Summary
The initial interaction of a pathogenic fungus with its host is complex and involves numerous metabolic pathways and regulatory proteins. Considerable attention has been devoted to proteins that play a crucial role in these interactions, with an emphasis on so‐called effector molecules that are secreted by the invading microbe to establish the symbiosis. However, the contribution of other types of molecules, such as glycans, is less well appreciated. Here, we present a random genetic screen that enabled us to identify 58 novel candidate genes that are involved in the pathogenic potential of the fungal pathogen Verticillium dahliae, which causes vascular wilt diseases in over 200 dicotyledonous plant species, including economically important crops. One of the candidate genes that was identified concerns a putative biosynthetic gene involved in nucleotide sugar precursor formation, as it encodes a putative nucleotide‐rhamnose synthase/epimerase‐reductase (NRS/ER). This enzyme has homology to bacterial enzymes involved in the biosynthesis of the nucleotide sugar deoxy‐thymidine diphosphate (dTDP)‐rhamnose, a precursor of L‐rhamnose, which has been shown to be required for virulence in several human pathogenic bacteria. Rhamnose is known to be a minor cell wall glycan in fungi and has therefore not been suspected as a crucial molecule in fungal–host interactions. Nevertheless, our study shows that deletion of the VdNRS/ER gene from the V. dahliae genome results in complete loss of pathogenicity on tomato and Nicotiana benthamiana plants, whereas vegetative growth and sporulation are not affected. We demonstrate that VdNRS/ER is a functional enzyme in the biosynthesis of uridine diphosphate (UDP)‐rhamnose, and further analysis has revealed that VdNRS/ER deletion strains are impaired in the colonization of tomato roots. Collectively, our results demonstrate that rhamnose, although only a minor cell wall component, is essential for the pathogenicity of V. dahliae.
Keywords: Agrobacterium tumefaciens‐mediated transformation (ATMT), attachment, carbohydrate, root colonization, tomato, UDP‐rhamnose, vascular wilt
Introduction
Vascular wilt diseases caused by soil‐borne pathogens are amongst the most devastating plant diseases worldwide. The fungus Verticillium dahliae causes vascular wilt diseases in over 200 dicotyledonous plant species, including important crops, such as eggplant, lettuce, olive, spinach and tomato (Fradin and Thomma, 2006; Klosterman et al., 2009). Triggered by root exudates, the melanized resting structures, microsclerotia, germinate and penetrate the roots through the root tips, wounds or sites of lateral root formation (Fradin and Thomma, 2006). After crossing the cortex, the hyphae of the fungus grow into the xylem vessels. The mycelium remains exclusively in these vessels and produces conidia which are transported acropetally with the water flow in the xylem throughout the plant. On senescence, microsclerotia are produced that are released into the soil on tissue decomposition (Wilhelm, 1955). Typical symptoms of plants infected with V. dahliae comprise stunting, wilting, chlorosis and vascular browning (Pegg and Brady, 2002).
Verticillium wilt diseases are difficult to control because of the longevity of the microsclerotia and the inability of fungicides to eliminate the fungus once it has entered the xylem tissues of the host plant (Fradin and Thomma, 2006; Wilhelm, 1955). Moreover, crop rotation is ineffective because of the broad host range of V. dahliae. The only effective control measure, soil fumigation, is expensive and has harmful environmental effects (Rowe et al., 1987). As a consequence, genetic resistance is presently preferred to control Verticillium wilt diseases. Importantly, Verticillium resistance has been described in several plant species, including crops (Bolek et al., 2005; Hayes et al., 2011; Schaible et al., 1951; Simko et al., 2004). From tomato (Solanum lycopersicum), the Ve1 Verticillium resistance gene has been cloned that mediates resistance to race 1 strains of V. dahliae that express the secreted Ave1 effector (Fradin et al., 2009; de Jonge et al., 2012). Putative Ve1 orthologues have been identified within and outside the Solanaceae family, suggesting that Ve1‐mediated Verticillium resistance may be widespread in plants (Thomma et al., 2011). Interestingly, Ave1 contributes to V. dahliae virulence on susceptible tomato plants, and homologues occur in a handful of taxonomically unrelated plant‐pathogenic fungi (de Jonge et al., 2012).
Various other virulence genes have been identified in V. dahliae, including members of the expanded LysM effector and NEP‐like (NLP) protein family (de Jonge et al., 2013; Klimes et al., 2015; Klosterman et al., 2011; Santhanam et al., 2013). Furthermore, genome analysis has revealed the relative abundance of particular families of cell wall‐degrading enzymes (CWDEs) when compared with other fungi (Klosterman et al., 2011). The importance of CWDEs during pathogenicity was demonstrated by the disruption of the CWDE regulator VdSNF1, which reduced V. dahliae virulence (Tzima et al., 2011). In addition, genes involved in nitrogen and amino acid metabolism were found to be important for establishment in the nutrient‐poor vascular system (Singh et al., 2010; Timpner et al., 2013). Finally, virulence genes have been described that are also involved in stress tolerance (Klimes et al., 2015).
The identification and subsequent characterization of novel genes involved in disease establishment are of fundamental importance to understand the interaction between V. dahliae and its hosts, and to potentially design novel strategies for disease control. An effective way to identify novel pathogenicity genes in plant‐pathogenic fungi is the use of random mutagenesis coupled with a rapid technique to screen the mutants for phenotypes. Agrobacterium tumefaciens is a Gram‐negative soil bacterium that can infect hosts as diverse as fungi, oomycetes and plants, and can be used to randomly insert a transfer DNA (T‐DNA) into the host genome. Over recent years, A. tumefaciens‐mediated transformation (ATMT) has been used to identify genes involved in disease development in various plant pathogens (Giesbert et al., 2012; Hüser et al., 2009; Jeon et al., 2007; Korn et al., 2015; Maruthachalam et al., 2011; Michielse et al., 2009; Munch et al., 2011; Ramos et al., 2013; Schumacher et al., 2014; Xu and Chen, 2013). In this study, we report the identification of pathogenicity and virulence genes in V. dahliae through ATMT. A collection of 900 transformants of V. dahliae was generated and tested for reduced disease development on susceptible tomato plants. We subsequently studied one of the candidates in detail, a gene that putatively encodes a biosynthetic gene involved in nucleotide sugar precursor formation, to reveal its role in pathogenicity.
Results
Identification of random V. dahliae transformants affected in aggressiveness
In this study, we generated 900 random T‐DNA insertion mutants of V. dahliae by transforming conidiospores through ATMT. All transformants were assessed for reduced potential to cause disease on tomato seedlings by root‐dip inoculation and scored for typical disease symptoms, including stunting, wilting, chlorosis and necrosis, for up to 14 days post‐inoculation (dpi). Seedlings that exhibited reduced Verticillium wilt symptoms when compared with plants inoculated with wild‐type V. dahliae were selected, and the corresponding V. dahliae mutants were retained for rescreening, leading to a set of 200 transformants. Subsequently, after calibration of the conidial concentration in the inoculum to 106 conidia/mL, all 200 transformants were reassessed for compromised ability to cause wilt disease on tomato plants for up to 21 dpi. Eventually, 80 transformants showed a reproducible defect in disease development when compared with the wild‐type progenitor strain (Fig. 1).
Figure 1.

Typical assay to identify transformants with reduced pathogenicity or virulence. Ten‐day‐old tomato seedlings were mock inoculated (M) or inoculated with conidiospores of wild‐type Verticillium dahliae (WT) or random T‐DNA insertion mutants. At 21 days post‐inoculation, the plants were scored for disease development by comparing plants inoculated with wild‐type V. dahliae with those inoculated with the T‐DNA insertion mutants. The sides (top) and tops (bottom) of plants inoculated with five T‐DNA insertion mutants that are impaired in aggressiveness are shown.
Isolation of T‐DNA flanking regions in selected V. dahliae transformants
Inverse polymerase chain reaction (iPCR) was carried out on the 80 selected transformants to determine the T‐DNA insertion sites. The genomic borders flanking the T‐DNA were amplified, sequenced and the resulting sequences were queried against the V. dahliae genome (Faino et al., 2015). In this manner, sequences flanking the T‐DNA were determined for 65 mutants, whereas T‐DNA flanking regions repeatedly could not be amplified from any of the borders of the remaining 15 mutants. Of the 65 flanking sequences, 12 sequences corresponded to the backbone of the vector that was used for ATMT, whereas 10 sequences could not unambiguously be assigned to a single genomic location. The remaining 43 sequences resulted in single genomic hits, allowing the determination of T‐DNA integration sites. In total, 14 insertions were found within predicted open reading frames (ORFs; Table 1), of which two transformants carried an insertion in the same ORF, whereas the remaining 29 insertions occurred in non‐coding regions. For these 29 insertions, the predicted genes flanking the T‐DNA were identified. In one case, we found that, for two transformants, the T‐DNA was integrated into the same non‐coding region, albeit at a slightly different location, resulting in the identification of the same flanking genes. In another case, the same candidate gene was identified twice in two transformants as a result of an insertion upstream and downstream of the gene. One flanking gene was only identified at 55,077 bp from the insertion site, for which we disqualified it as a candidate gene. In total, this resulted in the identification of 55 candidate genes from the 29 mutants with insertions in non‐coding regions.
Table 1.
List of candidate genes identified based on the T‐DNA insertion sites in the random mutants that showed compromised virulence on tomato.
| T‐DNA insertion within the CDS | ||||||
|---|---|---|---|---|---|---|
| Mutant | Old locus† | New locus‡ | Annotation | Expression§ | CDS¶ | |
| 78 | Not available | VDAG_JR2_Chr1g08240 | Glycoside hydrolase | + | – | |
| 73 | Not available | VDAG_JR2_Chr1g08240 | Glycoside hydrolase | + | – | |
| 407 | VDAG_00500 | VDAG_JR2_Chr1g19180 | Carboxypeptidase D | + | – | |
| 659 | VDAG_02200 | VDAG_JR2_Chr2g03300 | α/β‐Hydrolase fold family | – | – | |
| SD‐133 | VDAG_04418 | VDAG_JR2_Chr3g02140 | Ran GTPase‐activating protein 1 | + | – | |
| 444‐B | VDAG_08830 | VDAG_JR2_Chr3g07650 | Acetate kinase | + | – | |
| 211 | VDAG_06985 | VDAG_JR2_Chr4g06020 | Leucine‐rich repeat protein | + | – | |
| 375 | VDAG_07314 | VDAG_JR2_Chr4g10770 | Copper amine oxidase 1 | + | – | |
| 416 | VDAG_10292 | VDAG_JR2_Chr5g06230 | Conserved hypothetical protein | – | – | |
| 174 | VDAG_05684 | VDAG_JR2_Chr5g09640 | Hypothetical protein | + | – | |
| 657‐2 | VDAG_04769 | VDAG_JR2_Chr6g09500 | Phospho‐2‐dehydro‐3‐deoxyheptonate aldolase | + | – | |
| 367 | VDAG_06253 | VDAG_JR2_Chr7g00300 | Cyclic nucleotide‐binding protein | + | – | |
| 618 | VDAG_08151 | VDAG_JR2_Chr8g01560 | Hypothetical protein | + | – | |
| 48 | VDAG_08150 | VDAG_JR2_Chr8g01570 | CaiB/BaiF family enzyme | – | – | |
| T‐DNA insertion in intergenic region | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upstream direction | Downstream direction | |||||||||||
| Mutant | Dist.* | Old locus† | New locus‡ | Annotation | Expression§ | CDS¶ | Dist.† | Old locus | New locus‡ | Annotation | Expression§ | CDS¶ |
| 459‐1 | 2485 | VDAG_00838 | VDAG_JR2_Chr1g01770 | Predicted protein | + | – | 1784 | VDAG_00839 | VDAG_JR2_Chr1g01780 | Transcription initiation factor TFIID subunit 12 | + | – |
| 143 | 374 | VDAG_00843 | VDAG_JR2_Chr1g01820 | Not annotated | + | – | 100 | VDAG_00844 | VDAG_JR2_Chr1g01830 | Exosome complex exonuclease | + | – |
| SD‐102 | 270 | VDAG_00599 | VDAG_JR2_Chr1g20260 | Histidinol‐phosphate aminotransferase | + | – | 746 | VDAG_00600 | VDAG_JR2_Chr1g20270 | Acetyl esterase | + | – |
| 44 | 545 | VDAG_08030 | VDAG_JR2_Chr1g22770 | Deuterolysin metalloprotease | – | – | 669 | VDAG_08031 | VDAG_JR2_Chr1g22760 | SAFF domain containing protein | + | – |
| 11 | 100 | VDAG_08003 | VDAG_JR2_Chr1g23050 | F‐box protein | – | – | 3330 | VDAG_08988 | VDAG_JR2_Chr1g23040 | UMTA methyltransferase | + | – |
| SD‐107 | 165 | VDAG_07796 | VDAG_JR2_Chr1g25310 | Hydrolase | + | – | 1586 | VDAG_07797 | VDAG_JR2_Chr1g25300 | Briggsae CBR‐SMU‐2 protein | + | – |
| 182 | 470 | VDAG_09439 | VDAG_JR2_Chr2g01480 | SMC N‐terminal‐containing protein | + | – | 257 | VDAG_09440 | VDAG_JR2_Chr2g01470 | SGT1‐like protein | + | – |
| 525 | 895 | VDAG_02343 | VDAG_JR2_Chr2g01760 | Cell wall glucanase | + | 100 | 71 | VDAG_02344 | VDAG_JR2_Chr2g01750 | Phosphoribosylglycinamide formyltransferase | + | – |
| SD‐7 | 215 | VDAG_02014 | VDAG_JR2_Chr2g05250 | Myb‐like DNA‐binding protein | + | – | 1004 | VDAG_02015 | VDAG_JR2_Chr2g05240 | Zinc NFX1‐type‐containing protein | – | – |
| 540 | 622 | VDAG_01960 | VDAG_JR2_Chr2g05800 | KH domain containing protein | + | – | 336 | VDAG_01961 | VDAG_JR2_Chr2g05790 | DUF974 protein | + | – |
| 402 | 1788 | VDAG_01891 | VDAG_JR2_Chr2g06520 | Domain‐containing protein | + | 400 | 1318 | VDAG_01892 | VDAG_JR2_Chr2g06510 | Hypothetical protein | + | 100 |
| 417 | 88 | VDAG_01651 | VDAG_JR2_Chr2g09280 | F‐box protein | + | – | 731 | VDAG_01652 | VDAG_JR2_Chr2g09270 | Cytochrome p450 protein | – | – |
| 622 | 700 | Not available | VDAG_JR2_Chr3g02050 | Unknown | + | – | 101 | VDAG_04427 | VDAG_JR2_Chr3g02040 | AGC/AKT protein kinase | + | – |
| SD‐89 | 335 | VDAG_07703 | VDAG_JR2_Chr4g00600 | Transcriptional regulatory protein pro‐1 | – | – | 423 | VDAG_07704 | VDAG_JR2_Chr4g00580 | Pathway‐specific nitrogen regulator | + | – |
| SD‐78 | 1214 | VDAG_07011 | VDAG_JR2_Chr4g05750 | Subunit of Golgi mannosyltransferase complex | + | – | 1914 | VDAG_07012 | VDAG_JR2_Chr4g05740 | Fungal‐specific transcription factor | + | – |
| 414 | 4 | Not available | VDAG_JR2_Chr4g08330 | Myomesin | – | – | 1466 | Not available | VDAG_JR2_Chr4g08340 | UMTA methyltransferase | – | – |
| 584 | 51 | VDAG_09007 | VDAG_JR2_Chr4g08540 | Family protein | + | – | 1221 | VDAG_09008 | VDAG_JR2_Chr4g08550 | Zinc‐binding dehydrogenase | + | – |
| 534 | 737 | VDAG_09007 | VDAG_JR2_Chr4g08540 | Family protein | + | – | 535 | VDAG_09008 | VDAG_JR2_Chr4g08550 | Zinc‐binding dehydrogenase | + | – |
| 199 | 64 | VDAG_05141 | VDAG_JR2_Chr5g01900 | Low‐affinity potassium transport protein | – | – | 703 | VDAG_05142 | VDAG_JR2_Chr5g01910 | Sodium ion/H ion antiporter | + | – |
| 412 | 1647 | VDAG_09301 | VDAG_JR2_Chr5g03950 | Potassium sodium efflux p‐type fungal type | + | – | 1459 | VDAG_09302 | VDAG_JR2_Chr5g03940 | Glycoside hydrolase family 43 protein | + | 100 |
| 550 | 78 | VDAG_04591 | VDAG_JR2_Chr6g07500 | Calmodulin | + | – | 667 | VDAG_04592 | VDAG_JR2_Chr6g07510 | AP‐1 complex subunit γ‐1 | + | – |
| SD‐94 | 716 | VDAG_04810 | VDAG_JR2_Chr6g09950 | NADPH dehydrogenase | + | – | 1014 | VDAG_04811 | VDAG_JR2_Chr6g09960 | Hypothetical protein | – | 100 |
| 75 | 1060 | VDAG_09054 | VDAG_JR2_Chr6g10940 | Ubiquitin fusion degradation protein | + | – | 196 | VDAG_09055 | VDAG_JR2_Chr6g10930 | Ubiquitin fusion degradation protein | + | – |
| 374 | 1023 | VDAG_06275 | VDAG_JR2_Chr7g00090 | Maintenance of telomere capping 1 | + | – | 279 | VDAG_06276 | VDAG_JR2_Chr7g00080 | Major facilitator superfamily transporter | + | – |
| 389 | 56 | VDAG_06010 | VDAG_JR2_Chr7g02960 | NAD‐dependent epimerase dehydratase | + | – | 2363 | VDAG_06011 | VDAG_JR2_Chr7g02950 | Squalene synthetase | + | – |
| 438 | 977 | VDAG_10534 | VDAG_JR2_Chr7g06650 | CTP synthase | + | – | 55077 | VDAG_06595 | VDAG_JR2_Chr7g06640 | SAD1/UNC protein | + | – |
| 59 | 47 | VDAG_10192 | VDAG_JR2_Chr7g10460 | Benzoate 4‐monooxygenase cytochrome P450 | – | – | 1565 | VDAG_10193 | VDAG_JR2_Chr7g10450 | Cellulose‐binding family II | + | – |
| 646 | 246 | VDAG_10191 | VDAG_JR2_Chr7g10470 | Methyltransferase | + | – | 422 | VDAG_10192 | VDAG_JR2_Chr7g10460 | Benzoate 4‐monooxygenase cytochrome p450 | – | – |
| 575 | 466 | VDAG_03431 | VDAG_JR2_Chr8g09910 | Vegetative cell wall protein gp1‐like | – | – | 1040 | VDAG_03432 | VDAG_JR2_Chr8g09920 | DUF221 protein | + | – |
*Distance from the T‐DNA to the coding sequence (CDS).
†Locus corresponds to the Verticillium dahliae strain VdLs.17 assembly deposited at http://genome.jgi.doe.gov/programs/fungi/ (Klosterman et al., 2011).
‡Locus corresponds to the Verticillium dahliae strain JR2 assembly deposited at http://fungi.ensembl.org/ (Faino et al., 2015).
§Evidence for in planta expression.
¶Evidence for expression in the region between T‐DNA and the closest open reading frame (ORF). Distances from T‐DNA are indicated in base pairs (bp).
In order to try to determine which of the two genes flanking a particular insertion site in the non‐coding region is likely to cause reduced disease development, in planta expression of the flanking genes was assessed making use of previously generated RNAseq data of V. dahliae‐inoculated Nicotiana benthamiana plants (Faino et al., 2012; de Jonge et al., 2012). For 10 insertions, this revealed that only one of the two flanking genes is expressed during infection, disqualifying the 10 candidate genes that are not expressed. Thus, 45 candidate genes were considered from mutants with an insertion in the non‐coding region and 13 candidate genes were identified from mutants with an insertion in the ORF (Table 1). Subsequently, the 58 candidate genes were queried against the pathogen–host interaction (PHI) database that contains experimentally verified pathogenicity and virulence factors of plant and animal pathogens (Winnenburg et al., 2006, 2008); this resulted in the identification of homologues (E < 10−6) for only 19 of the 58 candidate genes (Table 2).
Table 2.
Homologues identified in the pathogen–host interaction (PHI) database among the 58 potential Verticillium dahliae pathogenicity genes.
| Old locus* | New locus† | Annotation | E‐value | Accession‡ |
|---|---|---|---|---|
| VDAG_00839 | VDAG_JR2_Chr1g01780 | Transcription initiation factor tfiid subunit 12 | 7E‐19 | PHI:2168 |
| VDAG_00844 | VDAG_JR2_Chr1g01830 | Exosome complex exonuclease | 5E‐12 | PHI:3472 |
| VDAG_01461 | VDAG_JR2_Chr1g08260 | Ring finger protein | 2E‐09 | PHI:1393 |
| VDAG_00500 | VDAG_JR2_Chr1g19180 | Carboxypeptidase D | 4E‐55 | PHI:901 |
| VDAG_09440 | VDAG_JR2_Chr2g01470 | SGT‐1‐like protein | 1E‐156 | PHI:1615 |
| VDAG_09439 | VDAG_JR2_Chr2g01480 | SMC N‐terminal‐containing protein | 2E‐10 | PHI:359 |
| VDAG_02343 | VDAG_JR2_Chr2g01760 | Cell wall glucanase | 9E‐30 | PHI:816 |
| VDAG_02014 | VDAG_JR2_Chr2g05250 | Myb‐like DNA‐binding protein | 1E‐105 | PHI:1540 |
| VDAG_01960 | VDAG_JR2_Chr2g05800 | KH domain containing protein | 3E‐22 | PHI:1662 |
| VDAG_04427 | VDAG_JR2_Chr3g02040 | AGC/AKT protein kinase | 0E+00 | PHI:3131 |
| VDAG_07704 | VDAG_JR2_Chr4g00580 | Pathway‐specific nitrogen regulator | 0E+00 | PHI:1960 |
| VDAG_07012 | VDAG_JR2_Chr4g05740 | Fungal‐specific transcription factor | 0E+00 | PHI:1954 |
| VDAG_09008 | VDAG_JR2_Chr4g08550 | Zinc‐binding dehydrogenase | 7E‐13 | PHI:2895 |
| VDAG_07314 | VDAG_JR2_Chr4g10770 | Copper amine oxidase 1 | 1E‐106 | PHI:2171 |
| VDAG_09301 | VDAG_JR2_Chr5g03950 | Potassium sodium efflux p‐type fungal type | 0E+00 | PHI:2098 |
| VDAG_04591 | VDAG_JR2_Chr6g07500 | Calmodulin | 4E‐20 | PHI:2640 |
| VDAG_06253 | VDAG_JR2_Chr7g00300 | Cyclic nucleotide‐binding protein | 2E‐10 | PHI:372 |
| VDAG_10193 | VDAG_JR2_Chr7g10450 | Cellulose‐binding family II | 6E‐13 | PHI:2388 |
| VDAG_10191 | VDAG_JR2_Chr7g10470 | Methyltransferase | 9E‐40 | PHI:2315 |
*Locus corresponds to the V. dahliae strain VdLs.17 assembly deposited at http://genome.jgi.doe.gov/programs/fungi/ (Klosterman et al., 2011).
†Locus corresponds to the V. dahliae strain JR2 assembly deposited at http://fungi.ensembl.org/ (Faino et al., 2015).
‡PHI database accession.
Analysis of a T‐DNA insertion site links VdNRS/ER to virulence
In random mutant 389, the T‐DNA was integrated 56 bp upstream of the coding sequence (CDS) of a nucleotide‐rhamnose synthase/epimerase‐reductase (NRS/ER) homologue VDAG_06010 (Table 1; Klosterman et al., 2011), which corresponds to VDAG_JR2_Chr7g02960 in the V. dahliae JR2 assembly deposited at http://fungi.ensembl.org/ (Faino et al., 2015). The first CDS downstream of the T‐DNA insertion site is found only at 2.4 kb, encoding a squalene synthetase (VDAG_06011 or VDAG_JR2_Chr7g02950), an enzyme that has been implicated in sterol and triterpene biosynthesis. However, considering the distance between the T‐DNA insertion and the CDS of this gene, it is unlikely that it is affected and causal to the reduced pathogenicity phenotype of the RM‐389 mutant. To assess whether any genes had been overlooked by the automated gene prediction in the region between the T‐DNA insertion and the squalene synthetase CDS, RNAseq reads from V. dahliae‐inoculated N. benthamiana (Faino et al., 2012; de Jonge et al., 2012) were queried. However, no reads were found to map to this region, suggesting that no in planta transcribed genes reside in this region. Finally, with real‐time PCR on genomic DNA, using the single copy Ave1 gene as a reference (de Jonge et al., 2012), it was determined that only a single T‐DNA insertion was present in the genome of the RM‐389 mutant.
Verticillium dahliae NRS/ER (VdNRS/ER) shows weak (17%) amino acid sequence similarity to the Salmonella enterica deoxy‐thymidine diphosphate (dTDP)‐rhamnose biosynthesis component rmlD, a bacterial protein involved in the last step in the conversion of dTDP‐glucose to dTDP‐rhamnose. In bacteria, the nucleotide sugar dTDP‐rhamnose is the precursor that facilitates the incorporation of L‐rhamnose into glycan, which has been implicated in the virulence of pathogenic bacteria such as Salmonella enterica, Vibrio cholerae and Streptococcus mutans (Giraud et al., 2000). Thus, considering all of the above, functional analysis of the VdNRS/ER gene was pursued.
VdNRS/ER is required for V. dahliae pathogenicity
Assessment of VdNRS/ER expression on various growth media (potato dextrose broth, Czapek‐Dox, Murashige and Skoog medium) revealed that the gene is expressed under these conditions (data not shown). Targeted replacement of VdNRS/ER by a hygromycin resistance cassette through homologous recombination was pursued in wild‐type V. dahliae. Several independent VdNRS/ER deletion strains were obtained, two of which (Δ6010‐1 and Δ6010‐2) were used for further analysis (Fig. S1, see Supporting Information). To assess the role of VdNRS/ER in V. dahliae vegetative growth and conidiospore production, radial growth and sporulation of the VdNRS/ER deletion strains on potato dextrose agar were assessed, together with the random mutant (RM‐389) and an ectopic transformant with a random T‐DNA insertion outside of the VdNRS/ER gene. This analysis showed that growth and conidiation were not markedly affected on VdNRS/ER deletion (Fig. 2).
Figure 2.
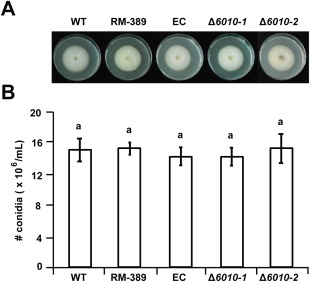
Targeted deletion of VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) does not impair growth and conidiogenesis. (A) Radial growth and colony morphology of wild‐type Verticillium dahliae (WT), random transformant RM‐389, an ectopic transformant (EC) and two VdNRS/ER deletion strains (Δ6010‐1 and Δ6010‐2) after 7 days of incubation on potato dextrose agar (PDA) medium at 22°C. (B) Average number of conidia produced after 7 days of growth on PDA medium based on two independent experiments. Letters indicate significant differences (P < 0.05) calculated using Student's t‐test.
Subsequently, the role of VdNRS/ER in V. dahliae pathogenicity was addressed. To this end, the expression of VdNRS/ER was assessed in wild‐type V. dahliae during infection of tomato plants in a time course harvested at regular intervals after inoculation, showing that VdNRS/ER is induced during in planta growth with a peak in expression at around 12 dpi (Fig. 3). Subsequently, the VdNRS/ER deletion strains were inoculated on tomato plants, alongside the random mutant (RM‐389) and an ectopic transformant, to investigate the role of VdNRS/ER in fungal virulence. Although inoculation of the plants with the wild‐type V. dahliae strain and the ectopic transformant resulted in the development of symptoms of Verticillium wilt disease that included stunting of the plants and wilting, inoculation with RM‐398 or the VdNRS/ER deletion strains did not result in disease development (Fig. 4A). The absence of disease symptoms was confirmed by measuring the surface area of the foliage of the plants that were inoculated with the various fungal strains. Plants that were inoculated with VdNRS/ER deletion strains developed similar foliage (canopy) areas to mock‐inoculated plants and plants inoculated with RM‐389 (Fig. 5A). In contrast, less foliage developed in plants that were inoculated with the wild‐type V. dahliae strain and the ectopic transformant (Fig. 5A). Thus, we conclude that VdNRS/ER is crucial for disease development by V. dahliae. To assess whether VdNRS/ER deletion strains retained the capability to colonize tomato plants, fungal outgrowth assays were performed by plating of stem sections harvested from the hypocotyls of the inoculated plants. Interestingly, although the ectopic transformants and the wild‐type strain extensively colonized the stems of tomato plants, no outgrowth was observed for the VdNRS/ER deletion strains and RM‐389, demonstrating that these genotypes were not able to colonize the xylem vessels of the inoculated tomato plants (Fig. 4A). The absence of tomato colonization by VdNRS/ER deletion strains was confirmed by real‐time PCR quantification of the fungal biomass (Fig. 5B). Importantly, pathogenicity was restored on complementation of the VdNRS/ER deletion strains with a genomic construct containing the wild‐type VdNRS/ER locus (Fig. S2, see Supporting Information). Thus, it can be concluded that VdNRS/ER is required for pathogenicity on tomato. Collectively, our findings thus suggest that the T‐DNA insertion 56 bp upstream of VDAG_06010 in random mutant 389 impaired the function of this gene, probably by impairing its expression in planta. However, as this mutant was no longer able to colonize tomato plants, the absence of gene expression during attempted host colonization could not be assessed.
Figure 3.
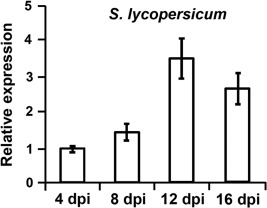
Expression of VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) during infection of Verticillium dahliae on tomato. Ten‐day‐old tomato (Solanum lycopersicum) cultivar MoneyMaker plants were root inoculated with V. dahliae, and plants were harvested at regular intervals from 4 to 16 days post‐inoculation (dpi). After RNA isolation and cDNA synthesis, real‐time polymerase chain reaction was performed to determine the relative expression levels of VdNRS/ER using the V. dahliae elongation factor 1‐α gene as a reference. Expression at 4 dpi is set to unity.
Figure 4.
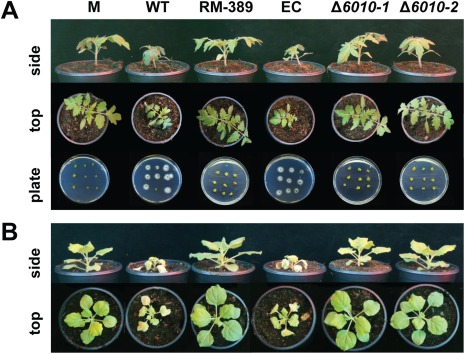
VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) is required for pathogenicity of Verticillium dahliae on tomato and Nicotiana benthamiana. (A) Top and side views of tomato cultivar MoneyMaker plants that were mock inoculated (M) or inoculated with wild‐type V. dahliae (WT), random transformant RM‐389, an ectopic transformant (EC) and two VdNRS/ER deletion strains (Δ6010‐1 and Δ6010‐2) at 14 days post‐inoculation (dpi). Fungal outgrowth at 7 days after plating of stem sections harvested at 14 dpi is shown at the bottom of the panel. (B) Top and side views of N. benthamiana plants inoculated as specified for (A).
Figure 5.
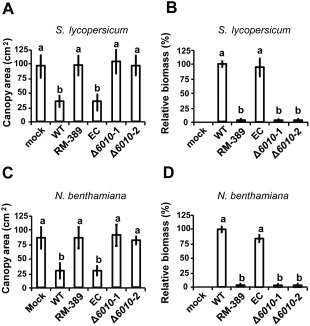
VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) is required for pathogenicity of Verticillium dahliae on tomato and Nicotiana benthamiana. Average canopy area of six tomato plants (A) and real‐time polymerase chain reaction (PCR) quantification of fungal biomass (B) at 14 days after mock inoculation (mock) or inoculation with wild‐type Verticillium dahliae (WT), random transformant (RM‐389), an ectopic transformant (EC) and two VdNRS/ER deletion strains (Δ6010‐1 and Δ6010‐2). Different letters indicate significant differences (P < 0.05). Average canopy area of six N. benthamiana plants (C) and real‐time PCR quantification of fungal biomass (D) on inoculation as specified for (A) and (B).
To investigate whether the observed role of VdNRS/ER in pathogenicity is confined to tomato only, or also extends to other host species, we tested the pathogenicity of the VdNRS/ER deletion strains on the solanaceous model plant N. benthamiana. Similar to tomato, targeted deletion of VdNRS/ER resulted in compromised pathogenicity, as these plants remained devoid of Verticillium wilt symptoms (Fig. 4B). Again, loss of pathogenicity was confirmed by measurement of the canopy surface area (Fig. 5C) and by real‐time PCR quantification of fungal biomass (Fig. 5D).
Absence of VdNRS/ER variation in a collection of V. dahliae strains
Allelic variation can be a sign of selection pressure and adaptation. To analyse potential VdNRS/ER diversity, we mined the genomes of 39 sequenced V. dahliae strains for the VdNRS/ER gene sequence (de Jonge et al., 2012) (L. Faino and B. P. H. J. Thomma, unpublished data). Intriguingly, only two single nucleotide polymorphisms (SNPs) were identified in two strains, each at a different position and both resulting in a synonymous substitution that does not affect the sequence of the encoded protein. We thus conclude that these substitutions do not affect the function of VdNRS/ER.
VdNRS/ER is not required for cell wall integrity and osmotic stress resistance
To examine the role of VdNRS/ER in cell wall integrity and osmotic stress resistance, the random mutant (RM‐389), VdNRS/ER deletion strains and ectopic transformant were grown on minimal agar medium supplemented with mannitol, sodium chloride, sorbitol or Congo red. After 7 days of growth, the colony diameter was measured, showing that mutant and wild‐type strains were equally sensitive to the cell wall stress reagents (Fig. 6). These findings suggest that VdNRS/ER is not involved in cell wall integrity or osmotic stress resistance during mycelium development.
Figure 6.
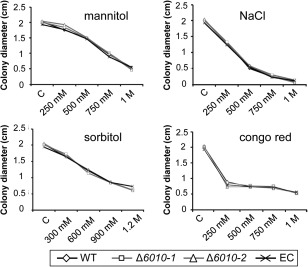
Targeted deletion of VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) does not affect cell wall integrity and osmotic stress resistance. Stress sensitivity assays were performed by placing a 5‐μL droplet (106 conidia/mL) of wild‐type Verticillium dahliae (WT), an ectopic transformant (EC) and two VdNRS/ER deletion strains (Δ6010‐1 and Δ6010‐2) on Czapek‐Dox medium (C) or Czapek‐Dox medium supplemented with Congo red (250 mm, 500 mm, 750 mm and 1 m), NaCl (250 mm, 500 mm, 750 mm and 1 m), mannitol (250 mm, 500 mm, 750 mm and 1 m) or sorbitol (300 mm, 600 mm, 900 mm and 1.2 m). The colony diameter was measured after 7 days of incubation at 22°C.
VdNRS/ER is required for tomato root colonization
To examine the role of VdNRS/ER in the initial stages of V. dahliae infection, the roots of 10‐day‐old tomato plants grown in a hydroponic solution were inoculated with conidiospores of the random mutant (RM‐389), the VdNRS/ER deletion strains and the ectopic transformant. After 5 days of incubation, the tomato roots were inspected using a binocular microscope. Our data show that wild‐type spores efficiently attached to the root surface, resulting in extensive fungal growth on the tomato roots (Fig. 7A). In contrast, little fungal growth was observed on roots inoculated with the random mutant (RM‐389) or the VdNRS/ER deletion strains (Fig. 7A). The reduced fungal biomass of the random mutant (RM‐389) and the VdNRS/ER deletion strains on the tomato roots was confirmed by quantitative PCR (Fig. 7B). Collectively, these data show that VdNRS/ER functions to contribute to successful colonization of roots by V. dahliae, potentially through root attachment.
Figure 7.
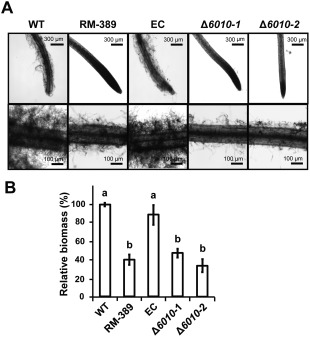
VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) is required for the colonization of tomato roots. Roots of 10‐day‐old tomato cultivar MoneyMaker seedlings were immersed in one‐fifth potato dextrose broth (PDB) containing 106 conidia/mL of wild‐type Verticillium dahliae (WT), random transformant (RM‐389), an ectopic transformant (EC) and two VdNRS/ER deletion strains (Δ6010‐1 and Δ6010‐2) for 72–96 h. (A) Roots were rinsed with water and photographed under a microscope. (B) Real‐time polymerase chain reaction quantification of fungal biomass on the roots. Different letters indicate significant differences (P < 0.05).
VdNRS/ER is a functional enzyme that converts uridine diphosphate‐4‐keto‐6‐deoxy‐glucose (UDP‐KDG) to UDP‐rhamnose
To examine whether VdNRS/ER is involved in the biosynthesis of UDP‐rhamnose and to determine whether this activated sugar is the precursor for the synthesis of rhamnose‐containing glycan, we examined the polysaccharide composition of V. dahliae grown in vitro. As mentioned previously, VdNRS/ER is expressed under various in vitro conditions. Thus, polysaccharides were extracted from conidiospores of wild‐type V. dahliae, the VdNRS/ER deletion strain and the complementation strain, and analysed by gas chromatography‐mass spectrometry (GC‐MS). In the samples derived from wild‐type V. dahliae and the complementation strain, a peak was detected that corresponds to rhamnose, which was absent in the VdNRS/ER deletion strain (Fig. 8). This finding confirms that VdNRS/ER is involved in rhamnose biosynthesis.
Figure 8.
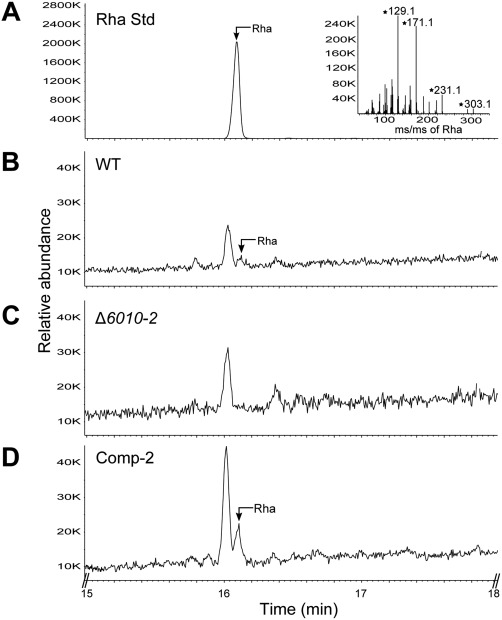
VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) deletion strains are depleted in uridine diphosphate (UDP)‐rhamnose. Gas chromatography‐mass spectrometry (GC‐MS) analysis of alditol acetate derivatives of the glycosyl residues from conidiospore polysaccharides. (A) Total ion count chromatogram of alditol acetate‐derived standard rhamnose (Rha) and mass spectrum (inset) showing fragmentation pattern fingerprints with m/z 129, 171, 231, 303. (B) Polysaccharides from wild‐type Verticillium dahliae (WT) conidiospores display a peak eluting at 16.10 min that has the same retention time as rhamnose standard. (C) VdNRS/ER deletion strain (Δ6010‐1) lacks a peak eluting at 16.10 min. (D) The VdNRS/ER deletion strain complemented with the native VdNRS/ER gene (Comp‐2) displays a peak eluting at 16.10 min that has the same retention time as the rhamnose standard.
If rhamnose‐containing glycans play a role during the early stages of host colonization, such as the attachment of conidiospores to the root, we anticipate that such glycans should be produced during conidiospore germination. Therefore, we determined the sugar composition of germinating conidiospores. Indeed, the sample derived from wild‐type V. dahliae comprised a clear rhamnose peak (Fig. 9A,B) that was absent in samples from the VdNRS/ER deletion strain (Fig. 9C). Taken together, these results indicate that VdNRS/ER plays a role in the formation of rhamnose‐containing glycan in germinating conidiospores during the early stages of host colonization.
Figure 9.
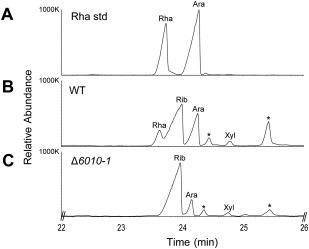
Uridine diphosphate (UDP)‐rhamnose is produced in germinating conidiospores. Gas chromatography‐mass spectrometry (GC‐MS) analysis of alditol acetate derivatives of the glycosyl residues from germinating conidiospore polysaccharides. (A) Total ion count chromatogram of alditol acetate‐derived standard rhamnose (Rha). (B) Polysaccharides of conidiospores from wild‐type Verticillium dahliae (WT) display a peak with the same retention time as the rhamnose standard. (C) VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) deletion strain (Δ6010‐1) does not display a rhamnose peak. Other peaks in the chromatogram correspond to ribose (Rib), arabinose (Ara) and xylose (Xyl). Asterisks indicate unidentified compounds that are not sugars.
In contrast with bacteria that produce dTDP‐rhamnose in a three‐step pathway, plants follow a two‐step pathway to synthesize UDP‐rhamnose (Oka et al., 2007; Watt et al., 2004). The genes involved in UDP‐rhamnose production have also been identified recently in other fungi, including the rice pathogen Magnaporthe oryzae and the broad‐host‐range pathogen Botrytis cinerea (Martinez et al., 2012). In B. cinerea, UDP‐glucose is converted by a 4,6‐dehydratase (BfDh) into an intermediate UDP‐KDG that is subsequently converted by a bifunctional 3,5‐epimerase/4‐reductase (BfER, i.e. NRS/ER) into UDP‐rhamnose (Martinez et al., 2012). Thus, we examined whether VdNRS/ER, like BfER, is a functional enzyme that can convert UDP‐KDG to UDP‐rhamnose. To this end, the dual enzyme activities of recombinant VdNRS/ER and BfDh were analysed by liquid chromatography‐mass spectrometry (LC‐MS) using UDP‐glucose (UDP‐Glc) as a substrate (Martinez et al., 2012). The first enzyme in the pathway is capable of converting UDP‐Glc to UDP‐KDG (Fig. 10B,D), as expected, and, with the addition of NRS/ER activity (VdNRS/ER), UDP‐KDG was converted to UDP‐rhamnose (Fig. 10C). The authenticity of UDP‐rhamnose was determined by its elution and by its mass and MS/MS ion fragmentation (see inset in Fig. 10C). The enzyme activity of recombinant VdNRS/ER unambiguously shows that VdNRS/ER is a bifunctional enzyme with 3,5‐epimerase and 4‐reductase activities (Fig. 10E). Thus, we conclude that deletion of VdNRS/ER contributed to the loss of UDP‐rhamnose formation and the lack of rhamnose‐containing glycans during spore germination and the early stages of infection.
Figure 10.
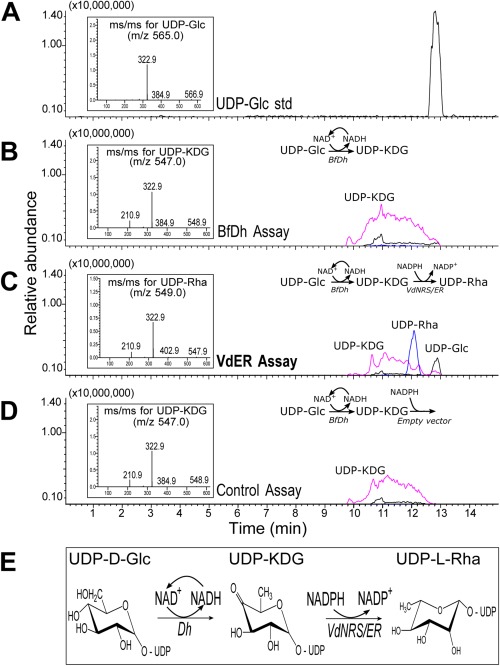
VdNRS/ER (NRS/ER, nucleotide‐rhamnose synthase/epimerase‐reductase) converts uridine diphosphate‐4‐keto‐6‐deoxy‐glucose (UDP‐KDG) to UDP‐rhamnose. (A) UDP‐glucose (UDP‐Glc) standard elutes from a HILIC (hydrophilic interaction liquid chromatography) column at 12.8 min and is detected by mass spectrometry (MS) with m/z 565 [M – H]–. The MS/MS of the parent ion (inset) gives m/z 323 and 385 diagnostic ion fragments, [UMP – H]– and [UDP – H2O – H]–, respectively. (B) HILIC separation of products from the enzymatic reaction with BfDh (4,6‐dehydratase) displays a peak with the same retention time as UDP‐KDG with diagnostic [M – H]– m/z 547 and MS/MS ion fragment with m/z 323. (C) HILIC separation of products from the dual enzymatic reaction with BfDh and VdNRS/ER displays a peak with the same retention time as UDP‐rhamnose (UDP‐Rha) with diagnostic [M – H]– m/z 549 and MS/MS of 403, 323, 210. (D) Dual enzymatic reaction with BfDh and an empty vector control displays a peak with the same retention time as UDP‐KDG with diagnostic [M – H]– m/z 547 and MS/MS ion fragment with m/z 323. (E) UDP‐rhamnose metabolic pathway in fungi showing the enzymes involved in the sequential conversion of UDP‐Glc to UDP‐Rha.
Discussion
Rhamnose is found in a variety of fungal glycoproteins and exopolysaccharides, such as rhamnomannans. In our study, on a random mutant library generated in the vascular wilt fungus V. dahliae, we show, for the first time, that rhamnose‐containing macromolecules are required for fungal pathogenicity through their contribution to the colonization of tomato roots. In random mutant 389, the T‐DNA was integrated 56 bp upstream of the coding region of a putative NRS/ER. Our biochemical analyses of cell wall polysaccharides (Figs 8 and 9) and enzyme activity (Fig. 10C) confirm that VdNRS/ER is involved in UDP‐rhamnose biosynthesis and show that this activated sugar is the precursor for rhamnose‐containing glycan.
VdNRS/ER shows similarity to the bacterial rmlD substrate‐binding domain, which is involved in the biosynthesis of dTDP‐rhamnose. In Gram‐negative bacteria, such as Salmonella enterica, Vibrio cholerae or Escherichia coli, L‐rhamnose is an important residue in the O‐antigen of lipopolysaccharides, which are essential for serum resistance and intestinal colonization (Chiang and Mekalanos, 1999). In mycobacteria, such as Mycobacterium tuberculosis, L‐rhamnose maintains the structural integrity of the cell wall through connection of the inner peptidoglycan layer to the arabinogalactan polysaccharides (Giraud et al., 2000). Moreover, disruption of dTDP‐rhamnose biosynthesis in the plant growth‐promoting rhizobacteria Azospirillum brasilense and Azorhizobium caulinodans results in reduced root attachment and colonization (Gao et al., 2001; Jofré et al., 2004). Furthermore, work on the foliar pathogen Colletotrichum graminicola, the causal agent of stalk rot of cereals and anthracnose of maize, has revealed the secretion of mucilaginous matrix by the fungus to promote the survival of fungal spores, the major component of which appeared to be high‐molecular‐weight glycoproteins containing rhamnose and mannose oligomers (Ramadoss et al., 1985). Subsequently, this matrix was proposed to allow for the rapid adhesion of even ungerminated conidia to the hydrophobic plant surface to ensure germination and appressorium formation and the establishment of an infection court (Nicholson and Kunoh, 1995). Interestingly, further rhamnose‐containing extracellular matrix is released on germ tube elongation to surround the hyphae and extend between them (Sugui et al., 1999). Finally, a recent study has identified two genes encoding UDP‐4,6‐glucose dehydratase (UG4,6‐Dh) and UDP‐4‐keto‐6‐deoxyglucose‐3,5‐epimerase‐4‐reductase (UDP4k6dG‐ER) enzymes from M. oryzae and B. cinerea involved in UDP‐rhamnose formation, although their contribution to pathogenicity was not investigated (Martinez et al., 2012). Based on our study, however, strong evidence is presented that fungal UG4,6‐Dh and U4k6dG‐ER enzymes are involved in the formation of UDP‐rhamnose, which is incorporated into rhamnose‐containing polysaccharides that are crucial for fungal pathogenicity. Interestingly, the plant colonization‐associated phenotypes observed thus far in our study and in those of others concern adhesion to various plant tissues, including roots, foliage and probably even the interior of xylem vessels, as V. dahliae is known to spread throughout the xylem by producing conidiospores that are carried with the water stream and that germinate once trapped, for instance at vessel ends (Fradin and Thomma, 2006). Arguably, the surfaces encountered under these conditions are highly divergent, for instance with respect to hydrophobicity, but also with respect to chemical composition. However, a full understanding of how rhamnose‐containing macromolecules contribute to plant surface adhesion requires the identification of the rhamnose‐containing macromolecules that are produced by the various plant‐associated microbes, and their potential molecular targets in the various host plants.
Here, we have shown that targeted deletion of VdNRS/ER contributes to the loss of V. dahliae pathogenicity on various host plants. The role of rhamnose‐containing macromolecules in cell wall integrity and oxidative stress tolerance in fungi is largely unknown. VdNRS/ER deletion strains were able to grow and sporulate without any visible defects, suggesting that rhamnose‐containing glycans, such as glycoproteins, polysaccharides or exopolysaccharides, are not essential for vegetative growth or sporulation despite the observation that VdNRS/ER is expressed on growth in vitro. In addition, the growth rate of VdNRS/ER knockout mutants was not affected in the presence of inducers of cell wall stress and osmotic stress, suggesting that the rhamnose‐containing glycans are not involved in functional cell wall structure and osmotic stress tolerance.
Based on the findings reported in this study, we propose a model to explain the contribution of UDP‐rhamnose to the virulence of V. dahliae during infection on host plants. In the cytoplasm, UDP‐glucose is converted into UDP‐rhamnose in two steps using VdUG4,6‐Dh and VdNRS/ER. The rhamnose of UDP‐rhamnose is then transferred to glycan(s). The nature of these glycans remains unknown, but they consist of NaOH‐solubilized cell wall polysaccharides and, potentially, glycoproteins. Presently, we cannot exclude the possibility that the rhamnose‐containing polysaccharide is not an extracellular polysaccharide (EPS), as shown to be the case in B. cinerea. We propose that the rhamnose‐containing glycan, produced during spore germination, plays a role in the attachment of spores to the root surface, which results in successful colonization of host plants. During host colonization, conidiospores are produced by V. dahliae in the xylem to facilitate the spread of the fungus throughout the host plant (Fradin and Thomma, 2006). Arguably, rhamnose then acts to facilitate conidiospore attachment to vessel ends and vessel walls in order to facilitate penetration into the next vessel elements. Thus, the production of conidiospores and subsequent germination is an ongoing process in repetitive cycles during disease progression, requiring continuous rhamnose biosynthesis. As VdNRS/ER deletion strains are unable to synthesize UDP‐rhamnose and unable to cause disease, inhibitors of UDP‐rhamnose synthesis may result in impaired pathogenicity and could present a novel strategy for disease control.
Experimental Procedures
Agrobacterium tumefaciens‐mediated transformation (ATMT)
The binary vector (pBHt2) harbouring the hygromycin B resistance gene (hph) under the control of the Aspergillus nidulans trpC promoter was used for ATMT (Mullins et al., 2001). This vector was introduced to A. tumefaciens strain SK1044 to transform conidia of race 1 V. dahliae strain JR2, as described previously (Faino et al., 2015; Santhanam, 2012). Briefly, A. tumefaciens was grown at 28 °C for 2 days in minimal medium supplemented with kanamycin (25 μg/mL). The A. tumefaciens cells were diluted to an optical density at 600 nm (OD600) of 0.15 in induction medium (IM), supplemented with 200 μm acetosyringone (AS). The cells were grown for an additional 6 h before mixing them with an equal volume of V. dahliae conidiospore suspension (106 conidia/mL); 200 μL of this mixture was plated onto a Hybond‐N+ filter placed on IM supplemented with 200 μm AS. The plates were incubated in the dark at room temperature for 48 h after which the filter was transferred onto a selection plate [potato dextrose agar (PDA) supplemented with 50 μg/mL of hygromycin B and 200 μm of cefotaxime]. After 10–14 days, individual transformants were transferred to 24‐well culture plates containing 1 mL of selection medium and incubated for 7–10 days. Spores from these cultures were stored in 30% glycerol at −80 °C until further analysis.
Plant inoculations
Pathogenicity assays were performed as described previously (Santhanam and Thomma, 2013). Briefly, individual transformants were subcultured in six‐well culture plates for 7–10 days. Subsequently, 10 glass beads (∼3 mm) and 3 mL of tap water were added to each of the wells and the plates were sealed. The conidiospores were released by shaking the plates for 15 min at 200 rpm on a reciprocal shaker, after which the sealant was removed. The roots of 10‐day‐old tomato (Solanum lycopersicum cv. MoneyMaker) seedlings were rinsed in water and dipped into the conidiospore suspension for 5 min. Seedlings were replanted in soil and scored for symptom development (wilting and stunting) up to 14 days. Seedlings that exhibited reduced Verticillium wilt symptoms when compared with wild‐type V. dahliae‐inoculated plants were identified and the corresponding mutants were retained for rescreening.
During rescreening, the infection assay was carried out essentially as described above, with the modification that the conidiospore concentration of the transformants was calibrated to 106 conidiospores/mL. The rescreening was repeated at least twice for each of the mutants that were retained in the initial screen.
Identification of T‐DNA insertion sites
Genomic sequences flanking the T‐DNA were isolated from the selected transformants with iPCR, as described previously (Meng et al., 2007; Santhanam, 2012). Essentially, 500 ng of genomic DNA were digested overnight with MspI or NcoI and heat‐inactivated at 65 °C for 20 min. Subsequently, 50 µL of ligation mix (10 µL of 10 × T4 DNA ligase buffer, 5 units of T4 DNA ligase and 38 µL of H2O) were added and incubated overnight at 15 °C. Next, the DNA was precipitated and dissolved in 50 µL of demineralized water. The genomic region flanking the T‐DNA was amplified in 50 µL of reaction mix using 2 µL of ligation product, 1 µL of each primer (Table S1, see Supporting Information), 1 × PCR buffer, 0.6 µL of deoxynucleoside triphosphate (dNTP) mix (10 mm), 0.8 units of GO Taq DNA polymerase and 35.25 µL of water. The cycling conditions consisted of one cycle for 2 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 55 °C and 3 min at 72 °C, and a final extension step for 10 min at 72 °C. PCR products were purified and sequenced. The sequences obtained were used as query to blast against the V. dahliae genome (http://fungi.ensembl.org/Verticillium_dahliaejr2/Info/Index) (Faino et al., 2015).
Targeted mutagenesis and complementation
To generate a VdNRS/ER deletion construct, sequences flanking the VdNRS/ER CDS were amplified from genomic DNA of V. dahliae strain JR2 using the primers KO‐6010‐LF with KO‐6010‐LR to amplify the left border, and KO‐6010‐RF with KO‐6010‐RR to amplify the right border, respectively (Table S1). The resulting amplicons were cloned into pRF‐HU2 as described previously (Frandsen et al., 2008). ATMT of V. dahliae was performed as described previously (Santhanam, 2012), and transformants were selected on PDA supplemented with 200 μg/mL of cefotaxime (Duchefa, Haarlem, the Netherlands) and 50 μg/mL of hygromycin (Duchefa). Homologous recombination was verified by PCR.
To generate a VdNRS/ER complementation construct, a 2404‐bp EcoRI/PacI fragment containing the VDAG_06010 (VDAG_JR2_Chr7g02960) CDS, with 1000‐bp upstream and 450‐bp downstream sequences, was amplified from V. dahliae strain JR2 genomic DNA, and cloned into binary vector pBT081 (Houterman et al., 2008). Complementation transformants were selected on PDA supplemented with 200 μg/mL of cefotaxime and 100 μg/mL of phleomycin (InvivoGen, San Diego, CA, USA).
Growth, conidiogenesis and stress assays
Radial growth was monitored by placing a 2‐μL droplet of a conidial suspension of 106 conidiospores/mL in the centre of PDA or Czapek‐Dox medium, incubated at 22 °C and the colony diameter was measured after 10 days. For the quantification of conidiospore production, 5 mL of water were added to the culture and a conidial suspension was prepared by gently rubbing the mycelium. A 10‐fold dilution of the conidial suspension was counted using a haemocytometer. Stress sensitivity assays were performed by placing a 2‐µL droplet with 106 conidiospores/mL of wild‐type V. dahliae, two VdNRS/ER deletion strains and an ectopic transformant in the centre of a Czapek‐Dox plate supplemented with Congo red (250 mm, 500 mm, 750 mm and 1 m), NaCl (250 mm, 500 mm, 750 mm and 1 m), mannitol (250 mm, 500 mm, 750 mm and 1 m) or sorbitol (300 mm, 600 mm, 900 mm and 1.2 m), and incubated at 22 ºC. Plates were photographed at 7 dpi and the colony diameter was measured using ImageJ software.
Root colonization assay
The root colonization assay was performed as described previously (Di Pietro et al., 2001). Briefly, the roots of 10‐day‐old tomato seedlings were rinsed in water and placed in Erlenmeyer flasks containing a suspension of 106 conidia/mL in one‐fifth potato dextrose broth (PDB), and incubated at 22 °C and 100 rpm. Fungal colonization of the root surface was observed macroscopically from 3 to 5 dpi. The experiments were performed three times with similar results.
Expression of recombinant VdNRS/ER
The following expression constructs were expressed in E. coli strain Bl21(DE3): pET‐6hSUMO‐VDAG_06010, containing a 6‐histidine‐small ubiquitin‐like modifier (6hSUMO) tag fused to the N‐terminus of V. dahliae VdNRS/ER (VDAG_06010); pET‐6hSUMO was used as an empty vector control (pET‐SUMO); pET28‐BfDH4.1 (Martinez et al., 2012) was used to express and purify the 6‐histidine‐tagged Botryotinia fuckeliana UDP‐Glc 4,6‐dehydratase (BfDH). The E. coli strains carrying the different constructs were cultured for 16 h at 37 °C in LB medium supplemented with 50 µg/mL kanamycin and 35 µg/mL chloramphenicol. On the following day, 5‐mL cultures were transferred to 245 mL of fresh LB liquid medium supplemented with the same antibiotics, and cell growth was resumed at 37 °C at 220 rpm until the cell density reached an OD600 of ∼0.6. Gene expression was then induced by the addition of isopropyl β‐d‐thiogalactoside (0.5 mm), and cultures were grown for an additional 20 h at 18 °C at 220 rpm. Cells were collected by centrifugation (6000 g for 10 min at 4 °C) and suspended in 10 mL of lysis buffer [50 mm Tris‐HCl, pH 7.5, 10% glycerol, 1 mm ethylenediaminetetraacetic acid (EDTA), 2 mm dithiothreitol (DTT), 0.5 mm phenylmethylsulfonylfluoride (PMSF), with or without 50 mm NaCl]. Cells were lysed in an ice bath with an S‐4000 sonicator (Misonix Inc., Farmingdale, NY, USA) equipped with a 1/8‐in microtip probe using 12 sonication cycles (10 s pulse; 20 s off). The lysed cells were centrifuged (6000 g for 10 min at 4 °C); the supernatant was supplemented with 1 mm DTT and centrifuged again (20 000 g for 30 min at 4 °C). The resulting supernatant was kept at −20 °C. Protein purification was carried out as described previously (Martinez et al., 2012).
Enzyme assays
The dual activity of recombinant enzymes BfDH and VdNRS/ER was analysed by LC‐MS in a total volume of 50 μL, containing 50 mm Tris‐HCl, pH 6.5, 0.25 mm UDP‐Glc, 0.5 mm NAD+, 0.5 mm NADPH, 5 µL of purified B. fuckeliana BfDH with or without the addition of 2 µL of VdNRS/ER. The reaction was incubated at 37 °C for 1 h and terminated by heating for 5 min at 95 °C. After the addition of chloroform and centrifugation (10 000 g for 4 min), an aliquot (20 μL) of the upper phase layer was removed and mixed with 38 μL of acetonitrile and 2 μL of 0.5 m ammonium acetate, pH 4.35. A portion (25 μL) of this mixture was analysed using HILIC (hydrophilic interaction liquid chromatography) and mass spectrometry analyses. This was performed using an liquid chromatography‐ mass spectrometry electrospray ionization‐ion trap‐time of flight (LC‐MS/MS IT‐TOF MS) system (Shimadzu Scientific Instruments, Columbia, MD, USA) operating in the negative ion mode with a Nexera, Shimadzu, Columbia, MD, USA UFPLC LC‐30AD pump (Shimadzu), autosampler (Sil30) and column heater (set to 37 °C). Nucleotide sugars were separated on an Accucore, Waltham, MA, USA 150‐amide HILIC column (150 × 4.6 mm2, 2.6 μm particle size, Thermo Scientific, Waltham, MA, USA) using 40 mm ammonium acetate, pH 4.3 (solvent A), and acetonitrile (solvent B) with the gradient conditions as in Li et al. (2015).
Isolation of polysaccharide and sugar analyses by GC‐MS
Conidiospores were harvested from 7‐day‐old V. dahliae cultures grown on PDA (Thermo Fisher Scientific Inc., Breda, the Netherlands). Germinating spores were prepared by inoculating 30 µL of conidiospores into 12.5 mL of Gamborg's Glc (2%) on a medium‐sized plate (9 cm). The plates were incubated for 66 h at room temperature under regular light to allow the spores to germinate. Polysaccharides were either extracted directly from conidospore samples or from germinating conidiospores (<100 µg) starting by incubation for 3 h at 80 °C whilst stirring with 5 mL of 1 m NaOH. After centrifugation (10 000 g, 4 °C, 10 min), the alkaline extract was mixed with an equal volume of 96% ethanol. Following centrifugation (10 000 g, 4 °C, 15 min), the pellet was resuspended in 10 mL of water and dialysed (3000 molecular weight cut‐off) against two 10‐L changes of deionized water. The dialysed solution was centrifuged (10 000 g, 4 °C, 15 min) and the water‐soluble supernatant alkaline water soluble (aWS) was collected and freeze–dried. The glycosyl residue compositions of these extracts were then determined.
An aliquot of each extract (1 mg) was supplemented with 10 µg inositol and hydrolysed with 1 mL of 2 m trifluoroacetic acid at 120 °C. The released monosaccharides were reduced to their alditols (York et al., 1986) and acetylated. The resulting alditol acetate derivatives were analysed by a GC‐MS system consisting of a gas chromatograph (Agilent 7890a; Agilent Technologies, Santa Clara, CA, USA) equipped with an EI‐MS detector (5975c). A 1‐µL sample or standard was injected via an Agilent 7693 autosampler injector port (Agilent Technologies) (set at 250 °C; 3 mL/min helium) in a split (1 : 50) mode into a non‐polar Equity‐1 capillary column (30 m × 0.25 mm i.d.; film thickness, 0.25 µm). The chromatography gas carrier was helium (1 mL/min) and the column oven temperature program used to separate the sugar derivatives was as follows: initial injection temperature of 60 °C for 1 min, followed by an increase of 27.5 °C/min to 170 °C, then of 4 °C/min to 235 °C, and a hold at 235 °C for 2 min. The column was then kept at 260 °C for 12 min, cooled to 60 °C and then kept at 60 °C for 1 min prior to the next sample injection. The temperature of the transfer line between the column ends to the mass spectrometer was 250 °C. Detection was achieved using EI‐MS operating with an electron impact ionization energy of 70 V with the temperature of the MS ion source set at 230 °C and that of the quad at 150 °C. MS data were collected in full scan monitoring mode in the m/z range 50–550 from 5 to 50 min. The spectra were analysed using Software MSD ChemStation D.02.00.275 (Agilent Technologies). Alditol acetate derivatives of monosaccharide standards (50 μg each of rhamnose, fucose, xylose, mannose and glucose, and 30 μg each of ribose, arabinose and galactose), supplemented with 10 μg inositol (internal standard), were prepared under the same conditions as the samples. For data analysis, sugar residue peaks were identified based on the retention times of standard monosaccharides and their characteristic mass spectra.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Verification of deletion and complementation strains by polymerase chain reaction (PCR). (A) Amplification of the T‐DNA flanking sequences from wild‐type Verticillium dahliae (w), 10 independent random transformants (lanes 1–10) and the marker (m). (B) Amplification of the hygromycin resistance gene (hph) and VdNRS/ER from 10 independent transformants (lanes 1–10) and wild‐type V. dahliae (w). The marker (m) is indicated. True deletion strains are marked with asterisks, and the ectopic transformant is marked with an arrow. (C) Amplification of the zeocin resistance gene (ble) and VdNRS/ER from five independent complementation strains (lanes 1–5), a VdNRS/ER deletion strain (*) and wild‐type V. dahliae (w). The marker (m) and true complementation strains (+) are indicated.
Fig. S2 Complementation of VdNRS/ER deletion strain restores pathogenicity on tomato. (A) Side and top views of tomato cultivar MoneyMaker plants inoculated with wild‐type Verticillium dahliae (WT), a VdNRS/ER deletion strain (Δ6010‐1) and two complementation strains (Comp‐1 and Comp‐2), or mock inoculated at 14 days post‐inoculation (dpi). (B) Fungal outgrowth at 7 days after plating of stem sections harvested at 14 dpi. (C) Average canopy area of six plants at 14 dpi as described for (A). Different letters indicate significant differences (P < 0.05).
Table S1 Primers used in this study.
Acknowledgements
B.P.H.J.T. was supported by a Vici grant from the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO). This research was funded in part by Binational Agriculture Research and Development fund (BARD) grant # IS 4501‐12 to M.B.P. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflicts of interest. The authors thank Hugo de Vries and Emilie Fradin for technical assistance, and Bert Essenstam and Henk Smid for excellent plant care.
References
- Bolek, Y. , El‐Zik, K.M. , Pepper, A.E. , Bell, A.A. , Magill, C.W. , Thaxton, P. M. and Reddy, O. U. K. (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. [Google Scholar]
- Chiang, S.L. and Mekalanos, J.J. (1999) rfb mutations in Vibrio cholerae do not affect surface production of toxin‐coregulated pili but still inhibit intestinal colonization. Infect. Immun. 67, 976–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A. , Garcia‐MacEira, F.I. , Meglecz, E. and Roncero, M.I. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Faino, L. , de Jonge, R. and Thomma, B.P.H.J. (2012) The transcriptome of Verticillium dahliae‐infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signal. Behav. 7, 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Datema, E. , van den Berg, G.C. , Janssen, A. , Wittenberg, A. H. and Thomma, B. P. H. J. (2015) Single‐molecule real‐time sequencing combined with optical mapping yields completely finished fungal genome. MBio, 6, e00936–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. and Thomma, B.P.H.J. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Juarez Ayala, J.C. , Castroverde, C.D. , Nazar, R.N. , Robb, J. , Liu, C. M. and Thomma, B. P. H. J. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen, R.J. , Andersson, J.A. , Kristensen, M.B. and Giese, H. (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , D'Haeze, W. , De Rycke, R. , Wolucka, B. and Holsters, M. (2001) Knockout of an azorhizobial dTDP‐L‐rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata . Mol. Plant–Microbe Interact. 14, 857–866. [DOI] [PubMed] [Google Scholar]
- Giesbert, S. , Schumacher, J. , Kupas, V. , Espino, J. , Segmuller, N. , Haeuser‐Hahn, I. , Schreier, P. H. and Tudzynski, P. (2012) Identification of pathogenesis‐associated genes by T‐DNA‐mediated insertional mutagenesis in Botrytis cinerea: a type 2A phosphoprotein phosphatase and an SPT3 transcription factor have significant impact on virulence. Mol. Plant–Microbe Interact. 25, 481–495. [DOI] [PubMed] [Google Scholar]
- Giraud, M.F. , Leonard, G.A. , Field, R.A. , Berlind, C. and Naismith, J.H. (2000) RmlC, the third enzyme of dTDP‐L‐rhamnose pathway, is a new class of epimerase. Nat. Struct. Mol. Biol. 7, 398–402. [DOI] [PubMed] [Google Scholar]
- Hayes, R.J. , McHale, L.K. , Vallad, G.E. , Truco, M.J. , Michelmore, R.W. , Klosterman, S. J. , Maruthachalam, K. and Subbarao, K. V. (2011) The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. Theor. Appl. Genet. 123, 509–517. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J. and Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathog. 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüser, A. , Takahara, H. , Schmalenbach, W. and O'Connell, R. (2009) Discovery of pathogenicity genes in the crucifer anthracnose fungus Colletotrichum higginsianum, using random insertional mutagenesis. Mol. Plant–Microbe Interact. 22, 143–156. [DOI] [PubMed] [Google Scholar]
- Jeon, J. , Park, S.Y. , Chi, M.H. , Choi, J. , Park, J. , Rho, H. S. , Kim, S. , Goh, J. , Yoo, S. , Choi, J. , Park, J. Y. , Yi, M. , Yang, S. , Kwon, M. J. , Han, S. S. , Kim, B. R. , Khang, C. H. , Park, B. , Lim, S. E. , Jung, K. , Kong, S. , Karunakaran, M. , Oh, H. S. , Kim, H. , Kim, S. , Park, J. , Kang, S. , Choi, W. B. , Kang, S. and Lee, Y. H. (2007) Genome‐wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 39, 561–565. [DOI] [PubMed] [Google Scholar]
- Jofré, E. , Lagares, A. and Mori, G. (2004) Disruption of dTDP‐rhamnose biosynthesis modifies lipopolysaccharide core, exopolysaccharide production, and root colonization in Azospirillum brasilense . FEMS Microbiol. Lett. 231, 267–275. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M. K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K. V. and Thomma, B. P. H. J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. , Kombrink, A. , van den Berg, G.C. , Yadeta, K.A. and Thomma, B. P. H. J. (2013) Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimes, A. , Dobinson, K.F. , Klosterman, S.J. and Thomma, B.P.H.J. (2015) Genomics spurs rapid advances in our understanding of the basic biology of vascular wilt pathogens in the genus Verticillium. Annu. Rev. Phytopathol. 53, 181–198. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman, S. J. , Subbarao, K. V. , Kang, S. , Veronese, P. , Gold, S. E. , Thomma, B. P. H. J. , Chen, Z. , Henrissat, B. , Lee, Y. H. , Park, J. , Garcia‐Pedrajas, M. D. , Barbara, D. J. , Anchieta, A. , de Jonge, R. , Santhanam, P. , Maruthachalam, K. , Atallah, Z. , Amyotte, S. G. , Paz, Z. , Inderbitzin, P. , Hayes, R. J. , Heiman, D. I. , Young, S. , Zeng, Q. , Engels, R. , Galagan, J. , Cuomo, C. A. , Dobinson, K. F. and Ma, L. J. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, M. , Schmidpeter, J. , Dahl, M. , Muller, S. , Voll, L. M. and Koch, C. (2015) A genetic screen for pathogenicity genes in the hemibiotrophic fungus Colletotrichum higginsianum identifies the plasma membrane proton pump pma2 required for host penetration. PLoS One, 10, e0125960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Hwang, S. , Ericson, J. , Bowler, K. and Bar‐Peled, M. (2015) Pen and Pal are nucleotide‐sugar dehydratases that convert UDP‐GlcNac to UDP‐6‐deoxy‐D‐GlcNac‐5,6‐ene and then to UDP‐4‐keto‐6‐deoxy‐L‐AltNac for CMP‐Pseudaminic acid synthesis in Bacillus thuringiensis . J. Biol. Chem. 290, 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, V. , Ingwers, M. , Smith, J. , Glushka, J. , Yang, T. and Bar‐Peled, M. (2012) Biosynthesis of UDP‐4‐keto‐6‐deoxyglucose and UDP‐rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana . J. Biol. Chem. 287, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthachalam, K. , Klosterman, S.J. , Kang, S. , Hayes, R.J. and Subbarao, K.V. (2011) Identification of pathogenicity‐related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens‐mediated T‐DNA insertional mutagenesis. Mol. Biotechnol. 49, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Patel, G. , Heist, M. , Betts, M. F. , Tucker, S. L. , Galadima, N. , Donofrio, N. M. , Brown, D. , Mitchell, T. K. , Li, L. , Xu, J. R. , Orbach, M. , Thon, M. , Dean, R. A. and Farman, M. L. (2007) A systematic analysis of T‐DNA insertion events in Magnaporthe oryzae . Fungal Genet. Biol. 44, 1050–1064. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Cornelissen, B.J. and Rep, M. (2009) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. Genome Biol. 10, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, E. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Munch, S. , Ludwig, N. , Floss, D. S. , Sugui, J. A. , Koszucka, A. M. , Voll, L. M. , Sonnewald, U. and Deising, H. B. (2011) Identification of virulence genes in the corn pathogen Colletotrichum graminicola by Agrobacterium tumefaciens‐mediated transformation. Mol. Plant Pathol. 12, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, R.L. and Kunoh, H. (1995) Early interactions, adhesion, and establishment of the infection court by Erysiphe graminis . Can. J. Bot. 73, 609–615. [Google Scholar]
- Oka, T. , Nemoto, T. and Jigami, Y. (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP‐D‐glucose to UDP‐L‐rhamnose conversion. J. Biol. Chem. 282, 5389–5403. [DOI] [PubMed] [Google Scholar]
- Pegg, G.F. and Brady, B.L. (2002) Verticillium Wilts. CABI publishing, Wallingford, UK.
- Ramadoss, C.S. , Uhlig, J. , Carbon, D.M. , Butler, L.G. and Nicholson, R.L. (1985) Composition of the mucilaginous spore matrix of Colletotrichum graminicola, a pathogen of corn, sorghum, and other grasses. J. Agric. Food Chem. 33, 728–732. [Google Scholar]
- Ramos, B. , Gonzalez‐Melendi, P. , Sanchez‐Vallet, A. , Sanchez‐Rodriguez, C. , Lopez, G. and Molina, A. (2013) Functional genomics tools to decipher the pathogenicity mechanisms of the necrotrophic fungus Plectosphaerella cucumerina in Arabidopsis thaliana . Mol. Plant Pathol. 14, 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, R.C. , Davis, J.R. , Powelson, M.L. and Rouse, D.I. (1987) Potato early dying: causal agents and management strategies. Plant Dis. 71, 482–489. [Google Scholar]
- Santhanam, P. (2012) Random insertional mutagenesis in fungal genomes to identify virulence factors In: Plant fungal pathogens. Methods in molecular biology, (Bolton MD, T. B., ed.). Totowa: Humana press, pp. 509–517. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. and Thomma, B.P.H.J. (2013) Verticillium dahliae Sge1 differentially regulates expression of candidate effector genes. Mol. Plant–Microbe Interact. 26, 249–256. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , van Esse, H. P. , Albert, I. , Faino, L. , Nurnberger, T. and Thomma, B. P. H. J. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Schaible, L. , Cannon, O.S. and Waddoups, V. (1951) Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology, 41, 986–990. [Google Scholar]
- Schumacher, J. , Simon, A. , Cohrs, K.C. , Viaud, M. and Tudzynski, P. (2014) The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea . PLoS Genet. 10, e1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko, I. , Haynes, K. G. , Ewing, E. E. , Costanzo, S. , Christ, B. J. and Jones, R. W. (2004) Mapping genes for resistance to Verticillium albo‐atrum in tetraploid and diploid potato populations using haplotype association tests and genetic linkage analysis. Mol. Genet. Genomics, 271, 522–531. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Braus‐Stromeyer, S. A. , Timpner, C. , Tran, V. T. , Lohaus, G. , Reusche, M. , Knufer, J. , Teichmann, T. , von Tiedemann, A. and Braus, G. H. (2010) Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross‐pathway control in the xylem. Appl. Microbiol. Biotechnol. 85, 1961–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui, J.A. , Kunoh, H. and Nicholson, R.L. (1999) Detection of protein and carbohydrate in the extracellular matrix released by Cochliobolus heterostrophus during germination. Mycoscience, 40, 11–19. [Google Scholar]
- Thomma, B.P.H.J. , Nurnberger, T. and Joosten, M.H.A.J. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpner, C. , Braus‐Stromeyer, S.A. , Tran, V.T. and Braus, G.H. (2013) The Cpc1 regulator of the cross‐pathway control of amino acid biosynthesis is required for pathogenicity of the vascular pathogen Verticillium longisporum . Mol. Plant–Microbe Interact. 26, 1312–1324. [DOI] [PubMed] [Google Scholar]
- Tzima, A.K. , Paplomatas, E.J. , Rauyaree, P. , Ospina‐Giraldo, M.D. and Kang, S. (2011) VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell‐wall degradation. Mol. Plant–Microbe Interact. 24, 129–142. [DOI] [PubMed] [Google Scholar]
- Watt, G. , Leoff, C. , Harper, A.D. and Bar‐Peled, M. (2004) A bifunctional 3,5‐epimerase/4‐keto reductase for nucleotide‐rhamnose synthesis in Arabidopsis. Plant Physiol. 134, 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, S. (1955) Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology, 45, 180–181. [Google Scholar]
- Winnenburg, R. , Baldwin, T. K. , Urban, M. , Rawlings, C. , Köhler, J. and Hammond‐Kosack, K. E. (2006) PHI‐base: a new database for pathogen host interactions. Nucleic Acids Res. 34, D459–D464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnenburg, R. , Urban, M. , Beacham, A. , Baldwin, T. K. , Holland, S. , Lindeberg, M. , Hansen, H. , Rawlings, C. , Hammond‐Kosack, K. E. and Kohler, J. (2008) PHI‐base update: additions to the pathogen host interaction database. Nucleic Acids Res. 36, D572–D576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. and Chen, W. (2013) Random T‐DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum . Mol. Plant–Microbe Interact. 26, 431–441. [DOI] [PubMed] [Google Scholar]
- York, W.S. , Darvill, A.G. , McNeil, M. , Stevenson, T.T. and Albersheim, P. (1986) Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118, 3–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Verification of deletion and complementation strains by polymerase chain reaction (PCR). (A) Amplification of the T‐DNA flanking sequences from wild‐type Verticillium dahliae (w), 10 independent random transformants (lanes 1–10) and the marker (m). (B) Amplification of the hygromycin resistance gene (hph) and VdNRS/ER from 10 independent transformants (lanes 1–10) and wild‐type V. dahliae (w). The marker (m) is indicated. True deletion strains are marked with asterisks, and the ectopic transformant is marked with an arrow. (C) Amplification of the zeocin resistance gene (ble) and VdNRS/ER from five independent complementation strains (lanes 1–5), a VdNRS/ER deletion strain (*) and wild‐type V. dahliae (w). The marker (m) and true complementation strains (+) are indicated.
Fig. S2 Complementation of VdNRS/ER deletion strain restores pathogenicity on tomato. (A) Side and top views of tomato cultivar MoneyMaker plants inoculated with wild‐type Verticillium dahliae (WT), a VdNRS/ER deletion strain (Δ6010‐1) and two complementation strains (Comp‐1 and Comp‐2), or mock inoculated at 14 days post‐inoculation (dpi). (B) Fungal outgrowth at 7 days after plating of stem sections harvested at 14 dpi. (C) Average canopy area of six plants at 14 dpi as described for (A). Different letters indicate significant differences (P < 0.05).
Table S1 Primers used in this study.


