Summary
Fsr1, a homologue of mammalian striatin, containing multiple protein‐binding domains and a coiled‐coil (CC) domain, is critical for Fusarium verticillioides virulence. In mammals, striatin interacts with multiple proteins to form a STRIPAK (striatin‐interacting phosphatase and kinase) complex that regulates a variety of developmental processes and cellular mechanisms. In this study, we identified the homologue of a key mammalian STRIPAK component STRIP1/2 (striatin‐interacting proteins 1 and 2) in F. verticillioides, FvStp1, which interacts with Fsr1 in vivo. Gene deletion analysis indicates that FvStp1 is critical for F. verticillioides stalk rot virulence. In addition, we identified three proteins, designated FvCyp1, FvScp1 and FvSel1, which interact with the Fsr1 CC domain via a yeast two‐hybrid screen. Importantly, FvCyp1, FvScp1 and FvSel1 co‐localize to endomembrane structures, each having a preferred localization in the cell, and they are all required for F. verticillioides stalk rot virulence. Moreover, these proteins are necessary for the correct localization of Fsr1 to the endoplasmic reticulum (ER) and nuclear envelope. Thus, we identified several novel components in the STRIPAK complex that regulates F. verticillioides virulence, and propose that the correct organization and localization of Fsr1 are critical for STRIPAK complex function.
Keywords: Fusarium verticillioides, stalk rot, striatin, STRIPAK, virulence
Introduction
The striatin family comprises a small group of proteins found in eukaryotic organisms, namely from filamentous fungi to mammals, but not in plants (Bartoli et al., 1999; Benoist et al., 2006; Castets et al., 2000; Pöggeler and Kück, 2004). Three well‐known family members in mammals are striatin, SG2NA and zinedin, all of which harbour a number of protein–protein interaction domains, i.e. a caveolin‐binding domain, a putative coiled‐coil (CC) domain, a Ca2+/calmodulin‐binding domain and a WD40‐repeat domain (Castets et al., 2000). These domains have been suggested to play critical roles in cellular functions by mediating interactions with other regulatory proteins (Castets et al., 1996; Moreno et al., 2000). Mammalian striatin homologues are involved in Ca2+‐dependent signalling in cells of the central nervous system and in endocytosis (Castets et al., 1996; Moreno et al., 2000). In addition, published reports have demonstrated that striatin forms a complex with kinases and phosphatases to regulate critical cellular mechanisms in other eukaryotic organisms (Beier et al., 2016; Bloemendal et al., 2012; Dettmann et al., 2013; Frey et al., 2015; Hwang and Pallas, 2014; Kean et al., 2011). Homologues of striatin in filamentous fungi are highly conserved and have a similar domain architecture to mammalian striatins, suggesting some common functionalities (Dettmann et al., 2013; Pöggeler and Kück, 2004; Shim et al., 2006; Wang et al., 2010). Fungal striatin homologues have been implicated in a variety of important biological functions, including cell cycle regulation, sexual development, hyphal fusion, conidiation and virulence.
Although striatin proteins have no intrinsic catalytic activity, it is recognized that they organize multiple signalling complexes, i.e. signalosomes, that spatially and temporally coordinate distinct and diverse cellular mechanisms (Benoist et al., 2006; Breitman et al., 2008; Goudreault et al., 2009; Qing et al., 2004). One of the first striatin‐interacting proteins discovered was phocein, an intracellular 26‐kDa protein (Baillat et al., 2001). A splicing variant of rat striatin‐3 (rSTRN3g) associates with oestrogen receptor‐a (ERa) in a ligand‐dependent manner (Tan et al., 2008). Subsequently, with additional insights obtained on striatin‐interacting proteins, Goudreault et al. (2009) defined a novel large multi‐protein assembly, referred to as the striatin‐interacting phosphatase and kinase (STRIPAK) complex, which includes protein phosphatase 2A catalytic (PP2Ac) and scaffolding A (PP2Aa) subunits. Striatin has been proposed to be a regulatory subunit of the PP2A complex, most likely belonging to the B''' family of PP2A regulatory (PP2Ar) subunits (Eichhorn et al., 2009; Goudreault et al., 2009; Moreno et al., 2000; Shin et al., 2013). The complex also contains the novel proteins STRIP1 and STRIP2 (striatin‐interacting proteins 1 and 2) (formerly FAM40A and FAM40B), the cerebral cavernous malformation 3 (CCM3) protein and members of the germinal centre kinase III family of Ste20 kinases. Recent studies have shown that striatin indeed forms complexes with phosphatases and kinases, and that these STRIPAK complexes play critical roles in devastating human disorders and diseases, including diabetes, autism, Parkinson's disease and cancer (Hwang and Pallas, 2014).
The STRIPAK complex is highly conserved in eukaryotes, and recent studies in filamentous ascomycetes, e.g. Sordaria macrospora and Neurospora crassa, have provided evidence that this complex plays important but also diverse roles in fungi. In S. macrospora, deletion mutations in STRIPAK complex components Δpro11, Δpro22, Δpro45, ΔSmmob3 and PP2Ac1 led to defects in fruiting body formation, vegetative growth and hyphal fusion (Beier et al., 2016; Bernhards and Pöggeler, 2011; Bloemendal et al., 2012; Nordzieke et al., 2015). Mutations in N. crassa STRIPAK components Δham‐2, Δham‐3 and Δpp2A led to similar defects to those observed in S. macrospora (Dettmann et al., 2013; Fu et al., 2011). Likewise, the striatin homologue StrA in Aspergillus nidulans has been shown to be important for fruiting body formation (Wang et al., 2010). In budding yeast, Far complex subunits show partial sequence similarity with the mammalian STRIPAK complex, and these are involved in the control of post‐mating cell fusion (Goudreault et al., 2009; Kemp and Sprague, 2003). A STRIPAK‐like complex in Schizosaccharomyces pombe has also been shown to be important for the control of septation and cytokinesis (Singh et al., 2011).
Our earlier studies have shown that striatin plays an important role in the plant‐pathogenic fungus Fusarium verticillioides (teleomorph: Gibberella moniliformis Wineland). We originally identified the gene encoding the striatin homologue, FSR1, through a forward genetic screen when investigating maize stalk rot pathogenicity in F. verticillioides (Shim et al., 2006). The knockout mutant (Δfsr1) revealed two important deficiencies: virulence and female fertility. When inoculated through artificial wounding, Δfsr1 colonized and advanced through the maize vascular bundle, but failed to develop stalk rot symptoms (W. B. Shim, unpublished data). Further characterization of Fsr1 has determined that the CC domain in the N‐terminus is essential for virulence, whereas the WD40 repeat in the C‐terminus is dispensable (Yamamura and Shim, 2008). The CC domain is known to mediate protein–protein interaction in eukaryotes, and we hypothesize that this interaction triggers further downstream cellular signalling directly associated with stalk rot pathogenesis. Homologues of striatin in filamentous fungi, e.g. Fsr1 in F. verticillioides, Str1 in A. nidulans, Str1 in Colletotrichum graminicola, Pro11 in S. macrospora and Ham‐3 in N. crassa, are highly conserved, suggesting some common functionalities (Dettmann et al., 2013; Pöggeler and Kück, 2004; Shim et al., 2006; Wang et al., 2010, 2016). However, we also predict that striatin/STRIPAK interacts with a unique set of proteins in different fungi and, in turn, regulates species‐specific functions through the modulation of protein localization, vesicular trafficking and phosphorylation (Benoist et al., 2006; Goudreault et al., 2009; Kean et al., 2011). In F. verticillioides, we have yet to characterize striatin‐interacting proteins that can help us to explain how Fsr1 regulates virulence. Therefore, our first aim was to test whether Fsr1 forms a complex with one of the key STRIPAK proteins, STRIP1/2 homologue, and to study its function. Subsequently, we sought to identify F. verticillioides proteins that interact with Fsr1 in vivo, and to characterize their role in stalk rot virulence through gene mutation and cellular localization studies.
Results
Functional characterization of F. verticillioides STRIP1/2 homologue FvSTP1
One of the key components of the eukaryotic STRIPAK complex is STRIP1/2, which was first reported in mammalian systems by Goudreault et al. (2009). We identified an F. verticillioides homologous locus FVEG_08004 (designated FvSTP1) with 2268 bp encoding a 705‐amino‐acid predicted protein with two functional motifs, pfam07923 and pfam11882 (Fig. S1, see Supporting Information), using the blastp algorithm. We generated a gene knockout mutant by homologous recombination to study its function (Fig. 1A). Hygromycin B phosphotransferase (HPH) was used as the selective marker, and homologous recombination was confirmed by polymerase chain reaction (PCR) (data not shown) and Southern blot (Fig. 1B). Notably, the ΔFvStp1 strain exhibited an almost identical phenotype to the FSR1 gene knockout mutant Δfsr1, i.e. reduced colony growth and fewer aerial mycelia, when grown on potato dextrose agar (PDA), V8 agar and defined medium agar (Fig. 1C). Therefore, we used the relative growth difference data (63.6% ± 3.1%) to normalize stalk rot and seedling rot assay results to accurately measure virulence.
Figure 1.
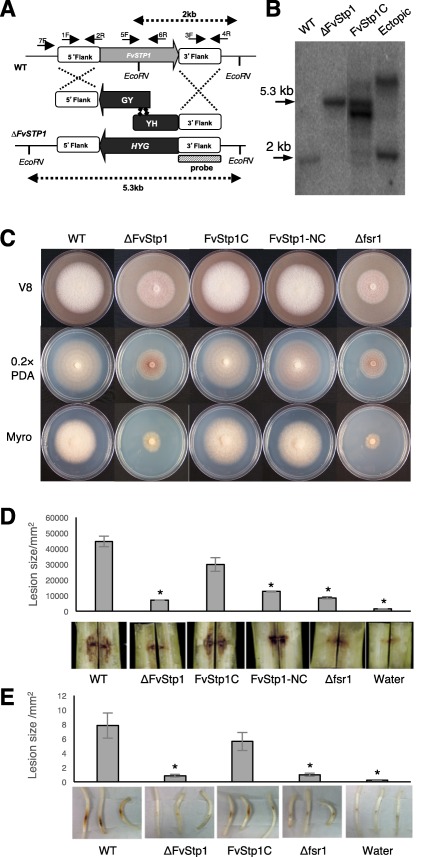
Functional characterization of F. verticillioides STRIP1/2 homolog FvSTP1. (A) Schematic representation of the homologous gene recombination strategy resulting in knockout mutant strain. Hygromycin phosphotransferase (HPH) was used as the selective marker. Arrows indicate primers used for PCR. HYG, hygromycin phosphotransferase gene, HY, HYG 5’ partial amplicon, YG, HYG 3’ partial amplification. (B) Southern analyses of wild‐type (WT), knockout mutant (DFvStp1), complementation (FvStp1C) and ectopic strains. 3’‐flanking region was used as a probe for Southern hybridization. Genomic DNA samples were digested with EcoRV. Anticipated band sizes before and after recombination are indicated above and under the scheme picture. (C) Vegetative growth of WT, DFvStp1, complementation strains (FvStp1C and FvStp1‐NC), and Dfsr1 were examined on V8, 0.2XPDA, and Myro agar plates. Strains were point inoculated with an agar block (0.5 cm in diameter) and incubated for 6 days at 25 °C under 14 h light/10 h dark cycle. (D) Eight‐week‐old B73 maize stalks were inoculated with 108/ml spore suspensions of fungal strains at the internodal region and incubated in a growth chamber for 10 days at 25 °C. Subsequently, maize stalks were split longitudinally to quantify the extent of the rot by Image J software. Three independent biological repetitions were performed. (E) Germinating B73 seedlings were inoculated with 108/ml spore suspension of fungal strains on mesocotyls. Lesion areas were quantified by Image J after 2‐week incubation. Asterisk above the column indicates statistically significant difference (P<0.05) analyzed by t‐Test.
In order to test whether the mutant phenotype is a result of a single gene mutation, we generated a complementation strain Fvstp1C by co‐transformation of ΔFvStp1 protoplasts with the wild‐type FvSTP1 gene (with its native promoter and terminator), together with the geneticin resistance gene. Southern analysis was also performed to confirm gene complementation (Fig. 1B). We also amplified and transformed the N. crassa ham‐2 gene, a STRIP1/2 homologue, into ΔFvStp1 protoplasts, together with the geneticin resistance gene, to generate a second complementation strain Fvstp1‐NC. PCR was performed to confirm this gene complementation (Fig. S2, see Supporting Information). On PDA, V8 agar and defined medium agar, FvSTP1 and its N. crassa homologue were able to restore the growth rate in the ΔFvStp1 mutant (Fig. 1C).
To test virulence, we inoculated internodes of B73 maize stalks (V10 to VT developmental stage) with spore suspensions of the wild‐type, ΔFvStp1, FvStp1C, FvStp1‐NC and Δfsr1 (negative control) strains and water (negative control). When the stalks were split open longitudinally after a 10‐day incubation, the ΔFvStp1 mutant showed significantly decreased levels of rot when compared with the wild‐type progenitor. Both ΔFvStp1 and Δfsr1 mutants showed greater than 90% reduction in virulence when analysed by the average stalk rot area (Fig. 1D). Gene complementation partially restored stalk rot virulence in the FvStp1C strain, up to approximately the 75% level when compared with the wild‐type progenitor. We also tested virulence on B73 maize seedlings, which showed outcomes that were very comparable with the stalk rot assays (Fig. 1E). These results suggest that FvSTP1 plays an important role in maize stalk rot virulence.
Fsr1–FvStp1 interaction in vivo confirmed by yeast two‐hybrid and split luciferase assays
To test the physical interaction between Fsr1 and FvStp1, we performed two independent assays: yeast two‐hybrid assay and split luciferase complementation assay. For yeast two‐hybrid assay, interaction between Fsr1 and FvStp1 was tested in Saccharomyces cerevisiae strain AH109. When yeast colonies were grown on Synthetic defined medium without Ade‐His‐Leu‐Trp (Clontech). amended with X‐alpha‐Gal, we observed a strong positive reaction (Fig. 2A). To further verify the physical interaction between Fsr1 and FvStp1 by split luciferase assay, we cloned FSR1 and FvSTP1 into pFNLucG and pFCLucH vectors, respectively (Kim et al., 2012), which were subsequently co‐transformed into F. verticillioides protoplasts. Fungal transformants co‐expressing FSR1‐nLuc and cLuc‐FvSTP1 showed strong luciferase activity, approximately 20‐fold higher than the negative controls (Fig. 2B). Two negative controls were used in this assay: one with two empty vectors (pFNLucG and pFCLucH) and the other with no vectors. As a positive control, we cloned F. verticillioides FCC1 (cyclin C) and FCK1 (cyclin‐dependent kinase) genes, which have been demonstrated to exhibit a strong in vivo interaction (Bluhm and Woloshuk, 2006), into pFNLucG and pFCLucH constructs. These two assays clearly demonstrated that Fsr1 physically interacts with FvStp1 in F. verticillioides.
Figure 2.
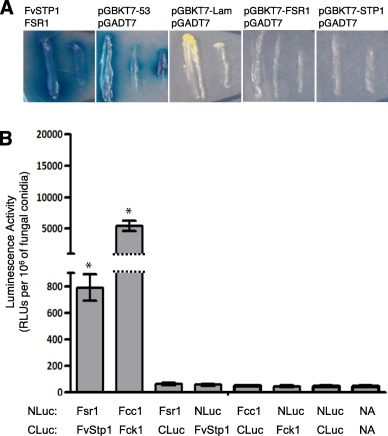
Verification of interaction between Fsr1 and FvStp1 interaction in vivo. (A) The pair of plasmids pGADT7‐FSR1 and pGBKT7‐FvSTP1 was used to test interaction by monitoring blue colonies that developed on Synthetic defined medium without Ade‐His‐Leu‐Trp (Clontech) amended with X‐alpha‐Gal. The pair of plasmids pGBKT7–53 and pGADT7 was used as a positive control. The plasmid pairs pGBKT7‐Lam/pGADT7, pGBKT7‐FSR1/pGADT7, and pGBKT7‐STP1/pGADT7 were used as negative controls. (B) Further verification of in vivo interactions between Fsr1 and FvStp1 was tested by split luciferase complementation assay. Fcc1/Fck1 was used as positive control, and six negative controls were tested (including no vector [NA]). Luminescence activity was obtained from three replicates of fungal inoculations and is presented as the number of RLUs (relative light units) per 106 fungal conidia. Asterisk above the column means statistically significant (P<0.05) analyzed by t‐Test.
Components of the putative F. verticillioides STRIPAK complex
Earlier studies in eukaryotic systems led us to hypothesize that Fsr1 regulates signalling in F. verticillioides by acting as a scaffolding protein and by interacting with kinases, phosphatases and/or transcription factors. Based on the knowledge reported for other eukaryotic organisms, e.g. Homo sapiens, Drosophila melanogaster, S. macrospora and N. crassa (Hwang and Pallas, 2014; Kück et al., 2016), we screened for putative STRIPAK components from the F. verticillioides genome (Table 1), in addition to striatin (Fsr1) and STRIP1/2 (FvStp1). As predicted in other organisms, protein phosphatase subunits are key components of STRIPAK. The homologues of germinal centre kinase were identified in S. macrospora (SmKIN3/24), N. crassa (Sid‐1) and F. verticillioides genomes (Beier et al., 2016; Frey et al., 2015; Heilig et al., 2013; Maerz et al., 2009). The functional role of this kinase has not been determined to date. Notably, Ccm3, a key component of mammalian STRIPAK known to interact with Stk24/Sok1, which is associated with vascular disease cerebral cavernous malformation, was not identified in fungal genomes. The study of STRIPAK components in mammalian systems and filamentous fungi led us to hypothesize that, although there are common core components, species‐specific, striatin‐interacting proteins can perform unique roles in different organisms.
Table 1.
STRIPAK (striatin‐interacting phosphatase and kinase) components in human (Homo sapiens, Hs), Sordaria macrospora (Sm), Neurospora crassa (Nc) and Fusarium verticillioides (Fv)
| Hs subunits 1 | Description | Sm subunits | Nc subunits | Fv subunits | Proposed function in filamentous fungi | References* |
|---|---|---|---|---|---|---|
| STRN1/3/4 | Striatin family (striatin/SG2NA/zinedin), PP2A regulatory subunit | Pro11 | Ham‐3 | FVEG_09767 (FSR1) | Vegetative growth (Sm, Nc, Fv), cell/hyphal fusion and sexual development (Sm, Nc), hyphal growth rate/density, female fertility, virulence (Fv) | 2–6 |
| PP2AAa/b | Protein phosphatase 2A structural subunit | PP2AA | PP2A‐A | FVEG_07097 | Hyphal growth, circadian clock (Nc) | 7,8 |
| PP2ACa/b | Protein phosphatase 2A catalytic subunit | PP2Ac1 | PP2A | FVEG_06252, FVEG_06432, FVEG_09543 (CPP1) | Vegetative growth, hyphal fusion and sexual development (Sm), vegetative growth, conidia production and fumonisin production (Fv) | 9,10 |
| STRIP1/2 | Striatin interacting protein 1/2 (FAM40A, FAM40B) | Pro22 | Ham‐2 | FVEG_08004 (FvSTP1) | Vegetative growth (Sm, Nc, Fv), hyphal fusion (Sm, Nc) and sexual development (Sm, Nc), female fertility, virulence (Fv) | 3,11–13 |
| MOB3 | Monopolar spindle‐one‐binder family 3, phocein | SmMob3 | Mob‐3 | FVEG_06088 | Vegetative growth, hyphal fusion and fruiting body development (Sm), vegetative cell fusion, fruiting body development (Nc) | 4,14 |
| SLMAP | Sarcolemmal membrane‐associated protein | Pro45 | Ham‐4 | FVEG_03887 | Sexual propagation and cell to cell fusion (Sm), vegetative growth, hyphal fusion and formation of protoperithecia (Nc), | 3,15 |
| MST3/4 | Germinal centre kinase III family (Stk24/Sok1) | SmKin3/24 | Sid‐1 | FVEG_05910, FVEG_07917, FVEG_08235 | Abnormal septum distribution and development (Sm), septum initiation and formation (Nc) | 16,17 |
| CCM3 | Cerebral cavernous malformation 3 | Not found | Not found | Not found | Not characterized to date | – |
*References: 1Hwang and Pallas (2013); 2Pöggeler and Kück (2004); 3Simonin et al. (2010); 4Bernhards and Pöggeler (2011); 5Shim et al. (2006); 6Yamamura and Shim (2008); 7Yatzkan et al. (1998); 8Yang et al. (2004); 9Beier et al. (2016); 10Choi and Shim (2008); 11Bloemendal et al. (2012); 12Xiang et al. (2002); 13This study; 14Maerz et al. (2009); 15Nordzieke et al. (2015); 16Heilig et al. (2013); 17Frey et al. (2015).
Identification of new Fsr1‐interacting proteins in F. verticillioides
Our previous study showed that the CC domain in the N‐terminus region of Fsr1 plays an important role in F. verticillioides virulence (Yamamura and Shim, 2008). Some of the key fungal STRIPAK subunits (Table 1), namely phocein/Mob3, are known to associate with striatin, primarily with the C‐terminal region (Bloemendal et al., 2012). Meanwhile, studies have also shown that the CC domain plays an important role in protein recruitment and structural assembly, which can influence overall STRIPAK function in mammalian systems (Chen et al., 2014; Gordon et al., 2011). One of our key aims was to identify F. verticillioides proteins that primarily interact with the Fsr1 CC motif and to test whether these proteins are involved in the regulation of stalk rot virulence. To do so, we first used the yeast two‐hybrid system to screen an F. verticillioides prey cDNA library with two bait constructs: FSR1 cDNA corresponding to the uninterrupted N‐terminal region (F1) and FSR1 cDNA devoid of the CC domain (F3). The prey library was prepared with F. verticillioides cDNAs generated from the fungus inoculated in corn stalk medium. We obtained 74 and 71 positive colonies when constructs F1 and F3, respectively, were used as bait (Table S1, see Supporting Information). We primarily aimed at proteins that are predicted to localize to the fungal endomembrane, i.e. in proximity to striatin homologues in fungal cells (Dettmann et al., 2013; Nordzieke et al., 2015; Wang et al., 2010).
From the screening (Fig. 3A), we selected three genes that showed interaction with the F1 construct [FVEG_00403 (FvCYP1, cyclophilin), FVEG_04097 (FvSCP1, sperm‐coating protein (SCP)‐like extracellular protein) and FVEG_12134 (FvSEL1, Sel1‐repeat protein)] and two genes that showed interaction with the F3 construct [FVEG_01920 (FvHEX1, Hex1‐like protein) and FVEG_11334 (FvPEX14, peroxisomal membrane anchor protein 14)]. The fact that FvHex1 and FvPex14 exhibit yeast two‐hybrid interaction with both the F1 and F3 constructs suggests that these two proteins do not require a CC domain for physical interaction with Fsr1. An additional description of these putative proteins is provided in Table S2 and Fig. S3 (see Supporting Information).
Figure 3.
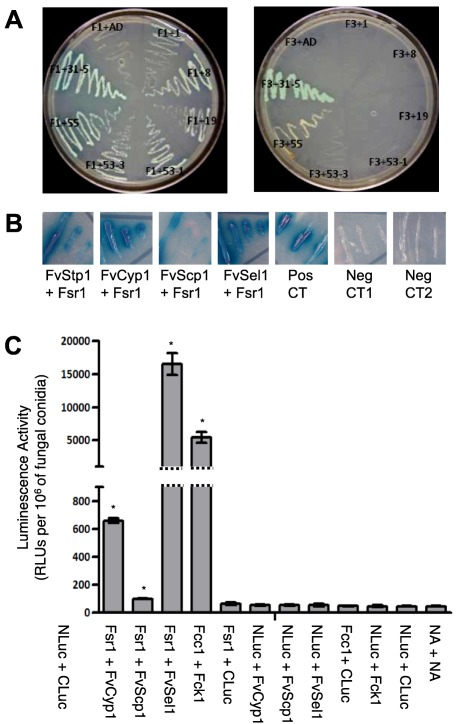
Identification of new Fsr1‐interacting proteins in F. verticillioides. (A) Bait F1 (N‐terminal Fsr1 with coiled coil motif) and Bait F3 (N‐terminal Fsr1 without coiled coil motif) were used for yeast‐two hybrid screening. Yeast strains bearing both the bait and prey plasmids were streaked on Synthetic defined medium without Ade‐His‐Leu‐Trp (Clontech) media amended with X‐alpha‐Gal. Prey strains, 1: HSP98, 8: SCP like extracellular protein, 19: Sel1 repeat protein, 53–1: Cyclophilin, 53–3: Cyclophilin, 55: Hex1, 31–5: Pex14. (B) Bait‐prey switch Y2H experiments were performed to verify performed interactions between Fsr1 and FvStp1 (positive control), FvCyp1, FvScp1, and FvSel1. The pair of plasmids pGBKT7–53 and pGADT7 was used as a positive control. The plasmid pairs pGBKT7‐Lam/pGADT7 (Neg CT1) and pGBKT7‐FSR1/pGADT7 (Neg CT2) were used as negative controls. (C) Further verification of in vivo interactions was tested by split luciferase complementation assay. Fcc1/Fck1 was used as positive control, and eight negative controls were tested (including no vector [NA]). Luminescence activity was obtained from three replicates of fungal inoculations and is presented as the number of RLUs (relative light units) per 106 of fungal conidia. Asterisk above the column means statistically significant (P<0.05) analyzed by t‐Test.
Functional characterization of FvCYP1, FvSCP1, FvSEL1, FvHEX1 and FvPEX14
To investigate the role of FvCYP1, FvSCP1, FvSEL1, FvHEX1 and FvPEX14 in F. verticillioides virulence, we generated gene knockout mutants following the established split‐marker protocol (Fig. S4, see Supporting Information). We also generated gene complementation strains using the corresponding wild‐type gene for each mutant. When we compared the vegetative growth of these mutants on synthetic media, the phenotypes of these mutants were very similar to those of the wild‐type progenitor, although ΔFvCyp1 and ΔFvSel1 exhibited slightly slower growth (Fig. 4A). However, when these strains were grown in synthetic liquid media, we did not see significant differences in fungal mass production (data not shown). ΔFvCyp1 strains produced less compact aerial mycelia when grown on synthetic media. Notably, the vegetative growth of ΔFvHex1 and ΔFvPex14 strains on V8 and PDA media were not significantly different from that of the wild‐type progenitor (Fig. S5, see Supporting Information).
Figure 4.
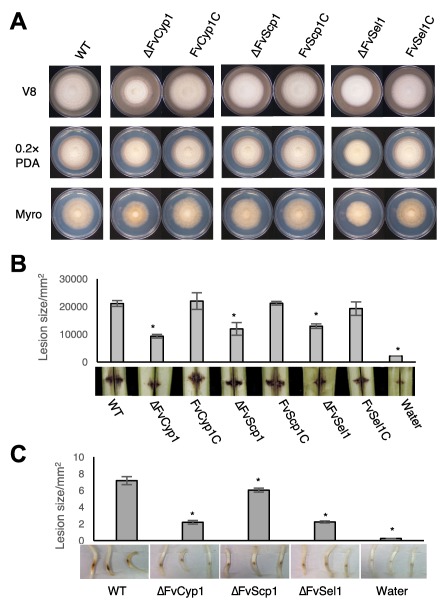
Functional characterization of FvCyp1, FvScp1, FvSel1. (A) Vegetative growth of WT, mutants and complementation strains were examined on V8, 0.2XPDA, and Myro agar plates. (B) Stalk rot pathogenicity assay was performed as described earlier. (C) Seedling rot pathogenicity assay was performed as described earlier.
Subsequently, we performed stalk rot assays with these mutants following the procedure described earlier (Shim et al., 2006). Conidial suspensions (1 × 108/mL) of wild‐type, mutants, complementation strains and water (negative control) were inoculated into maize stalks between internodal regions. After a 10‐day incubation, stalk rot symptoms generated by ΔFvCyp1, ΔFvScp1 and ΔFvSel1 strains were less severe than those formed by the wild‐type and complementation strains (Fig. 4B). In contrast, ΔFvHex1 and ΔFvPex14 showed no significant difference from the F. verticillioides wild‐type strain when stalk rot was tested (Fig. S5). When we quantified the lesion area distribution from longitudinally split stalks, the differences were significant, with the ΔFvCyp1 mutant showing the greatest difference when compared with the wild‐type strain (Fig. 4B). To further assess the loss of virulence in these mutants, we performed maize seedling rot assay as described previously by Christensen et al. (2014). Once again, ΔFvCyp1, ΔFvScp1 and ΔFvSel1 showed significantly reduced symptoms (Fig. 4C). These results strongly suggest that proteins that interact with the Fsr1 N‐terminus play important roles in F. verticillioides virulence.
To further verify protein–protein interaction, FvCyp1, FvScp1, FvSel1 and FvStp1 were subjected to bait–prey switch yeast two‐hybrid tests, and all showed positive results (Fig. 3B). Further confirmation of the physical interaction in vivo between the Fsr1 N‐terminus and FvCyp1, FvScp1 and FvSel1 came from split luciferase assay. Based on luminescence data, we concluded that Fsr1 interacts with FvCyp1, FvScp1 and FvSel1 in F. verticillioides. Notably, FvSel1 exhibited the highest level of interaction. Meanwhile, the average luminescence intensity between Fsr1 and FvScp1 was only slightly higher than the negative controls, but was statistically significant (P < 0.05) (Fig. 3C). Based on these results, we selected FvScp1, FvSel1 and FvCyp1 as Fsr1 N‐terminus‐interacting proteins for further characterization of their role in F. verticillioides virulence.
Cellular localization of Fsr1‐interacting proteins in F. verticillioides
After demonstrating the physical interaction between Fsr1 and FvCyp1, FvScp1 and FvSel1 in vivo, we next investigated the cellular localization of these proteins in F. verticillioides. In our earlier study, we used A. nidulans to visualize the subcellular localization of StrA, a homologue of Fsr1, in a living cell, and determined that StrA::eGFP co‐localizes to FM4–64‐stained endomembrane structures, probably the endoplasmic reticulum (ER) and nuclear envelope (Wang et al., 2010). FvCyp1, FvScp1 and FvSel1 are all predicted to be membrane‐associated proteins (The UniProt Consortium, 2017), and therefore we hypothesized that these interacting proteins would co‐localize to endomembrane structures in F. verticillioides. We transformed each F. verticillioides mutant with a construct that contains a green fluorescent protein (GFP)‐tagged target gene fused to Magnaporthe oryzae ribosomal protein 27 (RP27) promoter (Ribot et al., 2013; Zheng et al., 2007). FvCyp1‐GFP exhibited a pattern at or near the ER in mycelia (Fig. 5), and this association with ER was further tested by staining with ER‐Tracker™ red dye. We also observed that FvCyp1 was also distributed irregularly in the cytoplasm, perhaps with other endomembrane structures in the cell. FvScp1‐GFP and FvSel1‐GFP exhibited a slightly different localization pattern from FvCyp1: they were more concentrated near the nucleus, which was confirmed by staining with 4′,6‐diamidino‐2‐phenylindole (DAPI) (Fig. 5). However, FvSel1 localization also differed from FvScp1, as it showed irregular dispersion in the cytoplasm with tubular and punctate structures. These data suggest that, although FvCyp1, FvScp1 and FvSel1 co‐localize to endomembrane structures, most probably the nuclear membrane and ER, in F. verticillioides, they each hold unique localization sites in the cell when studied in more detail.
Figure 5.
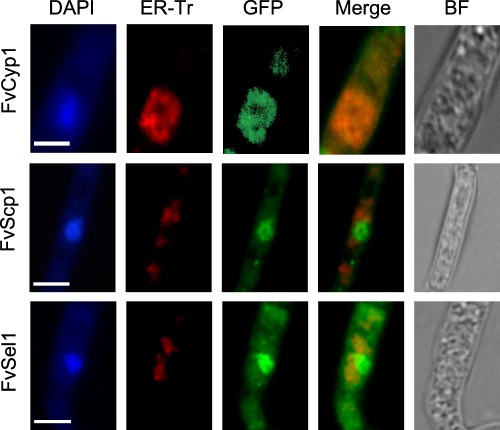
Cellular localization of Fvcyp1, Fvscp1, Fvsel1 proteins in F. verticillioides. FvCyp1‐GFP exhibited a pattern at or near ER in mycelia confirmed by ER‐tracker red (ER‐Tr). FvScp1‐GFP and FvSel1‐GFP exhibited a similar localization pattern near nucleus, which was confirmed by staining with DAPI. FM4–64 was used to stain the vacuole, septum, and vesicles. Scale bar = 5 μm.
Fsr1 localization is altered in F. verticillioides ΔFvCyp1, ΔFvScp1 and ΔFvSel1 mutants
After observing the impact of ΔFvCyp1, ΔFvScp1 and ΔFvSel1 mutations in F. verticillioides, we hypothesized that deletion of these genes, and thus the absence of these proteins, would negatively impact the cellular localization of Fsr1. Based on the fact that striatin serves as a critical component of the STRIPAK complex, incorrect localization of Fsr1 could lead to critical defects in STRIPAK‐mediated downstream cellular signalling. To test this hypothesis, we introduced the FSR1::GFP construct into Δfsr1, ΔFvCyp1, ΔFvScp1 and ΔFvSel1 mutants, and followed its localization in vivo. When FSR1::GFP was complemented into Δfsr1, GFP was largely associated with late endosome structures in mycelia, as detected by FM4‐64 staining. Surprisingly, Fsr1::GFP exhibited strong localization to vacuoles in ΔFvCyp1, ΔFvScp1 and ΔFvSel1 mutants (Fig. 6). Our study suggests that FvCyp1, FvScp1 and FvSel1 are necessary for the correct localization of Fsr1 to the ER in F. verticillioides and perhaps for the functional organization of the STRIPAK complex. Vacuoles are commonly known to play key roles in protein degradation, autophagy and cell death, and as a storage compartment for secondary metabolites (Xiao et al., 2009). We are not certain whether the localization shift seen in these mutants is a result of damaged Fsr1 or of the initiation of the autophagy process. Further study is warranted to answer these questions.
Figure 6.
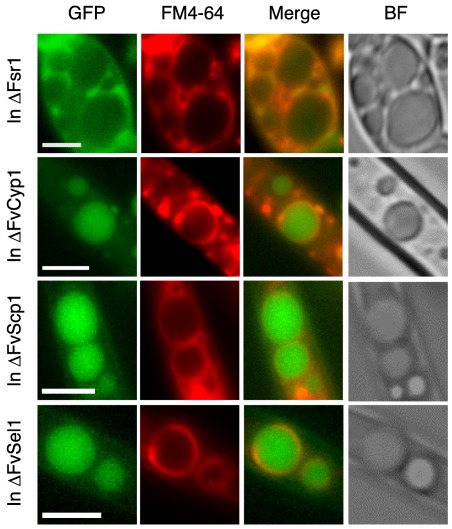
Fsr1 localization is altered in F. verticillioides ΔFvCyp1, ΔFvScp1, ΔFvSel1 mutants. Δfsr1::Fsr1‐GFP was associated with vacuolar membranes or late endosomes in mycelia, while Fsr1‐GFP exhibited a localization pattern inside vacuole/endomembrane in ΔFvCyp1, ΔFvScp1 and ΔFvSel1 mutants as confirmed by staining with FM4–64. Scale bar = 5 μm.
Discussion
Studies performed in eukaryotes, including filamentous fungi, strongly suggest that striatin homologues, as well as STRIPAK components, share highly conserved domain architectures. Although structural conservation suggests common functionalities, we are also learning that STRIPAK components in different organisms perform unique cellular functions by interacting with different subsets of kinases, phosphatases or transcription factors (Bloemendal et al., 2012; Dettmann et al., 2013; Qing et al., 2004; Shin et al., 2013). In the plant‐pathogenic fungus F. verticillioides, we demonstrated a role of striatin homologue Fsr1 in stalk rot virulence (Shim et al., 2006), and that the Fsr1 N‐terminus plays an essential role in the regulation of virulence (Yamamura and Shim, 2008). In this study, we characterized FvSTP1, the homologue of human STRIP1/2 protein, to confirm that Fsr1 indeed interacts with one of the main components of the STRIPAK complex and that F. verticillioides FvSTP1 is also necessary for virulence. Furthermore, with the understanding that the CC domain is critical for virulence, we aimed to identify F. verticillioides proteins that interact with Fsr1 in vivo to investigate their role in stalk rot virulence.
Recent studies have provided a better understanding on how striatins work with other interacting proteins to regulate cellular functions in fungi. In S. macrospora, the STRIP1/2 homologue Pro22, which interacts directly with the striatin homologue Pro11, plays a role in the regulation of septation, specifically during sexual development (Bloemendal et al., 2012). Neurospora crassa ham‐2 has also been characterized, which plays an important role in vegetative growth and hyphal fusion (Xiang et al., 2002). It has been hypothesized that ham‐2 functions in the mitogen‐activated protein (MAP) kinase pathway to regulate chemotropic polarized conidial growth and conidial germlings (Roca et al., 2005). In budding yeast, a STRIP1/2 homologue Far11, a component of the Far complex, is necessary for the maintenance of G1 phase cell cycle arrest required for sexual polarized growth (Kemp and Sprague, 2003). This evidence supports the idea that not only does the STRIP1/2 homologue exist, but it also performs distinct features in the maize pathogen F. verticillioides. Our yeast two‐hybrid and split luciferase data revealed that FvSTP1 interacts with Fsr1 in vivo, and subsequent molecular characterization demonstrated that FvSTP1 is important for F. verticillioides maize stalk virulence. The characterization of additional STRIPAK complex components (Table 1) can provide an improved understanding of how STRIPAK regulates important cellular functions, as demonstrated in S. macrospora and N. crassa (Beier et al., 2016; Bloemendal et al., 2012; Dettmann et al., 2013). The STRIPAK complex has been reported to be associated with multiple signalling pathways, e.g. the RhoA pathway, rapamycin (TORC) 2 pathway and cell wall integrity (CWI) MAP kinase pathway (Pracheil et al., 2012; Rispail and Di Pietro, 2009; Stockton et al., 2010; Zheng et al., 2010). Based on published reports, we hypothesized that Fsr1 functions together with putative phosphatases and kinases to regulate virulence. However, three F. verticillioides PP2A catalytic subunits (Table 1) did not show a detectable level of interaction with Fsr1 in F. verticillioides when tested by split luciferase assay. The identification of novel Fsr1‐interacting kinases and phosphatases will advance our understanding of STRIPAK‐mediated signalling mechanisms. However, this remains a challenging task because of the transient nature of these protein–protein interactions in fungal cells.
One of the key motivations for this study was to identify proteins that interact with the N‐terminus of Fsr1, namely with the CC domain, and to test whether these proteins play important roles in F. verticillioides virulence. Through our yeast two‐hybrid screen of the cDNA library generated from F. verticillioides‐infected maize stalks, we discovered three new STRIPAK‐interacting proteins: FvCyp1, FvScp1 and FvSel1. Notably, FvCyp1 has a well‐conserved cyclophilin‐like domain (CLD) of approximately 109 amino acids. Cyclophilin proteins are found across all organisms, with 16 in humans (Galat, 2003; Waldmeier et al., 2003), 29 in Arabidopsis thaliana (He et al., 2004) and eight in S. cerevisiae (Arevalo‐Rodriguez et al., 2004). All cyclophilins share a common CLD domain and are known to have peptidyl–prolyl cis–trans isomerase activity. As a result of their conserved nature and distribution throughout the cell, it is believed that cyclophilins perform a variety of fundamental cellular functions. Our FvCyp1 knockout mutant displayed significantly reduced virulence, similar to the Fsr1 mutant level, when tested on stalks and seedlings, suggesting a role in fungal virulence. In the plant‐pathogenic fungi Magnaporthe grisea and Botrytis cinerea, cyclophilin was shown to act as a virulence determinant during plant infection (Viaud et al., 2003). Cyclophilin proteins are also known to act as modulators of protein function in eukaryotes. For instance, mammalian cyclophilin A forms a complex with cyclosporin A and calcineurin to prevent the regulation of cytokine gene transcription (Fox et al., 2001). A mammalian Cyp40 has been shown to inhibit the activity of the transcription factor c‐Myb (Leverson and Ness, 1998). In budding yeast, cyclophilin Cpr1 is part of the Set3 complex that maintains histone‐deacetylase activity and is important for enabling the transcriptional events necessary during the switch from mitotic to meiotic cell division (Arevalo‐Rodriguez and Heitman, 2005; Arevalo‐Rodriguez et al., 2000; Pijnappel et al., 2001).
Two other proteins found in our yeast two‐hybrid screen, FvScp1 and FvSel1, also provide us with new insights into striatin function. FvScp1 is an SCP‐like extracellular protein, also called SCP/Tpx‐1/Ag5/PR‐1/Sc7 (SCP/TAPS), and all SCP/TAPS molecules share a common primary structure which can act as a Ca2+ chelator in various signalling processes (Cantacessi et al., 2009). SCP family members have been identified in various eukaryotes and belong to the cysteine‐rich secretory protein (CRISP) superfamily (Chalmers et al., 2008). The broader SCP family includes plant pathogenesis‐related protein 1 (PR‐1), CRISPs, mammalian cysteine‐rich secretory proteins and allergen 5 from vespid venom. FvSel1 was named after Sel1, a protein first identified in Caenorhabditis elegans that subsequently gave the name to the Sel1‐like repeat (SLR) solenoid protein family (Grant and Greenwald, 1996). SLR proteins are commonly found in bacteria and eukaryotes with an average frequency of 4.8 Sel1 repeats per protein, and are frequently involved in signal transduction pathways. FvSel1 has six Sel1 repeats, similar to the Hrd3 protein in S. cerevisiae; however, the lengths of the repeated sequences and the number of repetitions can vary in different prokaryotes and eukaryotes (Mittl and Schneider‐Brachert, 2007). Homologues of the Sel1 protein have been identified in several higher eukaryotes including humans, Drosophila melanogaster and A. thaliana (Mittl and Schneider‐Brachert, 2007; Zhang et al., 2015). In S. cerevisiae, there are at least three SLR proteins identified (Hrd1, Hrd3 and Chs4) with functional annotations. SLR proteins have been reported to be involved in the ER‐associated protein degradation (ERAD) in S. cerevisiae and A. thaliana under stress‐inducing conditions (Gardner et al., 2000; Kamauchi et al., 2005). Significantly, recent studies have shown that A. thaliana Sel1 is involved in RNA editing and splicing of plastid genes (Pyo et al., 2013; Zhang et al., 2015).
Although our study is the first to report in vivo interactions between STRIPAK and cyclophilin, SCP and SLR proteins in fungi, we still do not clearly understand how these proteins impact virulence activities. Based on the literature, it is reasonable to hypothesize that these proteins are involved in the regulation of STRIPAK complex assembly and therefore affect fungal development and virulence. However, questions still remain as to how these STRIPAK‐interacting proteins modulate cellular function in F. verticillioides, as well as other fungal pathogens. As striatin has been recognized as an endomembrane‐associated protein in eukaryotic cells, including in fungi (Castets et al., 2000; Okamoto et al., 1998; Pöggeler and Kück, 2004; Wang et al., 2010), we investigated the cellular localization of these Fsr1‐interacting proteins in F. verticillioides. We showed that FvCyp1 localizes to the ER membrane and perhaps plays a role in the recruitment of Fsr1 to the ER membrane. It is important to note that ER is a Ca2+ storage site in fungal cells (Bowman et al., 2009), and this result supports the idea that Fsr1 interacts with FvCyp1 to regulate Ca2+/calmodulin signalling pathways. Animal and human striatin homologues bind to calmodulin in a Ca2+‐dependent manner and are thought to be involved in neuron‐specific Ca2+ signalling events (Moreno et al., 2000). The localization of FvScp1 and FvSel1 was slightly different from that of FvCyp1 in that both were concentrated near the nuclear envelope rather than the ER. However, when we consider that the ER is adjacent to and encompasses the cell nucleus, we can conclude that these three proteins share localization with Fsr1 in F. verticillioides.
One of the surprising discoveries in this study was the altered cellular localization of Fsr1 in FvCyp1, FvScp1 and FvSel1 mutants. In wild‐type F. verticillioides, Fsr1 is localized to the endomembrane near the ER (Fig. 6), which is in agreement with the study performed in A. nidulans (Wang et al., 2010). However, in FvCyp1, FvScp1 and FvSel1 knockout mutants, Fsr1 failed to maintain its original localization and was seen inside the vacuoles (Fig. 6). Although we were not completely surprised by the fact the Fsr1 localization was altered in these mutants, we were perplexed to see very similar outcomes in all three mutants. One possible explanation is that Fsr1 proteins fail to translocate to the endomembrane for further processing without functional FvCyp1, FvScp1 and FvSel1 proteins, and that Fsr1 is shunted to the vacuole for degradation. Vacuoles typically store water, ions, secondary metabolites and nutrients, but also act as a key repository for waste products, excess solutes and toxic substances (Hatsugai et al., 2004; Martinoia et al., 2007). Vacuoles are also known to play key roles in cell death and autophagy (Xiang et al., 2013; Xiao et al., 2009). We can hypothesize that the degradation of Fsr1 in vacuoles results in the disruption of the STRIPAK complex and thus a loss of virulence. However, the reason for the incorrect localization is still not completely understood, and this mechanism remains to be further investigated.
Experimental Procedures
Fungal strains and culture media
Fusarium verticillioides strain 7600 (Fungal Genetics Stock Center, University of Missouri‐Kansas City, Kansas City, MO, USA) and all mutants generated in this study (Table S3, see Supporting Information) were cultured at 25 °C on V8 juice agar (200 mL of V8 juice, 3 g of CaCO3 and 20 g of agar powder per litre). Colony morphology was visually assayed after 6 days of growth on V8 agar, PDA (Difco, Detroit, MI, USA) and defined medium agar (1 g of NH4H2PO4, 3 g of KH2PO4, 2 g of MgSO4.7H2O, 5 g of NaCl, 40 g of sucrose and 20 g of agar powder per litre). For genomic DNA extraction, strains were grown in YEPD liquid medium (3 g of yeast extract, 10 g of peptone and 20 g of dextrose per litre) at 25 °C with shaking at 150 rpm.
Nucleic acid manipulation, PCR and transformation
Standard molecular manipulations, including PCR and Southern hybridization, were performed as described previously (Sagaram and Shim, 2007). Fungal genomic DNA was extracted using the OminiPrep genomic DNA extraction kit (G Biosciences, Maryland Heights, MO, USA). The constructs for the transformation of F. verticillioides were generated with a split‐marker approach described previously (Sagaram et al., 2007). Briefly, DNA fragments of 5′ and 3′ flanking regions of each gene were PCR amplified from wild‐type genomic DNA. Partial hygromycin B phosphotransferase (HPH) gene (HP‐ and ‐PH) fragments were amplified from the pBS15 plasmid. The 5′ and 3′ flanking region fragments were then fused with PH‐ and ‐HP fragments, respectively, by single‐joint PCR. The single‐joint PCR products were transformed into wild‐type fungal protoplasts. For complementation, respective wild‐type genes, driven by the native promoter, were co‐transformed with a geneticin‐resistant gene (GEN) into mutant protoplasts. All primers used in this study are listed in Table S4 (see Supporting Information). Fusarium verticillioides protoplasts were generated and transformed following standard protocols (Sagaram and Shim, 2007) with minor modifications. Murinase (2 mg/mL) was replaced with driselase (5 mg/mL) (Sigma, St Louis, MO, USA) in the protoplast digestion solution. Transformants were regenerated and selected on regeneration medium containing 100 µg/mL of hygromycin B (Calbiochem, La Jolla, CA, USA) and/or 150 µg/mL G418 sulfate (Cellgro, Manassas, VA, USA) as needed. Respective drug‐resistant colonies were screened by PCR and further verified by Southern analysis.
Yeast two‐hybrid experiments
Yeast two‐hybrid screening was performed using a Matchmaker™ Library Construction and Screening system (Clontech, Mountain View, CA, USA) following the manufacturer's suggested protocol. Total RNA was isolated from the wild‐type fungus grown in autoclaved corn stalk medium (2 g of pulverized B73 maize stalk in 100 mL of deionized water) for 5 days. Subsequently, an F. verticillioides cDNA prey library was generated by a BD‐SMART™ kit using oligo‐dT primer. Two bait vectors were constructed for the experiment: one with uninterrupted FvFSR1 N‐terminus cDNA (F1) and the other with the CC motif omitted from the FvFSR1 N‐terminus cDNA (F3). Both constructs were prepared from F. verticillioides genomic DNA, and all amplifications were carried out using Expand Long Template Polymerase (Roche, Indianapolis, IN, USA). All primers and amplification schemes are provided in Table S4.
The two bait vectors F1 and F3 were independently transformed into S. cerevisiae strain AH109 to generate the yeast bait strains BF12 and BF31, respectively, and were tested for autoactivation and toxicity. For the BF12 strain, the HIS3 background was controlled using 3AT10 (10 mm 3‐aminotriazole) in the medium. The F. verticillioides cDNA library was independently transformed into bait strains BF12 and BF31, and blue colonies that developed on SD‐Ade‐His‐Leu‐Trp medium amended with X‐alpha‐Gal were isolated and purified. Yeast plasmids were isolated from each positive colony and the prey inserts were PCR amplified using the primer pair 5′LD and 3′LD. The AD inserts were sequenced using T7 sequencing primer. After obtaining the sequence of putative Fsr1‐interacting proteins, we performed a bait–prey switch experiment. The coding sequence of each gene was amplified from the cDNA of F. verticillioides using the primers listed in Table S4 and cloned into pGBKT7 as the bait vectors. F1 and F3 were cloned into pGADT7 as the prey vectors. The pairs of yeast two‐hybrid plasmids were co‐transformed into S. cerevisiae strain AH109 following the standard protocol.
Split luciferase complementation assay
Plasmids used for split luciferase assay were generated following the method described by Kim et al. (2012). Briefly, the plasmids pUC19‐nLUC and pUC19‐cLUC carried N‐terminal (nLuc) and C‐terminal (cLuc) fragments of luciferase, respectively. The 35S promoter region was replaced in both plasmids with a fungal crp promoter. Fungus‐selectable GEN or HYG was introduced into these plasmids. The genes encoding the putative interacting proteins were amplified. Both PCR products were introduced into the SalI site of pFNLucG or the KpnI site of pFCLucH via an In‐FusionR HD Cloning Kit (Clontech). These two constructs were co‐transformed into F. verticillioides protoplasts. Fungal colonies were selected from regeneration medium with hygromycin and geneticin, and screened by PCR (data not shown). Fungal transformants carrying the constructed plasmids were used for further interaction studies. The strains carrying both empty vectors pFCLucH and pFNLucG were used as negative controls. The strains carrying FCC1 and FCK1 were used as positive controls. FCC1 and FCK1 are two strongly interacting proteins in F. verticillioides, as verified previously by yeast two‐hybrid assay (Bluhm and Woloshuk, 2006). All primers used are listed in Table S3.
The split luciferase complementation assays were performed as described previously (Kim et al., 2012) with minor modifications. Briefly, 96‐well plates filled with 170 μL of defined medium agar were inoculated with 10 μL of fungal conidia (108/mL) harvested from V8 agar medium, and incubated at 25 °C for 2 days before assay. When 10 μL of 15 mg/mL luciferin was added to the colonies, the light output was measured within 5 min after exposure to luciferin. Luciferase activity was measured using a GloMax 96 Microplate Luminometer (Promega, Madison, WI, USA). Luminescence activity was obtained from three replicates and is presented as the number of RLUs (relative light units) per 106 fungal conidia.
Maize infection assays
Stalk infection was performed on maize plants as described previously (Shim et al., 2006). B73 maize seeds (kindly provided by Dr Michael Kolomiets, Department of Plant Pathology and Microbiology, Texas A&M University, College Station, TX, USA) were planted in a glasshouse, and stalks (between V10 and VT stages) were inoculated with spores (1 × 108/mL) of fungal strains or water (negative control) between the nodal regions. Three inoculations were performed in the same plant on continuous nodal regions. Plants were maintained in a growth chamber with 70% humidity for 10 days at 28 °C with a 14‐h light/10‐h dark cycle and watered once. The stalks were split longitudinally in half with a scalpel to assess disease severity. At least three biological and three technical replicates were performed for each fungal strain.
Maize seedling rot pathogenicity assay was performed on 2‐week‐old maize inbred line B73 seedlings as described previously (Christensen et al., 2014) with minor modifications. Briefly, 1 × 108/mL spore suspensions in YEPD broth, together with a YEPD control, were inoculated on maize B73 mesocotyls. Plant mesocotyls were first slightly wounded by a syringe needle about 3 cm above the soil. A 5‐µL spore suspension was applied to the wound site. The seedlings were immediately covered with a plastic cover to create a high moisture environment suitable for infection and colonization. The seedlings were collected and analysed after a 2‐week growth period in the dark room. At least three biological and three technical replicates were performed for each fungal strain.
Construction of GFP fusion vectors and complementation
For in vivo localization, we generated GFP strains by introducing FvCyp1::GFP, FvScp1::GFP and FvSel1::GFP fusion constructs under their endogenous promoter. However, we were not successful in obtaining strains with good fluorescence signals. As an alternative, we redesigned our constructs under the control of the RP27 promoter, which has been successfully used in previous M. oryzae, F. graminearum and F. verticillioides protein localization studies (Bourett et al., 2002; Hou et al., 2015; Ribot et al., 2013). GFP was amplified from gGFP using the primers GFP‐F1/GFP‐R1, with five glycine–alanine repeat (GA‐5) sequences attached at the N‐terminus as a linker for GFP tagging at the gene's C‐terminus. Primers GFP‐F2/GFP‐R2 with five glycine–alanine repeat (GA‐5) sequences attached at the C‐terminus as a linker were used to amplify the GFP fragment for GFP tagging at the gene's N‐terminus. Primers RP27‐F and RP27‐R were used to amplify the RP27 promoter from the PET11 plasmid. The primers CYP‐F/CYP‐R, SCP‐F/SCP‐R and SEL‐F/SEL‐R were used to amplify gene fragments separately. For RP27::GFP::FvCyp1, the RP27::GFP fusion construct was generated first, and then fused with FvCyp1 by joint PCR. For RP27::FvScp1::GFP and RP27::FvSel1::GFP, each gene was first fused with GFP, and then the joint fragment was fused with RP27 by joint PCR.
For the Fsr1 altered localization study in Δfsr1, ΔFvcyp1, ΔFvscp1 and ΔFvsel1 mutants, the construct Rp27::GFP::Fsr1 was generated by fusion PCR as described previously. FSR1 was amplified using the primers FSR1‐F/FSR1‐R and fused with the Rp27::GFP construct. The generated construct together with the geneticin‐resistant cassette were co‐transformed into Δfsr1, ΔFvcyp1, ΔFvscp1 and ΔFvsel1 mutant protoplasts. Transformants were screened by PCR. All primers used are listed in Table S4.
Cytological assay
To visualize GFP strains, GFP strains were grown on minimal medium (MM) medium for 6 days at room temperature (Momany et al., 1999). An Olympus BX51 microscope (Olympus America, Melville, NY, USA) was used for observation with assistance from Dr Brian Shaw (Department of Plant Pathology and Microbiology, Texas A&M University, College Station, TX, USA). A detailed description of the features used for imaging from this microscope has been published previously (Sagaram et al., 2007). DAPI at a concentration of 10 ng/mL was used to stain hyphae for nuclear visualization. To visualize the cell endomembrane, hyphae were treated with 25 μm FM4–64 solution for 30 min before being observed under the microscope. To visualize ER, hyphae were treated with ER‐Tracker™ red dye (E34250, Invitrogen Carlsbad, CA, USA) at a final concentration of 1 μm for 20 min at 37 ºC before observation. Images were prepared for publication with Adobe Photoshop CS5.1 following a previously described protocol (Schultzhaus et al., 2015).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Yeast two‐hybrid (Y2H) screening results.
Table S2 Additional description of putative Fsr1‐interacting proteins.
Table S3 Fusarium verticillioides strains used in this study.
Table S4 Primers used in this study. The underlined sequences were for fusion purposes. The recognition site of the restriction enzyme is shaded in selected primer sequences.
Fig. S1 FvStp1 domain and sequence alignment.
Fig. S2 Neurospora crassa complementation transformation polymerase chain reaction (PCR).
Fig. S3 FvCyp1, FvScp1 and FvSel1 protein blast results.
Fig. S4 FvCYP1, FvSCP1 and FvSEL1 knockout strategies and Southern blot results.
Fig. S5 FvHex1 and FvPex14 characterization.
Acknowledgements
We thank Drs Ping He, Mike Kolomiets and Clint Magill (Texas A&M University, College Station, TX, USA) for providing valuable comments during our research and also for critical reading of the manuscript. We are also grateful to Drs Brian Shaw, Zach Schultzahaus and Libo Shan for providing valuable assistance in microscopy and luciferase assays. This work was supported in part by the National Research Initiative Competitive Grants Program of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (No. 2007‐35319‐18334).
References
- Arevalo‐Rodriguez, M. and Heitman, J. (2005) Cyclophilin A is localized to the nucleus and controls meiosis in Saccharomyces cerevisiae . Eukaryot. Cell, 4, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo‐Rodriguez, M. , Cardenas, M.E. , Wu, X.Y. , Hanes, S.D. and Heitman, J. (2000) Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3‐Rpd3 histone deacetylase. EMBO J. 19, 3739–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo‐Rodriguez, M. , Wu, X.Y. , Hanes, S.D. and Heitman, J. (2004) Prolyl isomerases in yeast. Front. Biosci. 9, 2420–2446. [DOI] [PubMed] [Google Scholar]
- Baillat, G. , Moqrich, A. , Castets, F. , Baude, A.S. , Bailly, Y. , Benmerah, A. and Monneron A. (2001) Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell, 12, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli, M. , Ternaux, J.P. , Forni, C. , Portalier, P. , Salin, P. , Amalric, M. and Monneron A. (1999) Down‐regulation of striatin, a neuronal calmodulin‐binding protein, impairs rat locomotor activity. J. Neurobiol. 40, 234–243. [PubMed] [Google Scholar]
- Beier, A. , Teichert, I. , Krisp, C. , Wolters, D.A. and Kück, U. (2016) Catalytic subunit 1 of protein phosphatase 2A is a subunit of the STRIPAK complex and governs fungal sexual development. Mbio, 7, e00870–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist, M. , Gaillard, S. and Castets, F. (2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris, 99, 146–153. [DOI] [PubMed] [Google Scholar]
- Bernhards, Y. and Pöggeler, S. (2011) The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora . Curr. Genet. 57, 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemendal, S. , Bernhards, Y. , Bartho, K. , Dettmann, A. , Voigt, O. , Teichert, I. , Seiler, S. , Wolters, D.A. , Pöggeler, S. and Kück, U. (2012) A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84, 310–323. [DOI] [PubMed] [Google Scholar]
- Bluhm, B.H. and Woloshuk, C.P. (2006) Fck1, a C‐type cyclin‐dependent kinase, interacts with Fcc1 to regulate development and secondary metabolism in Fusarium verticillioides . Fungal Genet. Biol. 43, 146–154. [DOI] [PubMed] [Google Scholar]
- Bourett, T.M. , Sweigard, J.A. , Czymmek, K.J. , Carroll, A. and Howard, R.J. (2002) Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet. Biol. 37, 211–220. [DOI] [PubMed] [Google Scholar]
- Bowman, B.J. , Draskovic, M. , Freitag, M. and Bowman, E.J. (2009) Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa . Eukaryot. Cell, 8, 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman, M. , Zilberberg, A. , Caspi, M. and Rosin‐Arbesfeld, R. (2008) The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. BBA Mol. Cell Res. 1783, 1792–1802. [DOI] [PubMed] [Google Scholar]
- Cantacessi, C. , Campbell, B.E. , Visser, A. , Geldhof, P. , Nolan, M.J. , Nisbet, A.J. , Matthews, J.B. , Loukas, A. , Hofmann, A. , Otranto, D. , Sternberg, P.W. and Gasser, R.B. (2009) A portrait of the “SCP/TAPS” proteins of eukaryotes – developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27, 376–388. [DOI] [PubMed] [Google Scholar]
- Castets, F. , Bartoli, M. , Barnier, J.V. , Baillat, G. , Salin, P. , Moqrich, A. , Bourgeois, J.P. , Denizot, F. , Rougon, G. , Calothy, G. and Monneron, A. (1996) A novel calmodulin‐binding protein, belonging to the WD‐repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 134, 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets, F. , Rakitina, T. , Gaillard, S. , Moqrich, A. , Mattei, M.G. and Monneron, A. (2000) Zinedin, SG2NA, and striatin are calmodulin‐binding, WD repeat proteins principally expressed in the brain. J. Biol. Chem. 275, 19 970–19 977. [DOI] [PubMed] [Google Scholar]
- Chalmers, I.W. , McArdle, A.J. , Coulson, R.M.R. , Wagner, M.A. , Schmid, R. , Hirai, H. and Hoffmann, K.F. (2008) Developmentally regulated expression, alternative splicing and distinct sub‐groupings in members of the Schistosoma mansoni venom allergen‐like (SmVAL) gene family. BMC Genomics, 9, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.C. , Shi, Z.B. , Zhang, W.Q. , Chen, M. , He, F. , Zhang, Z.Z. , Wang, Y. , Feng, M. , Wang, W. , Zhao, Y. , Brown, J.H. , Jiao, S. and Zhou, Z. (2014) Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2 : 2 heterotetrameric core of striatin‐interacting phosphatase and kinase (STRIPAK) complex. J. Biol. Chem. 289, 9651–9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.E. and Shim, W.B. (2008) Functional characterization of Fusarium verticillioides CPP1, a gene encoding a putative protein phosphatase 2A catalytic subunit. Microbiol, 154, 326–336. [DOI] [PubMed] [Google Scholar]
- Christensen, S.A. , Nemchenko, A. , Park, Y.S. , Borrego, E. , Huang, P.C. , Schmelz, E.A. , Kunze, S. , Feussner, I. , Yalpani, N. , Meeley, R. and Kolomiets, M.V. (2014) The novel monocot‐specific 9‐lipoxygenase Zmlox12 is required to mount an effective jasmonate‐mediated defense against Fusarium verticillioides in maize. Mol. Plant–Microbe Interact. 27, 1263–1276. [DOI] [PubMed] [Google Scholar]
- Dettmann, A. , Heilig, Y. , Ludwig, S. , Schmitt, K. , Illgen, J. , Fleissner, A. , Valerius, O. and Seiler, S. (2013) HAM‐2 and HAM‐3 are central for the assembly of the Neurospora STRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK‐1 in a MAK‐2‐dependent manner. Mol. Microbiol. 90, 796–812. [DOI] [PubMed] [Google Scholar]
- Eichhorn, P.J. , Creyghton, M.P. and Bernards, R. (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta, 1795, 1–15. [DOI] [PubMed] [Google Scholar]
- Fox, D.S. , Cruz, M.C. , Sia, R.A.L. , Ke, H.M. , Cox, G.M. , Cardenas, M.E. and Heitman, J. (2001) Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12‐FK506 in Cryptococcus neoformans . Mol. Microbiol. 39, 835–849. [DOI] [PubMed] [Google Scholar]
- Frey, S. , Reschka, E.J. and Pöggeler, S. (2015) Germinal center kinases SmKIN3 and SmKIN24 are associated with the Sordaria macrospora striatin‐interacting phosphatase and kinase (STRIPAK) complex. PLoS One, 10, e0139163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C. , Iyer, P. , Herkal, A. , Abdullah, J. , Stout, A. and Free, S.J. (2011) Identification and characterization of genes required for cell‐to‐cell fusion in Neurospora crassa . Eukaryot. Cell, 10, 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat, A. (2003) Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity targets–functions. Curr. Top. Med. Chem. 3, 1315–1347. [DOI] [PubMed] [Google Scholar]
- Gardner, R.G. , Swarbrick, G.M. , Bays, N.W. , Cronin, S.R. , Wilhovsky, S. , Seelig, L. , Kim, C. and Hampton, R.Y. (2000) Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, J. , Hwang, J. , Carrier, K.J. , Jones, C.A. , Kern, Q.L. , Moreno, C.S. , Karas, R.H. and Pallas, D.C. (2011) Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20‐like kinase Mst3. BMC Biochem. 12, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault, M. , D'Ambrosio, L.M. , Kean, M.J. , Mullin, M.J. , Larsen, B.G. , Sanchez, A. , Chaudhry, S. , Chen, G.I. , Sicheri, F. , Nesvizhskii, A.I. , Aebersold, R. , Raught, B. and Gingras, A.C. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin‐interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics, 8, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, B. and Greenwald, I. (1996) The Caenorhabditis elegans sel‐1 gene, a negative regulator of lin‐12 and glp‐1, encodes a predicted extracellular protein. Genetics, 143, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai, N. , Kuroyanagi, M. , Yamada, K. , Meshi, T. , Tsuda, S. , Kondo, M. , Nishimura, M. and Hara‐Nishimura, I. (2004) A plant vacuolar protease, VPE, mediates virus‐induced hypersensitive cell death. Science, 305, 855–858. [DOI] [PubMed] [Google Scholar]
- He, Z.Y. , Li, L.G. and Luan, S. (2004) Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134, 1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig, Y. , Schmitt, K. and Seiler, S. (2013) Phospho‐regulation of the Neurospora crassa septation initiation network. PLoS One, 8, e79464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, R. , Jiang, C. , Zheng, Q. , Wang, C.F. and Xu, J.R. (2015) The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in Fusarium graminearum . Mol. Plant Pathol. 16, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J.Y. and Pallas, D.C. (2014) STRIPAK complexes: structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 47, 118–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi, S. , Nakatani, H. , Nakano, C. and Urade, R. (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana . FEBS J. 272, 3461–3476. [DOI] [PubMed] [Google Scholar]
- Kean, M.J. , Ceccarelli, D.F. , Goudreault, M. , Sanches, M. , Tate, S. , Larsen, B. , Gibson, L.C. , Derry, W.B. , Scott, I.C. , Pelletier, L. , Baillie, G.S. , Sicheri, F. and Gingras, A.C. (2011) Structure–function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25 065–25 075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp, H.A. and Sprague, G.F. (2003) Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae . Mol. Cell Biol. 23, 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.K. , Cho, E.J. , Jo, S.M. , Sung, B.R. , Lee, S. and Yun, S.H. (2012) A split luciferase complementation assay for studying in vivo protein–protein interactions in filamentous ascomycetes. Curr. Genet. 58, 179–189. [DOI] [PubMed] [Google Scholar]
- Kück, U. , Beier, A.M. and Teichert, I. (2016) The composition and function of the striatin‐interacting phosphatases and kinases (STRIPAK) complex in fungi. Fungal Genet. Biol. 90, 31–38. [DOI] [PubMed] [Google Scholar]
- Leverson, J.D. and Ness, S.A. (1998) Point mutations in v‐Myb disrupt a cyclophilin‐catalyzed negative regulatory mechanism. Mol. Cell, 1, 203–211. [DOI] [PubMed] [Google Scholar]
- Maerz, S. , Dettmann, A. , Ziv, C. , Liu, Y. , Valerius, O. , Yarden, O. and Seiler, S. (2009) Two NDR kinase‐MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa . Mol. Microbiol. 74, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E. , Maeshima, M. and Neuhaus, H.E. (2007) Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58, 83–102. [DOI] [PubMed] [Google Scholar]
- Mittl, P.R.E. and Schneider‐Brachert, W. (2007) Sel1‐like repeat proteins in signal transduction. Cell Signal. 19, 20–31. [DOI] [PubMed] [Google Scholar]
- Momany, M. , Westfall, P.J. and Abramowsky, G. (1999) Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics, 151, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, C.S. , Park, S. , Nelson, K. , Ashby, D. , Hubalek, F. , Lane, W.S. and Pallas, D.C. (2000) WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin‐binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordzieke, S. , Zobel, T. , Franzel, B. , Wolters, D.A. , Kück, U. and Teichert, I. (2015) A fungal sarcolemmal membrane‐associated protein (SLMAP) homolog plays a fundamental role in development and localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria. Eukaryot. Cell, 14, 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T. , Schlegel, A. , Scherer, P.E. and Lisanti, M.P. (1998) Caveolins, a family of scaffolding proteins for organizing preassembled signaling complexes at the plasma membrane. J. Biol. Chem. 273, 5419–5422. [DOI] [PubMed] [Google Scholar]
- Pijnappel, W.W.M.P. , Schaft, D. , Roguev, A. , Shevchenko, A. , Tekotte, H. , Wilm, M. , Rigaut, G. , Séraphin, B. , Aasland, R. and Stewart, A.F. (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic‐specific repressor of the sporulation gene program. Genes Dev. 15, 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler, S. and Kück, U. (2004) A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot. Cell, 3, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracheil, T. , Thornton, J. and Liu, Z.C. (2012) TORC2 signaling is antagonized by protein phosphatase 2A and the Far complex in Saccharomyces cerevisiae . Genetics, 190, 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo, Y.J. , Kwon, K.C. , Kim, A. and Cho, M.H. (2013) Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 163, 1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing, L. , Pallas, D.C. , Surks, H.K. , Baur, W.E. , Mendelsohn, M.E. and Karas, R.H. (2004) Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor alpha. Proc. Natl. Acad. Sci. USA, 101, 17 126–17 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot, C. , Cesari, S. , Abidi, I. , Chalvon, V. , Bournaud, C. , Vallet, J. , Lebrun, M.H. , Morel, J.B. and Kroj, T. (2013) The Magnaporthe oryzae effector AVR1CO39 is translocated into rice cells independently of a fungal‐derived machinery. Plant J. 74, 1–12. [DOI] [PubMed] [Google Scholar]
- Rispail, N. and Di Pietro, A. (2009) Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant–Microbe Interact. 22, 830–839. [DOI] [PubMed] [Google Scholar]
- Roca, M.G. , Arlt, J. , Jeffree, C.E. and Read, N.D. (2005) Cell biology of conidial anastomosis tubes in Neurospora crassa . Eukaryot. Cell, 4, 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram, U.S. and Shim, W.B. (2007) Fusarium verticillioides GBB1, a gene encoding heterotrimeric G protein beta subunit, is associated with fumonisin B1 biosynthesis and hyphal development but not with fungal virulence. Mol. Plant Pathol. 8, 375–384. [DOI] [PubMed] [Google Scholar]
- Sagaram, U.S. , Shaw, B.D. and Shim, W.B. (2007) Fusarium verticillioides GAP1, a gene encoding a putative glycolipid‐anchored surface protein, participates in conidiation and cell wall structure but not virulence. Microbiology, 153, 2850–2861. [DOI] [PubMed] [Google Scholar]
- Schultzhaus, Z. , Yan, H.J. and Shaw, B.D. (2015) Aspergillus nidulans flippase DnfA is cargo of the endocytic collar and plays complementary roles in growth and phosphatidylserine asymmetry with another flippase, DnfB. Mol. Microbiol. 97, 18–32. [DOI] [PubMed] [Google Scholar]
- Shim, W.B. , Sagaram, U.S. , Choi, Y.E. , So, J. , Wilkinson, H.H. and Lee, Y.W. (2006) FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum . Mol. Plant–Microbe Interact. 19, 725–733. [DOI] [PubMed] [Google Scholar]
- Shin, J.H. , Kim, J.E. , Malapi‐Wight, M. , Choi, Y.E. , Shaw, B.D. and Shim, W.B. (2013) Protein phosphatase 2A regulatory subunits perform distinct functional roles in the maize pathogen Fusarium verticillioides . Mol. Plant Pathol. 14, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin, A.R. , Rasmussen, C.G. , Yang, M. and Glass, N.L. (2010) Genes encoding a striatin‐like protein (ham‐3) and a forkhead associated protein (ham‐4) are required for hyphal fusion in Neurospora crassa . Fungal Genet Biol. 47, 855–868. [DOI] [PubMed] [Google Scholar]
- Singh, N.S. , Shao, N. , McLean, J.R. , Sevugan, M. , Ren, L.P. , Chew, T.G. , Bimbo, A. , Sharma, R. , Tang, X. , Gould, K.L. and Balasubramanian, M.K. (2011) SIN‐inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates sin asymmetry in fission yeast. Curr. Biol. 21, 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton, R.A. , Shenkar, R. , Awad, I.A. and Ginsberg, M.H. (2010) Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207, 881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, B. , Long, X. , Nakshatri, H. , Nephew, K.P. and Bigsby, R.M. (2008) Striatin‐3 gamma inhibits estrogen receptor activity by recruiting a protein phosphatase. J. Mol. Endocrinol. 40, 199–210. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium . (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud, M. , Brunet‐Simon, A. , Brygoo, Y. , Pradier, J.M. and Levis, C. (2003) Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea . Mol. Microbiol. 50, 1451–1465. [DOI] [PubMed] [Google Scholar]
- Waldmeier, P.C. , Zimmermann, K. , Qian, T. , Tintelnot‐Blomley, M. and Lemasters, J.J. (2003) Cyclophilin D as a drug target. Curr. Med. Chem. 10, 1485–1506. [DOI] [PubMed] [Google Scholar]
- Wang, C.L. , Shim, W.B. and Shaw, B.D. (2010) Aspergillus nidulans striatin (StrA) mediates sexual development and localizes to the endoplasmic reticulum. Fungal Genet. Biol. 47, 789–799. [DOI] [PubMed] [Google Scholar]
- Wang, C.L. , Shim, W.B. and Shaw, B.D. (2016) The Colletotrichum graminicola striatin orthologue Str1 is necessary for anastomosis and is a virulence factor. Mol. Plant Pathol. 17, 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, L. , Etxeberria, E. and Van den Ende, W. (2013) Vacuolar protein sorting mechanisms in plants. FEBS J. 280, 979–993. [DOI] [PubMed] [Google Scholar]
- Xiang, Q.J. , Rasmussen, C. and Glass, N.L. (2002) The ham‐2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa . Genetics, 160, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H. , Chen, D.D. , Fang, Z. , Xu, J. , Sun, X.J. , Song, S. , Liu, J. and Yang, C. (2009) Lysosome biogenesis mediated by vps‐18 affects apoptotic cell degradation in Caenorhabditis elegans . Mol. Biol Cell. 20, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , He, Q. , Cheng, P. , Wrage, P. , Yarden, O. and Liu, Y. (2004) Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev, 18, 255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatzkan, E. , Szöor, B. , Fehér, Z. , Dombrádi, V. and Yarden O. (1998) Protein phosphatase 2A is involved in hyphal growth of Neurospora crassa . Mol Gen Genet., 259, 523–31. [DOI] [PubMed] [Google Scholar]
- Yamamura, Y. and Shim, W.B. (2008) The coiled‐coil protein‐binding motif in Fusarium verticillioides Fsr1 is essential for maize stalk rot virulence. Microbiology, 154, 1637–1645. [DOI] [PubMed] [Google Scholar]
- Zhang, H.D. , Cui, Y.L. , Huang, C. , Yin, Q.Q. , Qin, X.M. , Xu, T. , He, X.F. , Zhang, Y. , Li, Z.R. and Yang, Z.N. (2015) PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana . Photosynth. Res. 126, 311–321. [DOI] [PubMed] [Google Scholar]
- Zheng, W. , Chen, J.S. , Liu, W.D. , Zheng, S.Q. , Zhou, J. , Lu, G.D. and Wang, Z. (2007) A Rho3 homolog is essential for apressorium development and pathogenicity of Magnaporthe grisea . Eukaryot. Cell, 6, 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.J. , Xu, C. , Di Lorenzo, A. , Kleaveland, B. , Zou, Z.Y. , Seiler, C. , Chen, M. , Cheng, L. , Xiao, J. , He, J. , Pack, M.A. , Sessa, W.C. and Kahn, M.L. (2010) CCM3 signaling through sterile 20‐like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest. 120, 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Yeast two‐hybrid (Y2H) screening results.
Table S2 Additional description of putative Fsr1‐interacting proteins.
Table S3 Fusarium verticillioides strains used in this study.
Table S4 Primers used in this study. The underlined sequences were for fusion purposes. The recognition site of the restriction enzyme is shaded in selected primer sequences.
Fig. S1 FvStp1 domain and sequence alignment.
Fig. S2 Neurospora crassa complementation transformation polymerase chain reaction (PCR).
Fig. S3 FvCyp1, FvScp1 and FvSel1 protein blast results.
Fig. S4 FvCYP1, FvSCP1 and FvSEL1 knockout strategies and Southern blot results.
Fig. S5 FvHex1 and FvPex14 characterization.


