Summary
Receptor‐like kinases are membrane proteins that can be shared by diverse signalling pathways. Among them, the Arabidopsis thaliana FERONIA (FER) plays a role in the balance between distinct signals to control growth and defence. We have found that COK‐4, a putative kinase encoded in the common bean anthracnose resistance locus Co‐4, which is transcriptionally regulated during the immune response, is highly similar to the kinase domain of FER. To assess whether COK‐4 is a functional orthologue of FER, we expressed COK‐4 in the wild‐type Col‐0 and the fer‐5 mutant of Arabidopsis and evaluated FER‐associated traits. We observed that fer‐5 plants show an enhanced apoplastic and stomatal defence against Pseudomonas syringae. In addition, the fer‐5 mutant shows reduced biomass, smaller guard cell size, greater number of stomata per leaf area, fewer leaves, faster transition to reproductive stage and lower seed weight per plant than the wild‐type Col‐0. Except for the stomatal complex length and number of stomata, COK‐4 expression in fer‐5 lines partially or completely rescued both defence and developmental defects of fer‐5 to the wild‐type level. Notably, COK‐4 may have an additive effect to FER, as the expression of COK‐4 in Col‐0 resulted in enhanced defence and growth phenotypes in comparison with wild‐type Col‐0 plants. Altogether, these findings indicate that the common bean COK‐4 shares at least some of the multiple functions of the Arabidopsis FER kinase domain, acting in both the induction of plant growth and regulation of plant defence.
Keywords: Arabidopsis thaliana, Co‐4 locus, FERONIA, Phaseolus vulgaris, plant growth and development, plant immunity
Introduction
In order to resist pathogen invasion, plants induce innate immune responses that are often associated with a reduction in plant growth (Campos et al., 2016; Lozano‐Durán and Zipfel, 2015). This plant growth–defence balance is regulated by a crosstalk between different hormones and depends on both physiological and environmental factors (Huot et al., 2014; Züst and Agrawal, 2017). Pattern recognition receptors (PRRs) are often receptor‐like kinases (RLKs) that are important in self‐ and non‐self‐recognition. Some PRRs can be shared by different signalling pathways and work to fine tune plant responses to endogenous and exogenous stimuli; thus, PRRs play an important role in the regulation of growth–defence tradeoffs in plants (Belkhadir et al., 2014).
Amongst the subfamilies of RLKs, members of the Catharanthus roseus RLK1‐like (CrRLK1L) subfamily are known to be growth controlling elements (Nissen et al., 2016). FERONIA (FER), a member of the CrRLK1L subfamily in Arabidopsis thaliana (L. Heyhn.), is involved in pollen tube perception in the megagametophyte (Escobar‐Restrepo et al., 2007) and is a key regulator of cell expansion during vegetative growth (Guo et al., 2009). In roots, FER acts as a receptor for the secreted hormone peptide RALF1 (rapid alkalinization factor 1) and controls cell expansion (Haruta et al., 2014). In addition, FER can function in the crosstalk between plant hormones to regulate shoot growth. Brassinosteroid (BR) and ethylene signalling interact to balance hypocotyl growth through FER (Deslauriers and Larsen, 2010), whereas the crosstalk between abscisic acid (ABA) and auxin that enables normal shoot growth has FER as a convergent mediator (Yu et al., 2012).
Interestingly, Kessler et al. (2010) reported that a homozygous fer mutant is resistant to powdery mildew infection, and shows spontaneous cell death and high H2O2 production; thus, FER has been linked to plant defence responses. In addition, Keinath et al. (2010) observed enhanced accumulation of reactive oxygen species (ROS) in fer mutants, as well as enhanced flg22‐induced activation of mitogen‐activated protein kinases (MAPKs), constitutively closed stomata and low proliferation of the ΔavrPtoΔavrPtoB mutant strain of the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000, when compared with wild‐type plants. More recently, FER has been implicated in susceptibility to Fusarium oxysporum by recognizing a fungus‐produced RALF‐like molecule, which acts as a virulence factor that inhibits host immune responses (Masachis et al., 2016). Indeed, another Arabidopsis RALF peptide, RALF23, can dampen plant immune responses by recruiting FER and inhibiting the formation of the FLAGELLIN SENSING 2‐BRASSINOSTEROID INSENSITIVE 1/EF‐TU RECEPTOR‐ASSOCIATED KINASE (FLS2/EFR‐BAK1) immune complex (Stegmann et al., 2017).
We have found that COK‐4, a putative serine/threonine (Ser/Thr) kinase encoded within the anthracnose‐resistant locus Co‐4 from common bean (Melotto et al., 2004), is highly similar to members of the CrRLK1L family of proteins from various species, including the Arabidopsis FER and ANXUR (Oblessuc et al., 2015). Therefore, using genetic, physiological, phenotypic, molecular and in silico analyses, we sought to further understand the function of COK‐4. We first identified FER (At3g51550) as a putative orthologue of COK‐4 in Arabidopsis and genetically complemented the Arabidopsis fer‐5 mutant, as well as transformed Col‐0 control plants, with a COK‐4 open reading frame isolated from the common bean genotype G2333. We evaluated immune and growth responses in COK‐4‐expressing Arabidopsis lines. The results suggest that the kinase domain of FER shares similar functions to COK‐4, where both may act as negative regulators of immunity against Pst DC3000 and may be involved in the positive regulation of plant growth and development.
Results
COK‐4 is closely related to the CrRLK1L family in Arabidopsis
Previously, we have identified COK‐4 as being related to the kinase domain of protein members of the CrRLK1L family from various species (Oblessuc et al., 2015). In order to characterize the COK‐4 molecular function further, we searched the Arabidopsis genome for kinases that were most closely related to COK‐4. Phylogenetic analysis revealed that COK‐4 clusters with 15 of the 17 members of the CrRLK1L family in Arabidopsis (Fig. 1A). Amongst these CrRLK1L members, the kinase domain of FER showed the highest similarity to the whole COK‐4 protein, with 38% sequence identity based on protein sequence alignment analysis (Fig. S1A, see Supporting Information). No sequence matches were observed between COK‐4 and any other domain of all proteins used to create the phylogenetic tree (Figs S1A and S2, see Supporting Information). In addition, COK‐4 shares serine, threonine and tyrosine residues with FER in phosphorylation sites that were identified by both software prediction and experimental analyses (Fig. S1A). However, FER seems to contain more phosphorylation sites than COK‐4 (Fig. S1B). In addition, three‐dimensional (3D) protein structure modelling for both COK‐4 and FER proteins revealed that they share highly similar kinase domain structures (Fig. 1B). The hydrophobic domain (amino acids GSRFMSKQKQINVIVFWVIFVLLYELTHCH) of COK‐4 contains the previously predicted transmembrane region (Melotto and Kelly, 2001) and, interestingly, this region is not present in the FER kinase protein (Figs 1B and S1A).
Figure 1.
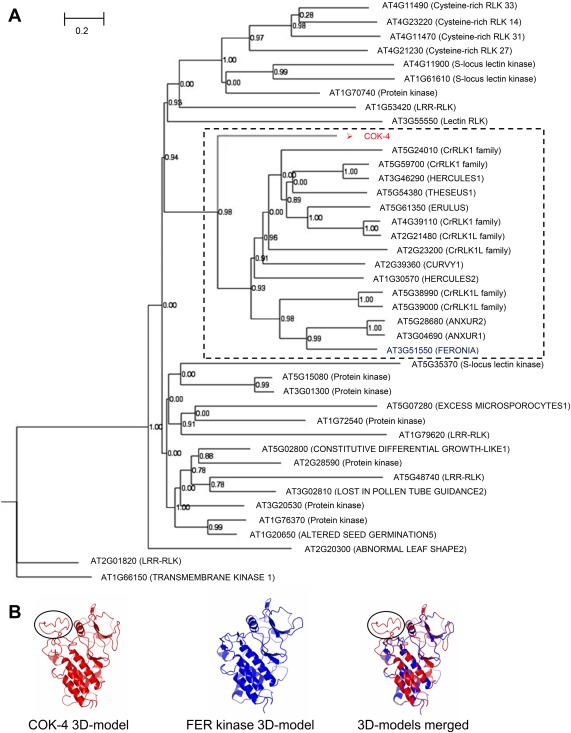
COK‐4 is highly similar to the Arabidopsis FERONIA (FER) kinase domain. (A) Phylogenetic tree showing the clustering of the predicted COK‐4 protein with 15 of the 17 Catharanthus roseus RLK1‐like (CrRLK1L) family members of Arabidopsis (broken square). The phylogenetic tree was obtained using the top 40 different protein kinases of Arabidopsis with significant alignment (threshold E‐value ≤ 3 × 10−33) with the predicted COK‐4 protein from the common bean line SEL 1308 [National Center for Biotechnology Information (NCBI) accession number AAF98554; Melotto and Kelly, 2001], using the maximum parsimony method from MAFFT software (Katoh and Standley, 2013). The nodes were confirmed by 1000 bootstraps. (B) Three‐dimensional (3D) protein structure of FER and COK‐4 kinase domains showing that both kinases have similar structures. The protein models were obtained with SWISS‐MODEL (https://swissmodel.expasy.org) and the alignment of the models was made using CCP4G v.2.10.6 (McNicholas et al., 2011). The black circles indicate the COK‐4 protein region that does not fit within the FER kinase domain structure. This region contains the transmembrane region of COK‐4 predicted by Melotto and Kelly (2001). LRR, leucine‐rich repeat; RLK, receptor‐like kinase.
FER and other members of the CrRLK1L family have similar kinase domains to each other, as well as to COK‐4 (Fig. S2; Kessler et al., 2014). Therefore, to further confirm that FER was the best candidate for having similar functions to COK‐4, we analysed the expression patterns of FER and the next two closest relatives of COK‐4, ANX1 and ANX2, based on protein phylogeny (Fig. 1A). Arabidopsis ANX1 and ANX2 are highly expressed, but not exclusively (Mang et al., 2017), in mature pollen, whereas FER shows ubiquitous expression across all plant tissues (Fig. S3A–C, see Supporting Information). Furthermore, in our experimental conditions, the knockout mutants anx1‐1 and anx2‐2 (Boisson‐Dernier et al., 2009) have the same reaction to dip inoculation of Pst DC3000 as the wild‐type Col‐0 plants (Fig. S3D,E). In addition, it is important to highlight that FER is the CrRLK1L member of Arabidopsis that has a more comprehensive description of its function in plant immunity (Keinath et al., 2010; Kessler et al., 2010; Masachis et al., 2016; Stegmann et al., 2017). Thus, based on phylogeny, phosphorylation sites, 3D protein modelling analyses and its function on the plant immune response, FER was selected to facilitate the functional analysis of COK‐4.
A functional FER contributes to stomatal opening and Arabidopsis susceptibility to Pst DC3000
In order to study the molecular function of COK‐4, we first assessed the immune response phenotypes associated with FER mutation using the knockdown fer‐5 mutant (Fig. S4A,B, see Supporting Information). Duan et al. (2010) first described this mutant as having shoot and root growth deficiencies. Here, we observed that fer‐5 has a constitutively smaller stomatal aperture width when compared with Col‐0 (Fig. 2A). When exposed to Pst DC3000, the fer‐5 stomatal aperture width decreases further within 2 h post‐inoculation (hpi) (Fig. 2B), indicating that stomatal defence is active in fer‐5. Interestingly, unlike Col‐0, fer‐5 stomata do not respond to coronatine produced by this bacterium and remain closed at 4 hpi (Fig. 2B).
Figure 2.
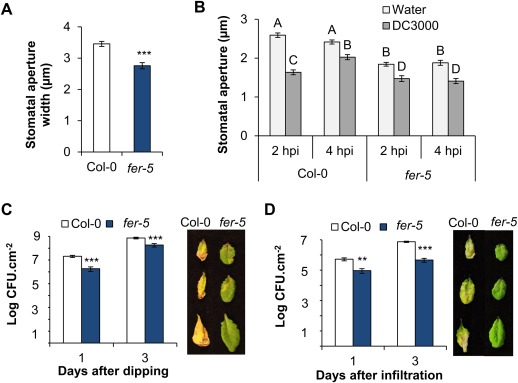
A functional FERONIA (FER) kinase domain contributes to stomatal opening and Arabidopsis susceptibility to Pseudomonas syringae pv. tomato (Pst) DC3000. (A) Stomatal aperture width in untreated Col‐0 and fer‐5 mature leaves. Results are shown as the mean (n = 120 ± SE) and statistical significance between the means was calculated with Student's t‐test (***P ≤ 0.001). (B) Stomatal aperture width in leaves inoculated with Pst DC3000 [1 × 108 colony‐forming units (CFU)/mL] at two time points: 2 and 4 h post‐inoculation (hpi). Results are shown as the mean (120 < n < 180 ± SE) and statistical significance amongst the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05). (C, D) Apoplastic bacterial population in leaves after dip inoculation with 1 × 108 CFU/mL (C) or vacuum infiltration with 1 × 106 CFU/mL (D) of Pst DC3000, 1 and 3 days after inoculation. Results are shown as the mean (n = 6 ± SE) (Student's t‐test; ***p ≤ 0.001 and **p ≤ 0.01). Photographs on the right were taken from various leaves at 3 days post‐inoculation.
Pathogenesis assays revealed that fer‐5 shows increased resistance to Pst DC3000 independent of the inoculation method. Either surface inoculation or vacuum infiltration of bacteria into the apoplast resulted in significantly smaller bacterial populations within fer‐5 leaves relative to Col‐0, as early as 1 day after inoculation (Fig. 2C,D). Furthermore, fer‐5 leaves showed no disease symptoms, whereas Col‐0 leaves showed chlorotic and necrotic spots as expected for this pathosystem (Fig. 2C,D). These results suggest that the FER kinase is required for plant susceptibility to Pst DC3000, as fer‐5 shows enhanced stomatal and apoplastic defences.
COK‐4 restores stomatal response and susceptibility to Pst DC3000 in fer‐5
We observed that fer‐5 stomata do not re‐open in response to Pst DC3000 and this plant is more resistant to bacterial infection (Fig. 2). Thus, we reasoned that these fer‐5 phenotypes could be complemented by expressing a kinase closely related to the FER kinase domain, such as COK‐4. To test this possibility, we first created two lines of Col‐0 and fer‐5 expressing the p35S‐GFP::COK‐4 construct (Col‐0/GFP::COK‐4 lines 8 and 14, and fer‐5/GFP::COK‐4 lines 10 and 12), as well as the empty vector as controls (Col‐0/GFP and fer‐5/GFP). Transient expression of these constructs in Nicotiana benthamiana, followed by western blot analysis, confirmed that the proteins were being expressed at the expected size (Fig. S5A, see Supporting Information). These constructs were then used to create stable Arabidopsis transgenic lines. Plant transformation was confirmed by imaging under fluorescence microscopy, and two independent transgenic lines for each construct were selected for further experimentation (Fig. 3).
Figure 3.
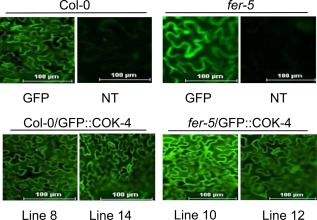
Transgenic lines expressing constructs driven by the 35S promoter. Fluorescence micrographs of Col‐0 and fer‐5 Arabidopsis leaves expressing either the green fluorescent protein gene (GFP) or the GFP::COK‐4 construct, as well as non‐transformed (NT) Col‐0 and fer‐5 leaves as a negative control. Two independent transgenic lines expressing GFP::COK‐4 were selected for further experimentation.
Col‐0 and fer‐5 plants expressing the green fluorescent protein gene (GFP) have the same stomatal response to Pst DC3000 as non‐transformed plants (Fig. S5B,C), and the expression of COK‐4 in both genetic backgrounds does not interfere with stomatal defence (i.e. the stomatal pores still close at 2 hpi) (Fig. 4A). Interestingly, expression of COK‐4 in fer‐5 plants restored the wild‐type stomatal response to Pst DC3000 and the stomatal pore re‐opened at 4 hpi in these transgenic plants (Fig. 4A). In addition, COK‐4‐expressing Col‐0 and fer‐5 lines supported increased bacterial apoplastic populations independent of the method of inoculation (dipping or vacuum infiltration) relative to the respective background plants expressing only GFP (Fig. 4B,C). Furthermore, fer‐5 lines expressing COK‐4 showed more severe symptoms in their leaves than in fer‐5/GFP (Fig. 4B,C). Altogether, these results suggest that the bean COK‐4 is a negative regulator of immunity and shares downstream partners in the same signalling pathway as the FER kinase during its action in plant defences against a bacterial pathogen.
Figure 4.
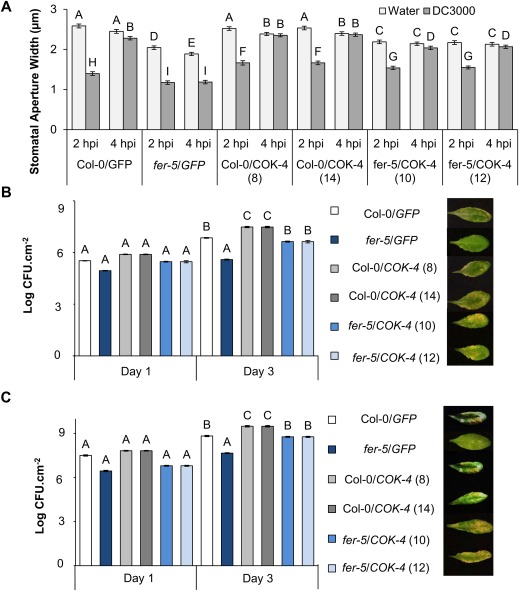
COK‐4 expression increases susceptibility to Pseudomonas syringae pv. tomato (Pst) DC3000. (A) Stomatal aperture width at 2 and 4 h post‐inoculation (hpi) with Pst DC3000 (1 × 108 CFU/mL). Results are shown as the mean (n = 240 ± SE). (B, C) Apoplastic bacterial population in leaves 1 and 3 days after dip inoculation with 1 × 108 CFU/mL (B) or vacuum infiltration with 1 × 106 CFU/mL (C) of Pst DC3000. Results are shown as the mean (n = 18 ± SE), and the statistical significance among the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05). It should be noted that some error bars are too small to appear at the graph scale. Photographs on the right were taken from various leaves at 3 days post‐inoculation.
COK‐4 complements some of the fer‐5 growth and development defects
fer‐5 stomata have constitutively smaller aperture widths than Col‐0 stomata (Fig. 2A), which prompted us to take other measures of the stomatal complex to determine whether FER interferes with stomatal development. We found that, in addition to the stomatal aperture width, the stomatal complex size, guard cell pair size and stomatal complex length in fer‐5 are also smaller than in the wild‐type plant (Fig. 5A–D). However, the fer‐5 mutant has a greater number of stomata per leaf area than Col‐0 (Fig. 5E). Interestingly, expression of the COK‐4 protein in the fer‐5 background partially rescued the stomatal width and stomatal complex size phenotypes, as indicated by ANOVA (Fig. 5A,B), and fully complemented the guard cell pair size (Fig. 5C). However, COK‐4 expression did not restore the stomatal complex length and number of stomata per leaf area (Fig. 5D,E). Expression of COK‐4 in Col‐0 plants did not result in altered phenotypes relative to Col‐0/GFP plants (Fig. 5A–E). Finally, the expression of FER increases during guard cell development, where mature guard cells express 1.9‐fold more FER than stomatal lineage cells (Arabidopsis eFBrowser) (Fig. 5G). These results suggest that FER functions in stomatal development and that some aspects of this function are impacted by COK‐4.
Figure 5.
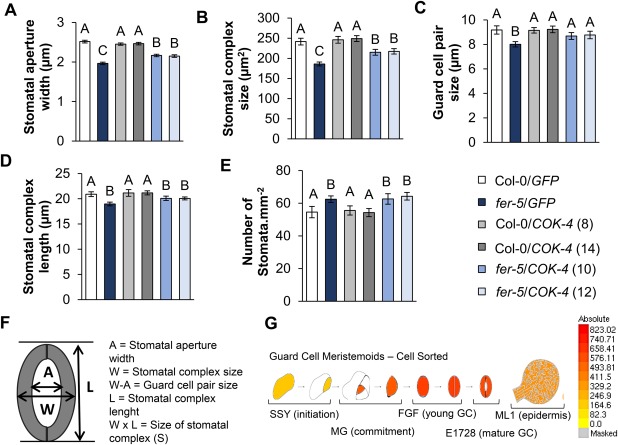
COK‐4 is required for normal stomatal development. Measurements were taken at 3 h after the lights were turned on in the morning: (A) stomatal aperture width; (B) stomatal complex size; (C) guard cell pair size; (D) stomatal complex length; (E) number of stomata per leaf area. Results are shown as the mean (n = 540 to 61 ± SE), and statistical significance amongst the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05). (F) Diagram representing the measurements taken from stoma‐forming guard cells. (G) Schematic representation of FERONIA (FER) gene expression during guard cell development in Arabidopsis adapted from eFBrowser.
We also noticed that fer‐5 rosettes are smaller overall than those of Col‐0 under the growth conditions set for the experiments (Fig. S6A, see Supporting Information; Duan et al., 2010). Thus, a series of measurements was undertaken in order to quantify this phenotypic difference. We verified significant differences in fresh and dry weights, number of leaves at maturity (4–5 weeks of age), days to bolting and seed weight between the fer‐5 mutant and Col‐0; however, no difference was observed in the number of days to emergence (Fig. S6B–G).
Next, we determined whether COK‐4 could alter Col‐0 growth and development, as well as complement the phenotypic defects of fer‐5 plants. Interestingly, Col‐0/COK‐4 lines showed enhanced growth, i.e. higher rosette fresh and dry weights and more leaves, than Col‐0/GFP (Fig. 6A–C), whereas Col‐0/GFP and Col‐0/COK‐4 showed the same average time to bolting and seed weight (Fig. 6D,E). Interestingly, fer‐5/COK‐4 plants showed higher average values for all phenotypes evaluated relative to fer‐5/GFP plants (Fig. 6A–E). However, although COK‐4 fully restored the number of leaves in the fer‐5 background (Fig. 6C), it partially rescued fresh and dry weights, days to bolting and seed weight (Fig. 6A,B,D,E). The number of days from sowing to seedling emergence did not change amongst the plant genotypes (Fig. 6F). The expression of GFP in fer‐5 and Col‐0 plants did not affect the overall appearance of the rosettes relative to those of non‐transformed plants, but an increase in rosette size was visible in COK‐4‐expressing lines (Fig. 6G). These findings suggest that the common bean COK‐4 is an up‐regulator of plant growth and functions in the same pathway as the Arabidopsis FER in controlling at least some aspects of plant growth and development.
Figure 6.
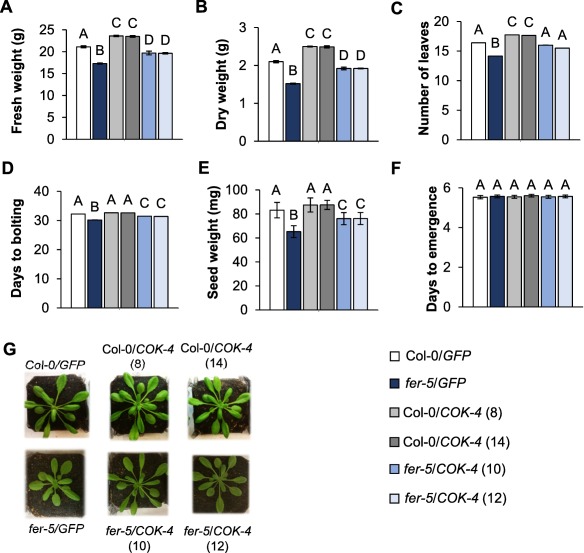
COK‐4 induces some aspects of plant growth and development. The graphs show the average fresh and dry weights of 4–5‐week‐old Arabidopsis rosettes (n = 30 ± SE) (A, B), number of leaves at maturity (4–5‐week‐old plants) (n = 20 ± SE) (C), days to bolting (n = 28 ± SE) (D), seed weight per plant (n = 5 ± SE) (E) and days to seedling emergence (n = 51 ± SE) (F). Statistical significance among the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05). (G) Representative photographs of 4–5‐week‐old plants used for each measurement. Note that error bars in (C) and (D) are too small to appear in the graphs.
Discussion
We have previously described the COK‐4 gene from Phaseolus vulgaris as a promising source of plant defence against a broad range of pathogens. For instance, the bacterium P. syringae pv. phaseolicola, the causal agent of halo blight disease in beans, down‐regulates a COK‐4‐like gene in an incompatible interaction with common bean (i.e. resistance), whereas the bacterial elicitor flagellin induces the expression of this gene (Oblessuc et al., 2015). In this study, we used the model plant Arabidopsis to further characterize the role of COK‐4 in plant immunity. We identified FER kinase as a putative orthologue of COK‐4 in Arabidopsis by protein alignment and 3D structure modelling analyses (Fig. 1). FER was first described as being responsible for pollen tube recognition during pollination (Escobar‐Restrepo et al., 2007). Its partners in this process are two ANXUR proteins (ANX1/2) which contribute to the growth of the pollen tube (Miyazaki et al., 2009). Although FER and ANX1/2 are closely related to each other and to COK‐4 (Figs 1 and S2), unlike ANX1/2, FER is highly expressed in all plant tissues, except pollen (Lindner et al., 2012). In addition, we observed that a lack of a functional FER, but not ANX1 or ANX2, resulted in altered plant defences against Pst DC3000 (Figs 2 and S2) when dip inoculated with high inoculum concentration [1 × 108 colony‐forming units (CFU)/mL]. However, in a different experimental set‐up (syringe infiltration with 5 × 104 CFU/mL), Mang et al. (2017) observed that anx1–2 and anx2‐2 leaves can support smaller bacterial populations than Col‐0 leaves. Moreover, ANX1 may act as a negative regulator of immunity by interfering with FLS2–BAK1 formation under flagellin elicitation (Mang et al., 2017). These recent studies indicate that other members of CrRLK1L could also be involved in plant immune responses.
Previously, the FER mutation in fer‐5 has been shown to cause reduction in shoot size, defects in root hairs, reduction in signalling response to auxin, partial insensitivity to RALF and hypersensitivity to ABA in relation to wild‐type plants (Duan et al., 2010; Haruta et al., 2014; Yu et al., 2012). However, this is the first study to report responses to pathogen infection in the fer‐5 mutant background. We observed altered stomatal and apoplastic immune responses in fer‐5 plants after inoculation with Pst DC3000, such as smaller stomatal aperture and an absence of stomatal re‐opening in response to this bacterium (Fig. 2). Seedlings of another FER mutant allele, named as fer, also exhibit smaller stomatal aperture in addition to higher accumulation of ROS (Keinath et al., 2010). Increased stomatal closure in response to ABA was observed in the fer‐4 mutant allele, and was caused, at least in part, by the higher accumulation of ROS in guard cells (Yu et al., 2012). Thus, it is possible that fer‐5 guard cells also accumulate high levels of ROS, and this may contribute to the constitutively smaller stomatal aperture in this mutant.
In addition, fer‐5 showed an increased resistance to Pst DC3000 (Fig. 2). Keinath et al. (2010) also observed that the FER mutation (fer) results in increased resistance to the Pst DC3000 ΔavrPtoΔavrPtoB bacterial strain, in relation to the wild‐type plant. These authors attributed this phenomenon, at least in part, to the enhanced fer mutant cell death response that could limit bacterial proliferation (Keinath et al., 2010). Moreover, this same FER mutant (fer) shows enhanced resistance to powdery mildew fungus infection (Kessler et al., 2010). As the fungal invasion of plant cells and pollen tube perception share some common features, it has been suggested that a functional FER probably enables fungal growth and plant colonization (Kessler et al., 2010). Lack of a functional FER in the fer‐4 mutant plant increases resistance to another fungus, F. oxysporum, in which the RALF‐like peptide produced by the fungus targets FER to suppress host immunity (Masachis et al., 2016). Altogether, these studies highlight the importance of FER during plant–pathogen interactions, in which FER‐mediated suppression of plant immunity is a common outcome with distinct pathogens.
Previously, COK‐4 has been described as a possible negative regulator of immunity in beans (Oblessuc et al., 2015). In this study, we observed that COK‐4 restores stomatal re‐opening and leaf susceptibility in response to Pst DC3000 in the Arabidopsis line fer‐5 expressing this common bean kinase (Fig. 4). Pst DC3000 produces the phytotoxin coronatine which can promote bacterial entrance into the leaf apoplast by inhibiting stomatal closure and inducing stomatal opening (Melotto et al., 2006; Panchal et al., 2016a). In addition, Pst DC3000 effectors, such as HopX1, HopF2 and HopM1, can prevent stomatal defence and/or induce stomatal re‐opening (Gimenez‐Ibanez et al., 2014; Hurley et al., 2014; Lozano‐Durán et al., 2014, Melotto et al., 2017). Therefore, FER of Arabidopsis and COK‐4 of beans could play a role in coronatine and/or effector signalling responses to promote disease. This hypothesis is further supported by the absence of chlorosis in fer‐5 leaves inoculated with Pst DC3000 (Figs 2 and 4), a well‐characterized symptom in response to coronatine (Melotto et al., 2006). Further studies on this issue would greatly assist the understanding of COK‐4 and FER function in the stomatal re‐opening pathway and disease progression.
Interestingly, Col‐0/COK‐4 lines support higher bacterial populations than do those of Col‐0/GFP. This finding suggests that COK‐4 expression may have an additive dosage effect on FER‐mediated regulation of plant immunity in response to Pst DC3000. Notably, the common bean genome has many copies of the COK‐4 gene that are highly similar, but not identical, to each other (Oblessuc et al., 2015). Therefore, it is possible that the encoded COK‐4 proteins work together to establish specific levels of disease reaction in different bean genotypes. This hypothesis deserves careful testing as FER, and probably COK‐4, may play a role in plant–pathogen interactions, acting as negative regulators of plant immunity.
High susceptibility to a coronatine‐deficient strain of Pst D3000 (COR–) has been observed in fer‐2 and fer‐4 knockout mutants (Stegmann et al., 2017), suggesting that FER could also promote plant immunity. FER is rapidly phosphorylated in the presence of flg22 (Benschop et al., 2007) and facilitates the formation of the FLS2–BAK1 complex at the cell membrane, thereby enabling the signal transduction that induces immune responses in the plant (Chinchilla et al., 2007; Stegmann et al., 2017). In addition, FER is a receptor of the small hormone peptide RALF, and RALF23–FER interaction inhibits the FER scaffold‐mediated formation of the FLS2–BAK1 complex. Therefore, FER can participate in both negative and positive regulation of plant innate immunity, depending on the perception of the hormone RALF23 (Stegmann et al., 2017). This highlights the importance of FER, and its malectin ectodomain, in the crosstalk between different signals in the plant. One cannot exclude that fer‐5 may be a dominant activating allele, unlike the loss‐of‐function alleles fer‐2 and fer‐4. The fer‐5 allele expresses the malectin domain, but not the kinase domain (Fig. S4), whereas fer‐2 and fer‐4 are complete knockout mutants (Deslauriers and Larsen, 2010; Duan et al., 2010). Recently, this phenomenon has been established for a mutant allele of another member of the CrRLK1L family, THESEUS1 (THE1). Similar to fer‐5, the mutant the1–4 also expresses the malectin domain, but not the kinase domain, of THE1 (Merz et al., 2017). Possibly, fer‐5 may possess a functional malectin domain that can perceive the extracellular environment and associate with signal transmitting partners in a stronger manner, because of the lack of the kinase domain, resulting in a dominant phenotype. This putative gain‐of‐function hypothesis is yet to be tested, and will probably depend on whether FER is the transducing element or whether it requires partners to transduce the perceived signal.
FER is required for plant responses to different hormones involved in the control of growth and development (Li et al., 2016). Similar to other members of the family, FER has been described as a modulator of plant development and is fundamental to cell growth (Deslauriers and Larsen, 2010; Duan et al., 2010; Guo et al., 2009; Haruta et al., 2014; Yu et al., 2012). For instance, fer‐2 seedlings are hyposensitive to the hormone BR and hypersensitive to ethylene, resulting in a small hypocotyl relative to wild‐type plants (Deslauriers and Larsen, 2010). BR signalling also regulates guard cell development by inhibiting stomatal formation in leaves (Zhu et al., 2013). Interestingly, we observed a reduction in stomatal aperture, guard cell complex size and guard cell length in fer‐5 leaves (Fig. 5), suggesting that FER may play a role in this hormonal control of stomatal development. In addition, COK‐4 expression partially complements these guard cell developmental defects of fer‐5, indicating that COK‐4 could be required for this process in beans. Therefore, FER and COK‐4 probably act in the same signalling pathways that enable guard cell development and stomatal movement.
FER can also work in the crosstalk between ABA and auxin, evidenced by the observation that fer‐4 and fer‐5 mutants are hypersensitive to ABA and hyposensitive to auxin, resulting in the repression of shoot growth (Yu et al., 2012). We also observed a reduction in fer‐5 shoot growth and other growth and developmental traits (Fig. 6). Notably, COK‐4 expression totally or partially rescued fer‐5 growth and developmental defects. Interestingly, the expression of COK‐4 in Col‐0 resulted in increased biomass (Fig. 6). Thus, the number of COK‐4 copies expressed in the genome may result in distinct biomass accumulation patterns in beans.
FER may be involved in specific cell‐type responses (Li et al., 2015; Liao et al., 2017), either as a result of the differential cell‐type expression pattern of its potentially 30 RALF‐like ligands (Haruta et al., 2014; Stegmann et al., 2017), or because of its opposite roles in ROS production in roots relative to guard cells (Yu et al., 2012). Although shoot growth and defence responses were enhanced in the presence of the COK‐4 protein in Col‐0 plants (Figs 4 and 6), Col‐0 plants with or without COK‐4 were indistinguishable with regard to guard cell development, stomatal movement, days to bolting and seed weight (Figs 5 and 6). This suggests that COK‐4 plays specific roles depending on the cell type, as previously hypothesized for FER. The putative kinase COK‐4 is a single domain protein, whereas FER has two functional domains: an extracellular malectin domain and an intracellular kinase domain (Li et al., 2016). Although it is unknown whether fer‐5 has a functional malectin domain, previous studies have shown that fer‐5 has a milder defective phenotype when compared with fer‐4, a FER mutant allele that lacks the expression of both protein domains (Duan et al., 2010; Haruta et al., 2014; Yu et al., 2012). Therefore, it is plausible to consider that COK‐4 requires the extracellular domain of FER to be fully functional. If so, COK‐4‐mediated signalling responses in guard cells and reproductive tissues could be limited by the similar FER–malectin expression pattern in both Col‐0 and Col‐0/COK‐4 plants. However, if COK‐4 does not require a FER–malectin domain to be functional in these cell types, the additive effect of COK‐4 expression in the Col‐0 background could be limited by other molecular partners that have the same expression levels in both Col‐0 and Col‐0/COK‐4 genotypes. In this scenario, it is also possible that COK‐4 may complement the function of the kinase domain of other CrRLK1L family members, as their kinase domains are functionally equivalent (Kessler et al., 2014). Moreover, a third hypothesis can be considered, in which the COK‐4 dosage is important in these cell types, and the expression of multiple copies of COK‐4 is necessary to fully complement the fer‐5 defects and enhance Col‐0 guard cell movement and development, as well as plant reproduction.
Our study provides evidence that the common bean COK‐4 acts in the same signalling pathway as the FER kinase in Arabidopsis, in which COK‐4 and the FER kinase domain are required for the regulation of both shoot growth/development and plant immunity (Fig. 7). Plant growth and defence share common signalling components (Huot et al., 2014; Züst and Agrawal, 2017). One of these components may be FER or COK‐4 which could function as a hub for the various plant responses to distinct environmental challenges to improve plant fitness. Continued studies on both FER and COK‐4 are necessary to pinpoint the steps in the molecular pathway(s) in which these proteins are involved during plant–pathogen interactions and growth responses.
Figure 7.
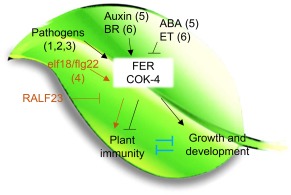
Working model for the possible roles of COK‐4 in the crosstalk between different signals in the control of plant shoot growth/development and immunity. The model is based on our results and previously published data: (1) Keinath et al. (2010); (2) Kessler et al. (2010); (3) Masachis et al. (2016); (4) Stegmann et al. (2017); (5) Yu et al. (2012); (6) Deslaurier and Larsen (2010). Orange arrows and bars indicate an alternative pathway for FERONIA (FER) function in immunity (Stegmann et al., 2017). Blue bars in the model indicate the possible existence of negative crossregulation of plant immunity and growth that is independent of FER/COK‐4. Arrows indicate positive regulation and bars indicate negative regulation. ABA, abscisic acid; BR, brassinosteroid; ET, ethylene; RALF, rapid alkalinization factor.
Experimental Procedures
In silico analyses
The phylogenetic tree was constructed using the top 40 unique protein kinases of Arabidopsis (Table S1, see Supporting Information) with significant alignment (threshold E‐value ≤ 3 × 10−33) with the predicted COK‐4 protein from the common bean line SEL 1308 [National Center for Biotechnology Information (NCBI) accession number AAF98554; Melotto and Kelly, 2001]. The Position‐Specific Iterated BLAST (PSI‐BLAST) program was used with the Arabidopsis thaliana database as query (Altschul et al., 1997, 2005). The phylogenetic tree was obtained using only the kinase domain of each protein with the maximum parsimony method from MAFFT software (Katoh and Standley, 2013). The nodes were confirmed by 1000 bootstraps.
The 3D protein models for FER and COK‐4 were obtained using SWISS‐MODEL software (Biasini et al., 2014). Both whole proteins were used for target–template alignment search with BLAST and HHBlits (Altschul et al., 1997, 2005; Remmert et al., 2012) against the SWISS‐MODEL template library. As a 3D structure is not available for the FER protein, the Pto protein kinase crystal structure (library id 2qkw.1.B; Xing et al., 2007) was used as a template to obtain the 3D models. Amongst the crystal protein structures available at the SWISS‐MODEL database, Pto has the highest sequence identity with both FER (58.97%) and COK‐4 (45.58%) proteins. The alignment between both protein models, COK‐4 and FER, was obtained using CCP4G v.2.10.6 software (McNicholas et al., 2011).
The amino acid sequence alignment was obtained using BioEdit software v.7.2.5 with the two sequence pairwise alignment method, allowing the ends to slide, or the ClustalW multiple alignment tool (Hall, 1999). The phosphorylation sites of FER and COK‐4 were predicted using PhosPhAt (Durek et al., 2010) and NetPhos 3.1 (Blom et al., 1999) software, respectively.
ANX1, ANX2 and FER expression in different Arabidopsis tissues was obtained using the developmental map from Arabidopsis eFBrowser software (http://bar.utoronto.ca; Winter et al., 2007). The expression of FER during Arabidopsis guard cell development was obtained using the Arabidopsis eFBrowser and data from Adrian et al. (2015).
Plant material
Arabidopsis thaliana (L. Heyhn.) wild‐type Columbia [Col‐0, Arabidopsis Biological Resource Center (ABRC) stock CS60000], the mutant fer‐5 (ABRC stock SALK_029056c) and the Col‐0 and fer‐5 lines expressing COK‐4 were sown in a 1 : 1 (v/v) mixture of growing medium (Redi‐earth plug and seedling mix, Sun Gro, Sacramento, CA, USA) and perlite. Plants were kept at 4 °C for 2 days for seed stratification, and transferred to a controlled environmental chamber set at 22 °C, 60% ± 5% relative humidity and 12‐h photoperiod under a light intensity of 100 μmol/m2/s.
The fer‐5 mutant has a T‐DNA insertion in the region that encodes the kinase domain at the end of its single exon. Thus, the malectin domain is expressed in this mutant, but the kinase domain is not (Fig. S4; Duan et al., 2010).
To express COK‐4 in Col‐0 and fer‐5 plants, the COK‐4 transcript was amplified from the common bean line G2333 using the primers listed in Table S2 (see Supporting Information). G2333 is the parental line of the breeding line SEL1308 that contains the Co‐42 locus (Melotto and Kelly, 2001). COK‐4 was cloned into the pB7WGF2 vector under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Karimi et al., 2002) to make the construct p35S‐GFP::COK‐4. This construct was transformed into the Agrobacterium tumefaciens strain C58C1 by the freeze–thaw method (Chen et al., 1994), which was used to transform Col‐0 and fer‐5 plants employing the floral dip method (Clough and Bent, 1998). Col‐0 plants and fer‐5 were also transformed with pB7WGF2 empty vector as controls. Transgenic lines were selected with BASTA (0.0114% glufosinate ammonium; Bayer®, Leverkusen, Germany) supplemented with 0.005% of Silwet L‐77 (Lehle Seeds, Round Rock, TX, USA). The expected transgene size was confirmed with western blotting of N. benthamiana leaves transiently expressing either GFP or GFP::COK‐4 protein, as described below.
Gene expression analysis
To assess the gene expression of both the malectin and kinase domains from FER in wild‐type Col‐0 plants and the fer‐5 mutant, leaves of each genotype were ground without any treatment. Total RNA was extracted using an RNeasy mini kit (Qiagen, Hilden, Germany) with in‐column DNAse treatment, according to the manufacturer's instructions. RNA was synthesized to cDNA using the Takara RNA PCR kit (AMV) (Clontech, Mountain View, CA, USA). End‐point reverse transcription‐polymerase chain reaction (RT‐PCR) (25 µL) was performed with 1 U DNA polymerase Gotaq® (Promega, Madison, WI, USA), 1 × enzyme buffer, 1.5 mm MgCl2, 200 µm deoxynucleoside triphosphate (dNTP), 1 µL (50 ng) of cDNA template and 200 nm of reverse and forward domain‐specific primers (Table S2). Reactions were carried out using the following cycling parameters: one cycle at 95 °C for 5 min, 40 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 10 min. Gene expression levels were normalized based on the expression of the housekeeping gene ACT2 (At3g18780; Table S2). Amplicons were visualized after gel electrophoresis using the UV light on a C300 imaging system (Azure Biosystems, Dublin, CA, USA).
Protein transient expression assay
Single colonies of A. tumefaciens strain C58C1 containing p35S‐GFP::COK‐4 or p35S‐GFP construct cloned into the binary vector pB7WGF2 were selected and cultured overnight at 28 ºC in Luria‐Bertani (LB) medium containing 100 µg/mL rifampicin, 10 µg/mL tetracycline and 100 µg/mL spectinomycin. Agrobacterium cells were harvested from overnight cultures, washed in infiltration buffer [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.8, 10 mm MgCl2, 0.2% sucrose, 300 μm acetosyringone] and suspended to an optical density at 600 nm (OD600)= 0.2 also in infiltration buffer. After 3 h of incubation in the dark, cultures were syringe infiltrated into mature leaf tissue of N. benthamiana. Infiltrated plants were returned to previous growing conditions (12‐h photoperiod and 25 ºC) and fluorescence was observed by microscopy at 2–3 days after inoculation. In addition, the transgene was detected using western blotting as described by Thines et al. (2007), with modifications. Briefly, after the extraction of soluble protein from leaves using protein buffer [100 mm dithiothreitol (DTT), 2% sodium dodecylsulfate (SDS), 50 mm tris(hydroxymethyl)aminomethane (Tris)‐HCl, pH 6.8, 10% glycerol], samples were run on a Mini‐protein® TGX Stain‐Free sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel (BioRad, Hercules, CA, USA). After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (PVDF‐Plus, Micron Separations Inc., Westborough, MA, USA). Protein transfer was performed in a Trans‐Bolt SD Cell (BioRad) set for 15 V for 20 min. The membrane was then incubated with the primary antibody (anti‐GFP, 1 : 50 000; Life Technologies, Carlsbad, CA, USA), overnight at 4 °C, on an orbital shaker (75 rpm). The secondary antibody (anti‐Rabbit IgG, 1 : 150 000; Life Technologies) was incubated for 2 h at room temperature. Membrane development was performed using chemiluminescent substrates (SuperSignal West Femto trial kit, Thermo Scientific, Rockford, IL, USA), detected with the CHEMI tool of a C300 imaging system (Azure Biosystems).
Microscopy
BASTA‐resistant plants and control non‐transformed plants were analysed for the presence of the GFP signal with a Nikon Eclipse 80i fluorescent microscope equipped with a Fluorescein IsoThioCyanate (FITC) filter (475–495 nm), Differential Interference Contrast (DIC) and long‐distance objectives, using NIS Elements Imaging software (Nikon Corporations, Shinagawa‐ku, Tokyo, Japan).
Pathogenesis assay
Pathogenesis assay was conducted using Pst DC3000, as described previously (Jacob et al., 2017; Katagiri et al., 2002). Briefly, plants were dip inoculated with 1 × 108 CFU/mL or vacuum infiltrated with 1 × 106 CFU/mL of bacterial suspension containing 0.03% or 0.004% of Silwet L‐77 (Lehle Seeds), respectively. The bacterial population in the leaf apoplast was determined on days 1 and 3 after infection using a serial dilution‐based method.
Stomatal bioassay
The stomatal response to the bacterium was assessed in 4–5‐week‐old plants as described previously (Montano and Melotto, 2017). As differences in stomatal movements could be attributed to developmental defects in mutants and transgenic plants, stomatal complex dimensions were also measured as follows: stomatal aperture width (A), stomatal complex width (W), guard cell pair size (W – A), length of the stomatal complex (L) and size of the stomatal complex (W × L), as described by Panchal et al. (2016b). The stomatal measurements were taken using non‐treated mature leaves at 3 h after the lights had been turned on in the morning.
Plant growth and developmental evaluations
Plant growth and developmental phenotypes were evaluated using several parameters. First, the fresh and dry weights of 35‐day‐old plant rosettes were measured following the procedure described by Abe et al. (2003). Briefly, rosettes were collected and immediately weighed for fresh weight, and the same rosettes were placed at 70 °C for 72 h to obtain the dry weight. Second, the number of true leaves produced by the apical meristem was recorded on bolted plants (around 35‐day‐old plants), following the description by Pouteau et al. (2004). Third, the bolting time was determined as the number of days from sowing to the elongation of the first floral stem at 0.1 cm height (Pouteau et al., 2004). Fourth, the seed weight was measured by collecting and weighing the seeds produced by each plant grown under standard conditions, as described above. Fifth, the days to seedling emergence were determined from the time of sowing to the germination of seedlings, i.e. when the cotyledons were visible. All measurements were obtained from three biological replicates (n = 5–51).
Statistical analyses
Statistical analyses were performed using Student's t‐test or analysis of variance (ANOVA) followed by Scott–Knott's test at 5% probability using InfoStat statistical software (Di Rienzo et al., 2014). All experiments were performed at least three times with similar results, where the data points represent the mean ± standard error (SE).
Author Contributions
PRO and MM conceived the project and designed all the research. RFA and PRO performed the experiments. RFA, PRO and MM analysed the data. MCGV and MM provided the materials. RFA, PRO and MM wrote the article.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Top 40 genes with protein sequence similarity to COK‐4 [National Center for Biotechnology Information (NCBI) accession number AAF98554] using the Position‐Specific Iterated BLAST (PSI‐BLAST) program and the Arabidopsis thaliana database (Altschul et al., 1997, 2005).
Table S2 Primer names and sequences.
Fig. S1 FERONIA (FER) and COK‐4 protein alignment and phosphorylation motif analyses. (A) Diagram showing the alignment between FER and COK‐4 proteins determined with the two sequence pairwise alignment function of BioEdit v.7.2.5. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated. The kinase domain residue alignment is shown below the diagram. COK‐4 does not align outside of the FER kinase domain. Kinase domain motifs were identified using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) and are highlighted in yellow (conserved domain cd14066: STKc_IRAK; FER, E‐value = 1.6 × 10−86; COK‐4, E‐value = 4.8 × 10−58). Black stars indicate predicted phosphorylation sites, and red stars indicate experimentally determined phosphorylation sites in the FER protein. Shared serine (S), threonine (T) and tyrosine (Y) between COK‐4 and FER are highlighted in green. The predicted transmembrane region of COK‐4, determined by Melotto and Kelly (2001), is underlined, and the region identified in the predicted COK‐4 three‐dimensional (3D) protein model with no overlap with the FER 3D protein (see Fig. 1) is shown in bold. (B) Predicted phosphorylation sites for COK‐4 and the FER kinase domain.
Fig. S2 Alignment between COK‐4 (GenBank: AAF98554.1) and the kinase domain of all 40 proteins used for phylogenetic analysis [see Table S1 (Supporting Information) for PSI‐BLAST results]. The alignment was obtained using the ClustalW multiple alignment tool of the BioEdit software v.7.2.5. It should be noted that COK‐4 has a single kinase domain that only aligns with the kinase domain of all proteins. The COK‐4 sequence is highlighted in yellow and its closest sequence (AT3G51550) is FERONIA (FER).
Fig. S3 Analyses of the Arabidopsis FERONIA (FER), ANXUR 1 and 2. (A–C) Diagrams depicting the developmental map of gene expression for ANX1, ANX2 and FER adapted from Arabidopsis eFBrowser. (D, E) Apoplastic bacterial population in leaves of Col‐0 and mutant plants after dip inoculation with 1 × 108 colony‐forming units (CFU)/mL of Pseudomonas syringae pv. tomato (Pst) DC3000, 1 and 3 days after inoculation. Results are shown as the mean (n = 6 ± SE), and statistical significance between Col‐0 and each mutant was performed using Student's t‐test (ns, non‐significant). The schematic representation below each graph shows the ANX1 and ANX2 gene structures adapted from TAIR (The Arabidopsis Information Resource) gene modelling. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated, as well as the position of the T‐DNA insertion present in the SALK_016179C (anx1‐1) and SALK_133057C (anx2‐2) lines. The dark blue box indicates the coding domain sequences, the light blue boxes indicate the untranslated regions (UTRs) of the genes and black lines represent the intron.
Fig. S4 Arabidopsis fer‐5 mutant lacks expression of the kinase domain. (A) Reverse transcription‐polymerase chain reaction (RT‐PCR) amplification of the malectin and kinase encoding regions of FERONIA (FER) of Col‐0 and the fer‐5 mutant. ACT2 was used as an RNA loading control. It should be noted that fer‐5 lacks the kinase domain. (B) Schematic representation of the FER gene structure adapted from TAIR (The Arabidopsis Information Resource) gene modelling. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated, as well as the position of the T‐DNA insertion present in the SALK_029056C line. The dark blue box indicates the coding domain sequence, the light blue boxes indicate the untranslated regions (UTRs) of the gene and the black lines represent the intron. The arrows indicate the region in which the primers used in (A) align with the FER gene.
Fig. S5 (A) Transient expression in Nicotiana benthamiana. Western blot analysis using anti‐green fluorescent protein (anti‐GFP) and total proteins extracted from leaves transiently expressing the indicated construct. The number above each gel lane indicates: 1, protein standard (Bio‐Rad®); 2, GFP::COK‐4 protein (69.2 kDa); 3, GFP protein (26.9 kDa). Fluorescence micrographs of N. benthamiana leaves showing the GFP signal in cells transformed with either GFP (control) or a GFP::COK‐4 construct. BF, bright field. (B, C) Col‐0 and fer‐5 have the same stomatal response to Pseudomonas syringae pv. tomato (Pst) DC3000 as the respective transgenic lines expressing GFP. (B) Col‐0 wild‐type (WT) and Col‐0/GFP (p35S‐GFP) and (C) fer‐5 mutant and fer‐5/GFP (p35S‐GFP). Plants were dip inoculated with 1 × 108 CFU/mL of Pst DC3000 and the stomatal aperture was assessed at 2 and 4 h post‐inoculation (hpi). Results are shown as the mean (n = 195–240 ± SE), and statistical significance amongst the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05).
Fig. S6 FERONIA (FER) affects plant growth and development. (A) Photograph of 3‐week‐old Col‐0 and fer‐5 plants grown under the conditions described in Experimental procedures. Note the fer‐5 mutant dwarf phenotype. (B, C) Graphs showing average fresh (B) and dry (C) weights of 4–5‐week‐old Arabidopsis rosettes. (D) Number of leaves at maturity (4–5 weeks of age). (E) Days to bolting. (F) Seed weight (n = 5 ± SE). (G) Days to seedling emergence. Data points represent the mean (5 < n < 51 ± SE), and statistical significance was calculated by Student's t‐test (***p ≤ 0.001; ns, non‐significant). Note that some error bars are too small and cannot be seen at the scale used in the graphs.
Acknowledgements
We thank Dr Nathan Pumplin for helpful discussions on this research and the Brazilian Funding Agency CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for a scholarship to RFA (award number BEX 3517/15‐2). The authors declare no conflicts of interest.
Contributor Information
Paula Rodrigues Oblessuc, Email: problessuc@ucdavis.edu.
Maeli Melotto, Email: melotto@ucdavis.edu.
References
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian, J. , Chang, J. , Ballenger, C.E. , Bargmann, B.O. , Alassimone, J. , Davies, K.A. , Lau, O.S. , Matos, J.L. , Hachez, C. , Lanctot, A. , Vatén, A. , Birnbaum, K.D. and Bergmann, D.C. (2015) Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self‐renewing population. Dev. Cell, 33, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Wootton, J.C. , Gertz, E.M. , Agarwala, R. , Morgulis, A. , Schaffer, A.A. and Yu, Y.K. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y. , Yang, L. , Hetzel, J. , Dangl, J.L. and Chory, J. (2014) The growth–defense pivot: crisis management in plants mediated by LRR‐RK surface receptors. Trends Biochem. Sci. 39, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop, J.J. , Mohammed, S. , O'Flaherty, M. , Heck, A.J.R. , Slijper, M. and Menke, F.L.H. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics, 6, 1198–1214. [DOI] [PubMed] [Google Scholar]
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , Kiefer, F. , Gallo, C.T. , Bertoni, M. , Bordoli, L. and Schwede, T. (2014) SWISS‐MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom, N. , Gammeltoft, S. and Brunak, S. (1999) Sequence‐ and structure‐based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 5, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Boisson‐Dernier, A. , Roy, S. , Kritsas, K. , Grobei, M. A. , Jaciubek, M. , Schroeder, J.I. and Grossniklaus, U. (2009) Disruption of the pollen‐expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development, 136, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, M.L. , Yoshida, Y. , Major, I.T. , Ferreira, D.O. , Weraduwage, S.M. , Froehlich, J.E. , Johnson, B.F. , Kramer, D.M. , Jander, G. , Sharkey, T.D. and Howe, G.A. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth–defense tradeoffs. Nat. Commun. 7, 12 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.H. , Leu, J.C. and Huang, T.C. (1994) Transport and hydrolysis of urea in a reactor–separator combining an anion‐exchange membrane and immobilized urease. J. Chem. Technol. Biotechnol. 61, 351–357. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nürnberger, T. , Jones, J.D. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deslauriers, S.D. and Larsen, P.B. (2010) FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant. 3, 626–640. [DOI] [PubMed] [Google Scholar]
- Di Rienzo, J.A. , Casanoves, F. , Balzarini, M.G. , Gonzalez, L. , Tablada, M. and Robledo, C.W. (2014) InfoStat versión 2014 Argentina: InfoStat Group, Facultad de Ciencias Agropecuarias, Universidad Nacioal de Córdoba. Available at http://www.infostat.com.ar [accessed 23 January 2018].
- Duan, Q. , Kita, D. , Li, C. , Cheung, A.Y. and Wu, H.M. (2010) FERONIA receptor‐like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA, 107, 17 821–17 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek, P. , Schmidt, R. , Heazlewood, J.L. , Jones, A. , Maclean, D. , Nagel, A. , Kersten, B. and Schulze, W.X. (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38, 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar‐Restrepo, J.M. , Huck, N. , Kessler, S. , Gagliardini, V. , Gheyselinck, J. , Yang, W.C. and Grossniklaus, U. (2007) The FERONIA receptor‐like kinase mediates male–female interactions during pollen tube reception. Science, 317, 656–660. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernández‐Barbero, G. , Chini, A. , Rathjen, J.P. and Solano, R. (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Li, L. , Ye, H. , Yu, X. , Algreen, A. and Yin, Y. (2009) Three related receptor‐like kinases are required for optimal cell elongation in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 106, 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Haruta, M. , Sabat, G. , Stecker, K. , Minkoff, B.B. and Sussman, M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science, 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant. 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, B. , Lee, D. , Mott, A. , Wilton, M. , Liu, J. , Liu, Y.C. , Angers, S. , Coaker, G. , Guttman, D.S. and Desveaux, D. (2014) The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One, 9, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, C. , Panchal, S. and Melotto, M. (2017) Surface inoculation and quantification of Pseudomonas syringae population in the Arabidopsis leaf apoplast. Bio. Protoc. 7, e2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inzé, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Katagiri, F. , Thilmony, R. and He, S.Y. (2002) The Arabidopsis thaliana–Pseudomonas syringae Interaction The Arabidopsis Book. American Society of Plant Biologists. Available at http://www.aspb.org/publications/arabidopsis [accessed 18 June 2015]. [DOI] [PMC free article] [PubMed]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath, N.F. , Kierszniowska, S. , Lorek, J. , Bourdais, G. , Kessler, S.A. , Shimosato‐Asano, H. , Grossniklaus, U. , Schulze, W.X. , Robatzek, S. and Panstruga, R. (2010) PAMP (pathogen‐associated molecular pattern)‐induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39 140–39 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, S.A. , Shimosato‐Asano, H. , Keinath, N.F. , Wuest, S.E. , Ingram, G. , Panstruga, R. and Grossniklaus, U. (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science, 330, 968–971. [DOI] [PubMed] [Google Scholar]
- Kessler, S.A. , Lindner, H. , Jones, D.S. and Grossniklaus, U. (2014) Functional analysis of related CrRLK1L receptor‐like kinases in pollen tube reception. EMBO Rep. 16, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Yeh, F.‐L. , Cheung, A.Y. , Duan, Q. , Kita, D. , Liu, M.‐C. , Maman, J. , Luu, E.J. , Wu, B.W. , Gates, L. and Jalal, M. (2015) Glycosylphosphatidylinositol‐anchored proteins as chaperones and co‐receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife, 4, e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Wu, H.M. and Cheung, A.Y. (2016) FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, H. , Tang, R. , Zhang, X. , Luan, S. and Yu, F. (2017) FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58, 1143–1150. [DOI] [PubMed] [Google Scholar]
- Lindner, H. , Müller, L.M. , Boisson‐Dernier, A. and Grossniklaus, U. (2012) CrRLK1L receptor‐like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15, 659–669. [DOI] [PubMed] [Google Scholar]
- Lozano‐Durán, R. and Zipfel, C. (2015) Trade‐off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. [DOI] [PubMed] [Google Scholar]
- Lozano‐Durán, R. , Bourdais, G. , He, S.Y. and Robatzek, S. (2014) The bacterial effector HopM1 suppresses PAMP‐triggered oxidative burst and stomatal immunity. New Phytol. 202, 259–269. [DOI] [PubMed] [Google Scholar]
- Mang, H. , Feng, B. , Hu, Z. , Boisson‐Dernier, A. , Franck, C. , Meng, X. , Huang, Y. , Zhou, J. , Xu, G. , Wang, T. , Shan, L. and He, P. (2017) Differential regulation of two‐tiered plant immunity and sexual reproduction by ANXUR receptor‐like kinases. Plant Cell, 1, tpc.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis, S. , Segorbe, D. , Turrà, D. , Leon‐Ruiz, M. , Fürst, U. , El‐Ghalid, M. , Leonard, G. , López‐Berges, M.S. , Richards, T.A. , Felix, G. and Di Pietro, A. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1, 16 043. [DOI] [PubMed] [Google Scholar]
- McNicholas, S. , Potterton, E. , Wilson, K.S. and Noble, M.E.M. (2011) Presenting your structures: the CCP4mg molecular‐graphics software. Acta Crystallogr. D: Biol. Crystallogr. 67, 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. and Kelly, J.D. (2001) Fine mapping of the Co‐4 locus of common bean reveals a resistance gene candidate, COK‐4, that encodes for a protein kinase. Theor. Appl. Genet. 103, 508–517. [Google Scholar]
- Melotto, M. , Coelho, M.F. , Pedrosa‐Harand, A. , Kelly, J.D. and Camargo, L.E.A. (2004) The anthracnose resistance locus Co‐4 of common bean is located on chromosome 3 and contains putative disease resistance‐related genes. Theor. Appl. Genet. 109, 690–699. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Zhang, L. , Oblessuc, P.R. and He, S.Y. (2017) Stomatal defense a decade later. Plant Physiol. 174, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz, D. , Richter, J. , Gonneau, M. , Sanchez‐Rodriguez, C. , Eder, T. , Sormani, R. , Martin, M. , Hématy, K. , Höfte, H. and Hauser, M.T. (2017) T‐DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J. Exp. Bot. 68, 4583–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, S. , Murata, T. , Sakurai‐Ozato, N. , Kubo, M. , Demura, T. , Fukuda, H. and Hasebe, M. (2009) ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19, 1327–1331. [DOI] [PubMed] [Google Scholar]
- Montano, J. and Melotto, M. (2017) Stomatal bioassay to characterize bacterial‐stimulated PTI at the pre‐invasion phase of infection In: Methods in Molecular Biology, Vol. 1578 (Shan L. and He P., eds), pp. 233–241. Springer Protocols, Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- Nissen, K.S. , Willats, W.G. and Malinovsky, F.G. (2016) Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci. 21, 516–527. [DOI] [PubMed] [Google Scholar]
- Oblessuc, P.R. , Francisco, C. and Melotto, M. (2015) The Co‐4 locus on chromosome Pv08 contains a unique cluster of 18 COK‐4 genes and is regulated by immune response in common bean. Theor. Appl. Genet. 128, 1193–1208. [DOI] [PubMed] [Google Scholar]
- Panchal, S. , Chitrakar, R. , Thompson, B.K. , Obulareddy, N. , Roy, D. , Hambright, W.S. and Melotto, M. (2016b) Regulation of stomatal defense by air relative humidity. Plant Physiol. 172, 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal, S. , Roy, D. , Chitrakar, R. , Price, L. , Breitbach, Z.S. , Armstrong, D.W. and Melotto, M. (2016a) Coronatine facilitates Pseudomonas syringae infection of Arabidopsis leaves at night. Front. Plant Sci. 7, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau, S. , Ferret, V. , Gaudin, V. , Lefebvre, D. , Sabar, M. , Zhao, G. and Prunus, F. (2004) Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Physiol. 135, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert, M. , Biegert, A. , Hauser, A. and Soding, J. (2012) HHblits: lightning‐fast iterative protein sequence searching by HMM‐HMM alignment. Nat. Methods, 9, 173–175. [DOI] [PubMed] [Google Scholar]
- Stegmann, M. , Monaghan, J. , Smakowska‐Luzan, E. , Rovenich, H. , Lehner, A. , Holton, N. , Belkhadir, Y. and Zipfel, C. (2017) The receptor kinase FER is a RALF‐regulated scaffold controlling plant immune signaling. Science, 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Winter, D. , Vinegar, B. , Nahal, H. , Ammar, R. , Wilson, G.V. and Provart, N.J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS One, 2, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, W. , Zou, Y. , Liu, Q. , Liu, J. , Luo, X. , Huang, Q. , Chen, S. , Zhu, L. , Bi, R. , Hao, Q. , Wu, J.W. , Zhou, J.M. and Chai, J. (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature, 449, 243–247. [DOI] [PubMed] [Google Scholar]
- Yu, F. , Qian, L. , Nibau, C. , Duan, Q. , Kita, D. , Levasseur, K. , Li, X. , Lu, C. , Li, H. , Hou, C. , Li, L. , Buchanan, B.B. , Chen, L. and Cheung, A.Y. (2012) FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA, 109, 14 693–14 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.Y. , Sae‐Seaw, J. and Wang, Z.Y. (2013) Brassinosteroid signalling. Development, 140, 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst, T. and Agrawal, A.A. (2017) Plant chemical defense indirectly mediates aphid performance via interactions with tending ants. Ecology, 98, 601–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Top 40 genes with protein sequence similarity to COK‐4 [National Center for Biotechnology Information (NCBI) accession number AAF98554] using the Position‐Specific Iterated BLAST (PSI‐BLAST) program and the Arabidopsis thaliana database (Altschul et al., 1997, 2005).
Table S2 Primer names and sequences.
Fig. S1 FERONIA (FER) and COK‐4 protein alignment and phosphorylation motif analyses. (A) Diagram showing the alignment between FER and COK‐4 proteins determined with the two sequence pairwise alignment function of BioEdit v.7.2.5. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated. The kinase domain residue alignment is shown below the diagram. COK‐4 does not align outside of the FER kinase domain. Kinase domain motifs were identified using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) and are highlighted in yellow (conserved domain cd14066: STKc_IRAK; FER, E‐value = 1.6 × 10−86; COK‐4, E‐value = 4.8 × 10−58). Black stars indicate predicted phosphorylation sites, and red stars indicate experimentally determined phosphorylation sites in the FER protein. Shared serine (S), threonine (T) and tyrosine (Y) between COK‐4 and FER are highlighted in green. The predicted transmembrane region of COK‐4, determined by Melotto and Kelly (2001), is underlined, and the region identified in the predicted COK‐4 three‐dimensional (3D) protein model with no overlap with the FER 3D protein (see Fig. 1) is shown in bold. (B) Predicted phosphorylation sites for COK‐4 and the FER kinase domain.
Fig. S2 Alignment between COK‐4 (GenBank: AAF98554.1) and the kinase domain of all 40 proteins used for phylogenetic analysis [see Table S1 (Supporting Information) for PSI‐BLAST results]. The alignment was obtained using the ClustalW multiple alignment tool of the BioEdit software v.7.2.5. It should be noted that COK‐4 has a single kinase domain that only aligns with the kinase domain of all proteins. The COK‐4 sequence is highlighted in yellow and its closest sequence (AT3G51550) is FERONIA (FER).
Fig. S3 Analyses of the Arabidopsis FERONIA (FER), ANXUR 1 and 2. (A–C) Diagrams depicting the developmental map of gene expression for ANX1, ANX2 and FER adapted from Arabidopsis eFBrowser. (D, E) Apoplastic bacterial population in leaves of Col‐0 and mutant plants after dip inoculation with 1 × 108 colony‐forming units (CFU)/mL of Pseudomonas syringae pv. tomato (Pst) DC3000, 1 and 3 days after inoculation. Results are shown as the mean (n = 6 ± SE), and statistical significance between Col‐0 and each mutant was performed using Student's t‐test (ns, non‐significant). The schematic representation below each graph shows the ANX1 and ANX2 gene structures adapted from TAIR (The Arabidopsis Information Resource) gene modelling. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated, as well as the position of the T‐DNA insertion present in the SALK_016179C (anx1‐1) and SALK_133057C (anx2‐2) lines. The dark blue box indicates the coding domain sequences, the light blue boxes indicate the untranslated regions (UTRs) of the genes and black lines represent the intron.
Fig. S4 Arabidopsis fer‐5 mutant lacks expression of the kinase domain. (A) Reverse transcription‐polymerase chain reaction (RT‐PCR) amplification of the malectin and kinase encoding regions of FERONIA (FER) of Col‐0 and the fer‐5 mutant. ACT2 was used as an RNA loading control. It should be noted that fer‐5 lacks the kinase domain. (B) Schematic representation of the FER gene structure adapted from TAIR (The Arabidopsis Information Resource) gene modelling. Coding regions for the kinase, transmembrane (TM) and two malectin domains are indicated, as well as the position of the T‐DNA insertion present in the SALK_029056C line. The dark blue box indicates the coding domain sequence, the light blue boxes indicate the untranslated regions (UTRs) of the gene and the black lines represent the intron. The arrows indicate the region in which the primers used in (A) align with the FER gene.
Fig. S5 (A) Transient expression in Nicotiana benthamiana. Western blot analysis using anti‐green fluorescent protein (anti‐GFP) and total proteins extracted from leaves transiently expressing the indicated construct. The number above each gel lane indicates: 1, protein standard (Bio‐Rad®); 2, GFP::COK‐4 protein (69.2 kDa); 3, GFP protein (26.9 kDa). Fluorescence micrographs of N. benthamiana leaves showing the GFP signal in cells transformed with either GFP (control) or a GFP::COK‐4 construct. BF, bright field. (B, C) Col‐0 and fer‐5 have the same stomatal response to Pseudomonas syringae pv. tomato (Pst) DC3000 as the respective transgenic lines expressing GFP. (B) Col‐0 wild‐type (WT) and Col‐0/GFP (p35S‐GFP) and (C) fer‐5 mutant and fer‐5/GFP (p35S‐GFP). Plants were dip inoculated with 1 × 108 CFU/mL of Pst DC3000 and the stomatal aperture was assessed at 2 and 4 h post‐inoculation (hpi). Results are shown as the mean (n = 195–240 ± SE), and statistical significance amongst the means, indicated by different letters above the bars, was calculated with analysis of variance (ANOVA) followed by Scott–Knott's test (P ≤ 0.05).
Fig. S6 FERONIA (FER) affects plant growth and development. (A) Photograph of 3‐week‐old Col‐0 and fer‐5 plants grown under the conditions described in Experimental procedures. Note the fer‐5 mutant dwarf phenotype. (B, C) Graphs showing average fresh (B) and dry (C) weights of 4–5‐week‐old Arabidopsis rosettes. (D) Number of leaves at maturity (4–5 weeks of age). (E) Days to bolting. (F) Seed weight (n = 5 ± SE). (G) Days to seedling emergence. Data points represent the mean (5 < n < 51 ± SE), and statistical significance was calculated by Student's t‐test (***p ≤ 0.001; ns, non‐significant). Note that some error bars are too small and cannot be seen at the scale used in the graphs.


