Summary
Plant natriuretic peptides (PNPs) have been implicated in the regulation of ions and water homeostasis, and their participation in the plant immune response has also been proposed. Xanthomonas citri ssp. citri contains a gene encoding a PNP‐like protein (XacPNP) which has no homologues in other bacteria. XacPNP mimics its Arabidopsis thaliana homologue AtPNP‐A by modifying host responses to create favourable conditions for pathogen survival. However, the ability of XacPNP to induce plant defence responses has not been investigated. In order to study further the role of XacPNP in vivo, A. thaliana lines over‐expressing XacPNP, lines over‐expressing AtPNP‐A and AtPNP‐A‐deficient plants were generated. Plants over‐expressing XacPNP or AtPNP‐A showed larger stomatal aperture and were more resistant to saline or oxidative stress than were PNP‐deficient lines. In order to study further the role of PNP in biotic stress responses, A. thaliana leaves were infiltrated with pure recombinant XacPNP, and showed enhanced expression of genes related to the defence response and a higher resistance to pathogen infections. Moreover, AtPNP‐A expression increased in A. thaliana on Pseudomonas syringae pv. tomato (Pst) infection. This evidence led us to analyse the responses of the transgenic plants to pathogens. Plants over‐expressing XacPNP or AtPNP‐A were more resistant to Pst infection than control plants, whereas PNP‐deficient plants were more susceptible and showed a stronger hypersensitive response when challenged with non‐host bacteria. Therefore, XacPNP, acquired by horizontal gene transfer, is able to mimic PNP functions, even with an increase in plant defence responses.
Keywords: AtPNP‐A, plant defence, plant natriuretic peptide, Pseudomonas syringae pv. tomato, XacPNP, Xanthomonas citri ssp. citri, Xanthomonas axonopodis pv. vesicatoria
Introduction
Natriuretic peptides (NPs) are a family of peptides involved in the regulation of salt and water homeostasis in vertebrates. The effects of NPs are mediated by their cognate signalling receptors. These receptors are guanylyl cyclases and, as a result of ligand binding, intracellular cyclic guanosine monophosphate (cGMP) concentrations increase (Potter, 2011). Plants possess heterologous proteins to animal NPs, named plant natriuretic peptides (PNPs), which are involved in a number of responses essential for plant homeostasis and growth (Gehring and Irving, 2003). Several activities have been observed for PNPs. Purified PNP application causes stomatal opening and the activation of membrane H+‐ATPase in leaves, transient elevation of cGMP levels in root tissue, increased osmoticum‐dependent volume in leaf protoplasts and modulation of ion influx (H+, K+ and Na+) across plasma membranes in roots (Ludidi et al., 2004; Maryani et al., 2003; Morse et al., 2004; Pharmawati et al., 1998a, 1998b, 1999; Suwastika and Gehring, 1999). The presence of a signal sequence in PNPs suggests that they are secreted into the plant apoplast; further, the immunodetection of PNPs in the conductive tissue of plants and the increase in cGMP levels as a result of PNP application have suggested that PNP function should be mediated via a receptor involving cGMP as a second messenger (Billington et al., 1997; Suwastika et al., 2000). In addition, in transiently transfected onion epidermal cells with a PNP signal peptide fused to green fluorescent protein (GFP), PNP‐GFP is located in the extracellular space (Wang et al., 2011) (for a review, see Gehring and Irving, 2013).
The most well‐characterized PNP of Arabidopsis thaliana is AtPNP‐A, encoded by the At2g18660 gene. Recently, a receptor for AtPNP‐A has been identified as a leucine‐rich repeat protein with guanylyl cyclase activity which allows cGMP‐dependent signalling (Turek and Gehring, 2016). Several efforts have been conducted to elucidate AtPNP‐A function, particularly during the stress response. A proteomic analysis of AtPNP‐A‐treated plant cells revealed that proteins involved in oxidation–reduction processes and in the response to salt stress were over‐represented. Consistently, it has been proposed that AtPNP‐A plays a key role in oxidation–reduction processes and in the response to salt stress (Turek et al., 2014). Further, AtPNP‐A is able to modulate its own expression, which enables the tuning of transcript and protein levels (Wang et al., 2011). It has also been reported that PNP signalling employs a systemic signal to alter photosynthesis and respiration, integrating the response to the whole plant (Ruzvidzo et al., 2011). In addition, a gene ontology analysis of AtPNP‐A and the 25 most expression‐correlated genes has revealed an over‐representation of genes associated with the plant defence response related to both abiotic and biotic stress, suggesting the involvement of this type of molecule in the plant defence response (Meier et al., 2008).
The phytopathogen Xanthomonas citri ssp. citri (Xcc) is the causal agent of the worldwide‐distributed citrus canker disease, and this bacterium possesses a gene coding for a PNP‐like protein, named XacPNP (XAC2654; Q8PJ87) (Gottig et al., 2008). Remarkably, no significant similarities between XacPNP and other bacterial proteins have been found to date, although homologues in phytopathogenic fungi have been found, i.e. Ave1 (avirulence on Ve1) present in Verticillium dahliae, Verticillium alboatrum, Colletotrichum higginsianum, Cercospora beticola and Fusarium oxysporum f. sp. lycopersici (de Jonge et al., 2012). Formerly, it has been proposed that XacPNP was acquired in an ancient lateral gene transfer event from plants (Nembaware et al., 2004), and evolutionary analysis of Ave1 proteins has shown that these fungal proteins are more related to plant PNPs than to XacPNP, also suggesting that Ave1 proteins have been horizontally acquired from plants (de Jonge et al., 2012).
Previous results have supported the hypothesis that XacPNP behaves as a PNP mimicking the host protein. The pure recombinant XacPNP protein elicits physiological responses in plants similar to AtPNP‐A (Gottig et al., 2008), such as an increase in the efficiency of photosynthesis in plant tissues infiltrated with this peptide (Garavaglia et al., 2010b). In line with these results, a comparative proteomic study of citrus leaves infected with wild‐type Xcc and a XacPNP deletion mutant revealed that photosynthetic proteins were under‐represented in the tissue infected with the mutant strain, and this could be reversed by the exogenous application of pure recombinant XacPNP or AtPNP‐A (Garavaglia et al., 2010a). During citrus infections, XacPNP is expressed in Xcc and serves to keep the plant tissue hydrated and in a healthier state, allowing the biotrophic pathogen to survive longer periods in the infected tissue, and this also occurs when the bacterial gene is replaced by the plant AtPNP‐A (Gottig et al., 2008). Therefore, the main known function for XacPNP, much like PNPs, is the modulation of plant homeostasis.
Despite the similar activities observed for XacPNP and AtPNP‐A, the observation that AtPNP‐A is co‐expressed with A. thaliana defence response genes led us to wonder whether these proteins contribute to increase the plant defence response, in addition to their homeostatic function. As little is known about the in vivo role of PNPs, in this article, we investigated the effect of these proteins in the defence response of A. thaliana. Accordingly, we analysed whether AtPNP‐A functions in vivo to modulate plant defence responses. Further, we hypothesized that the bacterial PNP may mimic PNPs in the response to biotic stress. Transgenic A. thaliana lines over‐expressing AtPNP‐A or bacterial XacPNP, as well as AtPNP‐A‐deficient plants, were obtained to evaluate these hypotheses. These plants were challenged with Pseudomonas syringae pv. tomato DC3000 (Pst) which has been widely used to infect A. thaliana seedlings (Yao et al., 2013) and, in addition, the non‐host interaction with Xanthomonas axonopodis pv. vesicatoria (Xav) was evaluated. These studies allowed us to gain a deeper insight into the function of these peptides in the plant defence response and to reveal that a bacterial PNP may be useful as a new candidate gene for crop improvement.
Results
Arabidopsis thaliana plants with modified expression of XacPNP and AtPNP‐A
To study whether XacPNP could trigger plant defence responses and to analyse the participation of AtPNP‐A in vivo in the defence response, A. thaliana plants with modified expression of these peptides were obtained. Transgenic Arabidopsis lines expressing XacPNP (named 35S:XacPNP) or AtPNP‐A (named 35S:AtPNP‐A) were obtained by transforming Col‐0 rdr6‐11 plants, as the background is deficient in silencing, allowing efficient over‐expression of these genes (Mecchia et al., 2013). The genes were cloned under the control of the 35S Cauliflower mosaic virus (CaMV) constitutive promoter in the binary vector pCHF3 (Fig. 1a). In addition, a mutant T‐DNA insertion line (SALK_000951C) with an insertion in the second intron of AtPNP‐A (atpnp‐A) (Alonso et al., 2003) and RNA interference (RNAi) lines with reduced levels of AtPNP‐A (AtPNP‐A‐RNAi) were characterized (Fig. 1a). To estimate the levels of XacPNP and AtPNP‐A expression, RNA was extracted from 10‐day‐old plants obtained in independent transformation events with 35S:XacPNP, 35S:AtPNP‐A or empty vector (EV) pCHF3, and reverse transcription‐polymerase chain reaction (RT‐PCR) and quantitative RT‐PCR (qRT‐PCR) assays were performed (Fig. S1, see Supporting Information). Figure 1b illustrates that 35S:XacPNP and 35S:AtPNP‐A lines show the highest expression. AtPNP‐A expression levels were similar between 35S:AtPNP‐A independent lines and at least seven‐fold higher than the expression of endogenous AtPNP‐A (Fig. 1b). 35S:XacPNP lines showed levels of expression of XacPNP of more than 100 times those of the endogenous AtPNP‐A in control plants (Fig. 1b). The expression of AtPNP‐A in several RNAi lines and in atpnp‐A lines (derived from SALK) was also determined and no transcripts were detected by RT‐PCR (Fig. S1). By qRT‐PCR, no peaks were detected in atpnp‐A lines, whereas, in RNAi lines, levels of expression of less than 3% relative to Col‐0 plants (with the empty pCHF3) were observed (Fig. 1b). The lines 35S:XacPNP 2, 35S:XacPNP 3, 35S:AtPNP‐A 1 and 35S:AtPNP‐A 3, with the highest expression levels, and atpnp‐A 2, atpnp‐A 4, AtPNP‐A‐RNAi7 and AtPNP‐A‐RNAi11 were chosen for further analyses of the roles of PNPs.
Figure 1.
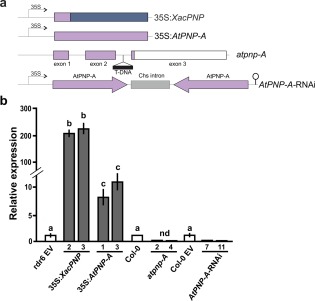
Construction of Arabidopsis thaliana lines with modified levels of plant natriuretic peptides (PNPs) and the analysis of PNP expression. (a) Schematic representation of the constructions used to obtain the transgenic lines. AtPNP‐A sequences are represented in light violet and XacPNP in blue. (b) Relative expression of PNPs analysed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of selected lines of transgenic plants and compared with the endogenous AtPNP‐A levels in control plants. rdr6 EV denotes Col‐0 rdr6‐11 bearing pCHF3, and Col‐0 EV denotes Col‐0 with pFGC5941 (EV, empty vector). nd, not detected. Values are the means of three biological replicates with three technical replicates each. Error bars indicate standard deviations. The data were analysed by analysis of variance (ANOVA) followed by Tukey's test. Bars with different letters are significantly different (P < 0.05).
PNPs increase stomatal opening
Phenotypic characterization of 35S:AtPNP‐A, 35S:XacPNP, AtPNP‐A‐RNAi and atpnp‐A lines showed no significant changes in morphology, root length, leaf area, rosette fresh weight and flowering time compared with their respective control lines or EV lines (Fig. S2, see Supporting Information). We have reported previously that treatment with recombinant XacPNP or AtPNP‐A results in a rapid and significant increase in stomatal conductance and aperture (Gottig et al., 2008). Thus, stomatal opening was measured in control and transgenic lines, as well as in controls treated with naphthalene acetic acid (NAA) and abscisic acid (ABA), which cause stomatal opening and closure, respectively. The stomatal aperture was increased significantly in PNP‐over‐expressing plants, whereas plants deficient in PNP showed reduced stomatal opening (P < 0.05) (Fig. 2).
Figure 2.
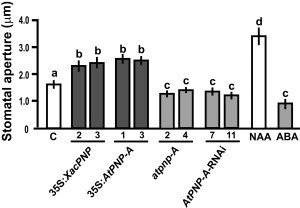
Plant natriuretic peptides (PNPs) promote stomatal aperture. Quantification of stomatal apertures in leaves of Arabidopsis thaliana transgenic lines. As control (C), Col‐0 and Col‐0 rdr6‐11 were analysed and showed similar apertures; thus, the average value was considered as the control. Naphthalene acetic acid (NAA) at 1 μm and abscisic acid (ABA) at 50 μm were used as controls of aperture and closure, respectively. Bars are the means of the apertures of 100 stomata, and the results are representative of three independent experiments. Error bars indicate standard deviations. The data were analysed by analysis of variance (ANOVA) followed by Tukey's test. Bars with different letters are significantly different (P < 0.05).
PNPs protect plants against saline and oxidative stress
In view of the proposed role of PNPs in the regulation of water and salt homeostasis, the response to saline stress was evaluated in the different A. thaliana lines. Plants were exposed to 150 mm NaCl at 1 week after germination. Experiments were performed in vitro in 0.5 X MS (Murashige and Skoog) agar because of the lack of reproducibility when they were performed in soil. Phenotypic characterization of the different lines in response to NaCl stress was evaluated. At 3 days post‐treatment, chlorosis and dehydration signals were evident mainly in atpnp‐A and AtPNP‐A‐RNAi plants, whereas 35S:XacPNP and 35S:AtPNP‐A lines showed better fitness (Fig. 3a). To further evaluate injury, the remaining chlorophyll was quantified, with 35S:XacPNP lines displaying about 70% remaining chlorophyll and 35S:AtPNP‐A lines showing about 50% compared with the untreated control (P < 0.05) (Fig. 3b). In the case of atpnp‐A and AtPNP‐A‐RNAi, on average, only 20% and 30% of remaining chlorophyll were observed relative to the control, respectively (P < 0.05) (Fig. 3b). Next, the behaviour of the different lines subjected to oxidative stress was evaluated. For this purpose, plants were challenged with the herbicide methyl viologen (MV), a superoxide‐generating agent (Youngman and Dodge, 1979). Two days after treatment, chlorotic lesions caused by the herbicide were observed. Chlorosis was greater in atpnp‐A and AtPNP‐A‐RNAi, whereas 35S:XacPNP and 35S:AtPNP‐A showed similar damage to the control lines (Fig. 3c). The amount of remaining chlorophyll was calculated, and 35S:XacPNP and 35S:AtPNP‐A plants showed slightly higher, but not significantly different (P < 0.05), levels of pigments compared with the controls. In contrast, PNP‐deficient lines showed almost three times less remaining chlorophyll than controls (P < 0.05) (Fig. 3d). Ion leakage was also quantified, and plants expressing XacPNP showed the lowest percentage of ion leakage, with a value of 18%, versus 38% observed in control plants (P < 0.05). The levels of ion leakage of 35S:AtPNP‐A plants were similar to those of control plants. PNP‐deficient plants were the most damaged, displaying values of 75%–82%, with controls about 40% (Fig. 3e).
Figure 3.
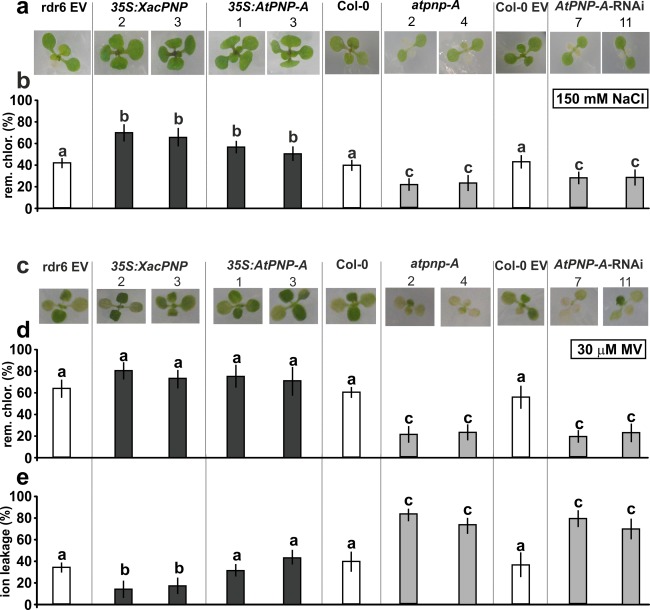
Plant natriuretic peptides (PNPs) protect Arabidopsis thaliana against saline and oxidative stress. (a) One‐week‐old plants were transplanted to 150 mm NaCl‐containing plates. Representative photographs are shown after 3 days of treatment. EV, empty vector. (b) Chlorophyll (rem. chlor.) was quantified at 3 days post‐treatment and is shown with reference to chlorophyll in the control treatment. (c) One‐week‐old plants were transplanted to 30 μm methyl viologen (MV)‐containing plates and representative photographs are shown after 2 days of treatment. (d) Chlorophyll was quantified at 2 days post‐treatment and is shown with reference to chlorophyll in the control. (e) Ion leakage quantification of leaves from the lines stated after 8 h of incubation with 30 μm MV. In (b), (d) and (e), values represent an average of 5–10 leaves per line and the experiments were repeated three times. Error bars are standard deviations. The data were analysed for statistical differences by one‐way analysis of variance (ANOVA) followed by Tukey's test. Bars with different letters are significantly different (P < 0.05).
XacPNP triggers A. thaliana defence responses
To analyse the participation of XacPNP in plant biotic stress triggering plant defence responses, A. thaliana Col‐0 leaves were infiltrated with the recombinant pure XacPNP protein and, at 6 h post‐inoculation (hpi), the transcript levels of several A. thaliana genes related to plant defence responses were analysed. RNA was extracted and real‐time qRT‐PCR was performed. Several defence marker genes were analysed: MITOGEN‐ACTIVATED PROTEIN KINASE 3 (MAPK3, At3g45640), MAP KINASE KINASE 4 (MKK4, At1g51660), WRKY30 transcription factor (WRKY30, At5g24110), GLUTATHIONE‐S‐TRANSFERASE 1 (GST1, At1g02930), PHENYLALANINE AMMONIA‐LYASE 1 (PAL1, At2g37040), PATHOGENESIS‐RELATED 1 (PR1, At2g14610) and PR5 (At1g75040). qRT‐PCR analysis showed that all transcripts were significantly (P < 0.05) more abundant in plants treated with XacPNP than in the 6 × His‐Trx‐infiltrated controls (His, histidine; Trx, thioredoxin), with a major up‐regulation for GST1, related to the oxidative stress response (Fig. 4a).
Figure 4.
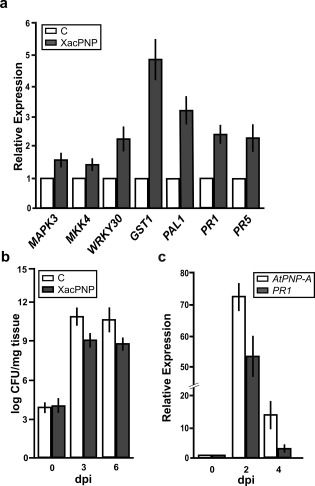
Induction of the defence response by XacPNP, and AtPNP‐A expression pattern, on Pseudomonas syringae pv. tomato (Pst) infection. (a) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of Arabidopsis thaliana genes related to the defence response (see text for gene definitions). RNA was extracted from leaves infiltrated with 5 µm XacPNP at 6 h post‐inoculation (hpi). Bars indicate the expression levels of the indicated gene relative to the control treatment. As control, 5 µm 6 × His‐Trx was used (His, histidine; Trx, thioredoxin). (b) Enhancement of the defence response mediated by XacPNP. Quantification of Pst growth at 0, 3 and 6 days post‐inoculation (dpi) in leaves pre‐infiltrated with 5 µm XacPNP and 6 × His‐Trx as control. CFU, colony‐forming unit. (c) qRT‐PCR of AtPNP‐A and PR1 in A. thaliana leaves infected with Pst at 0, 2 and 4 dpi. Values are the means of three biological replicates with three technical replicates each. Error bars indicate standard deviations. The data were analysed by analysis of variance (ANOVA) (P < 0.05).
To further evaluate whether XacPNP is involved in the defence response, the ability of this protein to enhance A. thaliana Col‐0 resistance to Pst was evaluated. Leaves were pre‐infiltrated with 5 µm XacPNP and 6 × His‐Trx as a control. The pre‐infiltrated leaves were then infiltrated with Pst at 5 × 106 colony‐forming units (CFU)/mL, and bacterial growth at 3 and 6 days post‐inoculation (dpi) was monitored. At both times analysed, XacPNP was able to induce defence responses, reducing significantly (P < 0.05) the population of bacteria by about two orders of magnitude (Fig. 4b).
AtPNP‐A expression is induced in A. thaliana on Pst infection
AtPNP‐A expression in leaves infected with Pst was quantified. Plants were infected with Pst at 5 × 106 CFU/mL and, at 0, 2 and 4 dpi, tissue samples were collected, RNA was extracted and expression levels of AtPNP‐A were quantified. As a marker of infection, the expression of PR1 was also evaluated. AtPNP‐A and PR1 expression showed a significant increase (P < 0.05) with a similar pattern in infected A. thaliana tissues relative to non‐infected tissues (Fig. 4c). Altogether, these results suggest that XacPNP can promote defence responses in A. thaliana and that AtPNP‐A also contributes to the plant defence response.
PNPs increase defence responses in both host and non‐host interactions
The response of A. thaliana lines with modified levels of PNPs to pathogens was evaluated. Flood inoculation with the virulent pathogen Pst at 1 × 106 CFU/mL was performed. This method allows reproducibility of the phenotypes observed and is more similar to natural infection than is inoculation by infiltration (Ishiga et al., 2011). The different lines showed similar infection phenotypes at 3 dpi, except for 35S:XacPNP lines which displayed less damaged tissue (Fig. 5a). To better characterize the infection process, in planta bacterial growth measurements were performed. At 3 and 5 dpi, PNP‐over‐expressing plants showed a decrease by one or two orders of magnitude in bacterial number compared with control lines, whereas PNP‐deficient lines exhibited an increased bacterial growth at both times, reaching populations of 1012 CFU/mg of tissue at 5 dpi, with control lines reaching only 1010 CFU/mg (Fig. 5b). To understand the differences in disease phenotypes and taking into consideration the increase in defence gene expression in recombinant XacPNP‐treated leaves (Fig. 4a), the expression of different genes related to the defence response was analysed at 24 hpi. 35S:XacPNP and 35S:AtPNP‐A plants showed a major induction of MKK4, WRKY30, PAL1 and PR1 genes relative to control plants, whereas, in PNP‐deficient lines, the opposite occurred (Fig. 5c).
Figure 5.
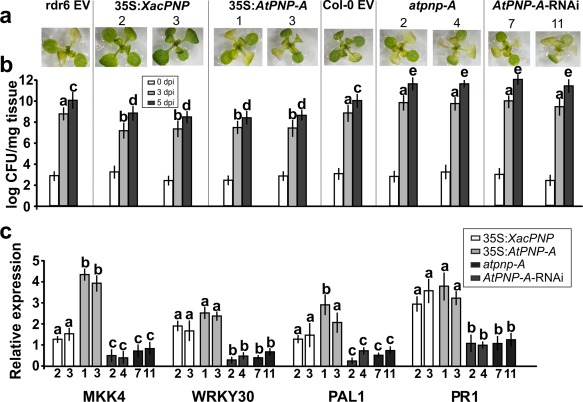
Plant natriuretic peptides (PNPs) protect Arabidopsis thaliana against Pseudomonas syringae pv. tomato (Pst). Two 3‐week‐old plants were flood inoculated with Pst. (a) Representative images of plants at 3 days post‐inoculation (dpi). Numbers represent independent lines. EV, empty vector. (b) Quantification of bacterial population size at the times stated. Bars indicate the mean bacterial population as log colony‐forming units (CFU)/mg tissue. An average of four plants was used for each line. The data were analysed by analysis of variance (ANOVA) followed by Tukey's test (P < 0.05). Bars with different letters are significantly different (P < 0.05). For the sake of clarity, letters for the population at 0 dpi were omitted. (c) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of A. thaliana genes related to the defence response at 24 h post‐inoculation (hpi). Numbers on the x‐axis represent independent transgenic lines. Bars indicate the gene expression levels, relative to infected control plants that were not included in the figure. In all cases, experiments were repeated three times and error bars indicate standard deviations. The data were analysed by analysis of variance (ANOVA) followed by Tukey's test. Bars with different letters are significantly different (P < 0.05). Comparisons were performed among different lines for each gene.
Inoculation with Xav at 5 × 107 CFU/mL was assayed. In this case, all the lines showed the typical defence response, known as the hypersensitive response (HR), with the symptoms observed being similar in 35S:XacPNP and 35S:AtPNP‐A plants compared with control lines at 24 hpi; however, atpnp‐A and AtPNP‐A‐RNAi plants showed more damaged tissue (Fig. 6a). This was also evidenced by the detection of hydrogen peroxide (H2O2) at 18 hpi, assessed by staining the plants with 3,3′‐diaminobenzidine (DAB). Microscopic observations (Fig. 6b) and quantification of DAB staining intensities (Fig. 6c) showed that, in PNP‐deficient lines, H2O2 production was significantly greater than in control lines, and, in PNP‐over‐expressing lines, this production was diminished (P < 0.05).
Figure 6.
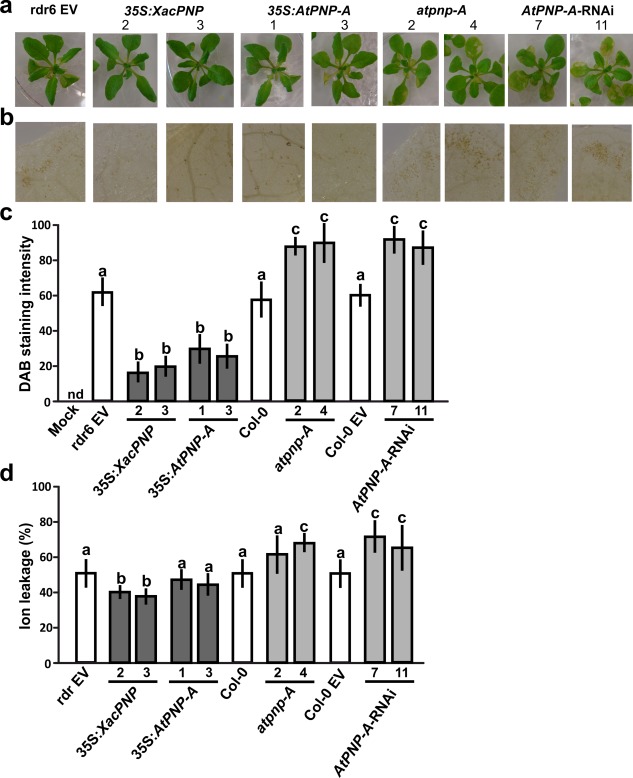
Plant natriuretic peptides (PNPs) alleviate hypersensitive response (HR) symptoms in the interaction of Arabidopsis thaliana with Xanthomonas axonopodis pv. vesicatoria (Xav). Four‐week‐old plants were flood inoculated with Xav. (a) Representative images of plants at 2 days post‐inoculation (dpi). Col‐0 and Col‐0 EV showed similar symptoms to Col‐0 rdr6 EV (rdr6 EV) and, because of the large amount of data, were not included. EV, empty vector. (b) 3,3′‐Diaminobenzidine (DAB) detection of H2O2 accumulation in leaves inoculated with Xav at 1 dpi. Representative photographs of DAB‐stained leaves are shown. (c) Quantification of DAB staining from the microscopic images as described in Experimental procedures. The means were calculated from five photographs from three independent experiments for each line. Error bars indicate standard deviations. Mock indicates treatment with sterile water and Silwett L‐77 0.025% (v/v). nd, not detected. The data were analysed by analysis of variance (ANOVA) followed by Tukey's test. Bars with different letters are significantly different (P < 0.05). (d) Ion leakage quantification at 18 h post‐inoculation (hpi). Values represent an average of four plants assayed for each line and the experiment was repeated three times. Error bars are standard deviations. The data were analysed by ANOVA followed by Tukey's test. Bars with different letters are significantly different (P < 0.05).
Finally, ion leakage was analysed in Xav‐treated plants and significant major damage was observed in atpnp‐A4 and AtPNP‐A‐RNAi lines, whereas, in 35S:XacPNP, ion leakage was lower than in control plants (P < 0.05). In the case of 35S:AtPNP‐A, no significant differences relative to the control were observed (Fig. 6d).
Discussion
It is known that PNPs modulate salt and water homeostasis (Gehring and Irving, 2013). In addition, the participation of AtPNP‐A in the plant defence response against pathogens has been proposed, taking into account that it is co‐expressed together with genes involved in this response (Meier et al., 2008; Turek et al., 2014). Moreover, the recently found AtPNP‐A receptor, which is a leucine‐rich repeat receptor‐like kinase and belongs to a family of proteins associated with plant defence responses to pathogens (Turek and Gehring, 2016), strengthens the idea that PNPs may be involved in the response to plant pathogens.
Previously, we have demonstrated that XacPNP and AtPNP‐A, expressed in Xac, are able to limit necrotic progress and to improve photosynthetic efficiency and water homeostasis during disease in citrus leaves (Garavaglia et al., 2010a; Gottig et al., 2008). In this work, we have revealed a role for PNPs in plant defence responses. The following observations indicate that PNPs play a role in plant defence responses: (i) pure XacPNP induces the expression of defence genes in Arabidopsis plants; (ii) pretreatment with recombinant XacPNP makes these plants more resistant to Pst; and (iii) AtPNP‐A shows a similar pattern of expression to PR1 on infection. Moreover, and for the first time, transgenic A. thaliana lines with modified levels of PNP expression were obtained to further evaluate the role of PNP in defence responses. These plants displayed different phenotypes with regard to stomatal aperture. Previously, we have observed that pure XacPNP or AtPNP‐A proteins are able to induce stomatal opening (Gottig et al., 2008). Accordingly, plants over‐expressing XacPNP or AtPNP‐A show an increase in stomatal aperture relative to control plants and PNP‐deficient lines show a clear decrease in stomatal aperture, providing support for the function previously observed for PNPs (Gottig et al., 2008). The active closure of stomata on bacterial contact is a widespread defence response in plants exposed to potential pathogens. Stomata play a role in the early phases of innate immunity; in particular, stomata are closed 1–2 h after bacterial recognition, preventing their entry into the host tissue (Melotto et al., 2006). However, Arabidopsis mutants that keep their stomata open in response to Pst, and thus do not respond as a barrier to bacterial invasion, do not show significantly higher susceptibility to the pathogen (Zeng et al., 2011). Accordingly, 35S:XacPNP and 35S:AtPNP‐A plants, although showing increased stomatal aperture to wild‐type plants, do not display increased susceptibility, and PNP‐deficient plants, although showing decreased stomatal aperture, do not show more resistance to Pst. These results suggest that the stomatal aperture of different plant lines does not determine bacterial entrance in this instance.
PNPs were also able to protect A. thaliana against saline stress, which is related to the known function of PNPs as regulators of K+, Na+ and H+ fluxes. In this work, another function has been attributed to PNPs, i.e. protection against oxidative stress. A proteomic analysis of PNP‐treated leaves showed an over‐representation of the gene ontology (GO) terms ‘oxidation–reduction process’, ‘translation’ and ‘response to salt stress’ (Turek et al., 2014). These results, together with our findings, indicate the participation of PNPs in the modulation of oxidative cell responses, in addition to their role in the regulation of ion fluxes.
In plant–pathogen interactions, challenge with Pst, which causes disease, revealed that transgenic plants over‐expressing XacPNP or AtPNP‐A were more resistant to this pathogen relative to control plants; bacterial growth was impaired, possibly through the increased expression of defence genes. In contrast, PNP‐deficient plants showed the opposite effect, being more susceptible to Pst. In the case of the non‐host response challenged with Xav, PNP‐deficient plants displayed stronger HR than both PNP‐over‐expressing and control plants. Although HR is related to the defence response, this stronger HR might be a consequence of larger tissue damage in the absence of PNP, as PNP has a role in tissue health maintenance. The role of this peptide as a molecule that maintains plant tissues in a healthier state and that also induces the defence response may be an adaptive reaction of the plant to fight against pathogens without the need to damage its own tissue. In general, 35S:XacPNP plants displayed a more noticeable phenotype than 35S:AtPNP‐A plants, and this may be a result of the higher levels of expression observed in the former. It has been shown that AtPNP‐A is able to modulate its own expression (Wang et al., 2011). Although a Col‐0 line impaired in gene silencing was used, and expression was under the control of the 35S CaMV promoter, AtPNP‐A may have been subjected to additional regulation, considering that XacPNP is a bacterial gene not encoded in the plant genome. It is worth mentioning that XacPNP is able to exert its function when infiltrated as a pure protein or when expressed by the plant. The recent finding of the AtPNP‐A receptor protein leads to the question of whether XacPNP interacts with this receptor and whether it has the same affinity as the Arabidopsis peptide. Both peptides share a conserved active site (Nembaware et al., 2004) and this region in AtPNP‐A is sufficient for interaction with the receptor to occur (Turek and Gehring, 2016); this suggests that they may have similar affinity for the receptor protein, and therefore the over‐expression of XacPNP, which is higher than the over‐expression of AtPNP‐A, may explain the more pronounced phenotypes observed in 35S:XacPNP plants. Our results reinforce the idea that XacPNP mimics its cognate protein in plants and, although it has been adapted to be expressed in the unique bacterium Xcc, it conserves its biological activity in its plant counterpart.
In summary, we have revealed a role for AtPNP‐A and bacterial XacPNP in biotic and abiotic stress responses, and we propose that PNPs are part of a concerted action to cope with the environmentally changing conditions to which plants must adapt, ranging from oxidative stress and salt stress to the challenge of a pathogen attack. Moreover, this work enhances our understanding of a bacterial‐acquired gene that mimics AtPNP‐A function in defence responses to plant pathogens.
Experimental Procedures
Plant material and growth conditions
Arabidopsis plants ecotype Columbia Col‐0 rdr6‐11 (At3g49500) (Mecchia et al., 2013) were used for the construction of plants over‐expressing PNPs, in order to achieve higher gene expression by avoiding gene silencing (Butaye et al., 2004). Arabidopsis plants ecotype Columbia (Col‐0) were used for the construction of AtPNP‐A‐RNAi experiments, and the Col‐0 (SALK_000951C) insertional mutant was purchased from the Arabidopsis Biological Resource Center, Columbus, OH, USA. Homozygous mutants from this line were isolated by PCR‐based genotyping using the gene‐specific PCR primers (AtPNPs, Table S1, see Supporting Information). Seeds were surface sterilized with ethanol 70% (v/v) and Tween‐20 0.1% (v/v) for 10 min, and spread on Murashige and Skoog (MS) basal medium plates. Seeds on plates were stratified for 2 days and then kept in growth chambers at 22 °C under fluorescent light, with 16 h : 8 h photoperiods. Seedlings were transferred to soil pots and kept in the same conditions until harvest.
Arabidopsis transformation
Arabidopsis thaliana transgenic plants over‐expressing XacPNP or AtPNP‐A and RNAi AtPNP‐A lines were obtained. AtPNP‐A was amplified from Col‐0 WT cDNA using 35S:AtPNP oligonucleotides (Table S1) and subcloned into KpnI‐BamHI of the pCHF3 binary vector under the control of the CaMV 35S promoter. For XacPNP over‐expression, the plant PNP signal peptide was fused to the XacPNP coding sequence. The signal peptide was amplified using PNP‐SP primers and XacPNP using 35S:XacPNP primers. Both PCR products were used as templates in a second round of amplification, and the final fusion product was subcloned into the pCHF3 binary vector under the control of the 35S CaMV promoter. For RNAi lines, AtPNP‐A was amplified using AtPNP‐A‐RNAi primers and the product was subcloned into AscI‐SwaI and XbaI‐BamHI sites from vector pFGC5941, which allows efficient silencing of the gene of interest (www.arabidopsis.org). The resulting constructs, pCHF3‐XacPNP, pCHF3‐AtPNP‐A and pFGC5941‐AtPNP‐A, were introduced into Agrobacterium tumefaciens GV3101 pMP90 strain by electroporation. Agrobacterium‐mediated plant transformation using floral dip was used to obtain transgenic lines in several independent transformations for each construct (Zhang et al., 2006). T1 seeds were selected on plates containing kanamycin when the vector pCHF3 was used, or by spraying the herbicide BASTA to soil‐grown plants when the vector pFGC5941 was used. Homozygous T3 or T4 seeds were obtained and several independent lines were analysed for each construct. For phenotype controls, Arabidopsis lines were obtained using both Col‐0 and Col‐0 rdr6‐11 plants transformed with empty pCHF3 and pFGC5941 vectors (EV).
RNA preparation, RT‐PCR and qRT‐PCR
Total RNA from leaves was isolated using TRIzol® reagent (Invitrogen, Grand Island, NY, USA), according to the manufacturer's instructions. RT‐PCRs and qRT‐PCRs were performed as described previously (Sgro et al., 2012), with the specific oligonucleotides detailed in Table S1. Values are the means of three biological replicates with three technical replicates each.
Stomatal opening assay
Leaves from 3–4‐week‐old plants were used. Individual leaves were incubated in 1 : 1 ethanol–KOH 5% (w/v) at 60 ºC for 30 min, and then washed and cleared using ethanol. Leaves were stained with 1% (w/v) safranin solution to increase contrast. NAA (1 μm) and ABA (50 μm) were used as controls. In this case, leaves were incubated for 2 h in 10 mm 2‐(N‐morpholino) ethanesulfonic acid (MES) and 10 mm KCl (pH 6.5), supplemented with each hormone. The pore widths of around 100–150 stomata were measured under the microscope with a calibrated ocular micrometer.
Saline and oxidative stress treatments
Seeds from the different A. thaliana lines were grown in 0.5 X MS (Murashige and Skoog) agar 0.8% for 7 days. They were then transplanted to MS plates as controls and to MS plates with the addition of NaCl (150 mm) to evaluate saline stress or MV (30 μm) to investigate oxidative stress.
Bacterial strains and growth conditions
Pst and Xav were grown at 28 °C in Luria–Bertani (LB) medium and Silva Buddenhagen (SB) medium (5 g/L sucrose, 5 g/L yeast extract, 5 g/L peptone and 1 g/L glutamic acid, pH 7.0), respectively, with constant agitation at 200 rpm on a rotating shaker.
Plant inoculation and in planta growth assays
Plant flood inoculation was performed as described previously (Ishiga et al., 2011). Col‐0 or Col‐0 rdr6‐11 plants were grown in Petri dishes containing 0.5 X MS (Murashige and Skoog) agar, vitamins and phytagel 0.3% (w/v) for 2 weeks. Bacterial cultures were diluted to the required concentrations in sterile water supplemented with Silwett L‐77 0.025% (v/v), and incubated with the plants for 3 min; then, the plates were drained and incubated in a growth chamber. Bacterial growth assays were performed by grinding previously weighed leaves in a 10 mm MgCl2 solution, diluted and plated in SB or LB plates to determine the fresh weight (CFU/mg). Experiments were repeated three times and plants from two plates were evaluated per bacterial strain in each experiment. For the analysis of Pst growth in leaves pre‐infiltrated with XacPNP, leaves were infiltrated with needleless syringes with 5 μm purified XacPNP and 5 μm 6 × His‐Trx as a control. Proteins were purified as described previously (Gottig et al., 2008). After 6 h, these leaves were infiltrated with a Pst suspension at 106 CFU/mL. Growth assays were performed from 10 infiltrated leaves for each treatment at the indicated times as detailed above.
Chlorophyll and ion leakage measurements
These experiments were performed as described previously (Dunger et al., 2012). Chlorophyll extraction was performed at 3 days post‐treatment after saline stress and at 2 days post‐treatment after oxidative stress. Values are expressed as the remaining percentage of chlorophyll compared with control infiltrated tissue. Ion leakage in Xav‐inoculated plants was performed at 18 h post‐treatment by placing leaf discs in 2 mL of water. In MV‐treated leaves, assay was performed at 8 h post‐treatment. Conductivity measurements were taken from each tube. The conductance of boiled tissue was measured and the values were used as representative of 100% of ion content (Dunger et al., 2005). The experiments were performed with three leaves and repeated three times.
DAB staining
Plants from the different lines were flood inoculated with Xav (107 CFU/mL) and, after 1 day, leaves were cut and the petioles were submerged in 0.1% (w/v) DAB solution and kept in the dark overnight. The leaves were cleared in ethanol, and observed and photographed in an optical microscope. DAB intensity was calculated from the digital photographs by the number of brown pixels relative to the total number of pixels covering the plant material, using Photoshop CS3 software (Adobe Systems Incorporated, San Jose, CA, USA). Average DAB measurements were calculated from at least five photographs from three independent experiments.
Statistical analysis
In all figures, bars show the mean of the data and error bars show the standard deviation. All data were analysed by one‐way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) test when stated.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of oligonucleotides used in this work.
Fig. S1 Analysis of the expression levels of plant natriuretic peptides (PNPs) in genetically modified plants. Reverse transcription‐polymerase chain reaction (RT‐PCR) (a) and quantitative RT‐PCR (qRT‐PCR) (b) performed on selected lines of transgenic plants with modified levels of PNPs. EV represents Col‐0 rdr6‐11 plants transformed with pCHF3 when the expression of either AtPNP‐A or XacPNP was analysed in 35S:XacPNP and 35S:AtPNP‐A lines (top), whereas EV represents Col‐0 plants transformed with pFGC5941 when the expression of AtPNP‐A was analysed in AtPNP‐A‐RNAi plants. C is the PCR‐positive control using Xanthomonas citri ssp. citri (Xcc) DNA as template. nd, not detected; tub, tubulin.
Fig. S2 Phenotypic analysis of plants with modified levels of plant natriuretic peptides (PNPs). Plants were grown in soil in similar conditions and the root length, fresh weight of rosettes and area of leaf 5 of 28‐day‐old plants were determined. The flowering time was established as the day on which the first flower appeared. No significant differences were observed. Ten plants were assayed for each line and the experiment was repeated twice. Error bars are standard deviations. The data were analysed for statistical differences by one‐way analysis of variance (ANOVA) (P < 0.05).
Acknowledgements
We thank Diego Aguirre (IBR‐CONICET) for plant technical assistance. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT2013‐0625) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (PIP2014‐2016). B.S.G., N.G. and J.O. are staff members and F.A.F. and C.G. are fellows of CONICET.
References
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Billington, T. , Pharmawati, M. and Gehring, C.A. (1997) Isolation and immunoaffinity purification of biologically active plant natriuretic peptide. Biochem. Biophys. Res. Commun. 235, 722–725. [DOI] [PubMed] [Google Scholar]
- Butaye, K.M. , Goderis, I.J. , Wouters, P.F. , Pues, J.M. , Delaure, S.L. , Broekaert, W.F. , Depicker, A. , Cammue, B.P. and De Bolle, M.F. (2004) Stable high‐level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39, 440–449. [DOI] [PubMed] [Google Scholar]
- Dunger, G. , Arabolaza, A.L. , Gottig, N. , Orellano, E.G. and Ottado, J. (2005) Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and nonhost plant responses. Plant Pathol. 54, 781–788. [Google Scholar]
- Dunger, G. , Garofalo, C.G. , Gottig, N. , Garavaglia, B.S. , Rosa, M.C. , Farah, C.S. , Orellano, E.G. and Ottado, J. (2012) Analysis of three Xanthomonas axonopodis pv. citri effector proteins in pathogenicity and their interactions with host plant proteins. Mol. Plant Pathol. 13, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia, B.S. , Thomas, L. , Gottig, N. , Dunger, G. , Garofalo, C.G. , Daurelio, L.D. , Ndimba, B. , Orellano, E.G. , Gehring, C. and Ottado, J. (2010a) A eukaryotic‐acquired gene by a biotrophic phytopathogen allows prolonged survival on the host by counteracting the shut‐down of plant photosynthesis. PLoS One, 5, e8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia, B.S. , Thomas, L. , Zimaro, T. , Gottig, N. , Daurelio, L.D. , Ndimba, B. , Orellano, E.G. , Ottado, J. and Gehring, C. (2010b) A plant natriuretic peptide‐like molecule of the pathogen Xanthomonas axonopodis pv. citri causes rapid changes in the proteome of its citrus host. BMC Plant Biol. 10, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, C.A. and Irving, H.R. (2003) Natriuretic peptides – a class of heterologous molecules in plants. Int. J. Biochem. Cell Biol. 35, 1318–1322. [DOI] [PubMed] [Google Scholar]
- Gehring, C. and Irving, H. (2013) Plant natriuretic peptides: systemic regulators of plant homeostasis and defense that can affect cardiomyoblasts. J. Invest. Med. 61, 823–826. [DOI] [PubMed] [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Daurelio, L.D. , Valentine, A. , Gehring, C. , Orellano, E.G. and Ottado, J. (2008) Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide‐like protein to modify host homeostasis. Proc. Natl. Acad. Sci. USA, 105, 18 631–18 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga, Y. , Ishiga, T. , Uppalapati, S.R. and Mysore, K.S. (2011) Arabidopsis seedling flood‐inoculation technique: a rapid and reliable assay for studying plant–bacterial interactions. Plant Methods, 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludidi, N. , Morse, M. , Sayed, M. , Wherrett, T. , Shabala, S. and Gehring, C. (2004) A recombinant plant natriuretic peptide causes rapid and spatially differentiated K+, Na+ and H+ flux changes in Arabidopsis thaliana roots. Plant Cell Physiol. 45, 1093–1098. [DOI] [PubMed] [Google Scholar]
- Maryani, M.M. , Morse, M.V. , Bradley, G. , Irving, H.R. , Cahill, D.M. and Gehring, C.A. (2003) In situ localization associates biologically active plant natriuretic peptide immuno‐analogues with conductive tissue and stomata. J. Exp. Bot. 54, 1553–1564. [DOI] [PubMed] [Google Scholar]
- Mecchia, M.A. , Debernardi, J.M. , Rodriguez, R.E. , Schommer, C. and Palatnik, J.F. (2013) MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech. Dev. 130, 2–13. [DOI] [PubMed] [Google Scholar]
- Meier, S. , Bastian, R. , Donaldson, L. , Murray, S. , Bajic, V. and Gehring, C. (2008) Co‐expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 8, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Morse, M. , Pironcheva, G. and Gehring, C. (2004) AtPNP‐A is a systemically mobile natriuretic peptide immunoanalogue with a role in Arabidopsis thaliana cell volume regulation. FEBS Lett. 556, 99–103. [DOI] [PubMed] [Google Scholar]
- Nembaware, V. , Seoighe, C. , Sayed, M. and Gehring, C. (2004) A plant natriuretic peptide‐like gene in the bacterial pathogen Xanthomonas axonopodis may induce hyper‐hydration in the plant host: a hypothesis of molecular mimicry. BMC Evol. Biol. 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmawati, M. , Billington, T. and Gehring, C.A. (1998a) Stomatal guard cell responses to kinetin and natriuretic peptides are cGMP‐dependent. Cell Mol. Life Sci. 54, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmawati, M. , Gehring, C. and Irving, H.R. (1998b) An immunoaffinity purified plant natriuretic peptide analogue modulates cGMP levels in the Zea mays root stele. Plant Sci. 137, 105–115. [Google Scholar]
- Pharmawati, M. , Shabala, S.N. , Newman, I.A. and Gehring, C.A. (1999) Natriuretic peptides and cGMP modulate K+, Na+, and H+ fluxes in Zea mays roots. Mol. Cell Biol. Res. Commun. 2, 53–57. [DOI] [PubMed] [Google Scholar]
- Potter, L.R. (2011) Natriuretic peptide metabolism, clearance and degradation. FEBS J. 278, 1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzvidzo, O. , Donaldson, L. , Valentine, A. and Gehring, C. (2011) The Arabidopsis thaliana natriuretic peptide AtPNP‐A is a systemic regulator of leaf dark respiration and signals via the phloem. J. Plant Physiol. 168, 1710–1714. [DOI] [PubMed] [Google Scholar]
- Sgro, G.G. , Ficarra, F.A. , Dunger, G. , Scarpeci, T.E. , Valle, E.M. , Cortadi, A. , Orellano, E.G. , Gottig, N. and Ottado, J. (2012) Contribution of a harpin protein from Xanthomonas axonopodis pv. citri to pathogen virulence. Mol. Plant Pathol. 13, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwastika, I.N. and Gehring, C.A. (1999) The plasma membrane H+‐ATPase from Tradescantia stem and leaf tissue is modulated in vitro by cGMP. Arch. Biochem. Biophys. 367, 137–139. [DOI] [PubMed] [Google Scholar]
- Suwastika, I.N. , Toop, T. , Irving, H. and Gehring, C. (2000) In situ and in vivo binding of natriuretic peptide hormones in Tradescantia multiflora . Plant Biol. 2, 1–3. [Google Scholar]
- Turek, I. and Gehring, C. (2016) The plant natriuretic peptide receptor is a guanylyl cyclase and enables cGMP‐dependent signaling. Plant Mol. Biol. 91, 275–286. [DOI] [PubMed] [Google Scholar]
- Turek, I. , Marondedze, C. , Wheeler, J.I. , Gehring, C. and Irving, H.R. (2014) Plant natriuretic peptides induce proteins diagnostic for an adaptive response to stress. Front. Plant Sci. 5, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.H. , Gehring, C. and Irving, H.R. (2011) Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 52, 837–850. [DOI] [PubMed] [Google Scholar]
- Yao, J. , Withers, J. and He, S.Y. (2013) Pseudomonas syringae infection assays in Arabidopsis. Methods Mol. Biol. 1011, 63–81. [DOI] [PubMed] [Google Scholar]
- Youngman, R.J. and Dodge, A.D. (1979) Mechanism of paraquat action: inhibition of the herbicidal effect by a copper chelate with superoxide dismutating activity. Z. Naturforsch. C, 34, 1033–1035. [PubMed] [Google Scholar]
- Zeng, W. , Brutus, A. , Kremer, J.M. , Withers, J.C. , Gao, X. , Jones, A.D. and He, S.Y. (2011) A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7, e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of oligonucleotides used in this work.
Fig. S1 Analysis of the expression levels of plant natriuretic peptides (PNPs) in genetically modified plants. Reverse transcription‐polymerase chain reaction (RT‐PCR) (a) and quantitative RT‐PCR (qRT‐PCR) (b) performed on selected lines of transgenic plants with modified levels of PNPs. EV represents Col‐0 rdr6‐11 plants transformed with pCHF3 when the expression of either AtPNP‐A or XacPNP was analysed in 35S:XacPNP and 35S:AtPNP‐A lines (top), whereas EV represents Col‐0 plants transformed with pFGC5941 when the expression of AtPNP‐A was analysed in AtPNP‐A‐RNAi plants. C is the PCR‐positive control using Xanthomonas citri ssp. citri (Xcc) DNA as template. nd, not detected; tub, tubulin.
Fig. S2 Phenotypic analysis of plants with modified levels of plant natriuretic peptides (PNPs). Plants were grown in soil in similar conditions and the root length, fresh weight of rosettes and area of leaf 5 of 28‐day‐old plants were determined. The flowering time was established as the day on which the first flower appeared. No significant differences were observed. Ten plants were assayed for each line and the experiment was repeated twice. Error bars are standard deviations. The data were analysed for statistical differences by one‐way analysis of variance (ANOVA) (P < 0.05).


