Summary
Pratylenchus penetrans is one of the most important species of root lesion nematodes (RLNs) because of its detrimental and economic impact in a wide range of crops. Similar to other plant‐parasitic nematodes (PPNs), P. penetrans harbours a significant number of secreted proteins that play key roles during parasitism. Here, we combined spatially and temporally resolved next‐generation sequencing datasets of P. penetrans to select a list of candidate genes aimed at the identification of a panel of effector genes for this species. We determined the spatial expression of transcripts of 22 candidate effectors within the oesophageal glands of P. penetrans by in situ hybridization. These comprised homologues of known effectors of other PPNs with diverse putative functions, as well as novel pioneer effectors specific to RLNs. It is noteworthy that five of the pioneer effectors encode extremely proline‐rich proteins. We then combined in situ localization of effectors with available genomic data to identify a non‐coding motif enriched in promoter regions of a subset of P. penetrans effectors, and thus a putative hallmark of spatial expression. Expression profiling analyses of a subset of candidate effectors confirmed their expression during plant infection. Our current results provide the most comprehensive panel of effectors found for RLNs. Considering the damage caused by P. penetrans, this information provides valuable data to elucidate the mode of parasitism of this nematode and offers useful suggestions regarding the potential use of P. penetrans‐specific target effector genes to control this important pathogen.
Keywords: pioneer effectors, plant‐parasitic, proline‐rich, root lesion nematode, transcriptome
Introduction
Root lesion nematodes (RLNs), namely Pratylenchus spp., are economically important pathogens that inflict damage and yield losses on a wide range of crops (Castillo and Vovlas, 2007). RLNs require an intimate association with their host to gain access to nutrients. Pratylenchus spp. are migratory endoparasitic nematodes that feed predominantly from the root cortical tissues, causing a reduction in root growth, accompanied by the formation of lesions, necrotic areas, browning and cell death (Castillo and Vovlas, 2007; Fosu‐Nyarko and Jones, 2016). In contrast with sedentary nematodes, such as cyst and root‐knot nematodes, which induce highly specialized and complex feeding structures (namely syncytia and giant cells, respectively), RLNs do not induce complex feeding structures (Fosu‐Nyarko and Jones, 2016). However, their mobility throughout their life cycle causes massive damage to the root system, predisposing the roots to secondary infections by other soil‐borne pathogens (Castillo and Vovlas, 2007).
One of the most important species of this genus is Pratylenchus penetrans because of its host range (nearly 400 species), including high‐value crops, such as grasses, forage crops and fruit trees (Castillo and Vovlas, 2007). Pratylenchus penetrans is an amphimictic species (Roman and Triantaphyllou, 1969), and all stages are vermiform and motile [except eggs and first‐stage juveniles (J1s)], capable of feeding both endoparasitically and ectoparasitically (Zunke, 1990). The life cycle of P. penetrans can range from 3 to 7 weeks depending on the environmental conditions (Mizukubo and Adachi, 1997), and thus several generations can develop during the life span of the crop.
Similar to other plant‐parasitic nematodes (PPNs), the successful infection of RLNs relies on the secretion of a repertoire of proteins with diverse parasitism‐related functions. These nematode‐secreted proteins (known as effectors) are crucial components in the outcome of the plant–nematode interaction by participating in the penetration and evasion of the host, with the consequent establishment of the nematode (Mitchum et al., 2013). In most Tylenchoidea, these nematode‐secreted effectors are primarily synthesized in three unicellular oesophageal glands (two subventral and one dorsal) and are ultimately secreted through the stylet, a hollow, protrusible, needle‐like structure (Hussey, 1989). These secretions can be delivered into different compartments of the host cells (e.g. apoplasm and cytoplasm), enabling nematode development and progression of the disease (Mitchum et al., 2013). In addition, proteins secreted by other nematode tissues, such as the hypodermis and amphids, can actively participate in different stages of host interaction (Mitchum et al., 2013). The invasion of roots by RLNs involves mechanical force of the stylet, pressure of the labial region and secretion of cell wall‐degrading enzymes (CWDEs) (Castillo and Vovlas, 2007). Despite their economic importance, the molecular mechanisms by which RLNs cause disease in plants are still largely unknown, but, similar to other plant pathogens, effector‐like proteins probably play an important role in their parasitic behaviour.
In this context, molecular studies have focused on the identification of nematode effector catalogues of different economically important PPNs. The majority of these studies have focused on sedentary plant parasites (e.g. cyst and root‐knot nematodes), showing that PPN effector repertoires can contain hundreds of proteins implicated in the establishment of a successful interaction (Mitchum et al., 2013). RLNs have long been considered as less specialized parasites, as they do not induce a specific feeding site, but rather feed on the contents of host cells they encounter during their destructive migration through the cortex of the root (Fosu‐Nyarko and Jones, 2016).
The availability of both genomic and transcriptomic datasets for several RLNs (Burke et al., 2015; Fosu‐Nyarko et al., 2016; Haegeman et al., 2011; Nicol et al., 2012), including P. penetrans (Denver et al., 2016; Mitreva et al., 2004; Vieira et al., 2015), has provided the opportunity to identify and catalogue putative candidate effectors. These studies have highlighted certain features of RLN effector repertoires, uncovering the presence of common effector genes often employed by other migratory and sedentary PPNs. A core set of candidate effectors has been identified, including a suite of genes encoding CWDEs, such as β‐1,4‐endoglucanases (GH5), pectate lyases (PL3), arabinogalactan endo‐1,4‐β‐galactosidases (GH53), xylanases (GH30) and expansin‐like genes (Vieira et al., 2015), often implicated in the softening and degradation of the plant cell wall (e.g. Smant et al., 1998). A few other genes or gene families frequently identified as part of the nematode–host secretome have also been recognized by these in silico analyses (Vieira et al., 2015), including, for example, fatty acid‐ and retinol‐binding proteins (FARs), transthyretin‐like proteins (TTLs), venom allergen‐like proteins (VAPs) and an array of diverse classes of putatively secreted proteases or genes involved in protection from host defences, such as reactive oxygen species (ROS). A prominent feature of these comparative analyses was the absence of transcripts encoding nematode effectors related to giant cell or syncytium formation by root‐knot and cyst nematodes, underlining the differences between sedentary nematode species and RLNs (Fosu‐Nyarko and Jones, 2016). Although efforts have been made to provide an exhaustive list of candidate effector genes of RLNs (Burke et al., 2015; Denver et al., 2016; Fosu‐Nyarko et al., 2016; Haegeman et al., 2011; Nicol et al., 2012; Vieira et al., 2015), a limited number have been experimentally validated or characterized. To date, only a handful of RLN effectors have been specifically localized in the oesophageal glands of P. thornei [e.g. one β‐1,4‐endoglucanase, one pectate lyase, one polygalacturonase, one glutathione‐S‐transferase and one VAP Fosu‐Nyarko and Jones, 2016], P. vulnus [e.g. two β‐1,4‐endoglucanases (Fanelli et al., 2014)] and P. zeae [e.g. one calreticulin, one β‐1,4‐endoglucanase and one SXP/RAL‐2 gene (Fosu‐Nyarko et al., 2016)].
In addition, the presence of predicted N‐terminal signal peptides and the absence of transmembrane domains have been used to mine the predicted secretomes of RLNs, complementing the list of candidate secreted proteins. A hallmark of RLNs transcriptome analyses, and, in particular, of P. penetrans, is the great proportion of transcripts encoding putative secreted proteins without a known function (Vieira et al., 2015). However, other putative effectors have been identified in the secretome of PPNs without having a classical signal peptide for secretion, suggesting alternative secretory pathways independent of the endoplasmic reticulum–Golgi network (Bellafiore et al., 2008; Dubreuil et al., 2007). Although the catalogue of effectors of species with distinct strategies of parasitism may share some common features, to date, a large portion of the newly identified pioneer effectors for other sedentary or migratory PPNs seem to be species‐ or genus‐specific (Bird et al., 2015). In this case, the number of predicted secreted proteins without functional annotation identified for RLNs, and, in particular, for P. penetrans (Vieira et al., 2015), could represent a powerful resource to identify novel, species‐specific, effectors.
Here, we combine spatially and temporally resolved next‐generation sequencing datasets of P. penetrans (Maier et al., 2013; Vieira et al., 2015) to catalogue effector genes, with special focus on the identification of novel effectors. We experimentally determine the spatial expression patterns of 38 nematode genes, revealing/validating gland cell expression for 22 candidate effectors. Furthermore, we combine in situ localization of effectors with available genomic data to identify a non‐coding motif enriched in the promoter regions of a subset of P. penetrans effectors, and thus a putative hallmark of spatial expression. In addition, we experimentally validate the temporal expression profile of candidate effectors during infection, further supporting their involvement in parasitism. Considering the detrimental effect caused by P. penetrans in a wide range of economically important crops, our results provide important information on the range of P. penetrans effector genes involved in the infection, and identify high‐priority candidates for gene targets in the control of this important plant pathogen.
Results
Candidate effector gene selection
To identify a more comprehensive list of P. penetrans effectors, we combined spatially and temporally resolved sequencing datasets. Although we expected considerable overlap between these approaches, they were nevertheless combined to safeguard against false negatives in each inherently imperfect approach. Based on a dataset of 1330 transcripts (Table S1, see Supporting Information) predicted to encode secreted proteins (i.e. presence of a signal peptide and no transmembrane domain) from the de novo transcriptome assembly of P. penetrans (Vieira et al., 2015), we ranked sequences by transcript abundance in: (i) 454 sequencing of a cDNA library generated from the oesophageal gland mRNA of P. penetrans (Maier et al., 2013); and (ii) Illumina RNA sequencing (RNAseq) of a nematode infection time course (Vieira et al., 2015).
The 454 gland cell reads were mapped to all P. penetrans transcripts in the transcriptome to identify sequences that may be expressed in these tissues. Using this approach, 85 of the 1330 transcripts encoding putatively secreted proteins were identified (Fig. S1A, see Supporting Information; Table S1). Amongst this list, we were able to re‐identify transcripts encoding homologues of known effectors, or genes relevant during nematode–host interaction, such as different classes of CWDEs, a calreticulin, a VAP, several TTLs and different proteases. Of the 85 transcripts, 40 sequences had no similarity to sequences in the non‐redundant (NR) database (blastx, e‐value < 10−5) (Fig. S1B). The Illumina RNAseq in planta infection time course reads were similarly mapped to all P. penetrans transcripts (Vieira et al., 2015) that putatively encode secreted proteins, and a total of 1286 of the 1330 transcripts were identified (Fig. S1; Table S1).
From these lists, a panel of candidate effectors was compiled to contain both those with similarity to previously characterized effectors and those that represented pioneer sequences (i.e. no known or annotatable function), because effector proteins are often evolutionarily diverse amongst different lineages of PPNs and are rarely similar to known proteins (Kikuchi et al., 2017). Thirty‐three candidates from this panel were similar to those described previously, for example various families of CWDEs, including β‐1,4‐endoglucanases (GH5), pectate lyases (PL3), xylanase (GH30), arabinogalactan endo‐1,4‐β‐galactosidase (GH53) and expansin‐like proteins (Table 1). Other candidates included homologues of known PPN genes with a putative participation in the suppression of plant defences, e.g. VAPs (Lozano‐Torres et al., 2012) and a calreticulin (Jaouannet et al., 2013), or genes commonly associated with nematode activity within the host, such as FARs (Iberkleid et al., 2013), TTLs (Lin et al., 2016), a glutathione peroxidase (Jones et al., 2004) and SXP/RAL‐2 proteins (Jones et al., 2000; Tytgat et al., 2005). A set of sequences encoding different classes of proteases and inhibitor‐like proteases was also included because of their potential participation in parasitism (Table 1). Although these types of proteins may play essential physiological roles (e.g. digestion), some proteases are secreted within the host tissues of both animal‐parasitic nematodes (APNs) and PPNs (Hewitson et al., 2009; Vieira et al., 2011), and are linked to putative roles in parasitism, such as suppression of the host immunity by APNs (Hewitson et al., 2009).
Table 1.
Summary of Pratylenchus penetrans gene transcripts with known annotation selected for in situ hybridization assays.
| Tylenchida | Aph. | Rhab. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. penetrans | Pratylenchidae | RKN | Cyst | Ang. | |||||||||||||||
| Transcript code | Gland dataset (n = 22) | In planta dataset (n = 33) | Pc | Pz | Pt | Pv | Rs | Mi | Mh | Gp | Gr | Dd | Bx | Ce | ANNOTATION – NR database | Top‐hit species | blast top hit e‐value | blast top hit score | Accession |
| Homologues of known effector or gene candidates with relevant annotation | |||||||||||||||||||
| Ppen15842_c0_seq1 | Yes | Yes | + | + | – | + | + | + | + | + | + | + | – | – | β‐1,4‐Endoglucanase | Pratylenchus penetrans | 0.00E+00 | 909.83 | BAB68522.1 |
| Ppen15605_c0_seq1 | – | Yes | + | + | – | + | + | + | + | + | + | + | – | – | β‐1,4‐Endoglucanase | Pratylenchus goodeyi | 3.00E‐165 | 478.79 | AJD14760.1 |
| Ppen16218_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | – | – | β‐1,4‐Endoglucanase | Pratylenchus coffeae | 1.90E‐88 | 289.66 | ABX79356.1 |
| Ppen13447_c0_seq1 | Yes | Yes | + | + | + | – | – | + | + | + | + | + | + | – | Pectate lyase | Heterodera glycines | 1.30E‐84 | 265.77 | ADW77534.1 |
| Ppen14256_c0_seq1 | Yes | Yes | + | – | – | – | – | + | + | + | + | + | + | – | Pectate lyase | Globodera pallida | 9.70E‐52 | 181.80 | AEA08853.1 |
| Ppen12533_c0_seq1 | Yes | Yes | + | + | – | + | – | + | + | + | + | + | + | – | Expansin‐like | Heterodera avenae | 2.20E‐40 | 150.21 | APC23320.1 |
| Ppen15554_c1_seq1 | Yes | Yes | + | + | – | + | + | + | + | + | + | + | + | – | Expansin‐like | Heterodera glycines | 1.90E‐60 | 207.61 | ADL29728.1 |
| Ppen9511_c0_seq1 | – | Yes | + | + | – | + | + | + | + | + | + | + | + | – | Expansin‐like | Heterodera glycines | 1.80E‐62 | 213.00 | ADL29728.1 |
| Ppen18759_c0_seq1 | Yes | Yes | + | – | – | – | – | – | – | + | + | – | – | – | Arabinogalactan endo‐1,4‐β‐galactosidase | Heterodera schachtii | 4.10E‐123 | 359.00 | ACY02855.1 |
| Ppen12597_c1_seq1 | Yes | Yes | + | + | – | – | + | + | + | – | – | – | – | – | Glucuronoarabinoxylan endo‐1,4‐β‐xylanase | Radopholus similis | 0.00E+00 | 549.67 | ABZ78968.1 |
| Ppen15229_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Calreticulin | Pratylenchus goodeyi | 0.00E+00 | 638.26 | AIW66697.1 |
| Ppen11632_c0_seq1 | Yes | Yes | + | + | + | + | – | + | + | + | + | + | + | + | Venom allergen | Globodera rostochiensis | 4.00E‐84 | 261.54 | AEL16453.1 |
| Ppen9526_c1_seq1 | – | Yes | + | + | + | – | – | + | + | + | + | + | + | + | Venom allergen | Globodera rostochiensis | 1.50E‐34 | 139.42 | AEL16453.1 |
| Ppen16493_c0_seq1 | Yes | Yes | + | + | – | + | – | + | + | + | + | + | + | + | Catalase | Ditylenchus destructor | 0 | 623.21 | AFJ15102.1 |
| Ppen16592_c0_seq1 | – | Yes | + | + | – | + | + | + | + | + | + | + | + | + | Glutathione peroxidase | Globodera rostochiensis | 5.10E‐137 | 399.44 | AHW98769.1 |
| Ppen14407_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Transthyretin‐like family | Radopholus similis | 1.80E‐81 | 250.75 | CAM84513.1 |
| Ppen11355_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Transthyretin‐like family | Ancylostoma duodenale | 3.90E‐61 | 197.98 | KIH61588.1 |
| Ppen12000_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Transthyretin‐like family | Ancylostoma duodenale | 1.20E‐60 | 196.82 | KIH61588.1 |
| Ppen14007_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Transthyretin‐like family | Meloidogyne javanica | 2.10E‐76 | 237.27 | AKU46811.1 |
| Ppen12895_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Fatty acid‐ and retinol‐binding protein | Pratylenchus penetrans | 6.00E‐27 | 112.46 | APT68073.1 |
| Ppen11068_c0_seq1 | – | Yes | + | + | – | + | + | + | + | + | + | + | + | + | Fatty acid‐ and retinol‐binding protein | Aphelenchoides besseyi | 1.20E‐48 | 168.70 | AOC59163.1 |
| Ppen12103_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | – | + | SXP RAL‐2 protein | Meloidogyne incognita | 1.20E‐44 | 159.46 | AAR35032.1 |
| Proteases and inhibitor proteases | |||||||||||||||||||
| Ppen15235_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Cathepsin L‐like cysteine protease | Ditylenchus destructor | 3.80E‐156 | 459.14 | ACT35690.1 |
| Ppen14741_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Cathepsin L | Ancylostoma ceylanicum | 3.00E‐90 | 287.73 | EYC42688.1 |
| Ppen15220_c0_seq1 | Yes | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Protein BMA‐NPA‐1 | Brugia malayi | 6.60E‐89 | 314.69 | CRZ25179.1 |
| Ppen16129_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Cysteine protease precursor | Onchocerca volvulus | 1.80E‐143 | 420.62 | AAC47348.1 |
| Ppen13948_c0_seq1 | – | Yes | + | + | + | + | + | + | + | + | + | + | + | + | Papain family cysteine protease | Oesophagostomum dentatum | 3.00E‐76 | 253.00 | KHJ95394.1 |
| Ppen16494_c0_seq1 | Yes | Yes | + | + | – | + | + | + | + | + | + | + | + | + | Fatty acid amide hydrolase | Strongyloides ratti | 1.80E‐94 | 312.38 | CEF68470.1 |
| Ppen16868_c0_seq1 | Yes | Yes | + | + | – | + | + | + | + | + | + | + | + | + | Aspartyl protease | Meloidogyne incognita | 0.00E+00 | 690.26 | ABC88426.1 |
| Ppen15876_c0_seq1 | Yes | Yes | + | – | – | – | + | + | + | + | + | + | + | – | Trypsin‐like serine protease | Heliconius melpomene | 5.10E‐17 | 90.12 | ADJ58583.1 |
| Ppen12385_c0_seq1 | Yes | Yes | + | – | – | – | – | – | + | + | – | – | + | – | Serine protease | Caligus rogercresseyi | 5.30E‐12 | 75.49 | ACO10196.1 |
| Ppen13849_c0_seq1 | Yes | Yes | + | – | – | + | – | + | + | – | – | + | + | + | Trypsin inhibitor‐like cysteine‐rich domain protein | Dictyocaulus viviparus | 6.10E‐12 | 70.86 | KJH50180.1 |
| Ppen11515_c0_seq1 | Yes | Yes | + | – | – | – | – | – | – | – | – | – | – | – | Trypsin inhibitor‐like cysteine‐rich domain protein | Necator americanus | 6.30E‐09 | 65.08 | ETN72713.1 |
blast searches were performed against sequences in the non‐redundant (NR) database at the National Center for Biotechnology Information (NCBI) for a putative annotation, and against specific nematode proteins or transcriptome datasets for the presence/absence of positive blast hits (e‐value cutoff of 1e‐5 and bitscore > 50) in the corresponding species.
Ang., Anguinidae; Aph., Aphelenchida; Bx, Bursaphelenchus xylophilus; Ce, Caenorhabditis elegans; Cyst, cyst nematodes (Heteroderidae); Dd, Ditylenchus destructor; Gp, Globodera pallida; Gr, G. rostochiensis; Mh, Meloidogyne hapla; Mi, M. incognita; Pc, Pratylenchus coffeae; Pt, P. thornei; Pv, P. vulnus; Pz, P. zeae; Rhab., Rhabditida; RKN, root‐knot nematode (Meloidogynidae); Rs, Radopholus similis.
To obtain a final list of 100 candidates, an additional set of 67 transcripts (pioneer sequences with unknown function) expressed in the gland cell dataset and/or the in planta time course data was chosen primarily based on the distribution of similar sequences across the phylum: 45 were apparently exclusive to P. penetrans and 22 had similar sequences in at least one other PPN species, but were absent from sequences of Caenorhabditis elegans (Table 2). Although we recognize that this pipeline will exclude effectors that have diversified from common ancestral genes, our goal was to identify whether P. penetrans carries novel effectors not derived from ancestral loci. It is important to note that, because of the incomplete nature of other RLN datasets, we cannot conclude that the 45 putatively P. penetrans‐specific pioneer sequences are truly absent from other RLNs.
Table 2.
Summary of Pratylenchus penetrans gene transcripts without functional known annotation selected for in situ hybridization assays.
| Tylenchida | Aph. | Rhab. | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. penetrans | Pratylenchidae | RKN | Cyst | ||||||||||||
| Transcript code | Gland dataset (n = 24) | In planta dataset (n = 67) | Pc (n = 18) | Pz (n = 11) | Pt (n = 4) | Pv (n = 5) | Rs (n = 3) | Mi (n = 9) | Mh (n = 10) | Gp (n = 5) | Gr (n = 4) | Dd (n = 5) | Bx (n = 1) | Ce (n = 0) | ANNOTATION – NR database |
| Ppen12587_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11402_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12898_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen3243_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13578_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13114_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen9482_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen14240_c2_seq1 | – | Yes | + | + | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13485_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen16416_c0_seq1 | Yes | Yes | + | + | + | + | – | + | + | – | – | – | – | – | – |
| Ppen8004_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12088_c0_seq1 | – | Yes | – | – | – | – | + | – | + | – | – | – | – | – | – |
| Ppen7984_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11964_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen17512_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11603_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11206_c0_seq1 | – | Yes | + | + | – | – | – | – | – | – | – | – | – | – | – |
| Ppen16605_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12016_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13388_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen14681_c0_seq1 | – | Yes | + | + | + | – | + | – | + | + | + | + | + | – | – |
| Ppen11135_c0_seq1 | – | Yes | + | – | – | + | – | + | – | – | – | – | – | – | – |
| Ppen3331_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen15637_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13090_c0_seq1 | – | Yes | + | + | – | – | – | + | + | + | + | – | – | – | – |
| Ppen14446_c0_seq1 | – | Yes | + | + | + | – | – | + | + | + | – | + | – | – | – |
| Ppen12616_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen14188_c0_seq1 | – | Yes | + | + | – | + | + | + | + | + | + | + | – | – | – |
| Ppen8861_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13037_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen18978_c0_seq1 | – | Yes | – | – | – | – | – | – | + | – | – | – | – | – | – |
| Ppen10370_c0_seq1 | – | Yes | + | + | + | – | – | – | – | – | – | – | – | – | – |
| Ppen10414_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen8129_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen9159_c0_seq1 | – | Yes | – | – | – | – | – | + | + | – | – | – | – | – | – |
| Ppen5003_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11094_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen15969_c0_seq1 | – | Yes | + | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen14399_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen10237_c0_seq1 | – | Yes | + | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen16124_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen9671_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11230_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen15066_c0_seq1 | Yes | Yes | + | + | + | – | – | + | + | + | + | + | – | – | – |
| Ppen14417_c0_seq1 | Yes | Yes | + | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen16557_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12211_c0_seq1 | – | Yes | – | – | – | + | – | – | – | – | – | – | – | – | – |
| Ppen12633_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12501_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11421_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen5669_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen18231_c0_seq1 | Yes | Yes | + | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13380_c0_seq1 | Yes | Yes | + | – | – | – | – | + | – | – | – | – | – | – | – |
| Ppen11641_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12399_c0_seq1 | – | Yes | + | + | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12805_c0_seq1 | – | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen18503_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen11677_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12366_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen8205_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen19523_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen13553_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12216_c0_seq1 | – | Yes | + | + | – | + | – | + | + | – | – | + | – | – | – |
| Ppen12120_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen16911_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen10194_c0_seq1 | Yes | Yes | + | – | – | – | – | – | – | – | – | – | – | – | – |
| Ppen12271_c0_seq1 | Yes | Yes | – | – | – | – | – | – | – | – | – | – | – | – | – |
blast searches were performed against sequences in the non‐redundant (NR) database at the National Center for Biotechnology Information (NCBI) for a putative annotation, and against specific nematode proteins or transcriptome datasets for the presence/absence of positive blast hits (e‐value cutoff of 1e‐5 and bitscore > 50) in the corresponding species.
n represents the total number of genes with a positive blast hit against the 67 genes of P. penetrans.
Aph., Aphelenchida; Bx, Bursaphelenchus xylophilus; Ce, Caenorhabditis elegans; Cyst, cyst nematodes (Heteroderidae); Dd, Ditylenchus destructor; Gp, Globodera pallida; Gr, G. rostochiensis; Mh, Meloidogyne hapla; Mi, M. incognita; Pc, Pratylenchus coffeae; Pt, P. thornei; Pv, P. vulnus; Pz, P. zeae; Rhab., Rhabditida; RKN, root‐knot nematode (Meloidogynidae); Rs, Radopholus similis.
In situ hybridization identifies specific genes to secretory organs of P. penetrans
In order to determine whether the selected genes of P. penetrans represent valid candidate effectors, in situ hybridization assays were performed on 100 candidates to determine their expression in the nematode tissues. In these analyses, a substantial number of homologues of PPN effectors were specifically expressed in the oesophageal glands of P. penetrans, which included transcripts encoding two β‐1,4‐endoglucanases (Ppen15842_c0_seq1 and Ppen16218_c0_seq1), two pectate lyases (Ppen13447_c0_seq1 and Ppen14256_c0_seq1), two expansin‐like proteins (Ppen12533_c0_seq1 and Ppen15554_c1_seq1), one xylanase (Ppen12597_c1_seq1), one arabinogalactan endo‐1,4‐β‐galactosidase (Ppen18759_c0_seq1), one VAP (Ppen11632_c0_seq1), one calreticulin (Ppen15229_c0_seq1), one FAR (Ppen12895_c0_seq1) and one SXP/RAL‐2 protein (Ppen12103_c0_seq1) (Fig. 1A–L, Table 1). Interestingly, transcripts encoding a catalase (Ppen16493_c0_seq1) are also localized to the oesophageal glands of P. penetrans (Fig. 1M).
Figure 1.
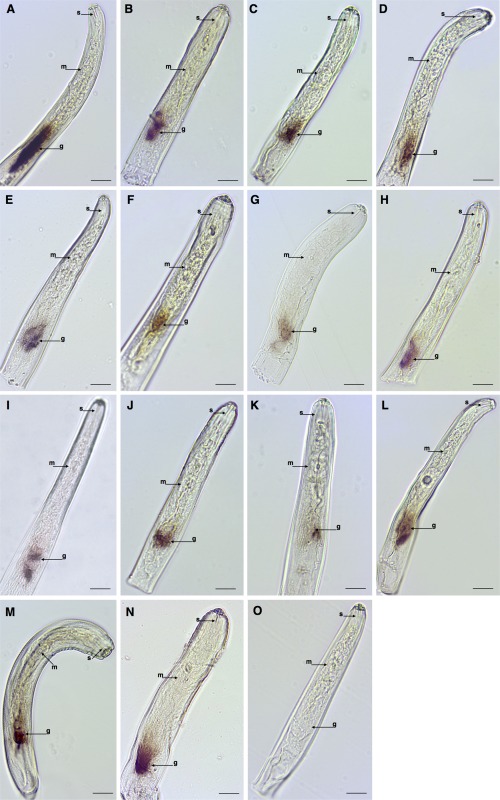
Detection of gene transcripts by in situ hybridization that encode genes with known annotation of Pratylenchus penetrans. (A, B) β‐1,4‐Endoglucanases. (C, D) Pectate lyases. (E, F) Expansin‐like. (G) Xylanase. (H) Arabinogalactan endo‐1,4‐β‐galactosidase. (I) Venom allergen‐like. (J) Calreticulin. (K) Fatty acid‐ and retinol‐binding protein. (L) SXP/RAL‐2. (M) Catalase. (N) Trypsin inhibitor‐like. (O) Example of a control image obtained using the sense probe (e.g. Ppen15842_c0_seq1). oesophageal glands; m, medium bulb; s, stylet. Bars, 20 µm.
Among the transcripts encoding different proteases, one was predicted to encode a putative trypsin inhibitor‐like protein (Ppen13849_c0_seq1), and was localized in the oesophageal glands of the nematodes (Fig. 1N). Remarkably, transcripts encoding two trypsin‐like serine proteases (Ppen15876_c0_seq1 and Ppen12385_c0_seq1) and a fatty acid amide hydrolase (Ppen16494_c0_seq1) were found to be predominantly expressed in the excretory duct of the excretory/secretory (E/S) system of P. penetrans (Fig. 2A–C) and, to our knowledge, these are the first genes ever found to be expressed in the E/S system of a RLN. In addition, transcripts encoding three other proteases (Ppen15235_c0_seq1, Ppen14741_c0_seq1 and Ppen13948_c0_seq1) were localized in the intestine of P. penetrans (Fig. 2D–F), probably associated with digestive processes of the nematode.
Figure 2.
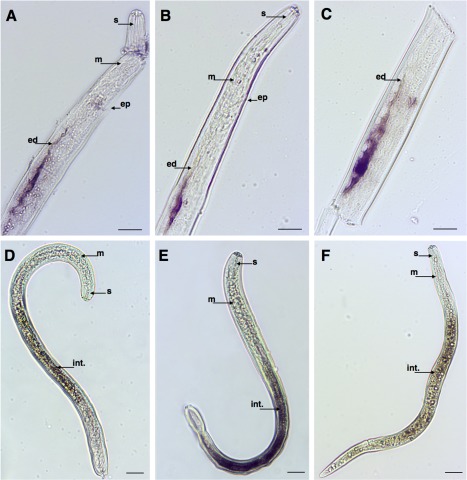
Detection of Pratylenchus penetrans gene transcripts by in situ hybridization that encode different proteases. (A) Fatty acid amide hydrolase. (B) Trypsin inhibitor‐like. (C) Serine protease. (D) Cathepsin L‐like cysteine protease. (E) Cathepsin L. (F) Papain family cysteine protease. ep, excretory pore; ed, excretory duct; int., intestine; m, medium bulb; s, stylet. Bars, 20 µm.
Of the pioneers (sequences of unknown function), eight candidates were specifically localized in the oesophageal glands (Ppen11402_c0_seq1, Ppen8004_c0_seq1, Ppen7984_c0_seq1, Ppen16605_c0_seq1, Ppen12016_c0_seq1, Ppen10370_c0_seq1, Ppen11230_c0_seq1 and Ppen15066_c0_seq1) of the nematode (Fig. 3A–H), increasing considerably the number of candidate parasitism‐related genes identified for this species. It is interesting to note that seven of the eight are, with reference to currently available datasets, unique to P. penetrans or to other RLNs (Table 2). Other relevant results amongst this set were a transcript localized to the amphids (Ppen13578_c0_seq1) (Fig. 3I), and two different transcripts localized along the hypodermis (Ppen9159_c0_seq1 and Ppen16557_c0_seq1) of the nematode (Fig. 3J,K). Although some genes expressed in the amphids and hypodermis have been shown to be relevant for the parasitism of other PPNs (Eves‐van den Akker et al., 2014; Iberkleid et al., 2013), we cannot exclude that they may be part of the ordinary development or physiology of the nematode.
Figure 3.
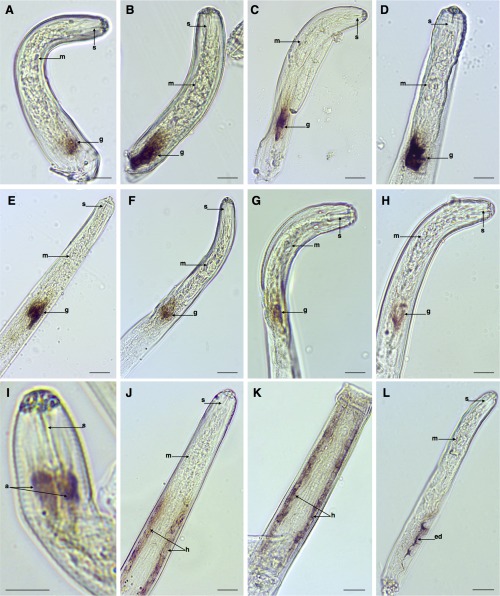
Detection of gene transcripts by in situ hybridization that encode genes with unknown predicted function of Pratylenchus penetrans. (A–H) Pioneer candidate effectors localized within the oesophageal glands (Ppen11402_c0_seq1, Ppen8004_c0_seq1, Ppen7984_c0_seq1, Ppen16605_c0_seq1, Ppen12016_c0_seq1, Ppen10370_c0_seq1, Ppen11230_c0_seq1, Ppen15066_c0_seq1). (I) Amphids (Ppen13578_c0_seq1). (J, K) Hypodermis (Ppen9159_c0_seq1 and Ppen16557_c0_seq1). (L) Excretory/secretory duct (Ppen16416_c0_seq1). a, amphids; ed, excretory duct; g, oesophageal glands; h, hypodermis; m, medium bulb; s, stylet. Bars, 20 µm.
In addition to the transcripts encoding proteases found within the E/S system, transcripts that encode a putatively secreted protein of unknown function (Ppen16416_c0_seq1) were found to be abundantly expressed in the E/S duct of different stages of P. penetrans (Fig. 3L). For the remaining candidates, in situ localization excluded their participation in parasitism (Fig. S2, see Supporting Information), or no signal was detected using the probes designed in this study (data not shown). As a control, the sense probe of each corresponding gene was used, and no hybridization signal was detected (e.g. Fig. 1O; for the remaining genes, data not shown).
Having a range of candidate effectors validated by in situ hybridization, we observed that, of the 22 effectors specifically expressed within the oesophageal glands, 17 were present within the gland transcriptome dataset, with a significant portion being highly abundant within the gland transcripts coding for putative proteins with a signal peptide and without a transmembrane domain (Fig. 4A). However, the 22 candidate effectors identified were each actively transcribed whilst the nematodes were in planta (Fig. 4B).
Figure 4.
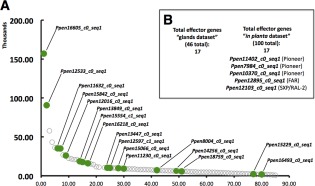
Relative abundance of transcripts encoding secreted proteins collected from the oesophageal glands of Pratylenchus penetrans. (A) Of the 46 genes selected, 17 genes were localized within the oesophageal glands. The annotation of each transcript can be found in Table 3. (B) Twenty‐two effector candidate genes (the previous 17 found within the “gland dataset” plus additional five) were detected in the in planta dataset.
Genetic characterization and annotation of gland cell‐expressed candidate effectors
Candidate effector‐encoding transcripts with spatial expression in the oesophageal glands were used for blastn searches (e‐value > 1e−10) against the low‐coverage genome skim assemblies of P. penetrans (Denver et al., 2016; I. A. Zasada, unpublished data, 2017) in order to identify their respective genomic sequences. These analyses allowed us to generate a preliminary prediction of the gene structure of the candidate effectors, and to substantiate the nematode origin of these genes, in particular for those often suggested to have been acquired via horizontal gene transfer (e.g. the CWDEs). This could not be determined for all candidates because the low‐coverage genomic skim is incomplete and highly fragmented; many P. penetrans transcripts were not present in their entirety (Fig. S3, see Supporting Information). Nevertheless, we could analyse possible gene structures for a subset of the candidates. Intron positions were determined by aligning the genomic DNA sequence to their corresponding transcripts. Most candidate effectors appear to be encoded by multi‐exon genes, with the number of exons varying from two to seven. The exon–intron boundaries of the majority are consistent with the canonical cis‐splicing GU‐AG rule.
The predicted protein sequences of all transcripts expressed within the glands were then used for InterPro scan, Pfam domain search and gene ontology (GO) term mapping to refine their annotation and to search for potential conserved domains using the Blast2GO suite (Table 3). A predicted function could be attributed to all annotated proteins, as the presence of Pfam domains was supported by relevant similarities with other characterized proteins within the NR database. Amongst the pioneers or sequences with unknown function localized within the oesophageal glands, only one candidate (Ppen15066_c0_seq1) showed low sequence identity to the Domain of Unknown Function‐DUF148 (PF02520.14 and IPR003677, e‐value of 4.9e−7) (Table 3).
Table 3.
Characterization of corresponding predicted protein sequences whose gene transcripts were specifically localized in the oesophageal glands of Pratylenchus penetrans
| PSORTII prediction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transcript code | Interpro accession | InterPro name | InterPro signatures | Protein (amino acid) | Domain position | Domain bit score | e‐value | Proline content (%) | Subcellular localization | Probability |
| Homologues of known effector genes or genes with relevant annotation | ||||||||||
| Ppen15842_c0_seq1 | IPR001547 | Glycoside hydrolase family 5 | PF00150 (PFAM) | 457 | 44–290 | 180.8 | 3.00E‐53 | 3.9 | Cytoplasmic | 60.9 |
| Ppen16218_c0_seq1 | IPR001547 | Glycoside hydrolase family 5 | PF00150 (PFAM) | 446 | 36–289 | 145 | 2.70E‐42 | 3.8 | Cytoplasmic | 47.8 |
| Ppen13447_c0_seq1 | IPR004898 | Pectate lyase catalytic | PF03211 (PFAM) | 260 | 19–236 | 214.1 | 1.50E‐63 | 1.2 | Nuclear | 56.5 |
| Ppen14256_c0_seq1 | IPR004898 | Pectate lyase catalytic | PF03211 (PFAM) | 264 | 20–257 | 86.4 | 1.70E‐24 | 3.8 | Nuclear | 73.9 |
| Ppen12533_c0_seq1 | IPR009009 | RlpA‐like protein double‐psi beta‐barrel domain | PF03330 (PFAM) | 180 | 56–73 | 28.8 | 8.70E‐07 | 5.6 | Nuclear | 34.8 |
| Ppen15554_c1_seq1 | IPR001919,IPR009009 | Carbohydrate‐binding type‐2 domain, RlpA‐like protein double‐psi beta‐barrel domain | PF00553 (PFAM), PF03330 (PFAM) | 323 | 25–118, 202–316 | 32 | 31.2 | 9.0E‐7, 2.1E‐8 | 6.8 | Nuclear | 39.1 |
| Ppen18759_c0_seq1 | IPR011683 | Glycosyl hydrolase family 53 | PF07745 (PFAM) | 336 | 27–283 | 296.7 | 1.80E‐88 | 3.6 | Cytoplasmic | 60.9 |
| Ppen12597_c1_seq1 | IPR033452,IPR033453 | Glycosyl hydrolase family 30 beta sandwich domain, glycosyl hydrolase family 30 TIM‐barrel domain | PF17189 (PFAM), PF02055 (PFAM) | 400 | 49–187 | 36.9 | 2.20E‐09 | 3.5 | Cytoplasmic | 47.8 |
| Ppen11632_c0_seq1 | IPR014044 | CAP domain | PF00188 (PFAM) | 212 | 35–174 | 59.2 | 1.30E‐20 | 2.4 | Nuclear | 39.1 |
| Ppen15229_c0_seq1 | IPR001580 | Calreticulin/calnexin | PF00262 (PFAM) | 412 | 23–333 | 206.9 | 9.80E‐123 | 5.6 | Endoplasmic reticulum | 55.6 |
| Ppen16493_c0_seq1 | IPR011614,IPR010582 | Catalase core domain, catalase immune‐responsive domain | PF00199 (PFAM), PF06628 (PFAM) | 512 | 44–425, 445–511 | 616.7 | 49.5 | 9.6E‐176, 9.5E‐14 | 7 | Cytoplasmic | 52.2 |
| Ppen13849_c0_seq1 | IPR002919 | Trypsin inhibitor‐like cysteine‐rich domain | PF01826 (PFAM) | 151 | 37–91 | 40.8 | 1.80E‐10 | 11.9 | Nuclear | 78.3 |
| Ppen12895_c0_seq1 | IPR008632 | Nematode fatty acid retinoid binding | PF05823 (PFAM) | 188 | 31–180 | 84.5 | 4.80E‐24 | 4.3 | Nuclear | 47.8 |
| Ppen12103_c0_seq1 | IPR003677 | Domain of unknown function DUF148 | PF02520 (PFAM) | 209 | 54 | 149 | 7.00E‐15 | 14.4 | Nuclear | 69.6 |
| Pioneer candidate effectors | ||||||||||
| Ppen11402_c0_seq1 | – | – | – | 79 | – | – | – | 6.3 | Cytoplasmic | 69.6 |
| Ppen8004_c0_seq1 | – | – | – | 92 | – | – | – | 23.9 | Nuclear | 65.2 |
| Ppen7984_c0_seq1 | – | – | – | 73 | – | – | – | 25.7 | Nuclear | 56.5 |
| Ppen16605_c0_seq1 | – | – | – | 102 | – | – | – | 22.5 | Nuclear | 43.5 |
| Ppen12016_c0_seq1 | – | – | – | 129 | – | – | – | 20.9 | Nuclear | 60.9 |
| Ppen10370_c0_seq1 | – | – | – | 101 | – | – | – | 13.9 | Nuclear | 39.1 |
| Ppen11230_c0_seq1 | – | – | – | 176 | – | – | – | 7.4 | Nuclear | 60.9 |
| Ppen15066_c0_seq1 | IPR003677 | Domain of unknown function DUF148 | PF02520 (PFAM) | 590 | 266–372 | 29.7 | 4.90E‐07 | 7.3 | Cytoplasmic | 69.6 |
Interestingly, we observed that most of the candidate pioneer effectors encoded an unusually high proportion of proline residues when compared with the other candidate secreted proteins selected for our analyses (Table 3). In one case, up to one‐quarter of the residues were prolines, whereas the average proline content of all predicted proteins of the transcriptome of P. penetrans is approximately 5.3% (Fig. 5). The five proline‐rich pioneer effectors were studied in more detail. Interestingly, on average, the proline content of these effectors is unevenly distributed across the predicted protein, and preferentially excluded from the first 20% (Fig. 5). This is in stark contrast with transcripts encoding putatively secreted proteins or, indeed, the predicted amino acid sequence of all other P. penetrans transcripts in the transcriptome (Fig. 5), suggesting that this trait is not a general feature of proteins/secreted proteins/effectors, but rather specific to this set. Although we cannot confirm that all the transcripts in the transcriptome are complete at their 5′ sequence, those that encode proteins with a predicted signal peptide are more likely to be complete, and are comparable with the proline‐rich effectors. The probability of randomly selecting five putatively secreted proteins that all exclude prolines from the first 20% of their open reading frame is empirically derived to be 2/250 (or P = 0.008). Furthermore, prolines are not randomly distributed across the proline‐rich 80% of the open reading frame, but are often present in pairs (position n + 1 to a proline) (Fig. 6). Prolines are also apparently more common in positions n + 3, n + 6 and n + 9 to another proline. This phenomenon does not appear to be a general feature of transcripts encoding proline‐rich proteins, as plotting those with >20% prolines (n = 145) does not generate the same pattern.
Figure 5.
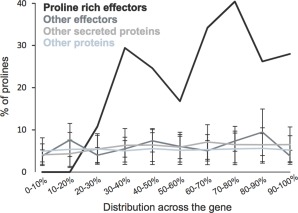
Prolines are preferentially excluded from the first 20% of proline‐rich pioneers. On average, the proline content of the proline‐rich effectors is non‐evenly distributed across the open reading frame, and preferentially excluded from the first 1%–20% (black). This is in stark contrast with all Pratylenchus penetrans predicted proteins (light blue), transcripts that encode putatively secreted proteins (light grey) and all transcripts expressed in the gland cells (dark grey). Five proteins were selected at random in each of 250 iterations. In each iteration, the average distribution of prolines in those five proteins was calculated. The means of all 250 iterations are shown, with error bars indicating the standard deviation.
Figure 6.
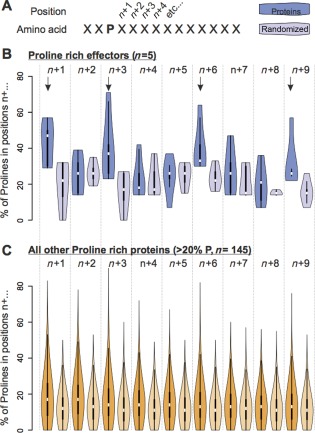
Distribution of prolines across proline‐rich pioneers and all other proline‐rich proteins predicted from the transcriptome of Pratylenchus penetrans. (A) For each proline (P), the probability of neighbouring positions (n + 1, n + 2, n + 3, etc.) also containing a proline was calculated. (B) For the proline‐rich effectors, positions n + 1, n + 3, n + 6 and n + 9 to a proline appear to be enriched for another proline (dark blue), when compared with the randomized primary amino acid sequence (purple). (C) No such enrichment is observed in any position for all other similarly proline‐rich proteins in the transcriptome dataset.
Putative promoter motifs associated with subventral gland expression
To determine whether the identified non‐coding promoter motifs are associated with gland cell expression in P. penetrans [as found previously for other PPNs (Eves‐van den Akker et al., 2016)], we identified the putative promoter regions of gland cell‐expressed transcripts in the available draft genome sequence (Denver et al., 2016). Given that this genome sequence was produced from a very low‐coverage skim, where possible, approximately 500 nucleotides of the 5′ sequence from the start codon were manually extracted based on blastn coordinates. The promoter regions of eight dorsal gland‐expressed transcripts and 14 subventral gland‐expressed transcripts were compared with a set of 28 promoters of transcripts not predicted to encode effectors (including those with experimentally verified non‐gland cell expression, e.g. egg, vulva region and amphids), using the differential motif discovery algorithm HOMER. The sequences of the identified promoter regions for the different candidate effector genes used are listed in Table S2 (see Supporting Information). A motif of the consensus sequence CAA[A|G|T|C]TG[T|G]C was identified as enriched in the subventral gland set (Figs 7A,B and S4, see Supporting Information). Given the nature of the genome skim assemblies for P. penetrans, and the consequent lack of gene calls, a global analysis of this motif's presence and frequency in P. penetrans promoters is not currently possible. However, we were able to show that the presence of this motif is not enriched in the sedentary PPNs Meloidogyne hapla and Globodera pallida (Fig. 7C,D), and multiple copies of the motif in the promoters of genes in these species cannot be used as a consistent predictor of secreted proteins, as was the case for the unrelated, but conceptually analogous, Dorsal Gland Box sequence of cyst nematodes (Eves‐van den Akker et al., 2016).
Figure 7.
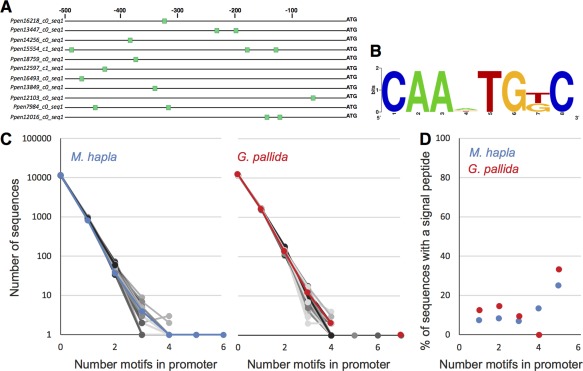
Identification of a non‐coding motif in the upstream region of the start codon associated with gland cell expression in Pratylenchus penetrans. (A) Each bar shows the distribution of the motif within 500 nucleotides upstream of the start codon. The annotation of each transcript can be found in Table 3. (B) Graphic representation of the consensus motif sequence. (C) In related plant‐parasitic nematodes with well‐annotated genomes available (Meloidogyne hapla and Globodera pallida), the number of promoter regions with multiple copies of this motif does not deviate from random. Normal promoter regions are shown in blue for M. hapla and red for G. pallida; 250 iterations of randomization of the sequence of each promoter region are shown in grey. (D) An increased number of motifs in the promoter region does not correlate with a greater chance of the corresponding gene encoding a predicted signal peptide in either species.
Expression of P. penetrans gland cell genes at different developmental stages
As most stages of P. penetrans are motile (with the exception of eggs and J1s), with the capacity to invade and migrate throughout the roots, we conducted semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analyses in order to detect transcripts at different nematode developmental stages [eggs, juveniles (J2–J4), adult females and adult males] (Fig. S5, see Supporting Information). Our results suggest that all motile stages are able to express the panel of effector genes described above. In some cases, the expression of some effectors could also be detected within the eggs, probably resulting from the non‐hatched second‐stage juveniles (J2s). The stage specificity of the different batches of cDNA was validated using the Pp‐18S rDNA gene as a constitutive gene (Fig. S5) and a pioneer gene (Ppen13485_c0_seq1) found to be specifically expressed in females (Fig. S5).
Expression profiles of P. penetrans effectors during infection in planta
To substantiate the involvement of the different effector candidates during root infection, quantitative RT‐PCR analyses were conducted to assess their transcription profiles at different time points after nematode infection. The time points were determined over a 10‐day infection time course in soybean hairy roots (Fig. 8). One day after inoculation (DAI), a mixture of juvenile and adult stages was observed feeding both ecto‐ and endoparasitically, with some nematodes reaching the inner layers of the roots (Fig. 8D). At this time, eggs were not observed within the root tissues. At 3 DAI, both juveniles and adult stages could be seen migrating and well established in different areas of the roots (Fig. 8E), whereas, at 7 DAI, a greater number of nematodes (including deposition of eggs by females) were observed within the inner layers of the roots (Fig. 8F). Consistent with the increased number of nematodes associated with the hairy roots, a discoloration of the roots could be observed in different areas parasitized by the nematodes (Fig. 8A–C).
Figure 8.
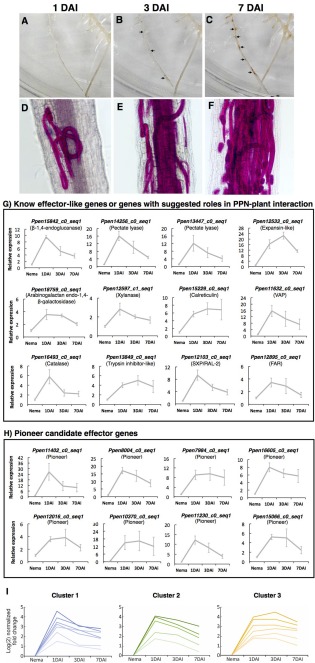
Expression profile of 20 Pratylenchus penetrans candidate effectors during the early time points of plant infection. (A–C) Symptom development of soybean hairy roots after P. penetrans infection at 1 (A), 3 (B) and 7 days after inoculation (DAI) (C), with arrows indicating root lesions. (D–F) Acid fuchsin staining of nematodes within soybean hairy roots at 1, 3 and 7 DAI, respectively. (G, H) The relative transcript expression value for each candidate effector gene was quantified by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) at 1, 3 and 7 DAI relative to the expression level of the 18S rDNA gene and using the transcription expression levels of nematodes not established within roots (Nema) as baseline. (I) The normalized expression values were used for clustering analysis, suggesting the occurrence of three expression clusters of the different candidate effectors.
We then established the expression profiles of 20 candidate effectors specifically expressed within the glands at 1, 3 and 7 DAI (Fig. 8G,H). For the control, RNA extracted from nematodes not yet established within the roots was used as the main reference. Most of the nematode effector genes were transcriptionally induced during infection and establishment of nematodes within roots. When individual levels of expression were compared, several of the pioneer candidate effectors were amongst the highest expressed transcripts during infection (e.g. Ppen11402_c0_seq1, Ppen8004_c0_seq1, Ppen10370_c0_seq1 and Ppen11230_c0_seq1), whereas transcripts encoding an expansin (Ppen12533_c0_seq1), two pectate lyases (Ppen14256_c0_seq1 and Ppen13447_c0_seq1), a VAP (Ppen11632_c0_seq1) and one β‐1,4‐endoglucanase (Ppen15842_c0_seq1) were amongst the top highly expressed genes with known annotation. The normalized expression values were then used for clustering analysis in order to visualize the expression patterns of the different candidate effectors. Three expression clusters were obtained when analysing 20 nematode candidate effectors according to their temporal expression levels (Fig. 8I). The profiles revealed that the expression of the majority of the transcripts tested peaked at 1 DAI, when nematodes became established within the host, followed by a consistent or decreased accumulation at 3 and 7 DAI, suggesting that this panel of effectors is likely to play an important role during the interaction of P. penetrans and the host.
Discussion
The purpose of this study was to identify and validate effector genes of P. penetrans, as very little is known about the infection mechanism adopted by this group of nematodes. Here, we provide novel insights into the catalogue of candidate effector genes of P. penetrans, covering different functional categories of known PPN effector genes, but also a large number of genes encoding proteins with unknown functions.
The expanded effector repertoire of P. penetrans, described herein, can be rationally subdivided into several apparently distinct functional groups based on sequence analysis. Consistent with previous findings for other PPNs, a significant number of genes encode different families of CWDEs or modifying enzymes (e.g. GH5, GH30, GH53, PL3 and expansin‐like proteins). We confirm that a subset of these is specifically expressed in the oesophageal glands of P. penetrans during infection. CWDEs are one of the few unifying features of PPN effector repertoires, and their similarity to bacterial or fungal genes, but absence in almost all other metazoans, implies acquisition by horizontal gene transfer (Danchin et al., 2010; Smant et al., 1998). The secretion of CWDEs by PPNs is hypothesized to facilitate penetration and migration through host tissue by softening or modifying the plant cell wall (e.g. Rosso et al., 1999; Smant et al., 1998; Wang et al., 1999). High cellulase and proteolytic enzyme activity has been found in P. penetrans homogenates (Morgan and McAllan, 1962), and the identification of these genes within the oesophageal glands suggests that these CWDEs might be secreted during parasitism.
Following the invasion of roots by plant pathogens, the activation of the plant immune system is considered to be a prominent feature (Jones and Dangl, 2006). The response of plants to RLNs is characterized by the dynamic expression of genes associated with defence pathways, including the production of secondary plant metabolites (Backiyarani et al., 2014; Yu et al., 2015; Zhu et al., 2014). The suppression of host defence responses is critical to successful colonization. In this context, VAPs are a conserved family of proteins through the Phylum and have been implicated in the suppression of host immunity (Gao et al., 2001; Lozano‐Torres et al., 2012, 2014). Globodera rostochiensis VAP1 (GrVAP1) has been shown to interact with the papain‐like cysteine protease Rcr3pim in tomato (Solanum lycopersicum L.), and this interaction perturbs the protease active site, resulting in increased plant susceptibility to the nematode (Lozano‐Torres et al., 2012), whilst silencing of this gene reduces nematode infectivity (Lozano‐Torres et al., 2014). Accordingly, overexpression of Hs‐VAP1 and Hs‐VAP2 increases infection by Heterodera schachtii (Lozano‐Torres et al., 2014). It will be interesting to explore whether VAPs in RLNs function similarly, and whether perturbation of their activity can be exploited to generate resistance towards RLNs as well.
There is increased evidence that PPNs harbour a significant number of genes that are involved in protection against host defences (Goverse and Smant, 2014). The effector repertoire of P. penetrans also includes a highly expressed catalase with a predicted N‐terminal signal peptide sequence. Catalases are found in most living organisms and provide protection against oxidative damage by the catalysis of ROS (Chelikani et al., 2004). An oxidative burst is one of the earliest defence responses to plant pathogen attack. The transient accumulation of ROS helps to defend the host from invading pathogens and can also act as a signalling molecule to trigger various other plant defence responses (Goverse and Smant, 2014). PPNs across the Phylum have apparently independently evolved a number of secreted proteins that may be involved directly or indirectly in the metabolism of host ROS [e.g. superoxidase dismutase, glutathione peroxidases, glutathione transferase (GST)] (Bellafiore et al., 2008; Espada et al., 2016; Jones et al., 2004). The resistance of some cultivars to RNLs has been linked to a strong capacity of plants to produce ROS, whereas, in susceptible varieties, a weaker production of ROS has been registered (Kathiresan and Mehta, 2005). It is interesting to note that secreted catalases have been proposed as virulence factors in pathogenic fungi, providing evidence that extracellular catalases could participate in the neutralization of ROS (Barek et al., 2015; Robbertse et al., 2003). The putative secretion of a catalase by P. penetrans is intriguing and, in this context, it will be interesting to analyse the role of this catalase through this nematode–plant interaction.
Proteases and protease inhibitors are present in the secretome of PPNs (e.g. Bellafiore et al., 2008; Shinya et al., 2013), and transcriptome analyses of P. penetrans have revealed a wide range of putatively secreted proteases/protease inhibitors for this species (Vieira et al., 2015). Although nematodes possess hundreds of protease encoding genes (Castagnone‐Sereno et al., 2011), only a portion of these will be ultimately secreted into the plant tissue, as suggested by the different proteases found within the intestine of P. penetrans. Likewise, protease inhibitors are highly abundant in the proteome of APNs (Hunt et al., 2017). These secreted proteases are known to participate in a wide spectrum of functions, including penetration and invasion of the host tissues (Zhu et al., 2014), acquisition of resources from the host and modulation of the host immune response (Balasubramanian et al., 2010; Hunt et al., 2017; Schwarz et al., 2015). In PPNs, the oesophageal gland cells are the major secretory tissues involved in effector delivery and host immune modulation (Mitchum et al., 2013). In APNs, the E/S system is considered to be the major component of the host immunomodulatory machinery (Hewitson et al., 2009). Of the panel of P. penetrans proteases studied, we specifically localized transcripts encoding a trypsin inhibitor‐like protein to the oesophageal gland cells, but, interestingly, also transcripts of several proteases to the E/S system. Given that a similarly specific expression pattern has been reported for two unrelated pioneer gene sequences of the plant‐parasitic Meloidogyne graminicola (Haegeman et al., 2013), the E/S system of PPNs may be more important in parasitism than previously appreciated, for migratory and sedentary PPNs alike.
Other candidate effectors expressed in the oesophageal glands of P. penetrans included a FAR gene and one gene of the SXP/RAL‐2 family. Both families are specific to nematodes. Similar to our results, transcripts of a FAR gene have been detected in the oesophageal glands of Bursaphelenchus xylophilus (Espada et al., 2016). Although the function of the FAR family members in PPNs is still relatively obscure, a correlation between the secretion of FAR‐1 by the hypodermis of cyst and root‐knot nematodes and host defence interaction has been established (Iberkleid et al., 2013; Prior et al., 2001). FAR‐1 binds a broad range of fatty acid precursors of the jasmonate signalling pathway [e.g. linolenic and linoleic acids (Prior et al., 2001)]. In P. penetrans, knockdown of FAR‐1 by plant‐mediated RNA interference (RNAi) resulted in a significant reduction of nematode propagation (Vieira et al., 2017a), consistent with a role in parasitism for this migratory species. Members of the SXP/RAL‐2 family are characterized by the presence of the Domain of Unknown Function‐DUF148 protein (Rao et al., 2000). Although their roles in pathogenicity have yet to be determined, silencing of an SXP/RAL2 gene in P. zeae resulted in a significant reduction in nematodes after the inoculation of carrot discs (Fosu‐Nyarko et al., 2016). The differential spatial expression, e.g. amphids or hypodermis of G. rostochiensis (Jones et al., 2000), oesophageal glands of P. zeae (Fosu‐Nyarko et al., 2016) and M. incognita (Tytgat et al., 2005), and our results, suggests multifaceted functions for this family.
In addition to the identification of conserved features between RLN and other PPN effectors, our results revealed eight new pioneer candidate effectors for P. penetrans. Most of these pioneer sequences are not annotatable in Pfam and identify no similar sequences by blast analyses in a panel of PPN genomes and transcriptomes across the Phylum. These apparently RLN‐specific effectors suggest an adaptation to the particular lifestyle of these species, or at least to P. penetrans. Attributing a function to such taxonomically restricted and apparently unique genes is challenging. Nevertheless, it is interesting to note that most of these pioneers are extremely proline‐rich (up to 25% of the primary amino acid sequence). Furthermore, prolines are not evenly distributed across this set of predicted proteins, but preferentially excluded from the first 20% and grouped into tandem arrays of proline pairs and/or triplets. Using the current datasets of P. penetrans, both of these phenomena appear to be specific features to these effectors. It is well documented that infection by RLNs induces the production and accumulation of tannin‐like deposits (Townshend et al., 1989; Vieira et al., 2017b). Tannins are astringent polyphenols induced on wounding and may contribute to the induced defence response (War et al., 2012). To counter this, many herbivores secrete tannin‐binding salivary proteins, which typically contain a high proportion of proline (Shimada, 2006). Whether P. penetrans proline‐rich pioneers function similarly remains to be tested.
The similarity amongst the effector genes of P. penetrans and other PPNs continues to support the idea of a parasitism strategy‐independent, ‘pan‐nematode’, effector repertoire (Bird et al., 2015). However, juxtaposed to this are the bewildering, and apparently species‐specific, pioneer effectors. The size of the effector repertoire seems to be correlated with the perceived ‘complexity’ of the nematode feeding strategy: a substantially larger number of effectors have been identified for sedentary nematodes (Abad et al., 2008; Danchin et al., 2010; Eves‐van den Akker et al., 2016; Thorpe et al., 2014), many of which are part of large multigene families (Eves‐van den Akker et al., 2016). The fact that RLNs do not induce the formation of a feeding site in planta presumably excludes a priori certain effectors involved in the formation of giant cells or syncytia (Fosu‐Nyarko and Jones, 2016), and may explain the apparently smaller number of effectors present in P. penetrans compared with other species. One constraint for the comprehensive identification of nematode effector repertoires lies in the relatively crude prediction pipelines. The strategies employed herein allowed us to identify a number of previously described and novel effectors for P. penetrans. Using these experimentally verified oesophageal gland cell‐expressed genes, we have identified a non‐coding promoter motif that appears to be associated with gland cell expression in P. penetrans [conceptually similar but sequence unrelated to the DOG box of Globodera effectors (Eves‐van den Akker et al., 2016)]. We anticipate that this motif may provide an additional useful criterion to expedite future effector prediction pipelines for this group of nematodes once complete and annotated genome sequences are available, and its accuracy can be validated.
Overall, we present a comprehensive set of candidate effectors of P. penetrans. We provide continued support for the presence of ‘common’ PPN effectors and implicate novel effectors in the parasitism process of RLNs. The unique composition and perhaps even delivery strategy of RLN effectors highlight the lack of knowledge for these species. This study provides an important prelude towards detailed functional analyses, and a platform for effector biology. Given the importance of effectors to parasitism, the expanded and novel effector repertoire of P. penetrans represents a series of new targets for the development of biotechnological alternatives to host resistance.
Experimental Procedures
Nematode collection and nematode extraction
Pratylenchus penetrans isolate (NL 10p RH) collected in Beltsville (MD, USA) was routinely multiplied in vitro in roots of corn (Zea mays cv. ‘Iochief’) growing in Murashige and Skoog (MS) medium agar plates. Nematodes were re‐cultured every 2 months onto new ex‐roots of corn and maintained in the dark at 25 °C.
Pratylenchus penetrans gene selection
Two distinct next‐generation sequencing datasets were used to identify a panel of putative effectors: (i) a subset of 1330 transcripts encoding for putatively secreted proteins from the de novo transcriptome assembly of P. penetrans, ranked according to normalized transcript abundance during root infection (Vieira et al., 2015); and (ii) a set of 454 reads derived from mRNA collected from the oesophageal glands of P. penetrans (Maier et al., 2013). These oesophageal gland cell reads were mapped to the 1330 transcripts encoding putatively secreted proteins using CLC Genomics v. 8 with default parameters. Relative transcript abundance was calculated based on RKPM values (reads per kilobase per million mapped reads).
blastp (e‐value cutoff of 1e−5 and bitscore > 50) was used to compare all 1330 putatively secreted proteins with sequences in the NR database and the proteomes of Clade 12 (van Megen et al., 2009) sedentary species [root‐knot nematodes Meloidogyne incognita (Abad et al., 2008) and M. hapla (Opperman et al., 2008) and cyst nematodes Globodera pallida (Cotton et al., 2014) and G. rostochiensis (Eves‐van den Akker et al., 2016)], Clade 12 migratory species [Ditylenchus destructor (Zheng et al., 2016) and Clade 10 B. xylophilus (Kikuchi et al., 2011)] and, finally, the Clade 9 free‐living species C. elegans (http://parasite.wormbase.org). Local tblastn searches were performed against the transcriptomes of additional Pratylenchidae species, namely P. coffeae (Haegeman et al., 2011), P. thornei (Nicol et al., 2012), P. vulnus [National Center for Biotechnology Information (NCBI) data], P. zeae (Fosu‐Nyarko et al., 2016) and the burrowing nematode Radopholus similis (Jacob et al., 2008).
RNA extraction and cDNA libraries
Total RNA was extracted from individual life stages [eggs, juveniles (J2–J4), adult females or males], or from a pool of mixed stages of P. penetrans, using the RNeasy Plant Mini kit (QIAGEN, GmbH, Hilden, Germany), following the manufacturer's instructions. RNA was treated with RNase‐free DNase (QIAGEN, GmbH, Hilden, Germany) before reverse transcription. The quantity and quality of the extracted RNA were assessed by an ND‐1000 NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and cDNA was synthesized using the iScript first‐strand synthesis kit (Bio‐Rad, Hercules, CA, USA) following the manufacturer's instructions.
In situ hybridization
Whole‐mount in situ hybridizations were performed in all stages of P. penetrans following the protocol of de Boer et al. (1998). Specific primers were designed to amplify a range of gene products varying from 170 to 300 nucleotides (Table S3, see Supporting Information) using the cDNA library produced from the mixed pool of P. penetrans stages. The resulting PCR products were used as template for the generation of sense and antisense DIG‐labelled probes, using a digoxigenin (DIG)‐nucleotide labelling kit (Roche, Indianapolis, IN, USA). Hybridized probes within the nematode tissues were detected using an anti‐DIG antibody conjugated to alkaline phosphatase and its substrate. Nematode sections were then observed using a Nikon Eclipse 5i light microscope (Melville, NY, USA).
Genetic characterization of P. penetrans candidate effectors
Focusing on a subset of candidate effectors with verified oesophageal gland cell expression in P. penetrans, additional in silico analyses were performed. Open reading frames were used to perform blastn searches (e‐value > 1e−10) against the low‐coverage genome skim of P. penetrans (Denver et al., 2016; I. A. Zasada, unpublished data, 2017). The most similar sequences were manually examined, and each transcript sequence was aligned to the respective genomic scaffold using MUSCLE (Edgar, 2004). Genomic sequences with >90% identities were submitted to FGENESH (www.softberry.com) for exon–intron prediction (Solovyev et al., 2006) and corresponding protein prediction. Gene schematics for predicted complete genes were generated with the Exon–Intron Graphic maker available at WormWeb.org. The protein sequences obtained from transcripts (transcriptome data) were then aligned to the respective genome predicted protein by MUSCLE (Edgar, 2004), and pairwise similarities were calculated using the software CLC Main Workbench v.9. SIGNALP v. 4.0 was used to confirm the presence/absence of protein signal peptide in the genome predicted proteins (Petersen et al., 2011). Proteins were scanned for InterPro scan and PFAM domain search using Blast2GO (Conesa et al., 2005) with default parameters. The PSORTII algorithm was used to predict the subcellular localization of the candidate effector protein sequences. Cysteine and proline contents were calculated for each predicted mature protein with CLC Main Workbench v.7.
Proline analyses
Proline distribution across all proline‐rich effectors, all other effectors, all other secreted proteins and all other proteins encoded in the transcriptome of P. penetrans was calculated. Proteins of interest were divided into 10 equal length fragments across their entire length (where possible), and the percentage of proline residues in each fragment was calculated using custom python script 1 (Script1_calculate_Proline_distributions.py, https://github.com/sebastianevda). The probability of randomly selecting five putatively secreted proteins that all exclude prolines from the first 20% of their open reading frame was empirically estimated to be 2/250 (or P = 0.008). To calculate the probability that residues adjacent to a proline in positions n + 1 to n + 9 are also a proline, custom python scripts 2 and 3 were used (Script2_calculate_next_letter_P_percent.py, Script3_calculate_next_letter_P_percent_random_250.py, https://github.com/sebastianevda).
Promoter analyses
To determine whether we were able to identify a non‐coding promoter motif that is descriptive of gland cell expression in P. penetrans, as for other PPNs (Eves‐van den Akker et al., 2016), we identified the putative promoter regions of gland cell‐expressed transcripts in the available draft genome sequence (Denver et al., 2016). Given that this genome sequence was produced from a very low‐coverage skim, and no gene calls are available, where possible, approximately 500 nucleotides of the 5′ sequence from the start codon were manually extracted based on blastn coordinates. The promoter regions of eight dorsal gland‐expressed transcripts and 14 subventral gland‐expressed transcripts were compared with a set of 28 promoters of transcripts not predicted to encode effectors (including those with experimentally verified non‐gland cell expression, e.g. egg, vulva region and amphids), using the differential motif discovery algorithm HOMER (Heinz et al., 2010). Instances of the motif were identified in FASTA sequences of promoter regions using the FIMO web server. The consensus sequences for the identified motifs were analysed using the WebLogo 3 program (http://weblogo.threeplusone.com/).
Developmental expression of candidate effectors at different nematode stages
The different nematode effectors of P. penetrans were amplified from the cDNA libraries generated for each nematode development stage (eggs, juveniles J2–J4, females and males) using the same primers as employed for the in situ hybridization protocol. Semi‐quantitative RT‐PCRs were conducted for transcript detection of each stage‐specific cDNA library, with the following PCR programme: 2 min at 94 °C; 38 cycles of 30 s at 94 °C, 30 s at 57 °C and 30 s at 72 °C; and one cycle of 72 °C for 10 min. The PCRs contained equal amounts of cDNA, 1 × PCR buffer, 1 U Taq polymerase (Invitrogen, Carlsbad, CA, USA) and 0.2 µm of each primer in a total solution of 50 µL. PCR products were separated by electrophoresis on a 1% agarose gel using TBE buffer [0.045 m Tris‐borate, 0.001 m ethylenediaminetetraacetic acid (EDTA), pH 8.0] and visualized using SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA, USA). The P. penetrans 18S rDNA gene, used as a control constitutive gene, and a pioneer gene specifically expressed in females were employed as controls of the different nematode stage cDNA library.
Plant inoculation and differential expression analyses of P. penetrans candidate effectors during infection in planta
Nematode sterilization and infection of soybean hairy roots followed the protocol described in Vieira et al. (2015). To follow the early steps of nematode infection, inoculated roots were stained with acid fuchsin following Byrd et al. (1983) from 1 to 10 DAI. Root tissues were then destained using a clearing solution (equal volumes of lactic acid, glycerol and distilled water) for 2–4 h at room temperature. After rinsing several times with tap water, roots containing nematodes were stored in acidified glycerol (five drops of 1.0 m HCl in 50 mL of glycerol) and observed using a Nikon Eclipse 50i light microscope.
To quantify the expression levels of P. penetrans candidate effector genes, total RNA was extracted from a pool of six infected soybean hairy root systems at 1, 3 and 7 DAI. Nematodes not yet established within the roots at 1 DAI were washed out from the medium and used for RNA extraction. The expression levels of transcripts from nematodes collected from the medium were used as baseline in comparison with the expression levels of transcripts from nematodes within the roots at the different time points. Specific primers were designed to amplify individual fragments of each candidate effector gene, and a 148‐bp fragment of the P. penetrans 18S rDNA gene was used as reference (Table S3). Quantitative real‐time RT‐PCR included 3.5 µL of SYBR green mix (Roche), 1 µL of 5 µm primers and 100 ng cDNA. Reactions were performed on a CFX96 Real‐time system machine (Bio‐Rad). The amplification reactions were run using the following programme: a hot start of 95 °C for 3 min, and then 40 cycles of 95 °C for 10 s and 60 °C for 30 s. After 40 cycles, a melt curve analysis or dissociation programme (95 °C for 15 s, 60 °C for 15 s, followed by a slow ramp from 60 to 95 °C) was performed to ensure the specificity (above 90%) of amplification. Three independent biological experiments were conducted by quantitative RT‐PCR, using three technical replicates for each independent experiment. Data analyses were performed using the CFX MANAGER v. 3 software (Bio‐Rad). The values of the relative normalized expression of each gene were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001), relative to the expression levels of the P. penetrans 18S rDNA gene and using the transcript expression levels of the non‐root established nematodes at 1 DAI as baseline.
Accession numbers
Raw RNAseq reads used in this publication are available under SRA accession PRJNA432986 and PRJNA304159. The predicted coding sequence (CDS) and corresponding predicted amino acid sequences of transcripts localized within the nematode tissues are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4h44313.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Summary of blast hit analyses of 1330 transcripts of Pratylenchus penetrans against the non‐redundant GenBank database and transcript quantification.
Table S2 List of transcripts and respective promoter sequences used for the identification of a non‐coding motif in the upstream region of the start codon associated with gland cell expression in Pratylenchus penetrans.
Table S3 List of primers.
Fig. S1 Distribution of transcripts encoding secreted proteins identified in different Pratylenchus penetrans datasets. (A) Venn diagram showing the number of nematode transcripts recovered from the nematode oesophageal glands versus the in planta datasets, when mapped against the full set of 1330 nematode transcripts encoding for predicted secreted proteins without transmembrane domains identified by the de novo assembly of the transcriptome of P. penetrans (Vieira et al., 2015). A complete description of the nematode transcripts is shown in Table S1. (B) Total number of annotated versus non‐annotated protein sequences by homology searches against the non‐redundant National Center for Biotechnology Information (NCBI) database of each nematode dataset.
Fig. S2 Detection of gene transcripts encoding pioneer genes of Pratylenchus penetrans by in situ hybridization in different nematode tissues. Whole‐mount in situ hybridization was performed using mixed stages of nematodes incubated with antisense probes (brown coloration) amplified from cDNA of P. penetrans. Transcripts of predicted pioneer genes localized in: (A) developing egg within the female (Ppen12587_c0_seq1); (B) surrounding the vulva region (Ppen13485_c0_seq1); (C) two dots‐like posterior to the medium bulb (Ppen14681_c0_seq1); (D) two dots‐like below the cuticle level (Ppen14446_c0_seq1); (E, F) testis region (Ppen14188_c0_seq1 and Ppen14399_c0_seq1, respectively). m, medium bulb; s, stylet. Bars, 20 µm.
Fig. S3 Prediction of gene structure of Pratylenchus penetrans candidate effectors with corresponding transcripts localized within the oesophageal glands. Only genes with complete genomic sequences obtained after blast analyses against the skim genome assemblies of P. penetrans were used. Exons are illustrated as black boxes and introns as black lines. Scale, 100 bases. FAR, fatty acid‐ and retinol‐binding gene; VAP, venom allergen‐like gene.
Fig. S4 Alignment of non‐coding promoter motif sequences associated with gland cell expression transcripts of Pratylenchus penetrans. Ppen12016_c0_seq1, pioneer; Ppen16493_c0_seq1, catalase; Ppen15554_c1_seq1, expansin‐like; Ppen14256_c0_seq1 and Ppen13447_c0_seq1, pectate lyases; Ppen12103_c0_seq1, SXP/RAL‐2; Ppen16218_c0_seq1, β‐1,4‐endoglucanase; Ppen18759_c0_seq1, arabinogalactan endo‐1,4‐β‐galactosidase; Ppen12597_c1_seq1, xylanase; Ppen13849_c0_seq1, trypsin inhibitor‐like; Ppen7984_c0_seq1, pioneer.
Fig. S5 Expression pattern of Pratylenchus penetrans effector candidate genes specifically localized in the oesophageal glands and detected by semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) in different nematode developmental stages. As positive control, each nematode developmental cDNA library [eggs, juveniles (J2–J4), females and males] was amplified using the primers of the 18S rDNA gene and a pioneer gene (Ppen13485_c0_seq1) specific to females. FAR, fatty acid‐ and retinol‐binding gene; VAP, venom allergen‐like gene.
Acknowledgements
The authors acknowledge the technical assistance of Sarah Shih. This material is based on work supported by the National Institute of Food and Agriculture, US Department of Agriculture, under grant number 2015‐67012‐22834. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. S.E.‐vdA. was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/M014207/1. This work was also supported by Hatch Act and State of Iowa funds.
References
- Abad, P. , Gouzy, J. , Aury, J.‐M. , Castagnone‐Sereno, P. , Danchin, E.G.J. , Deleury, E. , Perfus‐Barbeoch, L. , Anthouard, V. , Artiguenave, F. , Blok, V.C. , Caillaud, M.‐C. , Coutinho, P.M. , Dasilva, C. , De Luca, F. , Deau, F. , Esquibet, M. , Flutre, T. , Goldstone, J.V. , Hamamouch, N. , Hewezi, T. , Jaillon, O. , Jubin, C. , Leonetti, P. , Magliano, M. , Maier, T.R. , Markov, G.V. , McVeigh, P. , Pesole, G. , Poulain, J. , Robinson‐Rechavi, M. , Sallet, E. , Segurens, B. , Steinbach, D. , Tytgat, T. , Ugarte, E. , van Ghelder, C. , Veronico, P. , Baum, T.J. , Blaxter, M. , Bleve‐Zacheo, T. , Davis, E.L. , Ewbank, J.J. , Favery, B. , Grenier, E. , Henrissat, B. , Jones, J.T. , Laudet, V. , Maule, A.G. , Quesneville, H. , Rosso, M.‐N. , Schiex, T. , Smant, G. , Weissenbach, J. and Wincker, P. (2008) Genome sequence of the metazoan plant‐parasitic nematode Meloidogyne incognita . Nat. Biotechnol. 26, 909–915. [DOI] [PubMed] [Google Scholar]
- Backiyarani, S. , Uma, S. , Arunkumar, G. , Saraswathi, M.S. and Sundararaju, P. (2014) Differentially expressed genes in incompatible interactions of Pratylenchus coffeae with Musa using suppression subtractive hybridization. Physiol. Mol. Plant Pathol. 86, 11–18. [Google Scholar]
- Balasubramanian, N. , Toubarro, D. and Simões, N. (2010) Biochemical study and in vitro insect immune suppression by a trypsin‐like secreted protease from the nematode Steinernema carpocapsae . Parasite Immunol. 32, 165–175. [DOI] [PubMed] [Google Scholar]
- Barek, B. , Cordewener, J.H. , van der Lee, T.A. , America, A.H. , Mirzadi Gohari, A. , Mehrabi, R. , Hamza, S. , de Wilt, P.J. and Kema, G.H. (2015) Proteome catalog of Zymoseptoria tritici captured during pathogenesis in wheat. Fungal Genet. Biol. 79, 42–53. [DOI] [PubMed] [Google Scholar]
- Bellafiore, S. , Shen, Z. , Rosso, M.‐N. , Abad, P. , Shih, P. and Briggs, S.P. (2008) Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog. 4, e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, D.M. , Jones, J.T. , Opperman, C.H. , Kikuchi, T. and Danchin, E.G.J. (2015) Signatures of adaptation to plant parasitism in nematode genomes. Parasitology, 142, S71–S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer, J.M. , Yan, Y. , Smant, G. , Davis, E.L. and Baum, T.J. (1998) In‐situ hybridization to messenger RNA in Heterodera glycines . J. Nematol. 30, 309–312. [PMC free article] [PubMed] [Google Scholar]
- Burke, M. , Scholl, E.H. , Bird, D.M.K. , Schaff, J.E. , Colman, S.D. , Crowell, R. , Diener, S. , Gordon, O. , Graham, S. , Wang, X. , Windham, E. , Wright, G.M. and Opperman, C.H. (2015) The plant parasite Pratylenchus coffeae carries a minimal nematode genome. Nematology, 17, 621–637. [Google Scholar]
- Byrd, D.W. , Kirkpatrick, T. and Barker, K.R. (1983) An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143. [PMC free article] [PubMed] [Google Scholar]
- Castagnone‐Sereno, P. , Deleury, E. , Danchin, E.G.J. , Perfus‐Barbeoch, L. and Abad, P. (2011) Data‐mining of the Meloidogyne incognita degradome and comparative analysis of proteases in nematodes. Genomics, 97, 29–36. [DOI] [PubMed] [Google Scholar]
- Castillo, P. and Vovlas, N. (2007) Pratylenchus (Nematoda: Pratylenchidae): diagnosis, biology, pathogenicity and management In: Nematology Monographs and Perspectives, Vol. 6 (Hunt D. J. and Perry R. N., eds), p. 529 Leiden, Netherlands: Brill. [Google Scholar]
- Chelikani, P. , Fita, I. and Loewen, P.C. (2004) Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61, 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa, A. , Gotz, S. , Garcia‐Gomez, J.M. , Terol, J. , Talon, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cotton, J.A. , Lilley, C.J. , Jones, L.M. , Kikuchi, T. , Reid, A.J. , Thorpe, P. , Tsai, I. , Beasley, H. , Blok, V. , Cock, P.J.A. , Eves‐van den Akker, S. , Holroyd, N. , Hunt, M. , Mantelin, S. , Naghra, H. , Pain, A. , Palomares‐Rius, J.E. , Zarowiecki, M. , Berriman, M. , Jones, J.T. and Urwin, P.E. (2014) The genome and life‐stage specific transcriptomes of Globodera pallida elucidate key aspects of plant parasitism by a cyst nematode. Genome Biol. 15, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin, E.G.J. , Rosso, M.‐N. , Vieira, P. , de Almeida‐Engler, J. , Coutinho, P.M. , Henrissat, B. and Abad, P. (2010) Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. USA, 107, 17 651–17 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D.R. , Brown, A.M.V. , Howe, D.K. , Peetz, A.B. and Zasada, I.A. (2016) Genome skimming: a rapid approach to gaining diverse biological insights into multicellular pathogens. PLoS Pathog. 12, e1005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, G. , Magliano, M. , Deleury, E. , Abad, P. and Rosso, M.‐N. (2007) Transcriptome analysis of root‐knot nematode functions induced in the early stages of parasitism. New Phytol. 176, 426–436. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada, M. , Silva, A.C. , Eves van den Akker, S. , Cock, P.J. , Mota, M. and Jones, J.T. (2016) Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy. Mol. Plant Pathol. 17, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves‐van den Akker, S. , Lilley, C.J. , Jones, J.T. and Urwin, P.E. (2014) Identification and characterisation of a hyper‐variable apoplastic effector gene family of the potato cyst nematodes. PLoS Pathog. 10, e1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves‐van den Akker, S. , Laetsch, D.R. , Thorpe, P. , Lilley, C.J. , Danchin, E.G.J. , Da Rocha, M. , Rancurel, C. , Holroyd, N.E. , Cotton, J.A. , Szitenberg, A. , Grenier, E. , Montarry, J. , Mimee, B. , Duceppe, M.‐O. , Boyes, I. , Marvin, J.M.C. , Jones, L.M. , Yusup, H.B. , Lafond‐Lapalme, J. , Esquibet, M. , Sabeh, M. , Rott, M. , Overmars, H. , Finkers‐Tomczak, A. , Smant, G. , Koutsovoulos, G. , Blok, V. , Mantelin, S. , Cock, P.J.A. , Phillips, W. , Henrissat, B. , Urwin, P.E. , Blaxter, M. and Jones, J.T. (2016) The genome of the yellow potato cyst nematode, Globodera rostochiensis, reveals insights into the basis of parasitism and virulence. Genome Biol. 17, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanelli, E. , Troccoli, A. , Pousis, C. and De Luca, F. (2014) Molecular characterization and functional analysis of four β‐1,4‐endoglucanases from the root‐lesion nematode Pratylenchus vulnus . Plant Pathol. 63, 1436–1445. [Google Scholar]
- Fosu‐Nyarko, J. and Jones, M.G.K. (2016) Advances in understanding the molecular mechanisms of root lesion nematode host interactions. Annu. Rev. Phytopathol. 54, 253–278. [DOI] [PubMed] [Google Scholar]
- Fosu‐Nyarko, J. , Tan, C.H.J.‐A. , Gill, R. , Agrez, V.G. , Rao, U. and Jones, M.G.K. (2016) De novo analysis of the transcriptome of Pratylenchus zeae to identify transcripts for proteins required for structural integrity, sensation, locomotion and parasitism. Mol. Plant Pathol. 17, 532–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B. , Allen, R. , Maier, T. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2001) Molecular characterisation and expression of two venom allergen‐like protein genes in Heterodera glycines . Int. J. Parasitol. 31, 1617–1625. [DOI] [PubMed] [Google Scholar]
- Goverse, A. and Smant, G. (2014) The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 52, 243–265. [DOI] [PubMed] [Google Scholar]
- Haegeman, A. , Joseph, S. and Gheysen, G. (2011) Analysis of the transcriptome of the root lesion nematode Pratylenchus coffeae generated by 454 sequencing technology. Mol. Biochem. Parasitol. 178, 7–14. [DOI] [PubMed] [Google Scholar]
- Haegeman, A. , Bauters, L. , Kyndt, T. , Rahman, M.M. and Gheysen, G. (2013) Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 14, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S. , Benner, C. , Spann, N. , Bertolino, E. , Lin, Y.C. , Laslo, P. , Cheng, J.X. , Murre, C. , Singh, H. and Glass, C.K. (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson, J.P. , Grainger, J.R. and Maizels, R.M. (2009) Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 167, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, V.L. , Tsai, I.J. , Selkirk, M.E. and Viney, M. (2017) The genome of Strongyloides spp. gives insights into protein families with a putative role in nematode parasitism. Parasitology, 144, 343–358. [DOI] [PubMed] [Google Scholar]
- Hussey, R.S. (1989) Disease‐inducing secretions of plant‐parasitic nematodes. Annu. Rev. Phytopathol. 27, 123–141. [Google Scholar]
- Iberkleid, I. , Vieira, P. , de Almeida Engler, J. , Firester, K. , Spiegel, Y. and Brown Horowitz, S. (2013) Fatty acid‐ and retinol‐binding protein Mf‐FAR‐1 induces tomato host susceptibility to root‐knot nematodes. PLoS One, 8, e64586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, J. , Mitreva, M. , Vanholme, B. and Gheysen, G. (2008) Exploring the transcriptome of the burrowing nematode Radopholus similis . Mol. Gen. Genomics, 280, 1–17. [DOI] [PubMed] [Google Scholar]
- Jaouannet, M. , Magliano, M. , Arguel, M.J. , Gourgues, M. , Evangelisti, E. , Abad, P. and Rosso, M.‐N. (2013) The root‐knot nematode calreticulin Mi‐CRT is a key effector in plant defense suppression. Mol. Plant–Microbe Interact. 26, 97–105. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, J.T. , Smant, G. and Blok, V.C. (2000) SXP/RAL‐2 proteins of the potato cyst nematode Globodera rostochiensis: secreted proteins of the hypodermis and amphids. Nematology, 2, 887–893. [Google Scholar]
- Jones, J.T. , Reavy, B. , Smant, G. and Prior, A.E. (2004) Glutathione peroxidases of the potato cyst nematode Globodera rostochiensis . Gene, 324, 47–54. [DOI] [PubMed] [Google Scholar]
- Kathiresan, T. and Mehta, U.K. (2005) Effect of Pratylenchus zeae infection on the expression of isozyme activities in resistant and susceptible sugarcane clones. Nematology, 7, 677–688. [Google Scholar]
- Kikuchi, T. , Cotton, J.A. , Dalzell, J.J. , Hasegawa, K. , Kanzaki, N. , McVeigh, P. , Takanashi, T. , Tsai, I.J. , Assefa, S.A. , Cock, P.J.A. , Dan Otto, T. , Hunt, M. , Reid, A.J. , Sanchez‐Flores, A. , Tsuchihara, K. , Yokoi, T. , Larsson, M.C. , Miwa, J. , Maule, A.G. , Sahashi, N. , Jones, J.T. and Berriman, M. (2011) Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus . PLoS Pathog. 7, e1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, T. , Eves‐van den Akker, S. and Jones, J.T. (2017) Genome evolution of plant‐parasitic nematodes. Ann. Rev. Phytopathol. 55, 333–354. [DOI] [PubMed] [Google Scholar]
- Lin, B. , Zhuo, K. , Chen, S. , Hu, L. , Sun, L. , Wang, X. , Zhan, L.‐H. and Liao, J. (2016) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species‐scavenging system. New Phytol. 209, 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lozano‐Torres, J.L. , Wilbers, R.H.P. , Gawronski, P. , Boshoven, J.C. , Finkers‐Tomczak, A. , Cordewener, J.H.G. , America, A.H.P. , Overmars, H.A. , Klooster, J.W.V. , Baranowski, L. , Sobczak, M. , Ilyas, M. , van der Hoorn, R.A.L. , Schots, A. , de Wit, P.J.G.M. , Bakker, J. , Goverse, A. and Smant, G. (2012) Dual disease resistance mediated by the immune receptor Cf‐2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA, 109, 10 119–10 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Torres, J.L. , Wilbers, R.H. , Warmerdam, S. , Finkers‐Tomczak, A. , Diaz‐Granados, A. , van Schaik, C.C. , Helder, J. , Bakker, J. , Goverse, A. , Schots, A. and Smant, G. (2014) Apoplastic venom allergen‐like proteins of cyst nematodes modulate the activation of basal plant innate immunity by cell surface receptors. PLoS Pathog. 10, e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T.R. , Hewezi, T. , Peng, J. and Baum, T.J. (2013) Isolation of whole esophageal gland cells from plant‐parasitic nematodes for transcriptome analyses and effector identification. Mol. Plant–Microbe Interact. 26, 31–35. [DOI] [PubMed] [Google Scholar]
- van Megen, H. , van den Elsen, S. , Holterman, M. , Karssen, G. , Mooyman, P. , Bongers, T. , Holovachov, O. , Bakker, J. and Helder, J. (2009) A phylogenetic tree of nematodes based on about 1200 full‐length small subunit ribosomal DNA sequences. Nematology, 11, 927–950. [Google Scholar]
- Mitchum, M.G. , Hussey, R.S. , Baum, T.J. , Wang, X. , Elling, A.A. , Wubben, M. and Davis, E.L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Mitreva, M. , Elling, A.A. , Dante, M. , Kloek, A.P. , Kalyanaraman, A. , Aluru, S. , Clifton, S.W. , Bird, D. , Baum, T.J. and McCarter, J.P. (2004) A survey of SL1‐spliced transcripts from the root‐lesion nematode Pratylenchus penetrans . Mol. Gen. Genomics, 272, 138–148. [DOI] [PubMed] [Google Scholar]
- Mizukubo, T. and Adachi, H. (1997) Effect of temperature on Pratylenchus penetrans development. J. Nematol. 29, 306–314. [PMC free article] [PubMed] [Google Scholar]
- Morgan, G.T. and Mcallan, J.W. (1962) Hydrolytic enzymes in plant‐parasitic nematodes. Nematologica, 8, 209–215. [Google Scholar]
- Nicol, P. , Gill, R. , Fosu‐Nyarko, J. and Jones, M.G.K. (2012) De novo analysis and functional classification of the transcriptome of the root lesion nematode, Pratylenchus thornei, after 454 GS FLX sequencing. Int. J. Parasitol. 42, 225–237. [DOI] [PubMed] [Google Scholar]
- Opperman, C.H. , Bird, D.M. , Williamson, V.M. , Rokhsar, D.S. , Burke, M. , Cohn, J. , Cromer, J. , Diener, S. , Gajan, J. , Graham, S. , Houfek, T.D. , Liu, Q. , Mitros, T. , Schaff, J. , Schaffer, R. , Scholl, E. , Sosinski, B.R. , Thomas, V.P. and Windham, E. (2008) Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA, 105, 14 802–14 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. and Nielsen, H. (2011) SIGNALP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Prior, A. , Jones, J.T. , Blok, V.C. , Beauchamp, J. , McDermott, L. , Cooper, A. and Kennedy, M.W. (2001) A surface‐associated retinol‐ and fatty acid‐binding protein (Gp‐FAR‐1) from the potato cyst nematode Globodera pallida: lipid binding activities, structural analysis and expression pattern. Biochem. J. 356, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, K.V.N. , Eswaran, M. , Ravi, V. , Gnanasekhar, B. , Narayanan, R.B. , Kaliraj, P. , Jayaraman, K. , Marson, A. , Raghavan, N. and Scott, A.L. (2000) The Wuchereria bancrofti orthologue of Brugia malayi SXP1 and the diagnosis of bancroftian filariasis. Mol. Biochem. Parasitol. 107, 71–80. [DOI] [PubMed] [Google Scholar]
- Robbertse, B. , Yoder, O.C. , Nguyen, A. , Schoch, C. and Gillian Turgeon, B. (2003) Deletion of all Cochliobolus heterostrophus monofunctional catalase‐encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Mol. Plant–Microbe Interact. 16, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Roman, J. and Triantaphyllou, A.C. (1969) Gametogenesis and reproduction of seven species of Pratylenchus . J. Nematol. 1, 357–362. [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.‐N. , Favery, B. , Piotte, C. , Arthaud, L. , de Boer, J.M. , Hussey, R.S. , Bakker, J. , Baum, T.J. and Abad, P. (1999) Isolation of a cDNA encoding a β‐1,4‐endoglucanase in the root‐knot nematode Meloidogyne incognita and expression analysis during plant parasitism. Mol. Plant–Microbe Interact. 12, 585–591. [DOI] [PubMed] [Google Scholar]
- Schwarz, E.M. , Hu, Y. , Antoshechkin, I. , Miller, M.M. , Sternberg, P.W. and Aroian, R.V. (2015) The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection‐specific gene families. Nat. Genet. 47, 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T. (2006) Salivary proteins as a defense against dietary tannins. J. Chem. Ecol. 32, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Shinya, R. , Morisaka, H. , Kikuchi, T. , Takeuchi, Y. , Ueda, M. and Futai, K. (2013) Secretome analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS One, 8, e67377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smant, G. , Stokkermans, J.P.W.G. , Yan, Y. , de Boer, J.M. , Baum, T.J. , Wang, X. , Hussey, R.S. , Gommers, F.J. , Henrissat, B. , Davis, E.L. , Helder, J. , Schots, A. and Bakker, J. (1998) Endogenous cellulases in animals: isolation of β‐1,4‐endoglucanases from two species of plant‐parasitic nematodes. Proc. Natl. Acad. Sci. USA, 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, V. , Kosarev, P. , Seledsov, I. and Vorobyev, D. (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, P. , Mantelin, S. , Cock, P.J. , Blok, V.C. , Coke, M.C. , Eves‐van den Akker, S. , Guzeeva, E. , Lilley, C.J. , Smant, G. , Reid, A.J. , Wright, K.M. , Urwin, P.E. and Jones, J.T. (2014) Genomic characterisation of the effector complement of the potato cyst nematode Globodera pallida . BMC Genomics, 15, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townshend, J.L. , Stobbs, L. and Carter, R. (1989) Ultrastructural pathology of cells affected by Pratylenchus penetrans in alfalfa roots. J. Nematol. 21, 530–539. [PMC free article] [PubMed] [Google Scholar]
- Tytgat, T. , Vercauteren, I. , Vanholme, B. , De Meutter, J. , Vanhoutte, I. , Gheysen, G. , Borgonie, G. , Coomans, A. and Gheysen, G. (2005) An SXP/RAL‐2 protein produced by the subventral pharyngeal glands in the plant parasitic root‐knot nematode Meloidogyne incognita . Parasitol. Res. 95, 50–54. [DOI] [PubMed] [Google Scholar]
- Vieira, P. , Danchin, E.G.J. , Neveu, C. , Crozat, C. , Jaubert, S. , Hussey, R.S. , Engler, G. , Abad, P. , de Almeida Engler, J. , Castagnone‐Sereno, P. and Rosso, M.‐N. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, P. , Eves‐van den Akker, S. , Verma, R. , Wantoch, S. , Eisenback, J.D. and Kamo, K. (2015) The Pratylenchus penetrans transcriptome as a source for the development of alternative control strategies: mining for putative genes involved in parasitism and evaluation of in planta RNAi. PLoS One, 10, e0144674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, P. , Kamo, K. and Eisenback, J.D. (2017a) Characterization and silencing of the fatty acid‐ and retinol‐binding Pp‐far‐1 gene in Pratylenchus penetrans . Plant Pathol. 66, 1214–1224. [Google Scholar]
- Vieira, P. , Mowery, J. , Kilcrease, J. , Eisenback, J.D. and Kamo, K. (2017b) Characterization of Lilium longiflorum cv. ‘Nellie White’ infection with root lesion nematode Pratylenchus penetrans by bright‐field and transmission electron microscopy. J. Nematol. 49, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Meyers, D. , Yan, Y. , Baum, T.J. , Smant, G. , Hussey, R.S. and Davis, E. (1999) In planta localization of a β‐1,4‐endoglucanase secreted by Heterodera glycines . Mol. Plant–Microbe Interact. 12, 64–67. [DOI] [PubMed] [Google Scholar]
- War, A.R. , Paulraj, M.G. , Ahmad, T. , Buhroo, A.A. , Hussain, B. , Ignacimuthu, S. and Sharma, H.C. (2012) Mechanisms of plant defense against insect herbivores. Plant Signal Behav. 7, 1306–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Zeng, L. , Yan, Z. , Liu, T. , Sun, K. , Zhu, T. and Zhu, A. (2015) Identification of ramie genes in response to Pratylenchus coffeae infection challenge by digital gene expression analysis. Int. J. Mol. Sci. 16, 21 989–22 007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Peng, D. , Chen, L. , Liu, H. , Chen, F. , Xu, M. , Ju, S. , Ruan, L. and Sun, M. (2016) The Ditylenchus destructor genome provides new insights into the evolution of plant parasitic nematodes. Proc. Biol. Soc. 283, 20160942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Tang, S. , Tang, Q. and Liu, T. (2014) Genome‐wide transcriptional changes of ramie (Boehmeria nivea L. Gaud) in response to root‐lesion nematode infection. Gene, 552, 67–74. [DOI] [PubMed] [Google Scholar]
- Zunke, U. (1990) Observations on the invasion and endoparasitic behavior of the root lesion nematode Pratylenchus penetrans . J. Nematol. 22, 309–320. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Summary of blast hit analyses of 1330 transcripts of Pratylenchus penetrans against the non‐redundant GenBank database and transcript quantification.
Table S2 List of transcripts and respective promoter sequences used for the identification of a non‐coding motif in the upstream region of the start codon associated with gland cell expression in Pratylenchus penetrans.
Table S3 List of primers.
Fig. S1 Distribution of transcripts encoding secreted proteins identified in different Pratylenchus penetrans datasets. (A) Venn diagram showing the number of nematode transcripts recovered from the nematode oesophageal glands versus the in planta datasets, when mapped against the full set of 1330 nematode transcripts encoding for predicted secreted proteins without transmembrane domains identified by the de novo assembly of the transcriptome of P. penetrans (Vieira et al., 2015). A complete description of the nematode transcripts is shown in Table S1. (B) Total number of annotated versus non‐annotated protein sequences by homology searches against the non‐redundant National Center for Biotechnology Information (NCBI) database of each nematode dataset.
Fig. S2 Detection of gene transcripts encoding pioneer genes of Pratylenchus penetrans by in situ hybridization in different nematode tissues. Whole‐mount in situ hybridization was performed using mixed stages of nematodes incubated with antisense probes (brown coloration) amplified from cDNA of P. penetrans. Transcripts of predicted pioneer genes localized in: (A) developing egg within the female (Ppen12587_c0_seq1); (B) surrounding the vulva region (Ppen13485_c0_seq1); (C) two dots‐like posterior to the medium bulb (Ppen14681_c0_seq1); (D) two dots‐like below the cuticle level (Ppen14446_c0_seq1); (E, F) testis region (Ppen14188_c0_seq1 and Ppen14399_c0_seq1, respectively). m, medium bulb; s, stylet. Bars, 20 µm.
Fig. S3 Prediction of gene structure of Pratylenchus penetrans candidate effectors with corresponding transcripts localized within the oesophageal glands. Only genes with complete genomic sequences obtained after blast analyses against the skim genome assemblies of P. penetrans were used. Exons are illustrated as black boxes and introns as black lines. Scale, 100 bases. FAR, fatty acid‐ and retinol‐binding gene; VAP, venom allergen‐like gene.
Fig. S4 Alignment of non‐coding promoter motif sequences associated with gland cell expression transcripts of Pratylenchus penetrans. Ppen12016_c0_seq1, pioneer; Ppen16493_c0_seq1, catalase; Ppen15554_c1_seq1, expansin‐like; Ppen14256_c0_seq1 and Ppen13447_c0_seq1, pectate lyases; Ppen12103_c0_seq1, SXP/RAL‐2; Ppen16218_c0_seq1, β‐1,4‐endoglucanase; Ppen18759_c0_seq1, arabinogalactan endo‐1,4‐β‐galactosidase; Ppen12597_c1_seq1, xylanase; Ppen13849_c0_seq1, trypsin inhibitor‐like; Ppen7984_c0_seq1, pioneer.
Fig. S5 Expression pattern of Pratylenchus penetrans effector candidate genes specifically localized in the oesophageal glands and detected by semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) in different nematode developmental stages. As positive control, each nematode developmental cDNA library [eggs, juveniles (J2–J4), females and males] was amplified using the primers of the 18S rDNA gene and a pioneer gene (Ppen13485_c0_seq1) specific to females. FAR, fatty acid‐ and retinol‐binding gene; VAP, venom allergen‐like gene.


