Abstract
Objective
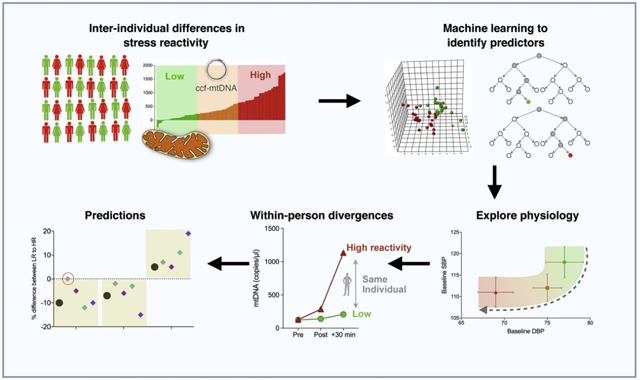
We have previously found that acute psychological stress may affect mitochondria and trigger an increase in serum mitochondrial DNA, known as circulating cell-free mtDNA (ccf-mtDNA). Similar to other stress reactivity measures, there are substantial unexplained inter-individual differences in the magnitude of ccf-mtDNA reactivity, as well as within-person differences across different occasions of testing. Here, we sought to identify psychological and physiological predictors of ccf-mtDNA reactivity using machine learning-based multivariate classifiers.
Method
We used data from serum ccf-mtDNA concentration measured pre- and post-stress in 46 healthy midlife adults tested on two separate occasions. To identify variables predicting the magnitude of ccf-mtDNA reactivity, two multivariate classification models, partial least-squares discriminant analysis (PLS-DA) and random forest (RF), were trained to discriminate between high and low ccf-mtDNA responders. Potential predictors used in the models included state variables such as physiological measures and affective states, and trait variables such as sex and personality measures. Variables identified across both models were considered to be predictors of ccf-mtDNA reactivity and selected for downstream analyses.
Results
Identified predictors were significantly enriched for state over trait measures (X2=7.03; p=0.008) and for physiological over psychological measures (X2=4.36; p=0.04). High responders were more likely to be male (X2=26.95; p<0.001) and differed from low-responders on baseline cardiovascular and autonomic measures, and on stress-induced reduction in fatigue (Cohen’s d=0.38-0.73). These group-level findings also accurately accounted for within-person differences in 90% of cases.
Conclusion
These results suggest that acute cardiovascular and psychological indices, rather than stable individual traits, predict stress-induced ccf-mtDNA reactivity. This work provides a proof-of-concept that machine learning approaches can be used to explore determinants of inter- and within-person differences in stress psychophysiology.
Keywords: psychological stress, mitochondria, ccf-mtDNA, stress reactivity, mitokine, machine learning
Graphical abstract
INTRODUCTION
Psychological stress triggers a coordinated multi-systemic stress response that increases cellular energy demand and involves mitochondrial recalibrations (Picard et al., 2018). While this stress response has evolved to promote adaptation and survival (Weiner, 1992), its chronic activation has been associated with increased disease risk (Cohen et al., 2007; Cohen et al., 2018; McEwen, 1998). To understand the basis for inter-individual differences in stress responses and the potential subsequent disease risk, efforts have been devoted to two main areas: i) mapping the biological consequences and cellular mechanisms of stress pathophysiology, and ii) identifying the psychobiological determinants at the origin of inter-individual and within-person differences.
Findings from pre-clinical and clinical studies suggest that psychological stress may affect the function and structural integrity of mitochondria (Cai et al., 2015; Liu and Zhou, 2012; Magariños et al., 1997), reviewed in (Picard and McEwen, 2018). Mitochondria are life sustaining organelles involved in energy production and intracellular signaling, and they contain their own genome – the circular mitochondrial DNA (mtDNA). Under some circumstances, the mtDNA can be released extracellularly, known as circulating cell-free mtDNA (ccf-mtDNA). Due to its bacterial origin, the mtDNA is immunogenic and triggers inflammation in model systems (West and Shadel, 2017; Zhang et al., 2010) – not unlike the pro-inflammatory state induced by acute psychological stress (Marsland et al., 2017). Physical stressors such as exercise, physical trauma (i.e., injury), and infection are also associated with both higher circulating levels of ccf-mtDNA and increased peripheral markers of inflammation (Boyapati et al., 2017; Nakahira et al., 2013; Stawski et al., 2017; Zhang et al., 2010), suggesting that ccf-mtDNA in humans is a physiologically meaningful signaling molecule.
Interestingly, in relation to psychological states, elevated ccf-mtDNA levels have also been described in cross sectional studies of psychiatric populations such as suicide attempters (Lindqvist et al., 2016) and patients with major depressive disorder (Lindqvist et al., 2018). Moreover, we recently showed that experimentally-induced acute psychological stress triggers a 2-3-fold increase in serum ccf-mtDNA within 30 minutes (Trumpff et al., 2018). Our findings are consistent with another recent study showing a rapid 1.7 fold increase in plasma ccf-mtDNA after induced psychological stress (Hummel et al., 2018), reinforcing initial evidence for a possible link between psychological states and ccf-mtDNA. The acute release of ccf-mtDNA, its ability to be transported through the blood, and its target effect via receptors on immune cells thus suggests a previously unrecognized “hormonal” function of ccf-mtDNA. However, little is known about what regulates its release.
To assess the factors contributing to ccf-mtDNA reactivity, classical statistical inference approaches (e.g., regression models) are not well-suited for the following reasons. First, we have no prior knowledge about the potential predictors of ccf-mtDNA reactivity and restricting our analyses to a specific set of variables would introduce substantial bias. Second, simultaneously considering multiple cardiovascular, autonomic, inflammatory, physiological, affective, psychosocial, and demographic variables introduces a problem of high-dimensionality where the number of variables approaches the sample size, for which inference-based statistical approaches are inadequate. In such case, independently testing each variable would also introduce the problem of multiple testing. Finally, our goal is to identify groups of variables (i.e., a pattern) capable of predicting ccf-mtDNA reactivity not only in this sample but also more generally in healthy women and men. But classical inference-based models are designed to maximize the fit to the current sample without regard to future generalizability. As such, traditional regression-based analyses would likely lead to data overfitting, with the natural potential drawback of limited replicability in future studies (for a review on this issue see (Yarkoni and Westfall, 2017)). In comparison, machine learning models are designed to find generalizable predictive patterns in the data (Bzdok et al., 2018). Machine learning is also an ideal data-driven approach to build prediction models from high-dimensionality datasets, which avoids fishing expeditions and P hacking (Simmons et al., 2011). In other fields such as the neurosciences, machine learning has successfully been used to identify predictive patterns of psychophysiological outcomes (Kragel et al., 2018). In the present study, we therefore opted for a machine learning approach to identify predictors of ccf-mtDNA reactivity.
To ensure the robustness of the identified predictors, two or more machine learning classifiers can be used, which leverages the overlap across models (Gromski et al., 2015). Two models in particular, partial least squares discriminant analysis (PLS-DA) (Wold et al., 2001) and Random Forest (RF) (Breiman, 2001) are mathematically distinct and represent complementary approaches that optimize variable selection when used conjointly (Menze et al., 2009). PLS-DA performs well to reduce high dimensional datasets, and RF is particularly good at eliminating irrelevant variables (Menze et al., 2009). Here, we therefore combined results of two machine learning-based classifiers, PLS-DA and RF, on data collected across two occasions of testing (Trumpff et al., 2018) to investigate psychological, behavioral, and physiological predictors of ccf-mtDNA reactivity induced by socio-evaluative stress in healthy adults.
METHOD
Participants
Data and samples of this study were obtained from the Vaccination and Immunity Project, a longitudinal study investigating the association of psychosocial, physiological and behavioral factors with antibody response to hepatitis B vaccination in a middle-age adult population (Carroll et al., 2011; Prather et al., 2009) and an analysis of the effects of stress reactivity on ccf-mtDNA levels in these samples (Trumpff et al., 2018). A total of 46 healthy middle-aged adult participants (28 men, 18 women, 88% Caucasian, 41-58 years old) were included in the present study. Participants having at least paired observations for task and +30 min at session 1 or session 2 (N=46) were included in the analyses, 64% completed both visits for a total of 74 observations. Informed consent was obtained in compliance with guidelines of the University of Pittsburgh Institutional Review Board.
Experimental stress procedure
The detailed study design is illustrated in Supplemental Information (Fig. S1) and has been previously reported (Carroll et al., 2011; Prather et al., 2009; Trumpff et al., 2018). In brief, participants attended two laboratory sessions scheduled 1 month apart. First, subjects completed a battery of questionnaires and their height and weight were measured to calculate body mass index (BMI; kg/m2). Then they were accompanied to a testing chamber where an intra-venous catheter was inserted into the antecubital fossa of one arm for the collection of blood samples. On the other arm, an occluding cuff was placed for automated measurement of heart rate (HR), systolic blood pressure (SBP) and diastolic blood pressure (DBP) (Critikon Dinamap 8100 Vital Signs Monitor, Tampa, FL). Participants were also fitted with a respiration strain gauge belt around the thorax and three electrocardiogram electrodes applied to each shoulder and the xiphoid process for the continuous assessment of respiration rate and heart rate, respectively. After instrumentation, participants rested for a 30-min period. Baseline blood pressure (BP) and HR were recorded every 90 s for the last 6 min of this rest period (4 readings). Then, 20 ml of blood was drawn and mood states (POMS) were assessed. Next, participants were asked to perform a public speaking task previously described in (Carroll et al., 2011; Prather et al., 2009; Trumpff et al., 2018). Mood states (POMS) were assessed and post-task blood samples were drawn immediately following completion of the task (20 ml) and again after the subject had rested quietly for 30 minutes (20 ml). At the second laboratory visit, the same procedure took place except that subjects were told that their “performance on the first speech task was slightly below average when compared with other participants’ speeches to avoid habituation. At both sessions, a similar increase in negative affect (anxiety, anger) and decrease in positive affect (calm, well-being) was observed (Trumpff et al., 2018). Physiological reactivity, including elevations in SBP, DBP, and HR were also similar across sessions (see Fig. S2).
EKG signals were digitized at a sampling rate of 1000Hz using CardioPro acquisition software (Thought Technology, Plattsburgh, NY). Time and frequency domain measures of heart rate variability (HRV) were continuously acquired. Time domain analyses provide the root mean of successive differences in interbeat intervals (RMSSD). Utilizing the Task Force guidelines (Force, 1996), spectral analyses was performed on the beat-to-beat intervals derived from the ECG data collection to obtain both low-frequency (LF) and high-frequency (HF) components using a Point Process statistical method (Weber et al., 1991).
Blood processing
Blood samples were allowed to clot, centrifuged at 1,000g for 10 minutes, and the serum was frozen at −80 °C until further processing. IL-6 levels were determined using a high sensitivity quantitative sandwich enzyme immunoassay kit (R & D Systems) as described previously (see (Carroll et al., 2011)) and log transformation was applied to normalize raw score distributions of the IL-6 values. Sample handling and processing have been described previously (Trumpff et al., 2018). In short, serum samples were centrifuged at 2,000 × g for 5 minutes, DNA was extracted from the supernatant, and circulating levels of mtDNA measured against a pooled standard curve by duplex quantitative real-time PCR (qPCR) with Taqman chemistry (Belmonte et al., 2016).
Salivary cortisol
Saliva samples were collected using oral swabs (Salimetrics) at the end of the baseline, task and recovery periods. Saliva samples were stored at −70 degrees Celsius and sent in batches to be analyzed by the Biopsychology Department at the Technical University of Dresden, Germany. Cortisol level was then determined using a time-resolved immunoassay with fluorometric end point detection (Dressendörfer et al., 1992).
Questionnaires
Mood states at baseline, task and +30 min were assessed using the Profile of Mood States (POMS) (McNair, 1971), with item selection based on Usala and Hertzog’s factor analysis of item loading (Usala and Hertzog, 1989). This questionnaire assesses current mood on a scale ranging from 0 (not at all) to 4 (extremely) and provides a score for anxiety (e.g., tense, nervous, uneasy, on edge), anger (e.g., angry, resentful, hostile), depression (e.g., sad, unhappy, depressed), calm (e.g., relaxed, comfortable, calm, at ease), fatigue (e.g., tired, sluggish, sleepy, fatigued, worn out), vigor (e.g., lively, energetic, full of pep), and well-being (e.g., cheerful, happy, pleased).
Depressive symptoms were assessed using the Beck depression inventory (BDI) (Beck et al., 1996) and state anxiety was assessed using the State-Trait Anxiety Inventory (STAI) (Spielberger et al., 2017). Perceived stress was measured using the Perceived Stress Scale (PSS) (Cohen et al., 1994). UCLA Loneliness Scale was used to measure loneliness (Russell et al., 1980). Negative and positive affect were measured using the Positive and Negative Affect Schedule (PANAS) scale (Watson et al., 1988). Personality measures included Goldberg’s Big-5 Factor Scales (Goldberg, 1992) and the Behavioral Inhibition Scale (BIS) (Carver and White, 1994). Participants completed a five items rumination measure assessing the extent of state rumination in response to the stressor, questions from the State Rumination Questionnaire – short form (Treynor et al., 2003) were adapted to ask about participant’s thoughts about the speech task.
The Paffenbarger physical activity index questionnaire was used to estimate physical activity level (usual kilocalories spent weekly) (Paffenbarger JR et al., 1978). Sleep quality was determined using the Pittsburgh Sleep Quality Index (PSQI) (Buysse et al., 1989). Participant’s age, gender, race and menopausal status were determined through self-report. The complete set of psychological and physiological variables tested is available as Supplementary Information.
Statistical analyses
Each participant visit (N=74) was treated as a unique observation. The change in ccf-mtDNA levels after stress was computed using the delta task to recovery (post-stress minus pre-stress). The distribution of response was then divided into tertiles to define low, medium, and high responder groups. Multivariate classification algorithms were trained to discriminate between the top tertile or “high responder”, and the bottom tertile “low responder”, using 56 physiological and psychological variables. Each variable was further classified as either trait or state to analyze the enrichment of specific types of variables in the final predictors.
Partial least squares discriminant analysis (PLS-DA) (Wold et al., 2001) and random forest (RF) (using 700 trees and 7 predictors) (Breiman, 2001) classification models were used to extract important variables predictive of group affiliation. Models were run using Metaboanalyst 3.0, an open source web software (Xia and Wishart, 2016). PLS-DA is a technique allowing the separation of two or more groups by relating two data matrices – X (the raw data used to separate the groups) and Y (the groups to be separated) by finding a linear subspace of the explanatory variables. This allows prediction of group affiliation (variable Y) based on a number of partial least square (PLS) components. The PLS components hereby describe the behavior of the groups as they span the mathematical subspace where the explanatory variables are projected (Wold et al., 2001). An advantage of this method is that it handles highly collinear and noisy datasets (Gromski et al., 2015). In comparison, RF is a machine learning algorithm that generates multiple decision trees to maximize the accurate grouping of individuals among groups. First, the total dataset is split into two groups: two-thirds of the data is used to generate a training set and one third to generate a test set. The procedure then uses bootstrapping of random sampling with replacement of individuals. The training set is used to build the decision trees, and the test set is used to define classification accuracy based on these trees (Breiman, 2001). An advantage of RF includes that it handles large datasets and is robust to over-fitting and outliers (Gromski et al., 2015).
The analytical plan was established prior to performing any analysis, including selection of the specific machine learning classifiers, focus on overlapping variables across models, and downstream univariate statistical comparisons only on overlapping variables. Participants with missing values for >20% of the variables were excluded (which included n=2 low responders and n=2 high responders). The variables predicting group affiliation were ranked by metrics reflecting the importance of each variable to predict group affiliation respectively - Variable of Importance Score (VIP) of the first component for PLS-DA; Mean Decrease Accuracy for RF. The top 15 variables were compared and only overlapping variables across both classifiers were identified as “predictors” and selected for further analysis. As a sensitivity analysis, to verify that our reactivity findings were not biased by baseline levels, we repeated the same analysis using baseline-adjusted delta variables and found similar results. For the continuous variables, group differences between low and high responders in the selected predictors were statistically assessed using non-parametric Mann-Whitney test for independent samples (missing values were kept as missing) and effect sizes were calculated as Hedges' g. The strength of the association between the selected predictors was assessed with Spearman’s rho. Group differences in categorical variables was tested using Chi Square. Continuous predictors were plotted in relation to each other using 2D scatterplots of the mean (± SEM) to visualize the extent of separation of low, medium and high responders and dose-response effects between predictors and ccf-mtDNA reactivity categories.
Finally, in a subset of participants classified as low responders at one session and high responders at the other (n=4), we tested if the identified predictors at the group level could “predict” when a given person would have a low or high ccf-mtDNA response. The percent difference in each predictor between the low and high responder groups (e.g., 5% higher heart rate in high responders than low responders) was compared to the percent difference between the “low” and “high” session for each person. Then we calculated the proportion of cases where group-level predictors correctly matched (i.e., “generalized”) within-person divergent responses across the two sessions. Additionally, we repeated the main analysis while excluding the four divergent responders from the total sample to ensure that the divergent responders were not driving the results of the main group-level analysis. Statistical analyses were performed using SAS statistical software 9.3 (SAS Institute Inc., Cary, NC, USA), SPSS (version 24) and Prism 7.0 (Graphpad).
Functional classification and enrichment analysis
Before any of the analyses, each variable was functionally classified as either a trait or state measure, and as either a physiological or psychological measure (all variables and their classification terms are listed in the Supplementary Information). We then performed an enrichment analysis to determine whether some categories of variables were over-represented in the final set of identified predictors. This enrichment analysis is a similar approach to those used on high dimensional omics data to define over- or under-representation of specific biological pathways (Huang et al., 2008). A Chi Square test was performed to compare the proportion of trait or state variables between the identified predictors compared to the total list of 56 variables initially used as input to the models.
RESULTS
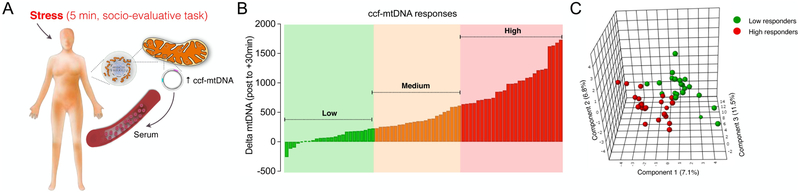
We previously showed that a brief socio-evaluative stress elevates serum ccf-mtDNA (Fig. 1A) (Trumpff et al., 2018), but noted that the magnitude of ccf-mtDNA response to stress varied substantially across participants. Here, we first separated the distribution of pre- to post-stress change in ccf-mtDNA (delta) into tertiles of responses (Fig. 1B). The bottom and top tertiles represent low and high responders, respectively. We then compared groups over a comprehensive and unbiased set of psychophysiological measurements, self-reported psychosocial measures, and demographic factors, for a total of 56 variables. The distribution of these variables is illustrated in Figure 2.
Fig. 1. Circulating cell-free mitochondrial DNA (ccf-mtDNA) in response to induced psychological stress and operationalization of low and high responders.
(A) Socio-evaluative stress induces a 2-3 fold elevation in serum ccf-mtDNA 30 minutes after stress. (B) Distribution of the ccf-mtDNA reactivity for all participants across both sessions (n=74 total visits). The distribution of ccf-mtDNA response was divided into tertiles to define low, medium, and high responder groups. (C) A partial least squares discriminant analysis (PLS-DA) model using 56 physio-psychological trait and state variables available in the study produces partial separation of the low and high responders, as shown in the 3D plot of the first three PLS-DA components. Each datapoint is a participant at one of the two visits.
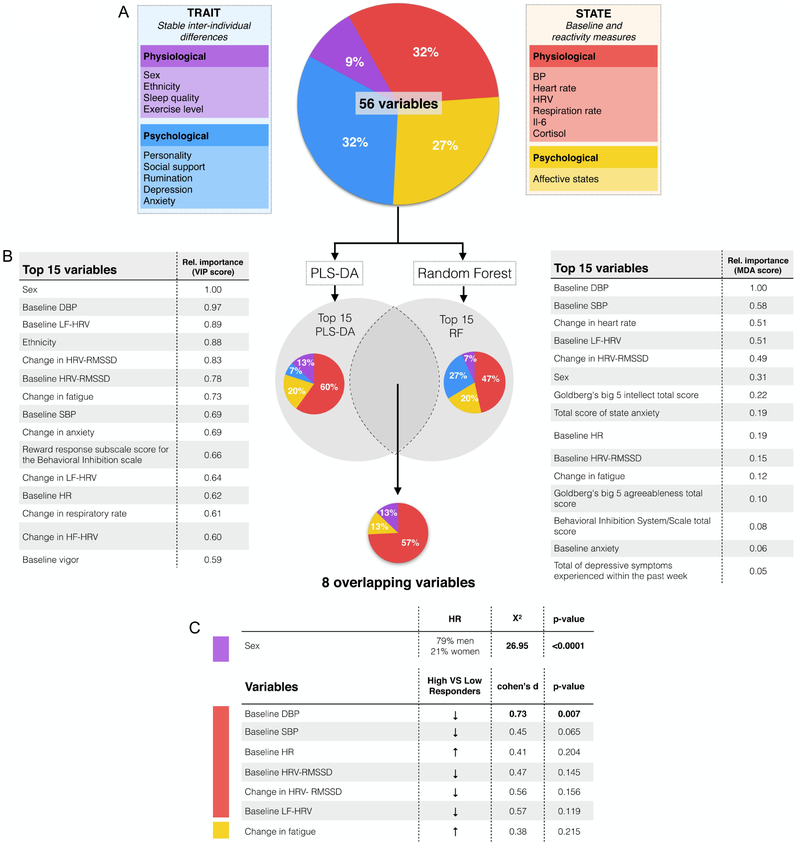
Fig. 2. Identifying predictors of ccf-mtDNA response to acute psychological stress using multivariate classification algorithms.
(A) 56 total variables a priori classified as stable trait and variable state were used. Partial least squares discriminant analysis (PLS-DA) and random forest (RF) classification models were trained to distinguish between low and high ccf-mtDNA responders. (B) Following our pre-established analytic plan, we identified the top 15 variables predicting group affiliation ranked by variable importance in projection (VIP) score for the first component in PLS-DA (left), and mean decrease accuracy (MDA) score from the RF model (right). (C) Overlapping variables across both classifiers. Metrics for group differences are shown for comparative purposes only (not for statistical inference), using non parametric independent samples Mann-Whitney test for the continuous variables. Chi square was performed for the categorical variable (sex). Effect sizes were calculated as Cohen’s d. Significant (p<0.05) results are in shown bold, n = 46 visits across low and high reactivity tertiles. Abbreviations: BP: Blood Pressure, DBP: Diastolic blood pressure, SBP: systolic blood pressure, HF-HRV: High frequency heart rate variability, HR: Heart rate, HRV: Heart rate variability, LF-HRV: low frequency heart rate variability, RMSSD: root mean squared successive differences, RF: Random Forest.
Common predictors of ccf-mtDNA responses identified by PLS-DA and RF classifiers
As illustrated by the physical separation of low and high responders in Fig. 1C, the PLS-DA model yielded a moderate level of discrimination between groups. This demonstrated that information contained within the 56 variables is sufficient to partially discriminate between individuals who release low or high levels of ccf-mtDNA upon psychological stress. The prediction accuracy metrics for PLS-DA were: accuracy = 0.62, R2 = 0.50, Q2 = −0.10 and for RF out of bag error = 0.478, reflecting relatively poor performance characteristics by each model separately. Therefore, the top 15 predictors from each model were ranked by their ability to discriminate between the groups and compared (Fig. 2B). The convergence of both models yielded a reduced set of 8 overlapping measures identified as predictors for subsequent analyses (Fig. 2C).
Physiological state characteristics are over-represented among predictors
The identified predictors included 1 trait and 7 state variables, representing a significant enrichment or over-representation for state characteristics (X2=7.03; p=0.008). This suggests that ccf-mtDNA reactivity may depend more on the current state of the person at the time of stress, rather than on stable individual traits. Moreover, 6 of the 7 state variables were classified as physiological, representing a significant enrichment for physiological over psychological characteristics (X2=4.36; p=0.04).
Among low responders, 60% were women and 40% were men. In contrast, among high responders only 21% were women and 79% were of men (X2=26.95; p<0.0001), establishing sex as the only trait predictor. Physiologically, high responders were characterized by lower baseline DBP (d=0.73 compared to low responders) and SBP (d=0.45), and higher HR (d=0.41). High responders also showed a lower baseline heart rate variability (HRV) measured as the root mean squared of successive differences (HRV-RMSSD, d= 0.47) and a lower baseline low frequency HRV (LF-HRV, d= 0.57). Whereas stress produced the expected reduction in HRV-RMSSD, this reduction was blunted in high responders (d=0.56). Finally, the only finding related to psychological factors showed that the stress-induced reduction in fatigue was more substantial in high responders than low responders (d=0.38). Inter-correlation between selected predictors as continuous variables are reported in Table 1. In sensitivity analyses (data not shown) where we repeated the same analysis with baseline-adjusted delta values for HR, BP, HRV and POMS, the delta HRV-RMSSD was selected by RF but not by PLS-DA, indicating that “change in HRV-RMSSD” may be a less robust predictor of ccf-mtDNA reactivity than baseline measures of cardiovascular function and change in fatigue. Given that sex was one of the predictors of ccf-mtDNA reactivity, we assessed if the selected predictors (n = 7) showed sex differences and found that baseline DBP was lower in men than in women, but other predictors did not show sex differences (see Table S1).
Table 1.
Inter-correlation between predictors of ccf-mtDNA reactivity identified by PLS-DA and RF classifiers
| Baseline DBP | Baseline SBP | Baseline heart rate | Baseline HRV-RMSSD |
Change in HRV-RMSSD |
Baseline LF- HRV | Change in fatigue | |
|---|---|---|---|---|---|---|---|
| Baseline DBP | 1.00 | ||||||
| Baseline SBP | 0.77** | 1.00 | |||||
| Baseline heart rate | 0.15* | 0.06 | 1.00 | ||||
| Baseline HRV-RMSSD | −0.09 | −0.10 | −0.53** | 1.00 | |||
| Change in HRV-RMSSD | 0.07 | 0.07 | 0.22** | −0.48** | 1.00 | ||
| Baseline LF-HRV | −0.067 | −0.13* | −0.30** | 0.54** | −0.45** | 1.00 | |
| Change in fatigue | −0.01 | −0.05 | 0.03 | 0.01 | 0.02 | −0.13* | 1.0 |
Values are Spearman’s rho coefficient. Significant results are in bold.
P < 0.05,
P < 0.01 level (2-tailed).
Abbreviations: SBP: systolic blood pressure, DBP: diastolic blood pressure, HRV: heart rate variability, LF-HRV: low frequency HR, HRV-RMSSD: HRV measured as root mean square of successive differences.
Exploratory analysis of stress induced ccf-mtDNA and changes in affect
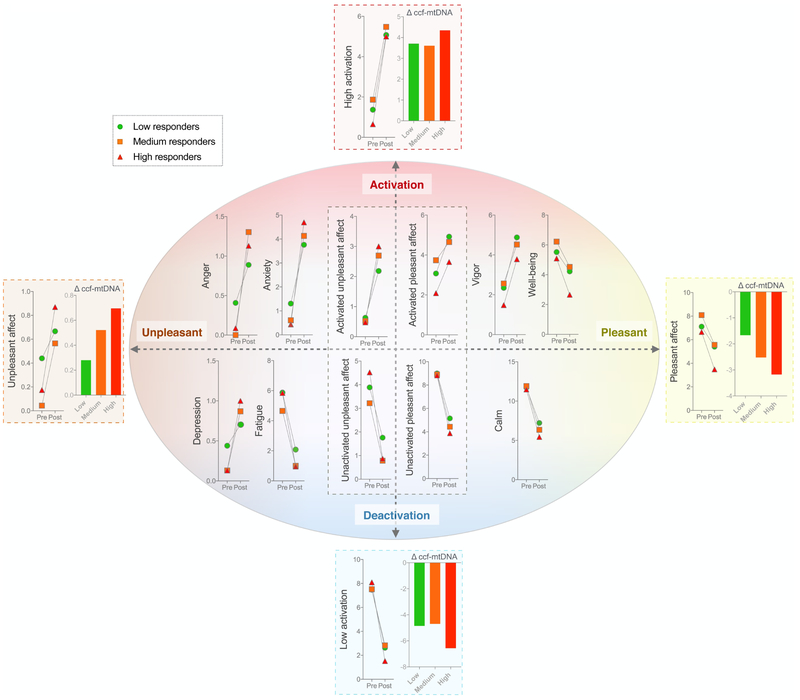
Because reduction in fatigue contributed to the discrimination of high and low responders, we conducted an exploratory analysis on the stress-induced changes in the valence and activation levels of different emotions. Using the circumplex model (Russell, 1980), changes in affective states were quantified using both individual POMS items and summary scores (McNair, 1971; Usala and Hertzog, 1989). Group differences in stress-induced changes in affective states between low, medium, and high responders were then assessed to explore potential dose-response relationships (Fig. 3). In 75% of cases, the high ccf-mtDNA responders tended to show greater changes in valence and activation of affects than the low responder group. For all summary scores, the changes in emotions were also greater among high ccf-mtDNA responders, providing preliminary evidence linking both emotional activation and valence to ccf-mtDNA reactivity.
Fig. 3. Stress-induced changes in affect valence and activation stratified by low, medium high ccf-mtDNA responders.
Timecourse of affective response before (pre) and after (post) the stressor for the low, medium and high ccf-mtDNA responders arranged according to the circumplex model of emotion. The histograms show the reactivity (delta, post to pre) quantified for each tertile. Affect ratings were obtained using the profile of mood states (POMS) instrument, which assesses immediate mood states. Shown at the four poles are composite indices for low/high activation, and pleasant/unpleasant emotions. The central box also includes composite scores. Note the opposite direction of effects along the activation and valence axes. (n=74 visits)
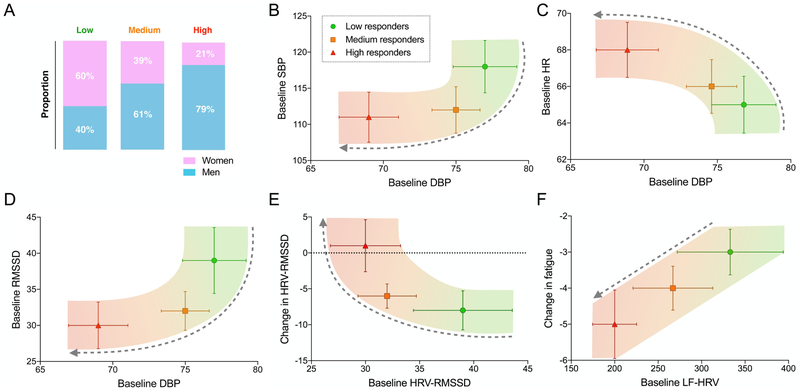
Predictors show dose-response progression from low, medium to high ccf-mtDNA responders
Next, predictors were plotted in pairs to visualize to what extent each predictor is able to separate low, medium and high responders (Fig. 4B-F). This showed that the averages for individual predictors among the low, medium and high responders groups occupied separate graphical spaces with minimal overlap in the confidence intervals. In all cases, predictors also exhibited gradual progression (linear or curvilinear) across low, medium and high responders, indicative of a dose-response association with ccf-mtDNA release. For example, Fig. 4B shows that the combination of low systolic and diastolic blood pressure strongly separates high responders from medium and low responders. The combination of baseline LF-HRV and change in fatigue (Fig. 3F) also creates a linear continuum that separates groups according to their ccf-mtDNA reactivity levels, suggesting that specific patterns of psychophysiological function are linked to mtDNA release.
Fig. 4. Visualizing predictors of ccf-mtDNA reveal dose-response patterns.
(A) Proportion of women and men stratified by low, medium, and high ccf-mtDNA responder groups. (B) Scatterplot of mean ± standard error of the mean (SEM) by tertiles of ccf-mtDNA reactivity for baseline SBP and DBP, (C) baseline DBP and HR, (D) baseline SBP and HRV-RMSSD, (E) baseline and change HRV-RMSSD, and (F) change in LF-HRV and change in fatigue. Dotted arrows show progression from low to high responders. (n = 74 total visits) Abbreviations: DBP: diastolic blood pressure, SBP: systolic blood pressure, LF-HRV: low frequency heart rate variability, HR: Heart rate, HRV: Heart rate variability, RMSSD: root mean squared successive differences.
Predictors account for within-individual divergent ccf-mtDNA reactivity profiles
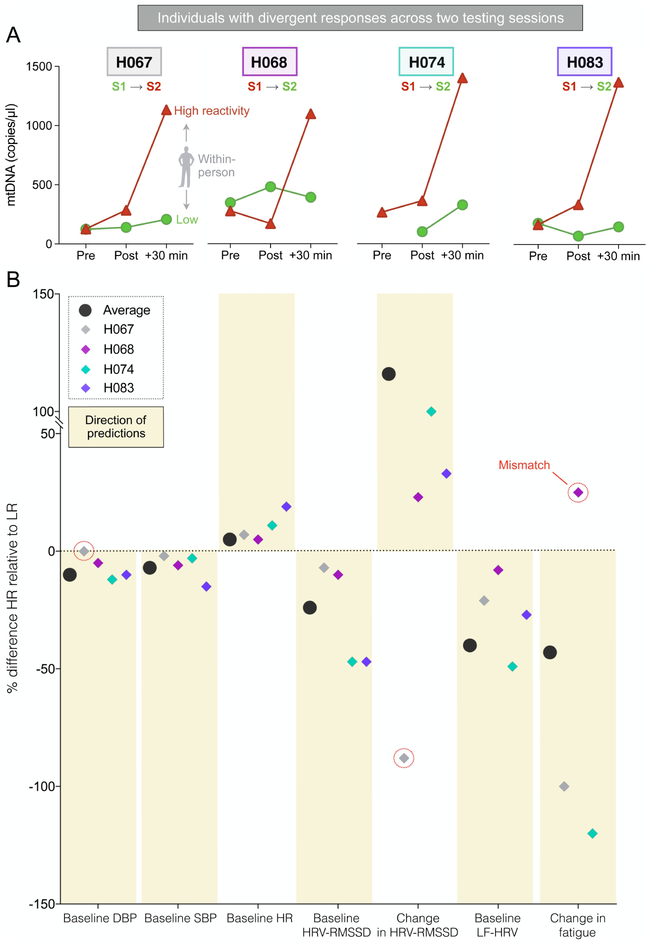
Since this study included two independent sessions, we had the opportunity to examine the within-person stability of these responses and their predictors. Of the 32 participants who completed both sessions, 4 (all males) were divergent responders, having been classified as low responders (bottom tertile) at one session and high responders (top tertile) at the other (Fig. 5A). Therefore, we reasoned that to the extent that the classification models and group differences shown in Figure 4 are robust and generalizable, the information about each variable should be sufficient to predict ccf-mtDNA release within a single individual. In other words, if the group-level differences detected by our algorithms are true predictors of ccf-mtDNA release, they should “generalize” and predict whether a given individual exposed to socio-evaluative stress on different occasions will exhibit a small or large ccf-mtDNA response.
Fig. 5. Individual profiles of divergent ccf-mtDNA responders and comparison with group-based predictions.
(A) Four participants (all males) exhibited low (green) and high (red) ccf-mtDNA responses on both occasions of testing and were thus identified as divergent responders. Shown are ccf-mtDNA levels before (Pre), immediately after (Post) and 30 min after (+30 min) the stressor for each participant. (B) Summary indicating the difference for each predictor between high and low ccf-mtDNA reactivity. Group-based findings for each predictor (black circle) establish the predicted direction of effects, shown as yellow boxes above or below the dotted line. For example, high responders are characterized by low baseline DBP and SBP, indicated by the black circles and boxes below 0. For each of the four divergent responders, the difference in each predictor between low and high reactivity sessions was plotted to assess the match in relation to group-based predictions. Individual data points going in the opposite direction than the group prediction (mismatch, 3 out of 28) are circled in red. Overall concordance between group-based predictions and individual divergent responses is 90%, compared to chance level (50%). Abbreviations: DBP: diastolic blood pressure, SBP: systolic blood pressure, LF-HRV: low frequency heart rate variability, HR: heart rate, HRV: heart rate variability, RMSSD: root mean squared successive differences.
To test this hypothesis, first, we calculated the direction of the change between low and high responder groups: participants released more ccf-mtDNA when they had lower baseline DBP, SBP, HRV-RMSSD, LF-HRV (negative percentage); and they released more ccf-mtDNA when they had higher baseline HR, lower stress-induced reduction of HRV-RMSSD, and higher decrease in fatigue (positive percentages). We then evaluated whether these patterns were conserved among each divergent responder for each predictor, for a total of 28 observations (Fig. 4B). Compared to chance level (50% accuracy), in 90% of cases the within-person change from low to high response sessions matched the predicted change based on the group findings. In particular, baseline SBP, HR, RMSSD and LF-HRV predicted the ccf-mtDNA response 100% of the time, suggesting their robustness. Since trait variables are stable within each person but state variables can change over time, these results further illustrated that ccf-mtDNA reactivity may depend more on the temporary states of the individual rather on stable traits.
DISCUSSION
Acute psychological stress may affect mitochondria and trigger an increase in ccf-mtDNA but substantial unexplained inter-individual differences exist. Here, we sought to identify predictors of ccf-mtDNA reactivity using machine learning-based multivariate classifiers. Using these converging methods, which handle high-dimensional datasets and generate more generalizable findings than traditional inference-based regression models, we have found that serum ccf-mtDNA reactivity is associated mostly with acute state measures of psychological and cardiovascular function.
It is established that physiological and emotional response to psychological stress shows considerable variability both across individuals and within the same individual tested on different occasions. Numerous factors are proposed to contribute to this response variability, including individual physiological and psychological traits and states. The magnitude of cardiovascular, immune and emotional responses to psychological stress vary by stable trait characteristics such as sex (Kajantie and Phillips, 2006), ethnicity (Busse et al., 2017), history of trauma (Carpenter et al., 2007), and other psychological (Chida and Hamer, 2008) and cognitive trait measures (Gaab et al., 2005), as well as behavioral factors such physical activity (Gröpel et al., 2018; Puterman et al., 2018) or sleep (Vargas and Lopez-Duran, 2017). To some extent, the magnitude of stress responses is correlated over time (Cohen et al., 2000), supporting the existence of underlying dispositional characteristics accounting for inter-individual differences. In addition to trait characteristics, a portion of the variance in stress reactivity is also attributable to the physiological and emotional state of the person at the time of testing. Manipulation of the individual’s physiological or psychological state prior to stress induction, using aerobic physical activity (Zschucke et al., 2015) or self-esteem training (Creswell et al., 2005; Sherman et al., 2009), can substantially attenuate neuroendocrine and cardiovascular responses to psychological challenge. These results are in line with our findings demonstrating that both trait and predominantly state variables account for the magnitude of stress reactivity.
Interestingly, we found that high ccf-mtDNA responders were ~4 times more likely to be men than women, suggesting a modulation of ccf-mtDNA reactivity by sex or gender. This is consistent with evidence of sex difference in stress reactivity, mitochondria biology, or metabolism. Previous work has shown that sex difference in response to psychological stress exist but the directionality of the findings are mixed, with men sometimes exhibiting stronger cortisol and cardiovascular reactivity (Chan et al., 2017; Goel et al., 2014; Juster et al., 2016). There are also sex differences in fundamental aspects of mitochondria biology such as respiratory capacity, reactive oxygen species production, or sensitivity to permeability transition, generally in the direction of increased mitochondrial vulnerability in males compared to females (Ventura-Clapier et al., 2017). Adult men and women also show significant differences in the circulating levels of mitochondria-related metabolites related to lipid synthesis and oxidation (Mittelstrass et al., 2011), and some mitochondrial disorders show sexual dimorphism with men being more susceptible than women (Rahman et al., 1996; Van Erven et al., 1987). Although more work is needed to confirm these sex differences in ccf-mtDNA reactivity, we speculate that greater vulnerability of male mitochondria could predispose them to the effects of acute stressors.
In relation to cardiovascular predictors, the high responder group had significantly lower baseline DBP and lower SBP (112/71 mmHg) than the low responder group (114/76 mmHg), although this difference is modest. The high responders also had a higher baseline HR and showed a greater decrease in fatigue after stress, indicating higher task activation. Regarding HRV, the high responders had lower baseline for both measures, LF-HRV and vagally-mediated HRV-RMSSD. The higher responder group also exhibited a reduced change in HRV-RMSSD following stress exposure, which, along with reduced basal HRV, paints a picture of lower cardio-vascular flexibility associated with ccf-mtDNA reactivity. At this point, the mechanistic connection between autonomic regulation and ccf-mtDNA, or mitochondrial function in general, remains unexplored in humans. But both reduced basal HRV and reduced change in stress-induced HRV have been associated with adverse health outcomes (Kemp et al., 2017), providing a potential future avenue of research to understand the cause and the downstream effects of stress-induced ccf-mtDNA levels.
Except for sex, all predictors identified were “state” variables, demonstrating that stress-induced ccf-mtDNA reactivity may vary within an individual over time. This prompted us to test whether the identified predictors of inter-individual variability between low to high ccf-mtDNA reactors were useful to predict divergent responders within-person. We found that group-based predictions matched individual divergent responses in the vast majority of the cases. This suggests that our approach was successful at isolating predictors of inter-individual variability, and that these generalize, albeit within the same sample, to within-person differences. Thus, although not definitive, this provides proof-of-concept evidence that machine learning approaches, such as multivariate classification algorithms using a heterogenous combination of psychological, behavioral, and physiological data can be useful to define features that distinguish experimental or naturally-occurring groups of individuals.
Classical inference approaches are well-suited to identify associations between variables in a given sample but have limited ability to predict future behavior (Bzdok et al., 2018). While up to now mostly used in the context of the “omics” fields such as metabolomics and genomics, and in computational neurosciences (Kragel et al., 2018), machine learning approaches and predictive analytics have demonstrated their utility in a variety of fields including human physiology and behavior (Yarkoni and Westfall, 2017). In general, results in the psychological and psychosocial sciences have been criticized for generating findings with particularly low replicability potential. This may be due to a combination of low effect sizes, overfitting, or the tendency for statistical models to take specific noise within a sample as signal. Machine learning does not solve these problems but minimizes their impact on the outcome. The present study illustrates how these approaches may contribute to may contribute to develop predictive models in psychoneuroendocrinology.
Some limitations of the current study must be considered. Although machine learning approaches handle more effectively a high variable-to-individual ratio than traditional inference-based models, the small sample size of this study remains a limitation. To address this issue we leveraged converging evidence of two distinct machine learning algorithms, which helped to refine and identify predictors (Gromski et al., 2015; Menze et al., 2009). Related points of limitation are the low performance metrics of each model individually most likely due to the low sample size, our inability to validate our model’s prediction accuracy on an entirely independent sample to provide definite estimates of the predictive accuracy for each predictor. Moreover, among the range of potential variables included here we identified a substantially greater number of physiological than psychological measures. This may reflect true physiological effects whereby baseline cardiovascular physiology is more closely linked to the mechanism underlying ccf-mtDNA release, as indicated above. Alternatively, objective cardiovascular indices have greater measurement precision relative to self-reported affective states, which have inherently more measurement error. Thus, we cannot rule out the possibility that the preferential enrichment of physiological measures among ccf-mtDNA predictors is in part due to selection bias introduced by measurement error that would particularly diminish the usefulness or strength of self-reported psychological measures among the classifiers.
To conclude, we provide a proof-of-concept that machine learning approaches can be used to explore the determinants of inter-individual and within-person differences in stress psychophysiology. In the context of the current study, our findings indicate that acute cardiovascular and psychological indices, rather than stable individual traits, are potential predictors of stress-induced serum ccf-mtDNA reactivity. This highlights a previously unappreciated degree of dynamic regulation of stress on mitochondria in humans, is consistent with the idea that the mitochondrial genome is a signalling molecule (i.e. mitokine), and points to potential links between psychological states, cardiovascular regulation, and ccf-mtDNA. Future investigations of mitochondrial psychobiology are needed to resolve the upstream mechanisms leading to ccf-mtDNA release and to understand the downstream short- and long-term health effects.
Supplementary Material
Highights.
Stress-included ccf-mtDNA levels increase shows substantial inter-individual variability
Within-person difference in ccf-mtDNA are observed across occasions of testing
Machine learning approaches can be used to identify predictors of ccf-mtDNA reactivity
Acute cardiovascular and psychological indices predict stress-induced ccf-mtDNA reactivity
Acknowledgments
Source of Funding
Support for this work was provided by NIH grants NR08237 (ALM), GM110424 (BAK), GM119793, MH113011, and the Wharton Fund (MP).
Acronyms
- BDI
Beck depression inventory
- BIS
Behavioral inhibition scale
- BMI
Body mass index
- BP
Blood pressure
- ccf-mtDNA
circulating cell free mitochondrial DNA
- DBP
Diastolic blood pressure
- HF
High frequency
- HR
Heart rate
- HRV
Heart rate variability
- LF
Low frequency
- mtDNA
Mitochondrial DNA
- PANAS
Positive and negative affect schedule
- POMS
Profile of mood states
- PLS
Partial least square
- PLS-DA
Partial least squares discriminant analysis
- PSS
Perceived stress scale
- PSQI
Pittsburgh sleep quality index
- qPCR
Quantitative real-time polymerase chain reaction
- RF
Random forest
- RMSSD
Root mean squared successive differences
- SBP
Systolic blood pressure
- STAI
State-trait anxiety inventory
- VIP
Variable of importance score
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: BAK declares significant financial interest in GSK unrelated to the current study. All the other authors declare no conflict of interest.
References
- Beck AT, Steer RA, Brown GK, 1996. Beck depression inventory-II. San Antonio: 78, 490–498. [Google Scholar]
- Belmonte FR, Martin JL, Frescura K, Damas J, Pereira F, Tarnopolsky MA, Kaufman BA, 2016. Digital PCR methods improve detection sensitivity and measurement precision of low abundance mtDNA deletions. Scientific reports 6, 25186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyapati RK, Tamborska A, Dorward DA, Ho G-T, 2017. Advances in the understanding of mitochondrial DNA as a pathogenic factor in inflammatory diseases. F1000Research 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, 2001. Random forests. Machine learning 45, 5–32. [Google Scholar]
- Busse D, Yim IS, Campos B, 2017. Social context matters: Ethnicity, discrimination and stress reactivity. Psychoneuroendocrinology 83, 187–193. [DOI] [PubMed] [Google Scholar]
- Buysse DJ, Reynolds CF III, Monk TH, Berman SR, Kupfer DJ, 1989. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry research 28, 193–213. [DOI] [PubMed] [Google Scholar]
- Bzdok D, Altman N, Krzywinski M, 2018. Points of significance: statistics versus machine learning. Nature Methods, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N, Chang S, Li Y, Li Q, Hu J, Liang J, Song L, Kretzschmar W, Gan X, Nicod J, 2015. Molecular signatures of major depression. Current Biology 25, 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter LL, Carvalho JP, Tyrka AR, Wier LM, Mello AF, Mello MF, Anderson GM, Wilkinson CW, Price LH, 2007. Decreased adrenocorticotropic hormone and cortisol responses to stress in healthy adults reporting significant childhood maltreatment. Biological psychiatry 62, 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JE, Low CA, Prather AA, Cohen S, Fury JM, Ross DC, Marsland AL, 2011. Negative affective responses to a speech task predict changes in interleukin (IL)-6. Brain, behavior, and immunity 25, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver CS, White TL, 1994. Behavioral inhibition, behavioral activation, and affective responses to impending reward and punishment: the BIS/BAS scales. Journal of personality and social psychology 67, 319. [Google Scholar]
- Chan JC, Houghton AB, Bale TL, 2017. Strained in Planning Your Mouse Background? Using the HPA Stress Axis as a Biological Readout for Backcrossing Strategies. Neuropsychopharmacology 42, 1749–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida Y, Hamer M, 2008. Chronic psychosocial factors and acute physiological responses to laboratory-induced stress in healthy populations: a quantitative review of 30 years of investigations. Psychological bulletin 134, 829. [DOI] [PubMed] [Google Scholar]
- Cohen S, Hamrick NM, Rodriguez MS, Feldman PJ, Rabin BS, Manuck SB, 2000. The stability of and intercorrelations among cardiovascular, immune, endocrine, and psychological reactivity. Annals of Behavioral Medicine 22, 171–179. [DOI] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, Miller GE, 2007. Psychological stress and disease. Jama 298, 1685–1687. [DOI] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermelstein R, 1994. Perceived stress scale. Measuring stress: A guide for health and social scientists, 235–283. [Google Scholar]
- Cohen S, Murphy ML, Prather AA, 2018. Ten Surprising Facts About Stressful Life Events and Disease Risk. Annual review of psychology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creswell JD, Welch WT, Taylor SE, Sherman DK, Gruenewald TL, Mann T, 2005. Affirmation of personal values buffers neuroendocrine and psychological stress responses. Psychological Science 16, 846–851. [DOI] [PubMed] [Google Scholar]
- Dressendörfer R, Kirschbaum C, Rohde W, Stahl F, Strasburger C, 1992. Synthesis of a cortisol-biotin conjugate and evaluation as a tracer in an immunoassay for salivary cortisol measurement. The Journal of steroid biochemistry and molecular biology 43, 683–692. [DOI] [PubMed] [Google Scholar]
- Force T, 1996. Heart rate variability: standards of measurements, physiological interpretation and clinical use. Circulation 93, 1043–1065. [PubMed] [Google Scholar]
- Gaab J, Rohleder N, Nater UM, Ehlert U, 2005. Psychological determinants of the cortisol stress response: the role of anticipatory cognitive appraisal. Psychoneuroendocrinology 30, 599–610. [DOI] [PubMed] [Google Scholar]
- Goel N, Workman JL, Lee TT, Innala L, Viau V, 2014. Sex differences in the HPA axis. Comprehensive Physiology 4, 1121–1155. [DOI] [PubMed] [Google Scholar]
- Goldberg LR, 1992. The development of markers for the Big-Five factor structure. Psychological assessment 4, 26. [Google Scholar]
- Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, Goodacre R, 2015. A tutorial review: Metabolomics and partial least squares-discriminant analysis-a marriage of convenience or a shotgun wedding. Analytica chimica acta 879, 10–23. [DOI] [PubMed] [Google Scholar]
- Gröpel P, Urner M, Pruessner JC, Quirin M, 2018. Endurance-and Resistance-Trained Men Exhibit Lower Cardiovascular Responses to Psychosocial Stress Than Untrained Men. Frontiers in psychology 9, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44. [DOI] [PubMed] [Google Scholar]
- Hummel E, Hessas E, Müller S, Beiter T, Fisch M, Eibl A, Wolf O, Giebel B, Platen P, Kumsta R, 2018. Cell-free DNA release under psychosocial and physical stress conditions. Translational Psychiatry 8, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juster R-P, Raymond C, Desrochers AB, Bourdon O, Durand N, Wan N, Pruessner JC, Lupien SJ, 2016. Sex hormones adjust “sex-specific” reactive and diurnal cortisol profiles. Psychoneuroendocrinology 63, 282–290. [DOI] [PubMed] [Google Scholar]
- Kajantie E, Phillips DI, 2006. The effects of sex and hormonal status on the physiological response to acute psychosocial stress. Psychoneuroendocrinology 31, 151–178. [DOI] [PubMed] [Google Scholar]
- Kemp AH, Koenig J, Thayer JF, 2017. From psychological moments to mortality: a multidisciplinary synthesis on heart rate variability spanning the continuum of time. Neuroscience & Biobehavioral Reviews. [DOI] [PubMed] [Google Scholar]
- Kragel PA, Koban L, Barrett LF, Wager TD, 2018. Representation, Pattern Information, and Brain Signatures: From Neurons to Neuroimaging. Neuron 99, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist D, Fernström J, Grudet C, Ljunggren L, Träskman-Bendz L, Ohlsson L, Westrin Å, 2016. Increased plasma levels of circulating cell-free mitochondrial DNA in suicide attempters: associations with HPA-axis hyperactivity. Translational psychiatry 6, e971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist D, Wolkowitz OM, Picard M, Ohlsson L, Bersani FS, Fernström J, Westrin Å, Hough CM, Lin J, Reus VI, 2018. Circulating cell-free mitochondrial DNA, but not leukocyte mitochondrial DNA copy number, is elevated in major depressive disorder. Neuropsychopharmacology, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhou C, 2012. Corticosterone reduces brain mitochondrial function and expression of mitofusin, BDNF in depression-like rodents regardless of exercise preconditioning. Psychoneuroendocrinology 37, 1057–1070. [DOI] [PubMed] [Google Scholar]
- Magariños AM, Verdugo JMG, McEwen BS, 1997. Chronic stress alters synaptic terminal structure in hippocampus. Proceedings of the National Academy of Sciences 94, 14002–14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland AL, Walsh C, Lockwood K, John-Henderson NA, 2017. The effects of acute psychological stress on circulating and stimulated inflammatory markers: a systematic review and meta-analysis. Brain, behavior, and immunity 64, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, 1998. Stress, adaptation, and disease: Allostasis and allostatic load. Annals of the New York Academy of Sciences 840, 33–44. [DOI] [PubMed] [Google Scholar]
- McNair DM, 1971. Profile of mood states instrument. Manual for the Profile of Mood States, 3–29. [Google Scholar]
- Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, Hamprecht FA, 2009. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC bioinformatics 10, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, Roemisch-Margl W, Polonikov A, Peters A, Theis FJ, 2011. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS genetics 7, e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Kyung S-Y, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, 2013. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS medicine 10, e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenbarger JR RS, Wing AL, Hyde RT, 1978. Physical activity as an index of heart attack risk in college alumni. American Journal of epidemiology 108, 161–175. [DOI] [PubMed] [Google Scholar]
- Picard M, McEwen BS, 2018. Psychological stress and mitochondria: A systematic review. Psychosom Med In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McEwen BS, Epel E, Sandi C, 2018. An energetic view of stress: Focus on mitochondria. Frontiers in neuroendocrinology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather AA, Carroll JE, Fury JM, McDade KK, Ross D, Marsland AL, 2009. Gender differences in stimulated cytokine production following acute psychological stress. Brain, behavior, and immunity 23, 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterman E, Weiss J, Lin J, Schilf S, Slusher A, Johansen KL, Epel ES, 2018. Aerobic exercise lengthens telomeres and reduces stress in family caregivers: A randomized controlled trial-Curt Richter Award Paper 2018. Psychoneuroendocrinology. [DOI] [PubMed] [Google Scholar]
- Rahman S, Blok R, Dahl HH, Danks D, Kirby D, Chow C, Christodoulou J, Thorburn D, 1996. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Annals of neurology 39, 343–351. [DOI] [PubMed] [Google Scholar]
- Russell D, Peplau LA, Cutrona CE, 1980. The revised UCLA Loneliness Scale: Concurrent and discriminant validity evidence. Journal of personality and social psychology 39, 472–480. [DOI] [PubMed] [Google Scholar]
- Russell JA, 1980. A circumplex model of affect. Journal of personality and social psychology 39, 1161. [DOI] [PubMed] [Google Scholar]
- Sherman DK, Bunyan DP, Creswell JD, Jaremka LM, 2009. Psychological vulnerability and stress: The effects of self-affirmation on sympathetic nervous system responses to naturalistic stressors. Health Psychology 28, 554. [DOI] [PubMed] [Google Scholar]
- Simmons JP, Nelson LD, Simonsohn U, 2011. False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological science 22, 1359–1366. [DOI] [PubMed] [Google Scholar]
- Spielberger CD, Gonzalez-Reigosa F, Martinez-Urrutia A, Natalicio LF, Natalicio DS, 2017. The state-trait anxiety inventory. Revista Interamericana de Psicologia/Interamerican Journal of Psychology 5. [Google Scholar]
- Stawski R, Walczak K, Kosielski P, Meissner P, Budlewski T, Padula G, Nowak D, 2017. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PloS one 12, e0178216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treynor W, Gonzalez R, Nolen-Hoeksema S, 2003. Rumination reconsidered: A psychometric analysis. Cognitive therapy and research 27, 247–259. [Google Scholar]
- Trumpff C, Marsland AL, Basualto-Alarcon C, Martin JL, Carroll JE, Sturm G, Vincent AE, Mosharov EV, Gu Z, Kaufman BA, 2018. Acute Psychological Stress Triggers Circulating Cell-Free Mitochondrial DNA. bioRxiv, 405886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usala PD, Hertzog C, 1989. Measurement of affective states in adults: Evaluation of an adjective rating scale instrument. Research on Aging 11, 403–426. [DOI] [PubMed] [Google Scholar]
- Van Erven P, Cillessen J, Eekhoff E, Gabreëls F, Doesburg W, Lemmens W, Slooff J, Renier W, Ruitenbeek W, 1987. Leigh syndrome, a mitochondrial encephalo (myo) pathy: a review of the literature. Clinical neurology and neurosurgery 89, 217–230. [DOI] [PubMed] [Google Scholar]
- Vargas I, Lopez-Duran N, 2017. Investigating the effect of acute sleep deprivation on hypothalamic-pituitary-adrenal-axis response to a psychosocial stressor. Psychoneuroendocrinology 79, 1–8. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Moulin M, Piquereau J, Lemaire C, Mericskay M, Veksler V, Garnier A, 2017. Mitochondria: a central target for sex differences in pathologies. Clinical Science 131, 803–822. [DOI] [PubMed] [Google Scholar]
- Watson D, Clark LA, Tellegen A, 1988. Development and validation of brief measures of positive and negative affect: the PANAS scales. Journal of personality and social psychology 54, 1063. [DOI] [PubMed] [Google Scholar]
- Weber E, Molenaar P, Van der Molen M, 1991. PSPAT: A program for spectral analysis of point events including a test for stationarity. [Google Scholar]
- Weiner H, 1992. Perturbing the organism: The biology of stressful experience. University of Chicago press, Chicago. [Google Scholar]
- West AP, Shadel GS, 2017. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nature Reviews Immunology 17, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Sjöström M, Eriksson L, 2001. PLS-regression: a basic tool of chemometrics. Chemometrics and intelligent laboratory systems 58, 109–130. [Google Scholar]
- Xia J, Wishart DS, 2016. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics 55, 14.10.11–14.10.91. [DOI] [PubMed] [Google Scholar]
- Yarkoni T, Westfall J, 2017. Choosing prediction over explanation in psychology: Lessons from machine learning. Perspectives on Psychological Science 12, 1100–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ, 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschucke E, Renneberg B, Dimeo F, Wüstenberg T, Ströhle A, 2015. The stress-buffering effect of acute exercise: Evidence for HPA axis negative feedback. Psychoneuroendocrinology 51, 414–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.