Abstract
Soil fauna play an essential role in the soil ecosystem, but they may be influenced by insecticidal Cry proteins derived from Bacillus thuringiensis (Bt) maize. In this study, a 2-year field trial was conducted to study the effects of transgenic cry1Ie maize, a type of Bt maize (Event IE09S034), on soil fauna, with the near-isogenic line non-Bt maize (Zong 31) as a control. The soil animals were collected with Macfadyen heat extractor and hand-sorting methods, respectively, and their diversity, abundance and community composition were calculated. Then, the effects of maize type, year, sampling time and soil environmental factors on the soil fauna were evaluated by repeated-measures ANOVA, redundancy analysis (RDA) and nonmetric multidimensional scaling (nMDS). Repeated-measures ANOVA showed that the diversity and abundance of the soil fauna were not affected by maize type, while they were significantly influenced by year and sampling time. Furthermore, for both the Macfadyen and hand-sorting methods, RDA indicated that soil fauna community composition was not correlated with maize type (Bt and non-Bt maize) but was significantly correlated with year, sampling time and root biomass. In addition, it was significantly related to soil pH according to the hand-sorting method. nMDS indicated that soil fauna community composition was significantly correlated with year and sampling time; however, it was not associated with maize type. In this study, we collected soil faunal samples according to the Macfadyen and hand-sorting methods and processed the obtained data with ANOVA, RDA, and nMDS in three ways, and our data indicate that transgenic cry1Ie maize (Event IE09S034) had no substantial influence on the diversity, abundance or community composition of the soil fauna.
Subject terms: Environmental economics, Biotechnology
Introduction
Since the first commercial genetically modified (GM) crop was cultivated in the United States in 19961, GM crops have been planted for 22 years2–4. In 2017, the total global planting area reached 189.8 million hectares5. With the spread of GM crops, environmental safety problems have caused wide public concern.
At present, the main method for researching the impacts of GM crops on environmental safety is to monitor biodiversity6,7 in the field. With this method, many studies have been conducted on the diversity of aboveground species communities8–12, but there has been less focus on belowground species communities. Even among the studies related to the belowground species community, most of them focused on soil microorganisms13–18. Recently, transgenic cry1Ab gene crops, a type of commercialized GM crop, have been studied and showed no impacts on soil organisms19–29. For example, transgenic cry1Ab rice had no significant effects on the residual decay- and decomposition-associated microbial community compositions in comparison to the non-Bt rice variety (Xiushui 11)16. Two types of cry1Ac/cpti transgenic rice (GM1 and GM2) also had no effects on the composition and abundance of bacterial and fungal communities in paddy soil during the growing season15. Bt hybrid cotton MECH-162 did not have adverse effects on both culturable and nonculturable microbial diversity according to the analysis of microbial community structure dynamics18. Mohammad et al. (2003) and Toschki et al. (2007) reported that both Bt maize and transgenic cry1Ab maize had no adverse effects on soil faunaus19,21. To date, no detrimental impacts of GM crops on belowground organisms have been found.
The Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS), isolated and cloned the anti-insect gene cry1Ie30, which encodes an insecticidal protein that does not show cross resistance with several other insecticidal proteins, Cry1Ab, Cry1Ac, Cry1Ah and Cry1F25,26,31,32, and shows certain virulence to the Asian corn borer (ACB) and cotton bollworm33. Therefore, the cry1Ie gene can be integrated with Bt genes, such as cry1Ah, to form stacked transgenic insect-resistant maize and thus to overcome the problems resulting from the high homology and interactive resistance of the single Bt gene maize type27,34. At present, the transgenic cry1Ie maize hybrid (Bt maize, Event IE09S034) is being tested for commercial production in China and has the potential to be commercialized.
Some studies have revealed that transgenic cry1Ie maize has no impact on the diversity of arthropod communities in the field35–37, while no study on the belowground soil fauna community has been reported until now.
In this study, we conducted field trials to compare the effects of transgenic cry1Ie (Bt maize, IE09S034) and non-Bt (Zong 31) maize hybrids on soil fauna, and the soil fauna were sampled according to two methods (the Macfadyen and hand-sorting methods). Then, we used the diversity and abundance parameters RDA and nMDS to analyse the soil fauna community and to evaluate the effects of the transgenic cry1Ie maize hybrid on soil fauna in the field. This study provides useful data for the commercialization of GM crops in the future.
Results
Soil fauna in Bt and non-Bt maize plots
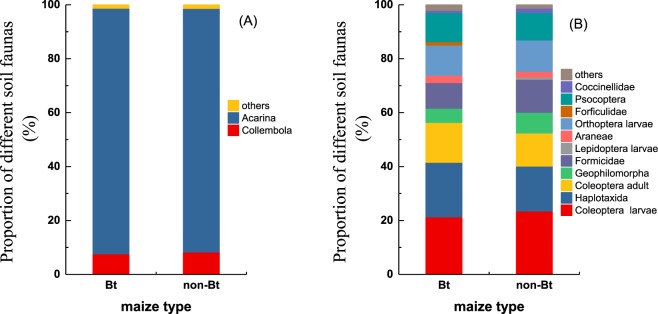
When the soil fauna community was extracted according to the Macfadyen method, in 2014 and 2015, a total of 20,133 and 21,777 soil animals were found in the Bt and non-Bt maize plots, respectively, and the animals in the two types of maize field both belonged to 11 species. In both years, the animals of Acarina accounted for the majority of the soil fauna in both Bt and non-Bt maize fields. The percentage of Acarina species was more than that of other species, which was 91.09 and 90.39% in the Bt and non-Bt maize fields, respectively, followed by Collembola species, which represented 7.59 and 8.25% of the animals in the Bt and non-Bt maize fields, respectively (Fig. 1A) and (Table 1).
Figure 1.
The taxon percentages of soil fauna found in Bt and non-Bt maize plots in 2-year field trials. Macfadyen method in 2014 and 2015 (A) hand-sorting method in 2014 and 2015. (B) This diagram illustrates every dominant soil fauna taxon with a proportion >1% and a total proportion of taxa <1% (others).
Table 1.
The abundance of soil animals captured in Bt and non- Bt maize fields by two years in Macfadyen and hand-sorting methods respectively.
| Method | Class | Order | Family | Bt | non-Bt |
|---|---|---|---|---|---|
| Macfadyen | Entognatha | Collembola | 1528 | 1796 | |
| Arachnida | Acarina | 18339 | 19685 | ||
| Insecta | Diplura | 22 | 19 | ||
| Insecta | Coleoptera larvae | 8 | 4 | ||
| Insecta | Psocoptera | 53 | 49 | ||
| Insecta | Hemiptera | Formicidae | 16 | 17 | |
| Insecta | Coleoptera | Staphylinidae | 1 | 2 | |
| Insecta | Coleoptera adult | 14 | 12 | ||
| Clitellata | Enchytraeidae | 40 | 47 | ||
| Clitellata | Haplotaxida | 101 | 144 | ||
| Chilopoda | Lithobiomorpha | 0 | 2 | ||
| Chilopoda | Geophilomorpha | 11 | 0 | ||
| Hand-sorting | Clitellata | Haplotaxida | 96 | 80 | |
| Chilopoda | Lithobiomorpha | 2 | 2 | ||
| Chilopoda | Geophilomorpha | 25 | 37 | ||
| Insecta | Coleoptera larvae | 101 | 113 | ||
| Insecta | Coleoptera adult | 70 | 59 | ||
| Insecta | Hemiptera | Formicidae | 45 | 59 | |
| Insecta | Lepidoptera larvae | 0 | 6 | ||
| Insecta | Araneae | 13 | 10 | ||
| Insecta | Orthoptera | Gryllidae | 2 | 0 | |
| Insecta | Orthoptera larvae | 53 | 56 | ||
| Insecta | Hemiptera | 4 | 0 | ||
| Insecta | Dermaptera | Forficulidae | 6 | 4 | |
| Insecta | Psocoptera | 51 | 49 | ||
| Insecta | Coleoptera | Coccinellidae | 6 | 8 |
When analysed according to the hand-sorting method, 474 and 481 soil animals were found in the Bt and non-Bt maize fields in 2014 and 2015, respectively. There were 13 and 12 species found in the Bt and non-Bt maize fields, respectively. In addition, the percentage of animals in each phylum was analysed. Coleoptera larvae, Haplotaxida, Coleoptera adults, Orthoptera larvae, and Psocoptera in the Bt maize plots accounted for 21.31, 20.25, 14.77, 11.18, and 10.76%, respectively, and the percentages of Coleoptera larvae, Haplotaxida, Coleoptera adults, Orthoptera larvae, Formicidae, and Psocoptera species in the non-Bt maize plots were 23.94, 16.63, 12.27, 11.64, 12.27, and 7.59%, respectively (Fig. 1B) and (Table 1).
Impacts of maize type, year and sampling time on soil fauna diversity and abundance
Repeated-measures ANOVA results showed that maize type had no significant effect on the Shannon-Wiener diversity index (H′), Simpson’s diversity index (D), Pielou’s evenness index (J), the number of species (S), or the total animal number (N) according to the Macfadyen and hand-sorting methods, indicating that transgenic cry1Ie maize did not influence the soil fauna. In addition, ANOVA showed that year, sampling time, and their interactions all had significant impacts on the H′, D, J, S and N of the soil fauna in the two types of maize plots when investigated according to the Macfadyen method, which indicated that the soil fauna was affected by year and sampling time. No effects of the interactions of year × maize type and year × maize type × sampling time on H′, D, J, S and N of soil fauna were detected for either method (Table 2).
Table 2.
Effects of year (2014 and 2015), maize type (Bt maize and non-Bt maize) and sampling time on soil fauna diversity and abundance, analyzed using a three-way unequally spaced repeated-measure ANOVA.
| Method | Variable | Diversity and abundance parameter | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon-Wiener index | Simpson’s diversity index | Pielou’s evenness index | Number of species | Abundance | |||||||
| (H′) | (D) | (J) | (S) | (N) | |||||||
| F | P | F | P | F | P | F | P | F | P | ||
| Macfadyen | Year | 13.37 | 0.006** | 22.59 | 0.001*** | 23.97 | 0.001*** | 10.13 | 0.013* | 9.00 | 0.017* |
| Maize type | 0.12 | 0.739 | 0.05 | 0.829 | 0.02 | 0.907 | 2.00 | 0.195 | 0.07 | 0.803 | |
| Sampling time | 4.69 | 0.004** | 2.88 | 0.038* | 3.91 | 0.011* | 4.10 | 0.009** | 15.41 | 0.003** | |
| Year × Maize type | 1.04 | 0.339 | 0.97 | 0.354 | 0.43 | 0.531 | 0.50 | 0.500 | 0.06 | 0.82 | |
| Year × Sampling time | 5.30 | 0.002** | 5.27 | 0.002** | 6.15 | 0.001*** | 3.27 | 0.020* | 19.43 | 0.002** | |
| Maize type × Sampling time | 0.49 | 0.743 | 0.36 | 0.834 | 0.80 | 0.536 | 0.31 | 0.867 | 0.06 | 0.831 | |
| Year × Maize type × Sampling time | 0.86 | 0.500 | 0.68 | 0.614 | 0.40 | 0.806 | 0.57 | 0.683 | 0.03 | 0.892 | |
| Mean ± SD (Bt maize) | 0.847 ± 0.131 | 0.325 ± 0.056 | 0.391 ± 0.060 | 4.833 ± 0.464 | 671.100 ± 159.636 | ||||||
| Mean ± SD (non-Bt maize) maize) | 0.818 ± 0.190 | 0.317 ± 0.083 | 0.387 ± 0.084 | 4.567 ± 0.654 | 725.900 ± 238.092 | ||||||
| Hand-sorting | Year | 0.06 | 0.821 | 0.20 | 0.668 | 0.09 | 0.768 | 0.04 | 0.850 | 0.58 | 0.468 |
| Maize type | 0.53 | 0.487 | 0.88 | 0.377 | 0.87 | 0.379 | 0.04 | 0.850 | 0.35 | 0.573 | |
| Sampling time | 1.07 | 0.387 | 0.60 | 0.667 | 0.37 | 0.828 | 1.80 | 0.153 | 1.62 | 0.193 | |
| Year × Maize type | 0.35 | 0.572 | 0.11 | 0.753 | 0.00 | 0.96 | 1.37 | 0.275 | 0.58 | 0.468 | |
| Year × Sampling time | 3.16 | 0.027* | 3.54 | 0.017* | 3.17 | 0.026* | 1.74 | 0.166 | 3.11 | 0.029* | |
| Maize type × Sampling time | 2.25 | 0.085 | 1.25 | 0.312 | 0.64 | 0.637 | 2.24 | 0.086 | 1.52 | 0.221 | |
| Year × Maize type × Sampling time | 1.02 | 0.411 | 0.96 | 0.441 | 0.95 | 0.450 | 0.45 | 0.771 | 0.89 | 0.482 | |
| Mean ± SD (Bt maize) | 2.046 ± 0.172 | 0.734 ± 0.061 | 0.828 ± 0.058 | 5.700 ± 0.504 | 17.000 ± 2.562 | ||||||
| Mean ± SD (non-Bt maize) | 2.142 ± 0.211 | 0.770 ± 0.052 | 0.858 ± 0.040 | 5.767 ± 0.733 | 16.100 ± 2.595 | ||||||
The values highlighted in bold are statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Repeated-measures ANOVA also showed that the root biomass (P = 0.258), soil water content (P = 0.2888) and soil pH (P = 0.067) in the Bt and non-Bt maize fields were not significantly different, indicating that the Bt and non-Bt maize had similar amounts of root biomass and that they had no significant effect on soil environmental factors, that is, soil water content and soil pH.
Effects of maize type and soil environment factors on soil fauna
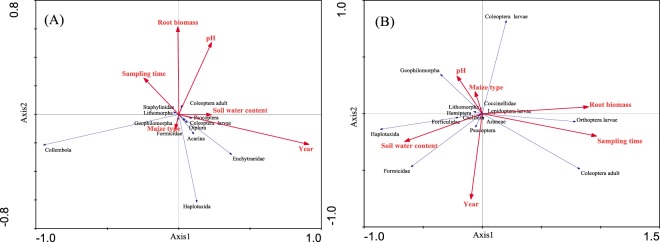
According to the Macfadyen and hand-sorting methods, year, sampling time and soil environmental variables together explained 25 and 32% of the total variability in the community composition of soil fauna collected according to the Macfadyen and hand-sorting methods, respectively, while maize type was not related to this variability (Table 3). For the Macfadyen method, axes 1 and 2 explained 49.7 and 75.0% of the variation in the species-environment relationship, respectively, with eigenvalues of 0.125 and 0.064 and species-environment correlations of 0.726 and 0.514, respectively (Fig. 2A). For the hand-sorting method, axes 1 and 2 explained 59.9 and 89.4% of variation in the species-environment relationship, respectively, with eigenvalues of 0.190 and 0.094 and species-environment correlations of 0.805 and 0.721, respectively (Fig. 2B).
Table 3.
Effects of maize type, sampling time, year and soil environment factors on the soil animals investigated according to the Macfadyen and hand-sorting methods in a Monte Carlo test of a RDA.
| Environmental factor | Macfadyen | Hand-sorting | ||||
|---|---|---|---|---|---|---|
| Variance explained (%) | F | P | Variance explained (%) | F | P | |
| Year | 11 | 7.17 | 0.0020** | 18 | 12.86 | 0.0020** |
| Sampling time | 5 | 3.46 | 0.0100** | 6 | 4.27 | 0.0020** |
| Root biomass | 5 | 3.44 | 0.0140* | 4 | 2.84 | 0.0080** |
| pH | 3 | 2.02 | 0.0660 | 3 | 2.17 | 0.0260* |
| Soil water content | 1 | 0.75 | 0.5700 | 1 | 0.52 | 0.8200 |
| Maize type | 0 | 0.32 | 0.9140 | 0 | 0.61 | 0.7500 |
| Total | 25 | 32 | ||||
The values highlighted in bold are statistically significant (*P < 0.05; **P < 0.01).
Figure 2.
Redundancy analysis (RDA) of the relationships among soil fauna community compositions and between soil fauna community compositions and environmental factors. (A) The soil fauna were collected according to the Macfadyen method; (B) the soil fauna were collected according to the hand-sorting method.
The relationship between soil fauna and maize type or other factors (year, sampling time, maize root biomass, soil water content and soil pH) was explored with the RDA method. The Monte Carlo test shows that when soil animal samples were collected according to the Macfadyen method, year (F = 7.17, P = 0.0020), sampling time (F = 3.46, P = 0.0100) and root biomass (F = 3.44, P = 0.0140) were significantly correlated with the composition of soil fauna. This result may have occurred because the different years and sampling times were associated with different soil temperatures and soil water contents, which influence the growth and development of soil fauna. In addition, root biomass is not only used as a type of food but also produces some substances that influence soil pH and the soil environment, so it also impacts the composition of soil fauna. When analysing the data from the hand-sorting method according to RDA, maize type did not impact the soil fauna, and soil water content (F = 0.52, P = 0.8200) was not significantly correlated with the composition of soil fauna. The soil fauna was highly related to soil pH (F = 2.17, P = 0.0260) when it was investigated according to the hand-sorting method, which was different from the result obtained according to the Macfadyen method. This may be because the hand-sorting method allows the capture of more soil animal species than the Macfadyen method (Table 3).
According to the Macfadyen method, the soil fauna of Collembola, Haplotaxida and Enchytraeidae were more easily affected by year, sampling time, maize root biomass, soil water content and pH than other soil animals. Among them, the number of Collembola was positively correlated with sampling time. This was due to Collembola reproduction increasing with increasing temperature. However, the number of Haplotaxida and Enchytraeidae were negatively correlated with sampling time, which might be due to changes in the soil water content (Fig. 2A). According to the hand-sorting method, the soil fauna of Coleoptera larvae, Geophilomorpha, Haplotaxida, Formicidae, Coleoptera and Orthoptera larvae were more easily affected by year, sampling time and environmental factors than other animals. Among them, Geophilomorpha, Haplotaxida, Formicidae, and Orthoptera larvae were all significantly negatively correlated with sampling time, while Coleoptera was significantly positively correlated with sampling time, which might have occurred because they have different suitable soil temperatures (Fig. 2B).
The similarity of soil fauna communities in Bt and non-Bt maize fields
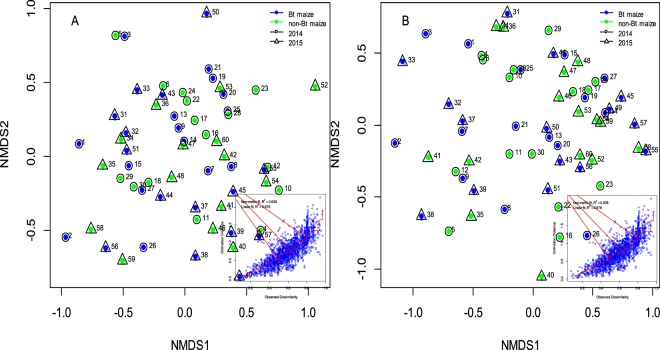
The soil fauna community structures in the Bt and non-Bt maize fields were further explored by nMDS. The distance between two sampling points was estimated using the pairwise Bray-Curtis similarity index. As shown in Fig. 3, the differences in the soil fauna composition among all animal samples were visualized with the nMDS plot, in which the samples were separated according to sampling time and year but not by maize type (Fig. 3A,B), which was substantiated by the more detailed analysis of similarity (ANOSIM). Significant correlations between soil fauna community composition and year or sampling time were found; however, no correlation between maize type and community composition was detected for soil fauna collected according to the Macfadyen and hand-sorting methods (Table 4).
Figure 3.
Non-metric multidimensional scaling (nMDS) plot of soil fauna community structure in different maize fields. Macfadyen method (A) and hand-sorting method (B) Circles with associated numbers from 1 to 30 indicate sampling points analysed in temporal order in 2014 (1–3: Bt maize at V0 stage, 4–6: non-Bt maize at V0 stage, 7–9: Bt maize at V3 stage, 10–12: non-Bt maize at V3 stage, 13–15: Bt maize at V6 stage, 16–18: non-Bt maize at V6 stage, 19–21: Bt maize at R1 stage, 22–24: non-Bt maize at R1 stage, 25–27: Bt maize at R6 stage, 28–30: non-Bt maize at R6 stage). Triangles with associated numbers from 31 to 60 indicate sampling points analysed in temporal order in 2015 (31–33: Bt maize at V0 stage, 34–36: non-Bt maize at V0 stage, 37–39: Bt maize at V3 stage, 40–42: non-Bt maize at V3 stage, 43–45: Bt maize at V6 stage, 46–48: non-Bt maize at V6 stage, 49–51: Bt maize at R1 stage, 52–54: non-Bt maize at R1 stage, 55–57: Bt maize at R6 stage, 58–60: non-Bt maize at R6 stage). The right side of the diagram represents Shepard’s stress plot.
Table 4.
Effects of maize type (Bt and non-Bt maize), sampling time and year on soil fauna community structure analysed according to the nMDS method.
| Correlation with nMDS structure | Macfadyen | Hand-sorting | ||
|---|---|---|---|---|
| R 2 | P | R 2 | P | |
| Sampling time | 0.29 | 0.001*** | 0.35 | 0.001*** |
| Year | 0.01 | 0.016* | 0.00 | 0.013* |
| Maize type | 0.00 | 0.953 | 0.00 | 0.601 |
The values highlighted in bold are statistically significant (*P < 0.05; ***P < 0.001).
Discussion
Transgenic crops can cause the retention of foreign gene expression products in soil through crop stubble, root exudates and pollen transmission38–41 and may cause changes in soil composition and content near the plant rhizosphere, which in turn affect the abundance and diversity of the soil fauna, ultimately posing a potential threat to the multiple functions of soil ecosystems42–44. At present, most related studies focus on the impacts on plants and surface animals, and only some specific orders, such as Collembola45–48, Haplotaxida28,29,49 and Enchytraeidae19,20,23. However, the overall structure and function of the soil fauna in the field are comprehensive.
In this study, soil fauna were investigated according to the Macfadyen and hand-sorting methods50–52. The combination of the two methods can comprehensively reflect changes in soil fauna. The two methods were successfully used to investigate the effects of different fertilization treatments on the soil fauna in the black soil area of Jilin Province, China52. In addition, the Shannon-Wiener index, Simpson’s index, Pielou’s evenness index, number of species and abundance have been successfully used to study the effects of transgenic crops on the arthropod community in the field35,36. Therefore, we used these five parameters to study the effects of transgenic cry1Ie maize IE09S034 on soil fauna. In this study, we analysed and compared the diversity of soil fauna in transgenic cry1Ie insect-resistant and non-transgenic control maize fields and concluded that the soil fauna were not significantly affected by the maize materials.
In recent years, RDA and nMDS analysis methods have been used for many types of ecological studies and evaluating the impact of transgenic crop cultivation on animal communities in the field53,54. This study also used the two methods to analyse the relationship between maize type and soil fauna composition and showed that transgenic maize did not have a significant impact on soil fauna.
Recently, as collembolans are a dominant animal group in crop fields and are sensitive to environmental changes55,56, they have been used as an indicator for evaluating the environmental safety of transgenic crop cultivation. For example, Mina45 et al. explored whether transgenic Bt cotton Mech162 endangered environmental safety by analysing its effects on the population structure of collembolans. Heckmann46 et al. found that transgenic Bt maize had no significant inhibitory effect on Protaphorura, and they concluded that the cultivation of Bt maize was not harmful to the environment. In addition, Chang47 et al. and Zhu48 et al. found that transgenic crops influenced the abundance of collembolans and concluded that the cultivation of transgenic crops was not safe for the environment. However, the effects of environmental factors on soil collembolans were not considered in the above studies. Collembolans are a dominant soil animal group, so they can be used as another indicator of the environmental safety of transgenic Bt maize. We considered the effects of environmental factors on collembolans through RDA and concluded that it is the environmental conditions (i.e., maize root biomass) but not maize type (transgenic or non-transgenic maize) that significantly affects the soil collembolan community.
According to the hand-sorting method, Haplotaxida, as a dominant group, was found to be an excellent index of the soil ecological environment due to its advantages in terms of promoting soil organic matter circulation and improving soil structure and fertility49. Some studies have shown that Haplotaxida can promote the decomposition of Bt proteins in soil and have no adverse effects on Haplotaxida themselves49. Zeilinger28 et al. only used four species of Haplotaxida to indicate whether transgenic cry1Ab and cry3Bb1 maize had effects on soil fauna. However, with the exception of the studied factor, the above studies related to Haplotaxida did not consider environmental effects. In this study, we concluded that Bt maize had no significant influence on the Haplotaxida community, which is more credible for the study considering the effects of environmental factors, such as maize root biomass and soil water content, identified through RDA.
As nMDS can be used to analyse the similarity of soil community structures57,58, Guo35,36 et al. used this method to study the effect of transgenic cry1Ie maize on the arthropod community in the field. In this study, we also used the nMDS ordination method to analyse the differences in soil fauna between transgenic and non-transgenic maize fields and illustrated that transgenic maize did not significantly affect the soil fauna58. This result was consistent with the RDA results. In this study, we used two sampling methods (Macfadyen and hand-sorting) and two analytical methods (RDA and nMDS) to explore whether transgenic cry1Ie maize (IE09S034) harmed the environment and concluded that the cultivation of transgenic cry1Ie insect-resistant maize did not influence the soil ecology. This result supports the popularization of transgenic cry1Ie maize (IE09S034) in spring maize production areas in China in the future.
Materials and Methods
Ethics statement
Field experiments were conducted using transgenic cry1Ie corn IE09S034 (insect resistant) in Jilin Province from 2013 to 2016, which was approved by the Ministry of Agriculture and Rural Affairs of China. In this study, no vertebrates were included, and none of the species are endangered or protected.
Materials and experimental design
Experimental materials
Bt maize (transgenic cry1Ie maize hybrid (IE09S034)) and near-isogenic non-Bt maize (Zong 31) were used in the study, which were provided by the Institute of Crop Sciences, CAAS. Field trials were conducted in 2014 and 2015 at the National Centre for Transgenic Plants Research and Commercialization/Jilin Academy of Agricultural Sciences in Gongzhuling (43°30′N, 124°49′E), Jilin Province, China. The soil at the experimental field is the typical black soil of northeast China, which contains 27.08 ± 0.07 g · kg−1 of organic matter, 77.54 ± 0.07 mg · kg−1 of alkaline nitrogen, 10.68 ± 0.07 mg · kg−1 of available phosphorus, and 154.10 ± 0.76 mg · kg−1 of available potassium. Before our experiments, only the non-GM hybrid Zhengdan 958 had been planted for the last three years. We sowed maize seeds on 7 May in 2014 and on 6 May in 2015. A randomized block design with three blocks was employed. Each plot was 10 m wide and 15 m long and contained 25 rows with 60-cm spacing. There were 40 plants in one row that were 25 cm apart. All plots were separated by 2 m bare borders. Both seeding and weeding were manually performed, and no chemical pesticides were applied during the entire growing period. Except for the lack of insecticide application, the maize was cultivated using the same agricultural management practices as normal.
Soil animal sampling
Macfadyen59 and hand-sorting60,61 methods were used to collect soil fauna before sowing (V0) and during four maize growing stages of the 3rd leaf (V3), elongation (V6), silking (R1), and physiological maturity (R6) stages in both years.
For the Macfadyen method, each time, we selected five sampling points from which to extract soil fauna from each plot. The five points were randomly selected every time in each plot. At each point, we collected soil samples from around the maize roots using a soil auger (15 cm in diameter, 10 cm in height). The soil samples were mixed, and 200 mL was collected with a measuring cylinder. Soil was collected within the maize row between two corn plants and approximately 12 cm away from each of the two plants. Each time, we took 30 soil samples in total from three replicates of the two maize types. The 200 mL sample was placed on a mesh with a mesh size of 20 on a Macfadyen extractor funnel, and the soil invertebrates were extracted for seven days at room temperature (25 °C). The soil invertebrates moved down through the funnel and dropped into a collection bottle below with 95% alcohol. The number and species of the collected animals were analysed at a magnification of 40 times with a microscope (Motic, China). Different soil animals were identified according to the keys of Yin62, Zhong63 and Zhang64. The numbers of soil animals collected five times at the five sites in one plot were summed for analysis.
In addition, we removed 5 g of soil from the 200 mL soil sample and dried it in an oven at 105 °C for the calculation of the actual water content.
Furthermore, 5 g of soil was mixed with an equal volume of 0.01 mol/L CaCl2, shaken for 5 minutes, and maintained under constant conditions for 2–24 h, and then the soil pH value was measured with a pH metre (Dynamica, UK).
To analyse the soil fauna in every plot with the hand-sorting method, only three of the above five sampling sites were randomly used because the sampling sites used in this method occupy a relatively large area and at each site two maize plants would be totally damaged each time. At each hand-sorting experimental site, we dug out a block of soil (0.5 m length × 0.5 m width × 0.5 m height) around the maize roots. The soil fauna were counted by the naked eye and collected with tweezers, the process of which was finished within 15 minutes. The total number of soil fauna in each plot was used as the soil fauna number per plot. The collected soil fauna were also stored in 95% ethanol for species identification and analysis. All roots of six maize plants in a plot destroyed by the hand-sorting method were collected, dried, weighed, and used as maize root biomass (g) per plot.
Statistical analysis
The changes in fauna occurrence, abundance, and diversity were analysed with the Data Processing System (DPS) version 2005 package (China)65. Three indices, the Shannon-Wiener index (H′), Simpson’s diversity index (D), and Pielou’s evenness index (J), were calculated as follows:
where Pi is the proportion of individuals belonging to the ith taxon in one plot66.
where Ni is the number of individuals in the ith taxon in one plot and N is the total number of individuals in one plot67.
where S is the number of faunal genera collected from one plot.
These three diversity indices were calculated for the Macfadyen and hand-sorting methods in 2014 and 2015 using the diversity index analysis module in the DPS.
Repeated-measures ANOVA (SPSS 23.0)68 was used to analyse the effects of every factor on soil fauna abundance and diversity, including maize type (Bt and non-Bt maize), year and sampling time. We also used repeated-measures ANOVA to analyse whether the environmental factors (maize root biomass, soil water content and soil pH) between Bt maize and non-Bt maize were different. For the above analysis, maize type (Bt and non-Bt maize), year and sampling time were included as fixed factors, and block and sampling points were considered as random factors.
A type of canonical analysis, redundancy analysis (RDA), was used to identify the factors influencing the soil fauna community by analysing the relationships among soil fauna and the relationships between the soil fauna and the soil environment69 and can be performed with Canoco and CanoDraw (Microcomputer Power, USA). Monte Carlo permutation tests (499 permutations) were performed to test the significance of the canonical axes of the RDA70,71. Additionally, an indirect ordination method, nonmetric multidimensional scaling (nMDS), was used to identify the effects of every influencing factor on the soil fauna community, which illustrated the similarity of the soil fauna samples through metric multidimensional scale analysis using the Bray-Curtis distance between sampling points57,58, and69. Analysis of similarity (ANOSIM) was used to calculate the distance between two sampling points according to the Bray-Curtis algorithm, and then all distances were sorted from small to large58. This analysis was conducted with the vegan package in R ver.3.2.3 (Auckland University, New Zealand)72–75.
Acknowledgements
We thank Professor Guoying Wang and Yunjun Liu from Institute of Crop Science, Chinese Academy of Agricultural Sciences, for providing all corn seeds. This project was supported by the Genetically Modified Organisms Breeding Major Project of China (2016ZX08011-003).
Author Contributions
Conceived and designed the experiments: C.F., F.W., B.W. and X.S. Performed the experiments: C.F., J.D. and J.Y. Analyzed the data: C.F. and X.S. Contributed materials: F.W., B.W., J.Y. and X.S. Wrote the paper: C.F. and X.S.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ishida Y, et al. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nature Biotechnology. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 2.James, C. Global status of commercialized biotech/GM crops: 2014. ISAAA Brief. 49 (ISAAA, Ithaca, NY, 2014).
- 3.Data gene of NY. ISAAA. Biological Technical Bulletin2, 102 (2015).
- 4.Wu, L. Global commercialization of GM crops for 20 years. Science news6 (2016).
- 5.International service for the acquisition of agri-biotech applications 2017 global trend of commercialization of biotechnology/GM crops. Chinese Biotechnology. 2018;38:1–8. [Google Scholar]
- 6.Song XY, et al. Strategy of environmental bio-safety assessment for transgenic plants. Journal of Biosafety. 2011;20:37–42. [Google Scholar]
- 7.Zuo J, et al. Progress on safety evaluation of transgenic corn. Maize Science. 2014;22:73–78. [Google Scholar]
- 8.Xu XL, et al. Field evaluation of effects of transgenic cry1Ab/cry1Ac, cry1C and cry2A rice on Cnaphalocrocis medinalis and its arthropod predators. Science China Life Sciences. 2011;54:1019–1028. doi: 10.1007/s11427-011-4234-2. [DOI] [PubMed] [Google Scholar]
- 9.Han Y, et al. Prey-mediated effects of transgenic cry2Aa, rice on the spider Hylyphantes graminicola, a generalist predator of nilapavarta lugens. Biocontrol. 2015;60:251–261. doi: 10.1007/s10526-014-9629-0. [DOI] [Google Scholar]
- 10.Xu, X. et al. Impacts of transgenic cry1Ab/cry1Ac rice Ganlü 1 on Cnaphalocrocis medinalis Guenée and Chilo suppressalis (walker) and their arthropod predators under field conditions. Journal of Plant Protection (2017).
- 11.Comas C, Lumbierres B, Pons X, Albajes R. No effects of Bacillus thuringiensis maize on non-target organisms in the field in southern europe: a meta-analysis of 26 arthropod taxa. Transgenic Research. 2014;23:135–143. doi: 10.1007/s11248-013-9737-0. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, et al. The cultivation of Bt corn producing Cry1Ac toxins does not adversely affect non-target arthropods. PloS One. 2014;9:e114228. doi: 10.1371/journal.pone.0114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei XD, et al. Field released transgenic papaya effect on soil microbial communities and enzyme activities. Journal of Environmental Sciences. 2006;18:734–740. [PubMed] [Google Scholar]
- 14.Griffiths BS, et al. A comparison of soil microbial community structure, protozoa and nematodes in field plots of conventional and genetically modified maize expressing the Bacillus thuringiensis, CryIAb toxin. Plant & Soil. 2005;275:135–146. doi: 10.1007/s11104-005-1093-2. [DOI] [Google Scholar]
- 15.Song YN, et al. Diversity of microbial community in a paddy soil with cry1Ac/cpti transgenic rice. Pedosphere. 2014;24:349–358. doi: 10.1016/S1002-0160(14)60021-7. [DOI] [Google Scholar]
- 16.Lu H, et al. Decomposition of Bt transgenic rice residues and response of soil microbial community in rapeseed–rice cropping system. Plant & Soil. 2010;336:279–290. doi: 10.1007/s11104-010-0476-1. [DOI] [Google Scholar]
- 17.Liu W, et al. Transgenic Bt rice does not affect enzyme activities and microbial composition in the rhizosphere during crop development. Soil Biology & Biochemistry. 2008;40:475–486. doi: 10.1016/j.soilbio.2007.09.017. [DOI] [Google Scholar]
- 18.Kapur M, et al. A case study for assessment of microbial community dynamics in genetically modified Bt cotton crop fields. Current Microbiology. 2010;61:118–124. doi: 10.1007/s00284-010-9585-6. [DOI] [PubMed] [Google Scholar]
- 19.Mohammad AA, et al. Effect of Bt corn for corn rootworm control on nontarget soil microarthropods and Nematodes. Environmental Entomology. 2003;32:859–865. doi: 10.1603/0046-225X-32.4.859. [DOI] [Google Scholar]
- 20.Saxena D, Stotzky G. Bacillus thuringiensis (Bt) toxin released from root exudates and biomass of Bt corn has no apparent effect on earthworms, nematodes, protozoa, bacteria, and fungi in soil. Soil Biol. Biochem. 2001;33:1225–1230. doi: 10.1016/S0038-0717(01)00027-X. [DOI] [Google Scholar]
- 21.Toschki A, Hothorn LA, ROß NM. Effects of cultivation of genetically modified Bt maize on epigeic arthropods (Araneae; Carabidae) Environmental Entomology. 2007;36:967–981. doi: 10.1603/0046-225X(2007)36[967:EOCOGM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Bhatti MA, et al. Field evaluation of the impact of corn rootworm (Coleoptera: Chrysomelidae) -protected Bt corn on ground-dwelling invertebrates. Environmental Entomology. 2005;34:1336–1345. doi: 10.1603/0046-225X(2005)034[1336:FEOTIO]2.0.CO;2. [DOI] [Google Scholar]
- 23.Neher DA, Muthumbi AW, Dively GP. Impact of coleopteran-active Bt corn on non-target nematode communities in soil and decomposing corn roots. Soil Biology and Biochemistry. 2014;76(Supplement C):127–135. doi: 10.1016/j.soilbio.2014.05.019. [DOI] [Google Scholar]
- 24.Höss S, et al. Effects of transgenic corn and Cry1Ab protein on the nematode, Caenorhabditis elegans. Ecotoxicology and Environmental Safety. 2008;70:334–340. doi: 10.1016/j.ecoenv.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Xu LN, et al. A proteomic approach to study the mechanism of tolerance to Bt toxins in Ostrinia furnacalis larvae selected for resistance to Cry1Ab. Transgenic Res. 2013;22:1155–1166. doi: 10.1007/s11248-013-9718-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang TT, et al. Inheritance patterns, dominance and cross-resistance of Cry1Ab-and Cry1Ac-selected Ostrinia furnacalis (Guenée) Toxins. 2014;6:2694–2707. doi: 10.3390/toxins6092694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L, et al. Cross-resistance of Cry1Ab selected Asian corn borer to other Cry toxins. Appl. Entomol. 2010;134:429–438. doi: 10.1111/j.1439-0418.2010.01517.x. [DOI] [Google Scholar]
- 28.Zeilinger AR, Andow DA, Zwahlen C, Stotzky G. Earthworm populations in a northern U.S. Cornbelt soil are not affected by long-term cultivation of Bt maize expressing Cry1Ab and Cry3Bb1 proteins. Soil Biology & Biochemistry. 2010;42:1284–1292. doi: 10.1016/j.soilbio.2010.04.004. [DOI] [Google Scholar]
- 29.Hönemann L, Nentwig W. Are survival and reproduction of Enchytraeus albidus (Annelida: Enchytraeidae) at risk by feeding on Bt-maize litter. European Journal of Soil Biology. 2009;45:351–355. doi: 10.1016/j.ejsobi.2009.03.001. [DOI] [Google Scholar]
- 30.Song FP, et al. Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl. Environ. Microbiol. 2003;69:5207–5211. doi: 10.1128/AEM.69.9.5207-5211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han HL, et al. Cross-resistance of Cry1Ac-selected Asian corn borer to other Bt toxins. Acta Phytophy. Sini. 2009;36:329–334. [Google Scholar]
- 32.He MX, et al. Selection for Cry1Ie resistance and cross-resistance of the selected strain to other Cry toxins in the Asian corn borer, Ostrinia furnacalis (Lepidoptera: Crambidae) Acta Phytophy. Sini. 2013;56:1135–1142. [Google Scholar]
- 33.Zhang YW, et al. Overexpression of a novel cry1Ie gene confers resistance to Cry1Ac-resistant cotton bollworm in transgenic lines of maize. Plant Cell Tiss. Org. 2013;115:151–158. doi: 10.1007/s11240-013-0348-5. [DOI] [Google Scholar]
- 34.Yang ZJ, et al. Studies on insect-resistant transgenic maize (Zea mays L.) harboring Bt cry1Ah and cry1Ie Genes. Journal of Agricultural Science and Technology. 2012;14:39–45. [Google Scholar]
- 35.Guo J, et al. Field trials to evaluate the effects of transgenic cry1Ie maize on the community characteristics of arthropod natural enemies. Scientific Reports. 2016;6:22102. doi: 10.1038/srep22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, et al. Effects of transgenic cry1Ie, maize on non-lepidopteran pest abundance, diversity and community composition. Transgenic Research. 2016;25:1–12. doi: 10.1007/s11248-015-9907-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G, Wan F, Lövei GL, Liu W, Guo J. Transmission of Bt toxin to the predator Propylaea japonica (Coleoptera: Coccinellidae) through its aphid prey feeding on transgenic Bt cotton. Environmental Entomology. 2006;35:143–150. doi: 10.1603/0046-225X-35.1.143. [DOI] [Google Scholar]
- 38.Gruber H, et al. Determination of insecticidal Cry1Ab protein in soil collected in the final growing seasons of a nine-year field trial of Bt-maize MON810. Transgenic Research. 2012;21:77–88. doi: 10.1007/s11248-011-9509-7. [DOI] [PubMed] [Google Scholar]
- 39.Olivergg K, et al. Constitutive expression of Cry proteins in roots and border cells of transgenic cotton. Euphytica. 2007;154:83–90. doi: 10.1007/s10681-006-9272-7. [DOI] [Google Scholar]
- 40.Baumgarte S, Tebbe CC. Field studies on the environmental fate of the Cry1Ab Bt-toxin produced by transgenic maize (MON810) and its effect on bacterial communities in the maize rhizosphere. Molecular Ecology. 2005;14:2539–2551. doi: 10.1111/j.1365-294X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 41.Saxena D, et al. Larvicidal Cry proteins from Bacillus thuringiensis are released in root exudates of transgenic B. thuringiensis corn, potato, and rice but not of B. thuringiensis canola, cotton, and tobacco. Plant Physiol Biochem. 2004;42:383–387. doi: 10.1016/j.plaphy.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Lu HH, et al. Soil microbial community responses to Bt transgenic rice residue decomposition in a paddy field. Journal of Soils & Sediments. 2010;10:1598–1605. doi: 10.1007/s11368-010-0264-9. [DOI] [Google Scholar]
- 43.Wei X, et al. Effects of transgenes insertion on pollen vigor and hybrid seed set of rice. Chinese Journal of Applied Ecology. 2005;16:115. [PubMed] [Google Scholar]
- 44.Hannula SE, Boer WD, Veen JAV. Do genetic modifications in crops affect soil fungi? a review. Biology & Fertility of Soils. 2014;50:433–446. doi: 10.1007/s00374-014-0895-x. [DOI] [Google Scholar]
- 45.Mina U, Chaudhary A, Kamra A. Effect of Bt cotton on enzymes activity and microorganisms in rhizosphere. Journal of Agricultural Science. 2011;3:1916–9752. doi: 10.5539/jas.v3n1p96. [DOI] [Google Scholar]
- 46.Heckmann LH, et al. Consequences for Protaphorura armata (Collembola: Onychiuridae) following exposure to genetically modified Bacillus thuringiensis (Bt) maize and non-Bt maize. Environmental Pollution. 2006;142:212–216. doi: 10.1016/j.envpol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Chang L, et al. Effect of transgenic cotton litter decomposition under elevated ozone concentration on soil Collembola. Chinese Journal of Aplied Entomology. 2014;51:1222–1229. [Google Scholar]
- 48.Zhu X, et al. Community structure and abundance dynamics of soil collembolans in transgenic Bt rice paddyfields. Acta Ecologica Sinica. 2012;32:3546–3554. doi: 10.5846/stxb201105170640. [DOI] [Google Scholar]
- 49.Schrader S, Münchenberg T, Baumgarte S, Tebbe CC. Earthworms of different functional groups affect the fate of the Bt-toxin Cry1Ab from transgenic maize in soil. European Journal of Soil Biology. 2008;44:283–289. doi: 10.1016/j.ejsobi.2008.04.003. [DOI] [Google Scholar]
- 50.Lu P, et al. Relationship between cropland soil arthropods community and soil properties in black soil area. Scientia Agricultura sinica. 2013;46:1848–1856. [Google Scholar]
- 51.Yang X, et al. Effect of straw-returning management on meso-micro soil fauna in a cultivated black soil area. Acta. Ecologica Sinica. 2017;37:2206–2216. doi: 10.1016/j.chnaes.2017.09.004. [DOI] [Google Scholar]
- 52.Lin YH, et al. Effect of long-term cultivation and fertilization on community diversity of cropland soil animals. Scientia Agricultura sinica. 2010;43:2261–2269. [Google Scholar]
- 53.Faith D, Minchin P, Belbin L. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio. 1987;69:57–68. doi: 10.1007/BF00038687. [DOI] [Google Scholar]
- 54.Lepš, J. & Šmilauer, P. Multivariate analysis of ecological Data Using CANOCO. (Cambridge University Press, Cambridge, UK, 2003).
- 55.Filser J. The role of Collembola in carbon and nitrogen cycling in soil. Pedobiologia. 2002;46:234–245. [Google Scholar]
- 56.Endlweber K, Scheu S. Effects of Collembola on root properties of two competing ruderal plant species. Soil Biol. Biochem. 2006;38:2025–2031. doi: 10.1016/j.soilbio.2006.01.004. [DOI] [Google Scholar]
- 57.Werley HH, et al. The NMDS: Abstraction tool for standardized, comparable, essential data[J] American Journal of Public Health. 1991;81:421–426. doi: 10.2105/AJPH.81.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 59.Macfadyen A. Improved funnel-type extractors for soil arthropods. Journal of Animal Ecology. 1961;30:171–184. doi: 10.2307/2120. [DOI] [Google Scholar]
- 60.Chen P. Methods of collection and investigation of soil animals. Chinese Journal of Ecology. 1983;3:46–51. [Google Scholar]
- 61.Cui, Z. D. Introduction of collection methods of soil animals. Chinese Journal of Zoology3 (1985).
- 62.Yin, W. Y. Chinese soil animal index map guide. Science Press (1998).
- 63.Zhong, J. M. Larval taxonomy. Agriculture press (1990).
- 64.Zhang, W. W. et al. A guide to the ecology of insects in China. Chongqing University Press (2011).
- 65.Tang, Q. Y. Dps data processing system: experimental design, statistical analysis and data mining. Science Press (2010). [DOI] [PubMed]
- 66.Shannon, C. E. & Weaver, W. The mathematical theory of communication. (University of Illinois Press, Chicago, IL, 1949).
- 67.Simpson EH. Measurement of diversit. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 68.George, D. & Mallery, P. SPSS for Windows step by step: A simple guide and reference. Computer Software, 357 (2003).
- 69.Jackson MM, Turner MG, Pearson SM, Ives AR. Seeing the forest and the trees: multilevel models reveal both species and community patterns. Ecosphere. 2012;3(art):79. [Google Scholar]
- 70.Rubinstein, R. Y. & Kroese, D. P. Simulation and the Monte Carlo method. Wiley Series in Probability and Mathematical Statistics. John Wiley & Sons, Inc., New York (1981).
- 71.Alday JG, Marrs RH. A simple test for alternative states in ecological restoration: the use of principal response curves. Appl. Veg. Sci. 2014;17:302–311. doi: 10.1111/avsc.12054. [DOI] [Google Scholar]
- 72.R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2015).
- 73.Li Y, et al. Consumption of Bt maize pollen containing cry1Ie does not negatively affect Propylea japonica (Thunberg) (Coleoptera: Coccinellidae) Toxins. 2017;9:108. doi: 10.3390/toxins9030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia HR, et al. The effects of Bt cry1Ie toxin on bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae) Scientific Reports. 2016;6:24664. doi: 10.1038/srep24664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pielou EC. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]