Summary
Verticillium dahliae is a phytopathogenic fungal pathogen that causes vascular wilt diseases responsible for considerable decreases in cotton yields. The lignification of cell wall appositions is a conserved basal defence mechanism in the plant innate immune response. However, the function of laccase in defence‐induced lignification has not been described. Screening of an SSH library of a resistant cotton cultivar, Jimian20, inoculated with V. dahliae revealed a laccase gene that was strongly induced by the pathogen. This gene was phylogenetically related to AtLAC15 and contained domains conserved by laccases; therefore, we named it GhLAC15. Quantitative reverse transcription‐polymerase chain reaction indicated that GhLAC15 maintained higher expression levels in tolerant than in susceptible cultivars. Overexpression of GhLAC15 enhanced cell wall lignification, resulting in increased total lignin, G monolignol and G/S ratio, which significantly improved the Verticillium wilt resistance of transgenic Arabidopsis. In addition, the levels of arabinose and xylose were higher in transgenic plants than in wild‐type plants, which resulted in transgenic Arabidopsis plants being less easily hydrolysed. Furthermore, suppression of the transcriptional level of GhLAC15 resulted in an increase in susceptibility in cotton. The content of monolignol and the G/S ratio were lower in silenced cotton plants, which led to resistant cotton cv. Jimian20 becoming susceptible. These results demonstrate that GhLAC15 enhances Verticillium wilt resistance via an increase in defence‐induced lignification and arabinose and xylose accumulation in the cell wall of Gossypium hirsutum. This study broadens our knowledge of defence‐induced lignification and cell wall modifications as defence mechanisms against V. dahliae.
Keywords: cell wall composition, defence‐induced lignification, GhLAC15, Gossypium hirsutum, Verticillium wilt resistance
Introduction
Allotetraploid upland cotton (Gossypium hirsutum L.) is an important cash crop and supplies renewable textile fibres as well as oilseed worldwide. Verticillium wilt, caused by the soil‐borne fungus Verticillium dahliae Kleb., stands out amongst the most serious biotic constraints affecting cotton (Bolek et al., 2005; Cai et al., 2009). The acreage of Verticillium wilt‐infected cotton fields in China is approximately 2.5 million hectares annually, which is equivalent to approximately 50% of the cotton planting area in the country and direct economic losses of approximately US$250–310 million (Li CH et al., 2015). Verticillium wilt has not been effectively controlled because of the long‐term existence of microsclerotia and its resting structures in host xylem vessels (Bell, 1992). No fungicide is available to cure commercial upland cotton once infected (Wang et al., 2016; Zhang et al., 2017).
A promising and environmentally friendly strategy to reduce the above losses is to enhance the immune system of plants via genetic engineering, and this is based on insights into the molecular mechanisms of interactions between plant and pathogen (Lacombe et al., 2010). Plants have evolved multiple systems to recognize pathogen attacks. The first layer involves the cell surface perception of conserved microbial components (i.e. pathogen‐/microbe‐associated molecular patterns) and pathogen‐generated plant signal molecules (damage‐associated molecular patterns) by pattern recognition receptors, and the subsequent activation of pattern‐triggered immunity or basal immunity (Boller and Felix, 2009; Zipfel, 2014). The second layer involves the perception of race‐specific pathogen effectors on the cell surface and in the cytosol by resistance (R) proteins, and the subsequent activation of effector‐triggered immunity or R‐mediated immunity (Jones and Dangl, 2006). The downstream responses triggered by these two innate immune strategies are partially overlapping, including the accumulation of secondary metabolites, phytohormones and reactive oxygen species, which activate the corresponding defence systems (De Vleesschauwer et al., 2014; Feng and Shan, 2014; Huang et al., 2011a,b; La Camera et al., 2004; Pieterse et al., 2009; Wu et al., 2014). More importantly, the pathogen elicitation of basal or R‐mediated immunity has been shown to activate the biosynthesis and deposition of lignin in cell wall appositions (CWAs) (Adams‐Phillips et al., 2010; Chezem et al., 2017; Kishi‐Kaboshi et al., 2010; Lee et al., 2001; Robertsen, 1986).
To protect against pathogen infection, plants have evolved multiple sophisticated defence mechanisms (Zhang et al., 2017). To overcome the barrier of the plant cell wall, phytopathogenic fungi secrete various cell wall‐degrading enzymes (CWDEs), such as cellulases, pectinase, hemicellulases, cutinase and protease. Most of these enzymes not only degrade cell wall components to obtain carbon sources for pathogen growth, but can also trigger multiple plant defence responses (Tayi et al., 2016). As lignin is a very difficult biopolymer to degrade because of the nature and heterogeneity of its linkages (Vanholme et al., 2010), it is regarded as a component of the defence response in plants (Miedes et al., 2014). For example, lignin has been reported to function as a defensive physical/chemical barrier to limit pathogen colonization or restrict pathogen growth (Bonello and Blodgett, 2003; Zhang et al., 2017). Thus, defence‐induced lignification is a conserved basal defence mechanism in the plant immune response against (hemi)biotrophic pathogens in a wide range of plant species (Baayen et al., 1996; Bhuiyan et al., 2009; Lange et al., 1995; Menden et al., 2007; Nicholson and Hammerschmidt, 1992; Siegrist et al., 1994; Smit and Dubery, 1997; Vance, 1980), and has been used as a biochemical marker of an activated immune response (Adams‐Phillips et al., 2010; Kishi‐Kaboshi et al., 2010). Previous studies manipulating lignin content and composition have primarily focused on the regulation of the monolignol pathway controlled by 11 enzymatic steps (Vanholme et al., 2013). However, genetic alterations in the monolignol pathway impact both plant growth and defence/resistance to hemibiotrophic pathogens (Bhuiyan et al., 2009), and cannot distinguish between cell wall lignification and defence‐induced lignification. For example, in Arabidopsis and tobacco (Nicotiana tabacum), loss or down‐regulation of phenylalanine ammonia lyase (PAL) leads to decreased basal immunity to the hemibiotrophic bacterial pathogen Pseudomonas syringae (Huang et al., 2010) and to the biotrophic viral pathogen Tobacco mosaic virus (Elkind et al., 1990; Pallas et al., 1996), respectively. In addition, PAL is also involved in the biosynthesis of the defence signal molecule salicylic acid (SA), which mediates local and systemic resistance to many (hemi)biotrophic pathogens (Sticher et al., 1997). Thus, it remains unclear whether the reduced resistance in PAL‐deficient plants is the result of significant reductions in lignin, SA or both. Recently, Hu et al. (2018) have reported that the overexpression of cotton GhLAC1 increases lignification and mediates jasmonic acid (JA) biosynthesis and the balance of the JA–SA defence response, resulting in a modulation of broad‐spectrum biotic stress tolerance. However, whether the improved resistance of transgenic plants is the result of significant increases in lignin, SA, JA or all three still remains unclear.
Lignin from angiosperms usually contains G and S units with low to trace amounts of H units (Davin and Lewis, 1992). S‐rich lignin is less condensed and more degradable (Skyba et al., 2013). G‐rich lignin is more cross‐linked and resistant to depolymerization than S‐rich lignin. The ratio of G to S units in lignin indicates the degree and nature of its polymeric cross‐linking (Chezem et al., 2017). G‐lignin should be a better defensive barrier against pathogen attack. For example, AtMYB15 mediates the defence‐induced synthesis of G‐lignin, which contributes to basal immunity in Arabidopsis (Chezem et al., 2017). However, several reports have shown inconsistent conclusions on the role of G‐rich lignin in basal immunity. For example, enhancing the G‐lignin level by genetic manipulation of the monolignol pathway in Arabidopsis led to a reduction, not an increase, in R‐mediated immunity to a hemibiotrophic bacterial pathogen (Goujon et al., 2003; Quentin et al., 2009). In wheat, a higher S‐lignin content is regarded as a cell wall biochemical trait related to Fusarium resistance (Lionetti et al., 2015). No genetic studies have been reported providing insights into the role of G‐ or S‐lignin in CWA‐associated defence against V. dahliae attack.
By scanning the transcriptomic data in the SSH library that reflects differential expression from Jimian20 root tissue inoculated with V. dahliae (Wang et al., 2008), we identified one expressed sequence tag (EST) that was homologous to the sequence of GaLAC (AY423714). The expression level of this EST, GhLAC, was strongly and significantly greater in V. dahliae‐infected plants than in plants receiving mock treatment, and the transcription level of GhLAC in resistant Jimian20 was significantly higher than that in susceptible Han208 at each time point (Wu et al., 2014a). Therefore, we carried out further related experiments to obtain insights into its function in this study. We found that GhLAC displayed a differential expression level between tolerant and susceptible cotton varieties, was involved in defence‐induced lignification and modulated Verticillium wilt resistance through an alteration in the G‐rich lignin level and cell wall composition.
Results
The GhLAC15 protein is phylogenetically related to the AtLAC15 laccase
Using the EST sequence combined with a previously established full‐length cDNA library platform (Zhang et al., 2010), we obtained the full‐length cDNA of GhLAC (GI: EU642559.1) from Jimian20, which contained 1701 bp in length. The genome sequences of GhLAC contained 2401 bp in length. To identify the homology between GhLAC and laccases from Arabidopsis, we blasted the amino acid sequence of GhLAC against those of 17 Arabidopsis laccase proteins. This phylogenetic analysis showed that the laccases could be divided into two groups. Some sequences (GhLAC, AtLAC14, AtLAC15 and PtLAC) were placed together in the same clade, whereas the others were placed within another clade. The GhLAC protein was especially closely related to the lignin‐specific AtLAC15 laccase; thus, we specifically renamed it GhLAC15 (Fig. S1, see Supporting Information). A search for amino acid sequence motifs of laccases showed that the GhLAC15 protein included one transmembrane domain, 13 N‐glycosylation sites, three copper oxidase‐like domains, one possible calmodulin‐binding region, one cell attachment sequence, one multicopper oxidase signature and other motif models (Fig. S2, see Supporting Information). A more detailed analysis based on the amino acid sequences was performed by multiple alignments. As shown in Fig. S2, despite the low sequence identity among these proteins, the domains of the cell attachment sequence (Arg‐Gly‐Asp, RGD), the multicopper oxidase signature (HCHLERHSSWGM) and three copper oxidase‐like domains were highly conserved. In addition, GhLAC15 was predominantly expressed in the roots, with significantly lower expression levels in stem and leaf (Fig. S3, see Supporting Information).
GhLAC15 displays different expression levels in tolerant and susceptible cotton
To investigate the expression levels of GhLAC15 amongst different Verticillium wilt‐resistant cottons, we used two types of varieties, one with a tolerant phenotype and one with a susceptible phenotype Table S2. As shown in Fig. S4 (see Supporting Information), primer efficiencies for GhLAC15 and GhActin were detected. The results indicated that GhLAC15 was more highly expressed in tolerant varieties than in susceptible varieties (Fig. 1). We further detected the abundance of lignin in the resistant variety Jimian20 and susceptible variety Han208, with higher GhLAC15 expression in the former than in the latter (Wu et al., 2014a), via basic fuchsin staining. As shown in Fig. 2, a greater area of stem tissue was stained with red (an indicator of lignification) in Jimian20 than in Han208, especially in interfascicular regions, and the basic fuchsin fluorescence intensity was also stronger in Jimian20 than in Han208. This difference suggested that the tolerant cotton accumulated more lignin or increased lignification on V. dahliae infection.
Figure 1.
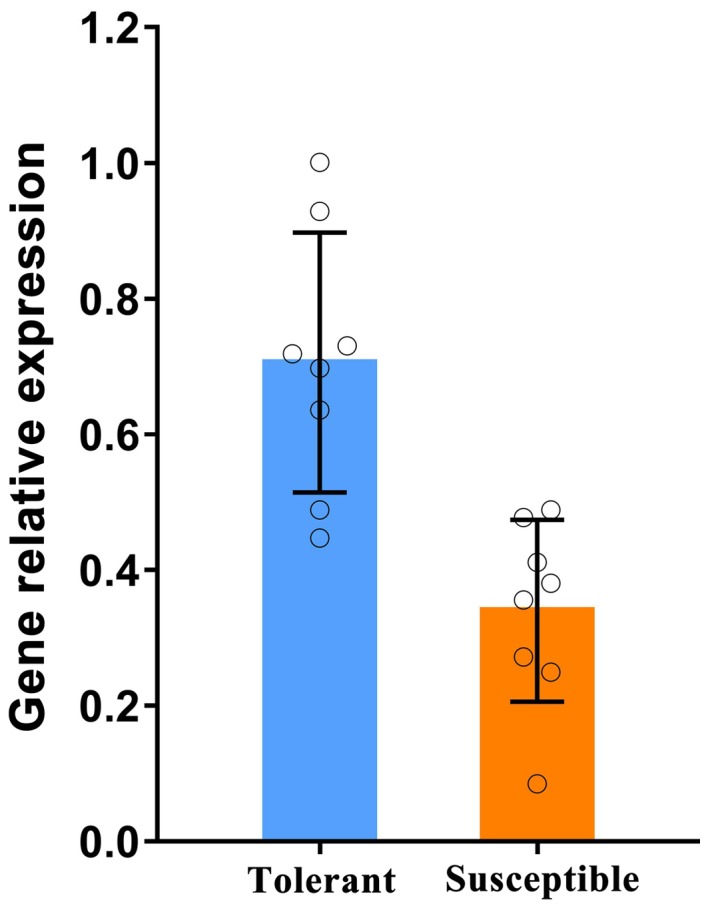
Expression of GhLAC15 in tolerant and susceptible cotton varieties, detected through quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). GhActin was used as an internal control. Tolerant and susceptible varieties included Tang mian7401, Zhong1421, Su yuan04‐3, Yu mian21, Jin mian20, Su mian22, CC28, Ku che96515 (resistant varieties), Jin zhou tui hua mian, 73‐782, Xu zhou1818, Ren dong67‐86, Xin lu zao28, 73‐184, Nong lin1 and Jun mian1 (susceptible varieties). Data are represented as average values with standard deviation (n = 8 varieties with three technical replicates). [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.
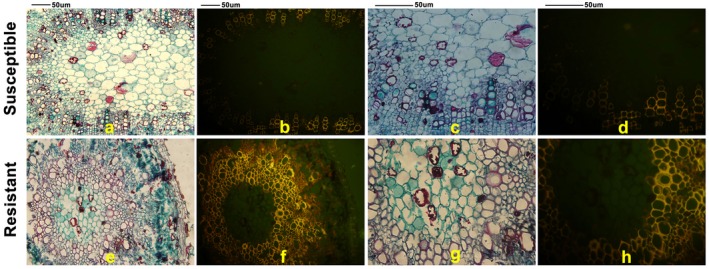
Basic fuchsin staining of cotton stem tissues. Images of susceptible variety cv. Han208 under white light (a, c) and fluorescent light (b, d). Images of resistant variety cv. Jimian20 under white light (e, g) and fluorescent light (f, h). Cross‐sections of stems from the same position of five seedlings were used for this assay. [Colour figure can be viewed at wileyonlinelibrary.com]
GhLAC15 protein is located in the cell wall
A GhLAC15 open reading frame (ORF) lacking the stop codon was ligated upstream of an enhanced green fluorescent protein (GFP) gene under the control of the constitutive promoter CaMV 35S. As shown in Fig. 3, the GhLAC15::GFP fusion protein was located in the cell wall of transgenic Arabidopsis and clearly exhibited fluorescence in the cell wall after being plasmolysed with 10% mannitol. As a control, Arabidopsis transformed with the GFP gene alone showed a very strong fluorescence signal, and the signal was uniformly and diffusely distributed in the cytosol and nucleus, as described by Cutler et al. (2000); fluorescence was absent in wild‐type (WT) Arabidopsis as expected. These results thus suggested that the GhLAC15 protein was localized to the cell wall.
Figure 3.
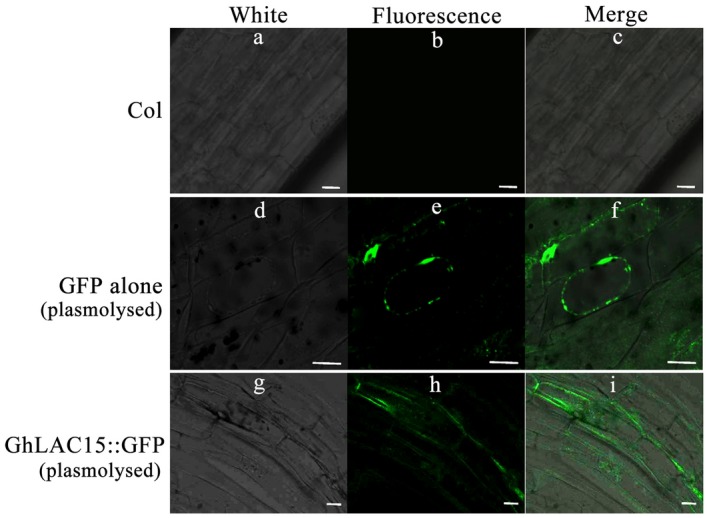
Subcellular localization of the GhLAC15::GFP fusion protein in the root cells of transgenic Arabidopsis. (a–c) Images of wild‐type (d–f) show GFP alone (plasmolysed) and (g–i) show GhLAC15::GFP (plasmolysed). Bar, 50 μm. [Colour figure can be viewed at wileyonlinelibrary.com]
GhLAC15 enhances the resistance of transgenic Arabidopsis against V. dahliae
To validate the function of the GhLAC15 gene, we first overexpressed GhLAC15 in Arabidopsis. Of the 16 independent T3 transgenic lines, three stable overexpressing lines selected via genome PCR and RT‐PCR analysis were inoculated with V. dahliae. Disease resistance was assessed by evaluation of the extent of leaf chlorosis and measurement of inflorescence heights and plant dry weights. Typical symptoms of vascular disease became evident in the infected WT plants, but were much less pronounced in transgenic plants at 20 days post‐inoculation (dpi) (Fig. 4a). Compared with WT, transgenic Arabidopsis exhibited significantly more resistance (Fig. 4b). The average inflorescence height of inoculated WT plants (155.3 cm) was significantly shorter than that of transgenic plants (199.5 cm), and the average dry weight of infected WT plants was lower than that of transgenic plants (Fig. 4c,d). The extent of leaf chlorosis also differed markedly between WT and transgenic plants. By comparison, disease symptoms developed more slowly and were less severe in transgenic plants than in WT (Fig. 4e). In detail, the percentage of chlorotic leaves in transgenic plants was nearly 50%, and only minor microsclerotium formation was observed in the vascular bundles (Fig. 5). However, more than 71% of WT leaves experienced chlorosis, and greater microsclerotium formation occurred in the vascular bundles of WT plants (Figs 4e and 5). Based on these symptoms, we concluded that GhLAC15 enhanced the Verticillium wilt resistance of transgenic Arabidopsis.
Figure 4.
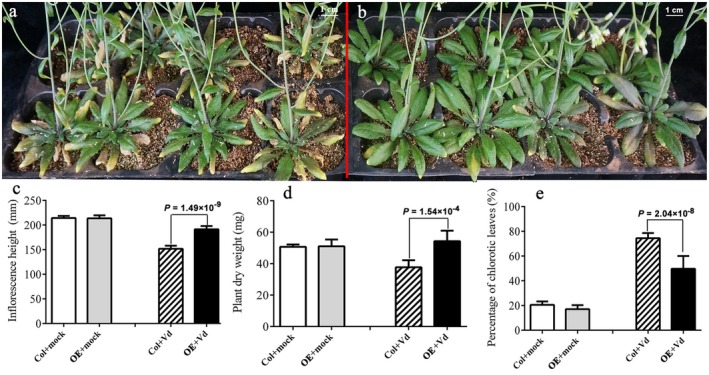
Overexpression of GhLAC15 in Arabidopsis improved resistance to Verticillium dahliae. (a, b) Four‐week‐old plants were inoculated by root dipping with 5 × 107 conidia/mL. Transgenic (b) and wild‐type (a) plants showed marked differences in Verticillium wilt resistance. Transgenic plants were more resistant than wild‐type plants. (c–e) Disease resistance was assessed by measurement of the inflorescence height (c) and plant dry weight (d), as well as the percentage of leaf chlorosis (e). The significance of differences was analysed with the t‐test (P < 0.01). Data are presented as average values with standard deviation (n = 39). Col, wild‐type Arabidopsis; OE, transgenic Arabidopsis. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 5.
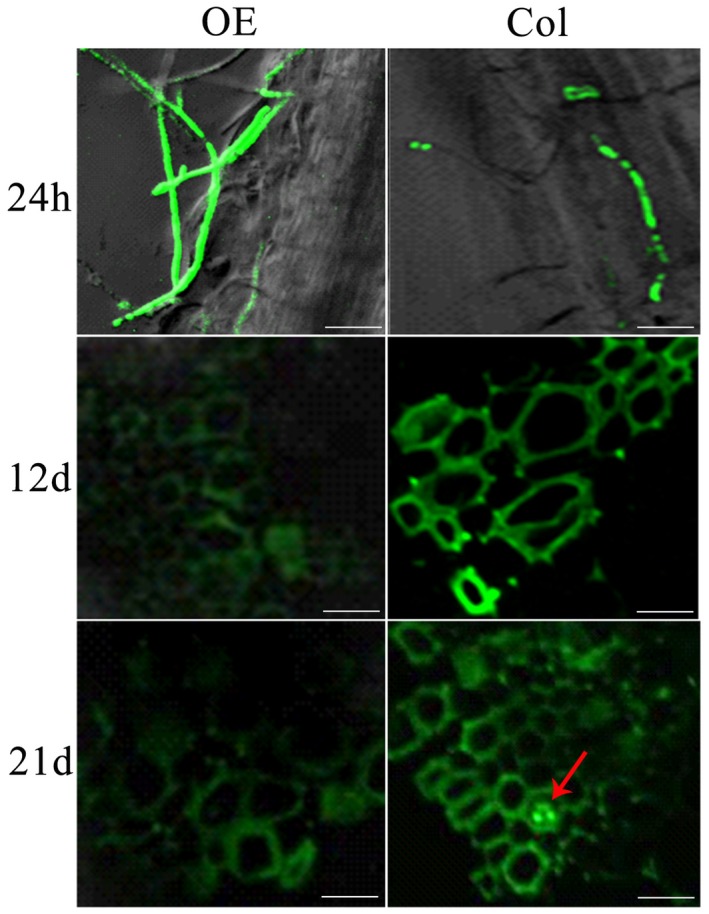
Progression of root colonization by green fluorescent protein (GFP)‐tagged isolate of Verticillium dahliae in transgenic Arabidopsis (OE) and wild‐type plants (Col). Representative images of conidiospores were taken at 24 h, 12 days and 21 days. The red arrow points to the accumulation of V. dahliae in the vascular bundle. Ten mature rosette leaves of each transgenic line were observed, and three transgenic lines were used for this assay. Bar, 20 µm. [Colour figure can be viewed at wileyonlinelibrary.com]
The GhLAC15 expression levels positively correlate with resistance against V. dahliae in G. hirsutum
Virus‐induced gene silencing (VIGS) is a promising approach in plant functional genomics and has been widely used to study gene function in cotton (Gao et al., 2011). Therefore, we employed VIGS to investigate the function of GhLAC15 in defence against V. dahliae infection. As shown in Fig. 6d, an albino phenotype appeared on newly developing true leaves and stems of plants infiltrated with agrobacteria carrying GhCLA1, indicating that the VIGS system worked efficiently under our experimental conditions. Meanwhile, quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analyses showed that the expression of GhCLA1 in VIGS plants was significantly lower than that in plants with the empty vector (Fig. 6e). When plants were challenged with V. dahliae, the down‐regulation of GhLAC15 expression resulted in reduced resistance to the pathogen (Fig. 6a–c). The disease index and rate of diseased plants were clearly greater in GhLAC15‐silenced plants than in non‐silenced plants (Fig. 6f,g). Larger numbers of fungal colonies were found in the roots of GhLAC15‐silenced plants, suggesting that the extent of fungal colonization was much more severe in GhLAC15‐silenced plants than in control plants (Fig. 6h).
Figure 6.
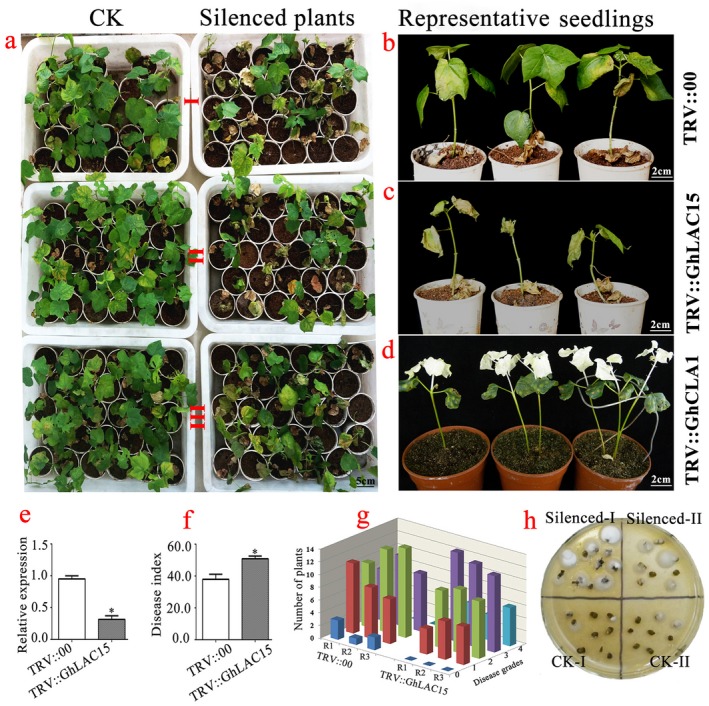
Effects of silencing of GhLAC15 on cotton susceptibility to Verticillium dahliae. Two weeks after infiltration, seedlings were inoculated with V. dahliae. (a) Responses of control (CK) (TRV::00) and silenced (TRV::GhLAC15) plants to the pathogen at 20 days post‐inoculation (dpi). Disease symptoms induced on CK and silenced plants with three experimental repeats. (b, c) Representative seedlings of CK and silenced plants after inoculation with V. dahliae at 20 dpi. (d) Seven‐day‐old cotton plants were infiltrated with Agrobacterium carrying TRV::GhCLA1. The photographs were taken at 2 weeks after infiltration. (e) Preliminary assay of the efficiency of virus‐induced gene silencing (VIGS) under our experimental conditions. (f, g) The disease index and rate of diseased plants were measured at 20 dpi. Error bars represent the standard deviation of three biological replicates (n = 36); asterisks indicate statistically significant differences, as determined by t‐test (P < 0.05). (h) Fifteen days after V. dahliae inoculation, surface‐sterilized hypocotyl sections prepared from CK and silenced plants were placed on agar medium at 7 dpi. The number of stem sections from which fungus grew represented the extent of fungal colonization. [Colour figure can be viewed at wileyonlinelibrary.com]
To clearly understand the relationship between changes in GhLAC15 expression and disease resistance in VIGS plants, the gene expression level and the corresponding disease resistance were investigated in 30 VIGS plants (Fig. S5, see Supporting Information). A reduction in GhLAC15 gene expression was positively correlated with aggravated susceptibility to V. dahliae. This result suggested that GhLAC15 played an important role in defence against V. dahliae in cotton.
We further investigated whether lignin deposition or monolignol composition was altered after silencing of GhLAC15. The histochemical staining results indicated that, at 4 dpi, hypocotyl sections from GhLAC15‐silenced plants exhibited lighter red staining of xylem vessel walls and parenchyma cell walls than control plants (Fig. 7a,b,d,e). In addition, the stem of GhLAC15‐silenced plants exhibited darker vascular browning (Fig. 7c,f). Monolignol composition analysis showed that both G‐ and S‐lignin levels, as well as the G/S ratio, decreased significantly after GhLAC15 was silenced in cotton (Table 1), which suggested that GhLAC15 functioned to modulate monolignol composition biosynthesis and the G/S ratio, thus regulating cotton Verticillium wilt resistance.
Figure 7.
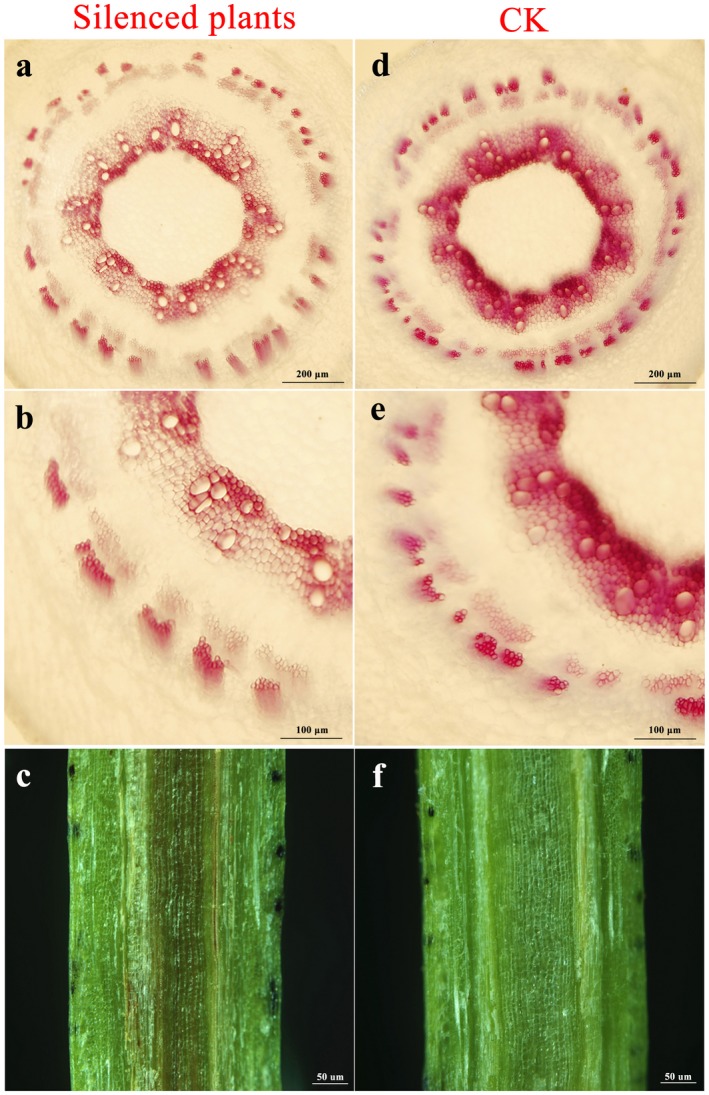
Determination of lignin in virus‐induced gene silencing (VIGS)‐treated cotton after inoculation with Verticillium dahliae. Light microscopy image of hypocotyl cross‐sections at 4 days post‐inoculation (dpi) after staining with phloroglucinol‐HCl to detect lignin in silenced plants (a, b) and control (CK) plants (d, e). At 20 dpi, tissue browning was rare in vascular bundles from CK plants (f), but severe in longitudinal sections from silenced plants (c). Cross‐sections of stem from the same position of five seedlings were used for the assays. [Colour figure can be viewed at wileyonlinelibrary.com]
Table 1.
Monolignol composition in cell walls from control (CK) and silenced cotton plants.
| Line | Thioacidolysis yield (μmol/g extract‐free sample) | G/S molar ratio | ||
|---|---|---|---|---|
| G | S | H | ||
| CK plant | 135.0 ± 6.9a | 72.6 ± 3.3a | 8.8 ± 0.6 | 1.86 ± 0.09a |
| Silenced plant | 87.1 ± 5.4b | 52.9 ± 4.0b | 9.3 ± 0.4 | 1.65 ± 0.02a |
The data are the mean ± standard error (SE) of three independent biological replications, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test).
GhLAC15 alters lignification and enzymatic hydrolysis in mature transgenic Arabidopsis stems
To confirm whether GhLAC15 functions in lignin polymerization, we determined the lignin content by measurement of both the Klason lignin (KL) and acid‐soluble lignin (ASL) concentrations of extract‐free stems. Transgenic plants exhibited higher KL content than the corresponding controls under V. dahliae stress or free (Table 2). Compared with mock plants, both WT and transgenic plants decreased in KL content after inoculation with V. dahliae. However, ΔKL in transgenic plants was 11.36%, significantly lower than that in WT plants (17.76%) (Table 2). ASL lignin contents were similarly low in all samples (within the range of 1%). The results indicated that GhLAC15 enhanced KL, which played a crucial role in disease resistance.
Table 2.
Lignin content of mature stems of Arabidopsis.
| Treatment | Line | Lignin content | |
|---|---|---|---|
| KL (%) | ASL (%) | ||
| Mock | Wild‐type | 21.12 ± 0.76b | 2.53 ± 0.24c |
| OE plant | 24.29 ± 0.07a | 3.17 ± 0.66b | |
| Inoculated | Wild‐type | 17.37 ± 1.20c | 2.94 ± 0.06b |
| OE plant | 21.50 ± 0.85b | 3.46 ± 0.00a | |
| ΔLignin | Wild‐type | -17.76% | +16.21% |
| OE plant | -11.36% | +9.15% | |
ASL, acid‐soluble lignin; KL, Klason lignin; OE, overexpressing. The data are the mean ± standard error (SE) of three independent biological replications, and different letters in the same column share a significant difference (P < 0.05) (Duncan’s multiple range test). ΔLignin = (Inoculated - Mock)/Mock.
Lignin content is known to be a factor in decreasing the cell wall susceptibility to enzymatic hydrolysis. Thus, we subjected the extract‐free samples to treatment with a commercial cellulose preparation to evaluate the saccharification potential of transgenic and WT plants. Consistent with their low KL level, WT plants lost an average of 29.76% of their weight when subjected to commercial cellulose treatment (cellulose and hemicellulase activities), which was much greater than that observed in transgenic plants. By contrast, transgenic plants were less easily hydrolysed than WT plants, with less weight loss (Fig. 8a). The amount of glucose (Glc) released by enzymatic hydrolysis was also significantly lower in transgenic plants (Fig. 8b). Thus, we concluded that the modified cell wall traits could function as a defence barrier to improve the resistance of Arabidopsis to V. dahliae infection.
Figure 8.
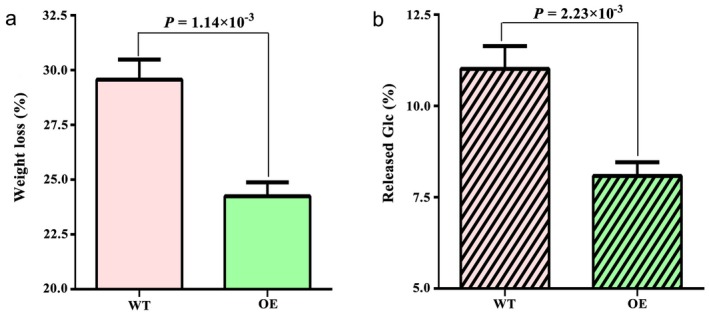
Enzymatic hydrolysis of mature stem from transgenic Arabidopsis. (a) Weight loss after enzymatic hydrolysis in wild‐type (WT) and overexpressing (OE) Arabidopsis. Data are presented as average values with standard deviation (n = 3 technical replicates). (b) Glucose (Glc) released after enzymatic hydrolysis of WT and OE Arabidopsis. The significance of differences was analysed with the t‐test. Data are presented as average values with standard deviation (n = 3 technical replicates). WT and OE samples displayed significantly different weight loss (P = 1.14 × 10−3) and amount of released Glc (P = 2.23 × 10−3). [Colour figure can be viewed at wileyonlinelibrary.com]
We further determined the impact of overexpression of GhLAC15 on lignin structure by thioacidolysis using entire mature inflorescence stems of 6‐week‐old plants (Table 3). The H thioacidolysis monomer will not be discussed further because of its small amounts recovered in all samples. As shown in Table 3, under normal conditions (free of V. dahliae), transgenic plants released significantly larger amounts of guaiacyl (G) monomers than WT, and equal amounts of syringyl (S) monomers as WT, indicating the crucial role of GhLAC15 in G‐lignin biosynthesis. On inoculation, the G monomer content was much higher in transgenic plants than in WT plants, even though there was a marked decrease in both transgenic and WT plants. The S monomer content in transgenic plants remained unchanged, whereas it decreased sharply in WT plants post‐inoculation with V. dahliae. Compared with its value in transgenic plants, the G/S ratio obviously decreased in WT after inoculation by V. dahliae (Table 3). The results suggested that GhLAC15 could maintain a relatively higher ratio of G‐units to S‐units in lignin, thus conferring improved Verticillium wilt resistance in transgenic plants (Xu et al., 2011).
Table 3.
Determination of the main G, S and H thioacidolysis monomers released by the lignin of extract‐free mature stems.
| Treatment | Line | Thioacidolysis yield (μmol/g extract‐free sample) | G/S molar ratio | |||
|---|---|---|---|---|---|---|
| G | S | H | Total | |||
| Mock | Wild‐type | 203.1 ± 0.0b | 74.0 ± 0.0a | 2.0 ± 0.0 | 252.2 ± 33.9b | 3.15 ± 0.05a |
| OE plant | 225.6 ± 0.0a | 77.2 ± 0.0a | 2.2 ± 0.0 | 314.0 ± 30.8a | 2.71 ± 0.21a | |
| Inoculated | Wild‐type | 62.0 ± 1.5d | 35.8 ± 0.2b | 1.3 ± 0.0 | 99.1 ± 1.8c | 1.64 ± 0.20b |
| OE plant | 176.4 ± 16.5c | 77.9 ± 11.7a | 1.9 ± 0.6 | 256.1 ± 28.9b | 2.28 ± 0.13a | |
| ΔLignin | Wild‐type | −69.5% | −51.6% | ‐ | −60.7% | ‐ |
| OE plant | −21.8% | +0.9% | ‐ | −18.4% | ‐ | |
The data are the mean ± standard error (SE) of three independent biological replications, and different letters in the same column share a significant difference (P < 0.05) (Duncan’s multiple range test). ΔLignin = (Inoculated − Mock)/Mock. OE, overexpressing.
GhLAC15 increases total lignin, arabinose (Ara) and xylose (Xyl) in transgenic Arabidopsis
The total lignin contents of the entire mature inflorescence stems were determined by the acetyl bromide method to further confirm the impact of the overexpression of GhLAC15 on lignin biosynthesis. The results indicated that the transgenic plants exhibited significantly higher total lignin content than WT whether under V. dahliae stress or free (Table 3). Compared with mock plants, total lignin content decreased in both WT and transgenic plants after inoculation by V. dahliae. However, ΔLignin in transgenic plants was 18.4%, significantly lower than that (60.7%) in WT plants (Table 3). Plant cell wall composition and features can greatly affect plant mechanical strength and resistance to pathogens and pests (Bonello and Blodgett, 2003; Mottiar et al., 2016; Naoumkina et al., 2010). Therefore, we analysed the predominant carbohydrate constituents of the cell wall in transgenic Arabidopsis, including Glc, Ara and Xyl. As shown in Table 4, both Ara and Xyl levels were significantly higher in transgenic Arabidopsis than in WT under V. dahliae stress. In contrast with WT, the Glc level showed a remarkable decrease in transgenic Arabidopsis. Thus, the overexpression of GhLAC15 changed the cell wall composition, leading to increases in Ara and Xyl content.
Table 4.
Detection of polysaccharide components and content in Arabidopsis.
| Line | Polysaccharide components of cell wall (%) | |||
|---|---|---|---|---|
| Ara content | Xyl content | Glc content | Ara + Xyl | |
| Wild‐type | 2.63 ± 0.03b | 8.87 ± 0.49b | 2.86 ± 0.27a | 11.62 ± 0.57b |
| OE plant | 3.33 ± 0.08a | 12.43 ± 0.10a | 1.70 ± 0.08b | 15.86 ± 0.00a |
Ara, arabinose; Glc, glucose; Xyl, xylose. OE, overexpressing. The data are the mean ± standard error (SE) of three independent biological replications, and different letters in the same column share a significant difference (P < 0.05) (Duncan’s multiple range test).
Discussion
The plant cell wall, composed of cellulose, hemicelluloses, lignin and pectic polysaccharides with minor structural proteins, is a dynamic structure that often determines the outcome of the interactions between plants and pathogens (Bellincampi et al., 2014; Xu et al., 2011). Lignin has been reported to be involved in multiple resistances, including cotton Verticillium wilt resistance (Bonello and Blodgett, 2003; Hu et al., 2018; Zhang et al., 2017). Previous studies have shown that plant laccases (LAC4, LAC11 and LAC17) are necessary and non‐redundant with peroxidase for lignin polymerization in Arabidopsis (Berthet et al., 2011; Zhao et al., 2013), and that they oxidatively polymerize monolignols (p‐coumaryl, coniferyl and sinapyl alcohols) into p‐hydroxyphenyl (H), guaiacyl (G) and syringyl (S) lignin units (Wang et al., 2015). In this study, we isolated a lignin laccase, GhLAC15, which affected Verticillium wilt resistance by increasing total lignin and KL levels and by modifying lignin composition and structure, as well as associated cell wall traits. These findings provide unprecedented insights into the defence mechanism against V. dahliae of cotton laccase genes.
The cell wall is a barrier that pathogens need to breach to colonize plant tissue. During infection, pathogens produce CWDEs, such as pectinases, xylanases and cellulases, to degrade cell wall polysaccharides to penetrate and colonize the host tissues (Wanyoike et al., 2002; Yang et al., 2012). The role of cell wall components in plant resistance has been reported in different species (Barros‐Riosa et al., 2011; Lionetti et al., 2015). New lines of evidence have suggested that the content and composition of cell wall polymers affect the susceptibility of cell walls to CWDEs and play important roles in the outcome of host–pathogen interactions (Blümke et al., 2014; Cantu et al., 2008; Pogorelko et al., 2013). Notably, the extent of cell wall degradation is often associated with the severity of diseases (King et al., 2011). Lignin is an important structural component involved in defence against invasive pathogens, making the cell wall more resistant to CWDEs and preventing the diffusion of pathogen‐produced toxins (Sattler and Funnell‐Harris, 2013; Zhang et al., 2017). In this study, we found that the overexpression of GhLAC15 increased significantly the lignin content of Arabidopsis, leading to improved Verticillium wilt resistance. Compared with mock plants, both transgenic and WT plants displayed decreased lignin content under V. dahliae stress. This may be a result of the degradation of plant cell walls by CWDEs produced by V. dahliae. However, the change in lignin amount in transgenic plants was much lower than that in WT plants. In other words, the cell wall of transgenic plants was more resistant to CWDEs produced by V. dahliae and probably maintained cell wall‐related integrity or exhibited higher plant mechanical strength.
Lignin is a factor in decreasing the cell wall susceptibility to enzymatic hydrolysis (Berthet et al., 2011). In this study, GhLAC15 transgenic plant stems were less easily hydrolysed. In a previous study, lac4‐2 and lac17 mutants showed low KL levels and much greater weight loss than WT plants when treated by commercial cellulose, and about one‐half of this loss was accounted for by Glc (Berthet et al., 2011). It has been reported that, during host and pathogen interactions, pathogens can degrade the host cell wall by self‐produced CWDEs, resulting in higher Glc levels (Yang et al., 2018). When we analysed the predominant carbohydrate constituents of cell walls in transgenic Arabidopsis, the Glc level was significantly lower in transgenic Arabidopsis than in WT; therefore, we concluded that cotton GhLAC15 altered the susceptibility of the Arabidopsis cell wall to enzymatic hydrolysis, which might be very important in defence against pathogen infection.
In this study, both Ara and Xyl levels were significantly higher in transgenic Arabidopsis than in WT under V. dahliae stress. The Ara level is a positive factor in rice lodging resistance and lignin level (Li F et al., 2015), and high concentrations of Xyl play an important role in defence against corn borers and are regarded as a defence mechanism of maize (Barros‐Riosa et al., 2011). Furthermore, the higher values for Ara and Xyl represent a high content of arabinose‐substituted xylan (arabinoxylan) (Barros‐Riosa et al., 2011). High concentrations of arabinoxylan in papillae effectively restrict pathogen penetration in barley (Chowdhury et al., 2014). Thus, we deduced that the increases in Ara and Xyl in Arabidopsis overexpressing GhLAC15 contributed to Verticillium wilt resistance.
Experimental Procedures
Growth of plant material and pathogen cultures
The G. hirsutum L. variety cv. Jimian20, which displays resistance to Verticillium wilt, was used in this study. Seeds of Jimian20 were surface disinfected in 0.5% sodium hypochlorite (NaOCl) for 5 min and then washed five times with sterile water. Any seed presenting internal fungal contamination was discarded. Seedlings of similar size were selected and cultivated in dishes containing sterile vermiculite under the following growth chamber conditions: 16‐h photoperiod, 30 ± 2 °C/25 ± 2 °C (day/night) and 75% relative humidity. Hoagland’s nutrient solution was added to the dishes every 4 days.
The highly aggressive defoliating V. dahliae strains Vd991 and Vd‐gfp77 were cultured on potato dextrose agar (PDA) for 7 days at 25 °C. The fungus was then inoculated into Czapek medium and cultured on a shaker at 120–140 g at 25 °C for another 3–4 days until the concentration of spores reached approximately 108–109 spores/mL. The suspension was adjusted to an approximate density of 107 conidia/mL with deionized water prior to use. Samples of seedlings inoculated with V. dahliae were used for RNA extraction, whereas seedlings inoculated with the gfp77 strain were used for observation of the progress of infection.
Gene cloning and bioinformatic analysis
Total RNA was extracted from cotton plants using an EASYspin Plus Plant RNA Kit (Aidlab Biotech) according to the manufacturer’s instructions. The purified RNA was used as a template to prepare cDNA with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa). A full‐length GhLAC15 coding sequence with KpnI and XbaI linkers was cloned using the primers LacF1 and LacR1 (Table S1, see Supporting Information).
Gene expression analysis
To determine the expression level of GhLAC15 in different cottons or tissues, the target samples were harvested, quickly frozen in liquid nitrogen, ground into a fine powder and stored at –80 °C until use. Total RNA isolation, cDNA synthesis and qRT‐PCR were performed as described previously (Zhang et al., 2017). The cotton GhLAC15‐specific primers LacF2/LacR2 (Table S1) were designed and included the 5′‐untranslated region (UTR) to discriminate the target GhLAC15 gene from other members of the gene family. Real‐time PCR was performed in triplicate with SYBR Premix Dimer Eraser (Perfect Real Time) (Dlian, China, TaKaRa). GhActin served as the reference gene to normalize the total amount of cDNA in each reaction. For each sample, cDNA was serially diluted in sterile water (1, 1 : 2, 1 : 4, 1 : 8, 1 : 16, 1 : 32, 1 : 64 and 1 : 128) to investigate the amplification efficiencies of the primers. Relative fold differences in mRNA abundance were defined by the mathematical model described by Livak and Schmittgen (2001). All experiments were performed with three technical replicates. The primers used for gene expression analysis are listed in Table S1.
Generation of transgenic Arabidopsis and disease assays
The full‐length ORF of GhLAC15 was amplified using forward (LacF3) and reverse (LacR3) primers (Table S1) that incorporated KpnI and XbaI cleavage sites, respectively. To prepare an overexpression (OE) construct, we inserted the GhLAC15 coding region of Jimian20 into the plant binary vector pCAMBIA1301S under the control of the CaMV 35S promoter. This vector was used to transform Agrobacterium tumefaciens strain GV3101 using a freeze–thaw method. Positive colonies were confirmed by PCR together with sequencing, and used for the genetic transformation of the Arabidopsis plants using the floral dip method (Clough and Bent, 1998). The identification of transformed seeds was based on hygromycin B screening and PCR detection with the primers LacF3/LacR3. Homozygous T3 seeds of transgenic lines were used for phenotypic analyses.
Fluorescence microscopy
Transgenic Arabidopsis seeds with the GhLAC15::GFP fusion were sterilized and germinated on Murashige and Skoog (MS) medium on a clean bench as described by Clough and Bent (1998). The roots of 7‐day‐old transgenic and WT Arabidopsis were plasmolysed with 10% mannitol (Komis et al., 2008). Specimen sections were observed using a fluorescence microscope (DM2500; Leica, Wetzlar, Germany).
VIGS in cotton and pathogen inoculation
Tobacco rattle virus (TRV)‐based VIGS was performed in cotton as described previously (Gao et al., 2011). The pTRV1, pTRV2 and pTRV2 derivatives harbouring specific regions of GhLAC15 were transformed into A. tumefaciens strain GV3101 by electroporation. The primers (LacF4/LacF4) used for GbLAC15 fragment amplification are listed in Table S1. Seven‐day‐old seedlings were transformed with a mixture (1 : 1, v/v) of Agrobacterium cultures harbouring pTRV1 with pTRV2 or its derivative plasmids. After completion of agro‐inoculation, the seedlings were washed with deionized water to remove excess agrobacterial inoculum and grown at 25 °C under a 16‐h/8‐h light/dark cycle in a controlled environmental chamber. After 2 weeks of cultivation, the plants were inoculated with V. dahliae. The experiments were performed with at least 36 seedlings per treatment and were repeated three times. The rate of diseased plants and the disease index were calculated as described previously (Zhang et al., 2016).
Lignin histochemical staining
Hypocotyl samples were taken at 3 dpi from 10 inoculated and 10 mock‐treated plants of each species. For histochemical analysis, 2‐cm‐long segments were excised and preserved in a mixture of acetic acid–formalin–ethanol (5 : 5 : 90, v/v/v). Tissues were transverse cross‐sectioned on a vibration microtome (VT 1000M; Leica). Lignin histochemistry was examined using Wiesner reagent (Pomar et al., 2004) or basic fuchsin staining. The cross‐sections were incubated for 10 min in a phloroglucinol solution (2 in 95% ethanol) or 95% ethanol (staining control), and then treated with 18% HCl for 5 min, and directly observed under bright‐field conditions with a fluorescence microscope (DM2500; Leica). Basic fuchsin staining of stem sections was performed according to the protocol of Kapp et al. (2015).
Determination of ASL and KL contents
Lignin chemical analysis was performed based on the above‐ground parts of Arabidopsis plants. Arabidopsis plants were collected and vacuum freeze‐dried for 48 h. Samples were evenly ground to pass through a 0.5‐mm sieve before an exhaustive solvent extraction, first with toluene–ethanol (2 : 1, v/v), then with ethanol and finally with water, as described previously (Berthet et al., 2011; Lapierre et al., 1995). The amounts of KL and ASL (spectrophotometric method) were determined by the Klason method with modifications (Dence, 1992). For each sample, 3 mL of 72% (w/w) sulfuric acid were added to 300 mg of the sample in a pressure tube. The samples were incubated in a water bath at 30 °C for 1.5 h with intermittent mixing using a glass rod. On completion of hydrolysis, the acid solution was diluted to a 4% concentration by the addition of 84 mL of water. The samples were mixed well by inverting the tube several times, and the Teflon caps were screwed on securely. The pressure tubes were then placed in an autoclave for 1 h using the ‘liquids’ setting (121 °C). The autoclaved hydrolysis solution was vacuum filtered. All remaining acid‐insoluble lignin was KL. The content was calculated as the weight percentage of the extract‐free wood and reported as the average of at least three independent determinations on the same sample. The filtrate was used to quantify ASL with a spectrophotometer. The maximal absorbance of the diluted solution was measured at 320 nm as described by Sluiter et al. (2013).
Determination of lignin content and composition
Mature stems were harvested for lignin analysis according to published procedures (Zhao et al., 2010). Lignin‐derived compounds were identified by analysis of their trimethylsilyl derivatives using gas chromatography/mass spectrometry. Lignin analyses were performed with three independent transformed lines and three biological replicates for lignin quantification and lignin composition. Statistical analysis was performed using the t‐test (P < 0.01 or P < 0.05).
Conflicts of Interest
The authors have declared that no competing interests exist.
Supporting information
Fig. S1 Phylogenetic analysis of laccase proteins. Phylogenetic relationship of GhLAC15, AtLAC proteins and two other plant laccases. The phylogenetic tree was constructed using DNAMAN6.0 Multiple Sequence Alignment programs. GhLAC15 is marked in red. The two letters preceding the protein name describe the organism from which the sequence was derived: At, Arabidopsis thaliana; Pt, Populus trichocarpa. LAC, laccase. AtLAC1 (NM_101674), AtLAC2 (NM_128470), AtLAC3 (NM_128574), AtLAC4 (NM_129364), AtLAC5 (NM_129597), AtLAC6 (NM_130222), AtLAC7 (NM_111756), AtLAC8 (NM_120181), AtLAC9 (NM_120182), AtLAC10 (NM_120197), AtLAC11 (NM_120404), AtLAC12 (NM_120621), AtLAC13 (NM_120795), AtLAC14 (NM_120972), AtLAC15 (NM_124184), AtLAC16 (NM_125281), AtLAC17 (NM_125395), PtLac (XM_002325536). The bar indicates the relative branch length. GhLAC15 and GhLAC17 laccases from Arabidopsis were used to build the tree. The branch length is proportional to the number of substitutions per site and represents evolutionary distance, as indicated by the scale bar.
Fig. S2 Multiple alignment of the deduced amino acid sequences of GhLAC15 and other plant laccases. The amino acid sequences of GhLAC15, GaLAC, PtLAC and Arabidopsis thaliana (AtLAC4 and AtLAC17) were aligned with ClustalW software. The consensus sequence of the cell attachment sequence (RGD) is marked by asterisks. The multicopper oxidase signature (HCHLERHSSWGM) is boxed. Three copper oxidase‐like domains are underlined. Dots represent gaps introduced to maximize similarities.
Fig. S3 Expression of GhLAC15 in different tissues (root, stem and leaf), tested through quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). GhActin was used as an internal control. Data are presented as average values with standard deviation (n = three technical replicates).
Fig. S4 Primer efficiencies for GhLAC15 and GhActin primer pairs.
Fig. S5 Correlation between GhLAC15 gene expression level and corresponding disease resistance in silenced seedlings.
Table S1 Primers used in this study.
Table S2 Cotton varieties used for GhLAC15 gene expression analysis.
Acknowledgements
This work was financially supported by the National Key Research and Development Program (2016YFD0101405), the Fund of the China Agriculture Research System (CARS18‐08), the Natural Science Foundation of Hebei Province (C2017204011) and Young Talents Support Program of Hebei Province.
References
- Adams‐Phillips, L. , Briggs, A.G. and Bent, A.F. (2010) Disruption of poly(ADP‐ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 152, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baayen, R.P. , Ouellette, G.B. and Rioux, D. (1996) Compartmentalization of decay in carnations resistant to Fusarium oxysporum f. sp. dianthi . Phytopathology, 86, 1018–1031. [Google Scholar]
- Barros‐Riosa, J. , Malvara, R.A. , Jung, H‐J.G. and Santiagoa, R. (2011) Cell wall composition as a maize defense mechanism against corn borers. Phytochemistry, 72, 365–371. [DOI] [PubMed] [Google Scholar]
- Bell, A.A. (1992) Verticillium wilt. Cotton Dis, Hillocks, R.J. (Ed.). CAB International, Oxon, UK. Pp. 87‐126. [Google Scholar]
- Bellincampi, D. , Cervone, F. and Lionetti, V. (2014) Plant cell wall dynamics and wall‐related susceptibility in plant–pathogen interactions. Front. Plant Sci. 5, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet, S. , Demont‐Caulet, N. , Pollet, B. , Bidzinski, P. , Cezard, L. , Le Bris, P. and Borrega, N. (2011) Disruption of LACCASE4 and 17 results in tissue‐specific alterations to lignification of Arabidopsis stems. Plant Cell, 23, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan, N.H. , Selvaraj, G. , Wei, Y. and King, J. (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümke, A. , Falter, C. , Herrfurth, C. , Sode, B. , Bode, R. , Schäfer, W. and Feussner, I. (2014) Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity‐related callose formation during wheat head infection. Plant Physiol. 165, 346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolek, Y. , El‐Zik, K.M. , Pepper, A.E. , Bell, A.A. , Magill, C.W. , Thaxton, P.M. and Reddy, O.U.K. (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bonello, P. and Blodgett, J.T. (2003) Pinus nigra–Sphaeropsis sapinea as a model pathosystem to investigate local and systemic effects of fungal infection of pines. Physiol. Mol. Plant Pathol. 63, 249–261. [Google Scholar]
- Cai, Y.F. , He, X.H. , Mo, J.C. , Sun, Q. , Yang, J.P. and Liu, J.G. (2009) Molecular research and genetic engineering of resistance to Verticillium wilt in cotton. Afr. J. Biotechnol. 8, 7363–7372. [Google Scholar]
- Cantu, D. , Vicente, A.R. , Labavitch, J.M. , Bennett, A.B. and Powell, A.L. (2008) Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci. 13, 610–617. [DOI] [PubMed] [Google Scholar]
- Chezem, W.R. , Memon, A. , Li, F.S. , Weng, J.K. and Clay, N.K. (2017) SG2‐type R2R3‐MYB transcription factor MYB15 controls defense‐induced lignification and basal immunity in Arabidopsis . Plant Cell, 29, 1907–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, J. , Henderson, M. , Schweizer, P. , Burton, R.A. , Fincher, G.B. and Little, A. (2014) Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei . New Phytol. 204, 650–660. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R. , Ehrhardt, D.W. , Griffitts, J.S. and Somerville, C.R. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin, L.B. and Lewis, N.G. (1992) Phenylpropanoid metabolism: biosynthesis of monolignols, lignans and neolignans, lignins and suberins In: Phenolic Metabolism in Plants (Stafford, H.A. and Ibrahim R.K., eds), pp. 325–375. New York: Plenum Press. [Google Scholar]
- Dence, C. (1992) Lignin determination In: Methods in Lignin Chemistry (Lin S.Y. and Dence C.W., eds), pp. 33–61. Berlin: Springer‐Verlag. [Google Scholar]
- De Vleesschauwer, D. , Xu, J. and Höfte, M. (2014) Making sense of hormone mediated defense networking: from rice to Arabidopsis . Front. Plant Sci. 5, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind, Y. , Edwards, R. , Mavandad, M. , Hedrick, S.A. , Ribak, O. , Dixon, R.A. and Lamb, C.J. (1990) Abnormal plant development and down‐regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia‐lyase gene. Proc. Natl. Acad. Sci. USA, 87, 9057–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, B. and Shan, L. (2014) ROS open roads to roundworm infection. Sci. Signal. 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X.Q. , Wheeler, T. , Li, Z.H. , Kenerley, C.M. , He, P. and Shan, L.B. (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon, T. , Sibout, R. , Pollet, B. , Maba, B. , Nussaume, L. , Bechtold, N. and Lu, F. (2003) A new Arabidopsis thaliana mutant deficient in the expression of O‐methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51, 973–989. [DOI] [PubMed] [Google Scholar]
- Hu, Q. , Min, L. , Yang, X.Y. , Jin, S.X. , Zhang, L. , Li, Y.Y. , Ma, Y.Z. , Qi, X.W. , Li, D.Q. , Liu, H.B. , Lindsey, K. , Zhu, L.F. and Zhang, X.L. (2018) Laccase GhLac1 modulates broad‐spectrum biotic stress tolerance via DAMP‐triggered immunity. Plant Physiol. 176, 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Gu, M. , Lai, Z. , Fan, B. , Shi, K. , Zhou, Y.H. and Yu, J.Q. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigard, J.A. and Donofrio, N.M. (2011a) HYR1‐mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7, e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigard, J.A. and Donofrio, N.M. (2011b) Suppression of plant‐generated reactive oxygen species is required for successful infection by the rice blast fungus. Virulence, 2, 559–562. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kapp, N. , Barnes, W.J. , Richard, T.L. and Anderson, C.T. (2015) Imaging with the fluorogenic dye Basic Fuchsin reveals subcellular patterning and ecotype variation of lignification in Brachypodium distachyon . J. Exp. Bot. 66, 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, B.C. , Waxman, K.D. , Nenni, N.V. , Walker, L.P. , Bergstrom, G.C. and Gibson, D.M. (2011) Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels, 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi‐Kaboshi, M. , Okada, K. , Kurimoto, L. , Murakami, S. , Umezawa, T. , Shibuya, N. , Yamane, H. , Miyao, A. , Takatsuji, H. , Takahashi, A. and Hirochika, H. (2010) A rice fungal MAMP responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis, G. , Galatis, B. , Quader, H. , Galanopoulou, D. and Apostolakos, P. (2008) Phospholipase C signaling involvement in macrotubule assembly and activation of the mechanism regulating protoplast volume in plasmolyzed root cells of Triticum turgidum . New Phytol. 178, 267–282. [DOI] [PubMed] [Google Scholar]
- La Camera, S. , Gouzerh, G. , Dhondt, S. , Hoffmann, L. , Fritig, B. , Legrand, M. and Heitz, T. (2004) Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198, 267–284. [DOI] [PubMed] [Google Scholar]
- Lacombe, S. , Rougon‐Cardoso, A. , Sherwood, E. , Peeters, N. , Dahlbeck, D. , van Esse, H.P. and Smoker, M. (2010) Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Lapierre, C. and Sandermann, H. Jr (1995) Elicitor‐induced spruce stress lignin: structural similarity to early developmental lignins. Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre, C. , Pollet, B. and Rolando, C. (1995) New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res. Chem. Intermed. 21, 397–412. [Google Scholar]
- Lee, S. , Sharm, Y. , Lee, T.K. , Chang, M. and Davis, K.R. (2001) Lignification induced by pseudomonads harboring avirulent genes on Arabidopsis. Mol. Cells, 12, 25–31. [PubMed] [Google Scholar]
- Li, C.H. , Feng, Z.L. , Li, Z.F. , Zhang, Z.G. , Shi, Y.Q. , Zhao, L.H. and Zhu, H.Q. (2015) In vitro sensitivity of Verticillium dahliae Kleb strains against some effective fungicides. China Cotton, 42, 16–18. [Google Scholar]
- Li, F. , Zhang, M. , Guo, K. , Hu, Z. , Zhang, R. , Feng, Y. and Yi, X. (2015) High‐level hemicellulosic arabinose predominantly affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol. J. 13, 514–525. [DOI] [PubMed] [Google Scholar]
- Lionetti, V. , Giancaspro, A. , Fabri, E. , Giove, S.L. , Reem, N. , Zabotina, O.A. , Blanco, A. , Gadaleta, A. and Bellincampi, D. (2015) Cell wall traits as potential resources to improve resistance of durum wheat against Fusarium graminearum . BMC Plant Biol. 15, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B.M. (2007) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry, 68, 513–520. [DOI] [PubMed] [Google Scholar]
- Miedes, E. , Vanholme, R. , Boerjan, W. and Molina, A. (2014) The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar, Y. , Vanholme, R. , Boerjan, W. , Ralph, J. and Mansfield, S.D. (2016) Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200. [DOI] [PubMed] [Google Scholar]
- Naoumkina, M.A. , Zhao, Q. , Gallego‐Giraldo, L. , Dai, X. , Zhao, P.X. and Dixon, R.A. (2010) Genome‐wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11, 829–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. [Google Scholar]
- Pallas, J.A. , Paiva, N.L. , Lamb, C. and Dixon, R.A. (1996) Tobacco plants epigenetically suppressed in phenylalanine ammonia‐lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 10, 281–293. [Google Scholar]
- Pieterse, C.M. , Leon‐Reyes, A. , van der Ent, S. and van Wees, S.C. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pogorelko, G. , Lionetti, V. , Bellincampi, D. and Zabotina, O. (2013) Cell wall integrity: targeted post‐synthetic modifications to reveal its role in plant growth and defense against pathogens. Plant Signal. Behav. 8, e25435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar, F. , Novo, M. , Bernal, M.A. , Merino, F. and Barcelo, A.R. (2004) Changes in stem lignins (monomer composition and cross linking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during Verticillium wilt in Capsicum annuum . New Phytol. 163, 111–123. [DOI] [PubMed] [Google Scholar]
- Quentin, M. , Allasia, V. , Pegard, A. , Allais, F. , Ducrot, P.H. , Favery, B. , Levis, C. , Martinet, S. , Masur, C. , Ponchet, M. , Roby, D. , Schlaich, N.L. , Jouanin, L. and Keller, H. (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 5, e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertsen, B. (1986) Elicitors of the production of lignin‐like compounds in cucumber hypocotyls. Physiol. Mol. Plant Pathol. 28, 137–148. [Google Scholar]
- Sattler, S.E. and Funnell‐Harris, D.L. (2013) Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front. Plant Sci. 4, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist, J. , Jeblick, W. and Kauss, H. (1994) Defense responses in infected and elicited cucumber (Cucumis sativus L.) hypocotyl segments exhibiting acquired resistance. Plant Physiol. 105, 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skyba, O. , Douglas, C.J. and Mansfield, S.D. (2013) Syringyl‐rich lignin renders poplars more resistant to degradation by wood decay fungi. Appl. Environ. Microbiol. 79, 2560–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter, A. , Hames, B. , Ruiz, R. , Scarlata, C. , Sluiter, J. , Templeton, D. and Crocker, D. (2013) NERL: determination of structural carbohydrates and lignin in biomass [EB]. Available at: https://www.nrel.gov/biomass/analytical_procedures.html [accessed on 15 March 2013].
- Smit, F. and Dubery, I.A. (1997) Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry, 44, 811–815. [Google Scholar]
- Sticher, L. , Mauch‐Mani, B. and Métraux, J.P. (1997) Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. [DOI] [PubMed] [Google Scholar]
- Tayi, L. , Maku, R. , Patel, H.K. and Sonti, R.V. (2016) Action of multiple cell wall‐degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol. Plant–Microbe Interact. 29, 599–608. [DOI] [PubMed] [Google Scholar]
- Vance, C.P. (1980) Lignification as a mechanism in disease resistance. Annu. Rev. Phytopathol. 18, 259–288. [Google Scholar]
- Vanholme, R. , Cesarino, I. , Rataj, K. , Xiao, Y.G. , Sundin, L. , Goeminne, G. , Kim, H. , Cross, J. , Morreel, K. , Araujo, P. , Welsh, L. , Haustraete, J. , McClellan, C. , Vanholme, B. , Ralph, J. , Simpson, G.G. , Halpin, C. and Boerjan, W. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science, 341, 1103–1106. [DOI] [PubMed] [Google Scholar]
- Vanholme, R. , Morreel, K. , Ralph, J. and Boerjan, W. (2010) Lignin biosynthesis. New Phytol. 187, 273–285.20642725 [Google Scholar]
- Wang, S.F. , Tian, H.Y. , Ma, Z.Y. , Zhang, G.Y. , Xuan, Z.L. , Wang, W.S. and Sun, Y.X. (2008) SSH library construction of upland cotton resistant cultivar under the stress of Verticillium dahliae . Cotton Sci. 1, 3–8. [Google Scholar]
- Wang, Y. , Bouchabke‐Coussa, O. , Lebris, P. , Antelme, S. , Soulhat, C. , Gineau, E. and Dalmais, M. (2015) LACCASE 5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 168, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liang, C. , Wu, S. , Zhang, X. , Tang, J. , Jian, G. , Jiao, G. , Li, F. and Chu, C. (2016) Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant, 9, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Wanyoike, M.W. , Kang, Z. and Heinrich, B. (2002) Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 108, 803–810. [Google Scholar]
- Wu, L.Z. , Wang, X.F. , Zhang, Y. , Li, X.H. , Zhang, G.Y. , Wu, L.Q. , Li, Z.K. and Ma, Z.Y. (2014) Function of acid insoluble lignin and GhLaccase in cotton resistance to Verticillium wilt. Acta Agron. Sin. 40, 1157–1163. [Google Scholar]
- Wu, S. , Shan, L. and He, P. (2014) Microbial signature‐triggered plant defense responses and early signaling mechanisms. Plant Sci. 228, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhu, L. , Tu, L. , Liu, L. , Yuan, D. , Jin, L. , Long, L. and Zhang, X. (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA‐Seq‐dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Y. , Liang, Y.B. , Qiu, D.W. , Zeng, H.M. , Yuan, J.J. and Yang, X.F. (2018) Lignin metabolism involves Botrytis cinerea BcG1‐induced defense response in tomato. BMC Plant Biol. 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Jensen, J.D. , Svensson, B. , Jorgensen, H.J.L. , Collinge, D.B. and Finnie, C. (2012) Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat. Mol. Plant Pathol. 13, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.Y. , Wang, X.F. , Zhang, G.Y. , Wu, L.Q. , Chi, J.N. , Li, Z.K. and Ma, Z.Y. (2010) ESTs analysis of suppression subtractive hybridization library from upland cotton resistant cultivar infected by Verticillium dahliae. Cotton Sci. 1, 17–22. [Google Scholar]
- Zhang, Y. , Wang, X.F. , Rong, W. , Yang, J. and Ma, Z.Y. (2016) Island cotton Enhanced Disease Susceptibility 1 gene encoding a lipase‐like protein plays a crucial role in response to Verticillium dahliae by regulating the SA level and H2O2 accumulation. Front. Plant Sci. 7, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X.F. , Rong, W. , Yang, J. , Li, Z.K. , Wu, L.Q. , Zhang, G.Y. and Ma, Z.Y. (2017) Histochemical analyses reveal that stronger intrinsic defenses in Gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae . Mol. Plant–Microbe Interact. 30, 984–996. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Gallego‐Giraldo, L. , Wang, H. , Zeng, Y. , Ding, S.Y. , Chen, F. and Dixon, R.A. (2010) An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula . Plant J. 63, 100–114. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Nakashima, J. , Chen, F. , Yin, Y.B. , Fu, C.X. , Yun, J.F. and Shao, H. (2013) LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis . Plant Cell, 25, 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2014) Plant pattern‐recognition receptors. Trends Immunol. 35, 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic analysis of laccase proteins. Phylogenetic relationship of GhLAC15, AtLAC proteins and two other plant laccases. The phylogenetic tree was constructed using DNAMAN6.0 Multiple Sequence Alignment programs. GhLAC15 is marked in red. The two letters preceding the protein name describe the organism from which the sequence was derived: At, Arabidopsis thaliana; Pt, Populus trichocarpa. LAC, laccase. AtLAC1 (NM_101674), AtLAC2 (NM_128470), AtLAC3 (NM_128574), AtLAC4 (NM_129364), AtLAC5 (NM_129597), AtLAC6 (NM_130222), AtLAC7 (NM_111756), AtLAC8 (NM_120181), AtLAC9 (NM_120182), AtLAC10 (NM_120197), AtLAC11 (NM_120404), AtLAC12 (NM_120621), AtLAC13 (NM_120795), AtLAC14 (NM_120972), AtLAC15 (NM_124184), AtLAC16 (NM_125281), AtLAC17 (NM_125395), PtLac (XM_002325536). The bar indicates the relative branch length. GhLAC15 and GhLAC17 laccases from Arabidopsis were used to build the tree. The branch length is proportional to the number of substitutions per site and represents evolutionary distance, as indicated by the scale bar.
Fig. S2 Multiple alignment of the deduced amino acid sequences of GhLAC15 and other plant laccases. The amino acid sequences of GhLAC15, GaLAC, PtLAC and Arabidopsis thaliana (AtLAC4 and AtLAC17) were aligned with ClustalW software. The consensus sequence of the cell attachment sequence (RGD) is marked by asterisks. The multicopper oxidase signature (HCHLERHSSWGM) is boxed. Three copper oxidase‐like domains are underlined. Dots represent gaps introduced to maximize similarities.
Fig. S3 Expression of GhLAC15 in different tissues (root, stem and leaf), tested through quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). GhActin was used as an internal control. Data are presented as average values with standard deviation (n = three technical replicates).
Fig. S4 Primer efficiencies for GhLAC15 and GhActin primer pairs.
Fig. S5 Correlation between GhLAC15 gene expression level and corresponding disease resistance in silenced seedlings.
Table S1 Primers used in this study.
Table S2 Cotton varieties used for GhLAC15 gene expression analysis.


