Summary
Puccinia striiformis f. sp. tritici (Pst) is an obligate biotrophic fungus that causes extensive damage in wheat. The pathogen is now known to be a heteroecious fungus with an intricate life cycle containing sexual and asexual stages. Orthologues of the STE12 transcription factor that regulate mating and filamentation in Saccharomyces cerevisiae, as well as virulence in other fungi, have been extensively described. Because reliable transformation and gene disruption methods are lacking for Pst, knowledge about the function of its STE12 orthologue is limited. In this study, we identified a putative orthologue of STE12 from Pst in haustoria‐enriched transcripts and designated it as PstSTE12. The gene encodes a protein of 879 amino acids containing three helices in the homeodomain, conserved phenylalanine and tryptophan sites, and two C2/H2‐Zn2+ finger domains. Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analyses revealed that the expression of PstSTE12 was highly induced during the early infection stages and peaked during haustorium formation and the pycniospore stage in the aecial host barberry. Subcellular localization assays indicated that PstSTE12 is localized in the nucleus and functions as a transcriptional activator. Yeast one‐hybrid assays revealed that PstSTE12 exhibits transcriptional activity, and that its C‐terminus is necessary for the activation of transcription. PstSTE12 complemented the mating defect in an α ste12 mutant of S. cerevisiae. In addition, it partially complemented the defects of the Magnaporthe oryzae mst12 mutant in plant infection. Knocking down PstSTE12 via host‐induced gene silencing (HIGS) mediated by Barley stripe mosaic virus (BSMV) resulted in a substantial reduction in the growth and spread of hyphae in Pst and weakened the virulence of Pst on wheat. Our results suggest that PstSTE12 probably acts at an intersection participating in the invasion and mating processes of Pst, and provide new insights into the comprehension of the variation of virulence in cereal rust fungi.
Keywords: mating, Puccinia striiformis f. sp. tritici, STE12, virulence
Introduction
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is a serious threat to worldwide wheat production and global food security (Kolmer, 2005). Yield losses of more than 90% in a single crop season caused by Pst have been reported in many countries (Chen, 2014; Line, 2002; Roelfs and Bushnell, 1985; Wellings, 2011). Despite the importance of Pst, the intricate life cycle has impeded the pursuit of the dissection of pathogenesis in this pathogen. As a strict obligate biotrophic fungus, Pst cannot survive without living tissue from cereal crops to provide nutrition (Eugenea et al., 2009). In addition, stripe rust fungi require an alternative host barberry to complete the macrocyclic sexual life cycle (Jin et al., 2010; Zhao et al., 2013). In general, barberry enables rust fungi to survive winter, provides initial inoculum to cereal crops, generates new races and confers diversification of rust populations (Zhao et al., 2016). The urediniospores are usually produced early in the next growing season and dispersed as air‐borne inoculum to produce new infections in vegetative tissues of the host plant. This cycle is repeated many times, often causing epidemics in the wheat crop (Eugenea et al., 2009; Kolmer, 2005). This complex life cycle, especially the sexual stage, has contributed to the high degree of virulence diversity in Pst, which can enhance its pathogenicity, increase its host range and generate new physiological races through sexual hybridization (Wang and Chen, 2013; Zheng et al., 2013). Therefore, sexual reproduction plays a critical role in Pst, as well as virulent urediniospores.
Previous studies have shown that many genes perform biochemical or physiological functions involved in sexual and asexual reproduction, including transcription factors (TFs), which interact with different DNA‐binding domains (Min et al., 2012; Son et al., 2011) and key conserved components of signal transduction pathways (Johnson, 1995; Urban et al., 2003; Yang et al., 2015; Zhou et al., 2011). Mitogen‐activated protein (MAP) kinase cascades, as essential signalling pathways, have attracted considerable research attention because of their broadly conserved roles in a variety of fungi, plants and mammals (Hamel and Ellis, 2012). In Saccharomyces cerevisiae, the filamentation invasion pathway and mating pathway are controlled by FUS3 and KSS1, which are MAP kinases (Madhani and Fink, 1997). Activation by mating pheromones occurs only in cells containing a STE12/DIG1/DIG2 complex. The FUS3‐ or KSS1‐mediated phosphorylation of STE12 relieves their inhibitory activity through two elementary proteins, DIG1 and DIG2 (Chou et al., 2006; Schamber et al., 2010). In contrast, the TEC1/STE12/DIG1 complex, containing the TF TEC1, is known to be involved in the activation of genes that determine filamentation formation (Chou et al., 2006). Although the activation of the two pathways depends on different activation mechanisms and components, they share the downstream TF STE12 as a central node to respond to mating as well as invasive growth (Madhani and Fink, 1997).
STE12‐like TFs showing similarities to yeast STE12 have been identified in many true filamentous fungi and play essential roles in sexual development and pathogenicity (Hoi and Dumas, 2010). These proteins contain two C‐terminus‐located, tightly linked, C2H2 zinc fingers, which are absent in yeast STE12. Currently, the role of the C2H2 domain is still unclear. Deletion of this domain does not impair DNA binding in vitro (Chang et al., 2004; Hoi et al., 2007), but, in vivo, the integrity of this domain is required for the function of the protein in Colletotrichum lindemuthianum and Magnaporthe oryzae (Hoi et al., 2007; Park et al., 2004). The saprophytic Neurospora crassa STE12‐like pp‐1 mutant shows a severe reduction in the growth rate and production of aerial hyphae, and fails to develop protoperithecia (Li et al., 2005). A similar phenotype has been observed for the MAP kinase mak‐2 mutant, suggesting that pp‐1 in N. crassa is controlled by this MAP kinase pathway. In hemibiotrophic fungal pathogens, C. lindemuthianum CLSTE12 may be involved in the production of cell surface proteins and host cell wall‐degrading enzymes (Hoi et al., 2007). In plant pathogens, M. oryzae STE12 orthologous genes are important for the development of specialized infectious structures, including appressoria, for the penetration of leaf surfaces and subsequent invasive growth. STE12‐deficient mutants are defective in penetration and either non‐pathogenic or strongly reduced in virulence (Kramer et al., 2009; Park et al., 2002). Similar functions for STE12 have also been documented in the basidiomycete yeast Cryptococcus neoformans, which is an important human fungal pathogen (Chang et al., 2001; Yue et al., 1999). Deletion of STE12 in both MATa and MATα strains affects the mating efficiency and markedly reduces the virulence in mice. From previous studies, almost all STE12 and STE12‐like factors play key roles in the regulation of fungal development and pathogenicity (Hoi and Dumas, 2010; Rispail and Pietro, 2010); however, their roles in sexual development cannot be ascribed for all fungi, e.g. the M. oryzae mst12 mutation has no effect on sexual development (Park et al., 2002).
Although STE12 orthologues have been reported to be involved in various cellular processes in many pathogens (Hoi and Dumas, 2010; Hoi et al., 2007; Rispail and Di, 2009; Schamber et al., 2010), the role of STE12 in Pst is still unknown. In a previous study, RNA sequencing (RNA‐seq) analysis indicated that sequences with homology to the STE12‐like gene (PST79_9215) were found in both haustoria and germinated spores of Pst and the encoded proteins with functions were related to TF activity (Garnica et al., 2013). Subsequently, three pheromone receptor (STE3) mating‐type genes, together with two allelic homeodomain (HD) pairs (HD1 and HD2), were identified in each dikaryotic Puccinia species. The HD proteins were active during mating through heterologous expression in Ustilago maydis, and host‐induced gene silencing (HIGS) of the HD and STE3 genes reduced infection in wheat (Cuomo et al., 2016). In this study, we report our findings on the characterized mating‐related gene from Pst, PstSTE12, which was identified by transcriptome sequencing. To clarify whether and how PstSTE12 participates in the mating and virulence of Pst, we performed sequence analysis, subcellular localization, transcriptional activation analyses, complementation of mutants of S. cerevisiae and M. oryzae, and PstSTE12 knockdown via HIGS. Our findings suggest that PstSTE12 is an important TF responsible for Pst virulence, and may be involved in the mating process, providing insights into virulence variation in cereal rust fungi.
Results
A highly induced gene in Pst haustoria is identified as a STE12 gene
To find novel pathogenicity factors and to identify haustoria‐enriched transcripts in Pst, we performed RNA‐seq analysis with haustoria isolated from wheat Suwon 11 (Su11) leaves infected with the virulent Pst isolate CYR32. One of the transcript sets that encodes a putative fungal TF was identified in the resulting RNA‐seq data. Mapping to the CYR32 genome (Zheng et al., 2013) showed that it encodes a putative STE12 involved in the MAP kinase pathway. We designated this gene PsSTE12. PstSTE12 has an open reading frame of 2637 bp encoding a protein of 879 amino acids and containing three conserved HD kinase domains. N‐terminal residues of PstSTE12 indicate significant identity to other STE12 proteins (Fig. S1A, see Supporting Information), and this region includes three conserved HDs from residues 106–199 of PstSTE12, based on the S. cerevisiae STE12 sequence. Saccharomyces cerevisiae STE12 contains the DNA recognition domain for the binding of pheromone response elements with HD‐Helix III (Vallim et al., 2000; Yuan and Fields, 1991). As shown in Fig. S1A, this helix exhibits high conservation amongst these fungi, and PstSTE12 contains the universally conserved tryptophan (residue 95) followed by the phenylalanine of Helix III believed to be essential for DNA binding. Amino acid identity over this region is 86%, 88%, 62%, 59%, 60%, 63% and 46% for Puccinia triticina, Puccinia graminis f. sp. tritici, Fusarium graminearum, C. neoformans, N. crassa, M. oryzae and S. cerevisiae, respectively. PstSTE12 shows highest similarity to STE12 orthologues from P. graminis f. sp. tritici and P. triticina, and three proteins show high conservation with other STE12 proteins in the HDs.
STE12 proteins from different pathogens were selected to study their relationships. The maximum‐likelihood (ML) phylogenetic analysis of PstSTE12 with orthologues from other fungi revealed that PstSTE12 displays higher similarity to STE12‐like proteins from basidiomycetes, compared with those from ascomycetes (Fig. S1B). The filamentous ascomycetes containing a C2H2 domain were clustered into one clade, whereas yeasts without this domain were clustered into another clade (Fig. S1B). blast analysis showed two C2/H2‐Zn2+ finger domains in the C‐terminus of PstSTE12 (Fig. S2, see Supporting Information).
PstSTE12 is highly induced in Pst during early infection stages in wheat and in pycniospores in barberry
To address the role of PstSTE12 during the infection stages of Pst with Su11, we compared the abundance of PstSTE12 in urediniospores, germ tubes, parasitic invasion hyphae, sporulation and pycniospores in barberry (Berberis shensiana) (Fig. 1). Values are expressed relative to an endogenous Pst reference elongation factor‐1 (EF‐1) gene, with the PstSTE12 transcripts in ungerminated urediniospores set to unity as the control (Cheng et al., 2015). The results showed that, once CYR32 contacted the leaf surface of Su11, the expression of PstSTE12 transcripts increased and gradually reached a maximum: an approximately 40‐fold increase at 36 h post‐inoculation (hpi) compared with that in the control. This occurred in the initial ‘parasitic/biotrophic’ stage during successful expansion of Pst into wheat. The transcript levels of PstSTE12 in the late ‘parasitic/biotrophic’ stage at 72–168 hpi and the ‘sporulation’ stage were down‐regulated compared with that of ungerminated urediniospores. In addition, the expression of PstSTE12 was induced four‐fold compared with the control during the pycniospore stage in the aecial host barberry [11 days post‐inoculation (dpi)] (Fig. 1), which is an alternative host for Pst (Jin et al., 2010; Zhao et al., 2016). The results suggest that, in addition to a role in sexual reproduction, PstSTE12 also participates in virulence.
Figure 1.
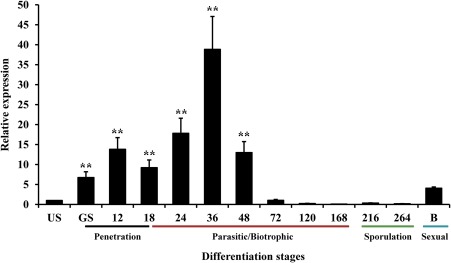
Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) assay of transcript profiles of PstSTE12 during Puccinia striiformis f. sp. tritici (Pst) infection stages. The relative transcript levels of PstSTE12 were calculated by the comparative Ct method compared with an endogenous standard elongation factor‐1 (EF‐1) gene. Pst infection of wheat can be divided into three major stages: ‘penetration’ (black), ‘parasitic/biotrophic’ (red) and ‘sporulation’ (green). Pycniospores in the alternative host berberis (Berberis shensiana) are marked in blue. US, urediniospores; GS, germinated urediniospores; 12–264 h, 12–264 h post‐inoculation with CYR32; B, infected Berberis shensiana. Data represent the mean of three biological replicates ± standard error (SE). Differences were assessed using Student's t‐test. Double asterisks indicate P < 0.01.
PstSTE12 shows transcriptional activation in yeast
To examine the transcriptional activity of PstSTE12, the fusion plasmids pBD‐PstSTE12 1–879, pBD‐PstSTE12 1–400 and the negative control pGBKT7 were separately transformed into yeast strain AH109 (Fig. 2). On selective dropout/–tryptophan (SD/–Trp) medium, clones of the three vectors transformed separately grew well, indicating the success of merging with the yeast strain (Fig. 2B). In contrast, only yeast cells containing the pBD‐PstSTE12 1–879 vector grew on the selective dropout/–tryptophan–histidine–adenine (SD/–Trp–His–Ade) medium (Fig. 2C), whereas yeast cells with the pBD‐PstSTE12 1–400 or negative control pGBKT7 vector did not survive. In the assay for the evaluation of α‐galactosidase activity, transformants containing the pBD‐PstSTE12 1–879 vector turned blue on SD/–Trp–His–Ade medium containing 5‐bromo‐4‐chloro‐3‐indolyl α‐d‐galactopyranoside (X‐α‐Gal) (Fig. 2C), suggesting that PstSTE12 effectively activated the transcription of His3 together with LacZ reporter genes. However, the C‐terminal deletion mutant of PstSTE12 lost the ability to activate the transcription of the reporter genes.
Figure 2.
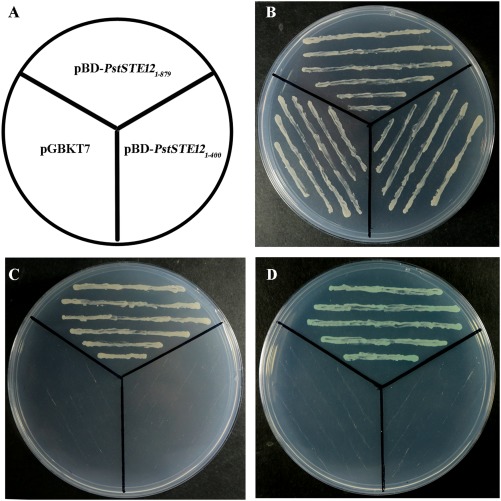
Transactivation analysis of the PstSTE12 protein in yeast. The fusion and control vectors were separately transformed into the yeast strain AH109. (A) The diagram indicates the corresponding vector for the assay. (B) The transformants were selected by growth on selective dropout/–tryptophan (SD/–Trp) medium at 30 °C for 3 days. (C) Transformants were streaked onto selective dropout/–tryptophan–histidine–adenine (SD/–Trp–His–Ade) medium. (D) The α‐galactosidase assay was conducted on SD/–Trp–His–Ade medium containing 5‐bromo‐4‐chloro‐3‐indolyl α‐d‐galactopyranoside (X‐α‐Gal).
PstSTE12 localizes to the nucleus of plant cells
To determine the subcellular localization of PstSTE12, we generated the fusion constructs pCaMV35S:PstSTE12‐GFP and pCAMBIA‐1302:PstSTE12‐GFP, which were transformed into Triticum aestivum protoplasts with polyethyleneglycol (PEG)‐calcium (Fig. 3A) or into Nicotiana benthamiana leaf cells by infiltration into Agrobacterium tumefaciens (Fig. 3B), respectively. When a PstSTE12‐green fluorescent protein (GFP) fusion protein was transiently expressed in T. aestivum protoplasts with the pCaMV35S:PstSTE12‐GFP construct, fluorescence was restricted to the nucleus, whereas GFP was distributed in the cytoplasm, cytomembrane and the nucleus of protoplasts in controls (Fig. 3A). When we transiently expressed the PstSTE12‐GFP fusion protein in N. benthamiana leaf cells, we found that the GFP fusion protein again only localized to the nucleus (Fig. 3B), whereas GFP was expressed throughout the cells in controls. These data indicate that PstSTE12 is constitutively expressed and localized to the nucleus.
Figure 3.
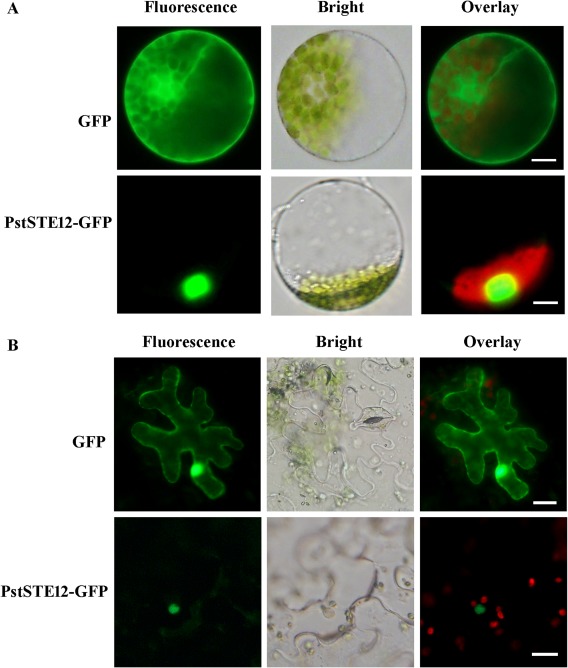
Subcellular localization of the PstSTE12 protein. All images were observed using a fluorescence microscope. (A) Green fluorescent protein (GFP) (control) and PstSTE12‐GFP fusion protein were expressed in wheat protoplasts following polyethyleneglycol (PEG)‐mediated transformation. Bar, 10 μm. (B) GFP and PstSTE12‐GFP fusion protein were expressed in Nicotiana benthamiana by transient agro‐infiltration assays. Bar, 20 μm. The fluorescence channel shows the localization of GFP and PstSTE12‐GFP (green). The bright channel shows the integrity of cells or protoplasts. An overlay of micrographs shows GFP and PstSTE12‐GFP (green) together with the chloroplasts (red).
Complementation of the S. cerevisiae ste12 (α) mutant with PstSTE12 restores mating efficiency
To verify whether PstSTE12 might possess a function similar to the yeast STE12 gene, we introduced PstSTE12 into a ste12 (α) mutant to test for complementation and a mutant rescue phenotype (Fig. S3, see Supporting Information). For the STE12 reporter, the mating efficiency was drastically reduced to approximately 0% in the ste12 mutants, whereas the mating efficiency in the wild‐type was 100% (Fig. 4A,B) (Hartwell, 1980; Yuan et al., 1993). Mating was assessed by quantitative assays. In accord with previous studies, our data showed that mating of the empty vector transformants occurred at an extremely low frequency (nearly 0%), consistent with the mutant type (Fig. 4). Meanwhile, the complemented strain, formed when the PstSTE12 gene was introduced into the ste12 mutant, mated with the wild‐type MATa, led to a recovery of 34% mated cells. Percentage data were calculated by dividing the mating efficiency of each construct by the mating efficiency of the wild‐type. Then, the ascospores were germinated in MacConkey medium and yeast cells were stained with phenol and methylene blue after growth for 5–6 days. Microscopic examination revealed blue conidia and red ascospores. The results indicated that both the yeast mutant ste12 (α) and the mutant carrying the empty vector failed to mate with MATa wild‐type cells (Fig. 4C). However, the wild‐type strains of yeast a/α and transformants of ste12/PstSTE12(α) mating with MATa wild‐type cells produced not only conidia, but also ascospores (Fig. 4C), confirming the successful mating with MATa. These results show that PstSTE12 can partially complement the ste12 mutation and suggest that it may be involved in the mating pathway in Pst.
Figure 4.
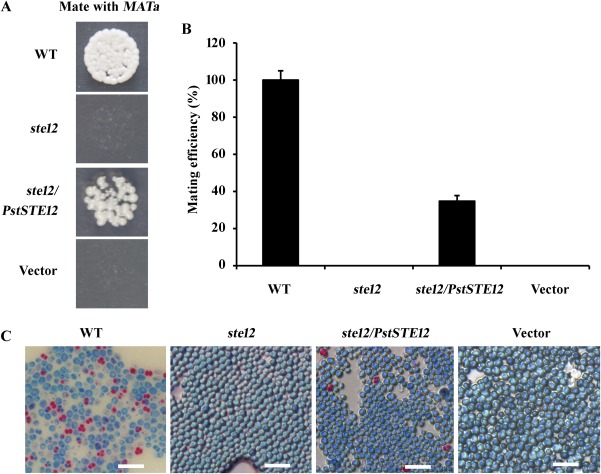
Functional characterization of the PstSTE12 transcription factor in the Saccharomyces cerevisiae ste12 mutant. (A) Patch mating assays for the ability of the wild‐type, mutant and transformant strains mated with the wild‐type MATa cells. PstSTE12 and empty vector were transformed into S. cerevisiae ste12 mutants. The strains were transferred onto a minimal plate spread with cells of the opposite mating type. (B) Quantitative assay of the mating efficiency. The mating efficiency of constructs was calculated as a percentage by dividing with the mating efficiency of wild‐type a/α. (C) Conidium and ascospore are labelled with blue and red, respectively. From left to right: the wild‐type strain of yeast MATa/MATα; ste12 (α) mutant mating with MATa wild‐type cells; transformants of ste12/PstSTE12 mating with MATa wild‐type; transformants carrying the empty vector mating with MATa wild‐type strains. Bar, 10 μm.
PstSTE12 partially complements the function of mst12 mutants in M. oryzae
The TF MST12 is required for infectious growth, but not appressorium formation, in M. oryzae (Park et al., 2002). To test whether PstSTE12 can functionally complement the mst12 mutant in M. oryzae, we transformed the PstSTE12 gene under the control of the strong constitutive RP27 promoter into the mst12 mutant (Park et al., 2002). Geneticin‐resistant transformants were isolated and verified by PCR to confirm the introduction of PstSTE12. Mature appressoria formed by the mst12 mutant were normal in size with typical morphology and melanization. Thus, appressorium formation was not significantly different between the transformants and wild‐type (Park et al., 2002; Zhou et al., 2011) (Fig. 5A). However, unlike appressoria formation, the mst12 mutant was defective in infection of barley and rice (Zhou et al., 2011). To determine whether PstSTE12 could recover the virulence of the mst12 mutant, 8‐day‐old barley seedlings of cultivar NB6 were drop inoculated with conidia. At 6 dpi, leaves inoculated with the wild‐type developed typical blast lesions (Fig. 5B). No lesions were observed on leaves inoculated with gelatin and the mst12 mutant also failed to cause lesions on barley leaves (Fig. 5B). Under the same conditions, transformants caused obvious lesions on barley leaves. These results indicate that PstSTE12 can partially complement the function of mst12 in plant infection.
Figure 5.
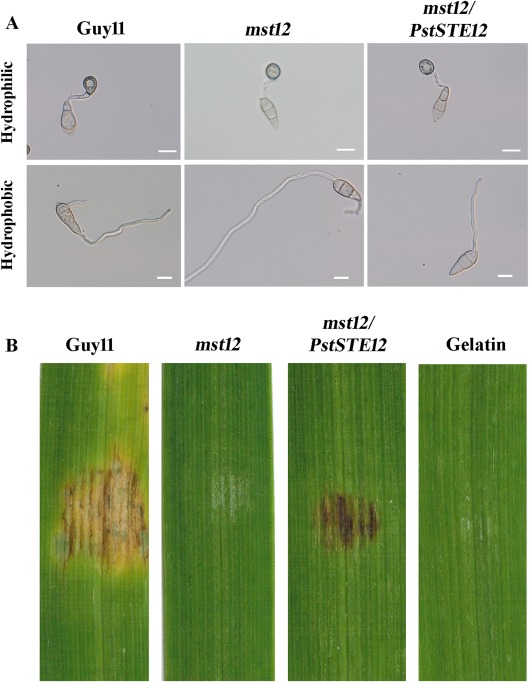
Complementation of the mst12 mutant with the pFL2‐PstSTE12 fusion construct. (A) Appressorium formation assays on hydrophobic and hydrophilic surfaces. Appressoria were formed by the wild‐type Guy11 and the mst12 mutant transformants on the hydrophobic (bottom) and hydrophilic (top) surfaces of Gelbond membranes at 24 h. Bar, 10 μm. (B) Barley infection assay. From left to right: barley leaves inoculated with conidia of Guy11, mst12 mutant, transformants and 0.25% gelatin. Typical phenotypes were photographed at 6 days post‐inoculation (dpi).
Silencing of PstSTE12 reduces virulence of Pst on wheat
To further characterize the function of PstSTE12 during the interaction between wheat and Pst, Barley stripe mosaic virus (BSMV) virus‐induced gene silencing (VIGS) was used in this study (Cheng et al., 2015; Yin et al., 2011). Two different ∼250‐bp fragments (BSMV:PstSTE12‐H and BSMV:PstSTE12‐B) were designed to specifically silence PstSTE12 (Fig. S4, Table S1, see Supporting Information). The assay was performed in wheat cv. Su11, which is susceptible to Pst isolate CYR32. All of the plants infected with virus, BSMV:γ, BSMV:PstSTE12‐H or BSMV:PstSTE12‐B, displayed mild chlorotic mosaic symptoms at 10 dpi, whereas mock‐inoculated plants were green and healthy (Fig. 6A). Subsequently, the fourth leaves were inoculated with urediniospores of Pst CYR32 on mock‐ and virus‐inoculated plants. After 14 days, very few uredinia were produced on leaves previously infected with BSMV:PstSTE12‐H or BSMV:PstSTE12‐B (Fig. 6B,C). In contrast, mock‐inoculated leaves and leaves infected with BSMV:γ inoculated with CYR32 produced numerous uredinia with abundant urediniospores (Fig. 6B,C). To determine the efficiency of knockdown by HIGS, transcript levels of PstSTE12 were detected at 48 and 120 hpi with the CY32 isolate. Compared with transcription levels of PstSTE12 in leaves inoculated with BSMV:γ, the expression of PstSTE12 was knocked down by 72% and 49% in leaves inoculated with BSMV:PstSTE12‐H at 48 and 120 hpi with the CYR32 isolate, respectively (Fig. 6D). Similarly, in leaves inoculated with BSMV:PstSTE12‐B, transcripts of PstSTE12 were reduced by 74% and 54% at 48 and 120 hpi with the CYR32 isolate, respectively (Fig. 6D). These findings indicate that, overall, the level of transcription of PstSTE12 is reduced in the invading fungus, and that this silencing probably influences the expression of PstSTE12 in Pst.
Figure 6.
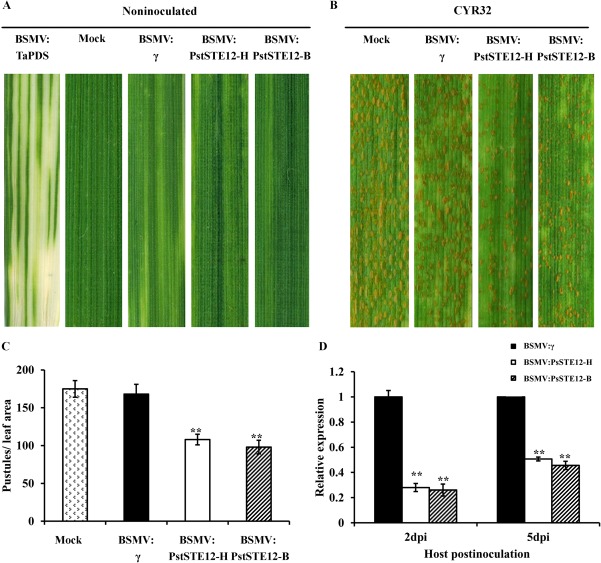
Silencing of PstSTE12 in the wheat–Puccinia striiformis f. sp. tritici (Pst) interaction using host‐induced gene silencing (HIGS) leads to reduced virulence. (A) No change in phenotype was observed in wheat leaves mock inoculated with FES buffer. Mild chlorotic mosaic symptoms were observed in wheat leaves inoculated with BSMV:γ, BSMV:PstSTE12‐H and BSMV:PstSTE12‐B. Photobleaching was evident on wheat leaves infected with BSMV:TaPDS. (B) Phenotypes of the fourth leaves of wheat plants inoculated with mock (FES buffer), BSMV:γ, BSMV:PstSTE12‐H and BSMV:PstSTE12‐B at 14 days post‐inoculation (dpi) with the virulent Pst isolate CYR32. (C) Quantification of uredinial density in silenced plants at 14 dpi with the CYR32 isolate. Values represent mean ± standard errors of three independent assays. Differences were assessed using Student's t‐tests. Double asterisks indicate P < 0.01. (D) Relative levels of PstSTE12 transcript abundance in knock‐down wheat leaves. RNA samples were isolated from the fourth leaves of wheat infected with BSMV:γ, BSMV:PstSTE12‐H and BSMV:PstSTE12‐B at 48 and 120 h post‐inoculation (hpi) with the CYR32 isolate. Values are expressed relative to an endogenous Pst reference elongation factor‐1 (EF‐1) gene, with the empty vector (BSMV:γ) set at unity. Values represent mean ± standard errors (error bars) of three independent samples. Differences were assessed using Student's t‐tests. Double asterisks indicate P < 0.01.
HIGS of PstSTE12 compromises fungal growth in the interaction between Pst and wheat
To determine how PstSTE12 is involved in Pst pathogenicity, we observed the cytological changes in PstSTE12‐silenced plants inoculated with Pst CYR32 at 48 hpi (Fig. 7A–C) and 120 hpi (Fig. 7D–F). The number of haustorial mother cells, as well as the number of hyphal branches, hyphal length and hyphal areas, were counted, as indicators of fungal penetration and expansion abilities, respectively (Fu et al., 2014; Tang et al., 2015). The numbers of haustorial mother cells and hyphal branches were not significantly different from controls at 48 hpi (Fig. 7G). However, the numbers of haustoria were significantly reduced in PstSTE12‐silenced plants (Fig. 7G). Moreover, hyphal lengths in PstSTE12‐silenced plants were shorter than those observed in BSMV‐γ‐treated leaves at 48 h (P < 0.01) (Fig. 7H). Consistent with this, hyphal expansion areas of Pst were strictly limited in PstSTE12‐silenced plants at 120 hpi. In contrast, control plants treated with BSMV:γ showed extensive colonization and the formation of secondary hyphae (Fig. 7D–F,I). These results indicate that PstSTE12 most probably contributes to fungal growth by participating in haustorium formation and hyphal development. Thus, PstSTE12 may also be involved in the invasion and pathogenicity of Pst.
Figure 7.
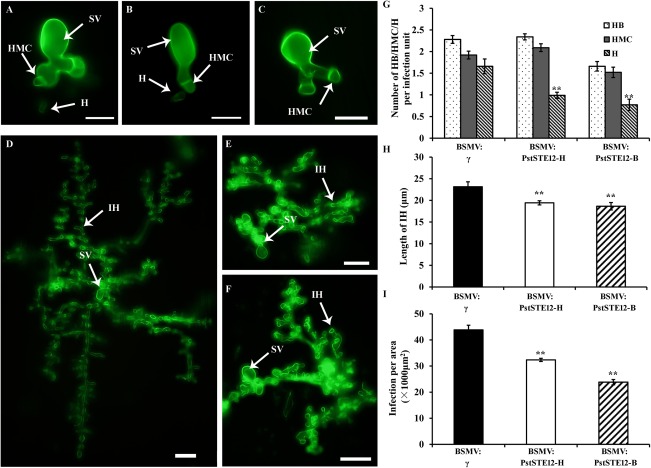
Histological observation of fungal growth in wheat infected with BSMV:γ and recombinant Barley stripe mosaic virus (BSMV) after inoculation with CYR32. (A–C) Fungal growth at 24 h post‐inoculation (hpi) in BSMV:γ‐infected plants (A), BSMV:PstSTE12‐infected plants (B) and BSMV:PstSTE12‐B‐infected plants (C). Bar, 20 μm. (D–F) Fungal growth at 120 hpi in BSMV:γ‐infected plants (D), BSMV:PstSTE12‐H‐infected plants (E) and BSMV:PstSTE12‐B‐infected plants (F). Bar, 50 μm. (G) The numbers of branches of infection hyphae (HB), haustorial mother cells (HMC) and haustoria (H) per infection unit were recorded at 48 hpi. (H) The length of infection hyphae (IH) in PstSTE12‐silenced plants at 48 hpi with the CYR32 isolate. The length of IH was measured from the substomatal vesicle to the apex of the longest infection hypha. (I) The infection unit area per infection unit was determined at 120 hpi. Values represent the means ± standard errors of three independent samples. Differences were assessed using Student's t‐tests. Asterisks indicate P < 0.05. Double asterisks indicate P < 0.01. SV, substomatal vesicle.
Discussion
The frequency of variation in cereal rust fungi often results in a rapid and sudden loss of resistance to Pst in cultivated wheat. Sexual reproduction is the primary process contributing to the origin of new races with combined virulence genes and diversification of stripe rust pathogens (Zhao et al., 2016). Although some virulence‐related genes have been described in Pst (Cheng et al., 2015; Liu et al., 2016; Tang et al., 2015), fewer studies have documented the genes that are involved in mating in Pst. A previous study has reported that the orthologue STE12, together with other components, contributes to the regulation of fertility in sexual crosses in heterothallic fungi, such as S. cerevisiae (Fields and Herskowitz, 1985; Hartwell, 1980; Rispail and Pietro, 2010). In this study, we identified and characterized PstSTE12, which shares structural features with STE12 orthologues in other fungi (Hoi and Dumas, 2010; Rispail and Pietro, 2010). The structural conservation of MAP kinase cascades between Pst and other fungal pathogens suggests a possible role of STE12 as a downstream target in different signalling pathways. In support of this hypothesis, we found that the expression of PstSTE12 restored the fertility in a ste12 mutant. Given the functional conservation properties of genes between different species, the recovery of the mating efficiency of the ste12 mutant complemented with PstSTE12 suggests that the function of PstSTE12 is conserved with yeast STE12. Possibly, PstSTE12 plays an important role in the mating pathway in Pst. Ustilago maydis is the most advanced model plant pathogen in basidiomycetes and lacks a STE12 orthologue (Brefort et al., 2005). A STE12 orthologue has been well characterized in the human pathogen model basidiomycete C. neoformans. In C. neoformans, CnSTE12α and CnSTE12a are required for haploid fruiting, and deletion of CnSTE12α or CnSTE12a reduces the mating frequency and virulence in mice (Chang et al., 2000, 2001). In this regard, it is of interest to note that we found that PstSTE12 was clearly expressed during the pycniospore stage in the aecial host barberry, a crucial stage of the reproduction process. The role of sexual reproduction in generating variation in virulence in Pst was not confirmed until the alternative host Berberis was discovered (Jin et al., 2010; Wang and Chen, 2013; Zhao et al., 2013). On alternative hosts, receptive hyphae are fertilized by pycniospores from other pycnia of the compatible mating type to produce dikaryotic mycelia, and then produce aecia containing one‐celled aeciospores with two nuclei (n + n) (Zhao et al., 2016). Thus, PstSTE12 may take part in the mating pathway by keeping the nuclei in communication during the dikaryotic stage, and contribute to virulence variation by generating new races through sexual reproduction.
The STE12 gene in S. cerevisiae encodes a protein that binds to the pheromone‐responsive element sequence in the nucleus, which is present in many different a‐ and α‐specific upstream regulatory sequences (Blackwell et al., 2007). As a TF, the PstSTE12 protein was predicted to contain a nuclear localization signal, and we confirmed that PstSTE12 was localized to the nucleus in heterologous (plant) systems. Cell surface receptors respond to the extracellular stimuli that induce a cascade of transduction signals, resulting in phosphorylation of various proteins positioned downstream in a variety of pathways, including the STE12 protein (Hamel and Ellis, 2012; Meng and Zhang, 2013; Rispail and Pietro, 2010). This rapid phosphorylation is associated with an increase in the ability of STE12 to stimulate transcription in the nucleus. We inferred that PstSTE12 may perform similar functions in Pst. The yeast one‐hybrid assay further showed that PstSTE12 has transcriptional activity and that the C‐terminal region (400–879 amino acids) is necessary for transactivation. Similarly, in S. cerevisiae, the removal of large C‐terminal regions of STE12 significantly reduced basal transcription. This cooperativity of STE12 to bind a single pheromone response element, such as that present in the STE2 gene, together with the binding of MCM1 to an adjacent site, requires the carboxyl‐terminal domain of STE12 (Kirkman‐Correia et al., 1993). These data suggest that PstSTE12 is a Pst TF, which may function as a transcriptional activator in the fungal nucleus.
STE12 positioned downstream of the MAP kinase pathway in S. cerevisiae also participates in filamentous invasive growth (Madhani and Fink, 1997; Rispail and Pietro, 2010). In addition, various orthologous STE12‐like proteins of pathogenic fungi, such as in M. oryzae, have been reported to regulate infection processes (Park et al., 2002; Zhou et al., 2011) and F. graminearum (Yang et al., 2015). In M. oryzae, germination and appressorium formation of the mst12 mutant were normal, but, in infection assays, the mst12 mutants appeared to be non‐pathogenic (Park et al., 2002). Our study showed that the expression of the PstSTE12 gene can partially restore function in the M. oryzae mst12 mutant. The PstSTE12‐transformed mutants showed restored function in appressorium penetration and invasive growth, suggesting that PstSTE12 plays a potential role in virulence in Pst. However, the phenotypes of the PstSTE12‐transformed mutants could not be entirely rescued, indicating that PstSTE12 is not fully functional in this ascomycete fungus, and also that there is specificity between different fungi. Pst, as an obligate parasitic pathogen, has a special and complex life style (Zheng et al., 2013), which is substantially different from that of M. oryzae. During evolution, sequence and structural variation in PstSTE12 may have enabled it to distinguish various environmental stimuli. This may reduce the functional efficiency of PstSTE12 in signal transduction in ascomycetes and account for the partial recovery of the phenotype. We also showed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) that the PstSTE12 gene is highly induced during Pst infection and functions as an important pathogenicity factor. PstSTE12 was clearly expressed during fungal colonization, haustorium formation, primary hyphal extension and sexual reproduction. A role in virulence was supported by further evidence via observations in HIGS experiments. Just as deletion of STE12 leads to defects in penetration and invasive growth in other fungi (Rispail and Di, 2009; Tsuji et al., 2003), silencing of PstSTE12 strongly reduces Pst virulence (Fig. 7). Silencing of PstSTE12 restricts Pst haustorium formation and development at 48 hpi, whereas the extension area of secondary hyphae and fungal colonization are significantly reduced at 120 hpi with the CYR32 isolate, suggesting that silencing of PstSTE12 at the early stages of infection suppresses further development at later stages. The growth of Pst was significantly limited, as indicated by the generation of a reduced number of uredinia on wheat leaves after 14 days. Our data suggest that PstSTE12 plays an important role in plant infection by regulating haustorium formation and fungal colonization. Collectively, our results, together with previous studies, provide strong circumstantial evidence suggesting that PstSTE12 plays a key role in the regulation of the response of infection signals lying downstream of a signal pathway network in Pst, and may be involved in the mating process. Therefore, future identification and characterization of rust fungi‐specific targets of PstSTE12 should result in a more complete understanding of the evolution of pathogenesis and virulence in Pst.
Experimental Procedures
Plant materials, strains and treatments
The wheat (T. aestivum) cultivar Suwon11 (Su11), which is highly susceptible to Pst isolate CYR32, was used in this study. Wheat was cultivated and inoculated by the procedures and conditions described previously (Kang et al., 2002). To investigate the PstSTE12 transcript profiles, samples were collected at 12, 18, 24, 36, 48, 72, 120, 168, 216 and 264 hpi during the CYR32 interaction with Su11, and in barberry (Berberis shensiana) leaves inoculated with CYR32 basidiospores at 11 dpi, which is the pycniospore stage in the aecial host barberry. After incubation at 4 °C for 6 h, germinated urediniospores were collected and frozen in liquid nitrogen for RNA extraction. The developmental stages of the Pst infection process analysed by qRT‐PCR were the same as those described previously (Cheng et al., 2015). Disease symptoms were photographed at 15 dpi. Three independent biological replicates were performed for each treatment.
RNA isolation and qRT‐PCR analysis
Total RNA was isolated using RNAiso Reagent (TaKaRa, Tokyo, Japan) according to the manufacturer's instructions. First‐strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) with an oligo(dT)18 primer as instructed. qRT‐PCR was performed on a CFX Connect Real‐Time System (Bio‐Rad, Hercules, CA, USA). EF‐1 was used as internal control to normalize the gene expression in Pst (Liu et al., 2016). Reactions were performed on a 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) under the conditions described by Cheng et al. (2015). The relative transcript expression was assessed by the 2–ΔΔ Ct method with three biological replicates (Livak and Schmittgen, 2001).
Isolation, cloning and sequence analysis of PstSTE12
PCR amplification of the PstSTE12 gene was performed using a CYR32‐inoculated Su11 cDNA sample as a template. The PCR products were purified and cloned into the pGEM‐T vector (Promega, Madison, Wisconsin, USA) and sequenced. Target P (http://www.cbs.dtu.dk/services/TargetP/) was used to predict the subcellular localization of PstSTE12. Multiple sequence alignments between homologous proteins from other fungi were compared using ClustalW (version 1.8) software and added shade by Boxshade online (http://www.ch.embnet.org/software/BOX_form.html). mega5 was used for phylogenetic analysis by the ML method. Some of the STE12 orthologues were obtained from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/), including Aspergillus nidulans (XP_659894.1), Aspergillus oryzae (KDE77588.1), Talaromyces marneffei (ABH09729.1), Setosphaeria turcica (AFS49719.1), Marssonina brunnea (EKD19163.1), Botrytis cinerea (ACJ06644.1), Sclerotinia borealis (ESZ98034.1), Colletotrichum lagenaria (BAC11803.1), C. lindemuthianum (CAD30840.2), Cryphonectria parasitica (ABE67104.1), M. oryzae (AAL27626.2), Trichoderma reesei (EGR50918.1), F. graminearum (XP_387486.1), Fusarium oxysporum (ACM80357.1), N. crassa (AAK14814.1), S. cerevisiae (KZV10819.1), Kluyveromyces lactis (AAA35270.1), P. triticina (PTTG_05606T0), P. graminis f. sp. tritici (EFP80971.2), Peronosclerospora sorghi (KNZ61697.1), Melampsora larici‐populina (EGF98701.1), Mixia osmundae (XP_014567023.1), Trichosporon asahii (EKC98332.1), Xanthophyllomyces dendrorhous (CED85281.1), Cryptococcus gattii (AAS92520.1), C. neoformans (AAN75715.1), Candida albicans (AAA64692.1) and Clavispora lusitaniae (AAD51741.1). The others were obtained from Ensembl Fungi (http://fungi.ensembl.org/index.html), including Trichosporon oleaginosus (KLT46070), Pisolithus microcarpus (KIK17142), Scleroderma citrinum (KIM53342), Hypholoma sublateritium (KJA26649), Laccaria amethystina (KIJ98362) and Laccaria bicolor (EDR03059).
Subcellular localization analysis
A pCAMBIA1302:PstSTE12‐GFP fusion was transformed into the A. tumefaciens strain GV3101 by electroporation. Individual colonies surviving after selection with kanamycin (50 μg/mL), rifampicin (30 μg/mL) and chloramphenicol (30 μg/mL) were verified by PCR with vector primers. The procedure for the infiltration of leaves was the same as that described previously (Cheng et al., 2015). Infiltrated plants were grown at 25 °C with a cycle of 16 h light and 8 h darkness. Tissue samples were collected after infiltration for 2 or 3 days and observed under an Olympus BX‐51 microscope (Olympus Corporation, Tokyo, Japan).
Triticum aestivum Su11 plants for protoplast transformation were grown in the glasshouse for 2–3 weeks. The plasmid pCaMV35S:PstSTE12‐GFP and the control plasmid pCaMV35S:GFP were separately transformed into T. aestivum protoplasts through PEG‐calcium transfection (Ito and Shinozaki, 2002; Li et al., 2015). After culture for 18–24 h, the GFP signals in transformed protoplasts were observed with a fluorescence microscope (Olympus BX‐51). All experiments were repeated three times, with each assay performed on at least three plants.
Transcriptional activation analysis in yeast
According to the manufacturer's instructions (Clontech, Tokyo, Japan), the reporter plasmids pBD‐PstSTE12 1–879 and pBD‐PstSTE12 1–400 and the negative control plasmid pGBKT7 were transformed into the yeast strain AH109 (Clontech), individually. The transformed yeast strains were cultured on SD/–Trp and SD/–Trp–His–Ade media at 30 °C, and subsequently photographed after 3 days. The α‐galactosidase activity assay was evaluated with X‐α‐Gal as a substrate according to the manufacturer's instructions.
Complementation of S. cerevisiae ste12 (α) mutants with PstSTE12
The yeast S. cerevisiae diploid mutant strains ste12 (BY4743; MATa/MATα; ura3Δ0/ura3Δ0; leu2Δ0/leu2Δ0; his3Δ1/his3Δ1; met15Δ0/MET15; LYS2/lys2Δ0; YHR084w/YHR084w::kanMX4) and wild‐type BY4741 (MATa; his3D1; leu2D0; met15D0; ura3D0) and BY4742 (MATα; his3D1; leu2D0; lys2D0; ura3D0) were purchased from the (EUROSCARF collection, Frankfurt, Germany). Tetrads of spores were arrested after plating on MacConkey medium at 30 °C for 5–7 days. The haploid mutant spores were identified as ste12 (MATα; ura3Δ0; leu2Δ0; his3Δ1; MET15; lys2Δ0; YHR084w::kanMX4) (data not shown), and separated haploid spores were transformed with pDR195‐PstSTE12 vectors by a lithium acetate method, as directed in the user's manual (Clontech). Quantitative mating assays were performed as described previously (Passmore et al., 1989; Sprague, 1991). Briefly, cultures with 2 × 106 cells of the transformants and 107 cells of the appropriate tester strain were mixed. The cells were plated on 1.0% yeast extract, 2.0% peptone, 2.0% glucose, 2.0% agar (YPD) plates and allowed to grow for 24 h at 30 °C, and then replica‐plated onto selective media. The number of mated cells was determined by their growth on selective media. The efficiency of mating was calculated from the number of cells that mated divided by the total number of cells of the transformants in the mating reaction. The mating efficiencies of wild‐type a and α strains were normalized to 100%, and the mating efficiencies of the other strains were expressed as a percentage of those of the wild‐type strains. The values are the means of two independent experiments. The staining assay of ascospores was performed on cultures grown for 5–6 days on MacConkey medium, and the cells were fixed with phenol and methylene blue by the method described previously (Lanchun et al., 2003).
Complementation of mutant mst12 by PstSTE12 in M. oryzae
For construction of the complementary expression vector in M. oryzae, the PCR products of the PstSTE12 gene amplified with primers STE12‐CF/CR (Table S1) were co‐transformed with XhoI‐digested pFL2 vector into S. cerevisiae strain XK1–25 (Bruno et al., 2004). The PstSTE12 gene was under the control of the strong constitutive RP27 promoter (derived from the M. oryzae ribosomal protein 27 gene). Then, the pFL2‐PstSTE12 construct was transformed into protoplasts of the M. oryzae ste12 deletion mutant. Geneticin‐resistant transformants were isolated and verified by PCR with primers PstSTE12‐CF/CR to confirm that the PstSTE12 gene had been integrated into the M. oryzae genome. Appressorium formation was assayed as described previously (Zhou et al., 2011). For plant infection investigation, conidia were resuspended to 105 conidia/mL in sterile distilled water. Eight‐day‐old barley seedlings of cultivar NB6 were used for infection assays (Zhou et al., 2011). Lesion formation was examined at 6 dpi.
Silencing by HIGS
Two cDNA fragments derived from different sites were used to silence PstSTE12 (Fig. S4, Table S1). Fragments contained no sequence similarity with known wheat genes by blast analysis of the public database from NCBI, indicating the specificity of the fragments. Capped in vitro transcripts were prepared from linearized plasmids containing the tripartite BSMV genome (Petty et al., 1990) using the RiboMAX™ Large‐Scale RNA Production System‐T7 and the Ribom7G Cap Analog (Promega), according to the manufacturer's instructions. The surface of the second leaf of two‐leaf wheat seedlings was inoculated with BSMV transcripts by mechanical rubbing, and followed by incubation at 25 °C. BSMV:TaPDS (TaPDS, the wheat phytoene desaturase) was used as a positive control (Holzberg et al., 2002), and plants inoculated with 1 × 77 mM glycine, 60 mM K2HPO4, 22 mM Na4P2O7 · 10H2O, 1% [wt/vol] bentonite, and 1% [wt/vol] celite (FES buffer) were used as a negative control (Mock). Ten days after virus inoculation, phenotypes were observed and photographed. The fourth leaves were further inoculated with CYR32 urediniospores and sampled at 48 and 120 hpi for RNA isolation and cytological observation. Phenotypes were recorded and photographed at 14 dpi with CYR32. The experiment was repeated at least three times.
Histological observations of fungal growth
Wheat leaves infected with BSMV were sampled at 48 and 120 hpi with Pst, and stained as described previously (Wang et al., 2007). Leaf segments excised from the inoculated leaves were fixed and decolorized in ethanol–trichloromethane (3 : 1, v/v) containing 0.15% (w/v) trichloroacetic acid for 3–5 days. The specimens were cleared in saturated chloral hydrate until the leaf tissue became translucent. Wheatgerm agglutinin conjugated to Alexa‐488 (Invitrogen, Carlsbad, CA, USA) was used to stain samples, as described previously (Ayliffe et al., 2011; Cheng et al., 2015). For each wheat leaf sample, 30–50 infection sites from three leaves were examined to record the number of haustorial mother cells and haustoria, hyphal branches and hyphal length, and the hyphal infection area. All microscopic examinations were performed with an Olympus BX‐51 microscope.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Multi‐sequence alignment and phylogenetic analysis of PstSTE12 and other orthologues of STE12 in other species. (A) Alignment showing the identity of STE12 homeodomains of Puccinia striiformis f. sp. tritici (Pst). Bold overlines mark the three helices of the homeodomain. Solid underline indicates the conserved tryptophan and phenylalanine residues of Helix III known to be essential for DNA binding. (B) Phylogeny of fungal STE12 orthologues. The phylogenetic tree was constructed using the maximum‐likelihood (ML) approach. The confidence level for the groupings was estimated using 500 bootstrap replicates. The numbers adjacent to the branch points indicate the percentage of replicates supporting each branch. Scale bars correspond to 0.1 amino acid substitutions.
Fig. S2 Two C2/H2‐Zn2+ finger domains of the C‐terminal region were predicted in PstSTE12 via the National Center for Biotechnology Information (NCBI).
Fig. S3 Tetrads were separated from the ascospores formed by the diploid mutant after cultivation in MacConkey medium. Four viable spores (A–D) were obtained from each of the tetrads (lanes 1, 3, 6–10 and 12) with some exceptions of two‐ or three‐spore tetrads as a result of random spore inviability (lanes 2, 4, 5 and 11).
Fig. S4 Sequence regions for the host‐induced gene silencing (HIGS) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) experiments in this study. Black arrows indicate the start codon, HIGS targeted sites, qRT‐PCR site and stop codon. The numbers in parentheses indicate the distance to the start codon (ATG). The sequences of the two domains are labelled in grey.
Table S1 Primers used for the assays in this study.
Acknowledgements
This study was supported by funds from the National Basic Research Program of China (No. 2013CB127700), the National Natural Science Foundation of China (No. 31371889) and the 111 Project from the Ministry of Education of China (No. B07049). The authors thank Professor Jinqiu Zhou (Institute of Biochemistry and Cell Biology, the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) for providing technical support on the tetrad separation of yeast, and Professor Larry Dunkle (USDA‐Agricultural Research Service at Purdue University, West Lafayette, IN, USA) for critical reading of the manuscript.
Contributor Information
Jun Guo, Email: guojunwgq@nwsuaf.edu.cn.
Zhensheng Kang, Email: kangzs@nwsuaf.edu.cn.
References
- Ayliffe, M. , Devilla, R. , Mago, R. , White, R. , Talbot, M. , Pryor, A. and Leung, H. (2011) Nonhost resistance of rice to rust pathogens. Mol. Plant–Microbe Interact. 24, 1143–1155. [DOI] [PubMed] [Google Scholar]
- Blackwell, E. , Kim, H.J.N. and Stone, D.E. (2007) The pheromone‐induced nuclear accumulation of the Fus3 MAPK in yeast depends on its phosphorylation state and on Dig1 and Dig2. BMC Cell Biol. 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort, T. , Müller, P. and Kahmann, R. (2005) The high‐mobility‐group domain transcription factor Rop1 is a direct regulator of prf1 in Ustilago maydis . Eukaryot. Cell, 4, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J.R. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryot. Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.C. , Wickes, B.L. , Miller, G.F. , Penoyer, L.A. and Kwon‐Chung, K.J. (2000) Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J. Exp. Med. 191, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.C. , Penoyer, L.A. and Kwon‐Chung, K.J. (2001) The second STE12 homologue of Cryptococcus neoformans is MATa‐specific and plays an important role in virulence. Proc. Natl. Acad. Sci. USA, 98, 3258–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.C. , Wright, L.C. , Tscharke, R.L. , Sorrell, T.C. , Wilson, C.F. and Kwon‐Chung, K.J. (2004) Regulatory roles for the homeodomain and C2H2 zinc finger regions of Cryptococcus neoformans Ste12αp. Mol. Microbiol. 53, 1385–1396. [DOI] [PubMed] [Google Scholar]
- Chen, X.M. (2014) Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 36, 311–326. [Google Scholar]
- Cheng, Y.L. , Wang, X.J. , Yao, J.N. , Voegele, R.T. , Zhang, Y.R. , Wang, W.M. , Huang, L.L. and Kang, Z.S. (2015) Characterization of protein kinase PsSRPKL, a novel pathogenicity factor in the wheat stripe rust fungus. Environ. Microbiol. 17, 2601–2617. [DOI] [PubMed] [Google Scholar]
- Chou, S. , Lane, S. and Liu, H.P. (2006) Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae . Mol. Cell. Biol. 26, 4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo, C.A. , Bakkeren, G. , Khalil, H.B. , Panwar, V. , Joly, D. , Linning, R. , Sakthikumar, S. , Song, X. , Adiconis, X. , Fan, L. , Goldberg, J.M. , Levin, J.Z. , Young, S. , Zeng, Q.D. , Anikster, Y. , Bruce, M. , Wang, M.N. , Yin, C.T. , McCallum, B. , Szabo, L.J. , Hulbert, S. , Chen, X.M. and Fellers, J.P. (2016) Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3 (Bethesda), 7, 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenea, M. , Kristian, K. and Mogenss, H. (2009) Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology, 99, 89–94. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Herskowitz, I. (1985) The yeast STE12 product is required for expression of two sets of cell‐type specific genes. Cell, 42, 923–930. [DOI] [PubMed] [Google Scholar]
- Fu, Y.P. , Duan, X.Y. , Tang, C.L. , Li, X.R. , Voegele, R.T. , Wang, X.J. , Wei, G.R. and Kang, Z.S. (2014) TaADF7, an actin‐depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici . Plant J. 78, 16–30. [DOI] [PubMed] [Google Scholar]
- Garnica, D.P. , Upadhyaya, N.M. , Dodds, P.N. and Rathjen, J.P. (2013) Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS One, 8, e67150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel, L.P. and Ellis, B.E. (2012) Mitogen‐activated protein kinase signaling in plant‐interacting fungi: distinct messages from conserved messengers. Plant Cell, 24, 1327–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L.H. (1980) Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 85, 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi, J.W.S. and Dumas, B. (2010) Ste12 and Ste12‐like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell, 9, 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi, J.W.S. , Herbert, C. , Bacha, N. , O'Connell, R. , Lafitte, C. , Borderies, G. , Rossignol, M. , Rougé, P. and Dumas, B. (2007) Regulation and role of a STE12‐like transcription factor from the plant pathogen Colletotrichum lindemuthianum . Mol. Microbiol. 64, 68–82. [DOI] [PubMed] [Google Scholar]
- Holzberg, S. , Brosio, P. , Gross, C. and Pogue, G.P. (2002) Barley stripe mosaic virus‐induced gene silencing in a monocot plant. Plant J. 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Ito, T. and Shinozaki, K. (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD‐finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Szabo, L.J. and Carson, M. (2010) Century‐old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology, 100, 432–435. [DOI] [PubMed] [Google Scholar]
- Johnson, A.D. (1995) Molecular mechanisms of cell‐type determination in budding yeast. Curr. Opin. Genet. Dev. 5, 552–558. [DOI] [PubMed] [Google Scholar]
- Kang, Z.S. , Huang, L.L. and Buchenauer, H. (2002) Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis . J. Plant Dis. Protect. 109, 25–37. [Google Scholar]
- Kirkman‐Correia, C. , Stroke, I.L. and Fields, S. (1993) Functional domains of the yeast STE12 protein, a pheromone‐responsive transcriptional activator. Mol. Cell. Biol. 13, 3765–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmer, J.A. (2005) Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 8, 441–449. [DOI] [PubMed] [Google Scholar]
- Kramer, B. , Thines, E. and Foster, A.J. (2009) MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea . Fungal Genet. Biol. 46, 667–681. [DOI] [PubMed] [Google Scholar]
- Lanchun, S. , Bochu, W. , Liancai, Z. , Jie, L. , Yanhong, Y. and Chuanren, D. (2003) The influence of low‐intensity ultrasonic on some physiological characteristics of Saccharomyces cerevisiae . Colloids Surf. B: Biointerfaces, 30, 61–66. [Google Scholar]
- Li, C.X. , Lin, H.Q. and Dubcovsky, J. (2015) Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 84, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Bobrowicz, P. , Wilkinson, H.H. and Ebbole, D.J. (2005) A mitogen‐activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa . Genetics, 170, 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line, R.F. (2002) Stripe rust of wheat and barley in North America: a retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Guan, T. , Zheng, P.J. , Chen, L.Y. , Yang, Y. , Huai, B.Y. , Li, D. , Chang, Q. , Huang, L.L. and Kang, Z.S. (2016) An extracellular Zn‐only superoxide dismutase from Puccinia striiformis confers enhanced resistance to host‐derived oxidative stress. Environ. Microbiol. 18, 4118–4135. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔ C T method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D. and Fink, G.R. (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science, 275, 1314–1317. [DOI] [PubMed] [Google Scholar]
- Meng, X. and Zhang, S. (2013) MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Min, K. , Shin, Y. , Son, H. , Lee, J. , Kim, J.C. , Choi, G.J. and Lee, Y.W. (2012) Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae . FEMS Microbiol. Lett. 334, 66–73. [DOI] [PubMed] [Google Scholar]
- Park, G. , Xue, C.Y. , Zheng, L. , Lam, S. and Xu, J.R. (2002) MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 15, 183–192. [DOI] [PubMed] [Google Scholar]
- Park, G. , Bruno, K.S. , Staiger, C.J. , Talbot, N.J. and Xu, J.R. (2004) Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53, 1695–1707. [DOI] [PubMed] [Google Scholar]
- Passmore, S. , Maine, G.T. , Elble, R. , Christ, C. and Tye, B.K. (1989) Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 204, 593–606. [DOI] [PubMed] [Google Scholar]
- Petty, I.T.D. , French, R. , Jones, R.W. and Jackson, A.O. (1990) Identification of barley stripe mosaic virus genes involved in viral RNA replication and systemic movement. EMBO J. 9, 3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail, N. and Di, P.A. (2009) Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant–Microbe Interact. 22, 830–839. [DOI] [PubMed] [Google Scholar]
- Rispail, N. and Pietro, A.D. (2010) The homeodomain transcription factor Ste12. Commun. Integr. Biol. 3, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfs, A.P. and Bushnell, W.R. (1985) The Cereal Rusts, Volume II: Diseases, Distribution, Epidemiology, and Control. Orlando, FL: Academic. [Google Scholar]
- Schamber, A. , Leroch, M. , Diwo, J. , Mendgen, K. and Hahn, M. (2010) The role of mitogen‐activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea . Mol. Plant Pathol. 11, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. , Seo, Y.S. , Min, K. , Park, A.R. , Lee, J. , Jin, J.M. , Lin, Y. , Cao, P.J. , Hong, S.Y. , Kim, E.K. , Lee, S.H. , Cho, A. , Lee, S. , Kim, M.G. , Kim, Y. , Kim, J.E. , Kim, J.C. , Choi, G.J. , Yun, S.H. , Lim, J.Y. , Kim, M. , Lee, Y.H. , Choi, Y.D. and Lee, Y.W. (2011) A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog. 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, G.F. (1991) Assay of yeast mating reaction. Methods Enzymol. 194, 77–93. [DOI] [PubMed] [Google Scholar]
- Tang, C.L. , Wei, J.P. , Han, Q.M. , Liu, R. , Duan, X.Y. , Fu, Y.P. , Huang, X.L. , Wang, X.J. and Kang, Z.S. (2015) PsANT, the adenine nucleotide translocase of Puccinia striiformis, promotes cell death and fungal growth. Sci. Rep. 5, 11 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, G. , Fujii, S. , Tsuge, S. , Shiraishi, T. and Kubo, Y. (2003) The Colletotrichum lagenarium Ste12‐like gene CST1 is essential for appressorium penetration. Mol. Plant–Microbe Interact. 16, 315–325. [DOI] [PubMed] [Google Scholar]
- Urban, M. , Mott, E. , Farley, T. and Hammond‐Kosack, K. (2003) The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4, 347–359. [DOI] [PubMed] [Google Scholar]
- Vallim, M.A. , Miller, K.Y. and Miller, B.L. (2000) Aspergillus SteA (sterile12‐like) is a homeodomain‐C2/H2‐Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36, 290–301. [DOI] [PubMed] [Google Scholar]
- Wang, C.F. , Huang, L.L. , Buchenauer, H. , Han, Q.M. , Zhang, H.C. and Kang, Z.S. (2007) Histochemical studies on the accumulation of reactive oxygen species ( and H2O2) in the incompatible and compatible interaction of wheat–Puccinia striiformis f. sp. tritici . Physiol. Mol. Plant Pathol. 71, 230–239. [Google Scholar]
- Wang, M.N. and Chen, X.M. (2013) First report of Oregon grape (Mahonia aquifolium) as an alternate host for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) under artificial inoculation. Plant Dis. 97, 839. [DOI] [PubMed] [Google Scholar]
- Wellings, C.R. (2011) Global status of stripe rust: a review of historical and current threats. Euphytica, 179, 129–141. [Google Scholar]
- Yang, C. , Liu, H.Q. , Li, G.T. , Liu, M.G. , Yun, Y.Z. , Wang, C.F. , Ma, Z.H. and Xu, J.R. (2015) The MADS‐box transcription factor FgMcm1 regulates cell identity and fungal development in Fusarium graminearum . Environ. Microbiol. 17, 2762–2776. [DOI] [PubMed] [Google Scholar]
- Yin, C. , Jurgenson, J.E. and Hulbert, S.H. (2011) Development of a host‐induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . Mol. Plant–Microbe Interact. 24, 554–561. [DOI] [PubMed] [Google Scholar]
- Yuan, Y.L. and Fields, S. (1991) Properties of the DNA‐binding domain of the Saccharomyces cerevisiae STE12 protein. Mol. Cell. Biol. 11, 5910–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.O. , Stroke, I.L. and Fields, S. (1993) Coupling of cell identity to signal response in yeast: interaction between the alpha 1 and STE12 proteins. Gene Dev. 7, 1584–1597. [DOI] [PubMed] [Google Scholar]
- Yue, C.L. , Cavallo, L.M. , Alspaugh, J.A. , Wang, P. , Cox, G.M. , Perfect, J.R. and Heitman, J. (1999) The STE12 alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans . Genetics, 153, 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, L. , Wang, Z.Y. , Chen, X.M. , Zhang, H.C. , Yao, J.N. , Zhan, G.M. , Chen, W. , Huang, L.L. and Kang, Z.S. (2013) Identification of eighteen Berberis species as alternate hosts of Puccinia striiformis f. sp. tritici and virulence variation in the pathogen isolates from natural infection of barberry plants in China. Phytopathology, 103, 927–934. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Wang, M.N. , Chen, X.M. and Kang, Z.S. (2016) Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Phytopathology, 54, 207–228. [DOI] [PubMed] [Google Scholar]
- Zheng, W.M. , Huang, L.L. , Huang, J.Q. , Wang, X.J. , Chen, X.M. , Zhao, J. , Guo, J. , Zhuang, H. , Qiu, C.Z. , Liu, J. , Liu, H.Q. , Huang, X.L. , Pei, G.L. , Zhan, G.M. , Tang, C.L. , Cheng, Y.L. , Liu, M.J. , Zhang, J.S. , Zhao, Z.T. , Zhang, S.J. , Han, Q.M. , Zhang, H.C. , Zhao, J. , Gao, X.N. , Wang, J.F. , Ni, P.X. , Dong, W. , Yang, L.F. , Yang, H.M. , Xu, J.R. , Zhang, G.Y. and Kang, Z.S. (2013) High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 5, 2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.Y. , Liu, W.D. , Wang, C.F. , Xu, Q.J. , Wang, Y. , Ding, S.L. and Xu, J.R. (2011) A MADS‐box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae . Mol. Microbiol. 80, 33–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Multi‐sequence alignment and phylogenetic analysis of PstSTE12 and other orthologues of STE12 in other species. (A) Alignment showing the identity of STE12 homeodomains of Puccinia striiformis f. sp. tritici (Pst). Bold overlines mark the three helices of the homeodomain. Solid underline indicates the conserved tryptophan and phenylalanine residues of Helix III known to be essential for DNA binding. (B) Phylogeny of fungal STE12 orthologues. The phylogenetic tree was constructed using the maximum‐likelihood (ML) approach. The confidence level for the groupings was estimated using 500 bootstrap replicates. The numbers adjacent to the branch points indicate the percentage of replicates supporting each branch. Scale bars correspond to 0.1 amino acid substitutions.
Fig. S2 Two C2/H2‐Zn2+ finger domains of the C‐terminal region were predicted in PstSTE12 via the National Center for Biotechnology Information (NCBI).
Fig. S3 Tetrads were separated from the ascospores formed by the diploid mutant after cultivation in MacConkey medium. Four viable spores (A–D) were obtained from each of the tetrads (lanes 1, 3, 6–10 and 12) with some exceptions of two‐ or three‐spore tetrads as a result of random spore inviability (lanes 2, 4, 5 and 11).
Fig. S4 Sequence regions for the host‐induced gene silencing (HIGS) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) experiments in this study. Black arrows indicate the start codon, HIGS targeted sites, qRT‐PCR site and stop codon. The numbers in parentheses indicate the distance to the start codon (ATG). The sequences of the two domains are labelled in grey.
Table S1 Primers used for the assays in this study.


