Abstract
With the increasing incidence and mortality of colorectal cancer (CRC), early and accurate diagnosis is of paramount priority to combat this cancer. Epigenetic alterations such as DNA methylation are innovative biomarkers for CRC, due to their stability, frequency, and accessibility in bodily fluids. In this study, blood samples were prospectively collected from patients before and after operation for CRC for determination of methylated septin 9 (mSEPT9) and compared to carcinoembryonic antigen (CEA). The sensitivity of using mSEPT9 methylation status for diagnosing CRC was significantly higher than using elevated CEA levels (73.2% vs 48.2%; p value < 0.001). The sensitivities of both tests increased with higher tumor staging (P = 0.004 and 0.04 respectively). Combined mSEPT9 and CEA had higher accuracy than single CEA or mSEPT9 (P = 0.009 and 0.532 separately). An increase in the methylation level of mSEPT9 detected in the post-operative samples was associated with a higher mortality rate (15.2% vs 1.8%; P = 0.024) and the presence of metastasis (27.3% vs 7.0%; P = 0.013). mSEPT9 was more sensitive than CEA for diagnosing CRC, and combined mSEPT9 and CEA was more accurate. After curative resection, detection of increased mSEPT9 methylation level may indicate adverse outcomes.
Subject terms: Gastroenterology, Colorectal cancer
Introduction
The incidence and mortality rates of colorectal cancer (CRC) has increased 10-fold globally, with an expected 60% increase by 20301. In Hong Kong, the incidence of CRC has surpassed lung cancer and emerged as the most common cancer. The mortality of CRC also ranked second, with crude rates of 35 and 22.8 per 100,000 men and women, respectively2. Due to the rapidly rising incidence and mortality of CRC, early and accurate detecting methods are of high importance. Currently, colonoscopy is the most direct method for detecting colorectal neoplasm. However, many people may not accept colonoscopy due to its potential risk and discomfort. In comparison, non-invasive blood test appears to be simple and may have a higher compliance as a screening test, and will permit frequent monitoring after diagnosis and treatment as well. As described in a German study, 63.4% (109/172) of patients refused screening colonoscopy, whereas 82.6% (90/109) of them chose blood test instead3.
Aberrant methylation is a regulatory mechanism of gene expressions which is commonly found in tumor suppressor genes in many cancers, including CRC4–6. Various epigenetic biomarkers have been identified in previous studies for the diagnosis as well as predicting prognosis of CRC7–9. Among various methylated genes, methylated septin 9 (mSEPT9) is found to have extremely high methylation levels in colorectal tumor tissues10 and has been studied for serological diagnosis of CRC with high sensitivity11–13. Detection of circulating mSEPT9 DNA in blood has been recently approved by the Food and Drug Administration (FDA) of the United States. This assay provided an alternative non-invasive test for the screening of CRC and is commercially available14. However, the utility of mSEPT9 for monitoring CRC patients after surgery has not yet been determined and there are few researches comparing carcinoembryonic antigen (CEA), a well-established tumor marker for CRC, with mSEPT9.
CEA is an oncofetal antigen generated from endodermal epithelial tumor cells in the alimentary tract15. The American Society of Clinical Oncology recommends that CEA to be measured every 3 months for at least 3 years in stage II and III CRC patients post-operatively16. Data are however insufficient to recommend other blood-based tumor markers for the monitoring of CRC patients at this stage.
The current study is a prospective study that aims to (1) evaluate the diagnostic accuracy of detecting mSEPT9 DNA in the blood of patients with different stages of colorectal neoplasm as compared to CEA and (2) to determine the role of mSEPT9, when compare to that of CEA, in monitoring CRC patients who have undergone curative resection of tumor.
Results
Participants
A total of 282 patients were included in the first part of this study for determination of diagnostic values of mSEPT9, including 117 patients with confirmed CRC, 45 patients with advanced adenoma, 50 patients with non-advanced adenoma and 70 patients with normal colonoscopy (Table 1). The percentage of male patients was 62.8% and the mean age of patients was 66.1 (SD 11.5) years. Among the 117 confirmed CRC patients, 98 patients who had serial blood taken before and after surgical resection were included in the second part of this study for characterization of the role of mSEPT9 on post-operative monitoring (Table 2). During the follow-up of up to 28 months (mean 18.80 ± 5.87), we observed 11 cases with recurrence, 15 cases with new metastases and 11 cases with CRC associated death (Fig. 1).
Table 1.
Patients’ characteristics.
| No.* | Mean age# (SD) |
Male (%) |
No. of tested mSEPT9** | No. of tested CEA** | |
|---|---|---|---|---|---|
| Colorectal Cancer | 117 | 67.3 (12.8) | 80 (68.4%) | 112 | 110 |
| Stage I | 20 | 67.2 (10.1) | 15 (75.0%) | 19 | 19 |
| Stage II | 47 | 69.6 (12.8) | 32 (68.1%) | 46 | 45 |
| Stage III | 35 | 66.1 (13.4) | 23 (65.7%) | 33 | 32 |
| Stage IV | 4 | 69.8 (8.3) | 3 (75.0%) | 4 | 4 |
| Unknown | 11 | 60.6 (15.7) | 7 (63.6%) | 10 | 10 |
| Colorectal Adenoma | 95 | 66.8 (9.0) | 58 (61.1%) | 92 | 90 |
| Advanced | 45 | 67.3 (8.6) | 31 (68.9%) | 43 | 40 |
| Non-advanced | 50 | 66.4 (9.5) | 27 (54.0%) | 49 | 50 |
| Normal | 70 | 63.1 (11.7) | 39 (55.7%) | 69 | 58 |
| Total | 282 | 66.1 (11.5) | 177 (62.8%) | 273 | 258 |
*No. means the number of cases.
#Age was calculated at the time of diagnosing.
**No. means the number of cases which had results of mSEPT9 (methylated septin 9) or CEA (carcinoembryonic antigen), and the corresponding sensitivity was shown in Fig. 2.
Table 2.
Characteristics of 98 colorectal cancer patients with serial follow up blood samples.
| Stage | I | II | III | IV | Unknown | Total |
|---|---|---|---|---|---|---|
| Number | 20 | 41 | 30 | 1 | 6 | 98 |
| Male (%) | 15 (75.0%) | 28 (68.3%) | 19 (63.3%) | 0 (0.0%) | 3 (50.0%) | 65 (66.3%) |
| Mean Age (SD) | 67.2 (10.1) | 69.9 (13.1) | 65.8 (13.7) | 66.0 | 54.0 (16.9) | 67.1 (13.3) |
| Location of tumor(s): | ||||||
| Proximal* | 4 | 12 | 4 | 0 | 0 | 20 |
| Distal | 16 | 28 | 26 | 1 | 6 | 77 |
| Synchronous | 0 | 1 | 0 | 0 | 0 | 1 |
*Proximal refers to lesion above the splenic flexure.
Figure 1.
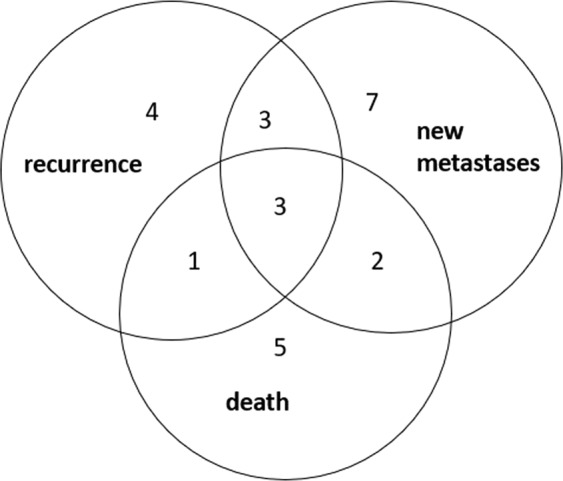
Number of cases with recurrence, new metastases and death during follow-up. Non-overlapped domain stands for colorectal cancer cases who had one poor prognostic outcome of recurrence, new metastases, and death; while the overlapped domain representing cases who had more than one.
Performance of mSEPT9 for diagnosis of colorectal neoplasm
In the first part of this study, we determined the sensitivities of mSEPT9 and CEA for patients with different colonoscopic diagnoses and tumor stages. The overall positive rates of mSEPT9 and CEA in the pre-operative blood samples of patients with confirmed CRC were 73.2% (95% CI, 65.0–81.1%) and 48.2% (95% CI, 38.1–57.3%; P < 0.001), respectively. There was a progressive increase in the detection rate for both mSEPT9 (P = 0.004) and CEA (P = 0.04) with higher tumor staging (Fig. 2). The positive rate of mSEPT9 increased from 52.6% in patients with stage I cancer to 100% in patients with stage IV cancer whereas the corresponding positive rate of CEA increased from 26.3% to 100% in these patients. There was a significant difference in the positive rates between mSEPT9 and CEA in stage II (P = 0.001) and stage III (P = 0.002) cancers, but not in patients with stage I (P = 0.184) and IV cancers. However, the sensitivities of both mSEPT9 and CEA were low (<27.6%) in patients with adenoma, both advanced and non-advanced, and there was no significant difference in positive rates between mSEPT9 and CEA for patients with adenoma. The overall specificity of mSEPT9 and CEA in colonoscopy-negative subjects was comparable (71.0% and 79.3%; P = 0.311). Table 3 compared the accuracy of mSEPT9, CEA and combination of the two for patients and controls who had both test results available. The combined results, when either one positive test is treated as positive, showed a higher accuracy than CEA (P = 0.009) or mSEPT9 (P = 0.532) alone. Positive CEA was more commonly found in proximal than distal CRC (72.2% vs 39.2%; P = 0.017), while positive mSEPT9 showed no significant difference between proximal and distal cancer (90.0% vs 69.4%; P = 0.085).
Figure 2.
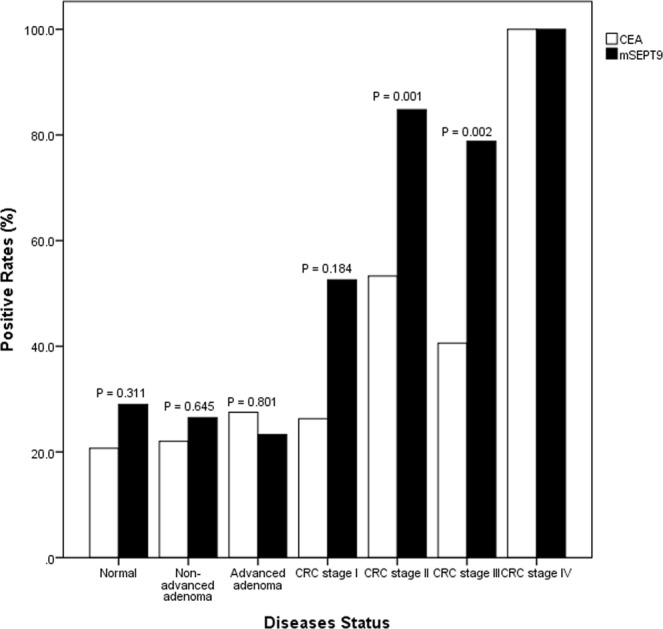
Positive rates of CEA and mSEPT9 in different patient groups. Sensitivities of carcinoembryonic antigen (CEA) and methylated septin 9 (mSEPT9) for detecting different stages of colorectal neoplasm; i.e. non-advanced adenoma (22.0% vs 26.5%; P = 0.645), advanced adenoma (27.5% vs 23.3%; P = 0.801), colorectal cancer (CRC): stage I (26.3% vs 52.6%; P = 0.184), stage II (53.3% vs 84.8%; P = 0.001), stage III (40.6% vs 78.8%; P = 0.002), and stage IV (100.0% vs 100.0%, p value is not available). In addition, the first pair of bars represented for false positive rates in participants with normal colonoscopy.
Table 3.
Accuracy of mSEPT9, CEA and the combination in detecting colorectal cancer.
| Tests | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| mSEPT9 | 71.8% (77/105) | 66.7% (38/57) | 71.0% (115/162) |
| CEA | 50.5% (53/105) | 78.9% (45/57) | 60.5% (98/162) |
| Combination# | 85.7% | 54.4% | 74.7% |
| P value | |||
| mSEPT9 vs CEA (2-sided) | 0.001 | 0.206 | 0.061 |
| mSEPT9 vs CEA (1-sided) | 0.001 | 0.103 | 0.030 |
| mSEPT9 vs Combination (2-sided) | 0.039 | 0.250 | 0.532 |
| mSEPT9 vs Combination (1-sided) | 0.020 | 0.125 | 0.266 |
| CEA vs Combination (2-sided) | 0.000 | 0.009 | 0.009 |
| CEA vs Combination (2-sided) | 0.000 | 0.005 | 0.004 |
#Combination refers to either mSEPT9 (methylated septin 9) or CEA (carcinoembryonic antigen) was positive among participants who had both tests done (n = 162).
Role of mSEPT9 and CEA in monitoring of cancer patients after surgery
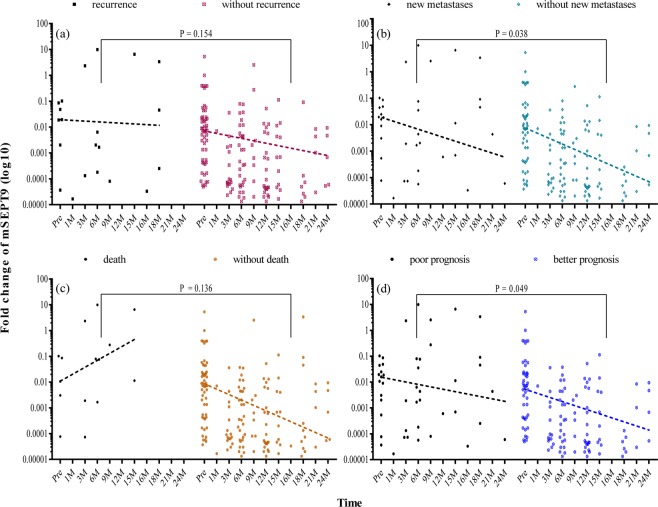
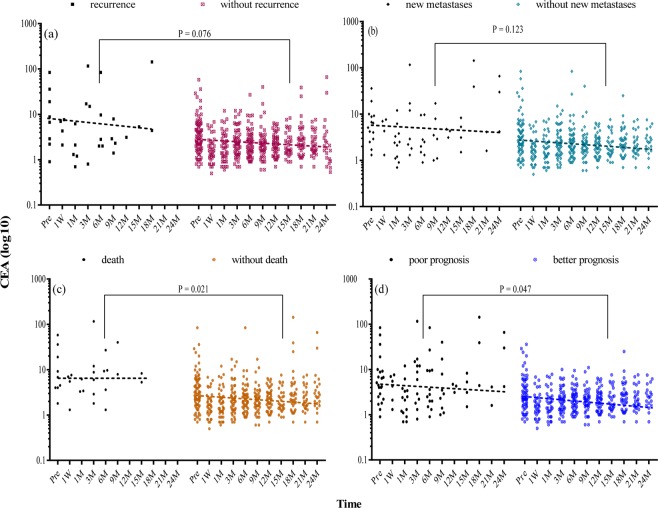
We also determined the changes in CEA and mSEPT9 in the post-operative blood samples of patients after resection of the tumor (Tables S1 amd S2). Overall, the proportion of patients with negative CEA was significantly higher than the proportion of patients with negative mSEPT9 at 6-months (71.8% vs 55.3%; P = 0.035) and 12-months (68.1% vs 48.1%; P = 0.028), especially for cases with non-advanced cancer (stage I/II) at 6-months (79.2% vs 55.6%; P = 0.013), and those without evidence of clinical recurrence at 6-months (72.9% vs 55.3%; P = 0.038) and 12-months (69.7% vs 49.0%; P = 0.033). This trend extends to 18-months and 24-months after operation although the difference did not reach statistical significance (Table 4). Among cancer patients with a positive mSEPT9 or CEA at baseline, 46.8% (mSEPT9) and 46.7% (CEA) turned negative at 6-months after operation (P = 1.0). These changes remain similar at 12-months, 18-months and 24-months (all P > 0.5). Figures 3 and 4 showed the distribution for different groups of mSEPT9 and CEA before and after operation. Patients with poor prognosis including recurrence, new metastases and death after operation had higher value of both mSEPT9 and CEA than those with better prognosis. When considering dynamic quantitative changes of mSEPT9 after operation, cases with increased methylation level had higher recurrence rate (15.2% vs 5.3%; P = 0.137), development of new metastases (27.3% vs 7.0%; P = 0.013) and higher mortality rate (15.2% vs 1.8%; P = 0.024). In the group of increased methylation level, patients had lower overall survival (P = 0.014) (Fig. 5) during the follow-up period.
Table 4.
Results of mSEPT9 and CEA in colorectal cancer patients after surgery.
| Cases | Time Point # | Positive mSEPT9 | Negative mSEPT9 | Positive CEA | Negative CEA | P Value * | |
|---|---|---|---|---|---|---|---|
| All patients after surgery | 6 M | 38 (44.7%) | 47 (55.3%) | 22 (28.2%) | 56 (71.8%) | 0.035 | |
| 12 M | 27 (51.9%) | 25 (48.1%) | 23 (31.9%) | 49 (68.1%) | 0.028 | ||
| 18 M | 15 (46.9%) | 17 (53.1%) | 18 (32.1%) | 38 (67.9%) | 0.180 | ||
| 24 M | 7 (63.6%) | 4 (36.4%) | 14 (34.1%) | 27 (65.9%) | 0.095 | ||
| Patients without recurrence | 6 M | 34 (44.7%) | 42 (55.3%) | 19 (27.1%) | 51 (72.9%) | 0.038 | |
| 12 M | 25 (51.0%) | 24 (49.0%) | 20 (30.3%) | 46 (69.7%) | 0.033 | ||
| 18 M | 11 (40.7%) | 16 (59.3%) | 15 (28.3%) | 38 (71.7%) | 0.316 | ||
| 24 M | 7 (63.6%) | 4 (36.4%) | 14 (34.1%) | 27 (65.9%) | 0.095 | ||
| Patients with recurrence | 6 M | 4 (50.0%) | 4 (50.0%) | 3 (37.5%) | 5 (62.5%) | 1.0 | |
| 12 M | 1 (50.0%) | 1 (50.0%) | 2 (40.0%) | 3 (60.0%) | 1.0 | ||
| 18 M | 4 (80.0%) | 1 (20.0%) | 3 (100.0%) | 0 (0.0%) | 1.0 | ||
| 24 M | 0 | 0 | 0 | 0 | NA | ||
mSEPT9, methylated septin 9; CEA, carcinoembryonic antigen; M, month.
#Time point was arranged as a time period, for example, 6 M concluded samples tested at 3 M and 6 M.
*P Value is the difference of negative percentages between mSEPT9 and CEA.
Figure 3.
Relative methylation levels of mSEPT9 in colorectal cancer at different time points. Relative methylation of mSEPT9 (line = median) in colorectal cancer (CRC) patients (a) with or without recurrence; (b) with or without new metastases; (c) with or without death; (d) with poor or better prognosis during follow-up after operation. Poor prognosis refers to the presence of any of the three mentioned prognostic factors, while better prognosis refers to none of them. The P value was calculated by paired t test through GraphPad Prism (version 7; GraphPad Software, San Diego, CA) and represented the significance level of the difference in each group as aforesaid. The dots represent the mean value of SEPT9 methylation level in three PCR reactions for each individual patient, while the dotted lines show the trend of the median methylation levels with time. Pre = pre-operation, M = months.
Figure 4.
Distribution of carcinoembryonic antigen in colorectal cancer at different time points. Levels of carcinoembryonic antigen (CEA) (line = median) in colorectal cancer (CRC) patients (a) with or without recurrence; (b) with or without new metastases; (c) with or without death; (d) with poor or better prognosis during follow-up after operation. Poor prognosis refers to the presence of any of the three mentioned prognosis, while better prognosis refers to none of them. The P value was calculated by paired t test through GraphPad Prism (version 7; GraphPad Software, San Diego, CA) and represented the significance level of the difference in each group as aforesaid. The dots represent individual CEA value while the dotted lines show the trend of the median CEA levels with time. Pre = pre-operation, M = months.
Figure 5.
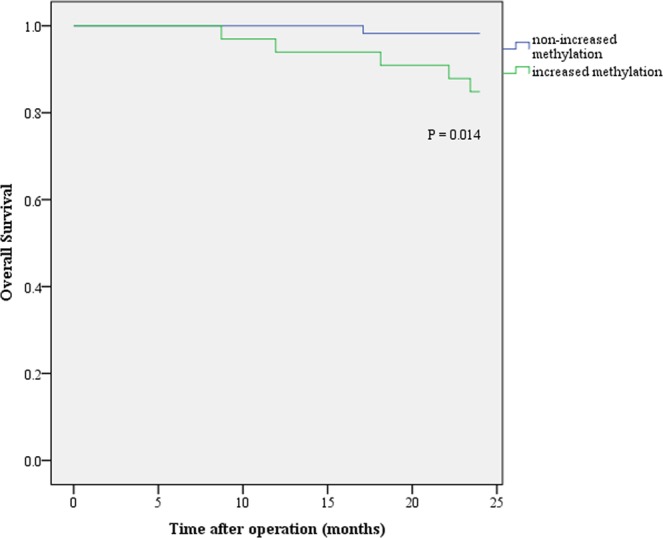
Overall survival in colorectal cancer patients after operation. The green and blue lines stand for the patient groups with and without increased methylation level after operation, respectively. Censor time is the death date or till the 24-month after surgery.
Discussion
The overall sensitivity of mSEPT9 for the diagnosis of CRC was found to vary from 48.2% to 95.6% and the specificity was 79.1 to 99.1% in previous reports17–21. The second generation of mSEPT9, however, was found to have a higher sensitivity (71.1 to 95.6%) and similar specificity21. In the largest multi-center prospective study that evaluated the role of mSEPT9 in screening asymptomatic subjects (PRESEPT study)17, the overall sensitivity of the first-generation assay was 48.2%, which increased from stage I (35%) to stage IV (77.4%) cancers. As yet, the study used duplicate rather than triplicate PCR reactions, which may lower the sensitivity. To date, there is no consensus on the diagnostic algorithm of mSEPT9 PCR assay, studies that defined ‘one out of three positive reactions’ as positive usually have higher sensitivities for cancer. The FDA also approved the commercially available SEPT9 assay based on the data from the reanalyzed PRESEPT trial that used the ‘one out of three positive PCR reactions’ as positive sample11,17, which was also adopted in this study.
Using the latest second-generation mSEPT9 assay, we found a significantly higher sensitivity of mSEPT9 than CEA for the diagnosis of CRC patients (73.2% vs 48.2%; P < 0.001), especially for patients with stage II and III cancer (both P < 0.01), but a slightly lower specificity for the former (P > 0.05). Toth et al. reported similar results, with respective sensitivities of 95.6% (88/92) and 51.8%(14/27), and specificities of 84.8% and 85.2% for mSEPT9 and CEA20. In another recent study, mSEPT9 was also shown to have higher diagnostic value than CEA for both sensitivity (61.8% vs 35.0%) and specificity (89.6% vs 62.6%)22. The sensitivity of plasma mSEPT9 for detecting CRC was highly tumor stage-dependent. As shown in Fig. 2, the sensitivity increased progressively from stage I to IV. This rising positive rate with higher tumor stage has also been found in other studies11–13. It is however interesting to note in our study that the sensitivity of mSEPT9 in detecting proximal and distal cancers varied less than CEA, which would be an advantage for mSEPT9. This point was also confirmed in a previous study, which showed similar sensitivity of mSEPT9 for both right- and left-sided CRC (94% and 96%, respectively)20. In our study, both mSEPT9 and CEA were not sensitive enough in identifying patients with adenomas, including advanced and non-advanced adenoma, with all positive rates of less than 27.6%. In contrast, using the same Epi proColon 2.0 assay, Song et al. reported a high positive rate of mSEPT9 in villous adenoma and adenoma with high-grade dysplasia (83.3% and 62.5%, respectively) though the positive rate for all adenomas was much lower (31.8%)13. In this study, we also found that when taking the combined result of mSEPT9 and CEA, there was a significantly higher sensitivity and accuracy than single CEA (both P < 0.01) or mSEPT9 (P = 0.039 and 0.532) for diagnosing CRC.
More importantly, we prospectively compared the performance of mSEPT9 with CEA as a blood-based biomarker in the post-operative surveillance of CRC patients. The overall negative rates of mSEPT9 are lower than CEA from 6-months post-operatively, especially in patients with non-advanced cancer and those with no tumor recurrence. Among cancer patients with a positive mSEPT9 or CEA before operation, only about half of them (46.8% and 46.7%, respectively) turned negative within 6-months after surgery, suggesting both markers may not be sensitive enough for monitoring early treatment response. On the other hand, both mSEPT9 and CEA levels tended to be higher among patients with poor prognostic factors including tumor recurrence, detection of new metastasis and death. We observed a subgroup of cancer patients who had increased in quantitative levels of mSEPT9 after operation, which were associated with significantly higher rates of new metastases and mortality (both P < 0.05). Tham et al. used an in-house quantitative mSEPT9 assay and found that increased methylation levels of SEPT9 one year after surgery was associated with CRC recurrence, while the change at 6-month after surgery did not have any correlation18. In addition, Fu et al. found that the CRC patients with continued positive mSEPT9 after operation had closer correlation with recurrences or metastases within 12-months than those whose positive mSEPT9 turned negative23.
Our study had some limitations. First, our sample size for the study may not be large enough for the stratified analysis of cancer patients with different stages. As we only selected patients who underwent curative resection, the number of patients with advanced cancer staging and hence, adverse clinical outcomes such as death or clinical recurrence was relatively low during the initial follow up period, which make it difficult to conclude the roles of these biomarkers in detecting recurrence. More studies with larger sample sizes would be needed to increase precision of the estimates. Although the increased methylation levels of mSEPT9 have been shown in this study to be associated with higher mortality and new metastasis, the low rates of conversion from positive to negative in mSEPT9 during early post-operative period, may imply that mSEPT9 is not sensitive enough during this period. Due to the qualitative nature of the commercially available mSEPT9, it may still be inferior to CEA which is a quantitative assay on monitoring of treatment response. There are, however, increasing doubts that CEA may not be the best surveillance biomarker for CRC recurrence24,25. Shinkins et al. showed that CEA was not a satisfactory predictor for CRC recurrence regardless of the cut-off threshold in a total of 582 CRC patients after curative resection24. The recommended threshold of 5 ug/L would miss half the recurrences (53.8%; 95% CI: 44.2–64.4%) while the threshold of 2.5 ug/L would get very high rate of false alarms (84.2%; 924/1097 referred).
In conclusion, we compared the performance of mSEPT9 with CEA both for the diagnosis of CRC and post-operative monitoring of CRC patients. We found that the second generation of commercially available plasma mSEPT9 assay was more sensitive than CEA for the diagnosis of CRC patients, which parallels with the increase in tumor staging. Combined mSEPT9 and CEA would result in even higher sensitivity than either CEA or mSEPT9 alone. Both mSEPT9 and CEA values showed increasing trend among patients with poor prognosis after operation. Moreover, increased methylation level of mSEPT9 after surgical resection may be able to identify a subgroup of patients with adverse outcomes. Further studies may be needed to identify a more sensitive way of detecting minor changes in circulating mSEPT9 in blood for the detection of early CRC recurrence after surgery.
Methods
Participants
This was a prospective study conducted in the Queen Mary Hospital of Hong Kong, a university teaching hospital. For determination of the diagnostic performance of mSEPT9 in subjects with different colorectal neoplasm ranging from non-advanced adenoma, advanced adenoma to carcinoma, we prospectively enrolled patients undergoing colonoscopy in the Endoscopy Center of the hospital. These included symptomatic patients who were referred for colonoscopy due to bowel symptoms or diagnostic work up of iron deficiency anemia as well as patients undergoing screening colonoscopy. We excluded patients with previous bowel resection, familial colorectal cancer syndrome, inflammatory bowel disease or diagnosis of any other malignancy in the past. After obtaining informed consent, venous blood was collected from patients prior to the colonoscopy. Plasma was separated by centrifugation and stored at −20 °C till further analysis. During colonoscopy, all polyps were removed for histological examination and lesions suspicious for colorectal cancer were biopsied. All histological samples were reviewed by experienced pathologists in the hospital. Patients were then classified into four groups according to the most advanced lesions found on colonoscopy as (1) adenocarcinoma, (2) advanced adenoma, (3) non-advanced adenoma, and (4) normal colonoscopy without any polyp or adenoma. Advanced adenoma was defined as a lesion with a diameter of 10 mm or above, with villous histology or the presence of high-grade dysplasia26.
To determine the role of mSEPT9 in the post-operative monitoring of CRC patients, we prospectively recruited patients who were newly diagnosed to have adenocarcinoma of colon or rectum and were scheduled for curative resection in our hospital. All patients had baseline blood samples taken immediately before surgery and then at three-monthly intervals for up to 24-month. The baseline demographic and tumor staging were retrieved for these patients. Tumor staging was classified as stipulated by the seventh edition of the American Joint Committee on Cancer TNM Classification27. All patients were then prospectively followed up in our colorectal clinic after operation. The charts were reviewed and the outcome of these patients were further verified by the centralized electronic patients’ record (ePR) of the Hospital Authority of Hong Kong which is a territory-wide health database that capture all clinical information of patients including deaths. Clinical relapse or recurrence was determined clinically by history, physical examination and relevant investigation findings. Imaging techniques including CT scan or PET-CT were arranged when clinically indicated to confirm the presence of distant or regional recurrence. Surveillance colonoscopy after surgical resection was performed in all patients according to current recommendation28.
Informed consent was obtained from all patients and the study protocol was approved by the Institutional Review Board of the Hospital Authority-Hong Kong West Cluster and University of Hong Kong (UW 12-489).
Detection of methylated SEPT9 and CEA
All plasma samples were coded with a unique sample collection number without patient identifier, final diagnosis or timing of blood sampling to ensure adequate blinding of laboratory staff involved in the testing of mSEPT9. mSEPT9 was determined by a commercially available assay (Epi proColon 2.0; Epigenomics AG, Berlin, Germany) in an independent laboratory (DiagCor, Hong Kong) authorized by the manufacturer. All samples were analyzed without patient information by real time PCR in triplicate and the mSEPT9 assay was considered positive when at least one of the three PCR reactions was positive17. Positive and negative controls were included in all PCR amplifications. Actin was used as reference, the results of SEPT9 methylation level was calculated using method similar to the 2(−∆∆CT) 29 with normalization to Actin.
CEA levels were determined by enzyme linked immunoassay in the Clinical Immunology Laboratory of our hospital. Abnormal or positive CEA was defined as a value above 3 ng/ml as recommended by the laboratory. Levels lower than this were considered negative in all analyses.
Statistical analysis
The sensitivity and specificity of mSEPT9 and CEA was computed with the 95% confidence intervals (CI). Chi-squared or Fisher Exact test was used to compare categorical data. All statistical analyses were two-sided, and a statistically significant difference was established when P < 0.05. All analysis was performed by the IBM SPSS Statistics software (version 21; IBM, USA) and GraphPad Prism (version 7; GraphPad Software, San Diego, CA).
Ethics approval and consent to participate
Informed consent was obtained from all patients and the study protocol was approved by the Institutional Review Board of the Hospital Authority Hong Kong West Cluster and University of Hong Kong (UW 12-489). In addition, all research was performed in accordance with the Declaration of Helsinki – Ethical Principles for Medical Research involving Human Subjects.
Supplementary information
Acknowledgements
We acknowledge all patients for participation in this study and the assistance offered by the hospital staff during the study period. This study was supported by the Health and Medical Research Fund of the Hong Kong SAR Government (Number 01121026). The sponsor had no role in the study’s design, conduct and reporting.
Author Contributions
Z. Ma performed the data analyses, data interpretation and writing of the manuscript. W.L. Law was involved in the study design, patient’s recruitment and critical review of the manuscript. E.K.O. Ng was responsible for analysis of blood samples. C.S.Y. Chan, K.S. Lau are responsible for blood sample collection and patient’s follow up logistics. Y.Y. Cheng provided conception and analysis of data, edit and review the manuscript. V. Shin and A. Kwong provided technical advice on study design and execution as well as critical review of the manuscript. W.K. Leung led and supervised the study, and critically reviewed the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46876-4.
References
- 1.Arnold M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Hong Kong Cancer Registry, H. A. Top Ten Cancers, http://www3.ha.org.hk/cancereg/topten.html (2015).
- 3.Adler A, et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC gastroenterology. 2014;14:183. doi: 10.1186/1471-230x-14-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goel A, Boland CR. Epigenetics of Colorectal Cancer. Gastroenterology. 2012;143:1442–1460.e1441. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204–1225.e1212. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juo YY, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Annals of Oncology. 2014;25:2314–2327. doi: 10.1093/annonc/mdu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung W, et al. Quantitative Detection of Promoter Hypermethylation in Multiple Genes in the Serum of Patients with Colorectal Cancer. American Journal of Gastroenterology. 2005;100:2274–2279. doi: 10.1111/j.1572-0241.2005.50412.x. [DOI] [PubMed] [Google Scholar]
- 8.Coppede F. Epigenetic biomarkers of colorectal cancer: Focus on DNA methylation. Cancer Lett. 2014;342:238–247. doi: 10.1016/j.canlet.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 9.Barault Ludovic, Amatu Alessio, Siravegna Giulia, Ponzetti Agostino, Moran Sebastian, Cassingena Andrea, Mussolin Benedetta, Falcomatà Chiara, Binder Alexandra M, Cristiano Carmen, Oddo Daniele, Guarrera Simonetta, Cancelliere Carlotta, Bustreo Sara, Bencardino Katia, Maden Sean, Vanzati Alice, Zavattari Patrizia, Matullo Giuseppe, Truini Mauro, Grady William M, Racca Patrizia, Michels Karin B, Siena Salvatore, Esteller Manel, Bardelli Alberto, Sartore-Bianchi Andrea, Di Nicolantonio Federica. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2017;67(11):1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Model F, et al. Identification and validation of colorectal neoplasia-specific methylation markers for accurate classification of disease. Mol Cancer Res. 2007;5:153–163. doi: 10.1158/1541-7786.Mcr-06-0034. [DOI] [PubMed] [Google Scholar]
- 11.Potter NT, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clinical chemistry. 2014;60:1183–1191. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 12.Jin P, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. Journal of Gastroenterology and Hepatology. 2015;30:830–833. doi: 10.1111/jgh.12855. [DOI] [PubMed] [Google Scholar]
- 13.Song L, et al. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017;9:599–610. doi: 10.2217/epi-2016-0146. [DOI] [PubMed] [Google Scholar]
- 14.Food and Drug Administration, U. S. Premarket Approval (PMA), https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPMA/pma.cfm?id=P130001 (
- 15.Gold P. Specific Carcinoembryonic Antigens Of The Human Digestive System. Journal of Experimental Medicine. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locker GY, et al. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. Journal of Clinical Oncology. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 17.Church TR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tham C, et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer. 2014;120:3131–3141. doi: 10.1002/cncr.28802. [DOI] [PubMed] [Google Scholar]
- 19.Grützmann R, et al. Sensitive Detection of Colorectal Cancer in Peripheral Blood by Septin 9 DNA Methylation Assay. PLOS ONE. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tóth K, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PloS one. 2012;7:e46000. doi: 10.1371/journal.pone.0046000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Scientific Reports. 2017;7:3032. doi: 10.1038/s41598-017-03321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie L, et al. Diagnostic value of methylated Septin9 for colorectal cancer detection. Frontiers in oncology. 2018;8:247. doi: 10.3389/fonc.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu, B. et al. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Disease markers2018 (2018). [DOI] [PMC free article] [PubMed]
- 24.Shinkins B, et al. The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: Results from the FACS trial. PLOS ONE. 2017;12:e0171810. doi: 10.1371/journal.pone.0171810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sørensen CG, Karlsson WK, Pommergaard H-C, Burcharth J, Rosenberg J. The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence - A systematic review. International journal of surgery (London, England) 2016;25:134–144. doi: 10.1016/j.ijsu.2015.11.065. [DOI] [PubMed] [Google Scholar]
- 26.Leung WK, et al. Detection of colorectal adenoma by narrow band imaging (HQ190) vs. high-definition white light colonoscopy: a randomized controlled trial. The American journal of gastroenterology. 2014;109:855–863. doi: 10.1038/ajg.2014.83. [DOI] [PubMed] [Google Scholar]
- 27.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Annals of Surgical Oncology. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 28.Kahi CJ, et al. Colonoscopy Surveillance After Colorectal Cancer Resection: Recommendations of the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2016;150:758–768.e711. doi: 10.1053/j.gastro.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.