Summary
Plant‐pathogenic microbes secrete effector molecules to establish themselves on their hosts, whereas plants use immune receptors to try and intercept such effectors in order to prevent pathogen colonization. The tomato cell surface‐localized receptor Ve1 confers race‐specific resistance against race 1 strains of the soil‐borne vascular wilt fungus Verticillium dahliae which secrete the Ave1 effector. Here, we describe the cloning and characterization of Ve1 homologues from tobacco (Nicotiana glutinosa), potato (Solanum tuberosum), wild eggplant (Solanum torvum) and hop (Humulus lupulus), and demonstrate that particular Ve1 homologues govern resistance against V. dahliae race 1 strains through the recognition of the Ave1 effector. Phylogenetic analysis shows that Ve1 homologues are widely distributed in land plants. Thus, our study suggests an ancient origin of the Ve1 immune receptor in the plant kingdom.
Keywords: Ave1 effector, leucine‐rich repeat, receptor‐like protein, RLP, Verticillium dahliae
Introduction
In order to activate immune responses to ward off invading microorganisms, plants employ immune receptors that detect pathogen(‐induced) ligands of various nature (Boller and Felix, 2009; Thomma et al., 2011). The recognition of such ligands by immune receptors results in the activation of defence responses, which are often accompanied by a hypersensitive response (HR), in which the necrosis of plant tissue surrounding the site of attempted penetration is activated to restrict further pathogen invasion.
Verticillium wilts are vascular wilt diseases caused by soil‐borne fungal pathogens that belong to the Verticillium genus. Verticillium dahliae is the most notorious species and can infect hundreds of dicotyledonous hosts (Fradin and Thomma, 2006; Inderbitzin et al., 2011). In tomato (Solanum lycopersicum), the Ve locus that confers race‐specific resistance against Verticillium has been characterized (Fradin et al., 2009; Kawchuk et al., 2001). This locus contains two closely linked and inversely oriented genes, Ve1 and Ve2, which encode extracellular leucine‐rich repeat receptor‐like proteins (eLRR‐RLPs) (Kawchuk et al., 2001; Wang G et al., 2008, 2010). Of these, only Ve1 was found to provide V. dahliae resistance in tomato (Fradin et al., 2009). Interestingly, interfamily transfer of Ve1 from tomato to Arabidopsis resulted in Verticillium resistance in the latter species (Fradin et al., 2011, 2014; Zhang et al., 2014), implying that the underlying immune signalling pathway is conserved (Fradin et al., 2011; Thomma et al., 2011).
Comparative genomics of V. dahliae race 1 and 2 strains identified the Ave1 effector that activates Ve1‐mediated immunity (de Jonge et al., 2012). Interestingly, Ave1 homologues were found in the bacterial plant pathogen Xanthomonas axonopodis pv. citri (XacPNP) and in the plant‐pathogenic fungi Colletotrichum higginsianum (ChAve1), Cercospora beticola (CbAve1) and Fusarium oxysporum f. sp. lycopersici (FoAve1), and these homologues were differentially recognized by Ve1 (de Jonge et al., 2012). During the optimization of an agroinfiltration assay in tobacco for the functional analysis of Ve1 signalling, we found that the expression of Ave1 in leaves of Nicotiana glutinosa triggered an HR, suggesting that this species contains an endogenous Ve1 allele (Zhang et al., 2013a). Indeed, inoculation experiments revealed that N. glutinosa is resistant to race 1 V. dahliae, whereas an Ave1 deletion strain was able to cause Verticillium wilt disease on these plants (Zhang et al., 2013a).
So far, several Ve1 homologues have been identified within and outside the Solanaceae family, such as SlVe1 from Solanum lycopersicoides (Chai et al., 2003), StVe1 from S. tuberosum (Simko et al., 2004a), StVe and StoVe1 from S. torvum (Fei et al., 2004; Liu et al., 2012), mVe1 from Mentha longifolia (Vining and Davis, 2009), Vr1 from Lactuca sativa (Hayes et al., 2011), VvVe from Vitis vinifera (Tang et al., 2016) and GbVe, Gbve1, Gbvdr5 and Gbvdr3 from Gossypium barbadense (Chen et al., 2016; Yang et al., 2014; Zhang et al., 2012, 2011). However, the functionality of these homologues against Verticillium wilt often remains obscure. Here, we describe the cloning and functional characterization of Verticillium wilt resistance genes from tobacco (N. glutinosa), potato (S. tuberosum), wild eggplant (S. torvum) and hop (Humulus lupulus), and demonstrate that particular Ve1 homologues govern resistance against V. dahliae race 1 strains through the recognition of the Ave1 effector.
Results
Isolation of NgVe1 from N. glutinosa
In our first attempt to clone the previously identified Ve1 homologue from N. glutinosa (Zhang et al., 2013a), a single cDNA fragment of ∼2800 bp was obtained using primers that were designed on the tomato Ve1 sequence (Table S1, see Supporting Information). To obtain the full‐length N. glutinosa Ve1 (NgVe1) transcript sequence, 3′ rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR) was performed, resulting in a single fragment of approximately 1200 bp. Likewise, a fragment of approximately 640 bp was amplified with 5′ RACE (Methods S1, see Supporting Information). The sequences of the three fragments were aligned to deduce the full‐length NgVe1 cDNA sequence. Subsequently, a pair of NgVe1‐specific primers (NgVe1‐F and NgVe1‐R; Table S1) was designed and amplicons amplified from N. glutinosa cDNA and genomic DNA were sequenced, indicating that both amplicons are identical (GenBank accession: KT895339) and that NgVe1 is an intronless gene.
The full‐length cDNA of NgVe1 is 3225 bp and contains a predicted translation initiation site (ATG) at nucleotide position 34 and a stop codon (TGA) at nucleotide position 3178, resulting in a single open reading frame of 3147 bp. The predicted NgVe1 protein comprises 1048 amino acids (GenBank accession: ALK26499) and shares an overall identity of 76% with tomato Ve1 (Fig. S1, see Supporting Information). Immunoblotting analysis using green fluorescent protein (GFP) antibody displayed clear signals for NgVe1‐GFP and Ve1‐GFP in transiently transformed tobacco leaves (Fig. S2, see Supporting Information).
Co‐expression of Ave1 and NgVe1 induces an HR in N. tabacum
Recently, an optimized agroinfiltration assay has been developed for Ve1‐mediated immune signalling in N. tabacum, revealing a swift HR on co‐expression of tomato Ve1 with V. dahliae Ave1 (Zhang et al., 2013a). To test the functionality of NgVe1, co‐expression with Ave1 on agroinfiltration in N. tabacum was performed. At 5 days post‐infiltration, the infiltrated leaves developed clear necrosis, and the HR induced on co‐expression of NgVe1 and Ave1 was as strong as the HR induced on co‐infiltration of tomato Ve1 and Ave1, for which the complete infiltrated areas became fully necrotic (Fig. 1A). In contrast, agroinfiltration of NgVe1 or Ave1 alone did not induce necrosis (Fig. 1A). These data strongly suggest that NgVe1 is a functional homologue of tomato Ve1.
Figure 1.

Expression of NgVe1 in tobacco mediates the Ave1‐triggered hypersensitive response (HR). (A) Co‐expression of NgVe1 and Ave1 in tobacco results in an HR. Photographs were taken at 5 days post‐infiltration and show representative leaves of at least three independent assays. As a positive control, HR was induced on co‐infiltration of Ve1 and Ave1. As a negative control, NgVe1 and Ave1 were expressed separately. (B) Ave1‐triggered HR, but not VdNLP1‐mediated cell death, is attenuated in NgVe1‐silenced Nicotiana glutinosa plants, whereas Avr9 does not trigger cell death.
Targeting of NgVe1 expression in N. glutinosa compromises Ave1‐induced HR, but not Verticillium resistance
To investigate the role of NgVe1 in N. glutinosa Verticillium resistance, we used virus‐induced gene silencing (VIGS). Tobacco rattle virus (TRV)‐based VIGS is a well‐established method for gene functional analysis in several Solanaceae species, and also for the investigation of Ve1‐mediated Verticillium resistance (Fradin et al., 2009; Senthil‐Kumar et al., 2007; Zhang et al., 2013a). In an attempt to establish VIGS in N. glutinosa, a 1 : 1 mixture of Agrobacterium tumefaciens cultures carrying pTRV1 and pTRV2::PDS to target the phytoene desaturase (PDS) gene was infiltrated into cotyledons of N. glutinosa plants. Visible photobleaching symptoms were observed in all agroinfiltrated N. glutinosa plants by 4 weeks post‐infiltration (Fig. S3A, see Supporting Information), albeit that a strongly varying degree of photobleaching was observed (Fig. S3A). Nevertheless, a recombinant TRV vector was designed to target NgVe1 expression (pTRV2::NgVe1). As a negative control, a construct (pTRV2::GUS) containing a fragment of the β‐glucuronidase (GUS) gene was used. At 4 weeks after TRV infection, mature leaves were agroinfiltrated to express Ave1, with VdNLP1 as a positive control (Santhanam et al., 2013) and the functionally and structurally unrelated effector Avr9 from the tomato leaf mould pathogen Cladosporium fulvum as a negative control (van Kan et al., 1991; Van der Hoorn et al., 2000). Agroinfiltration of Ave1 in N. glutinosa on GUS targeting resulted in a clear HR within 5 days, confirming that TRV infection did not compromise Ve1‐mediated HR (Fig. 1B). However, targeting of NgVe1 expression in N. glutinosa significantly compromised HR on expression of Ave1 (Fig. 1B). As expected, VdNLP1‐mediated cell death was not compromised on targeting of NgVe1 expression, whereas Avr9 expression never triggered HR (Fig. 1B).
To test the role of NgVe1 in Verticillium resistance, 3 weeks after TRV inoculation, plants were challenged with either V. dahliae race 1 strain JR2 (Faino et al., 2015) or a transformant from which the Ave1 gene had been deleted (V. dahliae JR2ΔAve1; de Jonge et al., 2012), and monitored for disease development (stunting, wilting, chlorosis and necrosis) up to 14 days post‐inoculation (dpi). As expected, no disease symptoms were observed in N. glutinosa plants on GUS targeting and subsequent mock inoculation or on inoculation with V. dahliae JR2, whereas the Ave1 deletion strain caused clear Verticillium wilt disease (Fig. S3B). However, unexpectedly, in repeated assays, no Verticillium wilt symptoms were observed on NgVe1 targeting and subsequent inoculation with V. dahliae (Fig. S3B). However, in line with the extremely variable photobleaching (Fig. S3A), assessment of the silencing efficiency revealed only a slight reduction in NgVe1 expression in NgVe1‐targeted N. glutinosa plants when compared with GUS‐silenced plants (Fig. S3C), and attempts to increase the silencing efficiency were unsuccessful. These results confirm previous observations that N. glutinosa is not very amenable to TRV‐based VIGS (Senthil‐Kumar et al., 2007; Zhang et al., 2013a) and, furthermore, suggest that the moderate silencing efficiency obtained in our experiments is sufficiently high to compromise NgVe1‐mediated HR, but insufficient to compromise NgVe1‐mediated resistance.
Expression of NgVe1 in Arabidopsis confers Verticillium resistance
As TRV‐based VIGS did not appear to be very suitable for gene functional analysis in N. glutinosa, we pursued other strategies to functionally characterize NgVe1. We have shown previously that interfamily transfer of tomato Ve1 to Arabidopsis results in resistance to race 1 Verticillium strains, providing a relatively fast method to assess Ve1 functionality (Fradin et al., 2011, 2014; Zhang et al., 2014). To further confirm functionality in Verticillium resistance, heterologous expression in Arabidopsis was obtained (Fig. S4, see Supporting Information). No obvious developmental alterations were observed in transgenic plants when compared with wild‐type plants (Fig. 2A) and, subsequently, three independent NgVe1‐transgenic lines (NgVe1‐1, NgVe1‐2 and NgVe1‐3), as well as transgenic plants expressing tomato Ve1 (Fradin et al., 2011) and non‐transgenic controls, were inoculated with V. dahliae JR2. Interestingly, like tomato Ve1‐expressing plants, NgVe1‐transgenic plants were clearly resistant to race 1 V. dahliae, as significantly fewer Verticillium wilt symptoms were observed when compared with non‐transgenic control plants (Fig. 2A,B). In contrast, NgVe1 and Ve1 transgenic plants were as diseased as non‐transgenic controls on inoculation with the Ave1 deletion strain (Fig. 2A,B). These data are further supported by fungal biomass quantifications which revealed significantly reduced fungal accumulation in NgVe1‐transgenic and Ve1‐expressing Arabidopsis plants for V. dahliae carrying Ave1, but not for the Ave1 deletion mutant, when compared with wild‐type Arabidopsis plants (Fig. 2C). Collectively, these data confirm that NgVe1 acts as a functional homologue of tomato Ve1 that recognizes race 1 V. dahliae.
Figure 2.
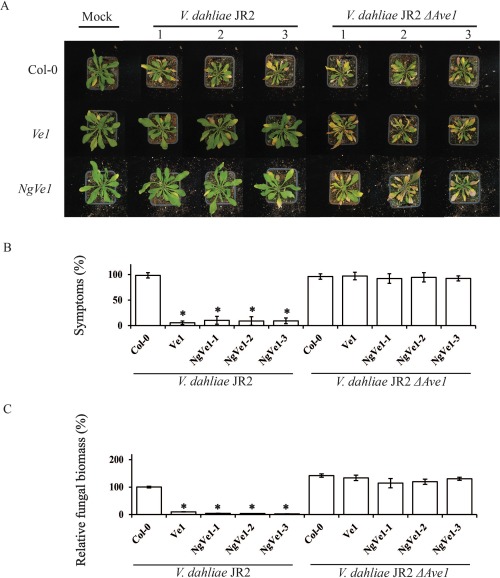
Expression of NgVe1 in Arabidopsis mediates resistance against race 1 Verticillium dahliae. (A) Typical appearance of non‐transgenic Arabidopsis and transgenic lines that constitutively express NgVe1 on mock inoculation or inoculation with V. dahliae strain JR2 or V. dahliae JR2 ΔAve1 at 21 days post‐inoculation (dpi). (B) Quantification of Verticillium wilt symptoms in Arabidopsis Col‐0 and transgenic plants at 21 dpi. Bars represent quantification of symptom development as the percentage of diseased rosette leaves with standard deviation, with Col‐0 (control) set to 100%. (C) Fungal biomass determined by quantitative real‐time polymerase chain reaction in Arabidopsis Col‐0 and transgenic plants at 21 dpi. Bars represent Verticillium internal transcribed spacer (ITS) transcript levels relative to AtRuBisCo (RuBisCo, ribulose‐1,5‐bisphosphate carboxylase/oxygenase) transcript levels (for equilibration) with standard deviation in a sample of five pooled plants. The fungal biomass in Col‐0 (control) is set to 100%. Three independent lines are shown (1, 2 and 3). Asterisks indicate significant differences when compared with Col‐0 (P < 0.05). Ve1 transgenic plants were used as a positive control. The data shown are representative of at least three independent experiments.
Cloning and functional analysis of Ve1 homologues from potato and wild eggplant
Ve gene homologues occur in the solanaceous species S. lycopersicoides (SlVe1; Chai et al., 2003) and the wild eggplant species S. torvum (StVe and StoVe1; Fei et al., 2004; Liu et al., 2012). Moreover, in tetraploid potato (S. tuberosum), a quantitative trait locus (QTL) for Verticillium resistance was identified using the tomato Ve1 gene as a probe. This locus was found to contain at least 11 genes, all putatively encoding LRR‐type receptor‐like proteins (Simko et al., 2004a). The tomato and potato genomes are highly collinear and the QTL locus was mapped to a region on potato chromosome 9 that is syntenic to the short arm of tomato chromosome 9 that carries Ve1 and Ve2 (Diwan et al., 1999; Simko et al., 2004b). Subsequently, this Verticillium resistance QTL locus was annotated and found to contain two predicted receptor‐like protein 12‐like genes [National Center for Biotechnology Information (NCBI): XM_006362308 and XM_006362309] in the genome sequence of S. tuberosum group Phureja DM1‐3 516 R44 (Xu et al., 2011). Here, the coding sequences (CDSs) of Ve gene homologues were amplified from cDNA of the heterozygous diploid potato breeding line S. tuberosum group Tuberosum RH 89‐039‐16 (Xu et al., 2011), sequenced and submitted to NCBI as StuVe1 and StuVe2 (GenBank accessions: KT946795 and KT946797) (Methods S1). The predicted StuVe1 and StuVe2 proteins are composed of 1053 and 1138 amino acids (GenBank accessions: ALK26501 and ALK26503), respectively, and share 87% and 84% identity with tomato Ve1 and 81% and 91% identity with tomato Ve2, respectively, and 82% identity with each other (Fig. S1).
To study the composition of the Ve locus in wild eggplant, the CDSs of Ve gene homologues were cloned from the cDNA of the Verticillium‐resistant S. torvum genotype Tuolubamu, sequenced and deposited at NCBI as StoVe1 and StoVe2 (GenBank accessions: KT946794 and KT946796) (Methods S1). The predicted StoVe1 and StoVe2 proteins comprise 1051 and 1135 amino acids (GenBank accessions: ALK26500 and ALK26502), respectively, and share 83% and 80% identity with tomato Ve1 and 81% and 85% identity with tomato Ve2, respectively, and 92% identity with each other (Fig. S1).
To check the functionality of StuVe1, StuVe2, StoVe1 and StoVe2, mature tobacco leaves were co‐infiltrated with a 1 : 1 mixture of A. tumefaciens cultures carrying Ave1 and the various Ve1 homologues. Intriguingly, agroinfiltration in at least three independent assays revealed that the expression of Ave1 together with StuVe1 or StoVe1 induced signs of a weak HR at 5 days post‐infiltration (Fig. 3A). However, when compared with the HR induced on co‐agroinfiltration of tomato Ve1 and Ave1, only a minor part of the infiltrated region developed necrosis (Fig. 3A). Agroinfiltration of Ave1 with StuVe2 or StoVe2 induced no such responses at all (Fig. 3A). Immunoblotting confirmed that the StuVe1, StuVe2, StoVe1 and StoVe2 fusion proteins were expressed (Fig. 3B).
Figure 3.
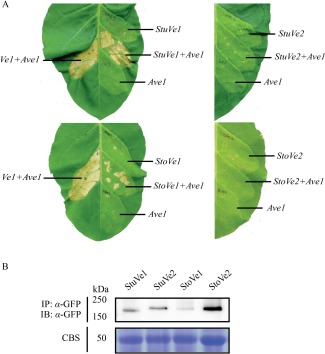
StuVe1 and StoVe1, but not StuVe2 and StoVe2, recognize Ave1 in Nicotiana tabacum. (A) Co‐expression of StuVe1 and StoVe1, but not StuVe2 and StoVe2, with Ave1 in tobacco induces signs of a relatively weak hypersensitive response (HR). Photographs were taken at 5 days post‐infiltration and show representative leaves of at least three independent assays. As a positive control, HR was induced on co‐infiltration of Ve1 and Ave1. As a negative control, StuVe1, StuVe2, StoVe1, StoVe2 and Ave1 were expressed separately. (B) Green fluorescent protein (GFP)‐tagged StuVe1, StuVe2, StoVe1 and StoVe2 proteins are detected in planta by immunopurification (IP) using GFP‐affinity beads, followed by immunoblotting (IB) using α‐GFP antibody. Coomassie blue staining (CBS) of the blot containing total protein extracts showed equal loading in each lane based on the 50‐kDa RuBisCo (ribulose‐1,5‐bisphosphate carboxylase/oxygenase) band.
As VIGS‐based gene silencing in potato genotype Tuberosum RH 89‐039‐16 and wild eggplant genotype Tuolubamu has not been established, we did not attempt VIGS‐based assays to test the role of these Ve1 homologues in Verticillium resistance. Rather, heterologous expression in Arabidopsis was pursued (Fig. S4). No developmental alterations were observed in transgenic plants when compared with Ve1‐expressing and wild‐type plants (Fig. 4A), and three independent transgenic lines expressing StuVe1, StuVe2, StoVe1 or StoVe2 were assayed for V. dahliae resistance. Intriguingly, despite the weak HR observed on agroinfiltration together with Ave1 in N. tabacum, StoVe1‐ and StuVe1‐expressing plants were clearly resistant to race 1 V. dahliae strain JR2, similar to Ve1‐transgenic plants (Fig. 4A,B). In contrast, StuVe2 and StoVe2 transgenes were as diseased as non‐transgenic controls (Fig. 4A,B). Importantly, all genotypes were equally susceptible to the V. dahliae Ave1 deletion mutant (Fig. 4A,B), suggesting that all of these Ve1 alleles recognize the Ave1 effector. The phenotypes correlated with the degree of V. dahliae colonization as determined by real‐time PCR (Fig. 4). Collectively, these data confirm that StuVe1 and StoVe1, but not StuVe2 and StoVe2, act as functional homologues of tomato Ve1 that confer resistance to race 1 V. dahliae.
Figure 4.
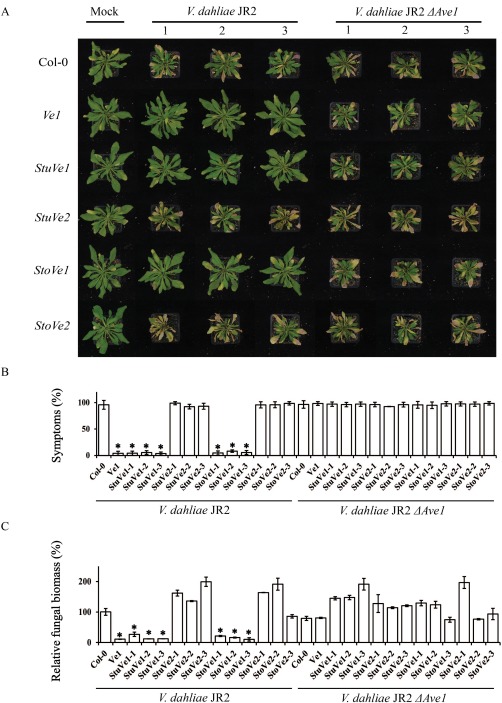
StuVe1 and StoVe1, but not StuVe2 and StoVe2, provide resistance against race 1 Verticillium dahliae in Arabidopsis. (A) Typical appearance of non‐transgenic Arabidopsis and transgenic plants that were engineered to express 35S‐driven StuVe1, StuVe2, StoVe1 or StoVe2 on mock inoculation or inoculation with V. dahliae JR2 or V. dahliae JR2 ΔAve1 at 21 days post‐inoculation (dpi). (B) Quantification of Verticillium wilt symptoms in Arabidopsis Col‐0 and transgenic plants at 21 dpi. Bars represent the quantification of symptom development as the percentage of diseased rosette leaves with standard deviation, with Col‐0 (control) set to 100%. (C) Fungal biomass determined by quantitative real‐time polymerase chain reaction in Arabidopsis Col‐0 and transgenic plants at 21 dpi. Bars represent Verticillium internal transcribed spacer (ITS) transcript levels relative to AtRuBisCo (RuBisCo, ribulose‐1,5‐bisphosphate carboxylase/oxygenase) transcript levels (for equilibration) with standard deviation in a sample of five pooled plants. The fungal biomass in Col‐0 (control) is set to 100%. Three independent lines are shown (1, 2 and 3). Asterisks indicate significant differences when compared with Col‐0 (P < 0.05). Ve1 transgenic plants were used as a positive control. The data shown are representative of at least three independent experiments.
HLVe1‐2A, but not HLVe1‐2B, recognizes Verticillium effector Ave1
Polygenic resistance to Verticillium spp. has also been described in several non‐solanaceous species, including hop, alfalfa, cotton and strawberry (Antanaviciute et al., 2015; Bolek et al., 2005; Jakse et al., 2013; Wang HM et al., 2008; Yang et al., 2008). Genetic resistance against Verticillium wilt in hop (Humulus lupulus) was introduced into breeding programmes from American wild hop (H. lupulus var. neomexicanus) and is still used today as the main resistance source (Darby, 2001). Genetic analysis identified a single significant QTL for this resistance, suggesting that Verticillium wilt resistance in hop is conferred by more than a single gene (Jakse et al., 2013; Majer et al., 2014). To investigate the presence of Ve‐like sequences in hop, Southern blotting with the tomato Ve1 gene as probe was performed, revealing low copy numbers of Ve‐like sequences in hop cultivars (Fig. S5 and Methods S1, see Supporting Information). With thermal asymmetric interlaced (TAIL)‐PCR (Terauchi and Kahl, 2000), several Ve‐like sequences were identified (Methods S1). Further analysis revealed two Ve1 alleles in the Verticillium‐resistant hop cultivar ‘Wye Target’, designated HLVe1‐2A (GenBank accession: KJ647426) and HLVe1‐2B (GenBank accession: KJ647427), which both encode 1039‐amino‐acid proteins (GenBank accessions: AIE39594 and AIE39595) sharing 52% and 51% identity with tomato Ve1 and Ve2, respectively, and 98% identity with each other (Fig. S1). To investigate the functionality of HLVe1‐2A and HLVe1‐2B in Verticillium resistance, co‐agroinfiltration with Ave1 in N. tabacum was performed. When mature tobacco leaves were co‐infiltrated with a 1 : 1 mixture of A. tumefaciens cultures carrying Ave1 and HLVe1‐2A, signs of a weak HR were observed at 5 days post‐infiltration with a minor part of the infiltrated region developing necrosis (Fig. 5A). However, in contrast, co‐expression of Ave1 and HLVe1‐2B in tobacco induced no such response, similar to the co‐agroinfiltration of Ve1, HLVe1‐2A and HLVe1‐2B with Avr9 (Fig. 5A). To test whether the failure of HLVe1‐2B to induce an HR was the result of the instability of the protein, the coding regions of HLVe1‐2A and HLVe1‐2B were cloned to generate C‐terminally GFP‐tagged expression constructs, and the stability of both proteins was verified by immunoblotting (Fig. 5B).
Figure 5.
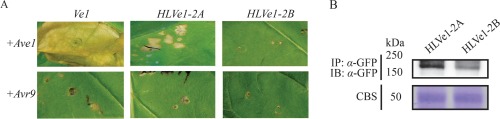
Co‐expression of HLVe1‐2A, but not HLVe1‐2B, with Ave1 in Nicotiana tabacum activates a hypersensitive response (HR). (A) HLVe1‐2A or HLVe1‐2B was transiently co‐expressed with Ave1 in N. tabacum. As a negative control, Avr9 was co‐expressed with Ve1 homologues. As a positive control, HR was induced on co‐expression of Ve1 and Ave1. (B) Green fluorescent protein (GFP)‐tagged HLVe1‐2A and HLVe1‐2B proteins are detected in planta by immunopurification (IP) using GFP‐affinity beads, followed by immunoblotting (IB) using α‐GFP antibody. Coomassie blue staining (CBS) of the blot containing total protein extracts showed equal loading in each lane based on the 50‐kDa RuBisCo (ribulose‐1,5‐bisphosphate carboxylase/oxygenase) band.
To further assess the role of HLVe1‐2A and HLVe1‐2B in resistance to V. dahliae, heterologous expression in Arabidopsis was pursued (Fig. S4). No phenotypic alterations were observed in plants that expressed HLVe1‐2A or HLVe1‐2B when compared with Ve1‐transgenic or non‐transgenic plants (Fig. 6A), and three independent transgenic lines for HLVe1‐2A and HLVe1‐2B were assayed for V. dahliae resistance. Interestingly, despite the weak HR observed on agroinfiltration together with Ave1 in N. tabacum, HLVe1‐2A‐expressing plants were clearly resistant to V. dahliae race 1 strain JR2 as few, if any, symptoms were observed (Fig. 6A,B). In contrast, HLVe1‐2B transgenic plants were as susceptible as non‐transgenic controls (Fig. 6A,B), and all genotypes were equally susceptible to the V. dahliae Ave1 deletion mutant (Fig. 6A,B). The phenotypes correlated with the level of V. dahliae biomass as determined by real‐time PCR (Fig. 6). Collectively, these data verify that HLVe1‐2A, but not HLVe1‐2B, acts a functional Ve1 homologue that provides resistance to race 1 V. dahliae.
Figure 6.
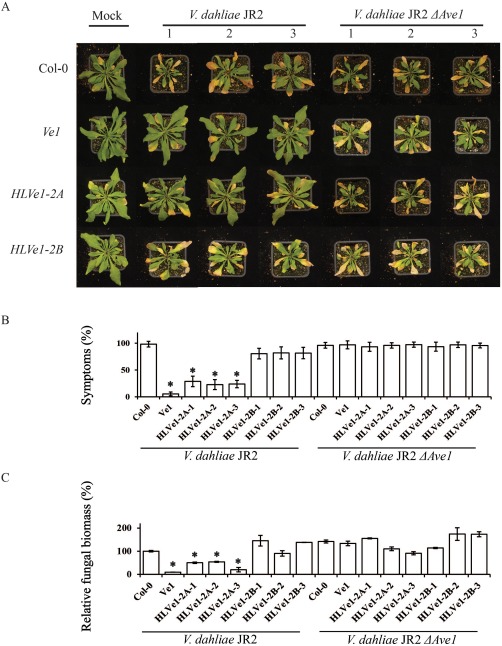
HLVe1‐2A, but not HLVe1‐2B, confers resistance to race 1 Verticillium dahliae in Arabidopsis. (A) Typical appearance of non‐transgenic Arabidopsis and transgenic lines that constitutively express HLVe1‐2A or HLVe1‐2B on mock inoculation or inoculation with V. dahliae strain JR2 or V. dahliae JR2 ΔAve1 at 21 days post‐inoculation (dpi). (B) Quantification of Verticillium wilt symptoms in Arabidopsis Col‐0 and transgenic lines at 21 dpi. Bars represent the quantification of symptom development as the percentage of diseased rosette leaves with standard deviation, with Col‐0 (control) set to 100%. (C) Fungal biomass determined by quantitative real‐time polymerase chain reaction in Arabidopsis Col‐0 and transgenic plants at 21 dpi. Bars represent Verticillium internal transcribed spacer (ITS) transcript levels relative to AtRuBisCo (RuBisCo, ribulose‐1,5‐bisphosphate carboxylase/oxygenase) transcript levels (for equilibration) with standard deviation in a sample of five pooled plants. The fungal biomass in Col‐0 (control) is set to 100%. Three independent lines are shown (1, 2 and 3). Asterisks indicate significant differences when compared with Col‐0 (P < 0.05). Ve1 transgenic plants were used as a positive control. The data shown are representative of at least three independent experiments.
Functional Ve1 homologues mediate the recognition of Ave1 homologues from multiple plant pathogens
We have shown previously that tomato Ve1 recognizes not only V. dahliae and V. albo‐atrum Ave1, but also homologues derived from F. oxysporum f. sp. lycopersici (FoAve1) and C. beticola (CbAve1) (de Jonge et al., 2012). To investigate whether the newly identified functional Ve1 homologues similarly recognize these Ave1 homologues, co‐expression of the five functional Ve1 homologues with a series of Ave1 homologues [FoAve1, CbAve1, ChAve1, CoAve1 (Colletotrichum orbiculare; Gan et al, 2013) and XacPNP (Gottig et al., 2008)] was performed (Fig. 7A). Co‐expression of the effector Avr9 (van Kan et al., 1991; Van der Hoorn et al., 2000) in combination with the Ve1 homologues was used as a negative control. To compare the HR induced on co‐expression of Ave1 homologues and functional Ve1 homologues in tobacco, HR development was measured by quantification of the leaf area that developed necrosis at 5 days post‐infiltration (Fig. 7B). Co‐expression of Ve1 with FoAve1 and CbAve1, but not with ChAve1, CoAve1 and XacPNP, in N. tabacum resulted in HR (Fig. 7). StuVe1 seems to recognize a wider panel of Ave1 homologues, as co‐expression with Ave1, FoAve1, CbAve1, ChAve1, CoAve1 and XacPNP induced HR (Fig. 7). For NgVe1 and StoVe1, co‐infiltration with Ave1 and FoAve1 induced HR, whereas infiltration with CbAve1, ChAve1, CoAve1 and XacPNP failed to induce HR (Fig. 7). Finally, the most narrow recognition spectrum is observed for HLVe1‐2A which recognizes none of the Ave1 homologues apart from that from V. dahliae (Fig. 7). These data demonstrate that the newly identified functional Ve1 homologues, similar to tomato Ve1, differentially recognize Ave1 homologues from different plant pathogens.
Figure 7.
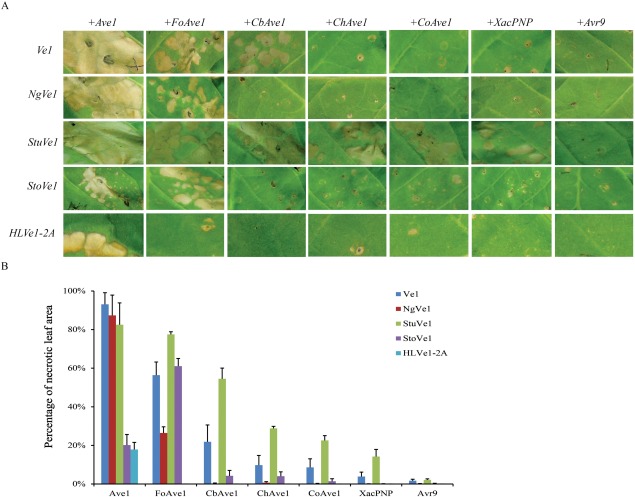
Distinct necrosis induced by Ave1 homologues through co‐expression with functional Ve1 orthologues in Nicotiana tabacum. (A) Co‐expression of functional Ve1 orthologues with Ave1 homologues (Ave1, FoAve1, CbAve1, ChAve1, CoAve1 and XacPNP) in N. tabacum. Expression of Avr9 in combination with Ve1 homologues is shown as a negative control. Leaves were photographed at 5 days post‐infiltration to visualize necrosis resulting from recognition by functional Ve1 homologues. (B) Quantification of necrosis resulting from the recognition of Ave1 homologues by functional Ve1 orthologues at 5 days post‐infiltration. Bars represent the average percentage of necrotic leaf area of infiltration zones with standard deviation.
Comparison of the protein sequences of Ve1 homologues
Tomato Ve1 is predicted to contain a signal peptide, an eLRR domain composed of two eLRR regions, separated by a non‐LRR island domain (also referred to as C1, C3 and C2, respectively; Figs S6 and S7, see Supporting Information), a transmembrane domain and a short cytoplasmic tail that lacks obvious motifs for intracellular signalling (Kawchuk et al., 2001; Wang et al., 2010; Zhang and Thomma, 2013). Alignment of the functional Ve1 protein sequences identified in this study clearly shows the typical eLRR‐RLP domain architecture (Fig. S6). All Ve1 homologues contain 37 eLRR repeats in two eLRR regions that are interrupted by a non‐LRR island domain (Fig. S6). Previously, we have determined that three eLRR regions are crucial for Ve1 functionality: eLRR_1 to eLRR_8, eLRR_20 to eLRR_23 and eLRR_32 to eLRR_37 (Zhang et al., 2014). A comparison of the functional Ve1 homologues studied here shows that the eLRR_1 to eLRR_8 (44.2% identity) and eLRR_20 to eLRR_23 (46.5% identity) regions of the C1 domain are only slightly more conserved than the eLRR_9 to eLRR_19 (40.2% identity) and eLRR_24 to eLRR_31 (45.0% identity) regions of the C1 domain (Figs S6 and S7). A similar comparison of the C1 domains among the non‐functional Ve1 homologues studied here shows that the eLRR_1 to eLRR_8 (50.2% identity) region of the C1 domain is slightly more conserved than the eLRR_9 to eLRR_19 (45.5% identity) region of the C1 domain, whereas the eLRR_20 to eLRR_23 (50.0% identity) region of the C1 domain is conserved to a similar extent to the eLRR_24 to eLRR_31 (50.3% identity) region of the C1 domain (Fig. S7). Further comparison among the functional Ve1 homologues shows that the C3 domain (eLRR_32 to eLRR_37, 48.4% identity) is more conserved than the C1 domain (43.3% identity), C2 domain (8.0% identity) and C‐terminal eLRR‐flanking domain (9.2% identity) (Figs S6 and S7). This result is consistent with a previous comparison of tomato eLRR‐RLPs, which showed that the C3 domain is more conserved than the C1 domain (Fradin et al., 2014). Finally, a comparison of the non‐functional Ve1 homologues studied here shows that the C1 domain (48.7% identity) and C3 domain (eLRR_32 to eLRR_37, 53.3% identity) are more conserved than the C2 domain (8.0% identity) and C‐terminal eLRR‐flanking domain (7.2% identity) (Fig. S7). Collectively, these findings do not point towards a particular conservation of the three LRR regions that were previously implicated in Ve1 functionality.
Phylogenetic analysis of Ve1 homologues in the plant kingdom
To determine the phylogenetic breadth among Ve1 homologues in plants, we systematically queried the available genomes of 41 plant species for the occurrence of Ve1 homologues. In these genomes, we identified 1361 bona fide Ve1 homologues, all of which occur in land plants (embryophytes) and none in green algae (Fig. 8A). To further analyse the phylogenetic relationship of tomato Ve1 and its close homologues, we used a neighbour‐joining phylogeny to guide the extraction of the tomato Ve1 clade and the relevant sister clades (Figs 8B and S8, see Supporting Information). These sequences were used to infer a refined maximum likelihood phylogeny containing 608 Ve1 homologues that encompass monocots and dicots. This phylogeny revealed a Ve1 orthologous group, defined at the last common ancestor of monocots and dicots, which contains all functional Ve1 homologues that have been described so far (Figs 8B and S8). The broad phylogenetic distribution, with homologues present in all land plants, establishes that Ve1 is an ancient immune receptor (Figs 8 and S8), and that the last common ancestor contained at least a single, but more likely several, Ve1‐like genes. Moreover, we inferred a Ve1 orthologous group that comprises both monocots and dicots and includes all functional Ve1 genes, suggesting the conservation of function within this group of genes.
Figure 8.
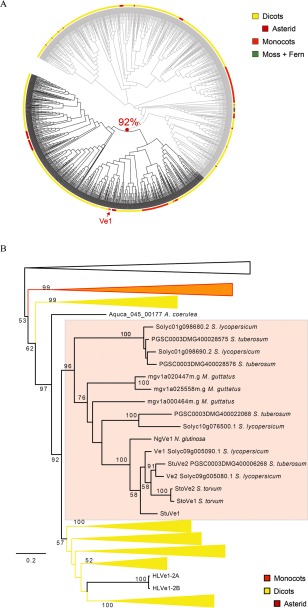
Phylogenetic analysis of Ve1 homologues indicates that they are widely distributed in land plants. (A) Phylogeny of Ve1 proteins from 41 plant species from the Phytozome database. Tomato Ve1 is indicated with a red arrow. (B) Maximum likelihood phylogenetic tree of selected Ve1 homologues (dark grey clades in A) displaying the tomato Ve1 clade and the relevant sister clades. The tomato Ve1 clade is indicated by the highlighted background. Official gene identifiers and species names are indicated. Bootstrap values are shown in the tree. The scale represents the branch length expressed as the relative number of amino acid substitutions.
Discussion
In this article, we describe the cloning and characterization of Ve1 homologues within and outside the Solanaceae family, and demonstrate that Ve1 homologues of tobacco (NgVe1), potato (StuVe1), wild eggplant (StoVe1) and hop (HLVe1‐2A) act as functional homologues of tomato Ve1 by providing resistance to race 1 V. dahliae, mediated through the recognition of the Ave1 effector, implying that functional Ve1 homologues are conserved across plant species within and outside the Solanaceae. We further show that all functional Ve1 proteins contain a conserved domain architecture with 37 eLRR repeats (Fig. S6). It has been determined previously that two regions of the C1 domain, namely eLRR_1 to eLRR_8 and eLRR_20 to eLRR‐23, are required for Ve1 functionality, probably because they contribute to ligand binding (Zhang et al., 2014). Here, these regions appear to be only slightly more conserved than other regions within the functional Ve1 homologues (Figs S6 and S7). In addition, the C3 domain (eLRR_32 to eLRR_37) has been shown to be critical for Ve1 functionality (Zhang et al., 2014; Fig. S6), potentially through interaction with common factors required for downstream signalling (Fradin et al., 2014; Fritz‐Laylin et al., 2005; Zhang and Thomma, 2013). As expected, this region is most conserved among the functional Ve1 homologues (Figs S6 and S7). Previously, Ve1 homologues from other plant species have also been associated with Verticillium wilt resistance, although conclusive evidence for a causal role in resistance has mostly been lacking. For example, Vining and Davis (2009) showed that the mint Ve1 homologue mVe1 associates with Verticillium wilt resistance. Genome‐wide analysis of disease resistance genes in lettuce, in combination with QTL mapping, showed that three Ve homologues, including Vr1, are located within a 100‐kb region on chromosome 9 that co‐segregates with resistance to race 1 V. dahliae (Christopoulou et al., 2015; Hayes et al., 2011). The grapevine Ve1 homologue VvVe has been shown to enhance defence against V. dahliae in N. benthamiana (Tang et al., 2016). Remarkably, cotton Ve1 homologues GbVe, Gbve1, Gbvdr5 and Gbvdr3 have been shown to confer Verticillium resistance on ectopic expression in Arabidopsis or cotton (Chen et al., 2016; Yang et al., 2014; Zhang et al., 2012, 2011), although this concerns V. dahliae isolates that do not carry the Ave1 effector gene, and thus Verticillium wilt resistance cannot be mediated by Ave1 recognition in these cases (Y. Song et al., unpublished data).
Previously, we have noted the absence of correlation between Ave1‐induced HR and resistance through Ve1, as treatment with Ave1 leads to HR in tomato and tobacco plants that express Ve1, but not in N. benthamiana or Arabidopsis, whereas Ve1‐transgenic Arabidopsis shows Ave1‐triggered resistance (de Jonge et al., 2012; Zhang et al., 2013a, 2013b). Similarly, in the present study, we observed robust resistance mediated by the S. torvum and hop Ve1 homologues StoVe1 and HLVe1‐2A, whereas only a weak HR was observed on co‐expression of Ave1. These findings suggest that the HR may occur as a consequence of Ve1/Ave1‐induced immune signalling under particular conditions, but is not required for V. dahliae resistance (Zhang et al., 2013b). This finding may also explain why we were unable to compromise NgVe1‐mediated V. dahliae resistance on VIGS in N. glutinosa, but were able to compromise Ave1‐mediated HR.
Phylogenetic analysis revealed that Ve1 homologues are widely distributed in phylogenetically distant plant species, implying an ancient origin of the Ve1 immune receptor. Nevertheless, this origin does not imply functionality in V. dahliae resistance. The most obvious example is the close tomato homologue Ve2 which, despite its homology, does not act as a functional V. dahliae resistance gene. Similarly, in this study, we identified the non‐functional Ve1 homologues StuVe2, StoVe2 and HLVe1‐2B. The Ve locus as it occurs in tomato, with two homologues Ve1 and Ve2, appears to originate before speciation, as clustered Ve1 family members also appear in potato and wild eggplant. Furthermore, a functional study addressing Ave1 recognition in the genus Nicotiana only identified Ave1 recognition, and thus the presence of a potentially functional Ve1 homologue, in the species N. glutinosa (Zhang et al., 2013a). Considering the extremely wide host range of V. dahliae, and the general occurrence of strains that carry Ave1, the question arises as to whether the ancient progenitor of the currently functional Ve1 orthologues already functioned as a V. dahliae resistance gene, and thus several species/homologues lost the capacity to recognize Ave1 as a result of adverse effects associated with Ve1 functionality, or whether several species/homologues evolved the capacity to recognize Ave1 after the occurrence of speciation events. The latter hypothesis would imply that Ave1 recognition evolved multiple times in the plant kingdom through parallel evolution. Our present data do not allow the verification or disqualification of either hypothesis.
Plants and animals employ germline‐encoded pattern recognition receptors (PRRs) to detect broadly distributed microbe‐associated molecular patterns (MAMPs) and to activate antimicrobial defence (Macho and Zipfel, 2014). We have noted previously that tomato Ve1 is an ancient pathogen receptor with traits of typical PRRs. This finding was based on the transferability of Ve1 across plant species and the observation that Ve1 resistance affected three fungal species: V. dahliae, V. albo‐atrum and F. oxysporum (Fradin et al., 2011; de Jonge et al., 2012; Thomma et al., 2011). We have now demonstrated that members of the Ve1 gene family in N. glutinosa, S. tuberosum, S. torvum and H. lupulus encode receptors that recognize Ave1 and are able to mediate V. dahliae resistance in Arabidopsis. As our findings are based on stable expression in a heterologous host, we realize that definitive evidence for the role of Ve1 homologues in disease resistance in the endogenous hosts from which the Ve1 homologues are derived needs to be provided through targeted gene deletion in these hosts, or stable expression in susceptible genotypes of these species. We also discovered that the functional Ve1 homologues have divergent recognition specificities, suggesting that some recognize an even wider spectrum of plant pathogens than tomato Ve1 (Fig. 7). Collectively, these findings mean that Ve1 has traits of a typical race‐specific resistance protein as well as of a typical PRR. Similarly, Arabidopsis RLP23 recognizes an epitope of Nep‐like effector proteins (NLPs) that are widely distributed among bacteria, fungi and oomycetes (Gijzen and Nürnberger, 2006) to induce immune responses (Albert et al., 2015). Thus, it is becoming apparent that MAMP receptor systems are more dynamic than generally appreciated and are conditioned in a similar manner to prototypical resistance genes (Albert et al., 2015; Cook et al., 2015; Shibuya and Desaki, 2015). Findings like these have inspired the proposal of the ‘Invasion Model’, which describes plant immunity as a surveillance system to detect invasion, in which host receptors, termed invasion pattern receptors (IPRs), detect either externally encoded or modified self‐ligands that indicate invasion, termed invasion patterns (IPs) (Cook et al., 2015).
Experimental Procedures
Plant growth conditions and manipulations
Plants were grown at 21°C/19°C during 16 h/8 h light/dark photoperiods in a climate chamber or glasshouse with a relative humidity of ∼75% and 100 W/m2 supplemental light when the light intensity dropped below 150 W/m2. Arabidopsis transformations were performed as described previously (Clough and Bent, 1998).
Isolation of Ve1 homologues
The isolation of Ve1 homologues from N. glutinosa, S. tuberosum, S. torvum and H. lupulus is described in Methods S1 and Tables S2–S4.
Binary over‐expression constructs and transient expression in planta
For NgVe1, StuVe1, StuVe2, StoVe1 and StoVe2 constructs, CDS regions were amplified from N. glutinosa, S. tuberosum and S. torvum cDNA, respectively, whereas the CDS regions of HLVe1‐2A and HLVe1‐2B were amplified from the corresponding plasmids (Table S1). The CDS fragments were cloned into pDONR207 using Gateway® BP Clonase® II Enzyme Mix (Invitrogen, Carlsbad, CA, USA). All pDONR207 clones were sequenced, and fragments were subsequently transferred to pEarleyGate 100, pSol2095 (C‐terminal GFP tag) (Zhang et al., 2013a) or pFAST_R02 as described previously (Shimada et al., 2010) using Gateway® LR Clonase® II Enzyme Mix (Invitrogen). Constructs for the constitutive expression of Ave1, FoAve1, CbAve1 and ChAve1 have been described previously (de Jonge et al., 2012). CoAve1 (Gan et al., 2013) and XacPNP (Gottig et al., 2008) were obtained by gene synthesis (Eurofins MWG Operon, Ebersberg, Germany), and subsequently recombined into the destination vector pSol2092 (Zhang et al., 2013a) to generate expression constructs pSol2092::CoAve1 and pSol2092::XacPNP. All constructs were transformed to A. tumefaciens strain GV3101 (pMP90) by electroporation.
Agrobacterium tumefaciens carrying expression constructs was infiltrated into tobacco plants as described previously (Zhang et al., 2013a). Agrobacterium tumefaciens cultures containing constructs to express Ave1 and Ve1 homologues were mixed in a 1 : 1 ratio and infiltrated into the leaves of 5–6‐week‐old tobacco plants. At 5 days post‐infiltration, photographs were taken, and necrosis was quantified using ImageJ to measure the area of necrosis as the percentage of the total infiltrated leaf area.
Protein extracts and immunoblotting
For the detection of GFP‐tagged Ve1 homologues, A. tumefaciens carrying the corresponding expression constructs was infiltrated into mature tobacco leaves as described previously (Zhang et al., 2013a). The co‐immunopurifications and immunoblotting were performed as described previously (Zhang et al., 2014).
Quantitative real‐time PCR and reverse transcription‐PCR (RT‐PCR)
The target specificity of the constructs TRV::GUS and TRV::NgVe1 was determined in TRV‐infected N. glutinosa plants. Four weeks after TRV inoculation, whole tobacco plants were collected, frozen in liquid nitrogen and stored at −80°C for total RNA isolation.
For the expression of Ve homologues in the corresponding transgenic plants, 2‐week‐old Arabidopsis seedlings were harvested and ground into a powder in liquid nitrogen. Total RNA extraction, cDNA synthesis and RT‐PCR were performed as described previously (Zhang et al., 2013b; Table S1). To analyse the expression of NgVe1 in TRV‐targeted N. glutinosa plants, quantitative real‐time PCR was conducted using the primers NgVe1‐F(qRT) and NgVe1‐R(qRT) with tobacco Actin as an endogenous gene (Table S1), employing the qPCR Core Kit for SYBR Green I (Eurogentec Nederland BV, Maastricht, the Netherlands) as described previously (Fradin et al., 2009).
Vigs
Constructs pTRV2::PDS and pTRV2::GUS were used as controls. To silence NgVe1 in N. glutinosa, the construct pTRV2::NgVe1 was generated. TRV vectors were agroinfiltrated as described previously (Liu et al., 2002; Zhang et al., 2013a). Briefly, cotyledons of 10–15‐day‐old N. glutinosa seedlings were infiltrated with 1 : 1 mixtures of pTRV1 and pTRV2 constructs. Photobleaching was observed at 4 weeks after agroinfiltration of pTRV2::PDS. For HR assays, 4 weeks after virus inoculations, mature leaves were agroinfiltrated to individually express Ave1, VdNLP1 (Santhanam et al., 2013) and Avr9 (van Kan et al., 1991; Van der Hoorn et al., 2000). VdNLP1 and Avr9 were used as positive and negative controls, respectively. Five days after agroinfiltration, leaves were examined for the development of HR. For Verticillium disease assays, 3 weeks after TRV infection, the TRV‐infected plants were inoculated with race 1 V. dahliae strain JR2 (Faino et al., 2015), the corresponding Ave1 deletion mutant (V. dahliae JR2 ΔAve1; de Jonge et al., 2012) and tap water as control. The inoculated plants were evaluated by the observation of disease symptoms at 14 dpi.
Disease assays
Verticillium dahliae was grown on potato dextrose agar at 22°C, and conidia were collected from 7–10‐day‐old plates and washed with tap water. Verticillium disease assays on N. glutinosa plants were performed as described previously (Zhang et al., 2013a). Briefly, 5‐week‐old plants were uprooted, the roots were rinsed in water, dipped for 3 min in a suspension of 106 conidiospores/mL water and transferred to soil. Verticillium disease assays on Arabidopsis, as well as Verticillium biomass quantification in infected Arabidopsis plants, were performed as described previously (Ellendorff et al., 2009).
Phylogenetic identification
To obtain the phylogenetic relationship of tomato Ve1 and its homologues, we identified similar sequences in 41 plant species acquired from Phytozome (v9.1) (Goodstein et al., 2012) and manually added Ve1 homologues of tobacco, potato, wild eggplant and hop. Sequence similarity was established using blast (version 322.28+), applying a conservative e‐value cut‐off of 1e‐50. To prevent spurious hits, we removed sequences in which the matching area was less than 75% or the ‘actual matching’ was less than 50% of either Ve1 or the subject. The matching area is defined as the area from the start position of the first segment to the end position of the last segment, and the ‘actual matching’ area is defined as the sum of the covered area by each individual segment. Moreover, sequences that deviate in length (<80% or >120% of the length of Ve1) or contain protein domains other than leucine‐rich repeats, as predicted by HMMER3 (version 3.0) (Finn et al., 2011) on a local PFAM database (version 27), were discarded. Subsequently, all protein sequences were aligned using MAFFT (version 7.047b) (Katoh et al., 2002) and the most consistent alignment (LINSI) was chosen using trimAl (version 1.2) (Capella‐Gutiérrez et al., 2009), after which the heuristic method of trimAl was applied to trim the alignment. This cleaned alignment was used to construct an initial phylogenetic tree employing quick tree (version 1.1; 1000 bootstraps). The clade of interest (with tomato Ve1) and surrounding sequences were manually gathered and realigned. The final phylogenetic tree was inferred using RAxML (version 7.6.3) (Stamatakis, 2006), with the gamma model of rate heterogeneity and the Whelan and Goldman amino acid substitution matrix.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Percentage of amino acid identity shared between nine Ve1 homologues. The highest percentage of homology between two Ve1 homologues is indicated in red. Dashes (‐) represent identical sequences.
Fig. S2. Stability of green fluorescent protein (GFP)‐fused NgVe1 protein in planta. Total protein extracts of transiently transformed leaf tissue were subjected to immunopurification (IP) using GFP‐affinity beads. Immunopurified proteins were subjected to sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblotted (IB) using α‐GFP antibody. Coomassie blue staining (CBS) of the blot containing total protein extracts showed equal loading in each lane based on the 50‐kDa RuBisCo (ribulose‐1,5‐bisphosphate carboxylase/oxygenase) band. GFP‐tagged Ve1 protein was used as a control.
Fig. S3. Tobacco rattle virus (TRV)‐mediated gene silencing in Nicotiana glutinosa. (A) Virus‐induced gene silencing of the phytoene desaturase (PDS) gene results in patchy photobleaching in leaves of N. glutinosa. Photographs were taken at 4 weeks after TRV::PDS infiltration, and show representative infected plants of at least three independent assays. (B) TRV::NgVe1‐inoculated plants show resistance to Verticillium dahliae strain JR2, but not V. dahliae JR2 ΔAve1. Nicotiana glutinosa plants were inoculated with a recombinant TRV targeting the β‐glucuronidase (GUS) gene as a control (TRV::GUS) or recombinant TRV targeting the NgVe1 gene (TRV::NgVe1). At 3 weeks after TRV infiltration, TRV‐inoculated plants were inoculated with either V. dahliae strain JR2 or V. dahliae JR2 ΔAve1 mutant. Photographs were taken 2 weeks after V. dahliae inoculation, and show representative inoculated plants of at least three independent assays. (C) Silencing efficiency was determined using quantitative real‐time polymerase chain reaction at 28 days post‐infiltration in TRV::NgVe1‐ and TRV::GUS‐inoculated N. glutinosa plants. Bars represent levels of NgVe1 transcripts relative to the transcript levels of N. glutinosa actin (for normalization) with standard deviation of a sample of three pooled plants. NgVe1 expression in TRV::GUS‐infected plants is set to unity.
Fig. S4. Expression of NgVe1, HLVe1‐2A, HLVe1‐2B, StuVe1, StuVe2, StoVe1 and StoVe2 in transgenic plants was detected by reverse transcription‐polymerase chain reaction (RT‐PCR). As an endogenous control, a fragment of the AtRuBisCo gene (RuBisCo, ribulose‐1,5‐bisphosphate carboxylase/oxygenase) was amplified from cDNA. For each construct, three independent transgenic lines are shown (1, 2 and 3). Water was used as a control.
Fig. S5. Southern blotting of seven hop cultivars with the tomato Ve1 gene as a probe reveals the presence of Ve‐like sequences in the hop genome. Seven hop cultivars are indicated: 1, ‘Wye target’; 2, ‘Fuggle’; 3, ‘Wye Challenger’; 4, ‘Savinjski Golding’; 5, ‘Aurora’; 6, ‘Celeia’; 7, ‘Yeoman’. Hop genomic DNA was digested with the restriction enzymes EcoRI, EcoRV and HindIII, separated on 0.8% agarose gel and blotted on nylon membranes with the 32P‐labelled tomato Ve1 gene sequence as a probe.
Fig. S6. Primary structure and protein sequence alignment of functional Ve1 proteins from tomato, tobacco, potato, wild eggplant and hop. The N‐terminal amino acids in the dashed frame denote the predicted signal peptides (SPs) of the functional Ve1 homologues. eLRR, extracellular leucine‐rich repeat (C1 domain and C3 domain); IS, non‐LRR island domain (C2 domain); AC, acidic domain; TM, transmembrane domain; CT, cytoplasmic domain. The locations of the predicted solvent‐exposed β‐sheet (xxLxLxx) on the concave surface of the receptor are indicated above the eLRR domains. Identical amino acid residues are highlighted in red, whereas conserved amino acid residues are highlighted in blue. Three consecutive eLRR regions required for the functionality of Ve1 are indicated by bold colour, whereas other regions that could not be implicated in Ve1 functionality are indicated in light colour. Ve1 homologue sequences can be found in the GenBank database using the following GenBank accessions: ACR33105 (Ve1), ALK26499 (NgVe1), ALK26501 (StuVe1), ALK26500 (StoVe1), AIE39594 (HLVe1‐2A).
Fig. S7. Percentage of amino acid identity shared between the C1 domain, C2 domain, C3 domain and C‐terminal extracellular leucine‐rich repeat (eLRR)‐flanking domain of the functional and non‐functional Ve1 homologues from tomato, tobacco, potato, wild eggplant and hop.
Fig. S8. An unrooted phylogenetic tree based on protein sequences of Ve1 homologues from the tomato clade and the relevant sister clades. The tomato Ve1 clade is indicated by the highlighted background. Official gene identifiers and species names are indicated. Bootstrap values are shown in the tree. The scale represents branch lengths expressed as the relative number of amino acid substitutions.
Table S1. Primers used for amplification, sequencing and expression of Ve1 homologues.
Table S2. Degenerative primers used for thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR) amplification of the flanking regions of putative hop Ve1 homologues.
Table S3. Primers used for the thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR)‐based isolation of hop Ve1 sequences.
Table S4. Amplification conditions used for thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR).
Methods S1. Isolation of Ve1 homologues.
Acknowledgements
Y.S. acknowledges a fellowship from the China Scholarship Council. B.P.H.J.T. and M.F.S. were supported by a Vici and a Veni grant, respectively, from the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO). B.J. and J.J. acknowledge financial support from the Slovenian Research Agency, Grant Number P4‐0077, and the support of A.M. by a PhD grant (No. 1000‐09‐310205). Bert Essenstam is acknowledged for excellent plant care.
References
- Albert, I. , Böhm, H. , Albert, M. , Feiler, C.E. , Imkampe, J. , Wallmeroth, N. , Brancato, C. , Raaymakers, T.M. , Oome, S. , Zhang, H. , Krol, E. , Grefen, C. , Gust, A.A. , Chai, J. , Hedrich, R. , Van den Ackerveken, G. and Nürnberger, T. (2015) An RLP23–SOBIR1–BAK1 complex mediates NLP‐triggered immunity. Nat. Plants, 1, 15140. [DOI] [PubMed] [Google Scholar]
- Antanaviciute, L. , Šurbanovski, N. , Harrison, N. , McLeary, K. , Simpson, D. , Wilson, F. , Sargent, D. and Harrison, R. (2015) Mapping QTL associated with Verticillium dahliae resistance in the cultivated strawberry (Fragaria×ananassa). Hortic. Res. 2, 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolek, Y. , El‐Zik, K.M. , Pepper, A.E. , Bell, A.A. , Magill, C.W. , Thaxton, P.M. and Reddy, O.U.K. (2005) Mapping of verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Capella‐Gutiérrez, S. , Silla‐Martínez, J.M. and Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y. , Zhao, L. , Liao, Z. , Sun, X. , Zuo, K. , Zhang, L. and Tang, K. (2003) Molecular cloning of a potential Verticillium dahliae resistance gene SlVe1 with multi‐site polyadenylation from Solanum licopersicoides . DNA Seq. 14, 375–384. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Kan, J. , Yang, Y. , Ling, X. , Chang, Y. and Zhang, B. (2016) A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae . Plant Physiol. Biochem. 98, 101–111. [DOI] [PubMed] [Google Scholar]
- Christopoulou, M. , Wo, S.R.‐C. , Kozik, A. , McHale, L.K. , Truco, M.‐J. , Wroblewski, T. and Michelmore, R. (2015) Genome‐wide architecture of disease resistance genes in lettuce. G3, 5, 2655–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cook, D.E. , Mesarich, C.H. and Thomma, B.P.H.J. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Darby, P. (2001) ‘Single gene traits in hop breeding’. Proceedings of the Scientific Commission of the International Hop Growers Convention, Canterbury, UK., (Seigner, E., ed.), 76–80. [Google Scholar]
- Diwan, N. , Fluhr, R. , Eshed, Y. , Zamir, D. and Tanksley, S. (1999) Mapping of Ve in tomato: a gene conferring resistance to the broad‐spectrum pathogen, Verticillium dahliae race 1. Theor. Appl. Genet. 98, 315–319. [Google Scholar]
- Ellendorff, U. , Fradin, E.F. , de Jonge, R. and Thomma, B.P.H.J. (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Datema, E. , van den Berg, G.C. , Janssen, A. , Wittenberg, A.H. and Thomma, B.P.H.J. (2015) Single‐molecule real‐time sequencing combined with optical mapping yields completely finished fungal genome. MBio, 6, e00936–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, J. , Chai, Y. , Wang, J. , Lin, J. , Sun, X. , Sun, C. , Zuo, K. and Tang K. (2004) cDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Seq. 15, 88–95. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Clements, J . and Eddy, S.R . (2011) HMMER web server: interactive sequence similarity searching. Nuc. Acids Res. 39, W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. and Thomma, B.P.H.J. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Ayala, J.C.J. , Castroverde, C.D. , Nazar, R.N. , Robb, J. , Liu C.‐M. and Thomma B.P.H.J. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Abd‐El‐Haliem, A. , Masini, L. , van den Berg, G.C. , Joosten, M.H. and Thomma, B.P.H.J. (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Rovenich, H. , Song, Y. , Liebrand, T.W. , Masini, L. , van den Berg, G.C. , Joosten, M.H. and Thomma, B.P.H.J. (2014) Functional analysis of the tomato immune receptor Ve1 through domain swaps with its non‐functional homolog Ve2. PLoS One, 9, e88208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz‐Laylin, L.K. , Krishnamurthy, N. , Tör, M. , Sjölander, K.V. and Jones, J.D. (2005) Phylogenomic analysis of the receptor‐like proteins of rice and Arabidopsis. Plant Physiol. 138, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, P. , Ikeda, K. , Irieda, H. , Narusaka, M. , O'Connell, R.J. , Narusaka, Y. , Takano, Y. , Kubo, Y. and Shirasu, K. (2013) Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197, 1236–1249. [DOI] [PubMed] [Google Scholar]
- Gijzen, M. and Nürnberger, T. (2006) Nep1‐like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- Goodstein, D.M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R.D. , Fazo, J. , Mitros, T. , Dirks, W. , Hellsten, U. and Putnam, N . (2012) Phytozome: a comparative platform for green plant genomics. Nuc. Acids Res. 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Daurelio, L.D. , Valentine, A. , Gehring, C. , Orellano, E.G. and Ottado, J. (2008) Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide‐like protein to modify host homeostasis. Proc. Natl. Acad. Sci. USA, 105, 18 631–18 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, R.J. , McHale, L.K. , Vallad, G.E. , Truco, M.J. , Michelmore, R.W. , Klosterman, S.J. , Maruthachalam, K. and Subbarao, K.V. (2011) The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. Theor. Appl. Genet. 123, 509–517. [DOI] [PubMed] [Google Scholar]
- Inderbitzin, P. , Davis, R.M. , Bostock, R.M. and Subbarao, K.V. (2011) The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS One, 6, e18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakse, J. , Cerenak, A. , Radisek, S. , Satovic, Z. , Luthar, Z. and Javornik, B. (2013) Identification of quantitative trait loci for resistance to Verticillium wilt and yield parameters in hop (Humulus lupulus L.). Theor. Appl. Genet. 126, 1431–1443. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao K.V. and Thomma, B.P.H.J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc . Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan, J.A. , Van den Ackerveken, G. and De Wit, P. (1991) Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant–Microbe Interact. 4, 52–59. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K.i. , and Miyata, T . (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nuc. Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk, L.M. , Hachey, J. , Lynch, D.R. , Kulcsar, F. , van Rooijen, G. , Waterer, D.R. , Robertson, A. , Kokko, E. , Byers, R. , Howard, R.J. , Fischer, R. and Prufer, D. (2001) Tomato Ve disease resistance genes encode cell surface‐like receptors. Proc. Natl. Acad. Sci. USA, 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Zhu, Y. , Xie, C. , Jue, D. , Hong, Y. , Chen, M. , Hubdar, A.K. and Yang, Q. (2012) Transgenic potato plants expressing StoVe1 exhibit enhanced resistance to Verticillium dahliae. Plant Mol. Biol. Rep. 30, 1032–1039. [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S. (2002) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Majer, A. , Javornik, B. , Cerenak, A. and Jakse, J. (2014) Development of novel EST‐derived resistance gene markers in hop (Humulus lupulus L.). Mol. Breed. 33, 61–74. [Google Scholar]
- Santhanam, P. , van Esse, H.P. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B.P.H.J. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. , Hema, R. , Anand, A. , Kang, L. , Udayakumar, M. and Mysore, K.S. (2007) A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus‐induced gene silencing. New Phytol. 176, 782–791. [DOI] [PubMed] [Google Scholar]
- Shibuya, N. and Desaki, Y. (2015) Immunity: one receptor, many pathogens. Nat. Plants, 1, 15149. [Google Scholar]
- Shimada, T.L. , Shimada, T. and Hara‐Nishimura, I. (2010) A rapid and non‐destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana . Plant J. 61, 519–528. [DOI] [PubMed] [Google Scholar]
- Simko, I. , Costanzo, S. , Haynes, K. , Christ, B. and Jones, R. (2004a) Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. [DOI] [PubMed] [Google Scholar]
- Simko, I. , Haynes, K. , Ewing, E. , Costanzo, S. , Christ, B. and Jones, R. (2004b) Mapping genes for resistance to Verticillium albo‐atrum in tetraploid and diploid potato populations using haplotype association tests and genetic linkage analysis. Mol. Genet. Genomics, 271, 522–531. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A . (2006) RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Tang, J. , Lin, J. , Yang, Y. , Chen, T. , Ling, X. , Zhang, B. and Chang, Y. (2016) Ectopic expression of a Ve homolog VvVe gene from Vitis vinifera enhances defense response to Verticillium dahliae infection in tobacco. Gene, 576, 492–498. [DOI] [PubMed] [Google Scholar]
- Terauchi, R. and Kahl, G. (2000) Rapid isolation of promoter sequences by TAIL‐PCR: the 5′‐flanking regions of Pal and Pgi genes from yams (Dioscorea). Mol. Gen. Genet. 263, 554–560. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Nürnberger, T. and Joosten, M.H. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A. , Laurent, F. , Roth, R. and De Wit, P.J. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr 9/Cf‐9‐induced and Avr 4/Cf‐4‐induced necrosis. Mol. Plant–Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Vining, K. and Davis, T. (2009) Isolation of a Ve homolog, mVe1, and its relationship to verticillium wilt resistance in Mentha longifolia (L.) Huds. Mol. Genet. Genomics, 282, 173–184. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Ellendorff, U. , Kemp, B. , Mansfield, J.W. , Forsyth, A. , Mitchell, K. , Bastas, K. , Liu, C.‐M. , Woods‐Tör, A. , Zipfel, C. , De Wit, P.J. , Jones, J.D.G. , Tör, M. and Thomma, B.P.H.J. (2008) A genome‐wide functional investigation into the roles of receptor‐like proteins in Arabidopsis. Plant Physiol. 147, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Fiers, M. , Ellendorff, U. , Wang, Z. , de Wit, P.J. , Angenent, G.C. and Thomma, B.P.H.J. (2010) The diverse roles of extracellular leucine‐rich repeat‐containing receptor‐like proteins in plants. Crit. Rev. Plant Sci. 29, 285–299. [Google Scholar]
- Wang, H.M. , Lin, Z.X. , Zhang, X.L. , Chen, W. , Guo, X.P. , Nie, Y.C. and Li, Y.H. (2008) Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J. Integr. Plant Biol. 50, 174–182. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Pan, S. , Cheng, S. , Zhang, B. , Mu, D. , Ni, P. , Zhang, G. , Yang, S. , Li, R. , Wang, J. , Orjeda, G. , Guzman, F. , Torres, M. , Lozano, R. , Ponce, O. , Martinez, D. , De la Cruz, G. , Chakrabarti, S.K. , Patil, V.U. , Skryabin, K.G. , Kuznetsov, B.B. , Ravin, N.V. , Kolganova, T.V. , Beletsky, A.V. , Mardanov, A.V. , Di Genova, A. , Bolser, D.M. , Martin, D.M. , Li, G. , Yang, Y. , Kuang, H. , Hu, Q. , Xiong, X. , Bishop, G.J. , Sagredo, B. , Mejía, N. , Zagorski, W. , Gromadka, R. , Gawor, J. , Szczesny, P. , Huang, S. , Zhang, Z. , Liang, C. , He, J. , Li, Y. , He, Y. , Xu, J. , Zhang, Y. , Xie, B. , Du, Y. , Qu, D. , Bonierbale, M. , Ghislain, M. , Herrera Mdel, R. , Giuliano, G. , Pietrella, M. , Perrotta, G. , Facella, P. , O'Brien, K. , Feingold, S.E. , Barreiro, L.E. , Massa, G.A. , Diambra, L. , Whitty, B.R. , Vaillancourt, B. , Lin, H. , Massa, A.N. , Geoffroy, M. , Lundback, S. , DellaPenna, D. , Buell, C.R. , Sharma, S.K. , Marshall, D.F. , Waugh, R. , Bryan, G.J. , Destefanis, M. , Nagy, I. , Milbourne, D. , Thomson, S.J. , Fiers, M. , Jacobs, J.M. , Nielsen, K.L. , Sønderkær, M. , Iovene, M. , Torres, G.A. , Jiang, J. , Veilleux, R.E. , Bachem, C.W. , de Boer, J. , Borm, T. , Kloosterman, B. , van Eck, H. , Datema, E. , Hekkert, B.T. , Goverse, A. , van Ham, R.C. and Visser, R.G. (2011) Genome sequence and analysis of the tuber crop potato. Nature, 475, 189–197. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Guo, W. , Li, G. , Gao, F. , Lin, S. and Zhang, T. (2008) QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 174, 290–298. [Google Scholar]
- Yang, Y. , Ling, X. , Chen, T. , Cai, L. , Liu, T. , Wang, J. , Fan, X. , Ren, Y. , Yuan, H. , Zhu, W. , Zhang, B. and Ma, D.‐P. (2014) A cotton Gbvdr5 gene encoding a leucine‐rich‐repeat receptor‐like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Rep. 33, 1–15. [Google Scholar]
- Zhang, B. , Yang, Y. , Chen, T. , Yu, W. , Liu, T. , Li, H. , Fan, X. , Ren, Y. , Shen, D. , Liu, L. , Liu, L. , Dou, D. and Chang, Y. (2012) Island cotton Gbve1 gene encoding a receptor‐like protein confers resistance to both defoliating and non‐defoliating isolates of Verticillium dahliae . PLoS One, 7, e51091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X. , Yang, S. , Chi, J. , Zhang, G. and Ma, Z. (2011) Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana . Plant Cell Rep. 30, 2085–2096. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. and Thomma, B.P.H.J. (2013) Structure–function aspects of extracellular leucine‐rich repeat‐containing cell surface receptors in plants. J. Integr. Plant. Biol. 55, 1212–1223. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Fradin, E. , de Jonge, R. , van Esse, H.P. , Smit, P. , Liu, C.M. and Thomma, B.P.H.J. (2013a) Optimized agroinfiltration and virus‐induced gene silencing to study Ve1‐mediated Verticillium resistance in tobacco. Mol. Plant–Microbe Interact. 26, 182–190. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Esse, H.P. , Damme, M. , Fradin, E.F. , Liu, C.M. and Thomma, B.P.H.J. (2013b) Ve1‐mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 14, 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Song, Y. , Liu, C.M. and Thomma, B.P.H.J. (2014) Mutational analysis of the Ve1 immune receptor that mediates Verticillium resistance in tomato. PLoS One, 9, e99511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Percentage of amino acid identity shared between nine Ve1 homologues. The highest percentage of homology between two Ve1 homologues is indicated in red. Dashes (‐) represent identical sequences.
Fig. S2. Stability of green fluorescent protein (GFP)‐fused NgVe1 protein in planta. Total protein extracts of transiently transformed leaf tissue were subjected to immunopurification (IP) using GFP‐affinity beads. Immunopurified proteins were subjected to sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblotted (IB) using α‐GFP antibody. Coomassie blue staining (CBS) of the blot containing total protein extracts showed equal loading in each lane based on the 50‐kDa RuBisCo (ribulose‐1,5‐bisphosphate carboxylase/oxygenase) band. GFP‐tagged Ve1 protein was used as a control.
Fig. S3. Tobacco rattle virus (TRV)‐mediated gene silencing in Nicotiana glutinosa. (A) Virus‐induced gene silencing of the phytoene desaturase (PDS) gene results in patchy photobleaching in leaves of N. glutinosa. Photographs were taken at 4 weeks after TRV::PDS infiltration, and show representative infected plants of at least three independent assays. (B) TRV::NgVe1‐inoculated plants show resistance to Verticillium dahliae strain JR2, but not V. dahliae JR2 ΔAve1. Nicotiana glutinosa plants were inoculated with a recombinant TRV targeting the β‐glucuronidase (GUS) gene as a control (TRV::GUS) or recombinant TRV targeting the NgVe1 gene (TRV::NgVe1). At 3 weeks after TRV infiltration, TRV‐inoculated plants were inoculated with either V. dahliae strain JR2 or V. dahliae JR2 ΔAve1 mutant. Photographs were taken 2 weeks after V. dahliae inoculation, and show representative inoculated plants of at least three independent assays. (C) Silencing efficiency was determined using quantitative real‐time polymerase chain reaction at 28 days post‐infiltration in TRV::NgVe1‐ and TRV::GUS‐inoculated N. glutinosa plants. Bars represent levels of NgVe1 transcripts relative to the transcript levels of N. glutinosa actin (for normalization) with standard deviation of a sample of three pooled plants. NgVe1 expression in TRV::GUS‐infected plants is set to unity.
Fig. S4. Expression of NgVe1, HLVe1‐2A, HLVe1‐2B, StuVe1, StuVe2, StoVe1 and StoVe2 in transgenic plants was detected by reverse transcription‐polymerase chain reaction (RT‐PCR). As an endogenous control, a fragment of the AtRuBisCo gene (RuBisCo, ribulose‐1,5‐bisphosphate carboxylase/oxygenase) was amplified from cDNA. For each construct, three independent transgenic lines are shown (1, 2 and 3). Water was used as a control.
Fig. S5. Southern blotting of seven hop cultivars with the tomato Ve1 gene as a probe reveals the presence of Ve‐like sequences in the hop genome. Seven hop cultivars are indicated: 1, ‘Wye target’; 2, ‘Fuggle’; 3, ‘Wye Challenger’; 4, ‘Savinjski Golding’; 5, ‘Aurora’; 6, ‘Celeia’; 7, ‘Yeoman’. Hop genomic DNA was digested with the restriction enzymes EcoRI, EcoRV and HindIII, separated on 0.8% agarose gel and blotted on nylon membranes with the 32P‐labelled tomato Ve1 gene sequence as a probe.
Fig. S6. Primary structure and protein sequence alignment of functional Ve1 proteins from tomato, tobacco, potato, wild eggplant and hop. The N‐terminal amino acids in the dashed frame denote the predicted signal peptides (SPs) of the functional Ve1 homologues. eLRR, extracellular leucine‐rich repeat (C1 domain and C3 domain); IS, non‐LRR island domain (C2 domain); AC, acidic domain; TM, transmembrane domain; CT, cytoplasmic domain. The locations of the predicted solvent‐exposed β‐sheet (xxLxLxx) on the concave surface of the receptor are indicated above the eLRR domains. Identical amino acid residues are highlighted in red, whereas conserved amino acid residues are highlighted in blue. Three consecutive eLRR regions required for the functionality of Ve1 are indicated by bold colour, whereas other regions that could not be implicated in Ve1 functionality are indicated in light colour. Ve1 homologue sequences can be found in the GenBank database using the following GenBank accessions: ACR33105 (Ve1), ALK26499 (NgVe1), ALK26501 (StuVe1), ALK26500 (StoVe1), AIE39594 (HLVe1‐2A).
Fig. S7. Percentage of amino acid identity shared between the C1 domain, C2 domain, C3 domain and C‐terminal extracellular leucine‐rich repeat (eLRR)‐flanking domain of the functional and non‐functional Ve1 homologues from tomato, tobacco, potato, wild eggplant and hop.
Fig. S8. An unrooted phylogenetic tree based on protein sequences of Ve1 homologues from the tomato clade and the relevant sister clades. The tomato Ve1 clade is indicated by the highlighted background. Official gene identifiers and species names are indicated. Bootstrap values are shown in the tree. The scale represents branch lengths expressed as the relative number of amino acid substitutions.
Table S1. Primers used for amplification, sequencing and expression of Ve1 homologues.
Table S2. Degenerative primers used for thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR) amplification of the flanking regions of putative hop Ve1 homologues.
Table S3. Primers used for the thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR)‐based isolation of hop Ve1 sequences.
Table S4. Amplification conditions used for thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR).
Methods S1. Isolation of Ve1 homologues.


