Abstract
A key metabolic adaptation of some species that face hypoxia as part of their life cycle involves an alternative electron transport chain in which rhodoquinone (RQ) is required for fumarate reduction and ATP production. RQ biosynthesis in bacteria and protists requires ubiquinone (Q) as a precursor. In contrast, Q is not a precursor for RQ biosynthesis in animals such as parasitic helminths, and most details of this pathway have remained elusive. Here, we used Caenorhabditis elegans as a model animal to elucidate key steps in RQ biosynthesis. Using RNAi and a series of C. elegans mutants, we found that arylamine metabolites from the kynurenine pathway are essential precursors for RQ biosynthesis de novo. Deletion of kynu-1, encoding a kynureninase that converts l-kynurenine (KYN) to anthranilic acid (AA) and 3-hydroxykynurenine (3HKYN) to 3-hydroxyanthranilic acid (3HAA), completely abolished RQ biosynthesis but did not affect Q levels. Deletion of kmo-1, which encodes a kynurenine 3-monooxygenase that converts KYN to 3HKYN, drastically reduced RQ but not Q levels. Knockdown of the Q biosynthetic genes coq-5 and coq-6 affected both Q and RQ levels, indicating that both biosynthetic pathways share common enzymes. Our study reveals that two pathways for RQ biosynthesis have independently evolved. Unlike in bacteria, where amination is the last step in RQ biosynthesis, in worms the pathway begins with the arylamine precursor AA or 3HAA. Because RQ is absent in mammalian hosts of helminths, inhibition of RQ biosynthesis may have potential utility for targeting parasitic infections that cause important neglected tropical diseases.
Keywords: biosynthesis, electron transport, Caenorhabditis elegans (C. elegans), hypoxia, ubiquinone, anthranilic acid, facultative anaerobe, helminths, kynureninase (kynu-1), rhodoquinone
Introduction
Adaptation to hypoxia is crucial for survival in several animal lineages (1). Such is the case with helminths (parasitic nematodes and platyhelminths), which are facultative anaerobes and live part of their life cycle under low-oxygen tension in the gastrointestinal tract of their vertebrate hosts. One of the key adaptations of these lineages is the use of an alternative electron transport chain (ETC)7 that allows them to harvest energy under hypoxic conditions (2–4). In the absence of oxygen, complex II of this alternative ETC functions in the opposite direction to the conventional ETC. To facilitate this reversal, fumarate is used as the final electron acceptor, and rhodoquinone (RQ) functions as the electron transporter. RQ differs from ubiquinone (Q), the electron transporter in the conventional ETC, by one substituent on the benzoquinone ring: RQ has a 2-amino substituent, whereas Q has a methoxy group in this position (Fig. 1A). RQ has a lower redox potential than Q (−63 mV versus 110 mV, respectively) (5, 6), enabling RQ to receive electrons from NADH through complex I and donate them to fumarate though complex II (Fig. 1B) (3, 7). In contrast to other fermentative pathways, the alternative ETC allows proton pumping and ATP synthesis through complex V, leading to higher efficiency in harvesting energy. This pathway, in which RQ is the signature metabolite, is also found in some bacteria and protists (1).
Figure 1.
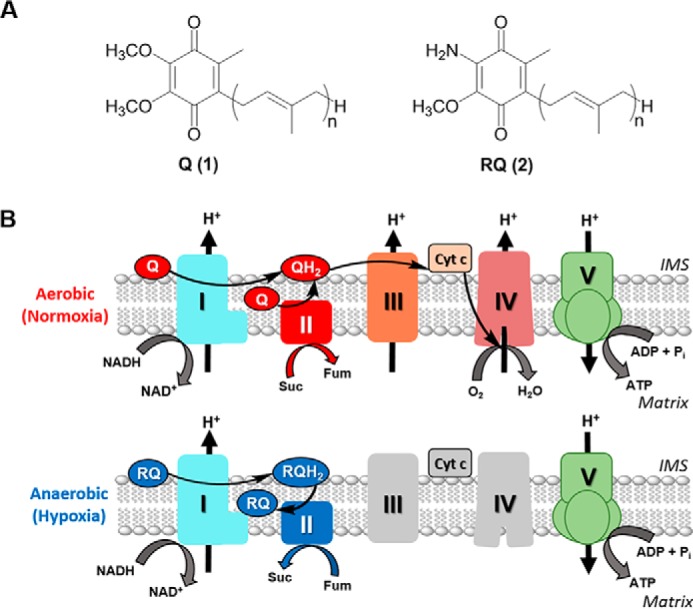
Structure and function of RQ and Q in the mitochondria of helminths. A, structures of ubiquinone (UQ or Q, compound 1) and RQ (compound 2), where n varies from 6 to 10 depending on species. B, Q and RQ are part of the mitochondrial ETC in normoxia and hypoxia, respectively. In normoxia, electrons from NADH and succinate are transferred to Q through complexes I and II, then from QH2 to cyt c through complex III, and finally from cyt c to O2 through complex IV. In hypoxia, the ETC functions with complexes I and II only. Electrons are transferred from NADH to RQ through complex I and then from RQH2 to fumarate through complex II. In both ETCs, a proton gradient across the inner membrane is generated and used to produce ATP through complex V. Suc, succinate; Fum, fumarate.
The biosynthetic pathway of RQ has been extensively studied in Rhodospirillum rubrum. In this organism, RQ biosynthesis requires Q as a precursor (8). Subsequently, it was discovered that the R. rubrum rquA gene is essential for RQ but not for Q biosynthesis (9). Despite a comprehensive study of R. rubrum knockout mutants, using aerobic versus anaerobic transcriptome data and comparative genomic analysis, no other genes besides rquA were identified to be essential for RQ biosynthesis (10). In parallel, it was shown that unicellular eukaryotes also possess a homolog of the rquA gene, most likely acquired by horizontal gene transfer (11). These studies indicate that rquA is the gene signature for RQ biosynthesis in bacteria and protists. More recently, the heterologous expression of R. rubrum rquA in two non-RQ–producing species, Escherichia coli and Saccharomyces cerevisiae, resulted in the in vivo conversion of native Q to synthetic RQ (12). Despite these advances, the biosynthesis of RQ in animals has not been elucidated, and the key genes involved have remained elusive.
RQ has been found in all helminths where it has been examined (7, 13). Importantly, RQ is also synthesized by the free-living nematode Caenorhabditis elegans (14), which faces hypoxia during development or as an environmental challenge. Studies in C. elegans have shown that Q is not a required precursor of RQ. Indeed, a KO strain in coq-7 (also known as clk-1) abolishes Q biosynthesis without affecting RQ biosynthesis (15, 16). Although helminths are not easily approachable, C. elegans is a formidable experimental organism (17) and has been used as a model for parasitic nematodes (18). In this study, we elucidate key steps in the RQ biosynthesis pathway using C. elegans. We demonstrate that the kynureninase KYNU-1 is essential for RQ biosynthesis, and based on RNAi experiments, we propose that RQ and Q have parallel pathways starting from different precursors. Because RQ is not synthesized or used by mammalian hosts, but is required for parasite survival, the RQ biosynthetic pathway is a unique target for antihelminthic drug design.
Results
KYNU-1 is essential for RQ biosynthesis in C. elegans
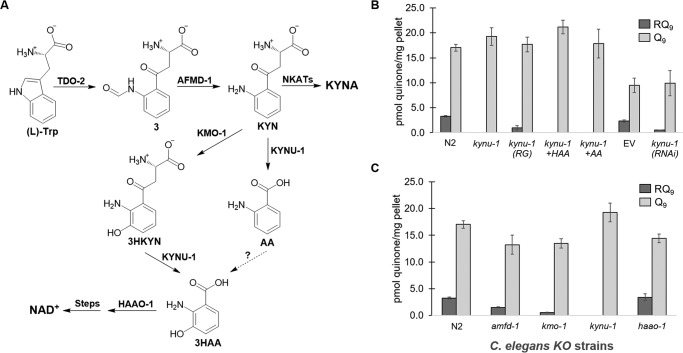
Because of the differences in RQ biosynthesis in R. rubrum and C. elegans, we reasoned that, in the case of animals, the amino group at position 2 of the benzoquinone ring (Fig. 1A) may be added at the beginning of RQ biosynthesis, rather than at the end. Because kynu-1 encodes a kynureninase that catalyzes the synthesis of two arylamines, anthranilic acid (AA) and 3-hydroxyanthranilic acid (3HAA), from l-kynurenine (KYN) and 3-hydroxy-l-kynurenine (3HKYN), respectively (Fig. 2A), we examined RQ biosynthesis in a kynu-1 KO strain. No trace of RQ was observed in the KO animals (Fig. 2B). In contrast, Q levels were not reduced in the KO. RNAi-mediated knockdown of kynu-1 in the C. elegans strain rrf-3(pk1426), which is hypersensitive to RNAi (19), exhibited a significant decrease in RQ levels (p < 0.001), with no decrease in Q, as compared with the empty vector (EV) control (Fig. 2B). The expression of the WT kynu-1 allele in the kynu-1 strain under the control of its own promoter rescued RQ biosynthesis (Fig. 2B). These results allow us to conclusively demonstrate that kynu-1 is essential for RQ biosynthesis and strongly suggest that AA or 3HAA is a key precursor for RQ biosynthesis. Intriguingly, supplementation experiments with AA or 3HAA did not restore RQ biosynthesis in the kynu-1 strain (Fig. 2B). These results indicate that RQ is synthesized de novo in C. elegans.
Figure 2.
Kynurenine pathway is essential for RQ biosynthesis. A, in the kynurenine pathway, l-tryptophan is first converted to l-formyl kynurenine (compound 3) by tryptophan 2,3-dioxygenase (TDO-2), which is then converted to KYN by an arylformamidase (AFMD-1). Kynurenine (KYN) is a branch point and can be converted to the following: 1) KYNA by kynurenine aminotransferases (NKATs); 2) 3HKYN by kynurenine 3-monooxygenase (KMO-1); or 3) AA by the kynureninase (KYNU-1). KYNU-1 also transforms 3HKYN to 3HAA, which has also been proposed to form from AA. Finally, 3HAA is converted to 2-amino-3-carboxymuconic semialdehyde (not shown) by 3-hydroxyanthranilic acid oxygenase (HAAO-1), which is then converted to NAD+. B, deletion of kynu-1 from N2 C. elegans abolished RQ biosynthesis. Overexpression of kynu-1 WT allele in the mutant kynu-1 strain restored RQ biosynthesis (RG). Supplementation with 3HAA and AA did not rescue RQ levels. The kynu-1 RNAi significantly reduced RQ levels compared with the EV control in rrf-3(pk1426) worms. C, amfd-1 and kymo-1 strains significantly reduced RQ levels, compared with N2, whereas the haao-1 strain had no effect. A full statistical analysis is given in Table S1.
Because KYN can be converted to 3HKYN by kynurenine 3-monooxygenase, KMO-1, we analyzed a kmo-1 mutant strain to discriminate whether AA or 3HAA is the RQ precursor. The kmo-1 strain had significantly reduced RQ levels compared with N2 (p < 0.001), but RQ biosynthesis was not completely abolished (Fig. 2C). The result is consistent with the fact that a hydroxyl substituent can be introduced at position 3 of the aromatic ring by kmo-1–dependent and kmo-1–independent routes (20, 21). Because KYNU-1 is required for the biosynthesis of both metabolites, this would explain the absolute requirement of this gene. Because both kynu-1 and kmo-1 belong to the kynurenine pathway (Fig. 2A), we analyzed a KO mutant in afmd-1, the gene that precedes kynu-1 in the pathway. afmd-1 encodes the kynurenine formamidase that converts l-formyl kynurenine to KYN. Surprisingly, the afmd-1 strain only reduced RQ levels to about half that of N2 (p < 0.001). We also analyzed a mutant strain in haao-1, which encodes a 3-hydroxyanthranilic acid oxygenase (HAAO-1) downstream of KYNU-1 in the pathway (Fig. 2A). This strain did not affect RQ biosynthesis (Fig. 2C). Thus, 2-amino-3-carboxymuconic semialdehyde is unlikely to be a precursor for RQ biosynthesis. Instead, RQ biosynthesis most likely branches from the AA or 3HAA precursors in the kynurenine pathway.
kynu-1 expression is restricted to the hypodermis of the worm
To assess the expression pattern of kynu-1 during the C. elegans life cycle, we generated and analyzed a translational reporter strain expressing GFP under the control of the kynu-1 promoter (IH25 strain). We detected expression of kynu-1 in the embryo early in the E lineage and epidermis. This pattern was maintained in the L1 stage, but starting at L2 through adulthood expression was only seen in the epidermis (Fig. 3 and Fig. S1). Because RQ is supposed to play a key role as part of an alternative ETC under hypoxia, we analyzed whether expression of kynu-1 was affected after exposure to 0.4% oxygen during 24 h in adult worms. We did not observe any obvious increase in GFP fluorescence nor a difference in the spatial expression of the reporter. This suggests that kynu-1 expression is not regulated under hypoxic conditions.
Figure 3.
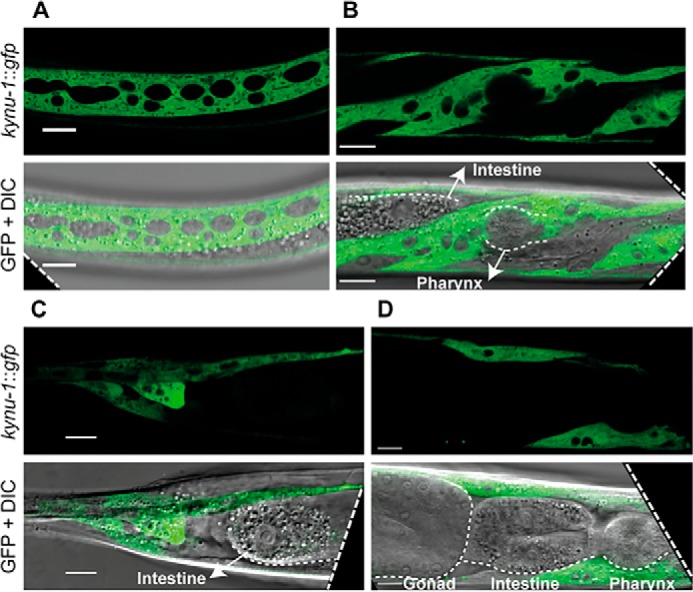
kynu-1 is expressed in the hypodermis with cytosolic localization. Confocal images of selected planes show head, middle body, and tail regions of Pkynu-1::kynu-1::gfp transgenic animal expression. A, middle body region. B, head of an L3 worm (lateral views). C, tail. D, head of an adult worm (lateral views). Pharynx, intestine, and gonad are indicated. Scale bar, 10 μm.
Enzymes involved in Q biosynthesis are also involved in RQ biosynthesis
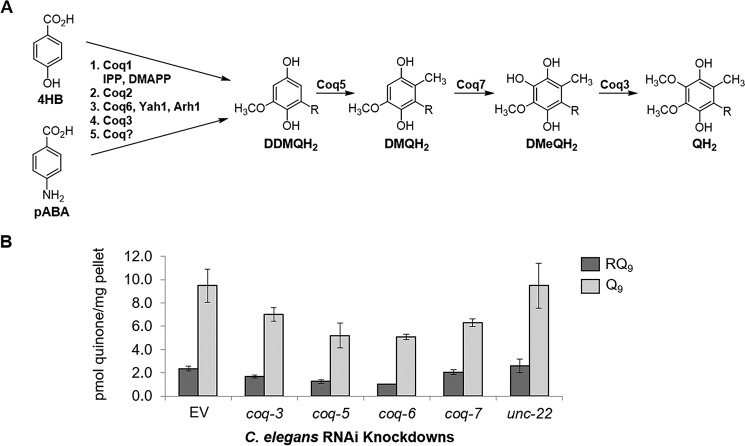
In the case of S. cerevisiae, Q can be synthesized from 4-hydroxybenzoic acid (4HB) or 4-aminobenzoic acid (pABA) in parallel pathways using common enzymes in most steps (Fig. 4A) (22, 23). Thus, we reasoned that some of the enzymes may also be involved in RQ biosynthesis. We performed RNAi knockdown assays of coq-3, coq-5, coq-6, and coq-7 genes. We found that coq-5 and coq-6 RNAi significantly decreased both Q and RQ levels compared with controls (p < 0.001), whereas coq-3 had a smaller effect on both (Fig. 4B and Table S1). As expected, coq-7 RNAi significantly decreased Q levels (p = 0.010) but not RQ. The mRNA levels for the silenced genes indicated efficient interference in all but the coq-3 RNAi samples (Fig. S2). These results clearly indicate that COQ-5 and COQ-6 are involved in both Q and RQ biosynthesis. Our results support the existence of parallel pathways that use several common enzymes to synthesize Q and RQ from different precursors.
Figure 4.
Biosynthesis of RQ shares common enzymes with the Q biosynthetic pathway. A, Q biosynthetic pathway in yeast can start from either 4HB or pABA. These pathways share the enzymes Coq1, Coq2, Coq6 (with Yah1 and Arh1), and Coq3. They merge at the common precursor demethyldemethoxyubiquinone (DDMQH2), which is converted to QH2 in three steps by Coq5, Coq7, and Coq3, respectively. B, RNAi strains of coq-3, coq-5, and coq-6 C. elegans show significant reduction of both RQ and Q, as compared with the EV and unc-22 controls (Table S1). RNAi of coq-7 significantly reduces Q levels, but RQ biosynthesis is unaffected (Table S1).
Discussion
The biosynthesis of RQ in animals has remained a puzzle for decades (24). In bacteria and protists, RQ is derived from Q, and rquA is the gene signature for its biosynthesis (9, 11). In contrast, in animals, RQ is not derived from Q, and no RQ-specific gene has been discovered. By analogy to the biosynthetic pathway of Q in yeast from pABA (22), we reasoned that the 2-amino substituent of RQ could be derived from an arylamine precursor. While this manuscript was in preparation, a different group independently reported the essential role of KYNU-1 for RQ biosynthesis, using the kynu-1 strain CB1003 (25). Our study was performed with the kynu-1 strain Tm4924. The genetic rescue of Tm4924 and RNAi experiments that we performed confirmed this finding. These results indicate that AA and/or 3HAA are RQ precursors. Consistent with this view, the strain used in this study has been previously reported to show increased levels of KYN and 3HKYN (21). Interestingly, supplementation with AA or 3HAA did not rescue RQ biosynthesis, suggesting the absence of transporters for uptake of these metabolites. The kmo-1 strain showed significantly reduced levels of RQ. Thus, whether AA or 3HAA or both are precursors of RQ is unclear. The kynu-1–dependent, kmo-1–independent biosynthesis of 3HAA has been postulated in several studies (20, 21), but to the best of our knowledge, no clear evidence regarding this reaction has been reported. In any case, the drastic decrease of RQ biosynthesis in the kmo-1 strain would suggest that 3HAA is an RQ precursor. The mutant strain in afmd-1, upstream of kynu-1, did not completely abolish RQ biosynthesis. This result would be explained if the afmd-1 strain is not a null-mutant or if KYN can be acquired from E. coli.
KYNU-1 expression was mostly restricted to the hypodermis, suggesting that the precursor of RQ is transported to other tissues. Two genes of the kynurenine pathway, tdo-2 and kmo-1, have also been found to be expressed almost exclusively in the worm hypodermis (26). KYN, AA, and 3HAA transport to other worm tissues is likely to be highly relevant because they are also precursors for other key metabolites, such as quinolinic acid and kynurenic acid. Interestingly, enigmatic deposits of fluorescent AA glycosyl esters are found in gut granules in dying worms (20). We found that the expression of kynu-1 was not up-regulated under hypoxic conditions. KYNU-1 may be constitutively expressed because it is also essential for de novo synthesis of NAD+ (27). An important conclusion of our study is that the kynurenine pathway is a complex metabolic hub, and that AA or 3HAA is a likely branch point for RQ biosynthesis.
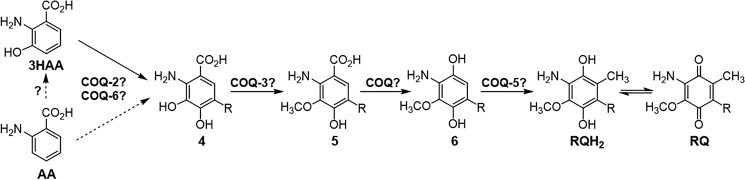
Our study reveals that from the kynurenine pathway branch point, Q and RQ biosynthesis in C. elegans make use of common enzymes, because coq-5 and coq-6 RNAi led to a significant decrease of both quinones. The fact that the enzymes involved in Q biosynthesis do not have strict substrate specificity is highlighted by the parallel pathways of Q biosynthesis in yeast that start from different precursors (23). In addition, neo-functionalization of Coq enzymes has been described for the COQ-5 bacterial ortholog (UbiE/MenG) in the biosynthesis of menaquinone (28). A scheme depicting a possible pathway for RQ biosynthesis in C. elegans is shown in Fig. 5, which utilizes COQ-2, COQ-3, COQ-5, and COQ-6. The order of these proposed steps will need to be determined.
Figure 5.
Proposed pathway for RQ biosynthesis in C. elegans. Either 3HAA or AA are proposed to be arylamine precursors to RQ. The Q biosynthetic enzymes, COQ-2 and COQ-6, may be used to form the common precursor, 2-amino-3,4-dihydroxy-5-nonaprenylbenzoic acid (compound 4). O-Methylation of compound 4 can be achieved using a S-adenosylmethionine–dependent methyltransferase, most likely COQ-3. The resulting compound 5 must be decarboxylated and hydroxylated, respectively, using the COQ enzyme(s), which would be analogous to those used in the Q biosynthetic pathway, to form the final 1,4-hydroquinone precursor to RQH2 (compound 6). The COQ-5 C-methyltransferase is proposed to catalyze the final methylation step to form RQH2, which can be oxidized to RQ. In C. elegans, R represents a tail with nine isoprenoid units (n = 9).
Our results highlight the existence of two independent evolutionary pathways for RQ biosynthesis. Interestingly, R. rubrum, and protists that synthesize RQ, lack the kynurenine pathway and use Q and RquA for the biosynthesis of RQ. In contrast, KYNU-1 is present in all helminths and in bivalves, suggesting that the kynurenine pathway has been co-opted for RQ biosynthesis. Our findings have practical applications for the identification of potential targets in the RQ biosynthetic pathway for antihelminthic drug development. Parasitic helminth infections have become a global health epidemic, and in the face of emerging drug resistance, new treatments are necessary to combat them (29). A key issue regarding future studies is to understand why mammals, and other animals that possess the kynurenine pathway, do not synthesize RQ. The discovery of key enzymatic steps that discriminate between Q and RQ precursors will be highly relevant to target a metabolic pathway that is essential for helminth survival within the mammalian host under hypoxic conditions, such as those found in the intestine.
Experimental procedures
C. elegans strains and culture conditions
The C. elegans strains used in this study are listed in Table S2. Transgenic lines were obtained according to Ref. 30. The pRF4 plasmid containing the injection marker rol-6(su1006) was co-injected with constructs containing Pkynu-1::kynu-1::gfp cloned into the pPD95.77 plasmid and injected into kynu-1(tm4924) animals. Independent transgenic lines were isolated and observed. The general methods used for culturing and maintenance of C. elegans are described in Ref. 31. All chemical reagents were purchased from Sigma. Chemical supplementation was carried out adding 10 mm AA or 10 mm 3HAA to NGM agar plates.
Reporter construct for expression and localization analysis
The expression pattern of kynu-1 was determined using GFP as a reporter. The translational constructs Pkynu-1::kynu-1::gfp included promoter (1.4 kb), exons, and introns of kynu-1 in-frame with the gfp coding sequence. Sequences were amplified by PCR using appropriate primers (Table S3) from N2 genomic DNA. The PCR products were cloned into the pPD95.77 vector that provides the unc-54 3′UTR. For the study of the kynu-1 expression pattern under hypoxic conditions, adult worms of the transgenic lines expressing the construct Pkynu-1::kynu-1::gfp were grown at 0.42% oxygen, 20 °C during 20 h in a C-Chamber incubator with a ProOx 110 oxymeter (Biospherix, Parish, NY). Worms were immediately mounted for visualization under the microscope. Animals were visualized under a confocal microscope Zeiss LSM 880 and images captured with the Zen black 2.3 software and processed with Fiji (32). Embryos were obtained by a transverse cut in a gravid adult (early stages) or picked directly from the plate (late embryonic stages).
RNAi assay
The expressions of the C. elegans kynu-1, coq-3, coq-5, coq-6, and coq-7 (clk-1) genes were interfered with by E. coli strain HT115 containing the plasmid pL4440 encoding the gene of interest (Table S4). Plasmids without an insert DNA (EV) or encoding unc-22 were used as controls. E. coli strains were grown overnight at 37 °C in LB plus ampicillin (50 μg/ml) and carbenicillin (30 μg/ml), followed by a 2-h outgrowth to obtain a cell density of 0.4–0.6 OD600 units. Each strain was seeded onto 20 NGM agar plates (150 μl per plate) plus ampicillin, carbenicillin, and 1 mm isopropyl 1-thio-β-d-galactopyranoside (to induce expression of dsRNA) and incubated for 48 h at 37 °C. RNAi was carried out by plating C. elegans rrf-3(pk1426), which were age-synchronized to the L1 stage, onto the seeded E. coli plates at 22 °C and grown for 7 days. Worms were washed from plates with M9 buffer, divided into aliquots for pelleting, and frozen at −80 °C until use.
Lipid extraction
For lipid extractions of C. elegans N2 or mutant strains, 4,000 synchronized L1s were grown on NGM plates at 20 °C to adulthood. For each experiment, ∼10,000 adult worms were harvested and washed several times with 18 megohm water to obtain pellets for extraction (∼100 mg). Lipid extraction of rrf-3 strains from RNAi assays was also performed using pellets containing ∼100 mg of worms, prepared from feeding plates as described above. Prior to extraction, 1000 pmol of Q3 internal standards was added to pellets, and then lipids were extracted using hexanes and ethanol as described previously (12).
RNA isolation and RT-quantitative PCR
RNA was extracted from ∼100-mg worm pellets using TRIzol reagent and the Zymo Quick-RNA MiniPrep kit and further purified using the Zymo RNA Clean and Concentrator Kit (Zymo Research, Irvine, CA). cDNA was prepared using the high-capacity RNA to cDNA kit (Applied Biosystems, Waltham, MA), and TaqMan gene expression assays (Table S4) were optimized and performed for each RNAi strain as described previously (10) using the endogenous control assay, cdc-42 (Ce02435138_g1).
LC-MS quantitation
LC-MS samples were prepared as described in Ref. 12. Standards were prepared and extracted at the following concentrations: Q3 (10 pmol/10 μl injection) and RQ9 (0.75, 1.5, 3.0, 4.5, or 6.0 pmol/10 μl injection). The RQ9 standard was isolated from Ascaris suum lipid extracts at Gonzaga University. In the absence of a standard, the quantity of Q9 was determined using a picomole conversion from the RQ9 standard curve and applying a RQ/Q response correction factor of 2.45 determined from RQ10/Q10 and RQ8/Q8 standard curves (12). Additional quinone-specific parameters are listed in Table S5. Samples were analyzed in triplicate and the picomole of quinone was determined from the standard curve and corrected for recovery of internal standard. Samples were normalized by milligrams of pellet mass.
Author contributions
P. M. R. B., L. R.-C., S. J. B., H. X., M. L. V., T. W. H., T. D. S., D. C. D., J. A. D., I. C., J. N. S., and G. S. investigation; P. M. R. B., L. R.-C., S. J. B., H. X., T. D. S., J. C. C., I. C., J. N. S., and G. S. methodology; P. M. R. B., L. R.-C., S. J. B., J. N. S., and G. S. writing-original draft; P. M. R. B., L. R.-C., S. J. B., H. X., M. L. V., T. W. H., D. C. D., J. A. D., I. C., J. N. S., and G. S. writing-review and editing; H. X., J. C. C., J. N. S., and G. S. conceptualization; I. C., J. N. S., and G. S. resources; J. N. S. and G. S. data curation; J. N. S. and G. S. formal analysis; J. N. S. and G. S. supervision; J. N. S. and G. S. funding acquisition; J. N. S. and G. S. project administration.
Supplementary Material
Acknowledgments
We thank Hugo Bisio, Cecilia Martínez, Gastón Risi, Drs. Lucía Otero, and Jorge Pórfido (Laboratorio de Biología de Gusanos) for helpful discussions on RQ biosynthesis, as well as Drs. Jeff Cronk and Kirk Anders from Gonzaga University, Dr. Catherine Clarke (UCLA), Drs. Andrew Roger, Courtney Stairs, and David Langelaan (Dalhousie University, Nova Scotia, Canada), and Dr. Gilles Basset and Ann Bernert (University of Florida, Gainesville). We thank Dr. Jennifer Watts (Washington State University) for RNAi clones and Alex Poppel and Fair Niven for their high school summer volunteer work at Gonzaga University. The Caenorhabditis Genetics Center is funded by National Institutes of Health Office of Research Infrastructure Program: P40 OD010440.
This work was supported by Agencia Nacional para la Innovación y la Investigación ANII FCE_1_2014_104366 (to G. S.), FOCEM (MERCOSUR Structural Convergence Fund) COF 03/11 (Institut Pasteur de Montevideo), Howard Hughes Medical Institute through the Undergraduate Science Education Program (to Gonzaga University), the Dr. Scholl Foundation (to J. N. S.), and the Agencia Nacional de Innovación e Investigación Post-doctoral Fellowship PD_NAC_2013_11008 (to I. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S7.
- ETC
- electron transport chain
- AA
- anthranilic acid
- 3HAA
- 3-hydroxyanthranilic acid
- 3HKYN
- 3-hydroxykynurenine
- KYN
- kynurenine
- KYNA
- kynurenic acid
- Q
- ubiquinone
- RQ
- rhodoquinone
- EV
- empty vector
- 4HB
- 4-hydroxybenzoic acid
- pABA
- para-aminobenzoic acid.
References
- 1. van Hellemond J. J., van der Klei A., van Weelden S. H., and Tielens A. G. M. (2003) Biochemical and evolutionary aspects of anaerobically functioning mitochondria. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 358, 205–215 10.1098/rstb.2002.1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamashita T., Ino T., Miyoshi H., Sakamoto K., Osanai A., Nakamaru-Ogiso E., and Kita K. (2004) Rhodoquinone reaction site of mitochondrial complex I, in parasitic helminth, Ascaris suum. Biochim. Biophys. Acta 1608, 97–103 10.1016/j.bbabio.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 3. Iwata F., Shinjyo N., Amino H., Sakamoto K., Islam M. K., Tsuji N., and Kita K. (2008) Change of subunit composition of mitochondrial complex II (succinate-ubiquinone reductase/quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol. Int. 57, 54–61 10.1016/j.parint.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 4. Müller M., Mentel M., van Hellemond J. J., Henze K., Woehle C., Gould S. B., Yu R. Y., van der Giezen M., Tielens A. G., and Martin W. F. (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 10.1128/MMBR.05024-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Erabi T., Higuti T., Kakuno T., Yamashita J., Tanaka M., and Horio T. (1975) Polarographic studies on ubiquinone-10 and rhodoquinone bound with chromatophores from Rhodospirillum rubrum. J. Biochem. 78, 795–801 10.1093/oxfordjournals.jbchem.a130968 [DOI] [PubMed] [Google Scholar]
- 6. Unden G., and Bongaerts J. (1997) Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320, 217–234 10.1016/S0005-2728(97)00034-0 [DOI] [PubMed] [Google Scholar]
- 7. Van Hellemond J. J., Klockiewicz M., Gaasenbeek C. P., Roos M. H., and Tielens A. G. (1995) Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J. Biol. Chem. 270, 31065–31070 10.1074/jbc.270.52.31065 [DOI] [PubMed] [Google Scholar]
- 8. Brajcich B. C., Iarocci A. L., Johnstone L. A., Morgan R. K., Lonjers Z. T., Hotchko M. J., Muhs J. D., Kieffer A., Reynolds B. J., Mandel S. M., Marbois B. N., Clarke C. F., and Shepherd J. N. (2010) Evidence that ubiquinone is a required intermediate for rhodoquinone biosynthesis in Rhodospirillum rubrum. J. Bacteriol. 192, 436–445 10.1128/JB.01040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lonjers Z. T., Dickson E. L., Chu T. P., Kreutz J. E., Neacsu F. A., Anders K. R., and Shepherd J. N. (2012) Identification of a new gene required for the biosynthesis of rhodoquinone in Rhodospirillum rubrum. J. Bacteriol. 194, 965–971 10.1128/JB.06319-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell A. R. M., Titus B. R., Kuenzi M. R., Rodriguez-Perez F., Brunsch A. D. L., Schroll M. M., Owen M. C., Cronk J. D., Anders K. R., and Shepherd J. N. (2019) Investigation of candidate genes involved in the rhodoquinone biosynthetic pathway in Rhodospirillum rubrum. PLoS ONE 14, e0217281 10.1371/journal.pone.0217281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stairs C. W., Eme L., Muñoz-Gómez S. A., Cohen A., Dellaire G., Shepherd J. N., Fawcett J. P., and Roger A. J. (2018) Microbial eukaryotes have adapted to hypoxia by horizontal acquisitions of a gene involved in rhodoquinone biosynthesis. Elife 7, e34292 10.7554/eLife.34292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernert A. C., Jacobs E. J., Reinl S. R., Choi C. C. Y., Roberts Buceta P. M., Culver J. C., Goodspeed C. R., Bradley M. C., Clarke C. F., Basset G. J., and Shepherd J. N. (2019) Recombinant RquA catalyzes the in vivo conversion of ubiquinone to rhodoquinone in Escherichia coli and Saccharomyces cerevisiae. BBA Mol. Cell. Biol. Lipids 1864, 1226–1234 10.1016/j.bbalip.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allen P. C. (1973) Helminths: comparison of their rhodoquinone. Exp. Parasitol. 34, 211–219 10.1016/0014-4894(73)90080-5 [DOI] [PubMed] [Google Scholar]
- 14. Takamiya S., Matsui T., Taka H., Murayama K., Matsuda M., and Aoki T. (1999) Free-living nematodes Caenorhabditis elegans possess in their mitochondria an additional rhodoquinone, an essential component of the eukaryotic fumarate reductase system. Arch. Biochem. Biophys. 371, 284–289 10.1006/abbi.1999.1465 [DOI] [PubMed] [Google Scholar]
- 15. Jonassen T., Larsen P. L., and Clarke C. F. (2001) A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc. Natl. Acad. Sci. U.S.A. 98, 421–426 10.1073/pnas.98.2.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyadera H., Amino H., Hiraishi A., Taka H., Murayama K., Miyoshi H., Sakamoto K., Ishii N., Hekimi S., and Kita K. (2001) Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 276, 7713–7716 10.1074/jbc.C000889200 [DOI] [PubMed] [Google Scholar]
- 17. Corsi A. K., Wightman B., and Chalfie M. (2015) A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 200, 378–407 10.1534/genetics.115.176099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bürglin T. R., Lobos E., and Blaxter M. L. (1998) Caenorhabditis elegans as a model for parasitic nematodes. Int. J. Parasitol. 28, 395–411 10.1016/S0020-7519(97)00208-7 [DOI] [PubMed] [Google Scholar]
- 19. Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., and Plasterk R. H. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317–1319 10.1016/S0960-9822(02)01041-2 [DOI] [PubMed] [Google Scholar]
- 20. Coburn C., and Gems D. (2013) The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid and the kynurenine pathway. Front. Genet. 4, 151 10.3389/fgene.2013.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Goot A. T., Zhu W., Vázquez-Manrique R. P., Seinstra R. I., Dettmer K., Michels H., Farina F., Krijnen J., Melki R., Buijsman R. C., Ruiz Silva M., Thijssen K. L., Kema I. P., Neri C., Oefner P. J., and Nollen E. A. (2012) Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc. Natl. Acad. Sci. U.S.A. 109, 14912–14917 10.1073/pnas.1203083109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marbois B., Xie L. X., Choi S., Hirano K., Hyman K., and Clarke C. F. (2010) para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838 10.1074/jbc.M110.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Awad A. M., Bradley M. C., Fernández-Del-Río L., Nag A., Tsui H. S., and Clarke C. F. (2018) Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 62, 361–376 10.1042/EBC20170106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawamukai M. (2018) Biosynthesis and applications of prenylquinones. Biosci. Biotechnol. Biochem. 82, 963–977 10.1080/09168451.2018.1433020 [DOI] [PubMed] [Google Scholar]
- 25. Del Borrello S., Lautens M., Dolan K., Tan K. H., Spensley A., Caudy A. A., and Fraser A. G. (2019) Identification of the pathway of rhodoquinone biosynthesis in C. elegans. bioRxiv 10.1101/627737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vohra M., Lemieux G. A., Lin L., and Ashrafi K. (2017) The beneficial effects of dietary restriction on learning are distinct from its effects on longevity and mediated by depletion of a neuroinhibitory metabolite. PLoS Biol. 15, e2002032 10.1371/journal.pbio.2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McReynolds M. R., Wang W., Holleran L. M., and Hanna-Rose W. (2017) Uridine monophosphate synthetase enables eukaryotic de novo NAD+ biosynthesis from quinolinic acid. J. Biol. Chem. 292, 11147–11153 10.1074/jbc.C117.795344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee P. T., Hsu A. Y., Ha H. T., and Clarke C. F. (1997) A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179, 1748–1754 10.1128/jb.179.5.1748-1754.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hotez P. J., Brindley P. J., Bethony J. M., King C. H., Pearce E. J., and Jacobson J. (2008) Helminth infection: the great neglected tropical diseases. J. Clin. Invest. 118, 1311–1321 10.1172/JCI34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mello C. C., Kramer J. M., Stinchcomb D., and Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.