Abstract
Conotruncal heart defects (CTDs) are among the most common and severe groups of congenital heart defects. Despite evidence of an inherited genetic contribution to CTDs, little is known about the specific genes that contribute to the development of CTDs. We performed gene-based genome-wide analyses using microarray-genotyped and imputed common and rare variants data from two large studies of CTDs in the United States. We performed two case-parent trio analyses (N = 640 and 317 trios), using an extension of the family-based multi-marker association test, and two case-control analyses (N = 482 and 406 patients and comparable numbers of controls), using a sequence kernel association test. We also undertook two meta-analyses to combine the results from the analyses that used the same approach (i.e. family-based or case-control). To our knowledge, these analyses are the first reported gene-based, genome-wide association studies of CTDs. Based on our findings, we propose eight CTD candidate genes (ARF5, EIF4E, KPNA1, MAP4K3, MBNL1, NCAPG, NDFUS1 and PSMG3). Four of these genes (ARF5, KPNA1, NDUFS1 and PSMG3) have not been previously associated with normal or abnormal heart development. In addition, our analyses provide additional evidence that genes involved in chromatin-modification and in ribonucleic acid splicing are associated with congenital heart defects.
Introduction
Congenital heart defects (CHDs) are the most common group of birth defects, with a prevalence of approximately 1% in live births [1]. CHDs are also the leading cause of birth defect related mortality [2] and account for the largest percentage of birth defect associated hospitalizations and hospitalization-associated costs [3]. In the United States, it is estimated that there are approximately 2.4 million CHD survivors (1.4 million adults, 1 million children) [4], the majority of whom will require lifelong cardiac care. Despite the impact on affected patients, their families, and the healthcare system, the causes of CHDs are not well defined [5].
There are many different CHD phenotypes, of which approximately one-third involve the cardiac outflow tracts and great arteries [6]–structures that develop from the cardiac neural crest and secondary heart field [7]. This subgroup of CHDs, collectively referred to as conotruncal heart defects (CTDs), includes some of the most severe and costly birth defects [3, 8]. In addition to their shared embryologic and anatomic basis, there is substantial evidence that the various CTD phenotypes (e.g. tetralogy of Fallot (TOF), truncus arteriosus) share common genetic underpinnings. For example, nationwide, population-based studies conducted in Norway and Denmark indicate that CTDs aggregate within families (recurrence risk ratios for CTDs in first-degree relatives: 9–12) [9, 10] and that affected relatives of patients with a CTD are at a higher relative risk for CTDs (sibling CTD recurrence risk ratio: 9.0, 95% confidence interval (CI) 4.0–20.0) than for other types of CHDs (sibling non-CTD, CHD recurrence risk ratio: 3.6, 95% CI 2.4–5.5) [9]. Further, there is evidence that, within affected relative-pairs, the specific type of CTD can differ. For example, among 28 CTD-affected siblings of patients with TOF, 17 also had TOF whereas 11 had a different CTD phenotype [11]. Additional evidence that the various CTD phenotypes share a common genetic basis is provided by the phenotypic characteristics of defined genetic syndromes. For example, in patients with the 22q11.2 deletion syndrome, the most common cardiac defects are CTDs, but the specific CTD phenotype (e.g. TOF, interrupted aortic arc) varies across patients with this deletion [12].
Studies of syndromes that include CTDs, such as the 22q11 deletion syndrome, have provided some clues regarding the specific genes that may be involved in determining the risk of CTDs (e.g. TBX1 [13]). In addition, studies of rare, presumably pathogenic, copy number variants [14–16], and inherited [17] and de novo [17, 18] single nucleotide variants have identified genes that may contribute to the risk of CTDs [18, 19]. Yet, most affected patients do not carry a confirmed or suspected rare, causative variant. Moreover, rare variants, in particular rare de novo variants, do not account for the observed increase in risk of CTDs among the relatives of affected patients.
Since rare, pathogenic variants are unlikely to fully account for the population prevalence or familial recurrence of CTDs, additional genetic mechanisms must also contribute to disease risk. While the involvement of more common variants that have more moderate impacts on CTD risk seems likely, genome-wide association studies (GWAS) [20–23] of common single nucleotide polymorphisms (SNPs) have identified only two genome-wide significant associations for CTDs (rs11065987, p = 7.7E-11 and rs7982677, p = 3.03E-11) [22]. However, given the huge number of variants evaluated in GWAS, the threshold for statistical significance is quite stringent (i.e. p<5E-08). Consequently, the lack of significant findings for CTDs may well reflect low study power rather than the lack of common, CTD-related genetic variants.
Gene-based GWAS provide an additional strategy for identifying disease-related genes, but, to our knowledge, there are no published gene-based GWAS for CTDs. Compared to SNP-based GWAS, gene-based studies have the advantage of a less stringent threshold for statistical significance (e.g. Bonferroni corrected p-value for 20,000 genes, 2.5E-06). In addition, SNP-based analyses generally exclude rare variants, due to low statistical power [24], whereas gene-based analyses can incorporate data from both common and rare variants [25] and, therefore, capture more genomic variation than SNP-based analyses. Given these advantages, we have undertaken gene-based analyses and meta-analyses using data from several large CTD datasets.
Materials and methods
Study subjects
The Children's Hospital of Philadelphia (CHOP)
Informed consent was obtained under a protocol approved by the Institutional Review Board for the protection of human subjects at CHOP. Adult subjects (parents or guardians) provided written consent for themselves and their minor children. Patients diagnosed with CTDs and their available parents of all races and ethnicities were recruited through the Cardiac Center at CHOP from 1992–2010 [21].
Patients with the following diagnoses were included in the study: TOF, persistent truncus arteriosus, D-transposition of great arteries (TGA), double outlet right ventricle, ventricular septal defects (conoventricular, posterior malalignment and conoseptal hypoplasia types), aortic-pulmonary window, interrupted aortic arch and isolated aortic arch anomalies. Cardiac diagnoses were confirmed using medical and operative reports as well as imaging (e.g., echocardiography, cardiac magnetic resonance imaging, cardiac catheterization) records. All potential patients were tested for the 22q11.2 deletion syndrome using fluorescence in situ hybridization and/or multiplex ligation-dependent probe amplification using standard techniques. Patients with a confirmed 22q11.2 deletion were excluded [26]. Patients with a clinically diagnosed chromosomal abnormality, single gene mutation, teratogenic syndrome or known maternal risk factor (e.g. diabetes, anticonvulsant use) were also excluded [21].
The CHOP patients were previously microarray genotyped in two phases. In the first phase, cases with any CTD phenotype and of any race and ethnicity, and their parents were genotyped to generate data for a case-parent trio study. In the second phase, only non-Hispanic Caucasian cases (based on self- or parental-reported race/ethnicity) were genotyped to generate data for a case-control study. Control data were obtained from existing microarray genotyped data from pediatric controls that were recruited during well child visits at CHOP [27].
Pediatric Cardiac Genomics Consortium (PCGC)
Informed consent was obtained from each participating individual or their parent or guardian in accordance with protocols approved by the Institutional Review Board of each participating institution. Patients with a CHD and their available parents of all races and ethnicities were recruited as part of the PCGC Congenital Heart Defect GEnetic NEtwork Study from 2010–2012 [18, 28, 29]. PCGC recruitment took place at five main clinical sites (including CHOP) and four satellite clinics. The patients recruited by PCGC through CHOP do not overlap with the CHOP patients described above. Participant information was collected through medical records, electronic case reports, and personal interviews. Our studies were restricted to include patients with a CTD (as described above) and without a clinically diagnosed chromosomal or genetic disorder.
Genetic methods
Blood samples were collected from each patient and pediatric control. When blood collection was scheduled in conjunction with a surgical procedure, the sample was collected prior to any blood transfusion. Blood or saliva samples were collected from available parents of patients. DNA extraction was performed using standard techniques.
Genome-wide microarray genotyping was performed at the CHOP Center for Applied Genomics. Samples collected at CHOP were genotyped using Illumina HumanOmni-2.5 or Illumina HumanHap550 (v2, v3), or 610 BeadChip platforms. Samples collected as part of the PCGC (including the PCGC samples collected at CHOP) were genotyped on the Illumina HumanOmni-1 or HumanOmni-2.5 platforms. Additional details regarding the CHOP and PCGC samples are provided elsewhere [18, 21, 27, 28].
Imputation and quality control (QC) procedures
The microarray genotyped data from CHOP and PCGC were imputed using Impute2 v2.3.0 and pre-phased haplotype data obtained from the 1000 Genomes Project (Phase-I integrated v3 variants set) as the reference [30]. Due to differences in the genotyping platforms, the CHOP and PCGC cohorts were imputed separately.
Standard QC procedures were performed for each dataset using PLINKv1.07 before and after imputation [31]. Before imputation, the array data were checked for strand and coding errors. Trios were removed if more than 1% of genotyped SNPs had Mendelian errors. Suspected duplicate samples were identified using pairwise identify-by-descent estimation and samples with pi-hat greater than 0.6 were removed. Samples with genotyping rates less than 95% were also removed. In addition, variants with minor allele frequency (MAF) less than 1%, genotyping rates less than 90%, or deviation from Hardy Weinberg Equilibrium (HWE) in controls (p<1E-05) were excluded, as were all non-autosomal variants.
After the pre-imputation exclusions, the CHOP data from different platforms (HumanOmni-2.5, HumanHap550K v2, 550K v3 and 610K) were combined and only those variants present on all platforms (N = 283,977 SNPs) were used for imputation. Similarly, the PCGC data from different Illumina platforms (HumanOmni-1 and HumanOmni-2.5) were combined and only those SNPs present on both platforms (N = 624,419 SNPs) were used for imputation. For each dataset, haplotypes were pre-phased using SHAPEIT2 v2.727 [32] and imputation was performed using Impute2 v2.3.0 [30]. A genotype was imputed only if the posterior probability value exceeded 0.9, the default calling threshold for Impute2. After imputation, we excluded variants with poor imputation quality (Impute2 information metric score <0.8), or genotyping rates less than 90%. Samples with genotyping rates less than 95% and all insertions or deletions were removed. For all case-control comparisons, variants were evaluated for deviation from HWE in the pediatric control group using the exact test [33] implemented in PLINK, and variants with p<1E-05 were excluded. Because we were interested in assessing both rare and common variants, the post-imputation QC procedures did not include restrictions based on MAFs.
Statistical analysis
Genome-wide gene-based analyses were conducted, as described below. Because the various gene-based approaches have different underlying assumptions, strengths and limitations, we used two different gene-based approaches, eFBAT-MM and SKAT-C, to optimize the probability of identifying CTD-related genes. All analyses included all autosomal RefSeq genes, defined by the transcription start-stop coordinates (Genome Reference Consortium Human genome build 37 or hg19 reference assembly) in the RefSeq gene records and we included variants that were 1kb upstream or downstream of each gene.
Family-based analyses
Data for case-parent trios ascertained through CHOP (CHOP-Trios) and PCGC (PCGC-Trios) trios were analyzed separately using an extension of the family-based multi-marker association test (eFBAT-MM) [34]. This test (i.e. eFBAT-MM) is a burden-type approach that collapses variant-level statistics over a gene or region to obtain a single p-value and makes the assumption that all associated variants in the gene or genetic region affect the phenotype in the same direction. The variants were weighted by the inverse of the MAF estimated from the parental genotypes. Meta-analysis of the gene p-values from the CHOP and PCGC trios was performed using Fisher’s combination of probability method [35].
Case-control analyses
For the present study, we formed two independent, case-control (CC) datasets using the microarray genotyped and imputed data from CHOP. The first dataset included the Caucasian subset of patients from the CHOP trios and an equal number of Caucasian pediatric controls (CHOP-CC1). The second dataset included a second set of Caucasian patients with a CTD and an equal number of Caucasian pediatric controls (CHOP-CC2). There was no overlap in the cases or the controls included in CHOP-CC1 and CHOP-CC2.
The two CHOP case-control datasets (CHOP-CC1 and CHOP-CC2) were analyzed separately using the sequence kernel association test for the combined effect of common and rare variants (SKAT-C) [36]. Using this approach, separate scores were calculated for rare and common SNPs and these scores were combined as a weighted sum to calculate the gene p-value. The SKAT-C recommended default parameters were used for variant weighting and analysis. To control for population stratification bias, only non-Hispanic Caucasian cases (based on self- or parental report) were included in the analyses. Since race and ethnicity were based on self-report (rather than ancestry informative genetic markers), each analysis was also adjusted for the first genotypic principal component. Genotypic principal component analyses were conducted in Golden Helix SVS8.1, using the default parameter settings (MAF-based allele classification, additive genetic model and data for each marker were normalized by its theoretical standard deviation under HWE) (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com). Meta-analysis of the gene p-values from the two case-control series was performed using Fisher’s combination of probability method [35]. Meta-analyses combining results from the eFBAT-MM and SKAT-C analyses were not performed, given the overlap in patients (i.e. the non-Hispanic Caucasian cases in the CHOP-Trios are the case group for CHOP-CC1) and the differences in the assumptions underlying the two analytic approaches.
For each of the family-based and case-control analyses, the genomic inflation factor (λ) was calculated (for the case-control analyses, λwas calculated using values that adjusted for the first genotypic principal component) and a quantile-quantile (Q-Q) plot was constructed to check for deviation of the genome-wide observed distribution of the test statistic from the expected null distribution. Genes with association p-values less than the Bonferroni-corrected p-value (based on the number of genes in each analysis) were considered genome-wide significant. Genes with p-values greater than the Bonferroni-corrected p-values but less than 1E-03 were considered to be suggestive of an association.
Gene-set enrichment analysis
Genes with p<0.01 in the eFBAT-MM or SKAT-C meta-analyses were evaluated together for gene-annotation enrichment using MetaCoreTM (Thomson Reuters, Life Science Research, https://portal.genego.com/metacore). A false-discovery rate (FDR) corrected p-value less than 0.05 was used to identify significant pathway maps and Gene Ontology (GO) processes. REVIGO was used for clustering GO terms based on p-values and semantic similarity score (simRel) [37]. The simRel scores range from 0 to 1 and we used a score threshold of 0.4 for filtering GO terms.
Gene annotation and prioritization
To prioritize genes with at least suggestive evidence of association with CTD (p<1E-3, in either the family-based or case-control meta-analysis), for future investigations, we considered: (1) whether the meta-analysis p-value for the gene was lower than the p-values in contributing datasets i.e. the evidence for association was stronger in the combined data than in either of the individual datasets; and, (2) gene expression levels, based on heart expression data from E9.5 and E14.5 mouse embryos [18]. For each gene with a meta-analysis p-value lower than the p-values for the contributing datasets, we annotated the variants that were included in our analyses, for location, function, MAF in the genome aggregation database [38], Combined Annotation Dependent Depletion (CADD) phred-scaled scores [39], Genome-Wide Annotation of VAriants (GWAVA) [40] and Genomic Evolutionary Rate Profiling scores [41]. Genes with meta-analysis p-values lower than the p-values from the contributing datasets, and with heart expression data in the top quartile at E9.5 or E14.5 were considered strong candidates for future investigations.
Results
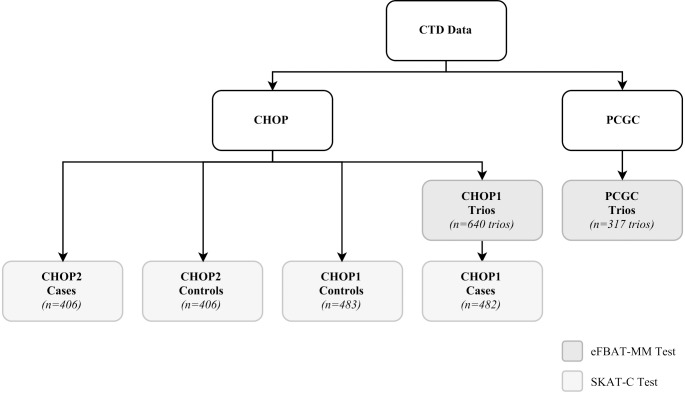
After QC exclusions, there were 640 CHOP trios and 317 PCGC trios for family-based analyses (Fig 1). In addition, there were 482 patients with CTD and 483 controls for CHOP-CC1, and 406 patients with CTD and 406 controls for CHOP-CC2. In both sets of trios, patients were predominantly Caucasian (Table 1). The two case-control datasets were restricted to Caucasians participants. In all groups, the most common heart defect was TOF.
Fig 1. Summary of conotruncal heart defects data cohorts.
The participants were recruited from the Children’s Hospital of Philadelphia (CHOP) and the Pediatric Cardiac Genomics Consortium (PCGC). CHOP-Trios and PCGC-Trios were analyzed using eFBAT-MM whereas SKAT-C was used to analyze the two case-control cohorts (CHOP-CC1 and CHOP-CC2). The cases in CHOP-CC1 are the Caucasian subset of cases in CHOP-Trios. One of 483 Caucasian cases was excluded during QC procedures prior to SKAT-C analysis.
Table 1. Characteristics of patients with conotruncal defects in the Children’s Hospital of Philadelpia (CHOP) and Pediatric Cardiac Genomics Consortium (PCGC) datasets.
| N (%) | ||||||
|---|---|---|---|---|---|---|
| CHOP-Trios / CHOP-CC1a (n = 640) |
CHOP-CC2 (n = 406) |
PCGC-Trios (n = 317) |
||||
| Race/ethnicity | ||||||
| Non-Hispanic Caucasian | 483 | (75.5) | 406 | (100.0) | 244 | (70.1) |
| Other | 157 | (24.5) | 0 | (0.0) | 73 | (29.9) |
| Sex | ||||||
| Male | 387 | (60.5) | 236 | (58.1) | 192 | (60.6) |
| Female | 253 | (39.5) | 170 | (41.9) | 125 | (39.4) |
| Conotruncal defect phenotype | ||||||
| Tetralogy of Fallot | 250 | (39.1) | 134 | (33.0) | 104 | (32.8) |
| D-transposition of the great arteries | 125 | (19.5) | 80 | (19.7) | 64 | (20.2) |
| Ventricular septal defects | 133 | (20.8) | 109 | (26.8) | 44 | (13.9) |
| Double outlet right ventricle | 66 | (10.3) | 25 | (6.2) | 46 | (14.5) |
| Isolated aortic arch anomalies | 30 | (4.7) | 22 | (5.4) | 7 | (2.2) |
| Persistent truncus arteriosus | 18 | (2.8) | 19 | (4.7) | 13 | (4.1) |
| Interrupted aortic arch | 11 | (1.7) | 10 | (2.5) | 9 | (2.8) |
| Other | 7 | (1.1) | 7 | (1.7) | 30 | (9.5) |
aThe cases used in CHOP-CC1 are the subset of the cases included in the CHOP-Trios (i.e. the non-Hispanic Caucasian cases, N = 483).
Gene-based GWAS of individual datasets
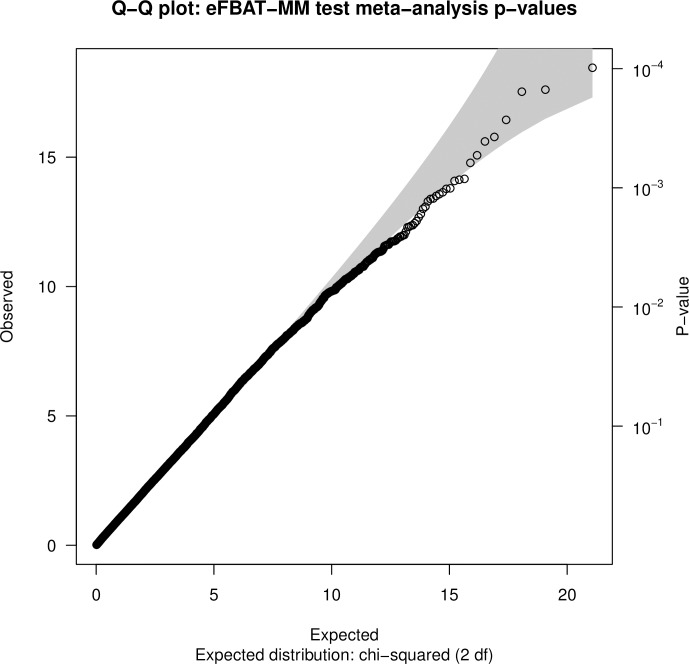
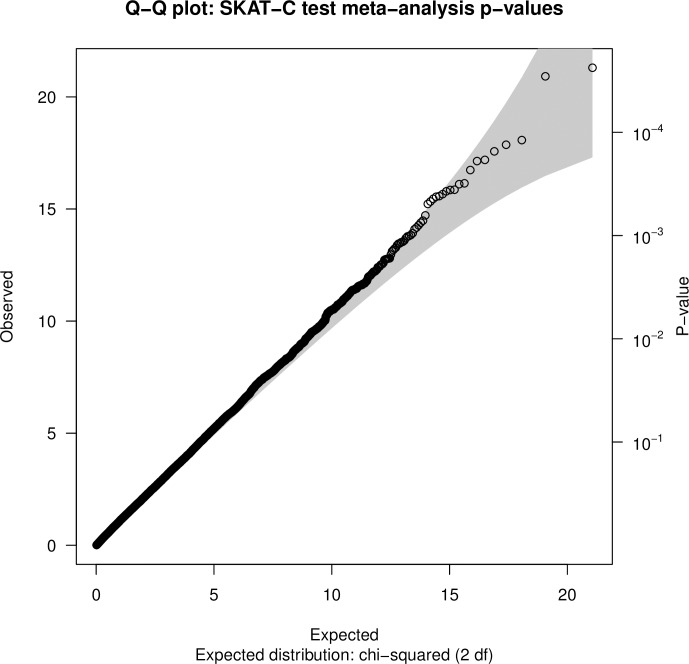
The number of variants and genes included in each analysis are summarized in Table 2. The genotype concordance for the imputation was >90%. The Q-Q plots (S1–S4 Figs) and genomic inflation factors (Table 2) provided little evidence for systematic bias in the observed p-values. No genome-wide significant associations were identified in the analyses of the individual datasets. The number of genes with suggestive evidence of association (p<1E-03) ranged from 13 to 27 (Table 2). There was no overlap across datasets or analyses with respect to the genes with suggestive evidence of association. Detailed genome-wide results for each analysis are included in Tables B-E in S1 File.
Table 2. Summary of eFBAT-MM and SKAT-C analyses and results.
| eFBAT-MM | SKAT-C | |||
|---|---|---|---|---|
| CHOP-Trios | PCGC Trios | CHOP-CC1 | CHOP-CC2 | |
| Total variants | 5,578,860 | 6,812,971 | 5,601,587 | 5,601,152 |
| Rare variantsa | 3,446,735 | 4,502,285 | 3,502,419 | 3,495,988 |
| Number of genes | 21,256 | 22,247 | 21,212 | 21,269 |
| Genomic inflation factor (λ) | 1.03 | 1.04 | 1.09 | 1.09 |
| Genes with p<1E-03b | 13 | 13 | 25 | 27 |
|
CBLN2 C22orf39 NCOA2 CEP95 DDX5 SQRDL SLMO2-ATP5E TRIP13 PADI3 RBM47 PRR14 ZC3H18 CREBZF |
MIR518C GOLGA2P9 POU6F2 LCE4A TMEM206 LOC100996349 ASAH2 MBNL1 IGFBPL1 RNF44 IRAK2 DDX59 ADGRA3 |
FAM225A ABCB4 STK33 BLOC1S6 MIR3916 LEXM DACT3 MIR99AHG TRIP10 LINC00620 DACT3-AS1 NPPC GPATCH1 AVPR1A FLNC NCAPG SHD ARAP3 FIGN NCAN SYMPK MIR548AA2 MIR548D2 BHMG1 FCGR3B |
PSMG3 NEXN FUBP1 PSMG3-AS1 DNAJB4 TSSC4 TSPAN10 MKX-AS1 MKX UHMK1 ATG9B MMP19 CRTAM NPLOC4 GRID2 COPZ2 PTPRT PYGL COA5 EEF1B2 APOPT1 GATC ACTL7B NDUFS1 LOC101927653 AACS EPHB4 |
|
a Variants with minor allele frequency <0.05
b Genes are listed by p-value (lowest to highest). Specific p-values are provided in Tables B-E in S1 File.
Meta-analysis
Separate meta-analyses were conducted using eFBAT-MM p-values for 21,170 genes that were analyzed in both the CHOP and PCGC trios, and from SKAT-C p-values for 21,077 genes that were analyzed in both the CHOP-CC1 and CHOP-CC2 case-control studies. The Q-Q plots (Figs 2 and 3) and genomic inflation factors provided little evidence for systematic bias in the observed p-values. No gene achieved genome-wide significance in either meta-analysis (Table F in S1 File provides p-values for all genes assessed in each meta-analysis). Suggestive evidence of association (p<1E-03) was obtained for 11 genes (8 protein coding, 2 pseudogenes, 1 RNA gene) in the trio-based meta-analysis (Table 3) and for 27 genes (23 protein coding, 4 RNA genes) in the case-control based meta-analysis (Table 4).
Fig 2. Quantile-quantile plot of eFBAT-MM test gene-level meta-analysis p-values.
Fig 3. Quantile-quantile plot of SKAT-C test gene-level meta-analysis p-values.
Table 3. Genes with suggestive evidence of association (p<1E-03) in the trio-based meta-analysis.
| CHOP-Trios (640 trios) |
PCGC-Trios (317 trios) |
Meta-analysis | ||||
|---|---|---|---|---|---|---|
| Gene | Function | Number of variants | p-valuea | Number of variants | p-valuea | p-value |
| POU6F2 | Protein coding | 2,662 | 8.9E-02 | 2,738 | 8.7E-05 | 9.8E-05 |
| MBNL1 | Protein coding | 534 | 3.3E-02 | 789 | 3.7E-04 | 1.5E-04 |
| SIGLEC11 | Protein coding | 37 | 4.1E-03 | 53 | 3.1E-03 | 1.6E-04 |
| GOLGA2P9 | Pseudogene | 38 | 9.8E-01 | 83 | 2.3E-05 | 2.7E-04 |
| MAP4K3 | Protein coding | 1,020 | 1.6E-03 | 1,063 | 2.1E-02 | 3.7E-04 |
| CBLN2 | Protein coding | 39 | 2.4E-04 | 52 | 1.5E-01 | 4.1E-04 |
| ROR1 | Protein coding | 1,585 | 2.4E-02 | 1,954 | 2.0E-03 | 5.3E-04 |
| LINC00207 | RNA gene | 26 | 2.0E-02 | 25 | 2.9E-03 | 6.2E-04 |
| KPNA1 | Protein coding | 317 | 5.4E-02 | 352 | 1.5E-03 | 8.4E-04 |
| GLIPR1 | Protein coding | 81 | 2.9E-02 | 104 | 2.9E-03 | 8.6E-04 |
| LOC100996349 | Pseudogene | 7 | 4.2E-01 | 15 | 2.0E-04 | 8.8E-04 |
a gene-level p-values from eFBAT-MM test
Table 4. Genes with suggestive evidence of association (p<1E-03) in the case-control based meta-analysis.
| CHOP-CC1 (482 CTD patients/483 controls) |
CHOP-CC2 (406 CTD patients/ 406 controls) |
Meta-analysis | ||||
|---|---|---|---|---|---|---|
| Gene | Function | Number of variants | p-valuea | Number of variants | p-valuea | p-value |
| PSMG3 | Protein coding | 41 | 6.5E-02 | 40 | 2.6E-05 | 2.4E-05 |
| FAM225A | RNA gene | 6 | 3.9E-05 | 7 | 5.3E-02 | 2.9E-05 |
| ABCB4 | Protein coding | 252 | 9.9E-05 | 272 | 9.5E-02 | 1.2E-04 |
| LEXM | Protein coding | 208 | 2.5E-04 | 185 | 4.2E-02 | 1.3E-04 |
| SHD | Protein coding | 28 | 6.9E-04 | 31 | 1.8E-02 | 1.5E-04 |
| NEXN | Protein coding | 150 | 5.1 E-01 | 162 | 3.0E-05 | 1.9E-04 |
| LOC100287036 | Protein coding | 12 | 6.3E-03 | 15 | 2.5E-03 | 1.9E-04 |
| PSMG3-AS1 | RNA gene | 29 | 1.8 E-01 | 28 | 1.1E-04 | 2.3E-04 |
| PRKD2 | Protein coding | 59 | 1.2E-03 | 67 | 2.3E-02 | 3.1E-04 |
| DACT3 | Protein coding | 16 | 3.1E-04 | 14 | 9.1E-02 | 3.2E-04 |
| EIF4E | Protein coding | 237 | 2.6E-02 | 271 | 1.2E-03 | 3.6E-04 |
| MKX-AS1 | RNA gene | 66 | 1.8 E-01 | 80 | 1.8E-04 | 3.6E-04 |
| MKX | Protein coding | 323 | 1.5 E-01 | 406 | 2.2E-04 | 3.7E-04 |
| RGS16 | Protein coding | 11 | 5.3E-03 | 11 | 6.7E-03 | 4.0E-04 |
| CHODL | Protein coding | 1,101 | 2.8E-02 | 1,396 | 1.3E-03 | 4.2E-04 |
| NCAPG | Protein coding | 76 | 6.8E-04 | 84 | 5.5E-02 | 4.2E-04 |
| DNAJB4 | Protein coding | 43 | 3.3 E-01 | 44 | 1.2E-04 | 4.4E-04 |
| FUBP1 | Protein coding | 79 | 8.1 E-01 | 91 | 5.2E-05 | 4.7E-04 |
| DACT3-AS1 | RNA gene | 14 | 4.7E-04 | 12 | 9.5E-02 | 4.9E-04 |
| STK33 | Protein coding | 769 | 1.1E-04 | 784 | 5.7 E-01 | 6.4E-04 |
| DCAF16 | Protein coding | 22 | 1.7E-03 | 28 | 4.0E-02 | 7.2E-04 |
| ACTL7B | Protein coding | 21 | 9.5E-02 | 20 | 7.5E-04 | 7.5E-04 |
| NDUFS1 | Protein coding | 181 | 8.6E-02 | 192 | 8.8E-04 | 8.0E-04 |
| FBXO47 | Protein coding | 66 | 2.2E-03 | 52 | 3.7E-02 | 8.4E-04 |
| ARF5 | Protein coding | 25 | 3.8E-03 | 24 | 2.2E-02 | 8.7E-04 |
| PYGL | Protein coding | 114 | 1.7 E-01 | 112 | 5.4E-04 | 9.5E-04 |
| SYMPK | Protein coding | 234 | 7.8E-04 | 251 | 1.2 E-01 | 9.9E-04 |
a gene-level p-values from SKAT-C
We identified genes that have previously been implicated in heart development and structural heart malformations (e.g. MBNL1, ROR1), and known disease-related genes (e.g. NEXN, dilated cardiomyopathy; NDUFS1, mitochondrial complex I deficiency). Several of the identified genes are also annotated to biological processes that are important during embryonic heart development including transcription (FUBP1, POU6F2, MKX), protein phosphorylation (MAP4K3, PRKD2, ROR1, STK33), positive regulation of the ERK1 and ERK2 cascade (ROR1, PRKD2), Wnt signaling (ROR1, DACT3), and cell adhesion (PRKD2, SYMPK).
Gene-set enrichment analysis
MetaCoreTM gene-set enrichment analysis was performed using genes with p<0.01 in the trio (195 genes) or case-control (246 genes) based meta-analyses (Table F in S1 File). We identified two significantly enriched pathways (FDR-corrected p<0.05): dynein-dynactin motor complex in axonal transport in neurons (FDR-corrected p = 0.02), and chromosome condensation in prometaphase (FDR-corrected p = 0.02) (Table G in S1 File). In addition, we identified 111 significantly enriched (FDR-corrected p<0.05) non-redundant (REVIGO-clustered) GO processes (Table H in S1 File). GO term clusters included processes relevant to heart defects including cellular response to hormone stimulus, angiogenesis and biological adhesion.
Gene annotations and prioritization
Of the 38 genes with suggestive evidence of association (p<1E-3) in either the family-based or case-control meta-analysis, 19 protein-coding and 2 RNA genes had a lower meta-analysis p-value than the p-values in contributing datasets i.e. the evidence for association was stronger in the combined data than in either individual dataset (Table 5). For the 21 genes with meta-analysis p-value less than the individual study p-values, the majority (95%) of variants included in the analyses were intronic (Table I in S1 File). Heart expression data from E9.5 and E14.5 mouse embryos [18] were available for 15 of the 19 protein-coding genes, of which eight (53%) were in the top quartile of expression at one or both time points (Table 5). We propose these eight genes (ARF5, EIF4E, KPNA1, MAP4K3, MBNL1, NCAPG, NDFUS1, PSMG3) as CTD candidate genes.
Table 5. Genes with suggestive evidence of association (p<1E-03) in either dataset and with a meta-analysis p-value that is lower than that obtained in either of the contributing analyses.
| Gene | Gene Name | Function | Gene-based Test | Dataset 1 p-valuea | Dataset 2 p-valuea | Meta-analysis p-value | Day 9.5b |
Day 14.5b |
|---|---|---|---|---|---|---|---|---|
| ARF5 | ADP-ribosylation factor 5 | Protein-coding | SKAT-C | 3.84E-03 | 2.17E-02 | 8.67E-04 | 95.3 | 88.8 |
| CHODL | Chondrolectin | Protein-coding | SKAT-C | 2.80E-02 | 1.32E-03 | 4.15E-04 | 5.8 | 10.4 |
| DCAF16 | DDB1 and CUL4 associated factor 16 | Protein-coding | SKAT-C | 1.70E-03 | 3.98E-02 | 7.18E-04 | No data | No data |
| EIF4E | Eukaryotic translation initiation factor 4E | Protein-coding | SKAT-C | 2.55E-02 | 1.24E-03 | 3.60E-04 | 81.7 | 76.4 |
| FAM225A | Family with sequence similarity 225 member 1 | RNA gene | SKAT-C | 3.86E-05 | 5.28E-02 | 2.87E-05 | -- | -- |
| FBXO47 | F-box only protein 47 | Protein-coding | SKAT-C | 2.17E-03 | 3.69E-02 | 8.36E-04 | 43.8 | 28.3 |
| GLIPR1 | Glioma pathogenesis-related protein 1 | Protein-coding | eFBAT-MM | 2.85E-02 | 2.88E-03 | 8.55E-04 | 57.6 | 34.9 |
| KPNA1 | Karyopherin alpha-1 | Protein-coding | eFBAT-MM | 5.37E-02 | 1.50E-03 | 8.41E-04 | 82.4 | 77.8 |
| LEXM (C1orf177) | Chromosome 1 open reading frame 177 | Protein-coding | SKAT-C | 2.52E-04 | 4.19E-02 | 1.32E-04 | 41.4 | 16.1 |
| LINC00207 | Long intergenic non-protein coding RNA 207 | RNA gene | eFBAT-MM | 2.01E-02 | 2.86E-03 | 6.17E-04 | -- | -- |
| LOC100287036 | Uncharacterized LOC100287036 | Protein-coding | SKAT-C | 6.34E-03 | 2.49E-03 | 1.90E-04 | No Data |
No data |
| MAP4K3 | Mitogen-activated protein kinase kinase kinase 3 | Protein-coding | eFBAT-MM | 1.56E-03 | 2.11E-02 | 3.74E-04 | 76.1 | 76.9 |
| MBNL1 | Muscleblind-like splicing regulator 1 | Protein-coding | eFBAT-MM | 3.29E-02 | 3.70E-04 | 1.50E-04 | 66.9 | 78.2 |
| NCAPG | Non-SMC condensin 1 complex subunit G | Protein-coding | SKAT-C | 6.79E-04 | 5.54E-02 | 4.21E-04 | 83.5 | 70.5 |
| NDUFS1 | NADH-ubiquinone oxidoreductase Fe-S protein 1 | Protein-coding | SKAT-C | 8.63E-02 | 8.79E-04 | 7.95E-04 | 94 | 97.1 |
| PRKD2 | Protein kinase D2 | Protein-coding | SKAT-C | 1.17E-03 | 2.31E-02 | 3.12E-04 | 48.1 | 66.9 |
| PSMG3 | Proteasome assembly chaperone 3 | Protein-coding | SKAT-C | 6.50E-02 | 2.55E-05 | 2.37E-05 | 79 | 54.1 |
| RGS16 | Regulator of G protein signaling | Protein-coding | SKAT-C | 5.31E-03 | 6.65E-03 | 3.97E-04 | 56.4 | 38.9 |
| ROR1 | Receeptor tyrosine kinase-like orphan receptor 1 | Protein-coding | eFBAT-MM | 2.42E-02 | 2.02E-03 | 5.33E-04 | 65.6 | 63.5 |
| SHD | SH2 domain-containing protein D | Protein-coding | SKAT-C | 6.86E-04 | 1.82E-02 | 1.53E-04 | 65.5 | 36.2 |
| SIGLEC11 | Sialic acid-binding immunoglobulin-like lectin 11 | Protein-coding | eFBAT-MM | 4.09E-03 | 3.10E-03 | 1.56E-04 | 36.2 | No data |
a When meta-analysis with p<1E-03 is FBAT, dataset 1 = CHOP Trios and dataset 2 is PCGC Trios. When meta-analysis with p<1E-03 is SKAT, dataset 1 = CHOP-CC1 patients with a CTD and controls and dataset 2 is CHOP-CC2 patients with a CTD and controls.
b Heart expression percentile rank [18].
Discussion
Our comprehensive genome-wide, gene-based analysis of common and rare variants identified 38 genes with suggestive evidence of association (meta-p<1E-3) with CTDs, as well as relevant biological pathways and processes that were significantly enriched (FDR-corrected p<0.05) among the genes with the most significant p-values in gene-based analyses. Based on both statistical evidence (i.e. the evidence for association was stronger in the meta-analysis than in any of the contributing studies) and gene expression data (top quartile of expression in mouse heart at E9.5 or E14.5) we propose eight genes (ARF5, EIF4E, KPNA1, MAP4K3, MBNL1, NCAPG, NDFUS1, PSMG3) as CTD candidate genes.
Four of the CTD candidate genes suggested by our work have not been associated with normal or abnormal heart development. These four genes are: ADP ribosylation factor 5 (ARF5), which encodes a GTP-binding protein involved in protein trafficking; karyopherin subunit alpha 1 (KPNA1), which functions in nuclear protein import; NADH:Ubiquinone oxidoreductase core subunit protein coding S1 (NDUFS1), which encodes the core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase, and; proteasome assembly chaperone 3 (PSMG3), which encodes a chaperone protein.
The known function of the remaining four candidate genes suggests that their altered expression could cause CHDs. Of these genes, the most significant association was observed for muscleblind-like splicing regulator 1 (MBNL1, eFBAT-MM meta-p = 1.5E-04). This gene encodes a CH3-type zinc finger protein (MBNL1) that is a key regulator of pre-RNA alternative splicing. Evidence that splicing regulators contribute to the etiology of CHDs is provided by the identification of a genome-wide, significant excess of damaging de novo and loss-of-function heterozygous mutations in another key splicing regulator, RBFOX2, in patients with CHDs [17].
Several additional lines of evidence also support a role for MBNL1 in cell differentiation and heart development. For example, MBNL1 and RBFOX2 appear to co-regulate the splicing changes that lead to the differentiation of pluripotent stem cells [42]. In addition, in the nucleotide repeat expansion disorder, myotonic dystrophy, reduced MBNL1 splicing activity (due to binding of MBNL1 protein to the expansion RNA) is thought to play a major role in determining the disease phenotype, which includes several cardiovascular abnormalities (conduction defects, arrhythmias, mitral valve prolapse) [43, 44]. There is also evidence that MBNL1 is involved in the fetal to adult transition in alternative splicing patterns in the heart [45], and that MBNL1 negatively regulates TGF-β signaling and the epithelial-mesenchymal transition in the endocardial cushions by restricting the timing and amount of TGF- β production in the atrioventricular canal and outflow tract endocardium [46, 47]. Mice null for MBNL1 protein present with abnormal heart valve development, regurgitation across both the in- and outflow valves, and ostium secundum septal defects [47].
Further evidence that genes involved in RNA splicing may be associated with CTDs is provided by our gene-set enrichment analyses. Specifically, genes mapping to the GO process ‘regulation of RNA splicing’ (GO:0043484) were significantly enriched (FDR-adjusted p = 0.03) among genes with association p<0.01 in our meta-analyses. In addition to MBNL1, seven genes mapping to this process (CLK3, DDX5, JMJD6, SRSF2, SRSF9 and TMBIM6) had meta-analysis p<0.01 (meta-analysis p-value range: 2E-03 to 8E-03) in either the family-based (i.e. eFBAT-MM) or case-control (i.e. SKAT-C) meta-analysis.
The second most significant of our proposed CTD candidate genes was eukaryotic translation initiation factor 4D (EIF4E, SKAT meta-p = 3.6E-04). The encoded protein, eIF4F, directs ribosomes to the mRNA 5’-cap and is a key factor in initiation of translation of many mRNAs [48]. Zhang et al. have presented evidence that eIF4E is involved in heart development via the p53-Rbm24 loop [49]. Specifically, they demonstrated that the multifunctional RNA-binding protein, Rbm24, prevents binding of eIF4E to p53 RNA, thereby repressing p53 translation and p53-dependent apoptosis. Further, they showed that mice deficient for Rbm24 develop endocardial cushion defects as a result of aberrant binding of eIF4E to p53 RNA resulting in overexpression of p53. Mutations in EIF4E have also been implicated as a cause of autism in humans [50], and enhanced eIF4E activity has been associated with autism-like phenotypes in animal models [51]. Hence, our finding adds EIF4E to the growing list of genes that may be related to both CHDs and neurodevelopment disabilities such as autism [17, 52].
Our study also identified the mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3) as a CTD candidate gene. The product of this gene is an upstream activator of the c-Jun-N-terminal kinase (JNK) signal transduction pathway, which is involved in several processes relevant to heart development (e.g. cell growth, differentiation and survival, apoptosis) [53]. Downstream effectors of JNK signaling relevant to heart development include the tumor suppressor/apoptosis gene, p53 (discussed above), and SMAD4. In a mouse model, disruption of Smad4 in neural crest cells resulted in multiple malformations including defects of the outflow tracts and ventricles [54]. Further, in humans, SMAD4 gain of function mutations cause Myhre syndrome, which includes CHD as a common (~2/3rds of patients) phenotypic finding [55]. There is also evidence that MAP4K3 is a central regulator of autophagy, a process that is critical for maintaining the supply of free amino acids for protein synthesis [56] that is required for embryonic growth and development.
Finally, our analyses identified non-SMC condensing I complex subunit G (NCAPG, SKAT meta-p = 4.2E-04) as a CTD candidate gene. The protein encoded by this gene forms part of the condensin complex, which is involved in mitotic chromatin condensation [57]. Further, our analyses indicated that genes in the MetaCore pathway map, “Chromosome condensation in prometaphase) were also significantly enriched (FDR-adjusted p = 0.02) among genes with association p<0.01 in our meta-analyses. In addition to NCAPG, a second member of the condensing complex, NCAPH (FBAT meta-p = 9.13E-03), and BAZ1B (FBAT meta-p = 5.62E-03), which is part of the WICH chromatin remodeling complex, had meta-analysis p<0.01. The involvement of chromatin-related genes, particularly H3K4me-H3K27me pathway genes, in CHD etiology has been suggested by studies of de novo mutations [18]. Our findings suggest that other classes of chromatin-modifiers may also contribute to CHDs.
We have previously conducted SNP-level (MAF≥0.05) GWAS using the same datasets as in the current gene-based analyses [20]. In our meta-analysis of the SNP-level results, we identified 36 variants with suggestive evidence of association (P≤1E-5). However, no association was genome-wide significant (P<5E-8). Further, none of the SNPs with suggestive evidence of association were located in, or within 1kb up or downstream of, the genes with suggestive evidence of association (Table 5) in the current, gene-based analyses. However, it should be noted that the SNP-level analyses were restricted to include only common variants and used slightly different configurations of the data. Specifically, in the SNP-level analyses we compared the cases used in CHOP-CC2 to all available CHOP pediatric controls (N = 2,976 controls), and the SNP-level meta-analysis was based on the combined results from CHOP-Trios, PCGC-Trios and the case-control analyses.
To our knowledge, this is the first gene-based genome-wide analysis of CTDs that is based on data for both common and rare variants. Because the various gene-based approaches have different underlying assumptions, strengths and limitations, we used two different gene-based approaches, eFBAT-MM and SKAT-C, to optimize the probability of identifying CTD-related genes. The family-based approach, eFBAT-MM, is robust to population stratification bias, but assumes that all variants within a gene have effects in the same direction and that the effect size is inversely proportional to the MAF. In contrast, the case-control approach, SKAT-C, is subject to stratification bias, but does not make assumptions about the direction of association. Therefore, SKAT-C is more powerful than eFBAT-MM when a large proportion of protective and neutral variants are present in a gene, and the converse is true when this proportion is small. Given the differences between the two methods, the lack of overlap in the genes identified by the two approaches is not particularly surprising.
Although the gene-based approaches used in our analyses had a lower multiple-testing burden than SNP-based GWAS, the criterion for achieving statistical significance (corrected p~2.5E-06) remained quite stringent. This, in combination with our relatively small sample sizes, suggests that associations with true CTD-related genes may have been missed in our analyses due to low study power. Further, the Q-Q plots for the eFBAT-MM analysis of individual datasets (Fig 2) indicate that this test may be too conservative, which would have also negatively impacted our power to detect a true association. Given these considerations, genes with suggestive evidence of association (meta-p<1E-03) and pathways and processes with FDR p<0.05 appear to be strong targets for further investigations of the genetic basis of CTDs.
In our analyses, we combined data across different CTD phenotypes, which could have obscured associations if the etiology of the individual phenotypes is distinct. For example, mutations in laterality genes (e.g. CFC1, FOXH1) have been observed in association with TGA [58, 59], suggesting that at least some cases of TGA might be more appropriately classified as laterality defects rather than CTDs. However, the preponderance of evidence suggests that the various CTD phenotypes share common genetic underpinnings. Studies of familial recurrence patterns, phenotypes of patients with known genetic syndromes (e.g. 22q11.2 deletion syndrome) and studies in animal models all indicate that the various CTD phenotypes share genetic risk factors. Moreover, studies of rare de novo and inherited variants in humans provide evidence that the genes involved in CHDs may be shared across even broader categories of defects. For example, Jin et al. reported genome-wide significant excess of damaging de novo and loss-of-function heterozygous mutations in seven genes among 2,871 patients with CHD. Of these seven genes, mutations in six were observed across broad CHD categories (i.e. CTDs, left-sided lesions and/or other CHDs) [17]. Hence, while studies of CTDs as a group might miss phenotype-specific associations, such studies appear to be appropriate for genes that contribute broadly to CHD risk and for genes that influence the spectrum of CTDs.
In summary, our genome-wide, gene-based analyses of common and rare variants identified enriched pathways and biological processes and candidate genes for CTDs. Our findings provide evidence for new CTD-related candidate genes, as well as support and expand on prior evidence implicating chromatin-related genes and splicing-regulators as determinants of CHD risk.
Conclusions
To our knowledge, this is the first study reporting the results of gene-based, genome-wide association studies for CTDs. The results of our study provide evidence for eight CTD candidate genes, of which four have previously been implicated in heart development and four are novel candidates. Thus, these findings add to our understanding of the complex, genetic etiology of CTDs, which may, in turn, enhance our ability to understand, predict and ultimately improve clinical outcomes for this patient population.
Supporting information
(XLSX)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
The Pediatric Cardiac Genomics Consortium author group consists of: Richard Kim from Children’s Hospital of Los Angeles, Los Angeles, California; Deepak Srivastava and Daniel Bernstein from J. David Gladstone Institutes, San Francisco, California; Daniel Bernstein from Stanford University, Stanford, California; Martina Brueckner and Richard Lifton from Yale School of Medicine, New Haven, Connecticut; Jane Newburger and Amy Roberts from Boston Children’s Hospital, Boston, Massachusetts; Christine Seidman and Jonathan Seidman from Harvard Medical School, Boston, Massachusetts; Sharon Tennstedt, Kimberly Dandreo and Julie Miller from New England Research Institutes, Watertown, Massachusetts; Angela Romano-Adesman from Steve and Alexandra Cohen Children's Medical Center, Hewlett, New York; Bruce Gelb from Icahn School of Medicine at Mount Sinai, New York, New York; Wendy Chung from Columbia University Medical School, New York, New York; George Porter from University of Rochester School of Medicine and Dentistry, Rochester, New York; Eileen C. King from Cincinnati Children’s Hospital, Cincinnati, Ohio; Elizabeth Goldmuntz from Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania; and Martin Tristani-Firouzi and H. Joseph Yost from University of Utah, Salt Lake City, Utah.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development [P01-HD070454 (EG)]; the National Heart, Lung, and Blood Institute [P50-HL74731 (EG)], including the Pediatric Cardiac Genomics Consortium (PCGC) [U01-HL098188 (Sharon Tennstedt, Kimberly Dandreo and Julie Miller from New England Research Institutes), U01-HL131003 (Eileen C. King from Cincinnati Children’s Hospital), U01-HL098147 (Jane Newburger and Amy Roberts from Boston Children’s Hospital), U01-HL098153 (EG), U01-HL098163 (Wendy Chung from Columbia University), U01-HL098123 (Bruce Gelb from Icahn School of Medicine at Mount Sinai), U01-HL098162 (Martina Breuckner, Richard Lifton from Yale University)]; the Cardiovascular Development Consortium [U01-HL098166 (Jonathan Seidman from Harvard Medical School)]; the National Human Genome Research Institute [U54-HG006504 (Richard Lifton from Yale University)]; and the National Center for Research Resources [M01-RR-000240 (EG), RR024134 (EG)], which is now the National Center for Advancing Translational Sciences [UL1-TR000003 (EG)]. Genome-wide microarray genotyping of The Children’s Hospital of Philadelphia (CHOP) cohorts was funded by an Institutional Development Fund to The Center for Applied Genomics from CHOP. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39(12):1890–900. 10.1016/s0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, Chen H, Correa A, Devine O, Mathews TJ, Honein MA. Racial differences in infant mortality attributable to birth defects in the United States, 1989–2002. Birth defects research Part A, Clinical and molecular teratology. 2006;76(10):706–13. 10.1002/bdra.20308 [DOI] [PubMed] [Google Scholar]
- 3.Arth AC, Tinker SC, Simeone RM, Ailes EC, Cragan JD, Grosse SD. Inpatient Hospitalization Costs Associated with Birth Defects Among Persons of All Ages—United States, 2013. MMWR Morbidity and mortality weekly report. 2017;66(2):41–6. 10.15585/mmwr.mm6602a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilboa SM, Devine OJ, Kucik JE, Oster ME, Riehle-Colarusso T, Nembhard WN, et al. Congenital Heart Defects in the United States: Estimating the Magnitude of the Affected Population in 2010. Circulation. 2016;134(2):101–9. 10.1161/CIRCULATIONAHA.115.019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khokha MK, Mitchell LE, Wallingford JB. An opportunity to address the genetic causes of birth defects. Pediatric research. 2017;81(2):282–5. 10.1038/pr.2016.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson TR. Conotruncal cardiac defects: a clinical imaging perspective. Pediatric cardiology. 2010;31(3):430–7. 10.1007/s00246-010-9668-y [DOI] [PubMed] [Google Scholar]
- 7.Kloesel B, DiNardo JA, Body SC. Cardiac Embryology and Molecular Mechanisms of Congenital Heart Disease: A Primer for Anesthesiologists. Anesth Analg. 2016;123(3):551–69. 10.1213/ANE.0000000000001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilboa SM, Salemi JL, Nembhard WN, Fixler DE, Correa A. Mortality resulting from congenital heart disease among children and adults in the United States, 1999 to 2006. Circulation. 2010;122(22):2254–63. 10.1161/CIRCULATIONAHA.110.947002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodwall K, Greve G, Leirgul E, Tell GS, Vollset SE, Oyen N. Recurrence of congenital heart defects among siblings-a nationwide study. American journal of medical genetics Part A. 2017;173(6):1575–85. 10.1002/ajmg.a.38237 [DOI] [PubMed] [Google Scholar]
- 10.Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120(4):295–301. 10.1161/CIRCULATIONAHA.109.857987 [DOI] [PubMed] [Google Scholar]
- 11.Peyvandi S, Ingall E, Woyciechowski S, Garbarini J, Mitchell LE, Goldmuntz E. Risk of congenital heart disease in relatives of probands with conotruncal cardiac defects: an evaluation of 1,620 families. American journal of medical genetics Part A. 2014;164A(6):1490–5. 10.1002/ajmg.a.36500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unolt M, Versacci P, Anaclerio S, Lambiase C, Calcagni G, Trezzi M, et al. Congenital heart diseases and cardiovascular abnormalities in 22q11.2 deletion syndrome: From well-established knowledge to new frontiers. American journal of medical genetics Part A. 2018;176(10):2087–98. 10.1002/ajmg.a.38662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow BE, McDonald-McGinn DM, Emanuel BS, Vermeesch JR, Scambler PJ. Molecular genetics of 22q11.2 deletion syndrome. American journal of medical genetics Part A. 2018;176(10):2070–81. 10.1002/ajmg.a.40504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak CCY, Chow PC, Liu APY, Chan KYK, Chu YWY, Mok GTK, et al. De novo large rare copy-number variations contribute to conotruncal heart disease in Chinese patients. NPJ Genom Med. 2016;1:16033 10.1038/npjgenmed.2016.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie HM, Werner P, Stambolian D, Bailey-Wilson JE, Hakonarson H, White PS, et al. Rare copy number variants in patients with congenital conotruncal heart defects. Birth Defects Res. 2017;109(4):271–95. 10.1002/bdra.23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nature genetics. 2009;41(8):931–5. 10.1038/ng.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature genetics. 2017;49(11):1593–601. 10.1038/ng.3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–3. 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nature genetics. 2016;48(9):1060–5. 10.1038/ng.3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agopian AJ, Goldmuntz E, Hakonarson H, Sewda A, Taylor D, Mitchell LE, et al. Genome-Wide Association Studies and Meta-Analyses for Congenital Heart Defects. Circulation Cardiovascular genetics. 2017;10(3):e001449 10.1161/CIRCGENETICS.116.001449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agopian AJ, Mitchell LE, Glessner J, Bhalla AD, Sewda A, Hakonarson H, et al. Genome-wide association study of maternal and inherited loci for conotruncal heart defects. PloS one. 2014;9(5):e96057 10.1371/journal.pone.0096057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordell HJ, Topf A, Mamasoula C, Postma AV, Bentham J, Zelenika D, et al. Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Human molecular genetics. 2013;22(7):1473–81. 10.1093/hmg/dds552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nature genetics. 2013;45(7):822–4. 10.1038/ng.2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. American journal of human genetics. 2008;82(1):100–12. 10.1016/j.ajhg.2007.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santorico SA, Hendricks AE. Progress in methods for rare variant association. BMC Genet. 2016;17 Suppl 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmuntz E, Clark BJ, Mitchell LE, Jawad AF, Cuneo BF, Reed L, et al. Frequency of 22q11 deletions in patients with conotruncal defects. Journal of the American College of Cardiology. 1998;32(2):492–8. 10.1016/s0735-1097(98)00259-9 [DOI] [PubMed] [Google Scholar]
- 27.White PS, Xie HM, Werner P, Glessner J, Latney B, Hakonarson H, et al. Analysis of chromosomal structural variation in patients with congenital left-sided cardiac lesions. Birth defects research Part A, Clinical and molecular teratology. 2014;100(12):951–64. 10.1002/bdra.23279 [DOI] [PubMed] [Google Scholar]
- 28.Pediatric Cardiac Genomics C, Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, et al. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circulation research. 2013;112(4):698–706. 10.1161/CIRCRESAHA.111.300297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang TT, Goldmuntz E, Roberts AE, Chung WK, Kline JK, Deanfield JE, et al. The Congenital Heart Disease Genetic Network Study: Cohort description. PloS one. 2018;13(1):e0191319 10.1371/journal.pone.0191319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5(6):e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature genetics. 2012;44(8):955–9. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. American journal of human genetics. 2005;76(5):887–93. 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De G, Yip WK, Ionita-Laza I, Laird N. Rare variant analysis for family-based design. PloS one. 2013;8(1):e48495 10.1371/journal.pone.0048495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher RAS. Statistical methods for research workers. Fourth ed—revised and enlarged ed: Edinburgh Oliver & Boyd; 1932. [Google Scholar]
- 36.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. American journal of human genetics. 2013;92(6):841–53. 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6(7):e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210. [Google Scholar]
- 39.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics. 2014;46(3):310–5. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nature methods. 2014;11(3):294–6. 10.1038/nmeth.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–13. 10.1101/gr.3577405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, et al. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480 10.1038/ncomms3480 [DOI] [PubMed] [Google Scholar]
- 43.Chaudhry SP, Frishman WH. Myotonic dystrophies and the heart. Cardiol Rev. 2012;20(1):1–3. 10.1097/CRD.0b013e31821950f9 [DOI] [PubMed] [Google Scholar]
- 44.Lee KY, Li M, Manchanda M, Batra R, Charizanis K, Mohan A, et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol Med. 2013;5(12):1887–900. 10.1002/emmm.201303275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105(51):20333–8. 10.1073/pnas.0809045105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeMasters KE, Blech-Hermoni Y, Stillwagon SJ, Vajda NA, Ladd AN. Loss of muscleblind-like 1 promotes invasive mesenchyme formation in endocardial cushions by stimulating autocrine TGFbeta3. BMC Dev Biol. 2012;12:22 10.1186/1471-213X-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coram RJ, Stillwagon SJ, Guggilam A, Jenkins MW, Swanson MS, Ladd AN. Muscleblind-like 1 is required for normal heart valve development in vivo. BMC Dev Biol. 2015;15:36 10.1186/s12861-015-0087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, et al. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115(6):739–50. 10.1016/s0092-8674(03)00975-9 [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Zhang Y, Xu E, Mohibi S, de Anda DM, Jiang Y, et al. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 2018;25(6):1118–30. 10.1038/s41418-017-0029-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neves-Pereira M, Muller B, Massie D, Williams JH, O'Brien PC, Hughes A, et al. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46(11):759–65. 10.1136/jmg.2009.066852 [DOI] [PubMed] [Google Scholar]
- 51.Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493(7432):371–7. 10.1038/nature11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nattel SN, Adrianzen L, Kessler EC, Andelfinger G, Dehaes M, Cote-Corriveau G, et al. Congenital Heart Disease and Neurodevelopment: Clinical Manifestations, Genetics, Mechanisms, and Implications. Can J Cardiol. 2017;33(12):1543–55. 10.1016/j.cjca.2017.09.020 [DOI] [PubMed] [Google Scholar]
- 53.Guo XX, An S, Yang Y, Liu Y, Hao Q, Tang T, et al. Emerging role of the Jun N-terminal kinase interactome in human health. Cell Biol Int. 2018;42(7):756–68. 10.1002/cbin.10948 [DOI] [PubMed] [Google Scholar]
- 54.Nie X, Deng CX, Wang Q, Jiao K. Disruption of Smad4 in neural crest cells leads to mid-gestation death with pharyngeal arch, craniofacial and cardiac defects. Developmental biology. 2008;316(2):417–30. 10.1016/j.ydbio.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin AE, Michot C, Cormier-Daire V, L'Ecuyer TJ, Matherne GP, Barnes BH, et al. Gain-of-function mutations in SMAD4 cause a distinctive repertoire of cardiovascular phenotypes in patients with Myhre syndrome. American journal of medical genetics Part A. 2016;170(10):2617–31. [DOI] [PubMed] [Google Scholar]
- 56.Hsu CL, Lee EX, Gordon KL, Paz EA, Shen WC, Ohnishi K, et al. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 2018;9(1):942 10.1038/s41467-018-03340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antonin W, Neumann H. Chromosome condensation and decondensation during mitosis. Curr Opin Cell Biol. 2016;40:15–22. 10.1016/j.ceb.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 58.Goldmuntz E, Bamford R, Karkera JD, dela Cruz J, Roessler E, Muenke M. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. American journal of human genetics. 2002;70(3):776–80. 10.1086/339079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Luca A, Sarkozy A, Consoli F, Ferese R, Guida V, Dentici ML, et al. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart. 2010;96(9):673–7. 10.1136/hrt.2009.181685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.