Abstract
Fine-tuning of flowering timing is crucial for plants to survive and leave offspring and depends on various endogenous and environmental factors. Here we report the identification of a vascular transcription factor, ANAC075, a putative regulator of VASCULAR-RELATED NAC-DOMAIN7 (VND7), as a negative regulator of flowering in Arabidopsis. Loss of function of ANAC075 causes the upregulation of floral integrator genes and early flowering under both long- and short-day conditions. ANAC075 promoter activity was detected in vascular tissues, including phloem. Previous reports suggested that ANAC075 is a transcriptional activator involved in the secondary cell wall formation, implying that the promotion of flowering time in anac075 mutants is caused by the disruption of flowering-time gene regulation in phloem and/or vascular tissue formation.
Keywords: flowering time regulation, NAC, secondary cell wall, transcription factor, vascular
Flowering-time regulation is controlled by various genetic, epigenetic, and environmental factors in plants. In the facultative long-day plant Arabidopsis, flowering-time regulation is achieved by interdependent pathways, including the photoperiod, autonomous, vernalization, and gibberellic acid (GA) pathways (Amasino and Michaels 2010; Berry and Dean 2015; Song et al. 2013). Various signals from these multiple pathways converge on the regulation of floral integrators, such as FLOWERING LOCUS T (FT; Kardailsky et al. 1999; Kobayashi et al. 1999), SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1; Lee et al. 2000; Samach et al. 2000), and TWIN SISTER OF FT (TSF; Kardailsky et al. 1999; Kobayashi et al. 1999; Michaels et al. 2005; Yamaguchi et al. 2005). These function as floral activators that induce the transition from vegetative to reproductive phase by regulating inflorescence identity genes. FT is a major component of florigen and acts as a mobile signal that is generated in the phloem of leaves and transported through the phloem to the shoot apical meristem, triggering the phase conversion (Corbesier et al. 2007; Takada and Goto 2003). FT transcription is directly activated by CONSTANS (CO), a B-box zinc finger transcription factor that accumulates particularly under long-day conditions (Samach et al. 2000; Song et al. 2012; Suárez-López et al. 2001; Valverde et al. 2004). The circadian regulation of CO expression and light-dependent regulation of CO stability and activity are keys to photoperiodic flowering. CO is expressed in vascular tissues of mature leaves and cotyledons as well as in the phloem and the protoxylem of stems. Weak expression has also been detected in the shoot apical meristem (An et al. 2004; Takada and Goto 2003).
TSF is the closest homolog of FT and its transcription is also activated by CO, in modes similar to those of FT (Yamaguchi et al. 2005). TSF and FT show similar expression profiles of diurnal oscillation and higher expression levels under long-day than those under short-day conditions (Yamaguchi et al. 2005). Interestingly, although overlapping roles between FT and TSF are suggested (Michaels et al. 2005; Yamaguchi et al. 2005), spatial expression patterns differ between these genes in young seedlings; FT is expressed in the phloem of leaves, whereas TSF is not expressed there but in the phloem of hypocotyls and near the shoot apical meristem (Takada and Goto 2003; Yamaguchi et al. 2005). TSF protein mobility and stability are lower than those of FT (Jin et al. 2015). These results suggest partially overlapping but distinct transcriptional regulation modes and roles of FT and TSF.
SOC1 encodes a MADS-box transcription factor and its transcription is activated by CO (Hepworth et al. 2002; Lee et al. 2000; Samach et al. 2000). FT also positively regulates SOC1 expression to promote flowering (Michaels et al. 2005; Yoo et al. 2005). The MADS-box transcription factors FLOWERING LOCUS C (FLC; Michaels and Amasino 1999; Sheldon et al. 1999) and SHORT VEGETATIVE PHASE (SVP; Hartmann et al. 2000), negative regulators of flowering, form a dimer that represses the transcription of floral activators (Fujiwara et al. 2008; Lee et al. 2007; Li et al. 2008; Mateos et al. 2015). FLC negatively regulates flowering by repressing the transcription of FT and TSF in the phloem and SOC1 in the shoot apical meristem (Hepworth et al. 2002; Lee et al. 2000; Michaels et al. 2005; Samach et al. 2000; Searle et al. 2006; Yamaguchi et al. 2005). SVP represses the expression of FT and TSF in leaves and SOC1 in the shoot apical meristem (Lee et al. 2007; Li et al. 2008; Jang et al. 2009).
Not only transcriptional regulation by transcription factors but also epigenetic regulation plays critical roles in flowering-time regulation. Among these, epigenetic FLC regulation is central to the vernalization-dependent flowering pathway (Berry and Dean 2015). LIKE HETEROCHROMATIN PROTEIN 1 (LHP1)/TERMINAL FLOWER 2 (TFL2), which is capable of binding H3K27me3 (Turck et al. 2007; Zhang et al. 2007a, b), binds to the FLC locus and maintains the repressed chromatin state (Mylne et al. 2006; Sung et al. 2006; Turck et al. 2007). LHP1 is expressed in the phloem tissues of leaves (Takada and Goto 2003). FT expression is also repressed by H3K27me3 and LHP1 (Jiang et al. 2008; Kotake et al. 2003; Turck et al. 2007; Zhang et al. 2007a, b).
Loss of function of JMJ14, a JmjC-domain containing histone demethylase, leads to early flowering under both long- and short-day conditions, and overexpression of JMJ14 (JMJ14-ox) leads to late flowering, indicating that JMJ14 functions as a floral repressor (Jeong et al. 2009; Lu et al. 2010; Ning et al. 2015; Searle et al. 2010; Yang et al. 2010). jmj14 under short days flowers considerably later than that under long days, suggesting that the mutants retain photoperiod sensitivity. This response is similar to that of mutants of FLC-dependent flowering pathways. JMJ14 is expressed in vascular tissues in young seedlings (Yang et al. 2010). The FLC level in jmj14 is not different from that in wild-type (WT), suggesting that JMJ14 functions independently of or downstream of FLC. Multiple floral integrator genes including FT and SOC1 show higher transcript levels in jmj14, suggesting that JMJ14 represses flowering by negatively regulating such gene expression. The mechanism by which JMJ14 represses flowering is controversial. Jeong et al. (2009) reported that JMJ14 directly demethylates H3K4, an active-mark modification, of the FT locus, but Lu et al. (2010) and Ning et al. (2015) indicated that FT locus is not the direct target of demethylation by JMJ14. JMJ14 is reported to mediate H3K4 demethylation of TSF loci as well (Yang et al. 2010). Interestingly, JMJ14 physically interacts with the NAC-family transcription factors ANAC050 and ANAC052 (Ning et al. 2015). Loss of function of ANAC050 and ANAC052 results in early flowering and overexpression of ANAC050 delays flowering, indicating that ANAC050 and ANAC052 function as floral repressors. More than half of the upregulated genes in jmj14 and ANAC050/ANAC052-RNAi plants overlapped, suggesting that JMJ14 and ANAC050/052 share common targets of repression.
Another JmjC-domain containing histone demethylase, EARLY FLOWERING 6 (ELF6), is also involved in flowering-time regulation as an H3K4 demethylase at the FT transcription initiation site (Jeong et al. 2009; Noh et al. 2004). Loss of function of ELF6 also causes early flowering under both long- and short-day conditions (Noh et al. 2004).
GA also affects flowering under both long- and short-day conditions. SOC1 and GAMYBs including MYB33 have been suggested to be involved in GA-mediated flowering promotion under short days (Achard et al. 2004; Gocal et al. 2001; Moon et al. 2003). MYB33 mRNA accumulates in the shoot apex by floral induction in response to GA treatment under short-day conditions or transition from short-day to long-day conditions (Gocal et al. 2001). Under long-day conditions, GA is required to increase the transcript levels of FT and TSF in vascular tissues (Porri et al. 2012). A recent report suggests that the SVP:FLC complex directly regulates GA metabolism-associated genes, also contributing to flowering-time regulation (Mateos et al. 2015). TEMPRANILLO (TEM) genes function as direct repressors of FT and GA4 biosynthetic genes GA3-oxidase1 and 2, suggesting that TEMs link the photoperiod and GA-dependent flowering pathways (Castillejo and Pelaz 2008; Osnato et al. 2012).
The factors described above and many others have been reported to be involved in flowering-time regulation. Many of them have been reported to be expressed and function in phloem, showing the importance of phloem for flowering-time regulation (Imaizumi and Kay 2006; Song et al. 2013).
In general, mature phloem accumulates secondary cell walls like those of vascular vessels and fiber cells in xylem, although it is difficult to observe the phloem secondary cell walls in Arabidopsis. The gene regulatory mechanism of secondary cell wall formation in xylem tissues has been well documented during the last decade. In particular, the important roles of NAC transcription factors have been clearly demonstrated. In vascular vessels, one specific clade of NAC transcription factors including VASCULAR-RELATED NAC-DOMAIN7 (VND7) have been shown to be master regulators of vascular vessel differentiation (Kubo et al. 2005). In xylem fiber cells, anther endothecium cells, and silique endodermal tissues, genes phylogenetically close to VND7, namely NAC SECONDARY CELL WALL THICKENING PROMOTING FACTOR1 (NST1), NST2, and NST3/SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), have been shown to encode master regulators of secondary cell wall formation (Mitsuda et al. 2005, 2007, 2008; Zhong et al. 2006). Overexpression of these transcription factors induced ectopic formation of secondary cell walls in various tissues (Kubo et al. 2005; Mitsuda et al. 2005, 2007; Zhong et al. 2006). Conversely, simultaneous loss of function of NST1 and NST3 induced loss of secondary cell walls in xylem fiber cells (Mitsuda et al. 2007; Zhong et al. 2007). In addition to these genes, the phylogenetically distinct NAC transcription factor ANAC075 has previously been characterized as an upstream regulator of VND7 (Endo et al. 2015). ANAC075 also restored the phenotype of an nst1 nst3 double mutant when it was expressed under the control of the NST3 promoter (Sakamoto and Mitsuda 2015). As expected, overexpression of ANAC075 induced ectopic secondary cell wall formation in various tissues (Endo et al. 2015; Sakamoto and Mitsuda 2015) through induced expression of VND7 (Endo et al. 2015). However, these experiments with ANAC075, using CaMV35S promoter and NST3 promoter, were somewhat artificial, so that the genuine function of ANAC075 in plants is still elusive.
In the present study, we identified ANAC075 as a negative regulator of flowering. Loss of function of ANAC075 caused early flowering under both long- and short-day conditions, associated with the upregulation of floral integrator genes. Possible roles and functional machineries of ANAC075 in flowering-time regulation are discussed.
Materials and methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used in this study were Columbia ecotype. anac075-1 (SALK_130117) and anac075-2 (SALK_132120) were obtained from the Arabidopsis Biological Resource Center. For flowering time analysis and expression analyses, seeds were soaked in water and kept in 4°C for 3–4 days and sown on soil. Plants were grown at 22°C under long-day (16-h light and 8-h dark) or short-day (8-h light and 16-h dark) conditions. For GUS assay, Arabidopsis T2 plants were grown on Murashige and Skoog medium containing 0.8% agar, 30 mg l−1 hygromycin, and 250 mg l−1 vancomycin.
Vector construction
For the construction of a promoter: GUS gene construct, a 2989-bp upstream region of ANAC075 coding region was amplified by polymerase chain reaction (PCR) with primer pairs shown in Supplementary Table 1. The amplified fragment was digested with AscI and BamHI and cloned into a pGUS_Ent vector (Mitsuda et al. 2007). The content of the resulting plasmid was then transferred to pBCKH T-DNA vector (Mitsuda et al. 2006) by Gateway LR clonase reaction (Thermo Fisher Scientific, MA, USA). ANAC075, ANAC052 or JMJ14 coding region without a stop codon was amplified by PCR from an Arabidopsis cDNA library fusing Gateway attB1 and attB2 sequences (Thermo Fisher Scientific) at the 5′ and 3′ ends, respectively, to prepare plasmids for constitutive expression of ANAC075-HA, ANAC052-HA, and JMJ14-FLAG. Primers used for PCR are shown in Supplementary Table 1. Each fragment was cloned into pDONR207 (Thermo Fisher Scientific) and introduced into the binary vectors pGWB414 (HA) and pGWB411 (FLAG, Nakagawa et al. 2007), respectively, using Gateway LR Clonase II (Thermo Fisher Scientific).
RNA extraction and qRT-PCR
RNA extraction and qRT-PCR were performed as described previously (Fujiwara et al. 2014a). Semi-quantitative RT-PCR was performed using the same cDNA used for the qRT-PCR. The gene-specific primers used for qRT-PCR and semi-quantitative RT-PCR are shown in Supplementary Table 1.
Immunoprecipitation assay
Transient protein co-expression in Nicotiana benthamiana and co-immunoprecipitation were performed as described previously (Fujiwara et al. 2014b) with some modifications as follows. α-DYKDDDDK tag antibody beads (Wako, Osaka, Japan) were used for immunoprecipitation. The blots were incubated with α-HA-HRP (1 : 1000; Wako) or α-DYKDDDDK-HRP (1 : 15,000; Wako).
Results
Loss of function of ANAC075 causes early flowering under both long- and short-day conditions
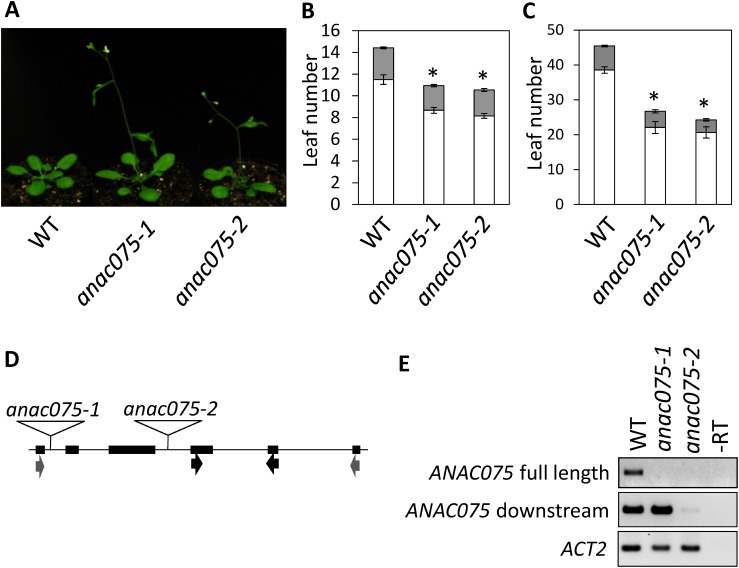
In an attempt to find novel transcription factors involved in flowering-time regulation, we found that a loss-of-function line of ANAC075 (At4g29230) flowered earlier than WT (Figure 1A–C). ANAC075 belongs to the NAC transcription factor family and has transactivation activity in plants (Sakamoto and Mitsuda 2015). These authors also reported that ANAC075 has the ability to restore the pendent stem phenotype of nst1 nst3 double loss-of-function plants, which lose the secondary cell wall when expressed by NST3 promoter and fused with VP16, suggesting that ANAC075 functions in secondary cell wall formation. However, its precise contribution to cell wall formation and function in flowering-time regulation has not been reported. Two independent T-DNA insertion lines SALK_130117 and SALK_132120, named as anac075-1 and anac075-2, respectively, showed significant shortening of the vegetative phase and flowered earlier than WT under both long- and short-day conditions (Figure 1A–D). The flowering phenotype of anac075-1 was slightly weaker than that of anac075-2 under short-day conditions (Figure 1C). To test the effect of T-DNA insertion on ANAC075 transcription in these two mutants, semi-quantitative RT-PCR analysis was performed using two primer sets located in the ANAC075 coding region (Figure 1D, E). For anac075-1, although no amplification was detected when primers were located upstream and downstream of the T-DNA, strong signal was detected when both primers were located downstream of the T-DNA. This result suggested that not the full-length but the 3′ region of ANAC075 was transcribed in this line and could be a leaky allele. No amplification was detected using both primer sets in anac075-2, suggesting that this is a null allele.
Figure 1. anac075 plants flowered early under long- and short-day conditions. (A) Representative WT, anac075-1 and anac075-2 plants grown under long-day conditions for 24 days. (B, C) Flowering time of WT, anac075-1, and anac075-2 plants under long- (B) and short-day conditions (C). Rosette (white bars) and cauline (gray bars) leaf numbers at the bolting stage were counted. n=12–20. Asterisks indicate significant differences compared with WT (p<0.05). (D) Positions of T-DNA insertions in the ANAC075 locus and primers used for semi-qRT-PCR. (E) Semi-qRT-PCR data using primers amplifying the full-length coding region (gray arrows shown in D) and partial fragment downstream of the T-DNA insertion sites (black arrows shown in D). ACTIN2 (ACT2) data is shown as an internal control.
Transcript levels of floral integrator genes were higher in anac075 mutants than those in WT
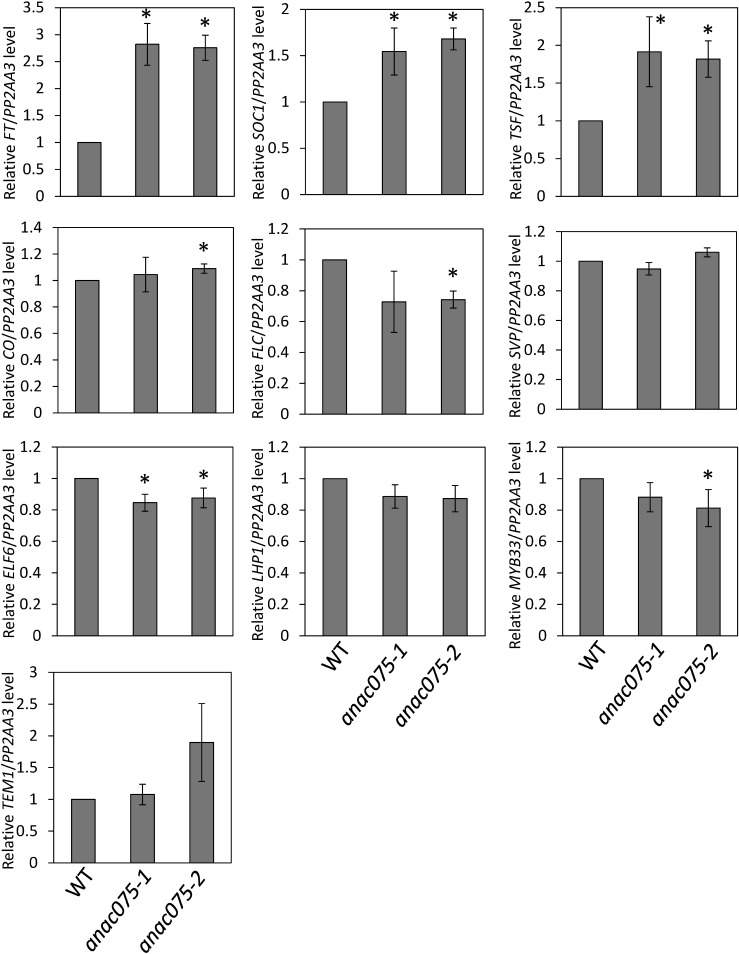
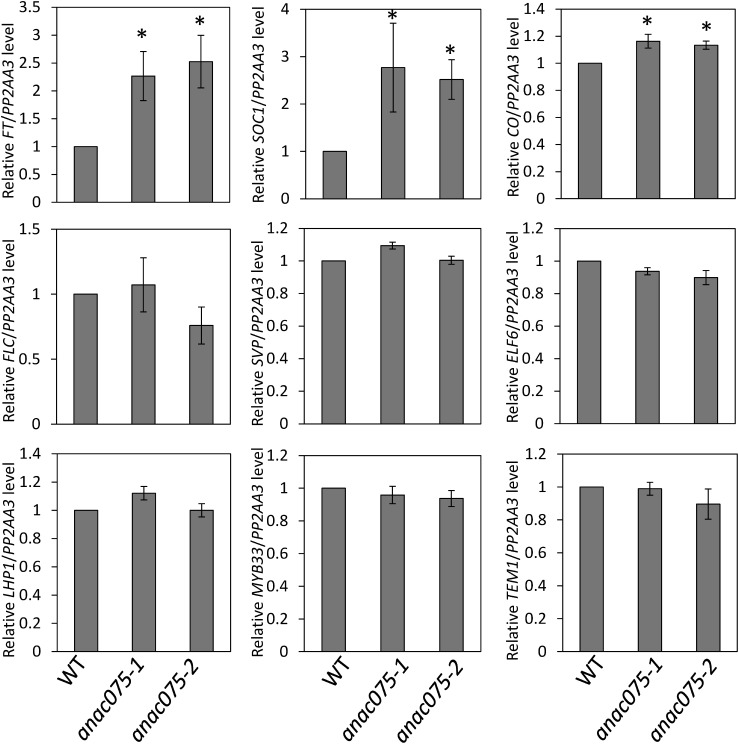
To identify the cause of the early flowering phenotype of anac075 mutants, we analyzed major flowering-time regulation-associated genes (Figures 2, 3). After cold treatment in water, WT, anac075-1, and anac075-2 seeds were sown on soil and grown under long- and short-day conditions. Aerial parts were harvested 7 and 14 days after sowing at ZT16 and ZT14 respectively, at times when WT plants showed high transcript levels of FT. The major floral integrator genes FT and SOC1 showed significantly higher transcript levels in anac075 mutants than in WT under both long- and short-day conditions, suggesting that the upregulation was the cause of the promotion of flowering. The upregulation of another floral integrator gene, TSF, was also detected under long-day conditions, whereas the expression level was too low to quantify under short-day conditions. This finding indicates that TSF upregulation also contributes to the early flowering phenotype of the anac075 mutants at least under long-day conditions. Given that the expression of these genes is known to be regulated by multiple floral regulation pathways, we also evaluated the transcript levels of major members of each pathway in which ANAC075 potentially functions. Under long-day conditions, transcript levels of FLC, ELF6, and MYB33 were slightly, but significantly, lower in the anac075 mutant than those in WT. In contrast, none of the genes tested showed significant downregulation in transcript level under short-day conditions. CO levels were slightly higher in the anac075 mutants under both long- and short-day conditions. These findings suggested that ANAC075 functions upstream of FT, SOC1, and TSF and that their upregulation may be partially caused by the up- and downregulation of upstream genes. However, it was not clear whether such small changes in the transcript levels of the potential upstream genes actually contribute to the upregulation of floral integrator genes and the strong early flowering phenotype. Taking these findings into account, we searched for other possible factors to explain the early flowering and differences in flowering time-associated genes in the anac075 mutants.
Figure 2. Transcript levels of flowering-time regulation-associated genes under long-day conditions. Transcript levels of flowering-time regulation-associated genes in WT, anac075-1 and anac075-2 were analyzed by qRT-PCR using aerial parts of seedlings grown under long-day conditions for 7 days and harvested at ZT16 (ZT: hours after the start of the light period) at the time at which the diurnal FT transcript level oscillation showed its maximum level in the WT. Mean values of four biological replicates are shown. Error bars represent±standard deviations. Asterisks indicate significant differences compared with WT (p<0.05).
Figure 3. Transcript levels of flowering-time regulation-associated genes under short-day conditions. Transcript levels of flowering-time regulation-associated genes in WT, anac075-1, and anac075-2 were analyzed by qRT-PCR using aerial parts of seedlings grown under short-day conditions for 14 days and harvested at ZT14 at the time at which diurnal FT transcript-level oscillation showed its maximum level in WT. Mean values of 4 biological replicates are shown. Error bars represent±standard deviations. Asterisks indicate significant differences compared with WT (p<0.05).
ANAC075 is expressed in the vascular tissues
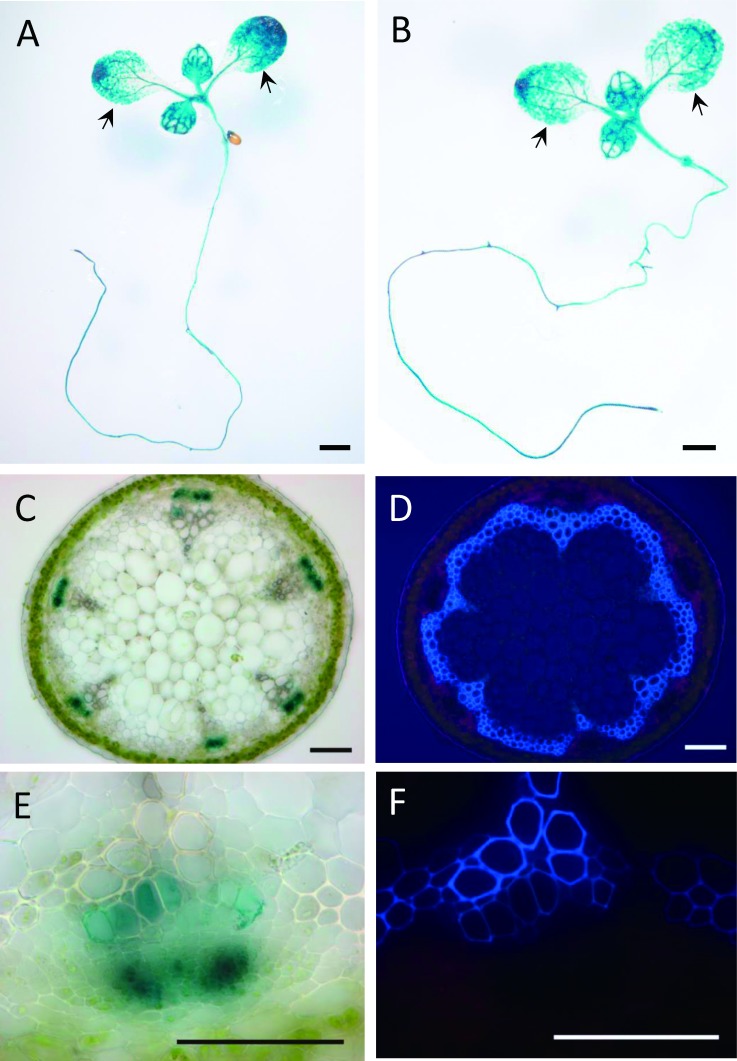
ANAC075 transcript levels are high in seeds, stems, and siliques (eFP browser; Winter et al. 2007). To further investigate the location of ANAC075 transcription, we generated transgenic plants expressing β-glucuronidase (GUS) driven by the ANAC075 promoter and performed histochemical analysis. Strong GUS signal was detected in the vascular tissue of leaves, cotyledons, hypocotyls, and roots in seven-day-old seedlings grown under long- and short-day conditions (Figure 4A, B). In adult plants, strong GUS staining was observed in the phloem of the stem, and a weak signal was also observed in xylem (Figure 4C–F). These observations suggest that ANAC075 is expressed in vascular tissues including phloem and xylem and that the early-flowering phenotype of anac075 mutants may be caused by the loss of ANAC075 function in the phloem.
Figure 4. ANAC075 is expressed in phloem and vessels. (A, B) GUS staining of 7-day-old ANAC075pro: GUS seedlings grown under long-day (A) and short-day (B) conditions. Black arrows indicate cotyledons. Bars represent 1 mm. (C–F) Cross section of inflorescence stem of ANAC075pro: GUS plant observed under normal light (C, E) and UV illumination (D, F) after GUS staining. Clear GUS staining was observed in phloem tissues in addition to newly differentiated vascular vessels. Blue autofluorescence in (D) and (F) indicates the presence of lignin. Bars represent 100 µm.
ANAC075 did not interact with a histone demethylase, JMJ14
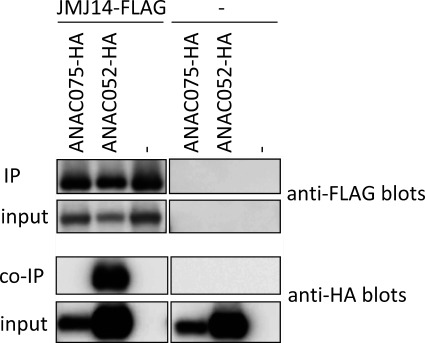
To further investigate the cause of the early-flowering phenotype of anac075, we searched for other potential factors that might be involved in it. Recent reports show that some NAC transcription factors are also involved in flowering-time regulation (Ning et al. 2015; Yoo et al. 2007). Among them, NAC050 and NAC052 were reported to function as negative regulators of flowering (Ning et al. 2015). Similarly to anac075 mutants, loss-of-function lines of NAC050/NAC052 flower early under both long- and short-day conditions. NAC050/052 physically associate with a histone demethylase, JMJ14, and activate the transcription of FLC, which indirectly represses FT expression. We hypothesized that ANAC075 also associates with JMJ14 and negatively regulates flowering by supporting the removal of H3K4me3 from the floral activator genes and/or disturbing the JMJ14 function on the floral repressor gene. A clear physical interaction between NAC052 and JMJ14 was detected by immunoprecipitation assay, as described above (Figure 5). However, interaction between JMJ14 and ANAC075 was not detected. This result suggests that the function of ANAC075 does not involve association with JMJ14 but with other unidentified factors.
Figure 5. Physical association between ANAC075 and JMJ14 was not detected. (A) FLAG-tagged JMJ14 (JMJ14-FLAG) was co-expressed with ANAC075-HA or ANAC052-HA in N. benthamiana leaves, and the protein complexes were immunoprecipitated using α-DYKDDDDK-tagged antibody beads. Inset, immunoprecipitated (IP) and co-immunoprecipitated (co-IP) samples were detected by α-DYKDDDDK or α-HA antibodies, respectively.
Discussion
We report that ANAC075 functions as a floral repressor that functions under both long- and short-day conditions. anac075 mutants flowered later under short-day than long-day conditions, suggesting that they still retain photoperiod sensitivity (Figure 1A–C) and that ANAC075 functions not only in the photoperiodic pathway but also in other pathways.
Although slight upregulation of CO and downregulation of FLC, ELF6, and MYB33 transcript levels were observed in anac075 mutants (Figures 2, 3), it was not clear whether such differences contributed to the clear early flowering phenotype and upregulation of floral integrator genes. It is possible that ANAC075 regulates transcription of a wide range of genes and that its loss of function causes early flowering as a result of a sum of multiple gene-expression changes.
ANAC075 itself showed transcriptional activation activity in a transient assay (Sakamoto and Mitsuda 2015), suggesting that ANAC075 functions as a transcriptional activator in the flowering-time regulation pathway. However, none of the flowering-time regulation-associated genes we tested showed clear reductions in their transcript levels in anac075 mutants, despite the clear early-flowering phenotype and upregulation of floral integrator genes (Figures 1–3). There are several possibilities for the interpretation of these results. One is that ANAC075 functions as a transcriptional activator and activates the function of genes that negatively regulate the expression of genes showing higher transcript levels in the anac075 mutant, such as FT, SOC1, and TSF. A second possibility is that ANAC075 actually functions as a transcriptional repressor, not an activator, in flowering-time regulation. ANAC075 may have an unknown transcriptional repression motif and/or represses expression of target genes by forming a protein complex with other proteins, such as transcriptional repressors and transcriptional corepressors. Indeed, differences in heterodimerization patterns of transcription factors change transcriptional activity and target genes in flowering-time regulation (Posé et al. 2012). It is also possible that ANAC075 affects the chromatin status of flowering-time regulation-associated genes, such as their histone modification status by associating with histone modifiers. JMJ14 was a possible partner of ANAC075 but we did not detect their association (Figure 5). Identification of proteins interacting with ANAC075 is a potential approach to elucidating the biological machinery of ANAC075 flowering repression.
ANAC075 shows high homology to SND2 and SND3 at the amino acid sequence level. Furthermore, their expression preferences are similar, according to public microarray databases. SND2 and SND3 are also suggested to be involved in secondary cell wall formation in xylem tissues (Hussey et al. 2011; Sakamoto and Mitsuda 2015; Zhong et al. 2008). In our previous study, fusion of SND2 and SND3 to the strong transcriptional activation domain VP16 but not SND2 and SND3 alone restored secondary cell wall formation in the nst1 nst3 double mutant when they were expressed by NST3 promoter like ANAC075 did (Sakamoto and Mitsuda 2015). In addition, constitutive expression of SND2-VP16 or SND3-VP16 induced ectopic formation of secondary cell walls as in ANAC075 (Sakamoto and Mitsuda 2015). Because SND2 and SND3 do not possess a long C-terminal part which ANAC075 does, it may be speculated that SND2 and SND3 are transcriptionally neutral factors that function as negative regulators, whereas ANAC075 appears to be a transcriptional activator in secondary cell wall formation. We accordingly propose that there is no gene redundancy between ANAC075 and SND2/3. In fact, their double and triple mutants did not show the enhancement of phenotype observed in anac075 mutants (data not shown).
It should be noted that anac075 mutants showed no recognizable phenotype associated with the secondary cell wall, suggesting that ANAC075 is not involved in secondary cell wall formation in plants. SOMBRERO, BEARSKIN1 (BRN1), and BRN2, which belong to the same clade as VND and NST transcription factors, are known to induce ectopic secondary cell wall formation when overexpressed, but their mutants show defects in root cap maturation and its detachment from root that are not associated with secondary cell walls (Bennett et al. 2010). ANAC075 could be another example to have a function not associated with secondary cell wall, even if its overexpression induces ectopic secondary cell wall formation. The long C-terminal region of ANAC075 may function not only as a transcriptional activation domain but also as a protein–protein interaction domain for flowering-time regulation. A search for proteins interacting with this region may lead to elucidation of the ANAC075 function and how it works. It is also possible that loss of function of ANAC075 affects phloem function, which affects the gene expression and trafficking of flowering-time regulation-associated proteins, RNA, and hormones. Precise analyses of spatial and temporal ANAC075 expression under both long- and short-day conditions will yield further information.
Acknowledgments
We thank Ms. Kanako Imokawa, Ms. Akiko Kuwazawa, Ms. Yuko Takiguchi, Ms. Yoshimi Sugimoto, and Ms. Miyoko Yamada for excellent technical assistance and other laboratory members for fruitful discussions. We also thank Prof. Tsuyoshi Nakagawa for kindly providing the pGWB414 and pGWB411 vectors and Arabidopsis Biological Research Center for providing anac075-1 (SALK_130117) and anac075-2 (SALK_132120) seeds. This work was partially supported by JSPS KAKENHI Grant number 26660296 to S.F.
Abbreviations
- ACT2
ACTIN2
- ANAC
Arabidopsis thaliana NAM/ATAF/CUC protein
- BRN
BEARSKIN
- CaMV
cauliflower mosaic virus
- CL
cauline leaf
- CO
CONSTANS
- ELF6
EARLY FLOWERING 6
- FLC
FLOWERING LOCUS C
- FT
FLOWERING LOCUS T
- GA
gibberellic acid
- GUS
β-glucuronidase
- IP
immunoprecipitation
- JMJ
JUMONJI
- LHP1
LIKE HETEROCHROMATIN PROTEIN 1
- NST
NAC SECONDARY CELL WALL THICKEING PROMOTING FACTOR
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse transcription PCR
- SND
SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CO 1
- SVP
SHORT VEGETATIVE PHASE
- TEM
TEMPRANILLO
- TFL2
TERMINAL FLOWER 2
- TSF
TWIN SISTER OF FT
- VND7
VASCULAR-RELATED NAC-DOMAIN7
- WT
wild-type
- ZT
zeitgeber time
Supplementary Data
References
- Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B (2010) SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22: 640–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Dean C (2015) Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J 83: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Endo H, Yamaguchi M, Tamura T, Nakano Y, Nishikubo N, Yoneda A, Kato K, Kubo M, Kajita S, Katayama Y et al. (2015) Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol 56: 242–254 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G et al. (2008) Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Kigoshi K, Mitsuda N, Suzuki K, Ohme-Takagi M (2014a) VP16 fusion efficiently reveals the function of transcriptional repressors in Arabidopsis. Plant Biotechnol 31: 123–132 [Google Scholar]
- Fujiwara S, Sakamoto S, Kigoshi K, Suzuki K, Ohme-Takagi M (2014b) VP16 fusion induces the multiple-knockout phenotype of redundant transcriptional repressors partly by Med25-independent mechanisms in Arabidopsis. FEBS Lett 588: 3665–3672 [DOI] [PubMed] [Google Scholar]
- Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, Li SF, Parish RW, Dennis ES, Weigel D et al. (2001) GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol 127: 1682–1693 [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA (2011) SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol 11: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Jeong JH, Song HR, Ko JH, Jeong YM, Kwon YE, Seol JH, Amasino RM, Noh B, Noh YS (2009) Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb Repressive Complex 2 Components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Jung HS, Chung KS, Lee JH, Ahn JH (2015) FLOWERING LOCUS T has higher protein mobility than TWIN SISTER OF FT. J Exp Bot 66: 6109–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 1286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Liu C, Cao X (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20: 387–390 [DOI] [PubMed] [Google Scholar]
- Mateos JL, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, Fornara F, Schneeberger K, Krajewski P, Coupland G (2015) Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol 16: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M (2006) Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J 4: 325–332 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M (2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56: 768–778 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Ning YQ, Ma ZY, Huang HW, Mo H, Zhao TT, Li L, Cai T, Chen S, Ma L, He XJ (2015) Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res 43: 1469–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Posé D, Yant L, Schmid M (2012) The end of innocence: Flowering networks explode in complexity. Curr Opin Plant Biol 15: 45–50 [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Mitsuda N (2015) Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol 2: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yang W, Jiang D, Jiang J, He Y (2010) A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62: 663–673 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE 2: e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007a) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE (2007b) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871 [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225: 1603–1611 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.