Summary
The cyclic adenosine monophosphate (cAMP) signalling pathway mediates signal communication and sensing during infection‐related morphogenesis in eukaryotes. Many studies have implicated cAMP as a critical mediator of appressorium development in the rice blast fungus, Magnaporthe oryzae. The cAMP phosphodiesterases, MoPdeH and MoPdeL, as key regulators of intracellular cAMP levels, play pleiotropic roles in cell wall integrity, cellular morphology, appressorium formation and infectious growth in M. oryzae. Here, we analysed the roles of domains of MoPdeH and MoPdeL separately or in chimeras. The results indicated that the HD and EAL domains of MoPdeH are indispensable for its phosphodiesterase activity and function. Replacement of the MoPdeH HD domain with the L1 and L2 domains of MoPdeL, either singly or together, resulted in decreased cAMP hydrolysis activity of MoPdeH. All of the transformants exhibited phenotypes similar to that of the ΔMopdeH mutant, but also revealed that EAL and L1 play additional roles in conidiation, and that L1 is involved in infectious growth. We further found that the intracellular cAMP level is important for surface signal recognition and hyphal autolysis. The intracellular cAMP level negatively regulates Mps1‐MAPK and positively regulates Pmk1‐MAPK in the rice blast fungus. Our results provide new information to better understand the cAMP signalling pathway in the development, differentiation and plant infection of the fungus.
Keywords: appressorium formation, cAMP level, MoPdeH/L, pathogenicity, phosphodiesterase activity
Introduction
Cyclic adenosine monophosphate (cAMP) acts as a widespread second messenger in the heterotrimeric G‐protein signalling pathway, which transmits extracellular signals to intracellular downstream signalling components (Daniel et al., 1998). The intracellular cAMP level depends on the balance between synthesis by adenylyl cyclases and degradation by cAMP phosphodiesterases (PDEases) (Colicelli et al., 1990; Sass et al., 1986). The cAMP signalling pathway plays critical roles in the regulation of the growth and differentiation of yeast and filamentous fungi, including the rice blast fungus, Magnaporthe oryzae (Bencina et al., 1997; D'Souza and Heitman, 2001; Durrenberger et al., 1998; Pan and Heitman, 1999; Ramanujam and Naqvi, 2010; Zhang et al., 2011a, 2011b), the causal agent of rice blast, which is the most destructive disease of cultivated rice worldwide (Talbot, 2003).
In M. oryzae, the cAMP signalling pathway and its components play vital roles in vegetative growth, asexual/sexual development, appressorium formation, cell wall integrity and plant infection (Choi and Dean, 1997; Lee and Dean, 1993; Ramanujam and Naqvi, 2010; Zhang et al., 2011a, 2011b). Intracellular cAMP levels are mainly regulated by the biosynthesis of adenylate cyclase MoMac1 and the hydrolysis of the PDEases MoPdeH and MoPdeL in M. oryzae (Ramanujam and Naqvi, 2010; Zhang et al., 2011a). Deletion of MoMAC1 results in defects in vegetative growth, asexual/sexual development, appressorium formation and penetration (Choi and Dean, 1997). Our previous study and that by Naqvi's laboratory have indicated that MoPdeH plays a major role in cAMP hydrolysis, and regulates surface signal recognition, conidiogenesis, cell wall integrity and plant infection; MoPdeL plays a minor role in cAMP hydrolysis, and only regulates conidial morphology in M. oryzae (Ramanujam and Naqvi, 2010; Zhang et al., 2011a).
MoPdeH and MoPdeL contain signature sequences of high‐ and low‐affinity cAMP PDEases (Hicks et al., 2005; Ramanujam and Naqvi, 2010; Zhang et al., 2011a). We named the conserved domains HD in MoPdeH, and L1 and L2 in MoPdeL. The HD domain is found in a superfamily of enzymes with predicted or known phosphohydrolase activity (Galperin et al., 2001). It also contains a conserved EAL (E, glutamine, A, alanine, L, leucine) domain, which is required for PDEase activity and negative regulation of biofilm formation in Yersinia pestis (Tamayo et al., 2005). In addition, it is also essential for cyclic diguanylate monophosphate (c‐di‐GMP) PDEase activity and virulence in Vibrio cholerae (Bobrov et al., 2005). Structure prediction has indicated that MoPdeH also possesses an EAL domain in the HD domain. These domains probably play conserved and critical roles in different organisms. Here, we characterized the HD, EAL, L1 and L2 domains separately and in chimeras. The results not only demonstrated the essential roles of HD and EAL for MoPdeH, but also yielded new findings with regard to the cAMP signalling pathway during development and virulence mediated by MoPdeH in the rice blast fungus.
Results
Overexpression of MoPdeL in ΔMopdeH partially rescued its intracellular cAMP level and appressorial formation on a hydrophilic surface
Previously, we have reported that MoPdeH is the major PDEase to degrade cAMP in M. oryzae, and is important for conidial morphology, infectious growth, cell wall integrity and surface signal recognition, whereas MoPdeL plays a minor role in cAMP degradation and is only involved in conidial morphology (Zhang et al., 2011a). To investigate whether MoPdeL could rescue the defects of the ΔMopdeH mutant, we expressed a fusion of MoPdeL with green fluorescent protein (MoPdeL‐GFP) in the ΔMopdeH mutant, and obtained a transformant designated MoPdeL which expressed MoPdeL‐GFP at a level 3.3‐fold higher than that of MoPdeL in the wild‐type. A high‐performance liquid chromatography (HPLC) assay revealed that the intracellular cAMP level in the MoPdeL transformant was reduced to 66.7% of that in the ΔMopdeH mutant, but was twice that in the wild‐type (Fig. 1A). Phenotypic analysis showed that most phenotypes of the MoPdeL transformant were similar to that of the ΔMopdeH mutant, with defects in hyphal autolysis, conidial morphology and production, and virulence (Fig. 1B,C). However, appressorial formation on a hydrophilic surface was partially rescued and was reduced to 35.3% in the MoPdeL transformant compared with 69.4% in the ΔMopdeH mutant (Fig. 1D). These results indicate that the intracellular cAMP level is critical for the development and virulence of the rice blast fungus. Different developmental stages require different cAMP levels to maintain normal physiological and biochemical activities. Hyphal cell wall integrity, conidial morphology and production, and virulence require lower cAMP levels than appressorial formation on a hydrophilic surface.
Figure 1.
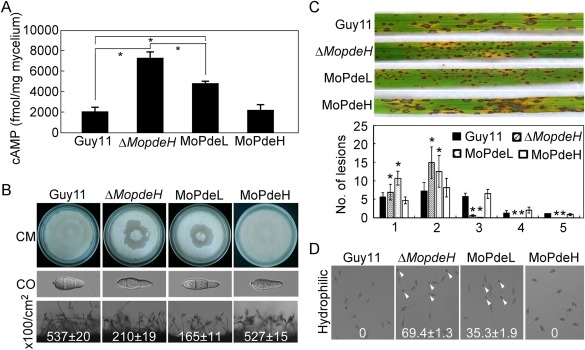
Phenotypic analysis of the MoPdeL overexpression strain. (A) Measurement of the intracellular cyclic adenosine monophosphate (cAMP) level in the indicated strains. (B) Mycelial autolysis (top panel), conidial morphology (middle panel) and conidiation (bottom panel) of the wild‐type Guy11, ΔMopdeH mutant, MoPdeL (MoPdeL overexpressed in ΔMopdeH mutant) and MoPdeH (ΔMopdeH mutant complemented transformant) strains. CO, conidium. (C) Rice spraying assay with conidial suspensions (5 × 104 spores/mL) prepared from the indicated strains, and lesion type statistical analysis: 0, no lesions; 1, dark‐brown pinpoint lesions; 2, 1.5‐mm brown spots; 3, 2–3‐mm lesions with brown margins; 4, eyespot lesions longer than 3 mm; 5, coalesced lesions infecting 50% or more of the leaf maximum size. Lesions were photographed and measured at 7 days post‐inoculation (dpi). The error bars indicate the standard deviation (SD) of three replicates. Asterisks indicate significant differences at P < 0.01. (D) Appressorium formation on hydrophilic surfaces photographed at 24 h post‐inoculation. White arrowheads indicate appressoria.
Structural identification and vector construction
Structural prediction (http://smart.embl-heidelberg.de/) revealed that the MoPdeH protein contains a phosphohydrolase domain (HD, amino acids 362–554) and a motif enriched in glutamine, alanine and leucine (EAL, amino acids 415–417). MoPdeL contains two PDEase domains (L1, amino acids 17–183; L2, amino acids 253–416). To investigate the contributions of these domains to MoPdeH and MoPdeL, we constructed domain mutation and chimeric vectors (Fig. 2A) and introduced them into the ΔMopdeH mutant. The resulting transformants were screened by GFP signals and the predicted size of the fusion proteins was confirmed by western blot analysis (Fig. S1, see Supporting Information). The final transformants MoPdeHEAL(AAA) (mutation of EAL to AAA), MoPdeHΔ HD (deletion of the HD domain), MoPdeHΔ HD+L1L2 (replacement of HD with both L1 and L2), MoPdeHΔ HD+L1 (replacement of HD with L1), MoPdeHΔ HD+L2 (replacement of HD with L2) and MoPdeH (full length) were used for phenotypic analysis. As all three transformants expressing each construct displayed the same phenotype (Figs S2–S5, see Supporting Information), only one transformant of each construct was presented in the main text.
Figure 2.
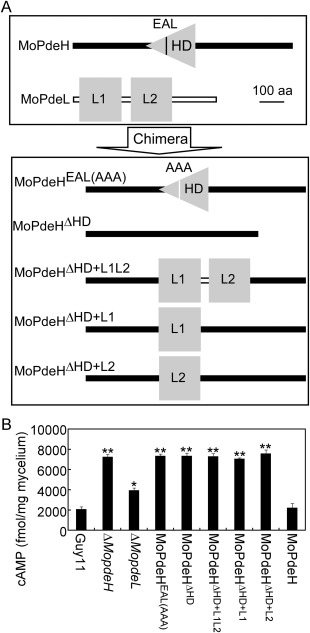
Strategy for domain mutation and chimera construction, and intracellular cyclic adenosine monophosphate (cAMP) measurement. (A) MoPdeHEAL(AAA), mutation of EAL to AAA in MoPdeH; MoPdeHΔ HD, deletion of HD domain in MoPdeH; MoPdeHΔ HD+L1L2 , replacement of HD with L1&L2 domain of MoPdeL; MoPdeHΔ HD+L1, replacement of HD with L1 domain of MoPdeL; MoPdeHΔ HD+L2, replacement of HD with L2 domain of MoPdeL. (B) Domain mutation and chimeric transformants, ΔMopdeH mutant and ΔMopdeL mutant, lead to increased accumulation of cAMP levels. Bar chart showing the quantification of intracellular cAMP in the mycelia of the above strains cultured for 2 days in complete medium. Two biological repetitions with three replicates were assayed. The error bars indicate the standard deviation (SD) of three replicates. Asterisks indicate significant differences at P < 0.01.
Intracellular cAMP level was not rescued in all transformants expressing domain mutation or chimeric vectors
We first measured the intracellular cAMP level of related transformants in comparison with the wild‐type Guy11, ΔMopdeH mutant, ΔMopdeL mutant and complement of the ΔMopdeH mutant. The results showed that MoPdeHEAL(AAA) and MoPdeHΔ HD exhibited the same cAMP level as the ΔMopdeH mutant, which was 3.5‐fold that of the wild‐type. Three chimeric transformants, MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1 and MoPdeHΔ HD+L2, also showed cAMP levels similar to that of the ΔMopdeH mutant, but much higher than that of the ΔMopdeL mutant (Fig. 2B). These results indicate that the HD domain is essential for cAMP hydrolysis activity of MoPdeH, and the EAL domain is indispensable for the function of the HD domain. However, the L1 and L2 domains, together or separately, are unable to replace the role of the HD domain in MoPdeH, and probably lose their phosphohydrolase activity in the chimera.
HD and EAL domains were indispensable for MoPdeH to maintain hyphal integrity and surface hydrophobicity
To investigate the roles of HD, EAL and chimeras in hyphal autolysis and surface hydrophobicity, the indicated strains were inoculated onto complete medium (CM) and sporulation medium (SDC) plates and cultured at 28 °C for 14 days. The results showed that only full‐length MoPdeH rescued the autolysis defect of the ΔMopdeH mutant on CM plates. The other transformants exhibited similar colony morphology to the mutant, with hyphal autolysis in the centre of the colonies. However, all transformants showed similar colony morphology to the wild‐type Guy11 on SDC plates under the same conditions (Fig. 3A). When inoculated with water droplets, domain mutation and chimeric transformants did not show an easily wettable phenotype, but were more readily wettable by detergent solutions, resulting in a soaked, wettable phenotype on aerial hyphae. The expression level of the hydrophobin gene, MPG1, was also significantly down‐regulated in all domain mutation and chimeric transformants (Fig. 3B), consistent with the ΔMopdeH mutant.
Figure 3.
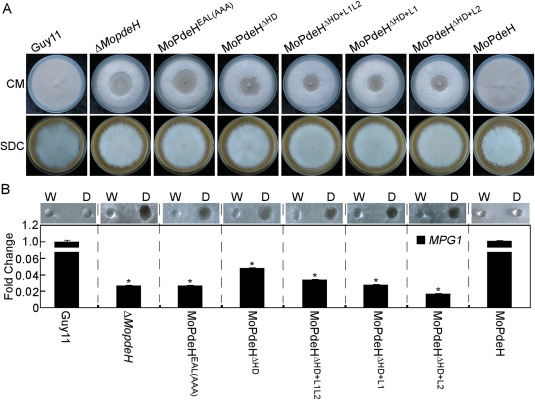
Defects of the indicated strains in mycelial autolysis and surface hydrophobicity. (A) The indicated strains were inoculated on complete medium (CM) and sporulation medium (SDC) plates for 14 days and photographed. (B) Ten microlitres of water (W) or detergent solution containing 0.02% sodium dodecylsulfate (SDS) and 5 mm ethylenediaminetetraacetic acid (EDTA) (D) were placed on the colony surfaces of the indicated strains cultured on CM plates for 14 days, and photographed after 5 min (top panel); quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of the expression of the MPG1 gene in the indicated strains is shown in the bottom panel. Error bars indicate the standard deviation (SD) of three replicates. Asterisks indicate significant differences at P < 0.01.
Conidial production and morphology were not restored in all domain mutation and chimeric transformants
The wild‐type Guy11, ΔMopdeH mutant and all transformants were inoculated onto SDC plates to promote conidiation. Compared with Guy11, MoPdeHEAL(AAA), MoPdeHΔ HD, MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1 and MoPdeHΔ HD+L2 transformants produced similar or fewer conidiophores and conidia when observed on glass slides (Fig. 4A). Further statistical analysis revealed that the conidial numbers of MoPdeHΔ HD, MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2 were similar to that of the ΔMopdeH mutant, which was 55% that of the wild‐type. MoPdeHEAL(AAA) and MoPdeHΔ HD+L1 produced many fewer conidia than the ΔMopdeH mutant, corresponding to 14% and 18% of the wild‐type, respectively (Table 1). Conidial morphology was also not restored in these transformants and they still produced thinner and elongated conidia, similar to those of the ΔMopdeH mutant (Fig. 4B). These results indicate that the HD domain is essential for the function of MoPdeH, and normal phosphohydrolase activity of MoPdeH is indispensable for conidiation in M. oryzae.
Figure 4.
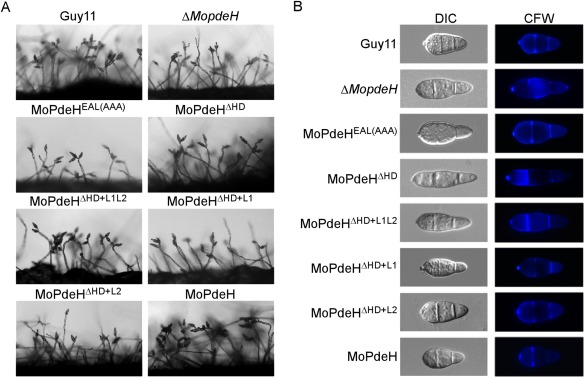
Conidial production and morphological observation of the indicated strains. (A) Conidial formation was observed under a light microscope after illumination for 14 h on a glass slide under fluorescent light and photographed. (B) Conidia of the wild‐type and transformants were stained with 1 mg/mL calcofluor white (CFW) for 5 min in the dark, and observed under a fluorescence microscope.
Table 1.
Phenotypic analysis of the indicated strains.
| Colony diameter (cm) † | Appressorium formation (%) § | Conidial size (μm) ¶ | |||||
|---|---|---|---|---|---|---|---|
| Strain | CM | SDC | Conidiation (×100/cm2) ‡ | Hydrophilic | Hydrophobic | Length | Width |
| Guy11 | 7.3 ± 0.1 | 5.5 ± 0.1 | 401.3 ± 27.8 | 0 | 99.2 ± 5.5 | 20.87 ± 0.59 | 9.78 ± 0.43 |
| ΔMopdeH | 7.3 ± 0.1 | 5.8± 0.2 | 221.5 ± 20.2* | 69.4 ± 1.3** | 98.8 ± 4.8 | 26.99 ± 0.42** | 8.94 ± 0.40 |
| MoPdeHEAL(AAA) | 7.1 ± 0.2 | 5.8 ± 0.1 | 56.7 ± 4.9** | 68.9 ± 0.8** | 99.6 ± 4.5 | 26.74 ± 1.34** | 8.76 ± 0.28 |
| MoPdeHΔHD | 7.3 ± 0.1 | 5.8 ± 0.2 | 237.6 ± 10.8* | 73.3 ± 3.1** | 99.5 ± 3.6 | 27.20 ± 0.06** | 8.37 ± 0.39 |
| MoPdeHΔHD+L1L2 | 7.2 ± 0.2 | 5.9 ± 0.2 | 169.4 ± 5.7** | 35.5 ± 5.3** | 97.5 ± 5.2 | 26.72 ± 0.78** | 8.20 ± 0.41 |
| MoPdeHΔHD+L1 | 7.2 ± 0.1 | 5.8 ± 0.1 | 73.8 ± 6.4** | 34.0 ± 2.0** | 98.6 ± 2.7 | 24.06 ± 0.36* | 8.88 ± 0.80 |
| MoPdeHΔHD+L2 | 7.1 ± 0.1 | 5.5 ± 0.1 | 211.6 ± 18.3* | 41.7 ± 4.8** | 99.0 ± 4.2 | 26.84 ± 1.51** | 8.33 ± 0.27 |
| MoPdeHHD | 7.2 ± 0.1 | 5.8 ± 0.1 | 367.9 ± 12.5* | 0 | 98.2 ± 2.3 | 21.15 ± 1.2 | 8.98 ± 0.81 |
| MoPdeH | 7.2± 0.2 | 5.8 ± 0.2 | 437.6 ± 32.7 | 0 | 99.3 ± 3.8 | 20.18 ± 0.53 | 9.34 ± 0.33 |
± standard deviation (± SD) was calculated from three repeated experiments and asterisks indicate statistically significant differences (P < 0.01).
†Colony diameter of the indicated strains on complete medium (CM) and sporulation medium (SDC) after 10 days of incubation at 28 °C.
‡Quantification of the conidial production of the indicated strains from SDC cultures.
§Appressorium formation on hydrophilic or hydrophobic surfaces at 24 h post‐inoculation (hpi).
¶Measurement of the conidial size of the indicated strains.
Appressorium formation on a hydrophilic surface was partially rescued in the MoPdeHΔHD+L1L2, MoPdeHΔHD+L1 and MoPdeHΔHD+L2 transformants
As the intracellular cAMP levels of domain mutation and chimeric transformants were much higher than those of the wild‐type Guy11, we examined appressorium formation on both artificial non‐inductive and inductive surfaces. After incubation for 24 h, all transformants, except Guy11 and the full‐length complemented strain, were able to form appressoria on non‐inductive surfaces (Fig. 5A). About 70% of conidia of MoPdeHEAL(AAA) and MoPdeHΔ HD formed melanized appressoria, which was similar to the rate of the ΔMopdeH mutant. However, the rates of appressorium formation by MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1 and MoPdeHΔ HD+L2 were 35.5%, 34.0% and 41.7%, respectively (Fig. 6A, Table 1). Similar to the wild‐type and the ΔMopdeH mutant, all transformants formed normal melanized appressoria on inductive surfaces (Fig. 5B, Table 1). Appressorial turgor on inductive surfaces did not differ markedly in all transformants compared with the wild‐type Guy11 and the ΔMopdeH mutant (Table S1, see Supporting Information).
Figure 5.
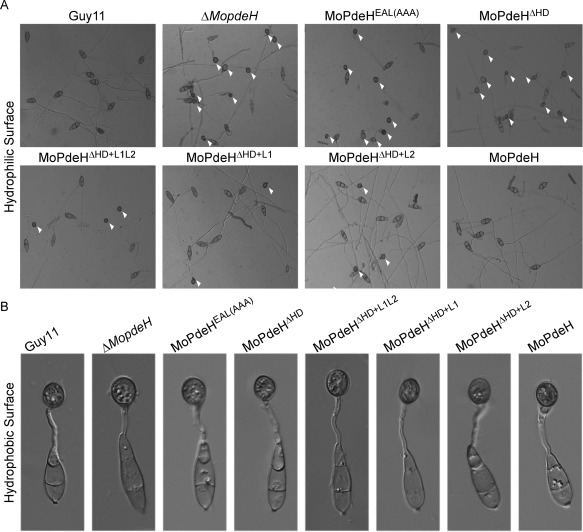
Appressorium formation of the indicated strains. (A) Conidia of the strains were incubated on a hydrophilic surface for 24 h and photographed. White arrowheads indicate appressoria. (B) Conidia of the strains were incubated on a hydrophobic surface for 24 h and photographed.
Figure 6.
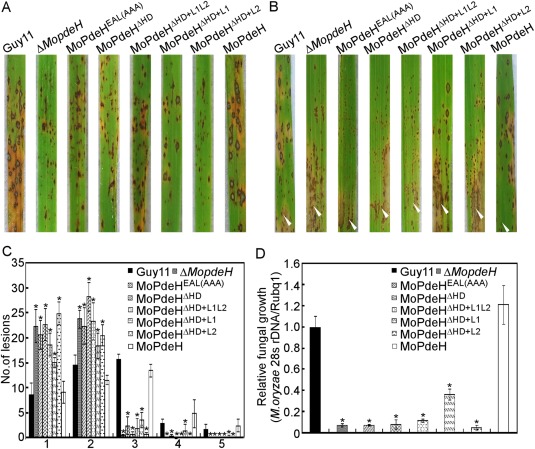
Defects of the indicated strains in pathogenicity. (A) Four millilitres of conidial suspension (5 × 104 spores/mL) of each strain were sprayed onto rice seedlings. Diseased leaves were photographed at 7 days post‐inoculation (dpi). (B) A conidial suspension (1 × 105 spores/mL) was injected into rice sheaths. Diseased leaves were photographed at 5 dpi. White arrowheads indicate injection sites. (C) Lesion type statistical analysis. Lesions were photographed and measured at 7 dpi. (D) The severity of disease was analysed by quantification of Magnaporthe oryzae genomic 28S rDNA relative to rice genomic Rubq1 DNA. Asterisks represent significant difference (P < 0.01).
Pathogenicity was partially rescued in the MoPdeHΔHD+L1 transformant
Conidial suspensions of the wild‐type Guy11, ΔMopdeH mutant and all transformants were sprayed or injected onto rice seedlings. At 7 days post‐inoculation (dpi), all domain mutation and chimeric transformants showed significantly reduced virulence, with tiny or fewer typical lesions compared with the numerous typical fusiform lesions of the wild‐type (Fig. 6A,B). Further lesion‐type scoring assays revealed that MoPdeHEAL(AAA), MoPdeHΔ HD, MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2 mainly produced type 1 and type 2 lesions, and almost no type 4 or type 5 lesions, similar to the ΔMopdeH mutant. However, compared with the ΔMopdeH mutant, MoPdeHΔ HD+L1 produced more type 3 and type 4 lesions, suggesting that it partially rescued the virulence defect of the ΔMopdeH mutant (Fig. 6C). Fungal biomass analysis in diseased leaves also confirmed the above conclusions (Fig. 6D).
Infectious hyphal growth was limited in all domain mutation and chimeric transformants
As all domain mutation and chimeric transformants caused tiny or fewer typical lesions, we supposed that they may have defects in infectious hyphal growth. To investigate this possibility, infectious growth was examined in rice leaf sheath epidermal cells. After 36 h of incubation, infectious hyphae of MoPdeHEAL(AAA), MoPdeHΔ HD, MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2 were limited to one cell with no or only one branch. Infectious hyphae of MoPdeHΔ HD+L1 were limited to one cell, but were longer than those of the former four transformants. In contrast, wild‐type Guy11 and MoPdeH formed extensive infectious hyphae spreading to neighbouring cells (Fig. 7A). Reactive oxygen species (ROS) generation and accumulation are obvious early features of plant–pathogen interactions (Liu et al., 2016; Wang et al., 2016), and the activation of ROS scavenging by phytopathogens is a mechanism for coping with plant defences to facilitate infection (Liu et al., 2016). As deletion of MoPdeH can elicit plant defence responses, we examined host‐derived ROS levels by staining with 3,3′‐diaminobenzidine (DAB) in rice cells infected by independent transformants. At 36 h post‐inoculation (hpi), 75%–90% of infected cells stained for ROS in the ΔMopdeH, MoPdeHEAL(AAA), MoPdeHΔ HD, MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2 strains. In contrast, only 20% of cells stained for ROS in MoPdeHΔ HD+L1 compared with 8% in the wild‐type (Fig. 7A,B). We also performed assays with diphenyleneiodonium (DPI) to inhibit the activity of plant NADPH oxidases, which are important for ROS generation in plants (Bolwell et al., 1998; Chen et al., 2014). After 36 h of incubation, the extension of infectious hyphae of ΔMopdeH, MoPdeHEAL(AAA), MoPdeHΔ HD, MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2 was restored to the levels of the wild‐type and MoPdeH, but not MoPdeHΔ HD+L1 (Fig. 8A). Additional infectious hyphal type statistical data were also consistent with the results of DAB and DPI assays (Fig. 7C). In addition, the expression levels of two rice defence‐related genes, PR1a and PBZ1, were strongly induced in all transformants except MoPdeHΔ HD+L1 at 48 hpi (Fig. 7D).
Figure 7.
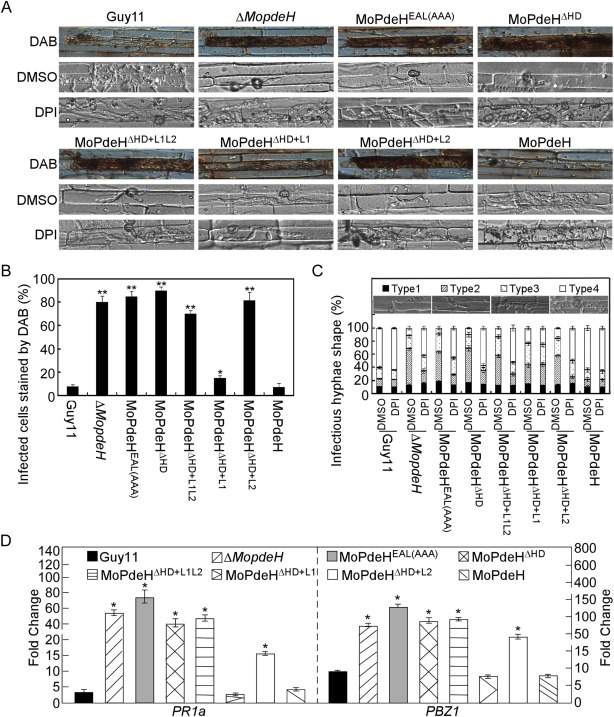
The indicated strains showed defects in infectious growth and induced a strong host defence response. (A) Conidial suspensions of the indicated strains treated with or without 0.5 µm diphenyleneiodonium (DPI) in dimethylsulfoxide (DMSO) were injected in rice leaf sheath and incubated for 36 h before staining with 3,3′‐diaminobenzidine (DAB) for 8 h and observed. (B) Percentage of cells with infectious hyphae stained by DAB. Data were analysed from three independent replicates. Asterisks represent significant difference (*P < 0.05, **P < 0.01). (C) Statistical analysis of the type of infectious hyphae treated with DPI or DMSO. DMSO treatment is a negative control. Three independent experiments were replicated. (D) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of the transcription levels of rice pathogenesis‐related (PR) genes at 48 h post‐inoculation (hpi). The average threshold cycle of triplicate reactions was normalized by the stable expression gene elongation factor 1α (Os03g08020) in Oryza sativa. Experiments were repeated three times with similar results. Asterisks represent significant difference (p < 0.01).
Figure 8.
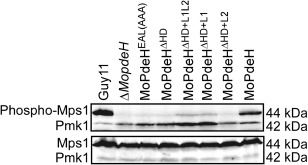
Defects of MoMps1 phosphorylation of the indicated strains. The total protein was harvested from mycelia cultured in liquid complete medium (CM) for 2 days. The phosphorylated MoMps1 was detected by binding of the antiphospho‐p44/42 antibody, with the Mpk1 antibody as a control.
All domain mutation and chimeric transformants showed defects in Mps1 phosphorylation
All transformants showed autolysis of hyphae and were defective in surface hydrophobicity, suggesting that they exhibited defects in cell wall integrity. In M. oryzae, the Mps1–mitogen‐activated protein kinase (MAPK) pathway is mainly involved in cell wall integrity and is important for appressorium penetration and infectious growth (Jeon et al., 2008; Xu et al., 1998; Yin et al., 2016). Therefore, we measured Mps1 phosphorylation levels in the transformants. The results revealed that the Mps1 phosphorylation level was almost abolished in all transformants, with only weak signals detected in MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L1. In contrast, strong signals were detected in the wild‐type Guy11 and MoPdeH strains (Fig. 8). Interestingly, compared with the wild‐type, the Pmk1 phosphorylation level was increased in all domain mutation and chimeric transformants (Fig. 8), indicating that the Pmk1‐MAPK signalling pathway was activated in these transformants.
HD domain of MoPdeH restored the ΔMopdeH mutant defects
Because MoPdeHΔ HD was unable to restore the defects of the ΔMopdeH mutant, we concluded that the HD domain was essential for the function of MoPdeH. To prove the role of the individual HD domain, the HD domain, driven by the native promoter of MoPdeH, was expressed in the ΔMopdeH mutant. The resulting transformants were confirmed by Western blot and the phenotype was analysed (Fig. S1). The results showed that MoPdeHHD rescued the autolysis, conidiation, conidial morphology, appressorium formation on a hydrophilic surface, virulence and intracellular cAMP level defects of the ΔMopdeH mutant (Fig. 9A–C, Table 1), indicating that the HD domain is sufficient for the full function of MoPdeH in M. oryzae.
Figure 9.
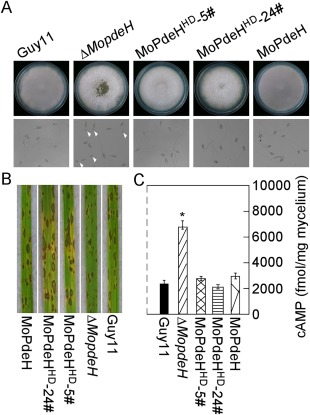
The HD domain rescued the defects of the ΔMopdeH mutant. (A) The indicated strains were inoculated on complete medium (CM) for 14 days and photographed (top panel). Conidia of the strains were incubated on a hydrophilic surface for 24 h and photographed (bottom panel). White arrowheads indicate appressoria. (B) Rice spraying assay with conidial suspensions (5 × 104 spores/mL) prepared from the indicated strains, and photographed at 7 days post‐inoculation (dpi). (C) Intracellular cyclic adenosine monophosphate (cAMP) level in the mycelia of the indicated strains cultured for 2 days in complete medium and quantified by high‐performance liquid chromatography (HPLC). The error bars indicate standard deviation (SD) of three replicates. Asterisk represents significant difference (P < 0.01).
HD and EAL domains were important for PDEase activity of MoPdeH in vitro
As the intracellular cAMP level was not rescued in all domain mutation and chimeric transformants, we speculate that phosphohydrolase activity was abolished in the domain mutation and chimera proteins. To test this possibility, we measured the phosphohydrolase activity by purifying the domain mutation and chimera proteins in vitro. It is known that cAMP generates AMP on hydrolysis by PDEase; AMP is then dephosphorylated by alkaline phosphatase, and the phosphate sensor can combine free inorganic phosphate (Pi), with increased fluorescence intensity (Brune et al., 1994). Our results showed that the samples treated with MoPdeH exhibited strong fluorescence intensity and those treated with MoPdeL showed relatively weak fluorescence intensity (52% of MoPdeH). However, the fluorescence intensity was decreased significantly in the samples treated with domain mutation and chimera proteins in comparison with MoPdeH and MoPdeL. The fluorescence intensity was decreased to 22% of that of MoPdeH in MoPdeHEAL(AAA), 11% in MoPdeHΔ HD, 27% in MoPdeHΔ HD+L1L2, 38% in MoPdeHΔ HD+L1 and 19% in MoPdeHΔ HD+L2 (Fig. 10). In addition, we also measured the phosphohydrolase activity of the individual HD, L1 and L2 domains, and found that the fluorescence intensity was 63% of MoPdeH in MoPdeHHD, 17% in MoPdeLL1L2, 38% in MoPdeLL1 and 5% in MoPdeLL2 (Fig. 10).
Figure 10.
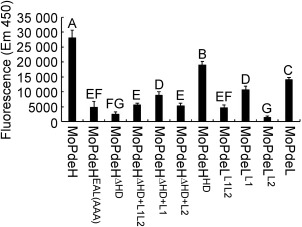
Detection of protein phosphodiesterase activity in vitro. The target proteins were expressed in Escherichia coli BL21‐CodonPlus (DE3) cells. The phosphate sensor was used to bind free inorganic phosphate (Pi), and fluorescence was read immediately at excitation (420 nm) and emission (450 nm). The error bars indicate the standard deviation of three replicates. Different letters indicate statistically significant differences (P < 0.01).
Suppressed adenylate cyclase in the ΔMopdeH mutant rescued its appressorium formation defect on a hydrophilic surface
In M. oryzae, adenylate cyclase plays a major role in intracellular cAMP biosynthesis, and deletion of its encoding gene MAC1 results in appressorium formation and infection defects, and decreased cAMP level (Choi and Dean, 1997, Zhou et al., 2012). To confirm whether the defects of ΔMopdeH relate directly to the intracellular cAMP level, we suppressed the adenylate cyclase activity by MDL‐12,330A, a type of adenylate cyclase inhibitor, in the wild‐type Guy11, ΔMopdeH mutant and complemented strain. The results showed that all three strains failed to form appressoria on both hydrophobic and hydrophilic surfaces on treatment with MDL‐12,330A; ΔMopdeH was even unable to produce germ tubes (Fig. 11A,B). Meanwhile, the ΔMopdeH mutant a showed similar growth rate to Guy11 and MoPdeH on treatment with MDL‐12,330A, which was remarkably decreased compared with the control (Fig. 11C). The intracellular cAMP level was also decreased significantly in Guy11 and the ΔMopdeH mutant on treatment with MDL‐12,330A. The treated Guy11 showed a similar cAMP level to the ΔMomac1 mutant, which was much lower than the control, and the treated ΔMopdeH showed 75% cAMP level compared with the control, but remained much higher than that of the wild‐type (Fig. 11D).
Figure 11.
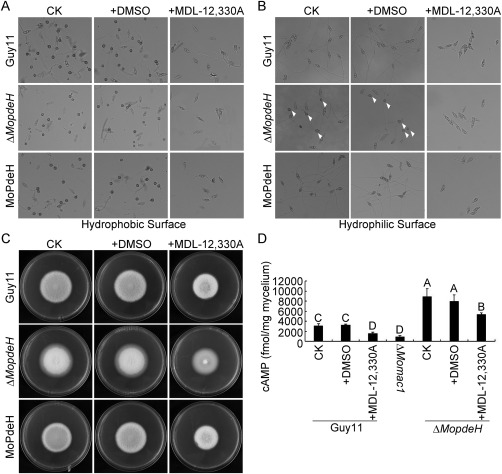
Suppressed adenylate cyclase in ΔMopdeH rescued its appressorium formation defect on a hydrophilic surface. (A, B) Conidia of the Guy11, ΔMopdeH mutant and complemented strain with 100 μm MDL‐12,330A and equal dimethylsulfoxide (DMSO) as a control were incubated on a hydrophobic or hydrophilic surface for 24 h and photographed. White arrowheads indicate appressoria. (C) Vegetative growth of the indicated strains on complete medium (CM) with or without MDL‐12,330A, and photographed at 7 days post‐inoculation (dpi). (D) Intracellular cAMP level in mycelia treated with or without MDL‐12,330A. The error bars indicate the standard deviation (SD) of three replicates. Different letters indicate statistically significant differences (P < 0.01).
Discussion
Here, we clarified the roles of major domains of MoPdeH and MoPdeL, separately or in chimeras, during development and virulence in M. oryzae. Overexpression of MoPdeL in the ΔMopdeH mutant decreased the intracellular cAMP level from 3.5‐ to 2.5‐fold that of the wild‐type, and only partially rescued appressorium formation on a hydrophilic surface, but did not rescue other defects. These results suggest that the cAMP level is critical for hyphal autolysis, conidial morphology and production, and infectious growth. It seems that a higher cAMP level is needed to disturb normal surface signal recognition than in other phenotypes in M. oryzae. Mutation HD and EAL domains of MoPdeH showed very similar intracellular cAMP levels and phenotypes to the ΔMopdeH mutant, whereas the expressed HD domain rescued the phenotypic defects of the ΔMopdeH mutant, indicating that HD and EAL domains are required for the PDEase activity of MoPdeH. This conclusion is supported by the in vitro PDEase activity data, which are also consistent with the findings in Y. pestis and V. cholerae, indicating that EAL is indispensable for PDEase activity (Bobrov et al., 2005; Tamayo et al., 2005). Surprisingly, all chimeric transformants showed high intracellular cAMP levels; this is a result of the defects in PDEase activity, which are similar to those in the ΔMopdeH mutant. One possible explanation is that the protein conformation may have been altered in the chimeras, thus affecting the PDEase activity. However, the homology modelling results revealed that MoPdeHΔ HD+L1 and MoPdeHΔ HD+L2 showed similar protein structures to MoPdeH (Fig. S6, see Supporting Information), and also showed similar PDEase activity to individual L1 or L2, suggesting that the phenotypic defects of MoPdeHΔ HD+L1 and MoPdeHΔ HD+L2 are a result of low PDEase activity, but not a change in protein structure. Although the protein structure of MoPdeHΔ HD+L1L2 was very close to that of MoPdeL (Fig. S6), indicating its change in protein conformation, this was defective in PDEase activity. In addition, all chimeric transformants also showed very similar phenotypes to the ΔMopdeH mutant, suggesting that these phenotypes may be closely related to the intracellular cAMP level in M. oryzae.
Our previous study indicated that the deletion of eight regulators of G‐protein signalling, MoRgs 1–8, resulted in increased intracellular cAMP levels and a series of phenotypic defects, including hyphal autolysis, appressorium formation, conidiation and virulence (Zhang et al., 2011b). In another study, we found that MoPdeH interacts with the cell wall integrity pathway Mck1‐Mkk1‐Mps1 (MAPK‐Mps1) (Jeon et al., 2008; Yin et al., 2016; Xu et al., 1998), and that the pathway positively modulates the expression of MoPdeH to affect intracellular cAMP levels (Yin et al., 2016). We also identified a protein phosphatase, MoYvh1, which functions upstream of MoPdeH and negatively regulates cAMP levels, conidial production, cell wall integrity and plant infection (Liu et al., 2016). These results indicate that a high intracellular cAMP level might have an important influence on asexual development, appressorium formation, cell wall integrity and virulence. In M. oryzae, the adenylate cyclase Mac1 mutant showed a very low intracellular cAMP level and was unable to form an appressorium on a hydrophobic surface (Choi and Dean, 1997; Zhou et al., 2012). In this study, suppressed adenylate cyclase in the ΔMopdeH mutant partially decreased its cAMP level, and blocked appressorium formation on a hydrophilic surface. This result supports the conclusion that the intracellular cAMP level might play an important role in surface recognition and appressorium formation. The cAMP pathway has also been reported to activate the Mst11‐Mst7‐Pmk1 pathway (MAPK‐Pmk1) and to be involved in appressorium formation and virulence (Park et al., 2006; Xu and Hamer, 1996; Zhang et al., 2011c; Zhao and Xu, 2007; Zhao et al., 2005). Together with the findings presented in this study, we conclude that the cAMP signalling pathway exhibits cross‐talk with the Mps1 and Pmk1 pathways; cAMP probably negatively regulates the Mps1 pathway and positively regulates the Pmk1 pathway in M. oryzae.
One visible phenotype is the abnormal conidial morphology of the ΔMopdeH mutant and chimeric transformants. However, the ΔMorgs1–8 and ΔMoyvh1 mutants showed no changes in conidial morphology, although they also showed high intracellular cAMP levels (Zhang et al., 2011b), indicating that other regulators or signalling pathways must exist to control conidial morphology, such as MoCom1, a transcription factor that is critical for conidial morphogenesis and pathogenicity in M. oryzae (Yang et al., 2010). In addition, not all cAMP level increases were associated with mutant hyphal autolysis defects, such as ΔMorgs2, ΔMorgs3, ΔMorgs5–8 and ΔMoyvh1 (Liu et al., 2016; Zhang et al., 2011b). These results also suggest that the cAMP level is not an essential factor for hyphal autolysis in M. oryzae. However, a review of published studies on M. oryzae indicated that a high, but not a low, intracellular cAMP level was more commonly associated with hyphal autolysis. Interestingly, the MoPdeHEAL(AAA) and MoPdeHΔ HD+L1 transformants produced fewer conidia than the ΔMopdeH mutant and other transformants, and only MoPdeHΔ HD+L1 showed a few typical lesions on rice leaves. These results suggest that the EAL and L1 domains are involved in an as yet unknown mechanism that regulates conidiation and infectious growth in M. oryzae. In addition, MoPdeHΔ HD+L1 transformants partially rescued certain phenotypic defects of the ΔMopdeH mutant; this is probably a result of its relatively high PDEase activity compared with MoPdeHΔ HD+L1L2 and MoPdeHΔ HD+L2. This is also consistent with the observation that the single L1 mutant displayed similar PDEase activity to MoPdeHΔ HD+L1.
Taken together, our findings provide new insights into the underlying cAMP regulatory mechanisms mediated by MoPdeH/L, which will help us to better understand the role of the cAMP signalling pathway during development and pathogenicity in the rice blast fungus and other phytopathogens.
Experimental Procedures
Strains and culture conditions
The M. oryzae Guy11 strain was used as the wild‐type; the ΔMopdeH and ΔMopdeL mutants were generated in a previous study (Zhang et al., 2011a) and used for protoplast transformation in this study. All strains were cultured on CM agar plates at 28 °C (Talbot et al., 1993). Mycelia collected from 2‐day‐old cultures in liquid CM were used to extract genomic DNA. Protoplast preparation and transformation were performed as described previously (Zhang et al., 2011b). Mycelial blocks were inoculated on SDC (100 g straw, 40 g corn powder, 15 g agar in 1 L of distilled water) agar plates to induce sporulation, as described previously (Zhang et al., 2011a).
Generation of the domain mutation, HD‐GFP, and chimera‐GFP or GST fusion constructs
To generate GFP fusion constructs, all domain mutation, individual domain and chimeric genes were driven by the native promoter of MoPDEH. The fragments were amplified by polymerase chain reaction (PCR) with primers and cloned into pYF11 plasmid using the yeast gap repair approach (Bruno et al., 2004). The resulting plasmids were confirmed by sequencing analysis to contain the in‐frame fusion constructs and transformed into the ΔMopdeH mutant. The resulting zeocin‐resistant transformants were screened by PCR or confirmed by the presence of GFP signals. To generate glutathione‐S‐transferase (GST) fusion constructs, full cDNA or fragments of the target genes were amplified by PCR with primers (Table S2, see Supporting Information), cloned into pGEX‐4T‐2 vector and sequenced, and transformed into Escherichia coli BL21‐CodonPlus for expression.
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis
To analyse the expression of pathogenesis‐related (PR) genes in the plants, total RNA was extracted from rice leaves inoculated with conidia at 24, 48 and 72 hpi. qRT‐PCR was performed on an ABI 7500 real‐time PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The stable expression ACTIN gene (MGG_03982) was used as an internal control. The experiment was repeated with three independent materials in three replicates. All primers used in this study are shown in Table S2.
Intracellular cAMP measurement and HPLC analysis
Two‐day‐old mycelia in liquid CM were harvested, quickly ground into powder with liquid nitrogen and lyophilized for 24 h. Intracellular cAMP extraction was performed following previously established procedures (Liu et al., 2007). The cAMP levels were quantified by HPLC analysis. The reactions were assessed as described previously (Liu et al., 2016).
Hyphal autolysis and surface hydrophobicity assays
For hyphal autolysis assay, mycelial blocks were inoculated on CM or SDC agar plates, cultured in the dark at 28 °C for 2 weeks and photographed. The experiments were repeated in triplicate. For surface hydrophobicity assay, 10 μL of sterile distilled water or detergent [with 0.02% sodium dodecylsulfate (SDS) and 5 mm ethylenediaminetetraacetic acid (EDTA)] were placed onto the surface of 14‐day‐old colonies and photographed.
Conidiation, appressorium formation and turgor assays
For conidiation, conidia were collected with 5 mL of distilled water from 10‐day‐old SDC cultures, filtered through three layers of lens paper and counted with a haemacytometer under a microscope. The conidial size was measured by a built‐in microscope ruler. For appressorium formation, conidia were resuspended in distilled water and the concentration was adjusted to 1 × 105 spores/mL. Conidial suspensions (20 μL/droplet) were placed on a hydrophobic or hydrophilic surface, and incubated in the dark at 28 °C. The appressorium formation rate was counted at 24 hpi under a microscope. More than 200 appressoria were counted for each strain and the experiments were repeated three times (Zhang et al., 2011a). For appressorium turgor assay, 1–4 m glycerol solution was used to measure the collapse of appressoria, as described previously (Liu et al., 2016; Zhang et al., 2009).
Pathogenicity, rice sheath penetration assay and ROS detection assay
Spraying assay and injection assay on rice seedlings were performed as described previously (Wang et al., 2016; Zhang et al., 2011a). For rice sheath penetration assay, 3‐week‐old rice seedlings (Oryza sativa cv. CO39) were inoculated with 1 × 105 spores/mL conidia, and kept in a chamber at 28 ºC with 90% humidity and in the dark for the first 24 h, followed by a 12‐h/12‐h light/dark cycle for 7 days. For microscopic observation of infectious hyphal growth in plant cells, detached rice sheaths were inoculated with 1 × 105 spores/mL conidia, and incubated at 28 ºC for 36 h. ROS staining with DAB (Sigma‐Aldrich, St. Louis, MO, USA) and DPI treatment assays were performed as described previously (Chen et al., 2014; Guo et al., 2010; Wang et al., 2016). All experiments were repeated three times, each with three replicates.
Detection of protein PDEase activity in vitro
GST, GST‐MoPdeH, GST‐MoPdeHEAL(AAA), GST‐MoPdeHΔ HD, GST‐MoPdeHΔ HD+L1L2, GST‐MoPdeHΔ HD+L1, GST‐MoPdeHΔ HD+L2, GST‐MoPdeHHD, GST‐MoPdeLL1L2, GST‐MoPdeLL1, GST‐MoPdeLL2 and GST‐MoPdeL were expressed in Escherichia coli BL21‐CodonPlus (DE3) cells (Stratagene, Cedar Creek, TX, USA). Samples were induced with 0.1 mm IPTG (isopropyl‐β‐d‐1‐thiogalactopyranoside) at 18 °C for 4 h. After centrifugation at 2500 g for 10 min, the precipitated cells were resuspended in lysis buffer [1 × phosphate‐buffered saline (PBS), 10% glycerol]. The target proteins were collected by sonication and further purified with glutathione sepharose beads incubated at 4 °C for 3 h. After centrifugation at 500 g for 5 min, glutathione sepharose beads were washed with washing buffer [20 mm Tris (pH 7.5), 0.25 mm NaCl, 2 mm EDTA, 2 mm ethyleneglycol‐bis(β‐aminoethylether)‐N,N'‐tetraacetic acid (EGTA)], eluted with washing buffer containing reduced glutathione (pH 8.0) (the elution contained 5% glycerol), concentrated at 4 °C and stored at −70 °C.
PDEase activity was detected through coupling with a phosphatase. The phosphate sensor is a phosphate‐binding protein modified with the N‐[2‐(1‐maleimidyl)ethyl]‐7‐(diethylamino)coumarin‐3‐carboxamide (MDCC) fluorophore (Brune et al., 1994). As a positive control, 2 × adenosine monophosphate (Amp) (Sigma A2252, St. Louis, MO, USA) and 2 × alkaline phosphatase (Calbiochem P/N 524545, San Diego, CA, USA) were incubated in phosphodiesterase enzymatic reaction buffer for 60 min at room temperature, followed by the addition of 2 × 1 μm phosphate sensor (Invitrogen P/N PV4406, Carlsbad, California, USA) in phosphate sensor detection buffer to each well. The plate was mixed and read immediately with excitation at 420 nm and emission at 450 nm. As a negative control, 2 × cAMP (Meilune P/N MB3159, Dalian, Liaoning, China), 2 × alkaline phosphatase and GST protein were incubated in phosphodiesterase enzymatic reaction buffer for 60 min at room temperature, followed by the addition of 2 × 1 μm phosphate sensor in phosphate sensor detection buffer to each well. The plate was mixed and read immediately. To ensure that the same amount of purified protein was used, we employed a Bradford Protein Assay Kit (Beyotime P0006, Songjiang district, Shanghai, China); 2 × cAMP, 2 × alkaline phosphatase and equal purified protein were incubated in phosphodiesterase enzymatic reaction buffer for 60 min at room temperature, followed by the addition of 2 × 1 μm phosphate sensor in phosphate sensor detection buffer to each well, and read immediately. All experiments were repeated three times, each with three replicates.
Homology modelling assay
Protein structural predictions were realized with the online platform SWISS‐MODEL (https://www.swissmodel.expasy.org/interactive/xBVTUG/templates/). The primary sequences of the proteins (MoPdeH, MoPdeHΔ HD, MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1, MoPdeHΔ HD+L2, MoPdeHHD, MoPdeLL1L2, MoPdeLL1, MoPdeLL2 and MoPdeL) were submitted and predicted (Arnold et al., 2006; Benkert et al., 2011; Biasini et al., 2014). The resulting protein structures were analysed by PyMOL.
Adenylate cyclase inhibition assay
MDL‐12,330A [N‐(cis‐2‐phenylcyclopentyl)azacyclotridecan‐2‐imine hydrochloride] (Sigma, M182–25MG) is a type of adenylate cyclase inhibitor (Guellaen et al., 1977; Hunt and Evans, 1980). The final concentration of MDL‐12,330A (100 μm) was added to CM medium or conidial suspensions to conduct vegetative growth, cAMP level measurement and appressorium formation assays on hydrophobic or hydrophilic surfaces. Equal dimethylsulfoxide (DMSO) was added as a control.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot analyses of the green fluorescent protein (GFP) expression of the indicated strains. Total proteins of the indicated strains were extracted from 2‐day‐old mycelia in liquid complete medium (CM). The antibody against GFP was used for analysis by western blot.
Fig. S2 Intracellular cyclic adenosine monophosphate (cAMP) measurement assays for additional transformants.
Fig. S3 Defects in mycelial autolysis and surface hydrophobicity of the additional transformants.
Fig. S4 Appressorium formation on hydrophilic surfaces of the additional transformants.
Fig. S5 Defects of the additional transformants in pathogenicity.
Fig. S6 Homology modelling of the indicated proteins. The target protein sequences of MoPdeH, MoPdeHΔ HD, MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1, MoPdeHΔ HD+L2, MoPdeHHD, MoPdeLL1L2, MoPdeLL1, MoPdeLL2 and MoPdeL were predicted by SWISS‐MODEL and the results were analysed by PyMOL.
Table S1 Appressorium turgor measurement of the indicated strains.
Table S2 Primers used in this study.
Acknowledgements
This study was supported by the Natural Science Foundation of China (Grant No. 31471736 to H.Z.), the Outstanding Youth Foundation of Jiangsu Province (Grant No. BK20160074 to H.Z.), the key program of the Natural Science Foundation of China (Grant No. 31530063 to Z.Z.) and the Agriculture Innovation Foundation of Jiangsu Province of China (Grant No. CX (15) 1054).
References
- Arnold, K. , Bordoli, L. , Kopp, J. and Schwede, T. (2006) The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics, 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Bencina, M. , Panneman, H. , Ruijter, G.J. , Legisa, M. and Visser, J. (1997) Characterization and overexpression of the Aspergillus niger gene encoding the cAMP‐dependent protein kinase catalytic subunit. Microbiology, 143, 1211–1220. [DOI] [PubMed] [Google Scholar]
- Benkert, P. , Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , Kiefer, F. , Gallo Cassarino, T. , Bertoni, M. , Bordoli, L. and Schwede, T. (2014) SWISS‐MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov, A.G. , Kirillina, O. and Perry, R.D. (2005) The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis . FEMS Microbiol. Lett. 247, 123–130. [DOI] [PubMed] [Google Scholar]
- Bolwell, G.P. , Davies, D.R. , Gerrish, C. , Auh, C.K. and Murphy, T.M. (1998) Comparative biochemistry of the oxidative burst produced by rose and French bean cells reveals two distinct mechanisms. Plant Physiol. 116, 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune, M. , Hunter, J.L. , Corrie, J.E. and Webb, M.R. (1994) Direct, real‐time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry, 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J.R. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryot. Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhai, S. , Zhang, H. , Zuo, R. , Wang, J. , Guo, M. , Zheng, X. , Wang, P. and Zhang, Z. (2014) Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae . Environ. Microbiol. 16, 788–801. [DOI] [PubMed] [Google Scholar]
- Choi, W. and Dean, R.A. (1997) The adenylate cyclase gene MAC1 of Magnaporthe grisea . Plant Cell, 9, 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli, J. , Field, J. , Ballester, R. , Chester, N. , Young, D. and Wigler, M. (1990) Mutational mapping of RAS‐responsive domains of the Saccharomyces cerevisiae adenylyl cyclase. Mol. Cell Biol. 10, 2539–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, P.B. , Walker, W.H. and Habener, J.F. (1998) Cyclic AMP signaling and gene regulation. Annu. Rev. Nutr. 18, 353–383. [DOI] [PubMed] [Google Scholar]
- D'Souza, C.A. and Heitman, J. (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25, 349–364. [DOI] [PubMed] [Google Scholar]
- Durrenberger, F. , Wong, K. and Kronstad, J.W. (1998) Identification of a cAMP‐dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis . Proc. Natl. Acad. Sci. USA, 95, 5684–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, M.Y. , Nikolskaya, A.N. and Koonin, E.V. (2001) Novel domains of the prokaryotic two‐component signal transduction systems. FEMS Microbiol. Lett. 203, 11–21. [DOI] [PubMed] [Google Scholar]
- Guellaen, G. , Mahu, J.L. , Mavier, P. , Berthelot, P. and Hanoune, J. (1977) RMI 12330 A, an inhibitor of adenylate cyclase in rat liver. Biochimica et biophysica acta, 484, 465–475. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Guo, W. , Chen, Y. , Dong, S. , Zhang, X. , Zhang, H. , Song, W. , Wang, W. , Wang, Q. , Lv, R. , Zhang, Z. , Wang, Y. and Zheng, X. (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 1053–1068. [DOI] [PubMed] [Google Scholar]
- Hicks, J.K. , Bahn, Y.S. and Heitman, J. (2005) Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A‐mediated negative feedback loop in Cryptococcus neoformans . Eukaryot. Cell, 4, 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, N.H. and Evans, T. (1980) RMI 12330A, an inhibitor of cyclic nucleotide phosphodiesterases and adenylate cyclase in kidney preparations. Biochim Biophys Acta, 613, 499–506. [DOI] [PubMed] [Google Scholar]
- Jeon, J. , Goh, J. , Yoo, S. , Chi, M.H. , Choi, J. , Rho, H.S. , Park, S.Y. , Kim, S. and Lee, Y.H. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae . Mol. Plant–Microbe Interact. 21, 525–534. [DOI] [PubMed] [Google Scholar]
- Lee, Y.H. and Dean, R.A. (1993) cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea . Plant Cell, 5, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Suresh, A. , Willard, F.S. , Siderovski, D.P. , Lu, S. and Naqvi, N.I. (2007) Rgs1 regulates multiple Galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 26, 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Qian, B. , Gao, C. , Huang, S. , Cai, Y. , Zhang, H. , Zheng, X. , Wang, P. and Zhang, Z. (2016) The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 29, 496–507. [DOI] [PubMed] [Google Scholar]
- Pan, X. and Heitman, J. (1999) Cyclic AMP‐dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae . Mol. Cell Biol. 19, 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, G. , Xue, C. , Zhao, X. , Kim, Y. , Orbach, M. and Xu, J.R. (2006) Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea . Plant Cell, 18, 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujam, R. and Naqvi, N.I. (2010) PdeH, a high‐affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae . PLoS Pathog. 6, e1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass, P. , Field, J. , Nikawa, J. , Toda, T. and Wigler, M. (1986) Cloning and characterization of the high‐affinity cAMP phosphodiesterase of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA, 83, 9303–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J. (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea . Annu. Rev. Microbiol. 57, 177–202. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J. , Ebbole, D.J. and Hamer, J.E. (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell, 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo, R. , Tischler, A.D. and Camilli, A. (2005) The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280, 33 324–33 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Yin, Z. , Tang, W. , Cai, X. , Gao, C. , Zhang, H. , Zheng, X. , Wang, P. and Zhang, Z. (2016) The thioredoxin MoTrx2 protein mediates ROS balance and controls pathogenicity as a target of the transcription factor MoAP1 in Magnaporthe oryzae . Mol. Plant Pathol. doi: 10.1111/mpp.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.R. and Hamer, J.E. (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea . Genes Dev. 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Xu, J.R. , Staiger, C.J. and Hamer, J.E. (1998) Inactivation of the mitogen‐activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA, 95, 12 713–12 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Zhao, X. , Sun, J. , Kang, Z. , Ding, S. , Xu, J.R. and Peng, Y.L. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 112–123. [DOI] [PubMed] [Google Scholar]
- Yin, Z. , Tang, W. , Wang, J. , Liu, X. , Yang, L. , Gao, C. , Zhang, H. , Zheng, X. , Wang, P. and Zhang, Z. (2016) Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen‐activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae . Mol. Plant Pathol. 17, 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhao, Q. , Liu, K. , Zhang, Z. , Wang, Y. and Zheng, X. (2009) MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol. Lett. 293, 160–169. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Liu, K. , Zhang, X. , Tang, W. , Wang, J. , Guo, M. , Zhao, Q. , Zheng, X. , Wang, P. and Zhang, Z. (2011a) Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae . PLoS One, 6, e17241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Tang, W. , Liu, K. , Huang, Q. , Zhang, X. , Yan, X. , Chen, Y. , Wang, J. , Qi, Z. , Wang, Z. , Zheng, X. , Wang, P. and Zhang, Z. (2011b) Eight RGS and RGS‐like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae . PLoS Pathog. 7, e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Xue, C. , Kong, L. , Li, G. and Xu, J.R. (2011c) A Pmk1‐interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae . Eukaryot. Cell, 10, 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. and Xu, J.R. (2007) A highly conserved MAPK‐docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea . Mol. Microbiol. 63, 881–894. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Kim, Y. , Park, G. and Xu, J.R. (2005) A mitogen‐activated protein kinase cascade regulating infection‐related morphogenesis in Magnaporthe grisea . Plant Cell, 17, 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.Y. , Zhang, H.F. , Li, G.T. , Shaw, B. and Xu, J.R. (2012) The cyclase‐associated protein Cap1 is important for proper regulation of infection‐related morphogenesis in Magnaporthe oryzae . PLoS Pathog. 8, e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot analyses of the green fluorescent protein (GFP) expression of the indicated strains. Total proteins of the indicated strains were extracted from 2‐day‐old mycelia in liquid complete medium (CM). The antibody against GFP was used for analysis by western blot.
Fig. S2 Intracellular cyclic adenosine monophosphate (cAMP) measurement assays for additional transformants.
Fig. S3 Defects in mycelial autolysis and surface hydrophobicity of the additional transformants.
Fig. S4 Appressorium formation on hydrophilic surfaces of the additional transformants.
Fig. S5 Defects of the additional transformants in pathogenicity.
Fig. S6 Homology modelling of the indicated proteins. The target protein sequences of MoPdeH, MoPdeHΔ HD, MoPdeHΔ HD+L1L2, MoPdeHΔ HD+L1, MoPdeHΔ HD+L2, MoPdeHHD, MoPdeLL1L2, MoPdeLL1, MoPdeLL2 and MoPdeL were predicted by SWISS‐MODEL and the results were analysed by PyMOL.
Table S1 Appressorium turgor measurement of the indicated strains.
Table S2 Primers used in this study.


