 10A). As before (32), the rate increased with the midazolam concentration (Figs. 9B and 10A). The P450 3A4 concentration was also increased in the presence of a fixed concentration of a midazolam (2.5 μm) (Fig. 10B), yielding increased rates as a function of P450 3A4 concentration. Combining the results of Fig. 10, C and D (from plots of single exponential fits of the data from Fig. 10, A and B, respectively), in Fig. 10E indicated discordance in the patterns of rate dependence, which is a pattern characteristic of conformational-selection (with fast pre-equilibrium steps) but not an induced-fit model, in which the second-order rate plots should be identical (53).
Figure 10.
Binding of midazolam and P450 3A4 as functions of concentration of each component. A, P450 3A4 (2 μm) was mixed with the indicated concentration of midazolam (color schemes match the indicated concentrations used). B, midazolam (5 μm) was mixed with the indicated concentration of P450 3A4 (dialyzed before use to remove glycerol). Color schemes match the indicated P450 3A4 concentrations used for mixing. C, plot of single-exponential rates (A) (fitted using KinTek Explorer) versus midazolam concentration. D, plot of single-exponential rates (from B) (fitted with KinTek Explorer) versus P450 3A4 concentration. E, combined data points from C and D (varying P450 3A4, ●; varying midazolam, □). The decreased signal/noise ratio with the higher P450 3A4 concentration used in B is due to the use of a 4-mm path length cell to reduce the absorbance of the Soret band.
Discussion
Several human P450s were examined regarding the kinetics of interaction with substrates, with the aim of developing models that are as simple as possible and judging whether induced-fit or conformational-selection dominates. Based on previous work (32, 33, 52) and new experimental studies, we conclude (Table 2) that (i) some systems are simple and can be represented by two states, (ii) most P450-substrate systems can be described by a conformational-selection model, (iii) some P450-substrate systems may be described by an induced-fit or a conformational-selection mechanism in the absence of more data, and (iv) some P450-substrate systems are still more complex and probably involve elements of both induced-fit and conformational-selection. The simple systems (item i and Table 2) may prove to be more complex upon further analysis.
Table 2.
Classification of human P450s in terms of binding modes
a Number of substrates analyzed.
Clearly many mammalian P450s show complex binding behavior, as judged by lack of increased binding rates with substrate concentration (e.g. Figs. 4, 5, 7, and 8). Even when conventional methods produce linear plots, further analysis may indicate more kinetic complexity (e.g. P450 2E1, Fig. 6). In previous studies with P450s, we concluded that initial encounters of substrates with P450s were fast (1–10 × 106 m−1 s−1) and that subsequent steps involved migration of the substrate to the vicinity of the heme prosthetic group to produce the spectral changes (31–33). This can be considered a type of induced-fit mechanism, or at least one that would appear to be in the kinetic analysis. However, in several cases a pure induced-fit mechanism could not fit the data (e.g. Figs. 4, 5, 7, and 8), and a conformational-selection model was more appropriate. An issue with a pure conformational-selection model for a P450 is that the initial conformational equilibrium (Fig. 2) should be independent of the substrate used, and the only major difference in the kinetics with different substrates should be in k−2, the koff rate constant (Fig. 2), in that kon (k−2) should be similar for different substrates.
In the review of the manuscript, one of the referees suggested that a possible explanation for the need to use biexponential fits for the substrate-binding traces was the existence of two noninterconverting enzyme conformations. We tried to fit some of the binding data with two E + S ⇆ ES equilibria. The model began to fit, but the amplitude was problematic and, as might be expected from the magnitude of the values of the slow exponential phases of binding (see several of the other figures, e.g. Figs. 5–9), the kon rate constants needed to be ≤104 m−1 s−1, which is unrealistic for a simple diffusion-limited reaction (see below) and would require the introduction of additional steps. Also, a model with two noninterconverting forms of the enzyme, in which one form bound the substrate but was spectrally silent, could not fit the data.
In our analysis, E′S (Fig. 2) has the H2O ligand at least partially removed and the heme iron atom at least partially in the high-spin state, giving the final spectra (1, 11–15), except in the case of P450 2E1 (Fig. 6). However, in principle it might be possible for the conformational equilibrium (E interconverting with E′) to be the result of a low-/high-spin equilibrium, i.e. in the absence of substrate both low- and high-spin iron might be present and only the high-spin form would bind the substrate. However, if this were the case then a P450 should exist in a mixed-spin state population (in the absence of ligand). This has been observed with some P450s, e.g. P450 2E1 (61), and P450 1A2 is isolated essentially only in the high-spin configuration (62) and cannot be used in binding studies of the type done here (31). However, this cannot be a general explanation for the kinetic observations presented here and elsewhere (52), because the analysis of P450 17A1 binding is consistent with a ratio of 0.4–0.7 (52) of two conformations, using the estimated forward and reverse rates of conformational change (termed kr and k−r in the nomenclature of Vogt and Di Cera (37)). When we analyzed our preparation of (unliganded) P450 17A1 using second-derivative analysis (63), the preparation was ≥95% low-spin (Fig. S4). Also, analysis of the data for P450 3A4 (Fig. 10), with the approach of Vogt and Di Cera (37), gives k−r = 1.1 s−1 and (kr + k−r) = 1.6 s−1, so kr/k−r = 0.5/1.1 = ∼0.5. However, our second derivative analysis showed that the preparation was ≥95% low-spin (Fig. S4) and this finding would not be consistent with the kr/k−r ratios or the rate constants used in the modeling.
The fits presented here are intended to employ the most minimal mechanisms and are admittedly less than perfect, with the goal of trying to identify main features. In the application of FitSpace software (KinTek Explorer) (64), the estimated rate constants could not be concluded to be highly constrained (data not shown) due to the lack of independent data sets to restrain the modeling. Another caveat of this work is that we have not extensively considered a large number of the substrates for the “drug-metabolizing” human P450s (e.g. P450s 2D6, 3A4, 2E1), which have many substrates.
The most generally appropriate value of a kon rate constant for an enzyme is uncertain. Although diffusion-limited values have been considered by some to be in the range of 108–109 m−1 s−1 (65), others have suggested lower values (105–107 m−1 s−1) (66) in the absence of intermolecular forces (e.g. charge). The values we used are in this range. Lower rates (e.g. ∼103–104 m−1 s−1 (20)) are often reported for surface plasmon resonance studies, but these are very compromised by surface artifacts (67).
In previous work with P450 3A4 we concluded that an induced-fit mechanism was an appropriate explanation for the multiphasic binding kinetics (33). This work was done before the theoretical framework for distinguishing mechanisms by concentration dependence of kinetics was published (37, 53). One argument against conformational-selection is that observed rates of binding are different and P450 3A4 ligands varied considerably (32, 33). However, it is possible that more than two conformational states may be involved (Fig. 2), and for instance, we might be using E, E′, and E″ in different cases with E″ being only a minor component or in slow equilibrium with the other forms (for binding certain ligands). The argument for induced-fit that we advanced in our 2007 report on P450 3A4 (33) was based on the results of a double-mixing experiment with indinavir followed by testosterone (Fig. 7 of Ref. 33). This experiment is complicated, in that the absorbance change (A390 increase) seen with binding testosterone is in the opposite direction of that seen with indinavir (A405 decrease) and at that time we did not resolve the spectra. We did observe a slow decrease in the amplitude of the testosterone response as a function of the time elapsed after indinavir was incubated (before the addition of testosterone). Also, as pointed out in the report (33), a conformational-selection model does not necessarily exclude silent steps. Furthermore, our more recent work with the dye/substrate Nile Red showed transient absorbance-silent (but fluorescent) changes with P450 17A1, an enzyme for which a conformational-selection mechanism was demonstrated (52).
Pearson et al. (20) also studied ketoconazole binding to P450 3A4 (Fig. S3), as well as itraconazole. As pointed out here, ketoconazole is both an inhibitor and a substrate, and the same applies to itraconazole. Pearson et al. (20) developed a model in which free P450 3A4 could bind ketoconazole in either of two modes. This possibility certainly cannot be ruled out and is not inconsistent with our own conclusions, except that the surface plasmon resonance method used (20) yielded kon rate constants of only 1–4 × 104 m−1 s−1, which are inconsistent with our own values in solution experiments. As pointed out earlier, surface plasmon resonance experiments involve bound molecules and are subject to surface artifacts (67).
Recently Montemiglio and co-workers (68) used a double-mixing approach to conclude that a bacterial P450, P450 OleP, utilizes a conformational-selection mechanism in binding 6-deoxyerythronolide B. As in the case of our earlier results (33), it is not clear that this particular mixing experiment proves conformational-selection. In the P450 3A4 experiment (33) the binding of indinavir was very tight (Kd 0.3 μm) and the P450 3A4 and indinavir concentrations were both 8 μm, so that essentially all indinavir would be complexed with the P450 if it bound before testosterone (33). In the P450 OleP study (68), the Kd was 5 μm and the 6-deoxyerythyronolide B and clotrimazole concentrations were 100 and 5–25 μm, respectively, so that there was competition for binding of these two ligands to P450 OleP. Nevertheless, the bulk of the other kinetic work by Montemiglio and co-workers (68) presents valid evidence for the involvement of conformational-selection for P450 OleP binding of 6-deoxyerythronolide B, and we do not dispute the overall conclusion.
The studies with the bacterial P450s OleP (68) and EryK (69) both implicate conformational-selection in the binding of substrates. In both cases the proteins are monomeric. We have not directly assessed the oligomeric state of any of our human P450s (except for P450 17A1, for which only about one-half is monomeric as judged by size exclusion chromatography (52)), and oligomerization is a general mode for mammalian P450s when they have been analyzed (70, 71). Changes in oligomerization, either in the degree of oligomerization or rearrangement within an oligomer, could be part of the process of conformational-selection and cannot be ruled out (72).
Bacterial P450cam (P450 101A1) is probably the most extensively characterized P450, from a biophysical standpoint (55). It is a monomeric enzyme and was the first P450 to be crystallized; numerous crystal structures of the enzyme at various steps in the catalytic cycle, with several ligands, are now available. There has been some controversy about the roles of open and closed forms in catalysis, particularly in the complexes with its accessory electron transfer partner putidaredoxin (73, 74). This seemingly simple P450 is also complex due to the issue of multiple ligand occupancy (74–77). Furthermore, conversion of high-spin iron back to low-spin occurs upon binding of the second molecule of camphor (substrate) (75, 77). Relevant to the present work is the multiplicity of X-ray crystal structures of ligand-free P450cam existing in both closed (78) and open (79) forms. NMR spectroscopy and molecular dynamic work also lead to the conclusion that ferric P450cam exists in an ensemble of conformations in the substrate-free form (80). However, we are not aware that any kinetic binding studies have been published on the binding of substrate as it relates to this phenomenon. We are also not aware if the presence of bound putidaredoxin affects the binding of any substrates to P450cam.
Another issue to consider is the complexity of other P450 reactions. For instance, the reduction of ferric P450 by NADPH-P450 reductase is often biphasic (but not always) (6). Complex explanations have been presented to account for this including membrane organization (81), flavin electron transfer (82), and spin state (83), although the latter cannot be general, e.g. case of P450 1A2 (6). One possibility is that ensembles of conformational forms of both free and substrate-bound P450 exist in equilibrium and are reduced at different rates, just as different ensembles bind ligands at different rates.
One point of discussion is that models for conformational-selection are generally restricted to two entities but that is probably not the limit, e.g. see Benkovic et al. (84). Multiple conformations may be relevant and, as mentioned earlier, be responsible for variations in the rates of binding of P450 3A4 (32, 33), even if the rates of conversion in the unliganded state are independent of the ligand. Realistically it is not useful to include more than two species in most efforts at kinetic modeling, in the absence of evidence that more conformations exist, due to the complexity. Another complicating issue is that we have built our models (Fig. 2) with the assumption that only one form of the enzyme can bind the substrate, but we cannot rule out the possibility that two (or more) forms both bind substrate (even yielding the expected spectral changes) and only one is poised for productive catalysis (20).
Another issue is that accessory proteins might modify the ligand binding behavior of a P450 (or other enzyme). In this regard, Scott (85) has reported that the presence of the iron-sulfur protein partner adrenodoxin enhances the binding of the substrate 11-deoxycosrticostereone to P450 11B2 by 7-fold, and we have repeated this finding.4 All microsomal P450s bind NADPH-P450 reductase and some also bind cytochrome b5. With human P450 17A1, which is known to interact with cytochrome b5 (86), we found that cytochrome b5 did not modify the binding of the substrate 17α-hydroxypregnenolone (52). No effect of NADPH-P450 reductase was seen in binding of substrates to P450s 1A2, 2A6, 2D6, and 3A4 (87). However, we have not systematically examined other P450s in this regard or for the effect of binding NADPH-P450 reductase or cytochrome b5 on the binding of substrates.
We have analyzed P450s with multiple substrates (Table 2) and concluded that conformational-selection was the dominant model in each case, as we did with P450 17A1 and seven steroids (52). In the case of P450 3A4 we analyzed data with four ligands (Figs. 9 and 10 and Figs. S1, S2, and S3). With P450 3A4 the patterns were rather consistent, but only in the case of the substrate midazolam did we extend the analysis to definitively corroborate conformational-selection (Fig. 10). To some extent the question of an induced-fit mechanism versus conformational-selection could be dependent upon the ligand. Although the phenomenon of conformational-selection should be independent of the ligand, the possibility exists that with a particular ligand an induced-fit phenomenon might be operative to the extent of overwhelming the overall nature of the observed kinetics. Another possibility, already mentioned, is the existence of more than two conformations and the preference of some to bind to a particular ligand.
Although we have done some analysis with nine human P450s in this and previous work (7, 30, 52), there are 48 other human P450s and we cannot comment on their behavior. With some of the mammalian P450s, the binding of substrates does not induce spectral changes (18, 62). Moreover, the weaker spectral changes seen with some of the P450-ligand associations are more difficult to analyze (e.g. Figs. 4, 7, and 8) and probably preclude detailed analysis of the type done in Fig. 10 by varying the protein concentration (53).
In conclusion, we analyzed a number of human P450s that we had interest in and were available in our laboratory. Some of the analyses were more difficult because of the weak spectral changes observed upon binding, but these did show the decrease in binding rates with increasing ligand concentration that is characteristic of conformational-selection (53). P450 3A4 binding rates increased with substrate concentrations but the detailed kinetic analysis with midazolam binding revealed a conformation selection mechanism due to the discordance of second-order rates of binding (Fig. 10E) (53). We conclude that many of the human P450s appear to bind their substrates via a conformational-selection mode.
Experimental procedures
Chemicals
Except for the synthesized chemical (see below), all others were purchased from Sigma and used without further purification.
Hexyl isonicotinate (isonicotinic acid hexyl ester)
Hexyl isonicotinate was synthesized by the condensation of isonicotinic acid with 1-hexanol using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and 4-dimethylaminopyridine in (CH3)2NCHO as described (17) in 66% yield: high resolution MS: calculated for C12H18NO2+ 208.1332, found 208.1341, Δ4.3 ppm. UV λmax (CH3OH) 275 nm, ϵ275 1,780 m−1 cm−1; 1H NMR (400 MHz, CD2Cl2), δ 0.91 (t, 3H, −CH3), 1.3–1.4 (m, 8H, -CH2-), 4.3 (t, 2H, -OCH2−), 7.85 (dd, 2H, H-3 (ring)), 8.75 (dd, 2H, H-2 (ring)).
Enzymes
Human P450s 2C8 (88), 2D6 (89), 2E1 (61), 3A4 (90, 91), 4A11 (27), and 21A2 (29) were expressed with C-terminal oligo-His tags (and slightly modified N-terminal amino acid sequences to improve expression) in Escherichia coli and purified to near electrophoretic homogeneity as described previously. All of these constructs have some truncation of the N terminus, but our work (27, 29, 61, 88–91) and that of others in the field all show what are expected to be full catalytic activities (92). With regard to the effect of truncation on conformation, it is difficult to answer completely, in that in only one case in which a crystal structure of a full-length mammalian P450 has been reported (93) and with all of the P450 structures the N-terminal tail is unstructured and has too much motion to be seen in the crystals (21).
Measurement of kinetics of binding
All measurements were made using an OLIS RSM-1000 stopped-flow spectrophotometer (On-Line Instrument Systems, Bogart, GA) in the rapid scanning mode with a 20 × 4 mm cell, 1.24 mm slits, and 600 line/500 nm gratings at 23 °C. In the cases where P450 heme absorbance was high (e.g. Fig. 10B, A418 > 1), a 4-mm path length cell (4 × 4 mm) was utilized. For collection time periods of ≤4 s, data were collected at 1000 scans/s. For time periods of ≥4 s, 62 scans/s were collected in the signal averaging mode. The wavelength range was 330–570 nm.
The general measurement mode involved mixing one syringe containing 2–4 μm P450 (in 100 mm potassium phosphate buffer, pH 7.4) with an equal volume of the same buffer containing varying concentrations of substrate or other ligand.
The data were saved as Excel files and most were converted to ΔAmax − Amin files (usually ΔA390–A418, except ΔA430–A410 with hexyl isonicotinate binding to P450 2E1). The resulting Excel files were corrected to ΔAt = 0 = 0 and saved as txt files for import into the KinTek Explorer program.
Kinetic modeling
All work was done with KinTek Explorer® software (Kintek, Snowshoe PA) using an Apple iMac OSX 10.13.6 system and Explorer Version 8.0 (2018) (54). txt files were imported directly into the program.
The general procedure involved an initial overall analysis a family of traces of ΔA versus time (varying substrate concentration), with a series of single exponential fits for each. The individual rates were plotted versus the substrate concentration. This analysis was followed by a series of biexponential fits of all traces and then plotting both rates (fast and slow phases) versus substrate concentration. From these plots a conclusion was reached whether the system followed a single 2-state or a more complex model, based on whether a plot of the apparent rate versus substrate concentration was linear.
Attempts were made to globally fit the data to either an induced-fit model (Model 1, Equations 1 and 2),
| (Eq. 1) |
| (Eq. 2) |
with E, P450; S, substrate; ES, initial substrate complex; E′S, final substrate complex and only E′S being observed (*), or to a conformational-selection model (Model 2, Equations 3 and 4),
| (Eq. 3) |
| (Eq. 4) |
with E and E′ being alternate conformational forms of P450; S being the substrate, and E′S being the only observed P450-substrate complex (*) (Fig. 2).
Individual rate constants for both forward and reverse steps and the ϵ (the extinction coefficient) were adjusted manually to obtain the most general fits with the various models, whereas attempting to hold (i) the kon rate (E + S → ES or E′ + S → E′S) ≥0.5 × 106 m−1 s−1 if possible and (ii) matching the maximum absorbance reached at the end of the reaction.
Author contributions
F. P. G. conceptualization; F. P. G. data curation; F. P. G. formal analysis; F. P. G. supervision; F. P. G. funding acquisition; F. P. G. validation; F. P. G., C. J. W., and T. T. N. P. investigation; F. P. G. visualization; F. P. G. writing-original draft; F. P. G. project administration; F. P. G., C. J. W., and T. T. N. P. writing-review and editing; C. J. W. and T. T. N. P. resources.
Supplementary Material
Acknowledgments
We thank S. M. Leddy and Prof. L. L. Furge for assistance in acquiring the previously data used in Fig. 4A (56), Dr. E. M. Isin for acquiring the data used in Fig. 9A and Figs. S1A, S2A, and S3A (32, 33), and K. Trisler for assistance in preparation of the manuscript.
This work was supported by National Institutes of Health Grants R01 GM118122 (to F. P. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
M. J. Reddish and F. P. Guengerich, unpublished results.
- P450
- cytochrome P450.
References
- 1. Ortiz de Montellano P. R. (2015) Substrate oxidation. in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano P. R., ed) 4th Ed., pp 111–176, Springer, New York [Google Scholar]
- 2. Rendic S., and Guengerich F. P. (2018) Human cytochrome P450 enzyme 5–51 as targets of drugs, natural, and environmental compounds: mechanisms, induction, and inhibition, toxic effects and benefits. Drug Metab. Rev. 50, 256–342 10.1080/03602532.2018.1483401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rendic S., and Guengerich F. P. (2015) Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem. Res. Toxicol. 28, 38–42 10.1021/tx500444e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guengerich F. P. (2018) Perspective: mechanisms of cytochrome P450-catalyzed oxidations. ACS Catalysis 8, 10964–10976 10.1021/acscatal.8b03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guengerich F. P., and Yoshimoto F. K. (2018) Formation and cleavage of C-C bonds by enzymatic oxidation-reduction reactions. Chem. Rev. 118, 6573–6655 10.1021/acs.chemrev.8b00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guengerich F. P., and Johnson W. W. (1997) Kinetics of ferric cytochrome P450 reduction by NADPH-cytochrome P450 reductase: rapid reduction in the absence of substrate and variations among cytochrome P450 systems. Biochemistry 36, 14741–14750 10.1021/bi9719399 [DOI] [PubMed] [Google Scholar]
- 7. Yun C.-H., Kim K. H., Calcutt M. W., and Guengerich F. P. (2005) Kinetic analysis of oxidation of coumarins by human cytochrome P450 2A6. J. Biol. Chem. 280, 12279–12291 10.1074/jbc.M411019200 [DOI] [PubMed] [Google Scholar]
- 8. Johnston W. A., Hunter D. J., Noble C. J., Hanson G. R., Stok J. E., Hayes M. A., De Voss J. J., and Gillam E. M. (2011) Cytochrome P450 is present in both ferrous and ferric forms in the resting state within intact Escherichia coli and hepatocytes. J. Biol. Chem. 286, 40750–40759 10.1074/jbc.M111.300871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remmer H., Schenkman J., Estabrook R. W., Sasame H., Gillette J., Narasimhulu S., Cooper D. Y., and Rosenthal O. (1966) Drug interaction with hepatic microsomal cytochrome. Mol. Pharmacol. 2, 187–190 [PubMed] [Google Scholar]
- 10. Schenkman J. B., Remmer H., and Estabrook R. W. (1967) Spectral studies of drug interaction with hepatic microsomal cytochrome P-450. Mol. Pharmacol. 3, 113–123 [PubMed] [Google Scholar]
- 11. Hildebrandt A., Remmer H., and Estabrook R. W. (1968) Cytochrome P-450 of liver microsomes: one pigment or many. Biochem. Biophys. Res. Commun. 30, 607–612 10.1016/0006-291X(68)90555-X [DOI] [PubMed] [Google Scholar]
- 12. Mitani F., and Horie S. (1969) Studies on P-450: V. on the substrate-induced spectral change of P-450 solubilized from bovine adrenocortical mitochondria. J. Biochem. (Tokyo) 65, 269–280 [PubMed] [Google Scholar]
- 13. Mitani F., and Horie S. (1969) Studies on P-450: VI. the spin state of P-450 solubilized from bovine adrenocortical mitochondria. J. Biochem. (Tokyo) 66, 139–149 10.1093/oxfordjournals.jbchem.a129129 [DOI] [PubMed] [Google Scholar]
- 14. Imai Y., Horie S., Yamano T., and Iizuka T. (1978) Molecular properties. in Cytochrome P-450 (Sato R., and Omura T., eds) pp. 37–135, Academic Press, New York [Google Scholar]
- 15. Sligar S. G., and Gunsalus I. C. (1976) A thermodynamic model of regulation: modulation of redox equilibria in camphor monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 73, 1078–1082 10.1073/pnas.73.4.1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng C. C., Pearson J. T., Rock D. A., Joswig-Jones C. A., and Jones J. P. (2010) The effects of Type II binding on metabolic stability and binding affinity in cytochrome P450 CYP3A4. Arch. Biochem. Biophys. 497, 68–81 10.1016/j.abb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Méard A., Fabra C., Huang Y., and Auclair K. (2012) Type II ligands as chemical auxiliaries to favor enzymatic transformations by P450 2E1. Chembiochem 13, 2527–2536 10.1002/cbic.201200524 [DOI] [PubMed] [Google Scholar]
- 18. Guengerich F. P. (1983) Oxidation-reduction properties of rat liver cytochromes P-450 and NADPH-cytochrome P-450 reductase related to catalysis in reconstituted systems. Biochemistry 22, 2811–2820 10.1021/bi00281a007 [DOI] [PubMed] [Google Scholar]
- 19. Isin E. M., and Guengerich F. P. (2008) Substrate binding to cytochromes P450. Anal. Bioanal. Chem. 392, 1019–1030 10.1007/s00216-008-2244-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pearson J. T., Hill J. J., Swank J., Isoherranen N., Kunze K. L., and Atkins W. M. (2006) Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry 45, 6341–6353 10.1021/bi0600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poulos T. L., and Johnson E. F. (2015) Structures of cytochrome P450 enzymes. in Cytochrome P450: Structure, Function, and Biochemistry (Ortiz de Montellano P. R., ed) 4th Ed., pp 3–32, Springer, New York [Google Scholar]
- 22. Schoch G. A., Yano J. K., Wester M. R., Griffin K. J., Stout C. D., and Johnson E. F. (2004) Structure of human microsomal cytochrome P450 2C8: evidence for a peripheral fatty acid binding site. J. Biol. Chem. 279, 9497–9503 10.1074/jbc.M312516200 [DOI] [PubMed] [Google Scholar]
- 23. Zhao B., Guengerich F. P., Bellamine A., Lamb D. C., Izumikawa M., Lei L., Podust L. M., Sundaramoorthy M., Kalaitzis J. A., Reddy L. M., Kelly S. L., Moore B. S., Stec D., Voehler M., Falck J. R., Shimada T., and Waterman M. R. (2005) Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J. Biol. Chem. 280, 11599–11607 10.1074/jbc.M410933200 [DOI] [PubMed] [Google Scholar]
- 24. Dabrowski M. J., Schrag M. L., Wienkers L. C., and Atkins W. M. (2002) Pyrene-pyrene complexes at the active site of cytochrome P450 3A4: evidence for a multiple substrate binding site. J. Am. Chem. Soc. 124, 11866–11867 10.1021/ja027552x [DOI] [PubMed] [Google Scholar]
- 25. Ekroos M., and Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 10.1073/pnas.0603236103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffin B. W., and Peterson J. A. (1972) Camphor binding by Pseudomonas putida cytochrome P-450: kinetics and thermodynamics of the reaction. Biochemistry 11, 4740–4746 10.1021/bi00775a017 [DOI] [PubMed] [Google Scholar]
- 27. Kim D., Cha G. S., Nagy L. D., Yun C.-H., and Guengerich F. P. (2014) Kinetic analysis of lauric acid hydroxylation by human cytochrome P450 4A11. Biochemistry 53, 6161–6172 10.1021/bi500710e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sohl C. D., and Guengerich F. P. (2010) Kinetic analysis of the three-step steroid aromatase reaction of human cytochrome P450 19A1. J. Biol. Chem. 285, 17734–17743 10.1074/jbc.M110.123711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pallan P. S., Wang C., Lei L., Yoshimoto F. K., Auchus R. J., Waterman M. R., Guengerich F. P., and Egli M. (2015) Human cytochrome P450 21A2, the major steroid 21-hydroxylase: structure of the enzyme·progesterone substrate complex and rate-limiting C-H bond cleavage. J. Biol. Chem. 290, 13128–13143 10.1074/jbc.M115.646307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson K. M., Phan T. T. N., Albertolle M. E., and Guengerich F. P. (2017) Human mitochondrial cytochrome P450 27C1 is localized in skin and preferentially desaturates trans-retinol to 3,4-dehydroretinol. J. Biol. Chem. 292, 13672–13687 10.1074/jbc.M116.773937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sohl C. D., Isin E. M., Eoff R. L., Marsch G. A., Stec D. F., and Guengerich F. P. (2008) Cooperativity in oxidation reactions catalyzed by cytochrome P450 1A2: highly cooperative pyrene hydroxylation and multiphasic kinetics of ligand binding. J. Biol. Chem. 283, 7293–7308 10.1074/jbc.M709783200 [DOI] [PubMed] [Google Scholar]
- 32. Isin E. M., and Guengerich F. P. (2006) Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 281, 9127–9136 10.1074/jbc.M511375200 [DOI] [PubMed] [Google Scholar]
- 33. Isin E. M., and Guengerich F. P. (2007) Multiple sequential steps involved in the binding of inhibitors to cytochrome P450 3A4. J. Biol. Chem. 282, 6863–6874 10.1074/jbc.M610346200 [DOI] [PubMed] [Google Scholar]
- 34. Sevrioukova I. F., and Poulos T. L. (2012) Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand. J. Biol. Chem. 287, 3510–3517 10.1074/jbc.M111.317081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gonzalez E., and Guengerich F. P. (2017) Kinetic processivity of the two-step oxidations of progesterone and pregnenolone to androgens by human cytochrome P450 17A1. J. Biol. Chem. 292, 13168–13185 10.1074/jbc.M117.794917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hammes G. G., Chang Y. C., and Oas T. G. (2009) Conformational-selection or induced-fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 10.1073/pnas.0907195106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vogt A. D., and Di Cera E. (2012) Conformational-selection or induced-fit? a critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902 10.1021/bi3006913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou H. X. (2010) From induced-fit to conformational-selection: a continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys. J. 98, L15–L17 10.1016/j.bpj.2009.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Changeux J. P., and Edelstein S. (2011) Conformational-selection or induced-fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chakraborty P., and Di Cera E. (2017) Induced-fit is a special case of conformational-selection. Biochemistry 56, 2853–2859 10.1021/acs.biochem.7b00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vogt A. D., Pozzi N., Chen Z., and Di Cera E. (2014) Essential role of conformational-selection in ligand binding. Biophys. Chem. 186, 13–21 10.1016/j.bpc.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davydov D. R., Halpert J. R., Renaud J.-P., and Hui Bon Hoa G. (2003) Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 312, 121–130 10.1016/j.bbrc.2003.09.247 [DOI] [PubMed] [Google Scholar]
- 43. Lampe J. N., Floor S. N., Gross J. D., Nishida C. R., Jiang Y., Trnka M. J., and Ortiz de Montellano P. R. (2008) Ligand-induced conformational heterogeneity of cytochrome P450 CYP119 identified by 2D NMR spectroscopy with the unnatural amino acid 13C-p-methoxyphenylalanine. J. Am. Chem. Soc. 130, 16168–16169 10.1021/ja8071463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Estrada D. F., Skinner A. L., Laurence J. S., and Scott E. E. (2014) Human cytochrome P450 17A1 conformational-selection: modulation by ligand and cytochrome b5. J. Biol. Chem. 289, 14310–14320 10.1074/jbc.M114.560144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petrunak E. M., Rogers S. A., Aubé J., and Scott E. E. (2017) Structural and functional evaluation of clinically relevant inhibitors of steroidogenic cytochrome P450 17A1. Drug Metab. Dispos. 45, 635–645 10.1124/dmd.117.075317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Porubsky P. R., Meneely K. M., and Scott E. E. (2008) Structures of human cytochrome P-450 2E1: insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J. Biol. Chem. 283, 33698–33707 10.1074/jbc.M805999200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Porubsky P. R., Battaile K. P., and Scott E. E. (2010) Human cytochrome P450 2E1 structures with fatty acid analogs reveal a previously unobserved binding mode. J. Biol. Chem. 285, 22282–22290 10.1074/jbc.M110.109017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bart A. G., and Scott E. E. (2018) Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands. J. Biol. Chem. 293, 19201–19210 10.1074/jbc.RA118.005588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walsh A. A., Szklarz G. D., and Scott E. E. (2013) Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J. Biol. Chem. 288, 12932–12943 10.1074/jbc.M113.452953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaur P., Chamberlin A. R., Poulos T. L., and Sevrioukova I. F. (2016) Structure-based inhibitor design for evaluation of a CYP3A4 pharmacophore model. J. Med. Chem. 59, 4210–4220 10.1021/acs.jmedchem.5b01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sevrioukova I. F., and Poulos T. L. (2010) Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl. Acad. Sci. U.S.A. 107, 18422–18427 10.1073/pnas.1010693107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guengerich F. P., Wilkey C. J., Glass S. M., and Reddish M. J. (2019) Conformational-selection dominates binding of steroids to human cytochrome P450 17A1. J. Biol. Chem. 294, 10028–10041 10.1074/jbc.RA119.008860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gianni S., Dogan J., and Jemth P. (2014) Distinguishing induced-fit from conformational-selection. Biophys. Chem. 189, 33–39 10.1016/j.bpc.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 54. Johnson K. A., Simpson Z. B., and Blom T. (2009) Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 10.1016/j.ab.2008.12.024 [DOI] [PubMed] [Google Scholar]
- 55. Mueller E. J., Loida P. J., and Sligar S. G. (1995) Twenty-five years of P450cam research: mechanistic insights into oxygenase catalysis. in Cytochrome P450-Structure, Mechanism, and Biochemistry (Ortiz de Montellano P. R., ed) 2nd Ed., pp. 83–124, Plenum, New York [Google Scholar]
- 56. Glass S. M., Leddy S. M., Orwin M. C., Miller G. P., Furge K. A., and Furge L. L. (2019) Rolapitant is a reversible inhibitor of CYP2D6. Drug Metab. Dispos. 47, 567–573 10.1124/dmd.118.085928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guengerich F. P., Kim D. H., and Iwasaki M. (1991) Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 4, 168–179 10.1021/tx00020a008 [DOI] [PubMed] [Google Scholar]
- 58. Bell L. C., and Guengerich F. P. (1997) Oxidation kinetics of ethanol by human cytochrome P450 2E1: rate-limiting product release accounts for effects of isotopic hydrogen substitution and cytochrome b5 on steady-state kinetics. J. Biol. Chem. 272, 29643–29651 10.1074/jbc.272.47.29643 [DOI] [PubMed] [Google Scholar]
- 59. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV protease. Anal. Biochem. 237, 260–273 10.1006/abio.1996.0238 [DOI] [PubMed] [Google Scholar]
- 60. Fitch W. L., Tran T., Young M., Liu L., and Chen Y. (2009) Revisiting the metabolism of ketoconazole using accurate mass. Drug Metab. Lett. 3, 191–198 10.2174/187231209789352085 [DOI] [PubMed] [Google Scholar]
- 61. Gillam E. M., Guo Z., and Guengerich F. P. (1994) Expression of modified human cytochrome P450 2E1 in Escherichia coli, purification, and spectral and catalytic properties. Arch. Biochem. Biophys. 312, 59–66 10.1006/abbi.1994.1280 [DOI] [PubMed] [Google Scholar]
- 62. Sandhu P., Guo Z., Baba T., Martin M. V., Tukey R. H., and Guengerich F. P. (1994) Expression of modified human cytochrome P450 1A2 in Escherichia coli: stabilization, purification, spectral characterization, and catalytic activities of the enzyme. Arch. Biochem. Biophys. 309, 168–177 10.1006/abbi.1994.1099 [DOI] [PubMed] [Google Scholar]
- 63. O'Haver T. C., and Green G. L. (1976) Numerical error analysis of derivative spectrometry for the quantitative analysis of mixtures. Anal. Chem. 48, 312–318 10.1021/ac60366a016 [DOI] [Google Scholar]
- 64. Johnson K. A., Simpson Z. B., and Blom T. (2009) FitSpace Explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 387, 30–41 10.1016/j.ab.2008.12.025 [DOI] [PubMed] [Google Scholar]
- 65. Fersht A. (1999) Structure and Mechanism in Protein Science. pp. 158–167, Freeman, New York [Google Scholar]
- 66. Schreiber G., Haran G., and Zhou H. X. (2009) Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 10.1021/cr800373w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson K. A. (2017) 12th New Enzymology Kinetics Workshop. in 12th New Enzymology Kinetics Workshop (Johnson K. A., ed) p. 23, KinTek, Austin, TX [Google Scholar]
- 68. Parisi G., Montemiglio L. C., Giuffre A., Macone A., Scaglione A., Giuffrè A., Cerutti G., Exertier C., Savino C., and Vallone B. (2019) Substrate-induced conformational change in cytochrome P450 OleP. FASEB J. 33, 1787–1800 10.1096/fj.201800450RR [DOI] [PubMed] [Google Scholar]
- 69. Savino C., Montemiglio L. C., Sciara G., Miele A. E., Kendrew S. G., Jemth P., Gianni S., and Vallone B. (2009) Investigating the structural plasticity of a cytochrome P450: three-dimensional structures of P450 EryK and binding to its physiological substrate. J. Biol. Chem. 284, 29170–29179 10.1074/jbc.M109.003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guengerich F. P., and Holladay L. A. (1979) Hydrodynamic characterization of highly purified and functionally active liver microsomal cytochrome P-450. Biochemistry 18, 5442–5449 10.1021/bi00591a029 [DOI] [PubMed] [Google Scholar]
- 71. French J. S., Guengerich F. P., and Coon M. J. (1980) Interactions of cytochrome P-450, NADPH-cytochrome P-450 reductase, phospholipid, and substrate in the reconstituted liver microsomal enzyme system. J. Biol. Chem. 255, 4112–4119 [PubMed] [Google Scholar]
- 72. Davydov D. R., Davydova N. Y., Sineva E. V., Kufareva I., and Halpert J. R. (2013) Pivotal role of P450-P450 interactions in CYP3A4 allostery: the case of α-naphthoflavone. Biochem. J. 453, 219–230 10.1042/BJ20130398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tripathi S., Li H., and Poulos T. L. (2013) Structural basis for effector control and redox partner recognition in cytochrome P450. Science 340, 1227–1230 10.1126/science.1235797 [DOI] [PubMed] [Google Scholar]
- 74. Colthart A. M., Tietz D. R., Ni Y., Friedman J. L., Dang M., and Pochapsky T. C. (2016) Detection of substrate-dependent conformational changes in the P450-fold by nuclear magnetic resonance. Sci. Rep. 6, 22035 10.1038/srep22035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Marden M. C., and Hoa G. H. (1987) P-450 binding to substrates camphor and linalool versus pressure. Arch. Biochem. Biophys. 253, 100–107 10.1016/0003-9861(87)90642-4 [DOI] [PubMed] [Google Scholar]
- 76. Yao H., McCullough C. R., Costache A. D., Pullela P. K., and Sem D. S. (2007) Structural evidence for a functionally relevant second camphor binding site in P450cam: model for substrate entry into a P450 active site. Proteins 69, 125–138 10.1002/prot.21508 [DOI] [PubMed] [Google Scholar]
- 77. Follmer A. H., Mahomed M., Goodin D. B., and Poulos T. L. (2018) Substrate-dependent allosteric regulation in cytochrome P450cam (CYP101A1). J. Am. Chem. Soc. 140, 16222–16228 10.1021/jacs.8b09441 [DOI] [PubMed] [Google Scholar]
- 78. Poulos T. L., Finzel B. C., and Howard A. J. (1986) Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry 25, 5314–5322 10.1021/bi00366a049 [DOI] [PubMed] [Google Scholar]
- 79. Lee Y. T., Wilson R. F., Rupniewski I., and Goodin D. B. (2010) P450cam visits an open conformation in the absence of substrate. Biochemistry 49, 3412–3419 10.1021/bi100183g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Asciutto E. K., Young M. J., Madura J., Pochapsky S. S., and Pochapsky T. C. (2012) Solution structural ensembles of substrate-free cytochrome P450cam. Biochemistry 51, 3383–3393 10.1021/bi300007r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Peterson J. A., Ebel R. E., O'Keeffe D. H., Matsubara T., and Estabrook R. W. (1976) Temperature dependence of cytochrome P-450 reduction: a model for NADPH-cytochrome P-450 reductase: cytochrome P-450 interaction. J. Biol. Chem. 251, 4010–4016 [PubMed] [Google Scholar]
- 82. Oprian D. D., Vatsis K. P., and Coon M. J. (1979) Kinetics of reduction of cytochrome P-450LM4 in a reconstituted liver microsomal enzyme system. J. Biol. Chem. 254, 8895–8902 [PubMed] [Google Scholar]
- 83. Backes W. L., Tamburini P. P., Jansson I., Gibson G. G., Sligar S. G., and Schenkman J. B. (1985) Kinetics of cytochrome P-450 reduction: evidence for faster reduction of the high-spin ferric state. Biochemistry 24, 5130–5136 10.1021/bi00340a026 [DOI] [PubMed] [Google Scholar]
- 84. Benkovic S. J., Hammes G. G., and Hammes-Schiffer S. (2008) Free-energy landscape of enzyme catalysis. Biochemistry 47, 3317–3321 10.1021/bi800049z [DOI] [PubMed] [Google Scholar]
- 85. Scott E. (2018) Cytochrome P450 CYP11B enzymes: ligand and adrenodoxin interactions. in 2018 International Meeting, 22nd Microsomes Drug Oxidations and 33rd Japanese Society Study of Xenobiotics, 1–5 October, Kanazawa, Japan [Google Scholar]
- 86. Katagiri M., Suhara K., Shiroo M., and Fujimura Y. (1982) Role of cytochrome b5 in the cytochrome P-450-mediated C21-steroid 17,20-lyase reaction. Biochem. Biophys. Res. Commun. 108, 379–384 10.1016/0006-291X(82)91877-0 [DOI] [PubMed] [Google Scholar]
- 87. Shimada T., Mernaugh R. L., and Guengerich F. P. (2005) Interactions of mammalian cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b5 enzymes. Arch. Biochem. Biophys. 435, 207–216 10.1016/j.abb.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 88. Tang Z., Martin M. V., and Guengerich F. P. (2009) Elucidation of functions of human cytochrome P450 enzymes: identification of endogenous substrates in tissue extracts using metabolomic and isotopic labeling approaches. Anal. Chem. 81, 3071–3078 10.1021/ac900021a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hanna I. H., Kim M. S., and Guengerich F. P. (2001) Heterologous expression of cytochrome P450 2D6 mutants, electron transfer, and catalysis of bufuralol hydroxylation: the role of aspartate 301 in structural integrity. Arch. Biochem. Biophys. 393, 255–261 10.1006/abbi.2001.2510 [DOI] [PubMed] [Google Scholar]
- 90. Gillam E. M., Baba T., Kim B. R., Ohmori S., and Guengerich F. P. (1993) Expression of modified human cytochrome P450 3A4 in Escherichia coli and purification and reconstitution of the enzyme. Arch. Biochem. Biophys. 305, 123–131 10.1006/abbi.1993.1401 [DOI] [PubMed] [Google Scholar]
- 91. Hosea N. A., Miller G. P., and Guengerich F. P. (2000) Elucidation of distinct ligand binding sites for cytochrome P450 3A4. Biochemistry 39, 5929–5939 10.1021/bi992765t [DOI] [PubMed] [Google Scholar]
- 92. Guengerich F. P., Gillam E. M., and Shimada T. (1996) New applications of bacterial systems to problems in toxicology. Crit. Rev. Toxicol. 26, 551–583 10.3109/10408449609037477 [DOI] [PubMed] [Google Scholar]
- 93. Ghosh D., Griswold J., Erman M., and Pangborn W. (2009) Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 457, 219–223 10.1038/nature07614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.