Summary
Fusarium culmorum is a soil‐borne fungal pathogen which causes foot and root rot and Fusarium head blight on small‐grain cereals, in particular wheat and barley. It causes significant yield and quality losses and results in the contamination of kernels with type B trichothecene mycotoxins. Our knowledge of the pathogenicity factors of this fungus is still limited. A transposon tagging approach based on the mimp1/impala double‐component system has allowed us to select a mutant altered in multiple metabolic and morphological processes, trichothecene production and virulence. The flanking regions of mimp1 were used to seek homologies in the F. culmorum genome, and revealed that mimp1 had reinserted within the last exon of a gene encoding a hypothetical protein of 318 amino acids which contains a ROGDI‐like leucine zipper domain, supposedly playing a protein–protein interaction or regulatory role. By functional complementation and bioinformatic analysis, we characterized the gene as the yeast Rav2 homologue, confirming the high level of divergence in multicellular fungi. Deletion of FcRav2 or its orthologous gene in F. graminearum highlighted its ability to influence a number of functions, including virulence, trichothecene type B biosynthesis, resistance to azoles and resistance to osmotic and oxidative stress. Our results indicate that the FcRav2 protein (and possibly the RAVE complex as a whole) may become a suitable target for new antifungal drug development or the plant‐mediated resistance response in filamentous fungi of agricultural interest.
Keywords: fungal pathogens, fungicide, Fusarium graminearum, Fusarium head blight, molecular target, transposon tagging, virulence genes
Introduction
Fusarium culmorum (W.G. Smith) Sacc., together with F. graminearum Schwabe and F. pseudograminearum O'Donnell & T. Aoki, are considered to be the most devastating fungal pathogens on small‐grain cereals (soft and durum wheat, barley, oat, rye and triticale). The interest in these species is justified by their role in the onset of two distinct diseases, namely foot and root rot (FRR, also known as ‘crown rot’) and Fusarium head blight (FHB) (Goswami and Kistler, 2004; Kazan et al., 2012; Scherm et al., 2013; Wagacha and Muthomi, 2007; Xu and Nicholson, 2009). FRR infection results in pre‐ or post‐emergence seedling death, brown discoloration on coleoptiles and the formation of whiteheads, leading to significant yield losses. However, yield and grain quality are particularly affected when these pathogens induce FHB, infecting the heads at anthesis and colonizing tissues until grain harvest. FHB infection causes contamination of the grain with mycotoxins, such as type B trichothecenes, i.e. sesquiterpene epoxides that are able to inhibit eukaryotic protein synthesis and induce apoptosis, and may play an important role as virulence factors.
Significant progress has been made during recent years towards a better understanding of the processes involved in FHB, especially in F. graminearum (Fu et al., 2013; Jiang et al., 2010; Kazan et al., 2012; Liu et al., 2014; Lysøe et al., 2011; Ma et al., 2013; Sperschneider et al., 2015). On the contrary, our knowledge on F. culmorum pathogenicity is still poorly understood and, although genes have been reported from whole‐genome sequencing projects (Moolhuijzen et al., 2013; Urban et al., 2016), only a few new potential fungicide targets in this species have been reported so far (Baldwin et al., 2010; Pasquali et al., 2013; Scherm et al., 2011; Skov et al., 2004).
With the aim to identify new F. culmorum genes playing a role in both FHB and FRR on durum wheat, we performed a transposon‐mediated mutagenesis approach based on the heterologous element mimp1 (Hua‐Van et al., 2000). mimp1 does not encode a transposase gene, but can be trans‐mobilized by the transposase of the impala element (Dufresne et al., 2007, 2008; Spanu et al., 2012).
Here, we report the identification of a new F. culmorum gene (FcRav2) by mimp1‐mediated insertional mutagenesis. The putative role of FcRav2 was determined in F. culmorum and, as a comparison, in F. graminearum by the analysis of the effect of deletion on the fungal phenotype.
Results
Molecular characterization of the mimp1‐tagged mutant R38
Polymerase chain reaction (PCR) and Southern blot analyses confirmed that the excision event in the revertant strain R38 was followed by the reinsertion of the mimp1/impala construct into a different genome site (not shown). Based on the left flanking sequence of the mimp1 element, blastn results revealed that mimp1 had reinserted at position 6542003 of the UK99 genome draft (LT598662.1) in the gene FCUL_11566.1 located in the fourth chromosome of F. culmorum (Fig. S1, see Supporting Information).
Characterization of the FcRav2 gene
FcRav2 (in F. graminearum, FgRav2, corresponding to Fg09428/FGSG_17209) has two introns and three exons, and codes for a hypothetical protein of 318 amino acids, with a molecular weight of 34.8 kDa and an isoelectric point of 6.43. The amino acid sequence contains a ROGDI leucine zipper domain (pfam10259), including a region of 30 amino acids with leucine repeats every seven or eight residues (Fig. S2, see Supporting Information). Homologous genes with unknown function were found in F. oxysporum (FOX_06114) and F. verticillioides (FVEG_03978) with sequence homologies of up to 83%, as well as in other filamentous fungi (Fig. S3, see Supporting Information). Orthology conservation was strongly confirmed (e‐value Eggnog e‐145 in the Class Leotiomycetes). Orthology classification from the National Center for Biotechnology Information (NCBI) allocated the gene within the Subclass Sordariomycetidae including the Neurospora crassa (NCU08091) and Magnaporthe oryzae (MGG_02604) ROGDI‐containing group. A lower level of conservation was found within the Class Saccharomycetes using both Eggnog and Inparanoid. As a result of the unique domain and homology along the overall protein length, we hypothesize that orthology also occurs with the Rav‐2 homologue in Saccharomyces cerevisiae, as well as with higher eukaryotes (Dawson et al., 2008). Protein localization is probably nuclear according to WoLF‐PSORT analysis (http://www.genscript.com/wolf-psort.html; Horton et al., 2007).
According to TMpred (http://www.ch.embnet.org/software/TMPRED_form.html; Hoffmann and Stoffel, 1993), the protein may contain a transmembrane domain (AA 178‐201) with its N‐terminus in the internal membrane region. The hypothetical structure obtained with RaptorX (Källberg et al., 2012) is reported in Fig. S4 (see Supporting Information).
Generation of FcRav2/FgRav2 deletion mutants in F. culmorum UK99 and F. graminearum PH1, and annotation confirmation
Seven hygromycin B‐resistant transformants were obtained from F. culmorum UK99, with five showing a distinct deletion and two being ectopic transformants (Fig. 1). One ectopic strain (FcB6) and two ΔFcRav2 strains (ΔFcRav2 B24, ΔFcRav2 B51) were used for all the following assays and characterizations. In addition, five FGSG_17209 deletion mutants were obtained from F. graminearum PH1, with ΔFgRav2 G8 and ΔFgRav2 G10 (Fig. 1) selected for further analyses.
Figure 1.
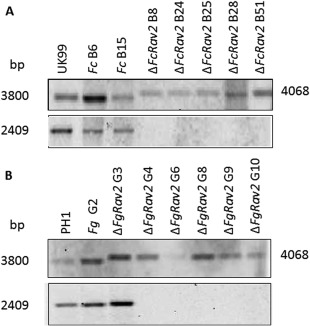
Southern blot analysis of Fusarium culmorum (A) and F. graminearum (B) transformants. Top: EcoRV‐digested DNAs were blotted and hybridized with a gene upstream probe obtained with primers 1F/2R (expected size for wild‐type and ectopic transformant, 3800 bp; expected size for deletion mutants, 4068 bp). Bottom: BglIII‐digested DNAs were blotted and hybridized with an internal FcRav2 gene upstream probe obtained with primers NF/NR (expected size for wild‐type and ectopic transformant, 2409 bp).
To verify the in silico annotation of the gene, the wild‐type strain UK99 was treated with 11 μmol of bafilomycin, which is known to inhibit specifically the V‐ATPase complex (Faraco et al., 2014). Indeed, the phenotype of the wild‐type treated with bafilomycin reproduced the same morphological features as the mutants, including colony morphology, hyphal hyperbranching and pigmentation (Fig. 2), allowing confirmation that at least part of the phenotypic effect of the FcRav2 mutation can be mimicked by a V‐ATPase‐inhibiting drug.
Figure 2.
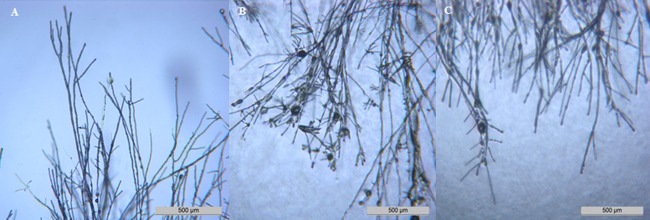
Comparison of Fusarium culmorum ΔFcRav2 mutant phenotype and in vitro effect of the vacuolar type H(+)‐ATPase inhibitor bafilomycin A1. (A) Fusarium culmorum wild‐type strain UK99 colony border after 48 h of growth on Czapek dox agar. (B) UK99 grown on Czapek dox agar containing 11 μmol of bafilomycin A1. (C) Fusarium culmorum mutant ΔFcRav2 B51 grown on Czapek dox agar. Microscopic detail (40×) shows hyperbranching and thickening of the hyphal tips in the ΔFcRav2 mutant and in the wild‐type exposed to bafilomycin A1.
Deletion of the FcRav2/FgRav2 gene involves significant changes in the physiological and metabolic profiles of F. culmorum and F. graminearum
All FcRav2 deletion mutants, as well as the revertant strain R38, showed significantly decreased growth compared with the recipient strains FcM7, F. culmorum UK99 and F. graminearum PH1 when coping with multiple osmotic stresses, whereas the ectopic transformant strain FcB6 displayed the same responsiveness as the F. culmorum wild‐type strain UK99 (Table 1; Figs S5–S8, see Supporting Information). The addition of 2 m sorbitol or 0.02% (w/v) sodium dodecylsulfate (SDS) led to more significant growth reductions than did 1 m NaCl. Fusarium graminearum was completely inhibited at 0.02% (w/v) SDS, and therefore PH1, ΔFgRav2 G8 and ΔFgRav2 G10 were characterized in a second test series on 0.01% (w/v) SDS, where deletion mutants displayed a significant growth reduction relative to the wild‐type PH1 (Table 1).
Table 1.
Phenotype assay of the strains used in this study.
| Strain | Sorbitol (2 m) | NaCl (1 m) | SDS (0.02%) | SDS (0.01%) | K2S2O8 (30 mm) | K2S2O8 (20 mm) | Tebuconazole (0.5 µg/mL) | Germinated conidia (%) |
|---|---|---|---|---|---|---|---|---|
| FcM7 | 2.30 ± 0.04 | 5.26 ± 0.02 | 2.14 ± 0.02 | N.D. | 2.56 ± 0.14 | N.D. | 5.26 ± 0.02 | 0.74 ± 0.01 |
| R38 | 0.84 ± 0.02** | 0.82 ± 0.02** | 1.14 ± 0.02** | N.D. | 0.70 ± 0.0** | N.D. | 1.58 ± 0.02** | 0.77 ± 0.01 |
| Fusarium culmorum UK99 | 2.98 ± 0.02 | 4.88 ± 0.05 | 2.78 ± 0.02 | N.D. | No growth | 4.87 ± 0.07 | 4.13 ± 0.14 | 0.30 ± 0.02 |
| FcB6 (ectopic) | 2.68 ± 0.10 | 5.02 ± 0.05 | 2.90 ± 0.03 | N.D. | No growth | 5.10 ± 0.06 | 4.33 ± 0.05 | 0.28 ± 0.01 |
| ΔFcRav2 B24 | 1.50 ± 0.07** | 1.40 ± 0.03 | 1.80 ± 0.03** | N.D. | No growth | 2.50 ± 0.0** | 2.77 ± 0.03** | 0.05 ± 0.01** |
| ΔFcRav2 B51 | 1.56 ± 0.05** | 1.80 ± 0.09 | 1.86 ± 0.02** | N.D. | No growth | 2.77 ± 0.03** | 3.13 ± 0.05** | 0.02 ± 0.01** |
| Fusarium graminearum PH1 | 4.02 ± 0.04 | 4.86 ±0.04 | No growth | 4.83 ± 0.03 | No growth | 5.00 ± 0.0 | 5.10 ± 0.10 | 0.34 ± 0.02 |
| ΔFgRav2 G8 | 1.08 ± 0.06** | 2.84 ± 0.05** | No growth | 3.26 ± 0.03** | No growth | 3.97 ± 0.07** | 2.08 ± 0.08** | 0.14 ± 0.01** |
| ΔFgRav2 G10 | 1.6 ± 0.09** | 3.78 ± 0.05** | No growth | 3.13 ± 0.08** | No growth | 3.70 ± 0.15** | 2.36 ± 0.05** | 0.15 ± 0.01** |
Colony growth was measured after 3 days of incubation at 25 °C on Czapek dox agar supplemented with 2 m sorbitol, 1 m NaCl, 0.01%–0.02% (w/v) sodium dodecylsulfate (SDS) (osmotic stress), 20–30 mm K2S2O8 (oxidative stress) or 0.5 µg/mL tebuconazole (fungicide sensitivity). Percentage spore germination was evaluated after 4 h of incubation in Czapek dox broth. Three independent tests were performed with three replicates for each test.
Results are expressed as colony diameter growth (cm) ± standard error. Significantly different from the control (**P < 0.01) based on Dunnett's test.
N.D., not determined.
Only F. culmorum strain FcM7 and its revertant R38 were able to grow in the presence of 30 mm K2S2O8, with R38 being significantly inhibited (over 70%) relative to FcM7. In the presence of 20 mm K2S2O8, all F. culmorum and F. graminearum deletion mutants were significantly inhibited relative to the respective wild‐type (Table 1).
Conidial germination was significantly impaired in the F. culmorum and F. graminearum deletion mutants relative to the wild‐type strains. In contrast, the germination capability of the revertant strain R38 was not reduced relative to that of the co‐transformant strain FcM7 (Table 1).
FcRav2 deletion dramatically reduced mycelium hydrophobicity: whereas, for both the wild‐type strain UK99 and the ectopic transformant FcB6, the time required to adsorb a 20‐µL drop of water was more than 15 min, in ΔFcRav2 B24 and ΔFcRav2 B51, the water droplet was immediately adsorbed on deposition (data not shown).
Tebuconazole sensitivity is significantly increased in the mimp1‐tagged mutant and in deletion mutants
The addition of 0.5 μg/mL of tebuconazole to Czapek dox agar produced a significant reduction in colony growth in all deletion mutants. In F. culmorum, growth of the mimp1‐tagged mutant R38 was decreased by 70% compared with the co‐transformant FcM7, whereas FcRav2 deletion resulted in 25%–35% growth inhibition on fungicide‐amended medium. Similarly, F. graminearum deletion mutants showed 50% growth reduction compared with the F. graminearum wild‐type strain PH1 in the presence of tebuconazole (Table 1).
FcRav2 gene plays a major role in sugar metabolism
Using a phenotype microarray approach, F. culmorum wild‐type strain UK99, its deletion mutant ΔFcRav2 B24 and the ectopic transformant strain FcB6 were further screened for their putative phenotype differences associated with low‐molecular‐weight carbon uptake and metabolism. The complete set of triplicate OD750 (optical density at 750 nm) readings recorded every 15 min from 0 to 72 h of incubation on 96 different carbon sources is reported in Table S1 (see Supporting Information).
Based on the analysis of the growth curves, the interval between 60 and 72 h was selected as the most informative to highlight differences in growth amongst the three tested strains. The heat map depicted in Fig. 3 reports the average OD750 readings recorded during the 60–72‐h interval on the 30 most differentiating carbon sources, listed in order of decreasing proportional growth difference between the deletion mutant ΔFcRav2 B24 and its wild‐type strain UK99.
Figure 3.

Phenetic heat map of carbon utilization patterns of Fusarium culmorum deletion mutant ΔFcRav2 B24 relative to its wild‐type strain UK99 and to ectopic transformant strain FcB6. Average OD750 (optical density at 750 nm) readings recorded during the 60–72‐h interval on the 30 most differentiating carbon sources are listed in order of decreasing proportional growth difference between ΔFcRav2 B24 and UK99.
The deletion mutant ΔFcRav2 B24 was impaired in its ability to catabolize different mono‐, di‐ and trisaccharide carbon sources (particularly l‐sorbose, d‐xylose, α‐methyl‐d‐glucoside, lactulose, l‐arabinose, l‐fucose, palatinose, d‐ribose, d‐mannose, d‐fructose, d‐raffinose, d‐melezitose), with percentage growth rates between 45% and 70% relative to the wild‐type UK99 and the ectopic strain FcB6 (Fig. 3). Moreover, the deletion mutant grew at a reduced rate on 2‐amino ethanol and on weak acids, such as bromosuccinic acid and β‐hydroxy‐butyric acid (Fig. 3).
FcRav2 influences the severity of crown rot and FHB on durum wheat
In a preliminary screening, the mimp1‐tagged mutant R38 was compared with the co‐transformant strain FcM7 and the wild‐type strain UK99 to evaluate the effect on seed germination. Abundant mycelium formation by all tested strains was observed after 24–48 h. However, only mutant R38 did not hamper the emergence of the primary root from the caryopsis, whereas FcM7 and the highly virulent strain UK99 killed 100% of the inoculated durum wheat seeds before germination (data not shown).
Glasshouse experiments further confirmed the role of FcRav2/FgRav2 in both crown rot and FHB. Insertion of the mimp1 transposable element or deletion of the FcRav2/FgRav2 gene in F. culmorum and F. graminearum caused a highly significant (P < 0.01) reduction in crown rot symptoms on durum wheat seedlings for all tested mutants, whereas the ectopic transformant FcB6 behaved similarly to the wild‐type reference strains UK99 and PH1, which caused disease incidences of 95 to 100, respectively (Table 2; Fig. S9, see Supporting Information).
Table 2.
Production of type B trichothecenes in vitro and in planta and virulence on durum wheat (cv. Iride) of the strains used in this study.
| Strain | DON production in vitro (ng/mL medium) † | Disease incidence (FRR) (0–100) | DON production in wheat seedlings (ng/g tissue) † | Disease incidence (FHB) (0–100) | DON production in wheat kernels (ng/g tissue) ‡ |
|---|---|---|---|---|---|
| Fusarium culmorum FcM7 | 3198.1 ± 155.5 | 61.9 ± 0 | 1092.9 ± 597.2 | 48.1 ± 15.1 | 5340.3 ± 4828.2 |
| R38 | 0** | 6.3 ± 5.5** | 0** | 4.7 ± 1.7** | 900.0 ± 435.9* |
| Fusarium culmorum UK99 | 21 085.6 ± 2727.4 | 95.0 ± 6.6 | 2219.4 ± 1324.1 | 57.8 ± 7.7 | 2598.0 ± 0 |
| FcB6 (ectopic) | 21 612.0 ± 1060.2 | 96.7 ± 3.8 | 1826.8 ± 289.3 | 77.8 ± 6.9 | 11 979.7 ± 1952.5** |
| ΔFcRav2 B24 | 24 044.4 ± 7431.9 | 22.5 ± 13.2** | 0** | 28.9 ± 7.0** | 1732.0 ± 433.0 |
| ΔFcRav2 B51 | 22 369.5 ± 642.7 | 4.2 ± 5.2** | 0** | 22.2 ± 8.4** | 2598.0 ± 1887.4 |
| Fusarium graminearum PH1 | 6336.2 ± 831.3 | 100 ± 0 | 1316.6 ± 447.9 | 44.4 ± 15.0 | N.D. |
| ΔFgRav2 G8 | 7439.6 ± 743.6 | 6.7 ± 5.8** | 0** | 0** | N.D. |
| ΔFgRav2 G10 | 2254.2 ± 349.8** | 4.2 ± 5.2** | 0** | 0** | N.D. |
Values are expressed as the mean of three or four replicates ± standard deviation.
Significantly different from the control strain (*P < 0.05, **P < 0.01) based on Dunnett's test.
FHB, Fusarium head blight; FRR, foot and root rot; N.D., not determined.
†Total concentrations of deoxynivalenol (DON) and its acetylated form (3‐ADON) were determined by liquid chromatography‐mass spectrometry (LC‐MS).
‡Total trichothecenes were measured by Rapid One Step Assay (ELISA lateral flow), designed for cereal flours.
In spray inoculation tests, FcRav2 inactivation by mimp1 insertion (R38) or deletion (ΔFcRav2 B51 and ΔFcRav2 B24) reduced FHB symptoms on durum wheat heads at 21 days post‐inoculation (dpi) to 10% and 5%–20% compared with F. culmorum control strains FcM7 and UK99, respectively (Table 2; Fig. S9). In contrast, the ectopic transformant FcB6 was not significantly affected in its virulence towards the host plant.
In F. graminearum, FgRav2 deletion mutants completely lost their pathogenicity, hence differing significantly from the wild‐type reference strain PH1 (Table 2).
Infection experiments showed a minimal, and insignificant, modulation of gene expression during the different stages of interaction with the plant. Indeed, expression profiles obtained in F. graminearum from array studies suggest that the gene modifies its expression according to the phenological stage of infection, but that these changes are dependent on various conditions and therefore show a high variability in experimental repetition in planta (Fig. 4).
Figure 4.
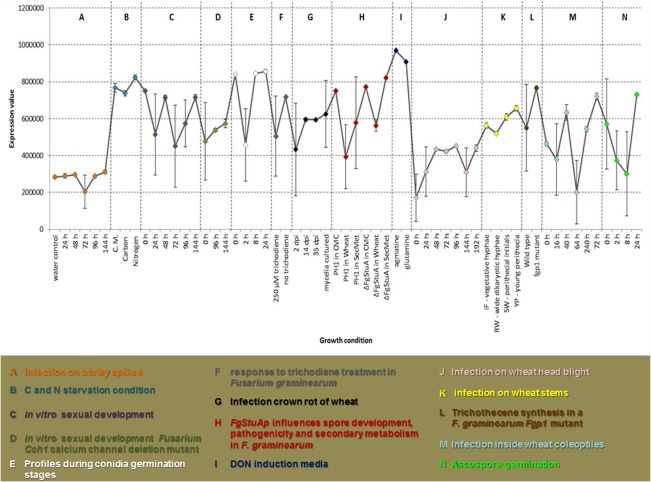
Expression patterns of different Affymetrix array experiments carried out on Fusarium graminearum in different environmental conditions obtained from the plexdb.org platform. The results are the average of three repetitions, with bars indicating standard error. CMC, carboxy‐methyl cellulose liquid medium; DON, deoxynivalenol.
FcRav2 is involved in type B trichothecene production
In in vitro experiments, only the F. culmorum mimp1‐tagged mutant R38 lost completely its ability to produce type B trichothecenes, whereas trichothecene production by the deletion mutants ΔFcRav2 B24 and ΔFcRav2 B51 did not differ significantly from that of the wild‐type strain UK99 or the ectopic transformant FcB6 (Table 2). In the case of F. graminearum, mutant ΔFgRav2 G8 was not significantly affected by gene deletion in its ability to produce deoxynivalenol (DON) and its acetylated form (3‐ADON), whereas trichothecene production by the mutant ΔFgRav2 G10 was reduced by approximately 70% (Table 2).
In accordance with the development of FRR symptoms, type B trichothecene levels reached 1000–2000 ng/g in seedling stem tissue infected by F. culmorum strains FcM7, UK99 and FcB6 and by F. graminearum strain PH1. In contrast, trichothecene mycotoxins were not detected in wheat seedlings on inoculation with the mimp1‐tagged mutant R38 or with deletion mutants of both F. culmorum UK99 and F. graminearum PH1 (Table 2).
When F. culmorum FcM7 was spray inoculated, multiple infection sites in the spikes resulted in high levels of trichothecene content in infected kernels, whereas the mimp1‐tagged mutant R38 produced approximately 15% mycotoxin compared with the co‐transformant strain FcM7 (Table 2). Infected kernels collected from spikes that had been spray inoculated with F. culmorum ectopic transformant FcB6 contained unusually high trichothecene content (approximately 12 000 ng/g tissue), whereas the wild‐type strain UK99 and both deletion mutants (ΔFcRav2 B24 and ΔFcRav2 B51) did not differ significantly in their DON production on kernel colonization (approximately 1700–2600 ng/g tissue; Table 2).
Discussion
Within the course of an extensive transposon‐mediated mutagenesis programme in the wheat pathogen F. culmorum (Spanu et al., 2012), a pathogenicity mutant was identified among over 2000 niaD revertants on minimal medium containing nitrate as the sole nitrogen source. In this mutant, the mimp1 transposable element was integrated within the last exon of the FcRav2 gene, encoding a hypothetical protein of 318 amino acids, and containing a ROGDI‐like leucine zipper domain known to have a regulatory role (pfam 10259). The ROGDI domain is conserved in eukaryotic genomes from Drosophila to yeasts and is mostly unique. Our protein presents a low level of similarity with the RAV2 gene from yeast (Seol et al., 2001), part of the RAVE complex involved in the assembly and disassembly of the V‐ATPase complex in yeast (Kane, 2006).
Based on the analysis of phenotype, expression and function, we hypothesize that, in F. graminearum and F. culmorum, the gene is also a rav2 homologue. As RAV2 gene homologues have not been investigated previously in other fungi, apart from Schizosaccharomyces pombe, where the gene is important for sugar response and calcium stress (Dawson et al., 2008), our work represents the first characterization of the role of RAV2 in filamentous fungi. More importantly, we showed that the protein affects the fitness of the fungus, including the ability to grow on different sugars, as well as the sensitivity to pH and fungicides, such as tebuconazole.
Treatment with the specific V‐ATPase inhibitor bafilomycin reproduced the same morphological features as in ΔFcRav2 mutants, including the hyphal hyperbranching observed on deletion of the vmaA1 gene, which encodes one of the subunits of the V‐ATPase in Aspergillus nidulans (Melin et al., 2004). The V‐ATPase has been shown to play a role in virulence in different fungi (Chen et al., 2013; Hilty et al., 2008; Patenaude et al., 2013). The impairment of pathogenicity observed in F. graminearum and F. culmorum confirms indirectly the putative function of FcRav2/FgRav2 as the homologue of RAV2, a controller of vacuolar and endosomal processing.
Further investigations are needed to experimentally confirm the role of the gene in filamentous fungi. Provided that the RAV2 gene may be present only in fungi (Parra et al., 2014), it appears now pivotal to focus on molecules able to interfere with RAV2 in planta in order to impair the toxigenic process, or to foster the activity of known fungicides, as suggested previously for human fungal pathogens (Hayek et al., 2014). For this reason, we propose that RAV2 in F. graminearum and F. culmorum may represent a new target for the development of integrative approaches against fungicide adaptation and resistance, or low‐sensitivity phenomena that are becoming important against different fungicides in Fusaria (Becher et al., 2011; Chen and Zhou, 2009; Dubos et al., 2011, 2013; Serfling and Ordon, 2014; Talas and McDonald, 2015).
FcRav2 appears to be deeply involved in a number of morphological and physiological traits, but no definite phenotypes were identified in Candida albicans in a large genomic screening of mutants (Noble et al., 2010). Spore germination and hydrophobicity are significantly altered in deletion mutants, as is resistance to various stresses: ΔFcRav2 inactivation results in increased sensitivity to osmotic and oxidative stress, and to the sterol biosynthesis inhibitor tebuconazole. The higher susceptibility to tebuconazole observed for the RAV2 mutant is a further confirmation of the importance of alternative mechanisms related to the extrusion of the compound in azole resistance. Zhang et al. (2010) have already hypothesized that the azole resistance mechanism also involves acidification of the medium.
Gene transcription seems to be influenced by azole treatment. Indeed, by analysis of the gene expression patterns of F. graminearum after treatment with sublethal concentrations of tebuconazole, Becher et al. (2011) found a decreased expression of FGSG_17209. This leads to the hypothesis that FGSG_17209 (FgRav2) may play an indirect role in controlling the expression of genes involved in compensation for azole stress, or that the gene is under‐expressed as growth is decreased.
A putative role of FcRav2 in stress resistance may also be inferred by the reduced growth of the deletion mutant on some of the tested carbon sources. For instance, l‐sorbose is a well‐known inhibitor of cell wall biosynthesis in fungi and shows a paramorphogenic action on N. crassa (Mishra and Tatum, 1972; Trinci and Collinge, 1973). Ethanolamine (2‐amino ethanol) is commonly used as a wood preservative (Humar et al., 2003). In addition, weak acids, such as bromosuccinic acid and β‐hydroxy‐butyric acid, may represent sources of stress on an impaired deleted mutant. These data suggest that FcRav2, as RAV2 in yeast, is involved in stress, including the stress to which F. culmorum is exposed when confronted with its victim plant.
The lower capacity of the FcRav2 deletion mutant to metabolize different mono‐, di‐ and trisaccharides in comparison with the wild‐type is also concordant with the hypothesis that it represents the RAV2 homologue. Indeed, the RAVE complex in yeast is important for the reassembly of the V‐ATPAse machinery on sugar starvation (Smardon et al., 2015). Our BIOLOG experiment confirmed the major role played by FcRav2 in sugar metabolism. This hypothesis is corroborated by expression data obtained from independent experiments, highlighting the under‐expression of FgRav2 in F. graminearum during glucose starvation (Fig. 4). In particular, among the sugars that determine the most significant differences in growth between the mutant and the wild‐type, xylose and arabinose belong to the pentose and glucuronate interconversion pathway (Fungipath), which is important in conditions of glucose starvation. It is noteworthy that RAV2 is overexpressed in S. cerevisiae exposed to high sugar stress (Erasmus et al., 2003), and that RAV2‐overexpressing Saccharomyces pastorianus bottom‐fermenting strains exhibit increased ethanol tolerance and increased fermentation rates in high‐sugar medium (Hasegawa et al., 2012).
As mentioned previously, the phenotype microarray analysis highlighted a significantly reduced ability of the FcRav2 deletion mutant to utilize 2‐amino ethanol as a carbon source. Interestingly, a behavioural screening of P‐transposon‐generated memory mutants coupled to DNA microarray analysis highlighted a Drosophila melanogaster ROGDI mutant and suggested its role in long‐term olfactory memory (Dubnau et al., 2003) and reduced ethanol tolerance (Berger et al., 2008). This supports indirectly the hypothesis that homologous functions are maintained by the gene with the ROGDI domain in higher eukaryotes, although further evidence is needed to corroborate such functional homology.
FcRav2/FgRav2 have a major impact on fungal virulence in F. culmorum and F. graminearum, respectively, and influence the severity of crown rot and FHB on durum wheat. This role was clearly demonstrated by different inoculation methods (contact between mycelium and caryopsis, spray inoculation of conidia), and was confirmed in F. graminearum.
Infection experiments with F. graminearum (Güldener et al., 2006; Lysøe et al., 2011) showed a minimal, and insignificant, modulation of gene expression during the different stages of interaction with the plant. Indeed, expression profiles suggest that the gene modifies its expression according to the phenological stage of infection, but that these changes are dependent on different conditions probably related to the variability in experimental repetitions in planta.
Less convincing was the implication of FcRav2 in mycotoxin production. The effect of FcRav2 disruption is probably dependent on the substrate: although the F. culmorum mimp1‐tagged mutant R38 lost completely its ability to produce type B trichothecenes in vitro, deletion mutants did not differ significantly from their wild‐type strains. In seedling stem tissue, mycotoxin biosynthesis was completely inhibited on FcRav2 deletion, whereas DON and 3‐ADON were still abundantly produced by deleted strains colonizing spray‐inoculated wheat spikes. This suggests that specific inducers of DON by‐passing FcRav2 regulation exist in the spike that are not able to activate the production of toxin in the stem or in vitro. Therefore, a plausible hypothesis is that some molecules in the spike may favour the synthesis of the toxin independent from FcRav2 control.
Interestingly, we observed a diverse DON production in vitro and in planta for some of the strains. This phenomenon has also been observed for the transcription factor in FgAtf1 (Nguyen et al., 2013), which is also a stress regulator (Lawrence et al., 2007), with opposite behaviour relative to FcRav2. To better elucidate the mechanisms that lead to high toxin contamination, there is a need to further characterize the specific molecules encountered by the fungus in planta.
In yeast, the lack of vacuolar acidification in the RAVE mutants is possibly compensated by other mechanisms of acidification of other compartments (Smardon et al., 2014). This would explain the relatively mild phenotype of the rav1Δ and rav2Δ mutants. We would anticipate that this acidification process is probably under the control of FgAtf1 in the HOG pathway. It is therefore plausible that the HOG pathway and the RAVE complex play two different, but complementary, roles in toxin production, linking toxin production and stress response pathways (Ponts, 2015). To further verify this hypothesis, we are currently investigating the cellular components of the assembly.
Expression profiles obtained from different array studies carried out on the F. graminearum PH1 strain confirm indirectly the homology of FcRav2 with the RAV‐2 gene from S. cerevisiae. Indeed, major oscillations of the gene (higher expression) are observed in agmatine medium, which is a strong toxin inducer (Gardiner et al., 2009a, b; Pasquali et al., 2016b). Given the need for vacuole assembly in conditions of high toxin production (toxisomes; Menke et al., 2013), it is plausible that vacuole acidification is strongly induced when high levels of toxins are produced.
From a more practical perspective, FcRav2 is proposed to be a suitable target for new antifungal drug development or in the plant‐mediated resistance response, given its pivotal role in affecting hydrophobicity, sugar metabolism, secondary metabolism, virulence and resistance to various stresses. Our work also confirms that untargeted methods for the identification of genes involved in fitness and pathogenicity are useful for genes that are not directly identified as homologous by normal orthology search programs. The further exploration of the mimp1 reinsertion mutant library in the future will possibly allow the identification of new genes involved in the fitness of the pathogen.
Experimental Procedures
Fungal strains and storage conditions
A mimp1‐mediated revertant (see below) strain collection was generated from the F. culmorum FcM7 co‐transformant strain, integrating a single copy of the niaD::mimp1 construct and the impalaE transposase gene under the constitutive control of the gpdA promoter (Dufresne et al., 2007; Spanu et al., 2012).
Homologous recombination experiments were carried out with F. culmorum wild‐type strain UK99 (Baldwin et al., 2010; kindly provided by Dr Kim Hammond‐Kosack, Rothamsted Research, Harpenden, Hertfordshire, UK) and with F. graminearum strain NRRL 31084 (syn. PH1; obtained from H. C. Kistler and deposited in the Luxembourg Microbial Culture Collection; Pasquali et al., 2016a).
All fungal strains were routinely cultured on potato dextrose agar (PDA; Sigma‐Aldrich, St. Louis, MO, USA). For long‐term storage, plugs colonized by mycelium were transferred to 50% (v/v) glycerol and stored at −80 °C.
Isolation of revertants obtained by the mimp1/impala double‐component system
The mimp1/impala revertant strains were selected on minimal medium agar supplemented with 50 µg/mL hygromycin (MMH50) containing sodium nitrate as sole nitrogen source (Spanu et al., 2012). Briefly, selection was performed with a phenotypic assay by inoculation of 10 µL of a spore suspension [106 colony‐forming units (CFU)/mL] of the co‐transformant strain FcM7 on MMH50 plates and by incubation at 25 °C for up to 1 month. First, nia+ colonies (referred to as ‘revertants’) appeared 15 days after inoculation on reacquisition of nitrate reductase function, and consisted of patches of aerial mycelium with a wild‐type phenotype. A single‐spore culture was obtained from each collected revertant strain.
Bioassay screening of the revertant strain collection
Single‐spore revertant strains were preliminarily screened for virulence during the first steps of kernel colonization using an in vitro bioassay described by Pasquali et al. (2013). Ten PDA‐mycelium plugs of each revertant bearing one durum wheat seed (cv. Simeto) were placed into a 90‐mm‐diameter sterile Petri dish and incubated at 25 °C for 3–5 days in the dark. Control assays included FcM7, F. culmorum strain UK99 and sterile PDA plugs. Inhibition of seed germination and kernel death were visually observed.
Molecular characterization of selected revertants
Excision of the mimp1 transposable element from the niaD gene was evaluated by Southern blot using a niaD‐specific probe, whereas a 120‐bp fragment of the mimp1 transposable element was used as probe to verify the reinsertion events (Table 3). Identification of the reinsertion site of mimp1 was achieved by Splinkerette‐PCR (Potter and Luo, 2010), according to the protocol described by Spanu et al. (2012). Flanking sequences were blasted to the F. culmorum genome (Urban et al., 2016) in order to identify mimp1 locations.
Table 3.
Primer sequences used in this study.
| Split‐marker recombination | Primer sequence |
|---|---|
| 1F‐FGSG17209 | 5‘‐AGTCCGCTTAAGACCCTCGAC‐3‘ |
| 2R‐FGSG17209 | 5‘‐TTGACCTCCACTAGCTCCAGCCAAGCCTAGTGAGGTGATGTTGAGATATGGA‐3‘ |
| 3F‐FGSG17209 | 5‘‐GAATAGAGTAGATGCCGACCGCGGGTTCCGTTGTAGTGGAAGCGACTATTATT‐3‘ |
| 4R‐FGSG17209 | 5‘‐GTTACACCAGTCTCGATACAACAT‐3‘ |
| Identification of mutants by PCR | |
| 1F‐FGSG17209 | 5‘‐AGTCCGCTTAAGACCCTCGAC‐3‘ |
| 4R‐FGSG17209 | 5‘‐GTTACACCAGTCTCGATACAACAT‐3‘ |
| NF‐FGSG17209 | 5‘‐ACTTACTAGCCATAACCTTGTCCA‐3‘ |
| NR‐FGSG17209 | 5‘‐AGACTTCCTGAACATCATTGCTATC‐3‘ |
| ITS1 | 5‘‐TCCGTAGGTGAACCTGCGG‐3‘ |
| ITS4 | 5‘‐TCCTCCGCTTATTGATATGC‐3‘ |
| Probes | |
| 1F‐FGSG17209 | 5‘‐AGTCCGCTTAAGACCCTCGAC‐3‘ |
| 2R‐FGSG17209probe | 5‘‐AAGCCTAGTGAGGTGATGTTGAGATATGGA‐3‘ |
| NF‐FGSG17209 | 5‘‐ACTTACTAGCCATAACCTTGTCCA‐3‘ |
| NR‐FGSG17209 | 5‘‐AGACTTCCTGAACATCATTGCTATC‐3‘ |
| Reinsertion events | |
| mi1 | 5‘‐TACAGTGGGATGCAATAAGTTTGAATAC‐3‘ |
| SacF | 5‘‐GGCTAGATCGAGCTCCCTCCTCAG‐3‘ |
| niaD144 | 5‘‐GTTCATGCCGTGGTCGCGC‐3‘ |
| niaD754r | 5‘‐AGTTGGAATGTCCTCGTCG‐3‘ |
Creation of deletion mutants and mutant screening
FcRav2 deletion mutants in F. culmorum strain UK99 and deletion mutants for the FGSG_17209 gene homologue in F. graminearum strain PH‐1 (FgRav2) were obtained by split‐marker recombination, as described by Breakspear et al. (2011). First, screening for deletion mutants and ectopic strains was carried out by PCR with the application of several primer combinations (Table 3; 1F‐FGSG17209/4R‐FGSG17209, NF‐FGSG17209/NR‐FGSG17209, NF‐FGSG17209/4R‐FGSG17209 and 1F‐FGSG17209/NR‐FGSG17209). The PCR mix consisted of 0.5 µm of each primer, 1 × Phusion® High‐Fidelity PCR Master Mix with HF Buffer (New England Biolabs, Ipswich, MA, USA) and 20 ng of DNA in a final volume of 20 µL. A subset of putative mutants was then analysed by Southern blot with two different probes: (i) gene upstream probe obtained with primers 1F/2R (916 bp); and (ii) internal gene probe NF/NR (456 bp) (Table 3). Probe labelling, hybridization and detection reactions were carried out using the Dig High Prime DNA labelling kit and detection starter II® (Roche Applied Science, Basel, Switzerland), following the manufacturer's protocol.
Phenotypic analysis of transposon‐tagged and deletion mutants of F. culmorum
The following strains were screened for altered growth under osmotic and oxidative stress or sensitivity to the fungicide tebuconazole: mimp1‐tagged F. culmorum revertant strain R38, co‐transformant strain FcM7, wild‐type F. culmorum strain UK99, deletion mutants ΔFcRav2 B24 and ΔFcRav2 B51, ectopic transformant strain FcB6, F. graminearum strain PH1, and deletion mutants ΔFgRav2 G8 and ΔFgRav2 G10. Assays were performed on Czapek dox agar (Oxoid Limited, Basingstoke, Hampshire, UK) and Czapek dox agar supplemented with 2 m sorbitol, 1 m NaCl, 0.01–0.02% (w/v) SDS (osmotic stress), 20–30 mm K2S2O8 (oxidative stress) or 0.5 µg/mL tebuconazole (fungicide sensitivity). Ten microlitres of a titrated conidial suspension (1 × 106 CFU/mL) were spotted on the centre of each plate (Ø = 60 mm). Colony diameter growth was measured after 3 days of incubation at 25 °C in the dark, and compared with the respective controls. Conidiogenesis and spore germination were evaluated by inoculating 150 mL of Czapek dox broth with 5 mL of conidial suspension (1 × 106 CFU/mL). At 0, 2, 4 and 8 h post‐inoculation (hpi) (25°C, 150 rpm), 100 conidia were examined with a haematocytometer using a light microscope (Olympus BX41, Hamburg, Germany). Three independent tests were performed for each assay, with three replicate plates for each test.
Mycelium hydrophobicity was evaluated according to Pasquali et al. (2013). Briefly, a 20‐μL droplet of sterile H2O was pipetted onto the surface of colonies grown on solid Vogel's medium (Vogel, 1956) for 5 days. The time (in seconds) needed to obtain complete droplet absorption by deletion mutants ΔFcRav2 B24 and ΔFcRav2 B51, and by the ectopic transformant strain FcB6, was compared with that of the wild‐type strain UK99.
The wild‐type F. culmorum strain UK99 and its deletion mutant ΔFcRav2 B24 were further screened for phenetic differences by the application of BIOLOG FF MicroPlates (Biolog Inc., Hayward, CA, USA). The plate panel composition, which contains 95 low‐molecular‐weight carbon sources, is available at the BIOLOG web site (http://www.biolog.com/products-static/microbial_identification_literature.php).
Spore suspensions were prepared in carboxy‐methyl cellulose liquid medium (CMC, Pasquali et al., 2013) and, after two subsequent washing steps with sterile distilled water, spores were suspended in ‘Inoculation fluid FF’ (Biolog Inc.) at a final concentration of 1 × 104 CFU/mL. One‐hundred microlitres of a spore suspension were pipetted into each of the 96 wells. The plates were incubated at 25 °C for 90 h and OD750 was recorded every 15 min using a Microarray Omnilog Reader (Biolog, Inc.). Samples were tested in triplicate.
Pathogenicity assay on durum wheat
The F. culmorum co‐transformant FcM7 and its revertant strain R38, wild‐type strain UK99, deletion mutants ΔFcRav21 B24 and ΔFcRav2 B51, the ectopic transformant strain FcB6, F. graminearum strain PH1, and deletion mutants ΔFgRav2 G8 and ΔFgRav2 G10 were tested for virulence in planta. Both FRR and FHB were evaluated.
Mycelium plugs, each bearing one durum wheat seed (Triticum durum cv. Iride, kindly provided by Unità di Ricerca per la Valorizzazione Qualitativa dei Cereali, CRA‐QCE, Rome, Italy), were placed in a plastic sowing pot and covered with sterile soil. Durum wheat seeds placed onto sterile PDA plugs (1 seed/plug) served as negative controls. FRR severity was assessed after 21 days of incubation at 25 °C in a glasshouse, and evaluated using the McKinney index (McKinney, 1923; Spanu et al., 2012). Three independent tests were carried out, each consisting of three replicates of 10 seedlings for each fungal strain.
The ability to cause FHB was tested on durum wheat (cv. Iride) on mid‐anthesis stage wheat heads. Spray inoculation was carried out with 4 mL of each spore suspension (1 × 105 spores/mL), covering the head until runoff. Inoculated heads were covered with a transparent plastic bag for 48 h to maintain high humidity conditions. Disease incidence was evaluated every 7 days and a final score was obtained at 21 dpi using the McKinney index. Each strain was tested in four replicates of five durum wheat heads each.
In vitro and in planta mycotoxin production
In vitro production of DON and its acetylated forms was determined in Vogel's medium (Vogel, 1956) as described by Pani et al. (2014), and expressed as ng/mL of culture filtrate.
The presence of trichothecenes in durum wheat seedlings (cv. Iride; three replicates of 10 seedlings each) was determined by the inoculation of seedlings at emergence with a conidial suspension (100 µL of 1 × 106 CFU/mL) of each strain. After 15 dpi, the basal portions of 10 seedlings were pooled, dried at 80 °C for 24 h, finely ground in a mortar and weighed. Samples were purified by MycoSep® 227 Trich+ columns (Romer Labs, Tulln, Austria), as described by the manufacturer, prior to liquid chromatography‐mass spectrometry (LC‐MS) analysis. Quantitative determinations were carried out as described previously (Pani et al., 2014), using a model HP 1100 LC‐MS detector (Agilent Technologies, Palo Alto, CA, USA).
The content of trichothecenes in durum wheat seeds (cv. Iride) harvested from the spray‐inoculated heads was determined using the ROSA® FAST5 DON assay (Rapid One Step Assay, Charm Sciences, Inc., Lawrence, MA, USA), according to the manufacturer's protocol. Total trichothecene content is expressed in ng/g plant tissue.
Growth on H(+)‐ATPase inhibitor bafilomycin
Fusarium culmorum wild‐type strain UK99 and its mutant ΔFcRav2 B51 were grown at 25 °C on Czapek dox agar amended with 6.7 μg/mL (equivalent to 11 μmol) of the vacuolar‐type H(+)‐ATPase inhibitor bafilomycin and on Czapek dox agar, respectively. After 48 h, colony diameters were measured and the phenotypes of the treated wild‐type and untreated mutant were compared.
Bioinformatic and statistical analysis
A one‐way analysis of variance, followed by multiple comparison using Dunnett's test, was performed on all data obtained from phenotypic, pathogenicity and trichothecene production assays using the Minitab® for Windows release 12.1 software.
Phenetic patterns were acquired with OmniLog‐OL_PM_FM/Kin 1.30 and OmniLog‐OL_PM_Par 1.30 software. Data were analysed separately by well with a mixed linear model that included the fixed effects of strain, sampling time, their interaction and the random effect of the replicates. The model was solved using PROC MIXED of SAS software (SAS Institute, 2008).
Sequences for alignment were obtained from FungiDB (http://FungiDB.org). Sequence analyses and expression profiles were analysed using CLC Main Workbench v 7.0. Expression profiles were obtained from the PLEXdb database (www.plexdb.org; Dash et al., 2012). Orthology was calculated with Eggnog 4.5 (Jensen et al., 2008) and Inparanoid 8 (Sonnhammer and Östlund, 2015). The putative protein structure was generated with RaptorX structure (raptorx.uchicago.edu).
Note by the authors
Whilst this article was under revision, a paper by Nguyen et al. (2017) showed the effect of deletion of the homologous FcRav2 in N. crassa, consisting of a decrease in growth of hyphae. The gene was selected as it has been proposed to be part of a set of candidate genes with a role in multicellular complexity, being conserved in complex eukaryotes. Our orthology search confirmed the work by Nguyen et al. (2017), suggesting that FcRAv2 homologues in Pezizomycotina may have many roles in governing cellular complexity that are partially lost in yeast. Moreover, the gene localization in N. crassa, shown by Nguyen et al. (2017), is consistent with our bioinformatic predictions, which suggest a cytoplasmic localization with a transmembrane domain.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 FcRav2 gene structure with sequence from ENSEMBLE modified to show the insertion site of mimp1 in the third exon of the gene. Untranslated region (UTR), introns and exons are reported.
Fig. S2 Protein domains identified in FcRav2 and the typical ROGDI leucine zipper pfam domain aligned to the FcRav2 amino acid sequence.
Fig. S3 Muscle alignment of FcRAV2 homologues.
Fig. S4 RaptorX putative three‐dimensional structure of the FcRAV2 protein and summarized results of prediction probabilities for modelling. Probably, only one part of the protein is correctly modelled, given the low GDT (Global Distance Test) value (<50).
Fig. S5 Phenotype of Fusarium culmorum mimp1 revertant R38, nit– recipient and M7 co‐transformant on different stress‐inducing media. SDS, sodium dodecylsulfate.
Fig. S6 Phenotype of Fusarium culmorum mimp1 revertant R38, nit– recipient and M7 co‐transformant on Czapek's medium amended with K2S2O8 (30 mm).
Fig. S7 Phenotype of Fusarium culmorum UK99, FcB6 (ectopic transformant), ΔFcRav2 B24 and ΔFcRav2 B51 on different stress‐inducing media after 10 days of growth at 25 °C.
Fig. S8 Phenotype of Fusarium graminearum PH1, ΔFgRav2 G8 and ΔFgRav2 G10 on different stress‐inducing media after 10 days of growth at 25 °C. SDS, sodium dodecylsulfate.
Fig. S9 Left: foot and root rot (FRR) symptoms on 21‐day‐old durum wheat cv. Iride seedlings mock inoculated (A) or infected with Fusarium culmorum mimp1 revertant R38 (B), co‐transformant M7 (C) or nit– recipient strain (D). Right: spikes of durum wheat cv. Iride mock inoculated (A) or infected with mimp1 revertant R38 (B), co‐transformant M7 (C) or nit– recipient strain (D) at 21 days post‐inoculation.
Table S1 Phenotypic growth raw data [OD750 (optical density at 750 nm) readings] of Fusarium culmorum wild‐type strain UK99, its deletion mutant ΔFcRav2 B24 and the ectopic transformant strain FcB6 recorded in triplicate every 15 min from 0 to 72 h of incubation on 96 different carbon sources. The complete set of data is available at: https://drive.google.com/file/d/0B2zMAlgHF40RWE5jX3lsTFdsY0k/view?usp=sharing.
Acknowledgements
This research was funded by the Ministry of University and Research (PRIN 2011 ‘Cell wall determinants to improve durum wheat resistance to Fusarium diseases’). B.S. acknowledges support from the University of Sassari (P.O.R. SARDEGNA F.S.E. 2007–2013, Obiettivo competitività regionale e occupazione, Asse IV Capitale umano, Linea di Attività l.3.1. ‘Identification of natural and natural‐like molecules inhibiting mycotoxin biosynthesis by Fusarium pathogens on cereals’). M.P. acknowledges the support of SUSTAIN COST action FA1208 on ‘Pathogen‐informed strategies for sustainable broad‐spectrum crop resistance’. Q.M. is grateful to Marie‐Jo Daboussi for her guidance in unveiling the marvellous world of fungal transposons.
This paper is dedicated to the memory of our friend and colleague Renato D'Ovidio.
Contributor Information
Matias Pasquali, Email: matias.pasquali@unimi.it.
Quirico Migheli, Email: qmigheli@uniss.it.
References
- Baldwin, T.K. , Urban, M. , Brown, N. and Hammond‐Kosack, K.E. (2010) A role for topoisomerase I in Fusarium graminearum and F. culmorum pathogenesis and sporulation. Mol. Plant–Microbe Interact. 23, 566–577. [DOI] [PubMed] [Google Scholar]
- Becher, R. , Weihmann, F. , Deising, H.B. and Wirsel, S.G.R. (2011) Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics, 12, 52 Available at DOI: 10.1186/1471-2164-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, K.H. , Kong, E.C. , Dubnau, J. , Tully, T. , Moore, M.S. and Heberlein, U. (2008) Ethanol sensitivity and tolerance in long‐term memory mutants of Drosophila melanogaster . Alcohol. Clin. Exp. Res. 32, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear, A. , Pasquali, M. , Broz, K. , Dong, Y. and Kistler, H.C. (2011) Npc1 is involved in sterol trafficking in the filamentous fungus Fusarium graminearum . Fung. Genet. Biol. 48, 725–730. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Liu, X. , Zhang, L. , Cao, H. , Lu, J. and Lin, F. (2013) Involvement of MoVMA11, a putative vacuolar ATPase c′ subunit, in vacuolar acidification and infection‐related morphogenesis of Magnaporthe oryzae . PLoS One, 8, e67804 Available at DOI: 10.1371/journal.pone.0067804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. and Zhou, M.G. (2009) Characterization of Fusarium graminearum isolates resistant to both carbendazim and a new fungicide JS399‐19. Phytopathology, 99, 441–446. [DOI] [PubMed] [Google Scholar]
- Dash, S. , Van Hemert, J. , Hong, L. , Wise, R.P. and Dickerson, J.A. (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 40, D1194–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, K. , Toone, W.M. , Jones, N. and Wilkinson, C.R.M. (2008) Loss of regulators of vacuolar ATPase function and ceramide synthesis results in multidrug sensitivity in Schizosaccharomyces pombe . Eukaryotic Cell, 7, 926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, J. , Chiang, A.S. , Grady, L. , Barditch, J. , Gossweiler, S. , McNeil, J. , Smith, P. , Buldoc, F. , Scott, R. and Certa, U. (2003) The staufen/pumilio pathway is involved in Drosophila long‐term memory. Curr. Biol. 13, 286–296. [DOI] [PubMed] [Google Scholar]
- Dubos, T. , Pasquali, M. , Pogoda, F. , Hoffmann, L. and Beyer, M. (2011) Evidence for natural resistance towards trifloxystrobin in Fusarium graminearum . Eur. J. Plant Pathol. 130, 239–248. [Google Scholar]
- Dubos, T. , Pasquali, M. , Pogoda, F. , Casanova, A. , Hoffmann, L. and Beyer, M. (2013) Differences between the succinate dehydrogenase sequences of isopyrazam sensitive Zymoseptoria tritici and insensitive Fusarium graminearum strains. Pestic. Biochem. Phys. 105, 28–35. [DOI] [PubMed] [Google Scholar]
- Dufresne, M. , Hua‐Van, A. , Abd el Wahab, H. , Ben M'barek, S. , Vasnier, C. , Teysset, L. , Kema, G.H.J. and Daboussi, M.J. (2007) Transposition of a fungal MITE through the action of a Tc1‐like transposase. Genetics, 175, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, M. , van der Lee, T. , Ben M'barek, S. , Xu, X. , Zhang, X. , Liu, T. , Waalwijk, C. , Zhang, W. , Kema, G.H. and Daboussi, M.J. (2008) Transposon‐tagging identifies novel pathogenicity genes in Fusarium graminearum . Fungal Genet. Biol. 45, 1552–1561. [DOI] [PubMed] [Google Scholar]
- Erasmus, D.J. , van der Merwe, G.K. and van Vuuren, H.J. (2003) Genome‐wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 3, 375–399. [DOI] [PubMed] [Google Scholar]
- Faraco, M. , Spelt, C. , Bliek, M. , Verweij, W. , Hoshino, A. , Espen, L. , Prinsi, B. , Jaarsma, R. , Tarhan, E. , de Boer, A.H. , Di Sansebastiano, G.P. , Koes, R. and Quattrocchio, F.M. (2014) Hyperacidification of vacuoles by the combined action of two different P‐ATPases in the tonoplast determines flower color. Cell Rep. 6, 32–43. [DOI] [PubMed] [Google Scholar]
- Fu, J. , Wu, J. , Jiang, J. , Wang, Z. and Ma, Z. (2013) Cystathionine gamma‐synthase is essential for methionine biosynthesis in Fusarium graminearum . Fungal Biol. 117, 13–21. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009a) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 46, 604–613. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009b) Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol. Plant–Microbe Interact. 22, 1588–1600. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Güldener, U. , Seong, K.Y. , Boddu, J. , Cho, S. , Trail, F. , Xu, J.R. , Adam, G. , Mewes, H.W. , Meehlbauer, G.J. and Kistler, H.C. (2006) Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta . Fungal Genet. Biol. 43, 316–325. [DOI] [PubMed] [Google Scholar]
- Hayek, S.R. , Lee, S.A. and Parra, K.J. (2014) Advances in targeting the vacuolar proton‐translocating ATPase (V‐ATPase) for anti‐fungal therapy. Front. Pharmacol. 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, S. , Ogata, T. , Tanaka, K. , Ando, A. , Takagi, H. and Shima, J. (2012) Overexpression of vacuolar H+‐ATPase‐related genes in bottom‐fermenting yeast enhances ethanol tolerance and fermentation rates during high‐gravity fermentation. J. Inst. Brew. 118, 179–185. [Google Scholar]
- Hilty, J. , Smulian, A.G. and Newman, S.L. (2008) The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages and virulence in a murine model of histoplasmosis. Mol. Microbiol. 70, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, K. and Stoffel, W. (1993) Tmbase – a database of membrane spanning proteins segments. Biol. Chem. H‐S. 374, 166. [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua‐Van, A. , Davière, J.M. , Langin, T. and Daboussi, M.J. (2000) Genome organization in Fusarium oxysporum: clusters of class II transposons. Curr. Genet. 37, 339–347. [DOI] [PubMed] [Google Scholar]
- Humar, M. , Petrič, M. , Pohleven, F. and Destpot, R. (2003) Upgrading of spruce wood with ethanolamine treatment. Eur. J. Wood Wood Prod. 61, 29–34. [Google Scholar]
- Jensen, L.J. , Julien, P. , Kuhn, M. , von Mering, C. , Muller, J. , Doerks, T. and Bork, P. (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Yang, J. , Fan, F. , Zhang, D. and Wang, X. (2010) The Type 2C protein phosphatase FgPtc1p of the plant fungal pathogen Fusarium graminearum is involved in lithium toxicity and virulence. Mol. Plant Pathol. 11, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg, M. , Wang, H. , Wang, S. , Peng, J. , Wang, Z. , Lu, H. and Xu, J. (2012) Template‐based protein structure modeling using the RaptorX web server. Nat. Prot. 7, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, P.M. (2006) The where, when, and how of organelle acidification by the yeast vacuolar H+‐ATPase. Microbiol. Mol. Biol. Rev. 70, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. , Gardiner, D.M. and Manners, J.M. (2012) On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 13, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.L. , Maekawa, H. , Worthington, J.L. , Reiter, W. , Wilkinson, C.R.M. and Jones, N. (2007) Regulation of Schizosaccharomyces pombe Atf1 protein levels by Sty1‐mediated phosphorylation and heterodimerization with Pcr1. J. Biol. Chem. 282, 5160–5170. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wang, J. , Xu, J. and Shi, J. (2014) FgIlv5 is required for branched‐chain amino acid biosynthesis and full virulence in Fusarium graminearum . Microbiology, 160, 692–702. [DOI] [PubMed] [Google Scholar]
- Lysøe, E. , Pasquali, M. , Breakspear, A. and Kistler, H.C. (2011) The transcription factor FgStuAp influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 54–67. [DOI] [PubMed] [Google Scholar]
- Ma, L.J. , Geiser, D.M. , Proctor, R.H. , Rooney, A.P. , O'Donnell, K. , Trail, F. , Gardiner, D.M. , Manners, J.M. and Kazan, K. (2013) Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. [DOI] [PubMed] [Google Scholar]
- McKinney, H.H. (1923) Influence of soil temperature and moisture on infection of wheat seedlings by Helminthosporium sativum . J. Agric. Res. 26, 195–217. [Google Scholar]
- Melin, P. , Schnürer, J. and Wagner, E.G.H. (2004) Disruption of the gene encoding the V‐ATPase subunit A results in inhibition of normal growth and abolished sporulation in Aspergillus nidulans . Microbiology, 150, 743–748. [DOI] [PubMed] [Google Scholar]
- Menke, J. , Weber, J. , Broz, K. and Kistler, H.C. (2013) Cellular development associated with induced mycotoxin synthesis in the filamentous fungus Fusarium graminearum . PLoS One, 8, e63077 Available at DOI: 10.1371/journal.pone.0063077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N.C. and Tatum, E.L. (1972) Effect of L‐sorbose on polysaccharide synthetases of Neurospora crassa . Proc. Natl. Acad. Sci. USA, 69, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolhuijzen, P.M. , Manners, J.M. , Wilcox, S.A. , Bellgard, M.I. and Gardiner, D.M. (2013) Genome sequences of six wheat‐infecting Fusarium species isolates. Genome Announc. 1, e00670‐13. Available at DOI: 10.1128/genomeA.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T.V. , Kröger, C. , Bönnighausen, J. , Schäfer, W. and Bormann, J. (2013) The ATF/CREB transcription factor Atf1 is essential for full virulence, deoxynivalenol production and stress tolerance in the cereal pathogen Fusarium graminearum . Mol. Plant–Microbe Interact. 26, 1378–1394. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.A. , Cissé, O.H. , Yun Wong, J. , Zheng, P. , Hewitt, D. , Nowrousian, M. and Jedd, G. (2017) Innovation and constraint leading to complex multicellularity in the Ascomycota. Nat. Commun. 8, 14 444 Available at 10.1038/ncomms14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, S.M. , French, S. , Kohn, L.A. , Chen, V. and Johnson, A.D. (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani, G. , Scherm, B. , Azara, E. , Balmas, V. , Jahanshiri, Z. , Carta, P. , Fabbri, D. , Dettori, M.A. , Fadda, A. , Dessì, A. , Dallocchio, R. , Migheli, Q. and Delogu, G. (2014) Natural and natural‐like phenolic inhibitors of type B trichothecene in vitro production by the wheat (Triticum sp.) pathogen Fusarium culmorum . J. Agric. Food Chem. 62, 4969–4978. [DOI] [PubMed] [Google Scholar]
- Parra, K.J. , Chan, C.Y. and Chen, J. (2014) Saccharomyces cerevisiae vacuolar H+‐ATPase regulation by disassembly and reassembly: one structure and multiple signals. Eukaryot. Cell, 13, 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali, M. , Spanu, F. , Scherm, B. , Balmas, V. , Hoffmann, L. , Hammond‐Kosack, K. , Beyer, M. and Migheli, Q. (2013) FcstuA from Fusarium culmorum controls wheat crown rot in a toxin dispensable manner. PLoS One, 8, e57429. Available at DOI: 10.1371/journal.pone.0057429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali, M. , Beyer, M. , Logrieco, A. , Audenaert, K. , Balmas, V. , Basler, R. , Boutigny, A.L. , Chrpová, J. , Czembor, E. , Gagkaeva, T. , González‐Jaén, M.T. , Hofgaard, I.S. , Köycü, N.D. , Hoffmann, L. , Lević, J. , Marin, P. , Miedaner, T. , Migheli, Q. , Moretti, A. , Müller, M.E.H. , Munaut, F. , Parikka, P. , Pallez‐Barthel, M. , Piec, J. , Scauflaire, J. , Scherm, B. , Stanković, S. , Thrane, U. , Uhlig, S. , Vanheule, A. , Yli‐Mattila, T. and Vogelgsang, S. (2016a) A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Front. Microbiol. 7, 406 Available at DOI: 10.3389/fmicb.2016.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali, M. , Cocco, E. , Guignard, C. and Hoffmann, L. (2016b) The effect of agmatine on trichothecene type B and zearalenone production in Fusarium graminearum, F. culmorum and F. poae . Peer J. 4, e1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude, C. , Zhang, Y. , Cormack, B. , Köhler, J. and Rao, R. (2013) Essential role for vacuolar acidification in Candida albicans virulence. J. Biol. Chem. 288, 26 256–26 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponts, N. (2015) Mycotoxins are a component of Fusarium graminearum stress‐response system. Front. Microbiol. 6, 1234 Available at DOI: 10.3389/fmicb.2015.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, C.J. and Luo, L. (2010) Splinkerette PCR for mapping transposable elements in Drosophila . PLoS One, 5, e10168 Available at DOI: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (2008) SAS/STAT Version 9.2. Cary, NC: SAS Institute Inc. [Google Scholar]
- Scherm, B. , Orrù, M. , Balmas, V. , Spanu, F. , Azara, E. , Delogu, G. , Hammond, T.M. , Keller, N.P. and Migheli, Q. (2011) Altered trichothecene biosynthesis in TRI6‐silenced transformants of Fusarium culmorum influences the severity of crown and foot rot on durum wheat seedlings. Mol. Plant Pathol. 12, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherm, B. , Balmas, V. , Spanu, F. , Pani, G. , Pasquali, M. and Migheli, Q. (2013) Fusarium culmorum: causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 14, 323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol, J.H. , Shevchenko, A. , Shevchenko, A. and Deshaies, R.J. (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V‐ATPase assembly. Nat. Cell Biol. 3, 384–391. [DOI] [PubMed] [Google Scholar]
- Serfling, A. and Ordon, F. (2014) Virulence and toxin synthesis of an azole insensitive Fusarium culmorum strain in wheat cultivars with different levels of resistance to fusarium head blight. Plant Pathol. 63, 1230–1240. [Google Scholar]
- Skov, J. , Lemmens, M. and Giese, H. (2004) Role of a Fusarium culmorum ABC transporter (FcABC1) during infection of wheat and barley. Physiol. Mol. Plant Pathol. 64, 245–254. [Google Scholar]
- Smardon, A.M. , Diab, H.I. , Tarsio, M. , Diakov, T.T. , Nasab, N.D. , West, R.W. and Kane, P.M. (2014) The RAVE complex is an isoform‐specific V‐ATPase assembly factor in yeast. Mol. Biol. Cell, 25, 356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon, A.M. , Nasab, N.D. , Tarsio, M. , Diakov, T.T. and Kane, P.M. (2015) Molecular interactions and cellular itinerary of the yeast RAVE (regulator of the H+‐ATPase of vacuolar and endosomal membranes) complex. J. Biol. Chem. 290, 27 511–27 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L. and Östlund, G. (2015) InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43, D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, F. , Pasquali, M. , Scherm, B. , Balmas, V. , Marcello, A. , Ortu, G. , Dufresne, M. , Hoffmann, L. , Daboussi, M.J. and Migheli, Q. (2012) Transposition of the miniature inverted‐repeat transposable element mimp1 in the wheat pathogen Fusarium culmorum . Mol. Plant Pathol. 13, 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Gardiner, D.M. , Thatcher, L.F. , Lyons, R. , Singh, K.B. , Manners, J.M. and Taylor, J.M. (2015) Genome‐wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol. Evol. 7, 1613–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talas, F. and McDonald, B.A. (2015) Significant variation in sensitivity to a DMI fungicide in field populations of Fusarium graminearum . Plant Pathol. 64, 664–670. [Google Scholar]
- Trinci, A.P.J. and Collinge, A. (1973) Influence of L‐sorbose on the growth and morphology of Neurospora crassa . Microbiology, 78, 179–192. [DOI] [PubMed] [Google Scholar]
- Urban, M. , King, R. , Andongabo, A. , Maheswari, U. , Pedro, H. , Kersey, P. and Hammond‐Kosack, K. (2016) First draft genome sequence of a UK Strain (UK99) of Fusarium culmorum . Genome Announc. 4, e00771–e00716. Available at DOI: 10.1128/genomeA.00771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, H.J. (1956) A convenient growth medium for Neurospora (Medium N). Microb. Genet. Bull. 13, 42–43. [Google Scholar]
- Wagacha, J.M. and Muthomi, J.W. (2007) Fusarium culmorum: infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Prot. 26, 877–885. [Google Scholar]
- Xu, X. and Nicholson, P. (2009) Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 47, 83–103. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.Q. , Gamarra, S. , Garcia‐Effron, G. , Park, S. , Perlin, D.S. and Rao, R. (2010) Requirement for ergosterol in V‐ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6 Available at DOI: 10.1371/journal.ppat.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 FcRav2 gene structure with sequence from ENSEMBLE modified to show the insertion site of mimp1 in the third exon of the gene. Untranslated region (UTR), introns and exons are reported.
Fig. S2 Protein domains identified in FcRav2 and the typical ROGDI leucine zipper pfam domain aligned to the FcRav2 amino acid sequence.
Fig. S3 Muscle alignment of FcRAV2 homologues.
Fig. S4 RaptorX putative three‐dimensional structure of the FcRAV2 protein and summarized results of prediction probabilities for modelling. Probably, only one part of the protein is correctly modelled, given the low GDT (Global Distance Test) value (<50).
Fig. S5 Phenotype of Fusarium culmorum mimp1 revertant R38, nit– recipient and M7 co‐transformant on different stress‐inducing media. SDS, sodium dodecylsulfate.
Fig. S6 Phenotype of Fusarium culmorum mimp1 revertant R38, nit– recipient and M7 co‐transformant on Czapek's medium amended with K2S2O8 (30 mm).
Fig. S7 Phenotype of Fusarium culmorum UK99, FcB6 (ectopic transformant), ΔFcRav2 B24 and ΔFcRav2 B51 on different stress‐inducing media after 10 days of growth at 25 °C.
Fig. S8 Phenotype of Fusarium graminearum PH1, ΔFgRav2 G8 and ΔFgRav2 G10 on different stress‐inducing media after 10 days of growth at 25 °C. SDS, sodium dodecylsulfate.
Fig. S9 Left: foot and root rot (FRR) symptoms on 21‐day‐old durum wheat cv. Iride seedlings mock inoculated (A) or infected with Fusarium culmorum mimp1 revertant R38 (B), co‐transformant M7 (C) or nit– recipient strain (D). Right: spikes of durum wheat cv. Iride mock inoculated (A) or infected with mimp1 revertant R38 (B), co‐transformant M7 (C) or nit– recipient strain (D) at 21 days post‐inoculation.
Table S1 Phenotypic growth raw data [OD750 (optical density at 750 nm) readings] of Fusarium culmorum wild‐type strain UK99, its deletion mutant ΔFcRav2 B24 and the ectopic transformant strain FcB6 recorded in triplicate every 15 min from 0 to 72 h of incubation on 96 different carbon sources. The complete set of data is available at: https://drive.google.com/file/d/0B2zMAlgHF40RWE5jX3lsTFdsY0k/view?usp=sharing.


