Plants appear to have modified the ancient and highly conserved eukaryotic energy sensor to better fit their unique lifestyle and more effectively cope with changing environmental conditions.
Abstract
Energy homeostasis is vital to all living organisms. In eukaryotes, this process is controlled by fuel gauging protein kinases: AMP-activated kinase in mammals, Sucrose Non-Fermenting1 (SNF1) in yeast (Saccharomyces cerevisiae), and SNF1-related kinase1 (SnRK1) in plants. These kinases are highly conserved in structure and function and (according to this paradigm) operate as heterotrimeric complexes of catalytic-α and regulatory β- and γ-subunits, responding to low cellular nucleotide charge. Here, we determined that the Arabidopsis (Arabidopsis thaliana) SnRK1 catalytic α-subunit has regulatory subunit-independent activity, which is consistent with default activation (and thus controlled repression), a strategy more generally used by plants. Low energy stress (caused by darkness, inhibited photosynthesis, or hypoxia) also triggers SnRK1α nuclear translocation, thereby controlling induced but not repressed target gene expression to replenish cellular energy for plant survival. The myristoylated and membrane-associated regulatory β-subunits restrict nuclear localization and inhibit target gene induction. Transgenic plants with forced SnRK1α-subunit localization consistently were affected in metabolic stress responses, but their analysis also revealed key roles for nuclear SnRK1 in leaf and root growth and development. Our findings suggest that plants have modified the ancient, highly conserved eukaryotic energy sensor to better fit their unique lifestyle and to more effectively cope with changing environmental conditions.
INTRODUCTION
A major challenge for all living organisms is the maintenance of energy homeostasis during growth and development and the continuous adaptation to changing environmental conditions and resource availability. In eukaryotic organisms, this is enabled by the highly conserved Animal AMP-activated kinase/yeast Suc Non-Fermenting1 kinase/plant SNF1-related kinase1 (AMPK/SNF1/SnRK1) protein kinases, which function as cellular fuel sensors, triggering the activation of catabolic reactions and repressing energy-consuming anabolic processes when energy supplies become limited. AMPK, SNF1, and SnRK1 function as heterotrimeric complexes with a catalytic α-subunit and regulatory β- and γ-subunits (Figure 1A; Supplemental Figure 1A; Hedbacker and Carlson, 2008; Hardie et al., 2012; Broeckx et al., 2016). The α-subunits consist of a highly conserved N-terminal Ser/Thr kinase domain and a large C-terminal regulatory domain, which mediates interaction with the β- and γ-subunits (Figure 1A; Supplemental Figures 1A and 1B). Phosphorylation of the kinase domain T-loop is an absolute prerequisite for AMPK/SNF1/SnRK1 kinase activity (Estruch et al., 1992; Hawley et al., 1996; Baena-González et al., 2007). The β-subunits act as complex scaffolds but also contribute to complex localization and substrate specificity. These subunits are characterized by a variable N-terminal domain, which is typically myristoylated; a carbohydrate-binding module (CBM); and a C-terminal domain, which binds to the α- and γ-subunits (Hedbacker and Carlson, 2008; Hardie et al., 2012; Broeckx et al., 2016). The prototypical γ-subunit links a divergent N terminus and a more recently identified pre-cystathionine β-synthase (CBS) domain (Viana et al., 2007; Ramon et al., 2013) with four CBS motifs that make up the two adenine nucleotide-binding Bateman domains, functioning as the energy (nucleotide charge) sensing module of the AMPK complex (Kemp, 2004; Scott et al., 2007; Xiao et al., 2011).
Figure 1.
Constitutive Regulatory Subunit-Independent Activity of the SnRK1 Catalytic α-Subunit.
(A) Space-filling homology models of the SnRK1α1β2βγ heterotrimeric complex, the FL (512-amino acid) SnRK1α1 subunit, and the truncated SnRK1α1 proteins, lacking part of (404-amino acid, 333-amino acid) or the entire regulatory domain (290-amino acid). The regulatory-α C-terminal domain (αCTD), α-linker, and ubiquitin-associated (UBA) domain are indicated in addition to the N-lobe, C-lobe, and catalytic cleft of the catalytic domain.
(B) Loss of SnRK1α1 interaction with the regulatory SnRK1β2 subunit upon truncation. CoIP of transiently coexpressed FL and truncated SnRK1α-HA with FLAG-tagged SnRK1β2 from Arabidopsis Col-0 leaf mesophyll protoplasts. Protein input and IP were visualized by immunoblot analysis using anti-HA and anti-FLAG epitope tag antibodies, as indicated.
(C) DIN6 promoter activity in leaf mesophyll protoplasts upon transient coexpression of FL and truncated SnRK1α1 subunits. Relative and normalized LUC reporter values are averages with sds, n = 6 biological repeats (independent protoplast transfections). One-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001. Protein expression was assessed by immunoblot analysis with anti-HA and anti-FLAG antibodies. RBCS staining with Coomassie Brilliant Blue R-250 was used as a protein loading control.
(D) qRT-PCR analysis of (induced) DIN1/SEN1 and (repressed) EXP10 SnRK1 target gene expression in leaf mesophyll protoplasts expressing the SnRK1α1 subunit. Values are averages with sds, n = 3 technical repeats. One-way ANOVA statistical analysis was performed in GraphPad Prism v7, ns = not significant.
(E) Phos-tag acrylamide-based (Wako Chemicals) mobility shift assay of (T175) T-loop phosphorylation of FL and truncated (404, 333, and 290 amino acids) SnRK1α1-HA proteins expressed in leaf mesophyll protoplasts. SnRK1α1 T175A was included as a negative control for T-loop phosphorylation. SnRK1α1 K48M is a kinase-dead (ATP binding site mutant) control. Arrows indicate phosphorylated protein bands. Immunoblot analysis was performed using anti-HA and anti-FLAG antibodies and RBCS staining with Coomassie Brilliant Blue R-250 as a protein loading control.
(F) Yeast mutant complementation. Growth of yeast snf1Δ and snf1Δsnf4Δgal83Δsip1Δsip2Δ (αβγ null) mutants expressing Snf1, SnRK1α1/KIN10, and SnRK1α2/KIN11 on fermentable Glc (Glc 2% [w/v]) and nonfermentable Glycerol (Gly 2% [v/v]-Ethanol (Eth; EtOH 3% [v/v]) medium. WT, wild type.
However, while the overall structure and function of this complex appear to be largely conserved, the diverse lifestyles of different types of eukaryotic organisms are also reflected in the molecular mechanisms of these complexes’ regulation. While AMPK and SNF1 are clearly regulated by adenine nucleotide charge, with AMP and/or ADP competing with ATP for γ-subunit binding and allosterically activating the kinase subunit through inhibiting T-loop dephosphoryation (Carling et al., 1989; Oakhill et al., 2011; Gowans et al., 2013), SnRK1 does not seem to be directly activated by AMP (Wilson et al., 1996; Sugden et al., 1999). More recently, Arabidopsis (Arabidopsis thaliana) SnRK1 was suggested to be both insensitive to AMP and ADP (consistent with amino acid substitutions that preclude binding) and resistant to T-loop dephosphorylation in vitro, indicating that plant SnRK1 indeed is an atypical member of the AMPK family (Emanuelle et al., 2015). Green plants also encode a unique hybrid βγ-subunit that has recruited an N-terminal CBM to the four-CBS γ-domain to function as the canonical plant γ-subunit (Lumbreras et al., 2001; Gissot et al., 2006; Ramon et al., 2013; Emanuelle et al., 2015) and a truncated β-subunit (SnRK1β3) that lacks the entire N-terminal extension and CBM, but still takes part in heterotrimeric complex formation (Gissot et al., 2004; Polge et al., 2008; Emanuelle et al., 2015).
More than other organisms, sessile and autotrophic plants depend on the ability to accurately monitor and adapt to changing environmental conditions that directly or indirectly affect energy supplies by photosynthesis, respiration, and carbon allocation. SnRK1 controls energy homeostasis and thereby plant growth and development as well as biotic and abiotic stress tolerance (Tsai and Gazzarrini, 2014; Broeckx et al., 2016; Hulsmans et al., 2016; Baena-González and Hanson, 2017; Wurzinger et al., 2018). Plants in general seem to prefer negative regulatory mechanisms, with many (notably hormone signaling) pathways that involve inactivation (and degradation) of repressor proteins, likely enabling more robust, reliable, and flexible signal integration and possibly also faster downstream responses (Muraro et al., 2013). Consistent with inactivation under energy-rich conditions and the release of inhibition (rather than direct activation) by energy depletion, sugar phosphates such as Glc-6-P and trehalose-6-P (T6P) were found to inhibit the plant SnRK1 kinase (Toroser et al., 2000; Zhang et al., 2009; Nunes et al., 2013). In addition, several more interacting proteins that negatively regulate SnRK1 signaling have been identified, including PLEIOTROPIC REGULATORY LOCUS1 (Bhalerao et al., 1999), SnRK1A Interacting Negative regulator proteins (Lin et al., 2014), and a family of small FCS-like zinc finger proteins (Jamsheer et al., 2018). SnRK1 was also found to restrict its own activity by sumoylation- and ubiquitination-mediated degradation, making for an efficient feedback mechanism to attenuate SnRK1 signaling and to avoid a persistent stress response (Crozet et al., 2016).
In this study, we determined that Arabidopsis SnRK1 harbors an active catalytic α-subunit, which translocates to the nucleus to trigger reprogramming of induced target gene expression under metabolic stress conditions. The myristoylated β-subunits can restrict nuclear translocation and SnRK1-induced gene expression. Analysis of transgenic Arabidopsis plants with an altered localization of the catalytic α-subunit confirmed the relevance and importance of this regulatory mechanism and reveals different nuclear and cytoplasmic functions of SnRK1 in plant growth and development.
RESULTS
The SnRK1 Catalytic α-Subunit Shows Regulatory Subunit-Independent Activity
Cell-autonomous SnRK1 signaling has been studied successfully via transient expression of epitope-tagged proteins in Arabidopsis leaf mesophyll protoplasts (Baena-González et al., 2007; Confraria and Baena-González, 2016). DARK INDUCED6 (DIN6)/ASPARAGINE SYNTHASE1 promoter activity and expression has been used as a direct target and physiologically relevant readout of SnRK1 activity (Baena-González et al., 2007; Dietrich et al., 2011). With its high N:C ratio, the amide Asp is preferentially synthesized under C-limiting stress conditions (Sieciechowicz et al., 1988; Lam et al., 1998; Baena-González et al., 2007). The DIN6 promoter is directly activated by heterodimers of SnRK1-phosphorylated C-class (bZIP63) and S1-class (bZIP11) basic region leucine zipper (bZIP) transcription factors (TFs; Mair et al., 2015). Whereas the AMPK/SNF1/SnRK1 kinases are generally believed to function as heterotrimeric complexes, overexpression of the catalytic SnRK1α subunit (encoded by SnRK1α1/KINASE10 [KIN10], SnRK1α2/KIN11, and SnRK1α3/KIN12 in Arabidopsis) is sufficient to confer high and specific SnRK1 activity, not only activating the DIN6 promoter, but also reprogramming the expression of >1,000 target genes in leaf cells (Baena-González et al., 2007). Using the same experimental setup, we found that progressive truncation of the SnRK1α1/KIN10 protein C-terminal regulatory domain down to the mere 290-amino acid catalytic domain abolished SnRK1 complex formation (interaction with the SnRK1β2 complex scaffold protein; Figures 1A and 1B; Supplemental Figure 1) but not SnRK1 signaling, as indicated by DIN6 promoter activity and RT-qPCR analysis of a set of established induced and repressed target genes (Figures 1C and 1D; Supplemental Figure 2; Baena-González et al., 2007). This suggests complex-independent activity of the catalytic subunit. Consistently, a Phos-tag mobility shift assay (Wako Chemicals) showed that the kinase domain T-loop (T175) of the transiently expressed full-length (FL) SnRK1α1 as well as its truncated versions were effectively phosphorylated (Figure 1E). Significantly reduced T-loop phosphorylation in the kinase-dead K48M mutant subunit indicates that this is largely dependent on SnRK1α1 kinase activity, most likely involving autophosphorylation.
We further analyzed the activity of the catalytic subunit by heterologous expression in yeast (Saccharomyces cerevisiae). As previously reported, SnRK1α1/KIN10 and SnRK1α2/KIN11 can complement the yeast snf1 mutant phenotype (Figure 1F; Supplemental Figure 3A; Alderson et al., 1991). However, unlike yeast Snf1 itself, heterologous expression of SnRK1α1 and SnRK1α2 also fully complemented the growth defect of an snf1Δsnf4Δgal83Δsip1Δsip2Δ quintuple αβγ mutant lacking all complex subunits on nonfermentable glycerol/ethanol medium (Figure 1F; Supplemental Figure 3A). This confirms the complex-independent activity of the Arabidopsis SnRK1α1 subunits. Conversely, transient overexpression of Snf1 did not induce SnRK1 target gene expression in leaf mesophyll protoplasts (Supplemental Figure 3B). Human AMPKα1 was unable to complement either yeast mutant or to activate the DIN6 promoter in leaf cells (Supplemental Figures 3A and 3B). These results confirm the notion that SnRK1 is an atypical AMPK/SNF1-related kinase with constitutive complex-independent catalytic activity, raising questions about the regulation of SnRK1 signaling in response to metabolic stress and energy supply and the exact role of the regulatory subunits.
The Regulatory β2 Subunit Inhibits SnRK1α-Induced Activation, But Not Repression, of Target Gene Expression
Although the SnRK1 catalytic subunit showed complex-independent activity, the conservation of the regulatory subunits and lethality of homozygous knockout of the SnRK1βγ subunit (Ramon et al., 2013; Gao et al., 2016) indicate that an intact heterotrimeric SnRK1 complex is required for normal plant function. To further explore the role of the regulatory subunits, we transiently coexpressed SnRK1β2 and SnRK1βγ with SnRK1α1 in leaf mesophyll protoplasts. Coexpression of the SnRK1β1 and SnRK1β2 subunits significantly suppressed SnRK1α1-induced activation of DIN6 promoter activity, while the truncated plant-specific SnRK1β3 subunit was less effective (Figure 2A). We focused subsequent analyses on the SnRK1β2 subunit, which suppressed SnRK1α1-induced promoter activity most efficiently. The activity of the truncated (290-amino acid) α-subunit was no longer significantly affected (Figure 2B), indicating that repression by coexpression of the β-subunit depends on the interaction of these subunits. Consistently, truncation of the SnRK1β2 CTD or CBM, which abolished interaction with the α-subunit (Supplemental Figure 4), also abolished the repression of SnRK1α1-induced promoter activation (Figure 2C). Coexpression of the β-subunit did not affect phosphorylation of the α-subunit T-loop (Figure 2D), suggesting that protein kinase activity itself was not affected. SnRK1β2 also appears to be phosphorylated. Coexpression of SnRK1βγ restored SnRK1α-induced gene expression (Figure 2E). This is consistent with our previous analyses. Transient RNAi-mediated silencing of the βγ-subunit significantly reduced SnRK1 responses, indicating that the βγ subunit is a positive regulator of these responses (Ramon et al., 2013).
Figure 2.
Inhibition of SnRK1α Target Gene Induction, But Not Repression, by the Regulatory SnR1β2 Subunit.
(A) DIN6 promoter activity (activation) in Arabidopsis leaf mesophyll protoplasts upon transient coexpression of SnRK1α1 with SnRK1β1, SnRK1β2, and SnRK1β3.
(B) DIN6 promoter activity in leaf mesophyll protoplasts upon transient coexpression of FL and truncated (290-amino acid) SnRK1α1 with SnRK1β2.
(C) DIN6 promoter activity in leaf mesophyll protoplasts upon transient coexpression of SnRK1α1 with FL SnRK1β2 and with SnRK1β2 lacking the C-terminal domain (−βCTD) or carbohydrate-binding module (−CBM).
(D) Phos-tag acrylamide-based (Wako Chemicals) mobility shift assay of (T175) T-loop phosphorylation of SnRK1α1-HA in leaf mesophyll protoplasts coexpressing SnRK1β2-FLAG. SnRK1α1 T175A-HA was included as a negative control. Arrows indicate phosphorylated protein bands of SnRK1α1 (black arrow) and SnRK1β2 (gray arrow).
(E) DIN6 promoter activity in leaf mesophyll protoplasts upon transient coexpression of SnRK1α1 with SnRK1β2, FL SnRK1βγ, and a truncated SnRK1βγ, lacking the N-terminal carbohydrate-binding module (−CBM).
(F) EXP10 promoter activity (repression) in leaf mesophyll protoplasts upon transient coexpression of SnRK1α1 and SnRK1β2. Relative and normalized LUC reporter values are averages with sds, n = 6 (A), (B), (E), (F), or three (C) biological replicates (independent protoplast transfections). One-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001; ns = not significant. Protein expression of HA- and FLAG-tagged proteins was assessed by immunoblot analysis with anti-HA and anti-FLAG antibodies, using RBCS staining with Coomassie Brilliant Blue R-250 as a protein loading control.
The effect of SnRK1 βγ is dependent on its N-terminal CBM (Figure 2E; Supplemental Figure 5A). Homology modeling of the plant SnRK1 complex suggested that the SnRK1βγ CBM can be positioned in the same space as the β-subunit CBM through its flexible linker, possibly affecting or competing for interaction with the catalytic domain (Supplemental Figures 5B and 5C; Broeckx et al., 2016). Surprisingly, SnRK1β2 coexpression only affected SnRK1α1-induced promoter activation, but not repression, as illustrated by the complete lack of effect on EXPANSIN10 (EXP10) promoter repression (Figure 2F). This was confirmed by RT-qPCR analysis of a number of known repressed target genes (Supplemental Figure 6), again indicating that protein kinase activity itself was not affected. To confirm the results of cellular assays in intact plants, we isolated a homozygous SnRK1β2 T-DNA knockout line (Supplemental Figure 7A). However, SnRK1β2 loss of function only slightly increased the SnRK1-mediated response of seedlings to sugar starvation (Supplemental Figure 7B) and did not affect the response of detached leaves to dark incubation (Supplemental Figure 7C). This points to functional redundancy with the other SnRK1β subunits.
The Regulatory SnRK1β2 Subunit Controls SnRK1α Localization
The regulatory β-subunits, with the exception of SnRK1β3, contain a long N-terminal extension that apparently lacks a specific secondary conformation and is absent from 3D structures and (homology) models (Supplemental Figures 8A to 8C; Broeckx et al., 2016). These subunits include an N-terminal myristoylation (N-MYR) site. In SnRK1β2, the MGNVNAR sequence matches the conserved MGNXX[ACGSTV][^DE] and MG[^DEFKRVWY]XX[ACGSTV][KR] motifs present in 60% of the n-myristoylome (Boisson et al., 2003), and the MGNVNAREE peptide was also experimentally shown to be myristoylated on Gly2 (Pierre et al., 2007). Gly2 N-MYR controls SnRK1β2 (and SnRK1β1) localization; the proteins mainly associate with membranes. Upon Gly2 to Ala (G2A) mutation, they localize to both the cytosol and nucleus (Pierre et al., 2007). We confirmed this via transient expression of green fluorescent protein (GFP) fusion proteins in leaf mesophyll protoplasts (Figure 3A). In addition, isolation and immunoblot analysis of the nuclear and cytoplasmic fractions of seedlings with specific antibodies confirmed that, unlike SnRK1α1, endogenous wild-type SnRK1β2 is excluded from the nucleus (Figure 3B). We therefore hypothesized that the β-subunits can restrict SnRK1-induced target gene activation by preventing nuclear localization of the catalytic α-subunit. Indeed, coexpression of SnRK1β2 in leaf mesophyll protoplasts led to the exclusion of SnRK1α1-GFP from the nucleus (Figure 3C). More detailed scanning of the nuclear area confirmed nuclear exclusion and possibly also nuclear membrane or perinuclear endoplasmic reticulum association of SnRK1α1 in those cells (Figure 3D).
Figure 3.
SnRK1β2 Controls the Subcellular Localization of SnRK1α1.
(A) Fluorescence microscopy analysis of the subcellular localization of SnRK1β2-GFP and SnRK1β2-G2A-GFP in leaf mesophyll protoplasts, 16 h after transfection. DIC, differential interference contrast image; G2A, Gly2 to Ala mutation (causing loss of N-MYR).
(B) Immunoblot analysis of nuclear (Nucl) and cytoplasmic (Cyto) fractions of endogenous SnRK1α1 and SnRK1β2 proteins in leaf mesophyll cells using specific anti-SnRK1α1 and anti-SnRK1β2 antibodies. Anti-Histone H3 antibodies and RBCS staining with Coomassie Brilliant Blue R-250 serve as controls for purity of the nuclear and cytoplasmic fractions, respectively. Ten percent of the cytoplasmic fractions and the complete nuclear fractions of samples were used for analysis.
(C) Fluorescence microscopy analysis of the subcellular localization of SnRK1α1-GFP in leaf mesophyll protoplasts upon coexpression of SnRK1β2 and SnRK1β2-G2A, 16 h after transfection. An SCF30-RFP nuclear reporter was also coexpressed. This reporter produces orange fluorescence in the nucleus. Colocalization with a nuclear GFP fusion protein produces a green to yellow color. Broken circles indicate the nucleus.
(D) Confocal image closeups of the nuclear areas of mesophyll protoplasts expressing SnRK1α1-GFP without and with SnRK1β2 coexpression in the absence of the SCF30-RFP nuclear reporter. Arrows indicate localization at/around the nuclear membrane. Localization studies have been repeated at least three times independently for each construct combination with consistent results. Representative pictures are shown.
This effect is dependent on the N-MYR of SnRK1β2, as coexpression of a G2A mutant protein did not alter SnRK1α1 localization (Figure 3C). In the DIN6-LUCIFERASE (LUC) reporter assay, the inhibition of the α-subunit by the SnRK1β2 subunit was only partly dependent on myristoylation. Truncation of the N-terminal 30 amino acids, which are also missing in a predicted short SnRK1β2 splice variant (Supplemental Figure 8C) and in SnRK1β3, further reduced the inhibitory effect significantly (Supplemental Figure 8D). This suggests that the N terminus plays an important additional regulatory role. Interestingly, the second (higher molecular weight) protein isoform produced by both endogenous (Figure 3B) and transiently overexpressed hemagglutinin (HA)-tagged SnRK1β2 (Figure 2) is most likely produced by additional posttranslational modification of the 30-amino acid N terminus. Finally, we confirmed the capacity of the SnRK1β2 MGNVNAREE N-MYR (βMYR) domain for nuclear exclusion (cytoplasmic retention) by fusing the domain to the G-BOX BINDING FACTOR5 (GBF5/bZIP2) TF (Supplemental Figure 8E). Nuclear exclusion (with apparent protein aggregation at the nuclear membrane or endoplasmic reticulum) was mediated by myristoylation, as G2A mutation of the βMYR domain restored its nuclear localization.
Metabolic Stress Triggers Nuclear Translocation of the SnRK1α Subunit
We then analyzed the effect of α-subunit localization on SnRK1 signaling in more detail by forcing its subcellular localization and isolating nuclear and cytoplasmic cell fractions. Increased nuclear localization of SnRK1α1 by N-terminal attachment of a simian virus 40 (SV40) nuclear localization sequence (NLS) led to increased activation of SnRK1 target promoter activity in leaf mesophyll protoplasts (Figure 4A; Supplemental Figure 9A). Conversely, nuclear exclusion by attachment of the SnRK1β2 βMYR motif significantly hampered SnRK1α1-induced activation of the target promoter (Figure 4A; Supplemental Figure 9A). The latter effect was lost with G2A mutation (Figure 4A). Consistent with the lack of effect of β-subunit coexpression on target gene repression (Figure 2F) and consistent with unaffected kinase activity, an increase or decrease in nuclear SnRK1α concentration did not affect target gene repression (Figure 4B). To confirm the effect of nuclear translocation, we also generated an SnRK1α1-glucocorticoid receptor fusion construct (Supplemental Figure 9B). In the absence of the dexamethasone (DEX) ligand, heteromerization with heat shock proteins and concomitant cytoplasmic retention (Schena et al., 1991) prohibited target promoter activation. The addition of DEX triggered nuclear translocation and target promoter activation (Supplemental Figures 9B and 9C). These results are consistent with a previous study noticing the effect of the nuclear localization of rice (Oryza sativa) SnRK1α on target gene induction (Cho et al., 2012).
Figure 4.
Effects of Altered SnRK1α1 and SnRK1β2 Localization on SnRK1 Target Gene Expression.
(A) DIN6 promoter activity in leaf mesophyll protoplasts upon transient expression of SnRK1α1 proteins with an SV40 NLS or SnRK1β2 (wild type or G2A mutant) nine-amino acid N-MYR (βMYR) domain.
(B) EXP10 promoter activity in leaf mesophyll protoplasts upon transient expression of SnRK1α1 proteins with an SV40 NLS or SnRK1β2 nine-amino acid N-MYR (βMYR) domain.
(C) DIN6 promoter activity in leaf mesophyll protoplasts upon transient coexpression of HA-tagged SnRK1α1 and NLS-SnRK1α1 proteins with SnRK1β2 and NLS-SnRK1β2 proteins. Relative and normalized LUC reporter values are averages with sds, n = 3 (A), (C), or 4 (B) biological replicates (independent protoplast transfections).
One-way ANOVA statistical analysis was performed in GraphPad Prism7, ****P < 0.0001; ***P < 0.001; **P < 0.01; ns = not significant. Protein expression was assessed by immunoblot analysis with anti-HA antibodies, using RBCS staining with Coomassie Brilliant Blue R-250 as a protein loading control.
Remarkably, while nuclear translocation of the α-subunit is both necessary and sufficient to trigger DIN6 target promoter activation, the repression of the target promoter EXP10 was unaffected by the nuclear localization of SnRK1α (Figure 4B, Supplemental Figure 9C). Consistent with a role for SnRK1β in cytoplasmic retention or nuclear exclusion of the catalytic subunit, activation of the DIN6 promoter by nucleus-enriched NLS-SnRK1α1 was not significantly affected by SnRK1β2 coexpression in protoplasts. Conversely, the use of a nucleus-targeted NLS-SnRK1β2 (lacking the MYR and the additional posttranslational modification) even further increased DIN6 promoter activation, most likely by increasing the nuclear localization of SnRK1α1 (Figure 4C).
We then analyzed the localization of endogenous SnRK1α1 in intact, growing Arabidopsis plants. No obvious changes were observed in α-subunit localization in adult (source) leaf tissue during the normal day/night cycle, which is consistent with the minor transient changes in SnRK1 target gene expression. However, extended night conditions that deplete transitory starch reserves (Usadel et al., 2008) triggered an enrichment of nuclear SnRK1α1 and a concomitant dramatic increase in expression of the DIN6 target gene (Figures 5A and 5B). We therefore assessed SnRK1α1 subunit localization in response to different metabolic stress conditions known to cause severe energy depletion and activate SnRK1 signaling (Baena-González et al., 2007). In leaf mesophyll protoplasts, transiently expressed SnRK1α1 protein was clearly enriched in the nuclear fraction after treatment with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a herbicide that inhibits photosynthesis by blocking the plastoquinone binding site of PSII or under hypoxic and dark conditions (Supplemental Figure 10). A similar nuclear translocation was observed for endogenous SnRK1α1 (Figure 5C). Importantly, endogenous SnRK1β2 did not translocate to the nucleus under these conditions (Figure 5C).
Figure 5.
Metabolic Stress-Induced Endogenous SnRK1α1 Translocation.
(A) qRT-PCR analysis of circadian DIN6 expression in Arabidopsis wild-type Col-0 plants grown for 4 weeks in soil (blue), and the effect on DIN6 expression of an extended (6 to 12 h) night (red). Values are averages with sds, n = 3 with three pooled leaves each.
(B) Immunoblot analysis of nuclear (N) and cytoplasmic (C) fractions of endogenous SnRK1α1 at different time points during the day–night cycle and in extended darkness using specific anti-SnRK1α1 antibodies.
(C) Immunoblot analysis of nuclear (N) and cytoplasmic (C) fractions of endogenous SnRK1α1 and SnRK1β2 in leaf mesophyll cells in control and metabolic stress conditions (6-h hypoxia, DCMU, and dark treatment). Anti-Histone H3 antibodies and RBCS staining with Coomassie Brilliant Blue R-250 serve as controls for purity of the nuclear and cytoplasmic fractions, respectively. Ten percent of the cytoplasmic fractions and complete nuclear fractions of the samples were used for analysis.
Altered SnRK1α Subunit Localization Affects Growth and Development as Well as Metabolic Stress Responses
To corroborate the physiological relevance of α-subunit localization, we generated transgenic lines uniquely expressing tagged wild-type, nuclear (SV40 NLS-fused), or cytoplasmic (βMYR-fused) SnRK1α1 in a homozygous snrk1α1 snrk1α2 (kin10 kin11) double knockout mutant background (Supplemental Figures 11A to 11E). As double knockout appears lethal, heterozygous T-DNA knockout plants were complemented with genomic SnRK1α1/KIN10 fragments containing a double FLAG-tag fused to the coding sequences (CDS), followed by selfing and selection for homozygous knockouts and tagged alleles. Initial analysis of three independent transgenic lines with altered α-subunit localization revealed consistent and distinct phenotypes, which were further quantified in representative lines. Enrichment in NLS-SnRK1α1 resulted in a more rounded leaf shape than wild-type and control (SnRK1α1 complemented, essentially SnRK1α2/kin11 mutant) plants, while plants with cytoplasmic SnRK1α1 retention (βMYR-SnRK1α1) produced narrower, dark green leaves with enhanced downward curvature (Figures 6A to 6C). When looking at the root system, we found that NLS-SnRK1α1 plants growing on one half Murashige and Skoog medium (0.5x MS, 0.5% Glc [w/v]) had significantly longer primary roots than wild-type and control plants, while root growth was impaired in βMYR-SnRK1α1 plants (Figures 7A and 7B). This correlated with the significantly increased and reduced root meristem size and number of meristem cells in NLS-SnRK1α1 and βMYR-SnRK1α1 plants, respectively (Figures 7C to 7E). In addition, NLS-SnRK1α1 plants had longer root hairs than wild-type and control plants on the same growth medium, whereas root hair growth was significantly impaired in βMYR-SnRK1α1 plants (Figures 7F and 7G), possibly due to altered auxin-induced ROOT HAIR DEFECTIVE6 LIKE2 (RSL2) basic helix-loop-helix TF-mediated signaling (Supplemental Figure 12).
Figure 6.
Altered SnRK1α1 Localization Affects Leaf Growth and Development.
(A) Distinct leaf shape phenotypes of 4-week–old soil-grown snrk1α1/snrk1α1 snrk1α2/snrk1α2 (kin10/kin10 kin11/kin11) Arabidopsis plants complemented with genomic βMYR-SnRK1α1 (left), SnRK1α1 (middle), and NLS-SnRK1α1 (right) constructs.
(B) Dissected rosettes of wild type and complemented lines. WT, wild type.
(C) Quantitative analysis of leaf shape. The leaf length:width ratio was determined using the software ImageJ for all rosette leaves of wild type and complemented lines. Values are averages with sds; n = 3 leaf series.
Figure 7.
Altered SnRK1 α1 Localization Affects Root Growth and Development.
(A) Primary root growth of wild-type (WT) and SnRK1α1 (α1), βMYR-SnRK1α1 (βMYR-α1), and NLS-SnRK1α1 (NLS-α1) complemented plants 10 d after sowing on 1/2 MS medium supplemented with 0.5% Glc. Representative seedlings are shown.
(B) Quantitative analysis of primary root length root length of 20 wild-type (WT) and complemented mutant seedlings 10 d after sowing on 1/2 MS medium supplemented with 0.5% Glc using the software ImageJ. Values are averages with sds. One-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001.
(C) Root meristems of 15 wild-type and complemented mutant seedlings 10 d after sowing on 1/2 MS medium supplemented with 0.5% Glc. Representative pictures are shown. WT, wild type.
(D) and (E) Root meristem size as quantified by (D) length and (E) number of cells in a single cell file. Values are averages with sds. One-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001, *P < 0.05. WT, wild type.
(F) Root hairs of complemented mutant seedlings 10 d after sowing on 1/2 MS medium supplemented with 0.5% Glc. Representative roots are shown.
(G) Distribution of root hair lengths of 20 wild-type or complemented mutant seedlings, measured using the software ImageJ. WT, wild type.
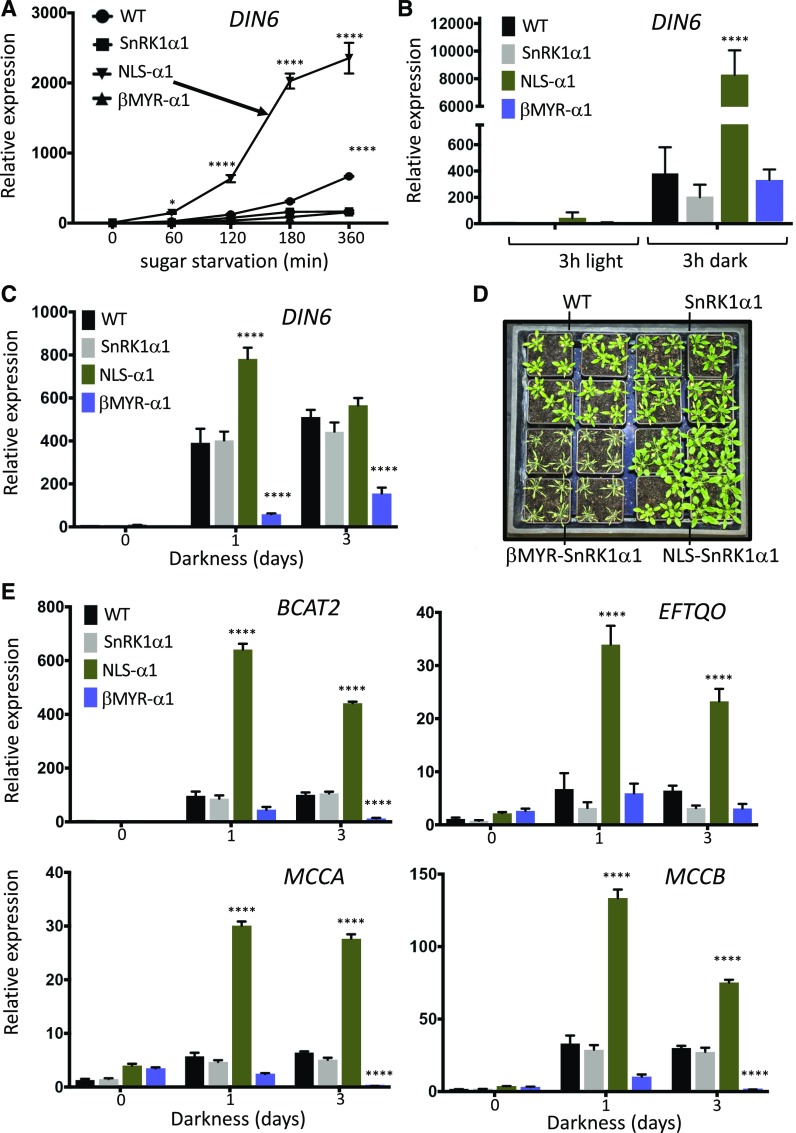
Finally, we analyzed the effect of SnRK1α localization on the metabolic stress response. To assess fast short-term responses in plants, we performed sugar starvation assays in seedlings. Whereas control and βMYR-SnRK1α1 seedlings showed a reduced or delayed induction of DIN6 target gene expression compared with wild-type plants upon removal of sugar from the growth medium, NLS-SnRK1α1 seedlings showed a significantly faster and stronger response (Figure 8A). A similar response was observed in detached rosette leaves upon dark incubation (Figure 8B). To assess longer-term effects on plant stress signaling and tolerance, we transferred soil-grown plants to complete darkness for several days. Whereas NLS-SnRK1α1 plants showed a stronger activation of DIN6 target gene expression than wild-type and control plants, βMYR-SnRK1α1 plants were significantly affected in their response (Figure 8C). The latter plants also perished after 3 d of dark incubation (Figure 8D).
Figure 8.
Altered SnRK1α1 Subcellular Localization Affects Metabolic Stress Responses.
(A) qRT-PCR analysis of DIN6 expression in wild-type (WT) and kin10/kin10 kin11/kin11 seedlings, complemented with SnRK1α1 (SnRK1α1), NLS-SnRK1α1 (NLS-α1), and βMYR-SnRK1α1 (βMYR-α1) at different time points after removal of sugar from the 1/2 MS growth medium (sugar starvation). Values are averages with sds, n = 3 biological repeats (15 pooled seedlings each). Two-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001.
(B) qRT-PCR analysis of DIN6 expression in leaves of wild-type (WT) and SnRK1α1-complemented kin10/kin10 kin11/kin11 plants 3 h after detachment and incubation in the light or dark. Values are averages with sds, n = 3 biological repeats (three pooled leaves each). Two-way ANOVA statistical analysis was performed in GraphPad Prism v7, ****P < 0.0001.
(C) qRT-PCR analysis of DIN6 expression in 4-week–old wild-type (WT) and SnRK1α1-complemented kin10/kin10 kin11/kin11 plants transferred to complete darkness for 3 d. Values are averages with sds, n = 3 biological replicates (three pooled leaves each). Two-way ANOVA statistical analysis was performed with GraphPad Prism v7, ****P < 0.0001.
(D) Phenotypes of wild-type (WT) and SnRK1α1-complemented kin10/kin10 kin11/kin11 plants after 3 d of complete dark incubation.
(E) qRT-PCR analysis of BCAT2, ETFQO, MCCA, and MCCB gene expression in 4-week–old wild-type (WT) and SnRK1α1-complemented kin10/kin10 kin11/kin11 plants transferred to complete darkness for 3 d. Values are averages with sds, n = 3 biological replicates (three pooled leaves each). Two-way ANOVA statistical analysis was performed with GraphPad Prism v7, ****P < 0.0001.
SnRK1 was recently shown to activate alternative mitochondrial metabolic pathways, such as branched-chain amino acid (BCAA) catabolism, through C/S1-class bZIP TF signaling to support respiration and ensure plant survival in extended darkness (Law et al., 2018; Pedrotti et al., 2018). Although NLS-SnRK1α1 plants showed enhanced induction of the BCAA catabolic genes BRANCHED CHAIN TRANSAMINASE2 (BCAT2), METHYLCROTONYL-COA CARBOXYLASE A (MCCA), MCCB, and ELECTRON-TRANSFER FLAVOPROTEIN:UBIQUINONE OXIDO-REDUCTASE (ETFQO), their induction was severely affected in βMYR-SnRK1α1 plants (Figure 8E). These results indicate that nuclear translocation of SnRK1α1 is required for the induction of alternative pathways to generate ATP from noncarbohydrate sources.
DISCUSSION
SnRK1 is a key sensor and regulator of plant energy homeostasis, but how it is activated by low energy stress is still unclear (Broeckx et al., 2016). AMPK and SNF1 are primarily regulated by complex-dependent α-subunit T-loop phosphorylation and (protection from) dephosphorylation. However, although T-loop phosphorylation is typically used as a measure of AMPK activity in animal cells, such a clear correlation is not always observed in plants (Baena-González et al., 2007; Fragoso et al., 2009; Coello et al., 2012; Rodrigues et al., 2013). Arabidopsis SnRK1α was also reported to be resistant to dephosphorylation in vitro (Emanuelle et al., 2015). Unlike mammalian AMPK and yeast SNF1, SnRK1 is also not allosterically activated by a relative increase in AMP and ADP levels (reduced nucleotide charge), which is consistent with the finding that a number of amino acid substitutions in the SnRK1βγ and SnRK1α subunits preclude efficient nucleotide binding and associated regulatory interactions (Ramon et al., 2013; Emanuelle et al., 2015). Moreover, the autoinhibitory domain in AMPK and SNF1 is lacking in the SnRK1α subunits, which feature a ubiquitin-associated domain instead (Broeckx et al., 2016). This results indicate that the SnRK1 catalytic α-subunits have complex-independent activity not affected by truncation of its regulatory domain. This effect is not an artifact of or limited to our experimental system, as upon heterologous expression in yeast, the plant α-subunits, unlike yeast Snf1 itself, did not require the yeast regulatory β- and γ-subunits for effective functional complementation. In addition, the FL and truncated SnRK1α proteins showed a similar, significant T-loop phosphorylation upon expression in leaf cells. Although upstream kinases (SnRK1 activating kinase1/Geminivirus Rep interacting kinase2 and SnRK1 activating kinase2/Geminivirus Rep interacting kinase1) have been identified that phosphorylate SnRK1 in vitro and in vivo (Kong and Hanley-Bowdoin, 2002; Shen and Hanley-Bowdoin, 2006; Shen et al., 2009; Crozet et al., 2010; Glab et al., 2017), the dramatic reduction in T-loop phosphorylation in catalytically dead K48M mutant SnRK1α1 suggests that this largely results from (complex-independent) autophosphorylation in leaf mesophyll protoplasts. Perhaps the upstream kinases are involved in the initial phosphorylation and activation of (newly synthesized) SnRK1α proteins.
The default activation state we observed is consistent with the identification of an increasing number of plant-specific negative SnRK1 regulators (Broeckx et al., 2016). Most notably, sugar-phosphates, such as Glc-6-P and T6P, were identified as potent allosteric inhibitors of SnRK1 activity (Toroser et al., 2000; Zhang et al., 2009; Nunes et al., 2013). This provides an alternative direct link with metabolic status and a straightforward mechanism for active SnRK1 repression under energy-rich conditions. Surprisingly, T6P was recently reported to directly bind to the catalytic SnRK1α subunits and to interfere with interaction with and T-loop phosphorylation by the upstream kinases (Zhai et al., 2018). However, the exact binding site has not been identified, and whether T6P and T-loop phosphorylation affect SnRK1α localization remains to be investigated. Our cellular assays also identified a regulatory role for the β-subunits in restricting target gene activation. T-loop phosphorylation and repressed target gene regulation were not affected, suggesting that catalytic activity itself is not regulated. The more limited effect of the truncated SnRK1β3 subunit suggested a key role for the variable N terminus with N-MYR. Myristoylation is known to mediate membrane association (Warden et al., 2001; Pierre et al., 2007), and confocal fluorescence microscopy confirmed that the SnRK1β1 and SnRK1β2 subunits restrict SnRK1α nuclear localization. Consistently, the nmt1-1 mutant, which is deficient in the activity of the main n-myristoyl transferase, showed increased SnRK1-specific activity in seedling extracts, which was attributed to the enrichment of the kinase in the soluble (nucleus and cytosol) fraction (Pierre et al., 2007). Myristoylation is typically a cotranslational modification, so it is not clear whether it can be regulated to provide a mechanism for SnRK1 signaling in response to stress or metabolic status.
We showed that different types of metabolic stress that deplete cellular energy levels (extended darkness, inhibition of photosynthesis, and respiration-inhibiting hypoxia) trigger nuclear translocation of endogenous SnRK1α1 and used the nuclear exclusion capacity of the βMYR domain in combination with the SV40 NLS as a tool to further analyze the physiological relevance of SnRK1α localization. Increasing nuclear levels (NLS-SnRK1α1) increased target promoter activation, while βMYR-SnRK1α1 (with reduced nuclear localization) was significantly less effective. DEX-inducible translocation confirmed that nuclear SnRK1α1 localization is sufficient for target gene induction.
Interestingly, SnRK1β2 did not localize or translocate to the nucleus and no longer repressed nuclear NLS-SnRK1α1, indicating that it must dissociate from the α-subunit under metabolic stress conditions. However, both SnRK1βγ and SnRK1β3 are localized to the cytoplasm and nucleus (Gissot et al., 2006; Bitrián et al., 2011; Gao et al., 2016). SnRK1 complex formation has been studied in vitro using coimmunoprecipitation (CoIP) assays with whole cell or tissue extracts, but more detailed analyses of interactions in different cellular locations will be needed to confirm that SnRK1β1/2-containing complexes are limited to the cytoplasm and that nuclear complexes exclusively contain the SnRK1β3 subunit. The yeast nonmyristoylated β-subunit Galactose Metabolism83 (Gal83) was also reported to relocalize to the nucleus with Snf1 under activating conditions (Vincent et al., 2001; Hedbacker et al., 2004a; Hedbacker and Carlson, 2008). In addition, the possibility of functional heterodimer formation with the hybrid SnRK1βγ subunit or SnRK1α monomers cannot be excluded. How the SnRK1α proteins translocate to the nucleus remains to be investigated, but they contain putative NLSs in their catalytic domains (Broeckx et al., 2016). These sequences may be exposed upon de-repression and dissociation from the β-subunits. Interestingly, a functional (leptomycin-sensitive) nuclear export signal in the C-terminal α-helix of the AMPKα subunits (with bulky hydrophobic amino acids) may also be conserved in the SnRK1α proteins (O’Brien et al., 2015).
Remarkably, the nuclear localization of SnRK1α is only required for induction and not repression of target gene expression. An increasing number of SnRK1-phosphorylated (nuclear) TFs (Broeckx et al., 2016) and chromatin interactions (Cho et al., 2012) are involved in transcriptional regulation, but how repressed SnRK1 targets are regulated is still poorly understood. Our findings suggest that this process involves cytoplasmic phosphorylation and retention of positive regulators and/or activation of (subsequently translocating) negative regulators of gene expression.
It is important to note that for immunoblot analyses, only 10% of the cytoplasmic fractions and entire nuclear fractions were used for analysis. Cytoplasmic SnRK1α1 levels far exceeded nuclear levels under control conditions and were still relatively high upon nuclear translocation or enrichment (stress, DEX treatment, SV40 NLS constructs). This is also consistent with SnRK1’s many cytoplasmic (enzyme) targets (Broeckx et al., 2016), with a possible role for the β-subunits in substrate specificity (Polge et al., 2008; Li et al., 2009).
Mutation of the MYR site only partially affected the repressive effect of SnRK2β, while deletion of the entire N-terminal 30-amino acid sequence reduced the effect to that of the SnRK1β3 subunit. This finding indicates that the N terminus has additional regulatory roles. Little is known about the structure of this 30-amino acid sequence, suggesting that it is flexible and rather unstructured, but it appears to be subject to additional posttranslational modification responsible for the second higher molecular weight SnRK1β2 signal in immunoblot analyses. It will be interesting to identify the exact type of modification and its role in SnRK1 signaling. Coexpression of the SnRK1βγ subunit also largely abolished the inhibition of SnRK1α by SnRK1β2. The βγCBM was recruited around the origin of the Viridiplantae and therefore must be involved in green plant-specific regulation (Ramon et al., 2013). Modeling suggested that the βCBM might interact with and/or compete with the βγCBM for binding to the α-subunit catalytic domain (Broeckx et al., 2016). In mammals, AMPK βCBM (S108) autophosphorylation and interaction with the catalytic domain protects the T-loop from dephosphorylation, and this interaction is disrupted by glycogen binding (Xiao et al., 2011, 2013; Li et al., 2015). AMPKβ S108 is conserved in plants (Broeckx et al., 2016), but whether starch or other carbohydrates bind to the plant SnRK1 CBMs is not yet fully resolved (Ávila-Castañeda et al., 2014; Emanuelle et al., 2015). A recent study reported maltose binding and activation of the complex involving the βγCBM (Ruiz-Gayosso et al., 2018).
Due to likely redundancy, future loss-of-function studies of SnRK1β will require the generation of double (and triple) knockouts or, in the case of lethality, transient or conditional/inducible knock-down. Analysis of SnRK1βγ function is also complicated by the lethality of knockouts due to a pollen hydration and germination defect (Ramon et al., 2013; Gao et al., 2016; Li et al., 2017). However, we were able to confirm the relevance of subcellular SnRK1α localization using transgenic plants uniquely expressing tagged NLS-SnRK1α1 and βMYR-SnRK1α1 proteins in their original gene context in an snrk1α1 snrk1α2 double mutant background. NLS-SnRK1α1 plants consistently showed a significantly faster and stronger transcriptional response to metabolic stress than the wild type, whereas βMYR-SnRK1α1 plants were severely affected in their response to more long-term energy depletion. Our results are also consistent with the notion that additional regulatory mechanisms repress and de-repress NLS-SnRK1α1-mediated signaling in response to metabolic status. βMYR-SnRK1α1 plants did not survive the severe carbon starvation of 3 d of dark incubation, which is consistent with an inability to transcriptionally induce BCAA catabolism as an alternative noncarbohydrate source for mitochondrial respiration and ATP generation (Pedrotti et al., 2018). Importantly, our approach of altering SnRK1α localization rather than expression (and activity) levels also revealed important functions for SnRK1 in shoot and root development, including the control of leaf shape, root meristem size, and root hair growth. Consistent with a growth-limiting function under low energy conditions, growth-related genes (such as EXP10) are typically repressed by SnRK1 (Baena-González et al., 2007). Our observation that repression requires cytoplasmic rather than nuclear SnRK1 may therefore partly explain the growth effects of altered SnRK1α localization. Identifying the exact mechanisms involved in SnRK1-mediated regulation of plant growth and development will require higher resolution studies using cell- and tissue-specific (e.g. stem cell niche defining factor) reporters and kinematic analyses of cell division and cell expansion. The transition between cell division and expansion along a basipetal gradient in Arabidopsis leaves is highly regulated, e.g. by the antagonistic microRNA396-GROWTH REGULATING FACTOR TF and miR319-class II TEOSINTE BRANCHED, CYCLOIDEA AND PCF (TCP) TF modules (Du et al., 2018; Maugarny-Calès and Laufs, 2018). Interestingly, miR319 and its TCP targets were found to mediate the carbon starvation response downstream of SnRK1 (Confraria et al., 2013), and TCP3 and TCP13 were picked up in a yeast two-hybrid screen with SnRK1α, identifying them as putative direct SnRK1 phosphorylation targets (Nietzsche et al., 2016). The altered leaf shape and curvature of NLS-SnRK1α1 and βMYR-SnRK1α1 plants may therefore be linked to an altered cell cycle arrest front shape (Baekelandt et al., 2018).
The processes affected in seedling roots are known targets of Target of rapamycin (TOR) kinase and auxin signaling, which also function downstream of SnRK1 (Xiong et al., 2013; Weiste and Dröge-Laser, 2014; Nukarinen et al., 2016; Weiste et al., 2017). An initial (low-resolution) RT-qPCR analysis identified a putative auxin-controlled molecular mechanism for SnRK1-mediated regulation of root hair growth (Supplemental Figure 12). Auxin-stimulated postmitotic root hair growth is mediated by the induction of the RSL2 and RSL4 basic helix-loop-helix TFs, which in turn activate the expression of genes that control cell growth through reactive oxygen species homeostasis and cell wall synthesis and remodeling, such as root hair-specific EXP7 (Yi et al., 2010; Mangano et al., 2018). Whole NLS-SnRK1α1 seedling roots showed increased expression of RSL2 and EXP7 (consistent with longer root hairs), whereas βMYR-SnRK1α1 roots showed reduced RSL2 expression (Supplemental Figure 12A). Sampling of entire root tissue most likely dilutes these effects. Posttranslational regulation may also be involved in the process. Local auxin synthesis via TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1, producing the indole 3-acetic acid precursor indole-3-pyruvate) was recently shown to mediate low Pi-induced root hair growth (Bhosale et al., 2018). We detected increased TAA1 expression levels in NLS-SnRK1α1 roots, although its expression was not reduced in βMYR-SnRK1α1 roots (Supplemental Figure 12B). However, root auxin levels are also controlled by biosynthesis in and transport from the shoot. Interestingly, TAA1 expression was significantly repressed in βMYR-SnRK1α1 seedling shoots (Supplemental Figure 12B), possibly contributing to the deficient root and root hair growth in these plants. More focused analyses (using auxin reporters) are needed to test this hypothesis.
In conclusion, our results show that SnRK1 regulation has evolved differently from that of analogous opisthokont (fungal and animal) energy sensing systems. Active suppression of (by default) activated SnRK1 and metabolic stress responses when carbon and energy supplies are abundant (and de-repression when they become depleted) likely represents a reliable strategy for helping autotrophic and sessile plants cope with rapidly changing conditions. The nuclear translocation of SnRK1 is an important regulatory mechanism in the energy stress response. Our analysis of transgenic plants with altered SnRK1α subunit localization also revealed important functions for nuclear and cytoplasmic SnRK1 signaling in the metabolic control of plant growth and development.
METHODS
The primers used for cloning, mutagenesis, confirmation of T-DNA insertion, and RT-qPCR are listed in Supplemental Data Set.
Homology Modeling
The Arabidopsis (Arabidopsis thaliana) heterotrimeric SnRK1 complex model was produced by combining individually modeled subunits (Broeckx et al., 2016). Subunits were modeled based on the 4RER crystal structure of the AMPK complex in the activated (phosphorylated) state (Xiao et al., 2013; Li et al., 2015), using SWISS-MODEL (https://swissmodel.expasy.org/; Biasini et al., 2014) with structural refinement by GalaxyWEB (http://galaxy.seoklab.org/; Ko et al., 2012). The complex was then assembled using the program PyMOL v1.3 Edu (https://pymol.org/edu/) and further refined using GalaxyRefineComplex (http://galaxy.seoklab.org/; Ko et al., 2012). Final figures were composed in PyMOL 1.3 Edu. The hybrid SnRK1βγ subunit was first modeled with the software RaptorX (http://raptorx.uchicago.edu/) based on structurally similar proteins (Källberg et al., 2012).
Plant Growth and Phenotyping
Arabidopsis seeds were vapor-sterilized and stratified at 4°C for 3 d before sowing. For leaf mesophyll protoplast isolation, Arabidopsis Columbia wild-type (Col-0) plants were grown under a 12-h light/12-h dark diurnal cycle at 21°C with 75-μE cool white fluorescent light (cat. no. F17T8/TL741/ALTO; Philips) for 4 weeks.
The SALK_037416 (snrk1β2), GABI_579_E09 (snrk1α1/kin10), and WiscDsLox320B03 (snrk1α2/kin11) T-DNA lines were obtained from the Arabidopsis Biological Resource Center (Ohio State University). Homozygous plants were selected on full MS medium including vitamins (cat. no. M0222; Duchefa Biochemie) with kanamycin, sulfadiazine, or glufosinate (Basta S; Bayer Crop Science) by PCR analysis and by immunoblot analysis.
For the seedling sugar starvation assays, 15 seeds were germinated in 1 mL of 0.5x MS medium supplemented with 0.5% (w/v) Glc in 6-well plates for each biological replicate. The plates were incubated under continuous cool white fluorescent light (65 μE) at 21°C for 6 d. After transfer to fresh 0.5× MS medium supplemented with 0.5% (w/v) Glc for one more day, the seedlings were washed, transferred to 0.5× MS medium without sugar, and sampled at the indicated time points after sugar removal by freezing in liquid nitrogen.
For the diurnal, extended night, and dark experiments, 4-week–old wild-type and transgenic plants were grown in a 12-h light/12-h dark diurnal cycle at 21°C with 75-μE cool white fluorescent light for 4 weeks and subjected to an extended night or continuous darkness. For each biological replicate, three adult source leaves from different plants were sampled at the indicated times by freezing in liquid nitrogen.
Leaf shape of soil grown plants (12-h light/12-h dark diurnal cycle at 21°C with 75-μE cool white fluorescent light) was determined 4 weeks after germination by measuring the leaf length/width ratio using the software ImageJ (U.S. National Institutes of Health). Root hair and primary root (meristem) size was determined for seedlings grown for 10 d on half-strength MS medium plates supplemented with 0.5% (w/v) Glc and placed vertically in a 12-h light/12-h dark diurnal cycle at 21°C with 75-μE cool white fluorescent light for 10 d. Roots were visualized using a Discovery model 8 Stereo microscope (Zeiss) with AxioVision v4.8.2 software (Zeiss), and lengths were quantified with the software ImageJ (with a 25-μm resolution for root hairs). Meristems were visualized by propidium iodide staining and fluorescence (confocal laser scanning) microscopy (FLUOVIEW FV1000 confocal laser scanning microscope; Olympus) with a 40× (1.3 oil) objective, focusing on the quiescent center. Root meristem size was determined based on both the number of cells in individual cortical cell files from the quiescent center up to where cell size doubled and the length (ImageJ).
Plasmid Construction
FL Arabidopsis SnRK1α1/KIN10, SnRK1α2/KIN11, SnRK1β1, SnRK1β2, SnRK1β3, SnRK1βγ, GBF5/bZIP2, yeast (Saccharomyces cerevisiae) SNF1, and human AMPKα1 CDS were PCR-amplified without the stop codon from Arabidopsis Columbia cDNA, yeast W303-1A genomic DNA, and human brain cDNA and inserted in the HBT95 expression vector with the 35SC4PPDK promoter (35S enhancer and maize [Zea mays] C4PPDK basal promoter) and nopaline synthase (NOS) terminator in-frame with a double HA, FLAG, or GFP tag (Sheen, 1996). N-terminal SV40 NLS and βMYR sequences were included in the forward cloning primers. The genomic SnRK1α1/KIN10 fragment was PCR-amplified from Arabidopsis Columbia genomic DNA, inserted in a pUC18 vector for modification (insertion of a C-terminal FLAG tag and addition of the N-terminal SV40 NLS and βMYR sequences), and subcloned in a pCB302-derived mini binary vector with a kanamycin resistance marker for transgenic plant selection (Xiang et al., 1999; Hwang and Sheen, 2001). Site-directed mutagenesis (point mutations, insertions, and deletions) was performed with PCR using complementary mutant primer pairs extending 12 to 15 nucleotides on either side of the modification. DpnI was used to digest the methylated template DNA. The DIN6/ASPARAGINE SYNTHASE1 (At3g47340) promoter-LUC reporter was described in Baena-González et al. (2007). The EXP10 (At1g26770) 2.5-kb promoter sequence was amplified from Arabidopsis Col-0 genomic DNA and cloned in a pUC18-based LUC vector. For yeast complementation, CDS were subcloned in a modified yeast pYX212 multicopy plasmid with a Hexose Transporter7 (HXT7) promoter and a Uracil requiring3 (URA3) marker (Vandesteene et al., 2010). All constructs were confirmed by sequencing (LGC Genomics).
Transient Expression in Leaf Mesophyll Protoplasts
Arabidopsis leaf mesophyll protoplast isolation and transfection were performed as described in Yoo et al. (2007). After polyethylene glycol (PEG)-Ca2+-mediated transfection, protoplasts were incubated in dim light for 6 h (LUC, β-Glucuronidase [GUS], and immunoblot analyses) or 12 h (GFP localization studies). For metabolic stress, cells were incubated under dark, hypoxic conditions (submergence in incubation buffer) or with 20-μM DCMU, Diuron D2425; Sigma-Aldrich). For DEX-induced nuclear translocation, 10-μM DEX (D1756; Sigma-Aldrich) was added to protoplasts for 4 h (2 h after transfection).
LUC and GUS Assays
For LUC activity measurements, protoplasts were lysed with 100-μL lysis buffer (25-mM Trip-Phosphate at pH 7.8, 2 mM DTT, 2-mM 1.2-diaminocylcohexane-n, n, n′,n′-tetra-acetic acid, 10% [v/v] glycerol, and 1% [v/v] Triton X-100). Twenty microliters of the cell lysate was dispensed into a luminometer tube and mixed with 100-μL LUC assay reagent (E1500 Kit; Promega). Luminescence was detected with a Lumat LB 9507 tube luminometer (Berthold Technologies). GUS activity of the UBQ-GUS control for transfection efficiency was measured with 5 μL of cell lysate in 45 μL of 10-mM 4-methylumbelliferyl-β-d-glucuronide solution (MUG, M-9130; Sigma-Aldrich). After 1 h incubation at 37°C, the reaction was stopped with 220 μL of 0.2-M Na2CO3, and fluorescence was measured with the GloMax-Multi+ Detection System (Promega).
Immunoblot Analyses
To detect transiently expressed and endogenous proteins in leaf mesophyll cells, 2 × 104 protoplasts were transfected with 20-μg DNA (CsCl-gradient–purified) and incubated for 6 h. A 2× quantity of loading buffer (120-mM Tris-HCl at pH 6.8, 5.4-M urea, 20% [v/v] glycerol, 4% [w/v] SDS, 5% [v/v] β-mercaptoethanol, and 0.5% [v/v] bromophenol blue) was added to the protoplast samples before loading on a 1.5-mm 10% acrylamide SDS-PAGE gel and protein separation in Tris-Gly running buffer (0.025-M Tris, 0.192-M Gly, and 0.1% [w/v] SDS at pH 8.5) at 60 Volts for 15 min and 160 Volts for 1 h. Proteins were then transferred to apolyvinylidene fluoride membrane (Immobilon-P; Millipore) with a semidry transfer system (Trans-Blot SD; Bio-Rad) in Tris-Gly buffer with 10% (v/v) methanol for 30 min at 20 Volts. After 1-h incubation with 5% (w/v) skimmed milk, the membrane was incubated with antibody in 1% (w/v) milk for 2 h at room temperature (horseradish peroxidase [HRP]-conjugated anti-HA antibody, 1/1,000 [50 μg/mL], cat. no. 12013819001, Roche; HRP-conjugated anti-FLAG antibody, 1/1,000 [1 μg/mL], cat. no. A8592, Sigma-Aldrich; primary anti-SnRK1β2 antibody, 1/500 [2 μg/mL], cat. no. AS09 462, Agrisera; and secondary goat-anti-rabbit IgG-HRP antibody, 1/10,000 [0.1 μg/mL], cat. no. AS09 602, Agrisera).
The membrane was washed three times intris(hydroxymethyl)aminomethane (Tris)-buffered saline (TBS) with polyethylene glycol sorbitan monolaurate (Tween 20; 50-mM Tris, 150-mM NaCl, and 0.05% [v/v] Tween 20), incubated with Pierce SuperSignal West Pico PLUS Chemiluminescent Substrate (cat. no. 34,577; Thermo Fisher Scientific) for 2 min, and exposed to film for a few seconds to several minutes. Ribulose bisphosphate carboxylase small chain (RBCS) staining of the blot with Coomassie Brilliant Blue R-250 was used as a protein loading control.
Phosphorylation Mobility Shift Assay
For the phosphorylation mobility shift assay, Phos-tag Acrylamide (Wako Chemicals) was added to the 1.5-mm 8% (w/v) acrylamide SDS-PAGE gel, as described by the manufacturer (cat. no. AAL-107; Wako Chemicals). Protoplast samples were prepared as described for “Immunoblot Analyses.” Proteins were separated in Tris-Gly running buffer (0.025-M Tris, 0.192-M Gly, and 0.1% [w/v] SDS at pH 8.5) at 30 mA until the bromophenol blue reached the bottom of the resolving gel. After electrophoresis, the gel was soaked twice in Tris-Gly transfer buffer containing 10% (v/v) methanol and 10-mM EDTA for 20 min each time with gentle agitation, followed by 10 min in transfer buffer without EDTA. Proteins were then transferred to a polyvinylidene fluoride membrane using a wet tank transfer system (Mini trans-blot cell; Bio-Rad) in Tris-Gly buffer with 10% (v/v) methanol for 2 h at 300 mA. Protein bands were visualized as described for “Immunoblot Analyses.”
Subcellular Localization
To observe the subcellular localization of SnRK1 subunits, 4 × 104 protoplasts were transfected with a total of 20-μg GFP-construct plasmid DNA and incubated for 6 to 16 h. GFP was visualized using confocal laser scanning microscopy (FV1000; Olympus) with a 40× (1.3 oil) objective.
CoIP Assays
For the CoIP assays, ∼2 × 105 Arabidopsis leaf mesophyll protoplasts were transfected with a total of 100 µg of DNA and incubated for 6 h. After harvest, the cells were lysed with 200-μl IP buffer (50-mM Tris-HCl at pH 7.5, 150-mM NaCl, 5-mM EDTA, 1% [v/v] Triton X-100, 0.5-mM DTT, and 1 tablet Complete Protease Inhibitor [cat. no. 04693159001; Roche]). A 20-μL aliquot was taken for the input control and the lysate was incubated for 3 h with 40-μL FLAG-conjugated agarose beads (anti-FLAG M2 Affinity Gel, cat. no. A2220; Sigma-Aldrich) prewashed three times with IP buffer at 4°C under gentle rotation. After incubation, the beads were washed five times with IP buffer. Eluted samples and input samples were subjected to immunoblot analysis using conjugated anti-HA and anti-FLAG antibodies.
Extraction and Fractionation of Proteins
To isolate nuclei from protoplasts, ∼4 × 105 Arabidopsis leaf mesophyll protoplasts were transfected with 150 μg of DNA and incubated for 6 h. After harvesting, cells were lysed with 300-μL lysis buffer (20-mM Tris-HCl at pH 7.0, 250-mM Suc, 25% (v/v) glycerol, 20-mM KCl, 2-mM EDTA, 2.5-mM MgCl2, 30-mM β-mercaptoethanol, 1% (v/v) Triton X-100, 0.5-mM spermidine, and 1× protease inhibitor cocktail) and kept on ice for 5 min. Lysate was then centrifuged at 600 relative centrifugal force (rcf; 2,000 rpm in an model no. 5702 centrifuge; Eppendorf) for 5 min. The cytoplasmic fraction was transferred to a new microcentrifuge tube while the nuclei pellet was washed three times with water without disrupting the nuclei. The nuclear pellet was resuspended in 20-μL resuspension buffer (20-mM Tris-HCl at pH 7.0, 25% (v/v) glycerol, 2.5-mM MgCl2, 30-mM β-mercaptoethanol, and 1× protease inhibitor cocktail). Five percent of the cytoplasmic fraction and the whole nuclear fraction were subjected to immunoblot analysis with anti-KIN10 antibody (Baena-González et al., 2007; 1:2,000 dilution) and conjugated anti-HA antibody (1:2,000 dilution). Anti-Histone H3 antibodies (1:3,000 dilution, cat. no. AS10710; Agrisera) and RBCS staining with Coomassie Brilliant Blue R-250 were used as controls for purity of the nuclear and cytoplasmic fractions, respectively.
To isolate nuclei from intact plants, leaves of soil-grown plants were ground in liquid nitrogen and homogenized with 3-mL lysis buffer. The homogenate was then filtered through a layer of wet Miracloth (cat. no. 475855; Millipore) and centrifuged at 1,400 rcf (3,000 rpm) for 10 min at 4°C to pellet the nuclei. The cytoplasmic fraction was transferred to a new tube, while the nuclear pellet was re-extracted twice with lysis buffer, discarding the supernatant. The nuclear pellet was washed in 1-mL resuspension buffer by pipetting, centrifuged at 1,400 rcf (3,000 rpm) for 10 min at 4°C, and the supernatant was discarded. Subsequently, the nuclear pellet was resuspended in 200-μL resuspension buffer. Twenty microliters of the cytoplasmic and nuclear fractions were subjected to immunoblot analysis.
RT-qPCR
For RT-qPCR quantification of gene expression in seedlings and transfected protoplasts, RNA extraction was performed with TRIzol Reagent (cat. no. 15,596,026; Thermo Fisher Scientific) according to the manufacturer’s instructions. One microgram of total RNA was used for reverse transcription with the Reverse Transcription System (A3500; Promega). qPCR was performed using the GoTaq qPCR Master Mix kit (cat. no. A6001; Promega) according to the manufacturer’s instructions in a total volume of 10 μL with 5-μL FAST SYBR GREEN buffer, 0.2 μL of each primer (10 μM), 2.5 μL of H20, 0.1 μL ofcarboxy-X-rhodamine (CXR) reference dye, and 2 μL of cDNA (5 ng/μL). The PCR program comprised an initial denaturation for 2 min at 95°C and amplification by 45 cycles of 3 s at 95°C and 30 s at 58°C in a StepOnePlus Real Time PCR system (Applied Biosystems). Expression levels were normalized to the expression of UBIQUITIN10, which is stable in the different tissues and metabolic stress conditions used (Baena-González et al., 2007).
Yeast Complementation
The yeast (Saccharomyces cerevisiae) strains MCY4908 (W303–1A snf1Δ10; Hedbacker et al., 2004b) and MCY5751 (W303–1A snf1Δ10 snf4Δ::hphMX gal83Δ::TRP1 sip1Δ::kanMX sip2Δ::kanMX; Momcilovic and Carlson, 2011) were used for growth defect complementation assays. Yeast transformation was performed using a LiAc/SS carrier DNA/PEG transformation protocol (Gietz and Schiestl, 2007). For the growth assays, cultures of the transformed strains were grown to exponential phase at 30°C on minimal synthetic defined medium without uracil (SD-ura) containing 2% (w/v) Glc and drop-assays were performed on SD-ura with 2% (w/v) Glc (control) or 2% (v/v) glycerol + 3% (v/v) ethanol. Transformants were streaked or spotted at OD600 1, and growth was analyzed after 3 d at 30°C. The assay was repeated three times with independent transformants with similar results.
Reproducibility and Statistics
All experiments were repeated at least three times with reproducible results. One-way and two-way analysis of variance (ANOVA) statistical analyses were performed using the program GraphPad Prism v7 (https://www.graphpad.com/scientific-software/prism/; Supplemental File).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: Arabidopsis SnRK1α1/KIN10 (At3g01090), SnRK1α2/KIN11 (At3g29160), SnRK1β1 (At5g21170), SnRK1β2 (At4g16360), SnRK1β3 (At2g28060), SnRK1βγ (At1g09020), GBF5/bZIP2 (At2g18160), UBIQUITIN10 (At4g05320), DIN1/SENESCENCE-ASSOCIATED PROTEIN1 (SEN1; At4g35770), EXP10 (At1g26770), SEN5 (At3g15450), DRM2 (At2g33830), PGPD14 (At5g22920), HDT1 (At3g44750), DWF4 (At3g50660), MYB75 (At1g56650), RSL2 (At4g33880), EXP7 (At1g12560), TAA1 (At1g70560), BCAT2 (At1g10070), MCCA (At1g03090), MCCB (At4g34030), ETFQO (At2g43400), yeast SNF1 (YDR477W), human AMPKα1 (AF100763.1).
Supplemental Data
Supplemental Figure 1. SnRK1 heterotrimeric complex and SnRK1α1 catalytic α-subunit domain structure and composition.
Supplemental Figure 2. Complex-independent regulation of target gene expression by SnRK1α1.
Supplemental Figure 3. Unique constitutive complex-independent activity of the SnRK1α1 subunit.
Supplemental Figure 4. SnRK1β2 subunit domain structure and truncation.
Supplemental Figure 5. SnRK1βγ subunit structure and truncation.
Supplemental Figure 6. SnRK1β2 does not affect SnRK1α1-mediated gene repression.
Supplemental Figure 7. Analysis of the SALK_037416 SnRK1β2 T-DNA knockout line.
Supplemental Figure 8. A role for the SnRK1β2 N-terminal variable extension and myristoylation.
Supplemental Figure 9. Effect of SnRK1α1 localization on target gene expression.
Supplemental Figure 10. Nuclear translocation of SnRK1α1 upon metabolic stress application.
Supplemental Figure 11. Development of transgenic lines with altered SnRK1α1 subcellular localization.
Supplemental Figure 12. Effect of SnRK1α1 localization on root hair growth- and auxin biosynthesis-associated gene expression.
Supplemental Data Set. Oligonucleotide primers used in this study.
Supplemental File. Statistical analysis results.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the Fund for Scientific Research - Flanders (grant FWO G0D2814N to the Rolland lab), the National Science Foundation (grant IOS-0843244 to the Sheen lab), the U.S. National Institutes of Health (grant R01 GM60493 to the Sheen lab), a Scientific Cooperation Project of the Fund for Scientific Research - Flanders and the National Research Foundation- Republic of Korea (grant NRF-2016K2A9A1A06922531), and the Korean Research Fellowship program of the Ministry of Science and information and communication technology through the National Research Foundation of Korea (fellowship 2017H1D3A1A03055171 to T.V.T.D.). The authors thank Elena Baena-González for the SnRK1α1/KIN10 and SnRK1α2/KIN11 antibodies, Marian Carlson for the yeast strains, the Arabidopsis Biological Resource Center (Ohio State University) for T-DNA mutant seeds, Tom Beeckman for advice on root meristem microscopy, Anja Vandeperre for excellent technical assistance, Hilde Verlinden for plant care, and Hannah Degroote, Jing Wen, and other former members of the lab for practical help and discussion.
AUTHOR CONTRIBUTIONS
M.R., J.S., and F.R. conceived the project; M.R. produced key constructs and transgenic plants, and performed initial analyses; T.V.T.D., T.B., S.H., N.C., and F.R. performed cellular assays and did plant characterization; T.B. performed the modeling; T.B., S.H., and N.C. collected statistics and developed the figures; F.R. wrote the article with input from all authors.
Disclaimer: The author Matthew Ramon is employed with the European Food Safety Authority (EFSA). However, the present article is published under the sole responsibility of the author and may not be considered as an EFSA scientific output. The positions and opinions presented in this article are those of the author alone and are not intended to represent the views or scientific works of EFSA. To learn about the views or scientific outputs of EFSA, please consult its website at http://www.efsa.europa.eu.
References
- Alderson A., Sabelli P.A., Dickinson J.R., Cole D., Richardson M., Kreis M., Shewry P.R., Halford N.G. (1991). Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl. Acad. Sci. USA 88: 8602–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila-Castañeda A., Gutiérrez-Granados N., Ruiz-Gayosso A., Sosa-Peinado A., Martínez-Barajas E., Coello P. (2014). Structural and functional basis for starch binding in the SnRK1 subunits AKINβ2 and AKINβγ. Front. Plant Sci. 5: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt A., Pauwels L., Wang Z., Li N., De Milde L., Natran A., Vermeersch M., Li Y., Goossens A., Inzé D., Gonzalez N. (2018). Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol. 178: 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Hanson J. (2017). Shaping plant development through the SnRK1-TOR metabolic regulators. Curr. Opin. Plant Biol. 35: 152–157. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert K., Bakó L., Okrész L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale R., et al. (2018). A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 9: 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., et al. (2014). SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrián M., Roodbarkelari F., Horváth M., Koncz C. (2011). BAC-recombineering for studying plant gene regulation: Developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 65: 829–842. [DOI] [PubMed] [Google Scholar]
- Boisson B., Giglione C., Meinnel T. (2003). Unexpected protein families including cell defense components feature in the n-myristoylome of a higher eukaryote. J. Biol. Chem. 278: 43418–43429. [DOI] [PubMed] [Google Scholar]
- Broeckx T., Hulsmans S., Rolland F. (2016). The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 67: 6215–6252. [DOI] [PubMed] [Google Scholar]
- Carling D., Clarke P.R., Zammit V.A., Hardie D.G. (1989). Purification and characterization of the AMP-activated protein kinase. Copurification of acetyl-CoA carboxylase kinase and 3-hydroxy-3-methylglutaryl-CoA reductase kinase activities. Eur. J. Biochem. 186: 129–136. [DOI] [PubMed] [Google Scholar]
- Cho Y.-H., Hong J.-W., Kim E.-C., Yoo S.-D. (2012). Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158: 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P., Hirano E., Hey S.J., Muttucumaru N., Martinez-Barajas E., Parry M.A., Halford N.G. (2012). Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 63: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confraria A., Baena-González E. (2016). Using Arabidopsis protoplasts to study cellular responses to environmental stress. Methods Mol. Biol. 1398: 247–269. [DOI] [PubMed] [Google Scholar]
- Confraria A., Martinho C., Elias A., Rubio-Somoza I., Baena-González E. (2013). miRNAs mediate SnRK1-dependent energy signaling in Arabidopsis. Front. Plant Sci. 4: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P., et al. (2016). SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 85: 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P., Jammes F., Valot B., Ambard-Bretteville F., Nessler S., Hodges M., Vidal J., Thomas M. (2010). Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J. Biol. Chem. 285: 12071–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K., Weltmeier F., Ehlert A., Weiste C., Stahl M., Harter K., Dröge-Laser W. (2011). Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Guan C., Jiao Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11: 1117–1134. [DOI] [PubMed] [Google Scholar]
- Emanuelle S., et al. (2015). SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 82: 183–192. [DOI] [PubMed] [Google Scholar]
- Estruch F., Treitel M.A., Yang X., Carlson M. (1992). N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S., Espíndola L., Páez-Valencia J., Gamboa A., Camacho Y., Martínez-Barajas E., Coello P. (2009). SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 149: 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.Q., Liu C.Z., Li D.D., Zhao T.T., Li F., Jia X.N., Zhao X.Y., Zhang X.S. (2016). The Arabidopsis KINβγ subunit of the SnRK1 complex regulates pollen hydration on the stigma by mediating the level of reactive oxygen species in pollen. PLoS Genet. 12: e1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (2007). Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 35–37. [DOI] [PubMed] [Google Scholar]
- Gissot L., Polge C., Bouly J.P., Lemaitre T., Kreis M., Thomas M. (2004). AKINbeta3, a plant-specific SnRK1 protein, is lacking domains present in yeast and mammals non-catalytic β-subunits. Plant Mol. Biol. 56: 747–759. [DOI] [PubMed] [Google Scholar]
- Gissot L., Polge C., Jossier M., Girin T., Bouly J.P., Kreis M., Thomas M. (2006). AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol. 142: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glab N., Oury C., Guérinier T., Domenichini S., Crozet P., Thomas M., Vidal J., Hodges M. (2017). The impact of Arabidopsis thaliana SNF1-related-kinase 1 (SnRK1)-activating kinase 1 (SnAK1) and SnAK2 on SnRK1 phosphorylation status: Characterization of a SnAK double mutant. Plant J. 89: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Gowans G.J., Hawley S.A., Ross F.A., Hardie D.G. (2013). AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 18: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G., Ross F.A., Hawley S.A. (2012). AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., Hardie D.G. (1996). Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271: 27879–27887. [DOI] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Hong S.P., Carlson M. (2004a). Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24: 8255–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Townley R., Carlson M. (2004b). Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24: 1836–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans S., Rodriguez M., De Coninck B., Rolland F. (2016). The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 21: 648–661. [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389. [DOI] [PubMed] [Google Scholar]
- Jamsheer K.M., Sharma M., Singh D., Mannully C.T., Jindal S., Shukla B.N., Laxmi A. (2018). FCS-like zinc finger 6 and 10 repress SnRK1 signalling in Arabidopsis. Plant J. 94: 232–245. [DOI] [PubMed] [Google Scholar]
- Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B.E. (2004). Bateman domains and adenosine derivatives form a binding contract. J. Clin. Invest. 113: 182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Park H., Heo L., Seok C. (2012). GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 40: W294–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.J., Hanley-Bowdoin L. (2002). A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.M., Hsieh M.H., Coruzzi G. (1998). Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J. 16: 345–353. [DOI] [PubMed] [Google Scholar]
- Law S.R., et al. (2018). Darkened leaves use different metabolic strategies for senescence and survival. Plant Physiol. 177: 132–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2015). Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 25: 50–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.D., Guan H., Li F., Liu C.Z., Dong Y.X., Zhang X.S., Gao X.Q. (2017). Arabidopsis SHAKER pollen inward K+ channel SPIK functions in SnRK1 complex-regulated pollen hydration on the stigma. J. Integr. Plant Biol. 59: 604–611. [DOI] [PubMed] [Google Scholar]
- Li X.F., Li Y.J., An Y.H., Xiong L.J., Shao X.H., Wang Y., Sun Y. (2009). AKINβ1 is involved in the regulation of nitrogen metabolism and sugar signaling in Arabidopsis. J. Integr. Plant Biol. 51: 513–520. [DOI] [PubMed] [Google Scholar]
- Lin C.-R., Lee K.W., Chen C.Y., Hong Y.F., Chen J.L., Lu C.A., Chen K.T., Ho T.H., Yu S.M. (2014). SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell 26: 808–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V., Albà M.M., Kleinow T., Koncz C., Pagès M. (2001). Domain fusion between SNF1-related kinase subunits during plant evolution. EMBO Rep. 2: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair A., et al. (2015). SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLife 4: e05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S., Denita-Juarez S.P., Marzol E., Borassi C., Estevez J.M. (2018). High auxin and high phosphate impact on RSL2 expression and ROS-homeostasis linked to root hair growth in Arabidopsis thaliana. Front. Plant Sci. 9: 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugarny-Calès A., Laufs P. (2018). Getting leaves into shape: A molecular, cellular, environmental and evolutionary view. Development 145: dev161646. [DOI] [PubMed] [Google Scholar]
- Momcilovic M., Carlson M. (2011). Alterations at dispersed sites cause phosphorylation and activation of SNF1 protein kinase during growth on high glucose. J. Biol. Chem. 286: 23544–23551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D., Byrne H.M., King J.R., Bennett M. (2013). Mathematical modelling plant signalling networks. Math. Model. Nat. Phenom. 8: 5–24. [Google Scholar]
- Nietzsche M., Landgraf R., Tohge T., Börnke F. (2016). A protein–protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Curr. Plant Biol. 5: 36–44. [Google Scholar]
- Nukarinen E., et al. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., Primavesi L.F., Patel M.K., Martinez-Barajas E., Powers S.J., Sagar R., Fevereiro P.S., Davis B.G., Paul M.J. (2013). Inhibition of SnRK1 by metabolites: Tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 63: 89–98. [DOI] [PubMed] [Google Scholar]
- O’Brien M., Kaplan-Levy R.N., Quon T., Sappl P.G., Smyth D.R. (2015). PETAL LOSS, a trihelix transcription factor that represses growth in Arabidopsis thaliana, binds the energy-sensing SnRK1 kinase AKIN10. J. Exp. Bot. 66: 2475–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakhill J.S., Steel R., Chen Z.P., Scott J.W., Ling N., Tam S., Kemp B.E. (2011). AMPK is a direct adenylate charge-regulated protein kinase. Science 332: 1433–1435. [DOI] [PubMed] [Google Scholar]
- Pedrotti L., et al. (2018). Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 30: 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre M., Traverso J.A., Boisson B., Domenichini S., Bouchez D., Giglione C., Meinnel T. (2007). N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19: 2804–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C., Jossier M., Crozet P., Gissot L., Thomas M. (2008). β-subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities; physical interaction with nitrate reductase specifically occurs via AKINβ1-subunit. Plant Physiol. 148: 1570–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M., Ruelens P., Li Y., Sheen J., Geuten K., Rolland F. (2013). The hybrid four-CBS-domain KINβγ subunit functions as the canonical γ-subunit of the plant energy sensor SnRK1. Plant J. 75: 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A., et al. (2013). ABI1 and PP2CA phosphatases are negative regulators of SNF1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25: 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gayosso A., Rodríguez-Sotres R., Martínez-Barajas E., Coello P. (2018). A role for the carbohydrate-binding module (CBM) in regulatory SnRK1 subunits: The effect of maltose on SnRK1 activity. Plant J. 96: 163–175. [DOI] [PubMed] [Google Scholar]
- Schena M., Lloyd A.M., Davis R.W. (1991). A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. USA 88: 10421–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.W., Ross F.A., Liu J.K., Hardie D.G. (2007). Regulation of AMP-activated protein kinase by a pseudosubstrate sequence on the gamma subunit. EMBO J. 26: 806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shen W., Hanley-Bowdoin L. (2006). Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142: 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Reyes M.I., Hanley-Bowdoin L. (2009). Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieciechowicz K.A., Joy K.W., Ireland R.J. (1988). The metabolism of asparagine in plants. Phytochemistry 27: 663–671. [Google Scholar]
- Sugden C., Crawford R.M., Halford N.G., Hardie D.G. (1999). Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 19: 433–439. [DOI] [PubMed] [Google Scholar]
- Toroser D., Plaut Z., Huber S.C. (2000). Regulation of a plant SNF1-related protein kinase by glucose-6-phosphate. Plant Physiol. 123: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A.Y.L., Gazzarrini S. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: The emerging picture. Front. Plant Sci. 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Bläsing O.E., Gibon Y., Retzlaff K., Höhne M., Günther M., Stitt M. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146: 1834–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L., Ramon M., Le Roy K., Van Dijck P., Rolland F. (2010). A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol. Plant 3: 406–419. [DOI] [PubMed] [Google Scholar]
- Viana R., Towler M.C., Pan D.A., Carling D., Viollet B., Hardie D.G., Sanz P. (2007). A conserved sequence immediately N-terminal to the Bateman domains in AMP-activated protein kinase γ-subunits is required for the interaction with the β-subunits. J. Biol. Chem. 282: 16117–16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O., Townley R., Kuchin S., Carlson M. (2001). Subcellular localization of the Snf1 kinase is regulated by specific β-subunits and a novel glucose signaling mechanism. Genes Dev. 15: 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden S.M., Richardson C., O’Donnell J. Jr., Stapleton D., Kemp B.E., Witters L.A. (2001). Post-translational modifications of the β-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem. J. 354: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste C., Dröge-Laser W. (2014). The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5: 3883. [DOI] [PubMed] [Google Scholar]
- Weiste C., Pedrotti L., Selvanayagam J., Muralidhara P., Fröschel C., Novák O., Ljung K., Hanson J., Dröge-Laser W. (2017). The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 13: e1006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.A., Hawley S.A., Hardie D.G. (1996). Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6: 1426–1434. [DOI] [PubMed] [Google Scholar]
- Wurzinger B., Nukarinen E., Nägele T., Weckwerth W., Teige M. (2018). The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 176: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]
- Xiao B., et al. (2011). Structure of mammalian AMPK and its regulation by ADP. Nature 472: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., et al. (2013). Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4: 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix–loop–helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhai Z., Keereetaweep J., Liu H., Feil R., Lunn J.E., Shanklin J. (2018). Trehalose 6-Phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A., Powers S.J., Schluepmann H., Delatte T., Wingler A., Paul M.J. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]