Summary
The phytohormone jasmonic acid (JA) and its derivatives, collectively referred to as jasmonates, regulate many developmental processes, but are also involved in the response to numerous abiotic/biotic stresses. Thus far, powerful reverse genetic strategies employing perception, signalling or biosynthesis mutants have broadly contributed to our understanding of the role of JA in the plant stress response and development, as has the chemical gain‐of‐function approach based on exogenous application of the hormone. However, there is currently no method that allows for tightly controlled JA production in planta. By investigating the control of the JA synthesis pathway in bacteria‐infected cotton (Gossypium hirsutum L.) plants, we identified a transcription factor (TF), named GhERF‐IIb3, which acts as a positive regulator of the JA pathway. Expression of this well‐conserved TF in cotton leaves was sufficient to produce in situ JA accumulation at physiological concentrations associated with an enhanced cotton defence response to bacterial infection.
Keywords: cotton; defence response; ERF transcription factor; jasmonate, Xanthomonas citri pv. malvacearum
Introduction
Phytopathogen attacks result in major agricultural problems. Diseases caused by pathogens in the field and at the post‐harvest stage are responsible for considerable losses, representing approximately one‐third of the world production of crop species. The annual cotton plant, providing fibres for numerous downstream industrial applications, is no exception. Bacterial blight triggered by Xanthomonas citri pv. malvacearum (Xcm) is the most damaging disease of cotton plants, affecting leaves and subsequently reducing cotton fibre yield (Delannoy et al., 2005). Although studies have been performed with the aim to genetically improve the resistance to Xcm (Essenberg et al., 2014), the molecular mechanisms underlying cotton resistance to Xcm remain poorly understood. In the cotton cultivar Réba B50, which contains the R genes B2B3, the avirulent Xcm strain (named race 18) induces effector‐triggered immunity (ETI; Cui et al., 2015) accompanied by a typical hypersensitive reaction (HR) phenotype. Several molecular traits associated with cotton HR have been characterized. These include the early production of reactive oxygen species (Martinez et al., 1998), enzymatic lipid peroxidation (Jalloul et al., 2002), the accumulation of salicylic acid (SA) correlated with the onset of systemic acquired resistance (Martinez et al., 2000), up‐regulation of peroxidase‐encoding genes (Delannoy et al., 2003), synthesis of antimicrobial compounds, such as flavonoids (Dai et al., 1996), as well as some phytoalexins belonging to the sesquiterpene family (Essenberg et al., 1990). More recently, jasmonic acid (JA) signalling has also been associated with cotton HR (Champion et al., 2009).
In addition to its functions in plant growth and development, JA is known to be involved in diverse physiological processes, such as programmed cell death (PCD), response to mechanical injury and plant–pathogen interactions (Wasternack and Hause, 2013). In response to stress, neo‐synthesis of JA is initiated in plastids by 13‐lipoxygenases (13‐LOX), which catalyse the oxidation of free and/or membrane‐esterified linolenic acid. The resulting fatty acid hydroperoxides are then sequentially processed by allene oxide synthase (AOS) and allene oxide cyclase (AOC) to produce oxo‐phytodienoic acid (OPDA). OPDA is, in turn, converted into JA by reduction and three rounds of β‐oxidation, the latter steps of which invoke acyl‐coenzyme A oxidases (ACX) probably localized to peroxisomes. This metabolic pathway, known as the octadecanoic pathway, can be completed in the cytoplasm, where JA is conjugated to the amino acid isoleucine (Ile) to form JA‐Ile, which is bioactive (Fonseca et al., 2009; Lee et al., 2008). Of note, OPDA can also be esterified on structural glycerolipids (namely arabidopsides) and may be mobilized directly to produce JA under stressful conditions (Kourtchenko et al., 2007).
Transcriptional regulation of JA‐responsive genes relies on a core protein complex, the main components of which are the F‐box protein CORONATINE INSENSITIVE1 (COI1), the JASMONATE ZIM DOMAIN (JAZ) transcriptional repressors and transcription factors (TFs) (Pauwels and Goossens, 2011). Basically, in the absence of JA‐Ile, target gene expression is constitutively repressed as a result of TF and JAZ protein interaction within the TOPLESS (TPL)‐NOVEL INTERACTORS OF JAZ (NINJA) repressor complex, which prevents TFs from binding to promoters (Pauwels et al., 2010). When JA‐Ile is produced, JAZ proteins are recruited by COI1 through indirect interaction with the hormonal conjugate, ubiquitinated and rapidly degraded in a proteasome‐dependent manner (Larrieu et al., 2015). Freeing TFs from their constraint thus results in the transcriptional activation of JA‐responsive genes, thereby launching the JA signalling cascade and downstream physiological responses.
JA perception and signalling have been well documented. However, it is still unclear how JA synthesis is activated in plants. Only a few regulatory components have been identified thus far (Ellis and Turner, 2001; Ellis et al., 2002; Ito et al., 2007; Schommer et al., 2008; Seo et al., 1995, 1999). Here, we report that a novel TF, namely GhERF‐IIb3, which belongs to the ethylene‐response factor (ERF) family, is rapidly and transiently up‐regulated in response to the avirulent Xcm strain. Using protoplast assays, we have shown that this TF is localized to the nucleus and is able to transactivate the Arabidopsis AOC2 promoter in a sequence‐specific manner. Furthermore, we also demonstrate that this TF is sufficient to bring about JA accumulation in planta to enhance the defence response to bacterial pathogens, such as Xcm.
Results
Identification of genes potentially involved in jasmonate biosynthesis in cotton plants
Before examining the expression of genes coding for octadecanoic enzymes in response to the bacterium Xcm, we first searched for the corresponding coding sequences in dedicated genomic databases (see Experimental procedures). Highly similar orthologues of AtAOS, AtAOC2 and AtACX1a were identified using Arabidopsis thaliana primary amino acid sequences as queries (Table S1, see Supporting Information). To validate our candidate genes, their expression was quantified in wounded cotton cotyledons using a haemostat to promote JA accumulation in planta. Wounded tissues were then harvested over a time period of 24 h and the expression of GhAOS‐like, GhAOC2‐like and GhACX1a‐like genes was quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). As expected, wounding triggered a marked increase in JA content preceded by a transient accumulation of its precursor OPDA, as evidenced by liquid chromatography‐mass spectrometry (LC‐MS) measurement (Fig. 1A). A close correlation between the kinetic induction of the three candidate genes and that of the octadecanoic metabolites was also obtained in wounded tissues (Fig. 1B).
Figure 1.
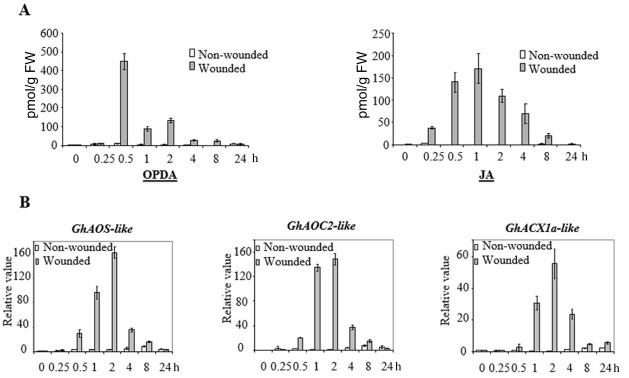
Wounding promotes both the up‐regulation of jasmonic acid (JA) biosynthesis‐related gene expression and oxo‐phytodienoic acid (OPDA)/JA accumulation in cotton cotyledons. (A) Twelve‐day‐old cotton plants were wounded four times with a haemostat. Damaged cotyledons were harvested at the indicated time points after wounding. OPDA/JA levels were determined by liquid chromatography‐mass spectrometry (LC‐MS). (B) Expression of cotton ALLENE OXIDE SYNTHASE‐like (GhAOS‐like), ALLENE OXIDE CYCLASE 2‐like (GhAOC2‐like) and ACYL‐COENZYME A OXIDASE‐like 1a (GhACX1a‐like) genes in damaged cotyledons was quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Results in (A) and (B) are averages of three biological replicates; error bars indicate standard deviation (n > 12).
To further reinforce these findings, the expression of these genes was quantified in JA‐treated cotyledons, because genuine JA biosynthesis genes are known to undergo a positive transcriptional regulatory loop (Browse, 2009). In accordance with their postulated function, all three identified cotton genes exhibited a significant induction at 2 h following application of 50 µm JA compared with solvent‐treated controls. Moreover, these genes, like others that respond to JA (Van der Does et al., 2013), were not regulated by 50 μm SA treatment (Fig. S1, see Supporting Information).
On the basis of these results, GhAOS‐like, GhAOC2‐like and GhACX1a‐like gene expression, together with metabolite levels, were followed as hallmarks for the potential activation of the octadecanoic pathway in further analysis in cotton plants.
Jasmonate and its precursor OPDA specifically accumulate during cotton HR
GhAOS‐like, GhAOC2‐like and GhACX1a‐like expression profiles were determined by qRT‐PCR. The cotton cultivar Reba B50, carrying the B2B3 blight resistance genes, was used for this purpose. Although this cultivar shows a resistance phenotype to Xcm race 18, characterized by a rapid and localized HR, it develops disease symptoms following inoculation with Xcm race 20 (Innes, 1983). During the incompatible interaction, steady‐state levels of GhAOS‐like, GhAOC2‐like and GhACX1a‐like transcripts increased significantly at 8 and 16 h post‐inoculation (hpi) when compared with those of untreated control and compatible tissues (Fig. 2A). Moreover, this stimulation of gene expression appeared to be specific to HR‐developing cotyledons.
Figure 2.
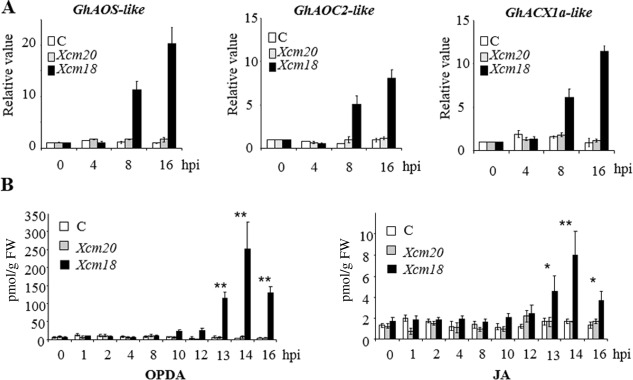
Coordinated induction of jasmonic acid (JA) biosynthesis‐related gene expression and metabolite accumulation on challenge with the avirulent strain of Xanthomonas citri pv. malvacearum (Xcm). (A) Time course expression of JA biosynthesis‐related genes cotton ALLENE OXIDE SYNTHASE‐like (GhAOS‐like), ALLENE OXIDE CYCLASE 2‐like (GhAOC2‐like) and ACYL‐COENZYME A OXIDASE‐like 1a (GhACX1a‐like) in response to Xanthomonas virulent (Xcm20) and avirulent (Xcm18) strains. Gene expression was measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Results are averages; error bars indicate standard deviation (n > 12). (B) Time course of oxo‐phytodienoic acid (OPDA) and JA production in response to Xcm infection. Results are averages; error bars indicate standard deviation (n > 12). Asterisks denote values significantly different from the control (C) (Student's test: *P < 0.05; **P < 0.01). All experiments (A, B) were repeated at least three times with similar results. hpi, h post‐inoculation.
To corroborate the transcriptional analysis, foliar concentrations of OPDA and JA on Xcm challenge were then measured. Consistent with the expression kinetics of the three putative octadecanoic genes, both OPDA and JA specifically accumulated in response to Xcm race 18, reaching an optimum at 14 hpi (Fig. 2B). Altogether, our data suggest a role for OPDA and JA in signalling events associated with resistance to Xcm.
The AtORA47 orthologous gene, GhERF‐IIb3, is specifically up‐regulated in HR‐developing cotton cotyledons
The data described above prompted us to investigate the molecular mechanisms responsible for the regulation of JA synthesis in pathogen‐challenged cotton plants. Recent work has pointed out that the ERF TF OCTADECANOIC‐RESPONSIVE ARABIDOPSIS APETALA2 (AP2)/ERF DOMAIN 47 (AtORA47) is a positive regulator of AtLOX3 and AtAOC2 expression on wounding and biotic stress in A. thaliana leaves (Walley et al., 2007). ERF proteins form a TF family mainly involved in stress responses and secondary metabolite production. Interestingly, it was shown that AtORA47 is able to interact directly with the two gene promoters, mediating their transcriptional transactivation (Pauwels et al., 2008; Zarei et al., 2011). Although it was not experimentally established that AtORA47 promotes JA production in the latter studies, we assumed that this TF and its relatives could do so and literally ‘switch on’ the octadecanoic pathway in plants. We thus carried out a phylogenetic analysis of AtORA47 relative proteins and found that they formed a distinct group within the ERF family (Fig. 3A), previously reported as group IIb (Champion et al., 2009). The clustering strength of group IIb was also supported by the presence of the conserved specific motifs of group II, dubbed CMII‐1 and CMII‐3 (Fig. 3B; Nakano et al., 2006). Among the identified putative TFs, GhERF‐IIb3 originating from the cotton plant Gossypium hirsutum exhibited an enhanced expression specific to Xcm race 18 infection characterized by a biphasic induction pattern (Fig. 3C). In addition, the GhERF‐IIb3 gene, like its A. thaliana counterpart AtORA47, was responsive to JA, whereas it did not respond to SA (Fig. S2, see Supporting Information; Pauwels et al., 2008). Combined with the data from Fig. 2, this last set of results indicates that GhERF‐IIb3 may be responsible for JA production during Xcm‐induced HR by positively regulating octadecanoic pathway genes.
Figure 3.
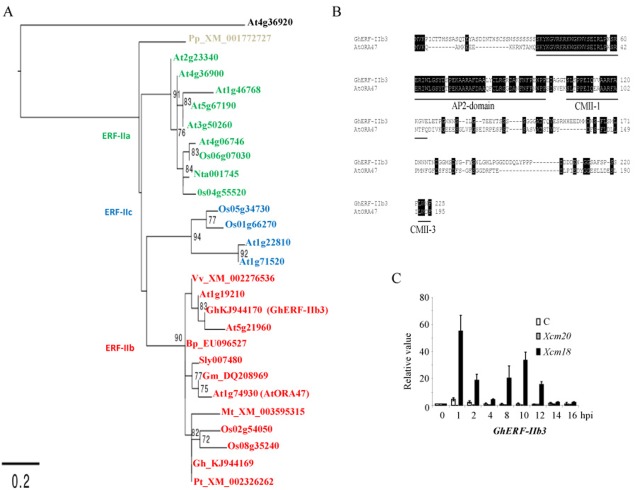
Identification of an ethylene‐response factor (ERF)‐related transcription factor induced during Xanthomonas citri pv. malvacearum (Xcm)‐induced hypersensitive reaction (HR) in cotton plants. (A) A maximum likelihood tree representing relationships among ERF proteins from Arabidopsis thaliana (At), Oryza sativa (Os), Medicago truncatula (Mt), Solanum lycopersicum (Sly), Nicotiana tabacum (Nt), Glycine max (Gm), Vitis vinifera (Vv), Populus trichocarpa (Pt), Broussonetia papyrifera (Bp), Physcomitrella patens (Pp) and Gossypium hirsutum (Gh) plants is rooted to the APETALA2 (AP2) domain R1 (At4g36920). Bootstrap values from 100 replicates were used to assess branch support. Bootstrap values over 70 are shown. The classification by Nakano et al. (2006) is indicated by coloured accession numbers. (B) Alignment of the Gossypium hirsutum GhERF‐IIb3 with the Arabidopsis ERF transcription factor AtORA47. Underlined sequences represent the AP2 domain and the two ERF group IIb‐specific conserved motifs CMII‐1 and CMII‐3. Strictly conserved residues are highlighted in black. Numbers on the right of the alignment indicate the amino acid position for ERF transcription factors. (C) Expression levels of GhERF‐IIb3 in response to Xanthomonas virulent (Xcm20) and avirulent (Xcm18) strains, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The results are averages; error bars indicate standard deviation (n > 12). These experiments were repeated at least three times with similar results.
Nuclear GhERF‐IIb3 proteins activate AtAOC2 promoter in plant cells
Given that GhERF‐IIb3 is a putative TF, it should present a typical nuclear localization. This hypothesis was challenged using a co‐localization approach. The GhERF‐IIb3 protein fused to green fluorescent protein (GFP) at its C‐terminal end was co‐expressed with the nuclear DsRed‐tagged NtKIS1a protein in A. thaliana leaf protoplasts and imaged by fluorescence microscopy. As shown in Fig. 4A, both protein combinations exhibit a round‐shaped fluorescence pattern consistent with a nuclear localization. In addition, GhERF‐IIb3‐associated fluorescence shows exact overlap with that of the nuclear marker NtKIS1 in good agreement with our working hypothesis.
Figure 4.
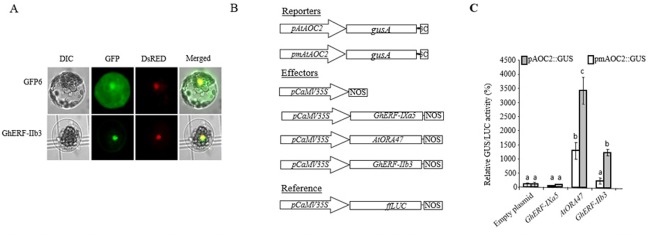
Transient overexpression of Xanthomonas citri pv. malvacearum (Xcm)‐inducible nuclear transcription factor (TF) GhERF‐IIb3 transactivates the AtAOC2 promoter in Arabidopsis thaliana cells. (A) Subcellular localization of the TF green fluorescent protein (GFP) fusions GhERF‐IIb3‐GFP in A. thaliana leaf protoplasts. GhERF‐IIb3‐GFP and NtKISa‐DsRED constructs were transformed simultaneously in protoplasts. The differential interference contrast (DIC) images are shown on the left, epifluorescence microscopic images are shown in the middle and merged images are shown on the right. (B) Schematic representation of the constructs used for promoter transactivation assays in A. thaliana protoplasts. The reporter construct consisted of the uidA gene driven by the AtAOC2 promoter [or AtAOC2 mutated promoter (pmAtAOC2) as described by Zarei et al. (2011)]. The effector constructs consisted of an expression vector carrying the CaMV35S promoter upstream of the GhERF‐IIb3, AtORA47 or GhERF‐IXa5 cDNAs. Empty effector vector was used as a negative control. The firefly luciferase (LUC) gene fused to the CaMV35S promoter served as a reference gene to correct for differences in transformation and protein extraction efficiencies. The firefly luciferase coding sequence (ffLUC) driven by the promoter CaMV35S was used as an internal control. (C) AtAOC2 promoter transactivation by the transcription factors GhERF‐IIb3 and AtORA47 in plant protoplasts. Bars represent average GUS/LUC ratios from triplicate experiments (±standard deviation) expressed relative to the vector control set at 100% (n = 6). Letters above the bars indicate distinct statistical groups as calculated by one‐way analysis of variance (ANOVA) (P < 0.01).
The TF activity of GhERF‐IIb3 was next tested using a promoter transactivation assay in A. thaliana leaf protoplasts. Protoplasts were co‐transformed with a tripartite vector system comprising: (i) a reporter plasmid carrying the AtAOC2 promoter‐driven β‐d‐glucuronidase (GUS)‐encoding uidA gene; (ii) an effector plasmid carrying AtORA47, GhERF‐IXa5 or GhERF‐IIb3 genes under the control of the CaMV35S promoter; and (iii) a reference plasmid carrying the firefly luciferase gene. The relative enzymatic activity of the reporter protein was monitored in vitro, and normalized to constitutively expressed luciferase activity (Fig. 4B). Although empty vector and the expression of the unrelated GhERF TF, which belongs to group IX (e.g. GhERF‐IXa5), had minimal or no effect on GUS activity, AtORA47 and, to a lesser extent, GhERF‐IIb3 were clearly able to transactivate the AtAOC2 promoter in A. thaliana cells (Fig. 4C). Similar results were also obtained when these two TFs were expressed in cotton protoplasts (Fig. S3, see Supporting Information), indicating that GhERF‐IIb3 may share with AtORA47 a regulatory binding site(s) to transactivate the AtAOC2 promoter. Indeed, in the A. thaliana protoplast assay, the use of an AtAOC2 promoter mutated in the GCC‐like box responsible for AtORA47 binding drastically diminished the GUS activity induced by both the A. thaliana and cotton proteins (Fig. 4C).
Altogether, these data point to GhERF‐IIb3 and AtORA47 as potent nuclear transcriptional activators of genuine JA biosynthesis genes in plant cells, as well as a common mode of action for both TFs.
Overexpression of GhERF‐IIb3 TFs induces OPDA and JA accumulation in cotton cotyledons
Next, we examined whether transient expression of GhERF‐IIb3 in cotton cotyledons allowed for the accumulation of OPDA/JA. An efficient Agrobacterium‐mediated expression system was successfully developed for this gain‐of‐function approach. Cotyledons of 10‐day‐old cotton plants were transformed with GhERF‐overexpressing constructs, using as vector the virG‐modified LBA1119 Agrobacterium strain (virGN54D), which improves T‐DNA transfer frequency (Fig. S4, see Supporting Information; Fits et al., 2000). Overexpression of the GhERF‐IIb3 gene resulted in substantial up‐regulation of GhAOS‐like, GhAOC2‐like and GhACX1a‐like expression (Fig. 5A). This increase in gene expression was also observed when the A. thaliana AtORA47 TF‐encoding gene was ectopically expressed in cotton cotyledons (Fig. S5A, see Supporting Information). Remarkably, the expression induction of the three putative JA biosynthesis genes previously identified by GhERF‐IIb3 and AtORA47 was accompanied by OPDA/JA accumulation in transformed tissues (Figs 5B and S5B), suggesting that the control of the octadecanoic pathway by AtORA47 and GhERF‐IIb3 TFs could be conserved in higher plants.
Figure 5.
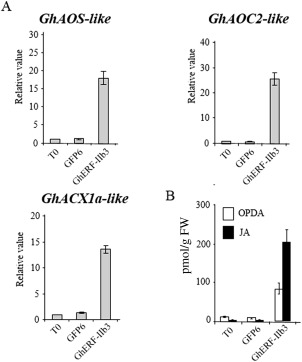
Overexpression of GhERF‐IIb3 promotes oxo‐phytodienoic acid (OPDA) and jasmonic acid (JA) accumulation in cotton plants. (A) Activation of JA‐responsive marker gene expression in cotton cotyledons overexpressing GhERF‐IIb3. (B) OPDA and JA contents in cotton cotyledons overexpressing GhERF‐IIb3. Ten‐day‐old Gossypium hirsutum plants were transformed with Agrobacterium tumefaciens carrying GhERF‐IIb3 or GFP6. At 2 days post‐transformation, gene expression and oxylipin content were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and liquid chromatography‐mass spectrometry (LC‐MS), respectively. Averages ± standard deviation (n = 24) were calculated from biological triplicates. GFP, green fluorescent protein.
Overexpression of GhERF‐IIb3 confers resistance to bacterial blight disease in cotton plants
Having established a system that permits the in planta accumulation of OPDA/JA at the flick of the GhERF‐IIb3 TF switch, we next investigated the role of the hormone and its precursor in Xcm–cotton plant interactions. Two days following agroinfiltration, cotton cotyledons overexpressing GhERF‐IIb3 were inoculated with the virulent (race 20) bacterial strain. Surprisingly, although control leaves expressing GFP6 showed typical disease symptoms characterized by spreading yellowing zones, GhERF‐IIb3‐transformed tissues presented HR‐like lesions reminiscent of the resistance phenotype (Fig. 6A). At 12 days post‐inoculation (dpi), GhERF‐IIb3‐induced hypersensitive‐like cell death resulted in 60% water loss (Fig. 6B), associated with a strong decrease in foliar bacterial concentration that was not observed in control tissues expressing GFP6 (Fig. 6C). Consistent with the increased OPDA/JA levels in cotyledons, this phenotype was accompanied by the up‐regulation of GhAOS‐like, GhAOC2‐like and GhACX1a‐like genes, as well as the up‐regulation of additional genes responsive to JA, including two TF genes GhERF‐IXa1 and GhERF‐IXa2 and the LOX gene GhLOX1, a hallmark of cotton hypersensitive cell death (Fig. S6, see Supporting Information). Likewise, ectopic expression of AtORA47 in cotton cotyledons also phenocopied the resistance response following virulent strain (race 20) inoculation that normally triggers disease (Fig. S7, see Supporting Information).
Figure 6.
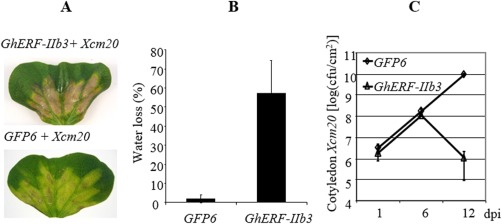
Overexpression of GhERGF‐IIb3 induces bacterial blight resistance in cotton plants. (A) Cotton cotyledons overexpressing GhERF‐IIb3 or GFP6 as a control were inoculated at 108 colony‐forming units (cfu)/mL with the virulent Xanthomonas citri pv. malvacearum (Xcm) strain (race 20). Symptom photographs at 12 days post‐inoculation (dpi). Note the hypersensitive reaction (HR)‐like symptoms in GhERFIIb3‐expressing cotyledons compared with typical yellow disease symptoms in GFP6 controls. (B) Cell death intensity at 12 dpi using water loss as proxy. (C) Kinetic evolution of cotyledon bacterial content. All experiments (B, C and D) were repeated at least three times with similar results. Averages ± standard deviation (n = 12) were calculated from biological triplicates. GFP, green fluorescent protein.
Altogether, these results point to a positive control of hypersensitive cell death by GhERF‐IIb3 and OPDA/JA in cotton plants challenged with Xcm.
Discussion
To date, the molecular mechanisms controlling JA production remain poorly understood. Here, we have identified the TF GhERF‐IIb3 as a novel positive regulator of JA synthesis in cotton. In this work, we have established that: (i) GhERF‐IIb3 is rapidly and transiently up‐regulated in response to the avirulent Xcm strain (Fig. 3C); (ii) this change in gene expression takes place before the activation of the three identified JA biosynthesis gene candidates and the increase in oxylipin metabolite concentrations (Fig. 2A,B); and (iii) overexpression of GhERF‐IIb3 leads to OPDA and JA overaccumulation in uninfected cotton cotyledons (Fig. 5B). In addition to a role for JA and its precursor in defence mechanisms associated with cotton HR, these data suggest that GhERF‐IIb3 proteins can activate the octadecanoic pathway in a linear model, where an as yet uncharacterized Xcm‐generated signal induces TF expression, causing OPDA and JA to accumulate. In line with this model, it is expected that genuine octadecanoic genes whose expression is driven by GCC‐like motif‐containing promoters will be directly regulated by the GhERF‐IIb3 TF, as indicated by the transactivation promoter assay (Fig. 4). Nonetheless, it cannot be ruled out that OPDA could be released from arabidopside‐like lipids, sustaining part of the Xcm‐dependent JA production. Moreover, given that GhAOS‐like, GhAOC2‐like, GhACX1a‐like and GhERF‐IIb3 genes are responsive to the exogenous application of JA, it is tempting to speculate that the ERF TF also contributes to the positive feedback amplification loop that operates in JA signalling, although this assumption requires further investigation.
Remarkably, we have also demonstrated that overexpression of GhERF‐IIb3 in cotton cotyledons converts a compatible into an incompatible interaction (Fig. 6). In other words, plants which are normally susceptible to the virulent race 20 of Xcm become resistant to the bacterium when they accumulate OPDA and JA. This resistant phenotype presents the two main characteristics of HR: cell death lesions localized to the infection site and low foliar bacterial growth a few days post‐inoculation. An executioner of cotton HR‐associated cell death, namely GhLOX1 (Jalloul et al., 2002; Marmey et al., 2007), is also transcriptionally up‐regulated in this context (Fig. S6). Altogether, these results strongly suggest that the increase in OPDA/JA levels recorded in response to the avirulent race 18 of Xcm could be responsible for hypersensitive cell death initiation within the infected zone by activating the 9‐LOX‐encoding gene GhLOX1. As proposed previously (Jalloul et al., 2002), the GhLOX1‐dependent lipid peroxidation could contribute to dismantling of the cellular structure during cotton HR. Our previous work lends support to this scenario. We have indeed shown that: (i) methyl jasmonate‐pretreated cotton cotyledons exhibit a strong accumulation of 9‐LOX activity accompanied by GhLOX1 gene expression; and (ii) infiltration of the pretreated cotyledons with LOX substrates, i.e. polyunsaturated fatty acids (PUFAs), triggers hypersensitive‐like symptoms in the absence of pathogens (Marmey et al., 2007). Hence, overexpression of the GhERF‐IIb3 TF could prime HR through the constitutive activation of the 9‐LOX gene, allowing hypersensitive cell death to be launched in the presence of the compatible race 20 of Xcm. If so, this situation implies the onset of lipid‐hydrolysing activities acting upstream of GhLOX1, as 9‐LOX preferentially oxidizes free PUFA. The patatin‐like protein GhPat1 has been suggested previously to form an enzymatic duo with GhLOX1 during cotton immunity, and therefore represents a good candidate for the release of the substrates of 9‐LOX (Cacas et al., 2009).
In the literature, contradictory results have been published with regard to the function of JA in the control of PCD. On the one hand, methyl jasmonate treatment of wild‐type A. thaliana plants has been reported to inhibit ozone‐induced cell death, a process which shares features with hypersensitive cell death (Tuominen et al., 2004). One such hormonal treatment also restricts lesion spread in the ozone‐sensitive mutant radical‐induced cell death1 (Overmyer et al., 2000). On the other hand, the development of HR‐like symptoms triggered by the mycotoxin fumonisin B1 requires JA to develop (Asai et al., 2000). Likewise, JA promotes singlet oxygen‐dependent cell death in flu mutants altered for chlorophyll anabolism (Danon et al., 2005). Furthermore, OPDA has been genetically proven to repress foliar necrotic lesions in flu mutants. In the cotton–Xcm pathosystem, it is difficult to draw conclusions on the respective roles of OPDA and JA in cell death propagation at the edge of infected areas and in naïve systemic tissues, as both molecules accumulate on GhERF‐Iib3 expression. Nonetheless, our results clearly establish that products of the octadecanoic pathway are invoked following biotrophic bacterial attack. This unexpectedly extends previous studies suggesting synergistic effects of ethylene and JA in the onset of defence against necrotrophic fungi, as well as antagonism between SA and JA in response to the biotrophic pathogen Pseudomonas syringae in A. thaliana (Pieterse et al., 2012).
Beyond academic research, the control of in situ JA production represents a major challenge not only to restrict pest infection in the field, but also for many additional horticultural and biotechnological applications (Wasternack, 2014). However, despite the fact that several JA regulatory components have been identified as aforementioned, there is currently no experimental system that allows us to tightly control JA production at the spatiotemporal level. In addition, manipulation of the rate‐limiting step of the octadecanoic pathway is not as straightforward as expected. The failure to significantly increase endogenous JA levels when AOS and AOC genes are overexpressed in distinct plant species illustrates the difficulty of this task (Laudert et al., 2000; Stenzel et al., 2003). Here, we report a novel alternative that potentially can be used to manipulate the octadecanoic pathway in situ. Based on sequence similarities and phylogenetic analysis (Fig. 3A,B and Table S3, see Supporting Information), many plant models, including monocotyledonous plants, such as rice (Oryza sativa), and dicotyledonous plants, such as tobacco (Nicotiana benthamiana), alfalfa (Medicago truncatula), tomato (Solanum lycopersicum) and grapevine (Vitis vinifera), contain AtORA47 sequence orthologues unrooted in group IIb, suggesting that the expression of GhERF‐IIb3, AtORA47 or endogenous AtORA47‐related TFs in all of these plant species could allow us to ‘switch on’ JA synthesis on demand.
Experimental Procedures
Biological materials, plant infection and treatments
Cotton cultivar G. hirsutum cv. Reba B50, which carries B2B3 blight resistance genes, was used in the present study. This cultivar is resistant to race 18 of the bacterium Xcm (Xcm18), but develops disease symptoms in response to race 20 (Xcm20) (Innes, 1983). Plants were grown and infected with Xcm strains as described previously (Jalloul et al., 2002). Xcm18‐triggered hypersensitive cell death symptoms were visually assessed and estimated by quantification of the water loss (Rustérucci et al., 1999). Bacterial counting assays were performed at 1, 6 and 12 dpi of the Xcm20 strain. Three distinct sets of serial dilutions were made for nine infected cotyledons. Twelve‐millimetre‐diameter discs were punched out from inoculated cotyledons, disinfected with 70% ethanol for 2 min, rinsed twice and ground in 5 mL of sterile water. The resulting suspension was serially diluted in water, and 100 μL of the dilutions were plated on Luria‐Bertani Glucose agar plates before being incubated at 28 ºC until colony counting. Cotyledons from 12‐day‐old cotton plants were wounded four times with a haemostat. Damaged cotyledons were harvested in liquid nitrogen after wounding for real‐time qRT‐PCR and high‐performance (HP)LC‐MS. Arabidopsis thaliana accession Columbia‐0 and G. hirsutum cv. Reba B50 were used to generate protoplasts from rosette leaves and cotyledons, respectively (Yoo et al., 2007).
Sequence analysis
GhERF and coding sequences of JA biosynthesis gene homologues were retrieved from the TIGR cotton database (http://plantta.tigr.org/search.shtml) using the TblastN tool, as described by Champion et al. (2009). Amino acid sequence alignments and phylogenetic analysis were performed using phylogeny.fr with default settings. The tree was edited with FigureTree (http://tree.bio.ed.ac.uk/software/Figuretree/).
Gene expression analysis
Transcript levels of cotton JA biosynthesis gene candidates, GhLOX1, GhERF‐IIb3 and GhERF‐IXa, were quantified by qRT‐PCR with MX3005P (Stratagene, LES ULIS Cedex, France), as described by Champion et al. (2009). Relative expression levels of reporter and target genes were determined based on the 2–ΔΔ C t method (Livak and Schmittgen, 2001) using cotton Actin2 (AY305724) as internal control (Champion et al., 2009; Li et al., 2005). Primers for qRT‐PCR analysis are listed in Table S2 (see Supporting Information).
Plasmid constructs
Cotton plants were transiently transformed with Gateway binary vector pMDC32 (for overexpression) (Curtis and Grossniklaus, 2003) by agroinfiltration. Reverse primers were designed to amplify cDNA with or without the stop codon for each coding sequence AtORA47 (At1g74930) and GhERF‐IIb3 (accession number KJ944170) from A. thaliana or cotton plants treated by JA for 1 h (see Table S2 for primer sequences). As control, GFP6 was amplified using pMDC84 as template in PCR. All PCR products were first cloned into pENTR‐D TOPO (Invitrogen, VILLEBON SUR YVETTE Cedex), and were then recombined into the binary plasmids pMDC32 or pMDC43 using LR Clonase (Invitrogen). Plasmid constructs were confirmed by sequencing. Plasmids were electroporated into Agrobacterium tumefaciens LBA1119m containing pBBRvirGN54D plasmid and used for cotton cotyledon transformation.
For the transactivation promoter assay, A. thaliana and cotton protoplasts were co‐transformed with plasmids carrying AtORA47, GhERF‐IXa5 (accession number KJ944171) or GhERF‐IIb3 and the reference plasmid p2rL7 carrying the Renilla reniformis luciferase gene and a version of the AtAOC2 promoter‐uidA fusion. Arabidopsis thaliana genomic DNA was used as template for the amplification of a 323‐bp‐long fragment of the AtAOC2 promoter using the specific primers listed in Table S2. The mutated promoter derivative pAOC2m was generated using a mutated version of the primer used to amplify the wild‐type sequence of pAOC2 (see Table S2 for the primer sequence). PCR products were cloned into pENTR‐D TOPO (Invitrogen), and were recombined into pMDC162 (Curtis and Grossniklaus, 2003).
Plant transformation
Agrobacterium‐mediated transient expression of cotton cotyledons was performed as described by Sparkes et al. (2006). Agrobacterium tumefaciens strains were grown for 18 h at 29 ºC with mild shaking. The agrobacterial density was estimated spectrophotometrically at 600 nm. When in the exponential phase [optical density (OD) between 0.6 and 0.8], bacteria were sedimented (3000 g for 20 min at 4 ºC) and the pellet was resuspended in infiltration medium (10 mm MgSO4, 200 μm acetosyringone and 20 mm MES, pH 5.5) to reach a final OD of 0.5. Agroinfiltration was carried out on 10‐old‐day cotton cotyledons. Each cotyledon was inoculated using a needle‐less syringe.
Quantification of OPDA and JA contents
Leaf samples harvested from cotton plants were ground with a mortar and pestle in liquid nitrogen. The resulting powder (250 mg/sample) was extracted with 1 mL of methanol containing 100 ng of deuterated JA (d6Ja) and 100 ng of deuterated OPDA (d5OPDA) as internal standards. Methanolic extracts were centrifuged at 5000 g for 10 min at 4 ºC and the supernatant was recovered in glass tubes. Pellets were re‐extracted three times, and methanolic extracts were pooled and dried at 40 ºC under a nitrogen flux. Dried residues were resuspended in 5 mL of 100 mm phosphate buffer, pH 7.8, containing 5% (w/v) NaCl. Suspensions were then washed three times with 2.5 mL of hexane and the organic phases were discarded. The pH of the clean pooled aqueous phases was adjusted to pH 1.4 using 5 M HCl. The aqueous phases were extracted three times using 2.5 mL of chloroform. Pooled chloroform phases were finally dried at 40 ºC under a nitrogen flux and dry residues were stored at −80°C until use. Samples were resuspended before LC‐MS analysis in 100 µL of methanol (injected volume, 20 µL).
The quantification of JA and OPDA was performed using a Waters 1515 liquid chromatograph coupled to a Waters ZQ mass spectrometer controlled by the software MassLynk V5.0 5 (MicroMass, Cary, NC, USA). The LC column used was an XTerra MS C18, 3.5 μm, 100 × 2.1 mm (Waters, Milford, MA, USA) and gradient elution was performed. A binary solvent was utilized, consisting of water with 15 mm formic acid (eluent A) and methanol (eluent B). The initial conditions were 40% A at a flow rate of 0.25 mL/min kept constant for 2 min. A gradient elution was introduced to change the chromatographic conditions to 60% A at 14 min to flush the analytical column, followed by restoration of the column to the initial conditions at 16 min, followed by equilibrium for 2 min. Under these conditions, the retention time of JA and its deuterated form was 9.3 min, and the retention time of OPDA and its deuterated form was 16.5 min. Both JA and OPDA were detected and quantified in negative electron spray ionization (ESI) mode. The analysis conditions for MS were as follows: vaporization temperature, 120 ºC; source temperature, 450 ºC; capillary voltage, 2.5 kV; cone voltage, 20 V. The quantification was carried out in the ‘selected ion monitoring’ mode using the following characteristic ions: 209 m/z ion for JA; 215 m/z ion for deuterated JA; 291 m/z ion for OPDA; and 295 m/z ion for deuterated OPDA. The amounts of endogenous compounds were calculated from the signal ratios of the unlabelled over the stable isotope‐containing mass fragments observed in both analysis channels.
Protoplast transfection, reporter enzyme assays and fluorescence microscopy
The protoplasts of cotton cotyledons and A. thaliana leaves were produced and transformed using the protocol of Yoo et al. (2007). Co‐transformations of pAOC2‐uidA or pAOC2m‐uidA with both the p2rL7 plasmid and one of the effector plasmids carrying AtORA47, GhERF‐IXa5 or GhERF‐IIb3 were carried out. An empty pMDC32 expression vector was used as an effector control. Protoplasts were transformed using polyethylene glycol as described previously (Yoo et al., 2007) with the three constructs in a ratio of 2 : 2 : 6 (GUS : LUC : effector plasmid). The protoplasts were collected 18 h after transformation and frozen in liquid nitrogen. GUS and luciferase (LUC) activities were measured as described previously (Zarei et al., 2011). In each sample, GUS and LUC activities were taken into account to correct for the differences in efficacy of transformation and protein extraction. The mean GUS/LUC ratio was expressed relative to that of the control empty vector.
In order to determine the subcellular localization of the GhERF‐IIb3 TF, protoplasts were co‐transformed with the plasmids, allowing the expression of GFP6‐tagged candidate proteins and NtKIS1a‐DsRFP (Jasinski, 2002), and then incubated in the dark for at least 16 h at 25°C. Fluorescence patterns were examined by fluorescence microscopy (Zeiss (Marly‐le‐Roi, France) Axio Imager A1 epifluorescence microscope coupled to a Zeiss Colibri LED illumination system and a QImaging Retiga‐SRV CCD camera).
Statistics
Data collected from protoplast transactivation experiments were subjected to a one‐way analysis of variance (ANOVA) in order to determine whether there were significant differences in AtAOC2 promoter activity between treatments. Means were compared using Tukey post‐hoc tests for pairwise comparisons (P < 0.01).
Conflict of Interest
The authors have no conflicts of interest to declare.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Genes encoding potential enzymes of jasmonate biosynthesis expressed in response to hormonal treatments in cotton. Expression of GhAOS‐like, GhAOC2‐like and GhACX1a‐like in response to JA and salicylic acid (SA) treatments, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Twelve‐day‐old cotton plants were treated for 2 h with 50 mm JA, 50 mm SA or a combination of the solvents dimethyl sulfoxide (DMSO; 0.1%) and sodium phosphate, pH 7 (0.5 mm) as control (C). Results are averages of three biological replicates; error bars indicate standard deviation (n > 12).
Fig. S2 Expression analysis of GhERF‐IIb3 gene in response to solvent (C, control), jasmonic acid (JA) and salicylic acid (SA) treatments. This experiment was carried out as described in Fig. S1.
Fig. S3 Both transcription factors AtORA47 and GhERF‐IIb3 transactivate the AtAOC2 promoter in cotton protoplasts. Protoplasts were produced and transformed using a protocol adapted from Yoo et al. (2007). β‐Glucuronidase (GUS) and luciferase (LUC) activity tests were performed as described previously (Zarei et al., 2011). Averaged GUS/LUC ratios were expressed relative to the control vector. Three independent experiments ± standard deviation (n = 6).
Fig. S4 Optimization of T‐DNA transfer to cotton cotyledon cells using Agrobacterium tumefaciens LBA1119 virGN54D bacterial vector. (A) Comparison of the transformation efficiency over time in agroinfiltrated cotyledons using two distinct bacterial strains. Agrobacterium tumefaciens strains LBA1119 and LBA1119 virGN54D were both transformed with the pCAMBIA1300::uidA‐intron and further used for the transformation of 10‐day‐old cotton cotyledons, as described in Experimental procedures. The virG‐modified LBA1119 was chosen because it is known to improve the T‐DNA transfer frequency (Fits et al., 2000). β‐Glucuronidase (GUS) activity was followed for 2 days for both strains. For each strain, 24 cotyledons were agroinfiltrated. GUS activities are reported as mean ± standard deviation (SD). Experiments were repeated at least three times with similar results. 4‐methylumbelliferone (MU) (B) Quantitative analysis of GUS activity until 6 days post‐inoculation (dpi) with LBA1119 virGN54D. Twenty‐four cotyledons were agroinfiltrated. GUS activity is reported as mean ± SD. The experiment was repeated twice with similar results. (C) Histochemical analysis of agroinfiltrated cotton cotyledons. The left half of the cotyledon was infiltrated with the empty bacterial strain as control, whereas the right half was infiltrated using the pCAMBIA1300‐uidA‐intron plasmid containing the virG‐modified LBA1119 strain. The photograph (2 dpi) shows the presence of indigo crystals over the whole region that had been transformed by pCAMBIA1300‐GUS‐intron.
Fig. S5 Overexpression of AtORA47 increases the gene expression of the jasmonic acid (JA)‐biosynthesis marker genes and endogenous oxo‐phytodienoic acid (OPDA)/JA levels in cotton. (A) Expression of GhAOS‐like, GhAOC2‐like and GhACX1a‐like genes in cotton cotyledons overexpressing AtORA47 or β‐glucuronidase‐encoding uidA genes. (B) OPDA and JA contents in cotton cotyledons overexpressing AtORA47 or GUS‐encoding uidA genes. Ten‐day‐old Gossypium hirsutum plants were transformed with Agrobacterium tumefaciens carrying either the AtORA47 or uidA construct. At 2 days post‐transformation, gene expression and oxylipin content were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and liquid chromatography‐mass spectrometry (LC‐MS), respectively. Averages ± standard deviation (n = 12) were calculated from biological triplicates.
Fig. S6 Gene expression of jasmonic acid (JA)‐responsive and defence marker genes in transgenic cotton overexpressing GhERF‐IIb3 and inoculated by the virulent Xcm20 strain. Relative transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) at 1 day post‐inoculation with Xcm20 in control tissues expressing GFP6‐ and in GhERF‐IIb3‐overexpressing tissues. Means ± standard deviation (n = 12). Experiments were repeated at least twice with similar results. GFP, green fluorescent protein; Xcm, Xanthomonas citri pv. malvacearum.
Fig. S7 Transient ectopic expression of AtORA47 in cotton cotyledons confers resistance to virulent Xanthomonas citri pv. malvacearum (Xcm) (race 20) bacteria. Cotton cotyledons overexpressing AtORA47 or GFP6 were inoculated at 108 colony‐forming units (cfu)/mL and the bacterial content was determined over time as described in Experimental procedures. Means ± standard deviation (n = 12). Experiments were repeated twice with similar results. GFP, green fluorescent protein.
Table S1 Jasmonic acid (JA) biosynthesis gene candidates ALLENE OXIDE SYNTHASE (AOS), ALLENE OXIDE CYCLASE (AOC) and ACYL‐COENZYME A OXIDASE (ACX) from Gossypium hirsutum share a significant degree of sequence conservation with their Arabidopsis thaliana counterparts.
Table S2 List of primers used in this study.
Table S3 List of APETALA2 (AP2) domains used in phylogenetic analysis.
Acknowledgements
This work was supported by the grant ‘Maturation de projet innovant’ from the Institut de Recherche pour le Développement (IRD) of France.
We thank J. Aribi for technical assistance with the IRD glasshouse work; C. Wasternack for providing the deuterated JA and OPDA forms; A. S. Petitot for providing the Agrobacterium tumefaciens strains and the plasmid pCAMBIA1300‐GUS‐intron and J. Memelink for providing the plasmid pBBRvirGN54D.
The institutes Institut National de la Recherche Agronomique (INRA) and IRD have filed a patent on behalf of the inventors P.D., M.N. and A.C. on the use of AtORA47 to switch on the overproduction of JA in transgenic plants and improved resistance to pathogenic agents.
References
- Asai, T. , Stone, J.M. , Heard, J.E. , Kovtun, Y. , Yorgey, P. , Sheen, J. and Ausubel, F.M. (2000) Fumonisin B1‐induced cell death in Arabidopsis protoplasts requires jasmonate‐, ethylene‐, and salicylate‐dependent signaling pathways. Plant Cell, 12, 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. [DOI] [PubMed] [Google Scholar]
- Cacas, J.L. , Marmey, P. , Montillet, J.L. , Sayegh‐Alhamdia, M. , Jalloul, A. , Rojas‐Mendoza, A. , Clérivet, A. and Nicole, M. (2009) A novel patatin‐like protein from cotton plant, GhPat1, is co‐expressed with GhLox1 during Xanthomonas campestris‐mediated hypersensitive cell death. Plant Cell Rep. 28, 155–164. [DOI] [PubMed] [Google Scholar]
- Champion, A. , Hebrard, E. , Parra, B. , Bournaud, C. , Marmey, P. , Tranchant, C. and Nicole, M. (2009) Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas . Mol. Plant Pathol. 10, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, G.H. , Nicole, M. , Andary, C. , Martinez, C. , Bresson, E. , Boher, B. , Danielson, J.F. and Geiger, J.P. (1996) Flavonoids accumulate in cell walls, middle lamellae and callose‐rich papillae during an incompatible interaction between Xanthomonas campestris pv. malvacearum and cotton. Physiol. Mol. Plant. Pathol. 49, 285–306. [Google Scholar]
- Danon, A. , Miersch, O. , Felix, G. , Camp, R.G.L. and Apel, K. (2005) Concurrent activation of cell death‐regulating signaling pathways by singlet oxygen in Arabidopsis thaliana . Plant J. Cell Mol. Biol. 41, 68–80. [DOI] [PubMed] [Google Scholar]
- Delannoy, E. , Jalloul, L.A. , Assigbetsé, K. , Marmey, P. , Geiger, J.P. , Lherminier, J. , Daniel, J.F. , Martinez, C. and Nicole, M. (2003) Activity of class III peroxidases in the defense of cotton to bacterial blight. Mol. Plant–Microbe Interact. 16, 1030–1038. [DOI] [PubMed] [Google Scholar]
- Delannoy, E. , Lyon, B.R. , Marmey, P. , Jalloul, A. , Daniel, J.F. , Montillet, J.L. , Essenberg, M. and Nicole, M. (2005) Resistance of cotton towards Xanthomonas campestris pv. malvacearum . Annu. Rev. Phytopathol. 43, 63–82. [DOI] [PubMed] [Google Scholar]
- Ellis, C. and Turner, J.G. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell, 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C. , Karafyllidis, I. , Wasternack, C. and Turner, J.G. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell, 14, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essenberg, M. , Grover, P.B., Jr. and Cover, E.C. (1990) Accumulation of antibacterial sesquiterpenoids in bacterially inoculated Gossypium leaves and cotyledons. Phytochemistry, 29, 3107–3113. [Google Scholar]
- Essenberg, M. , Bayles, M.B. , Pierce, M.L. and Verhalen, L.M. (2014) Pyramiding B genes in cotton achieves broader but not always higher resistance to bacterial blight. Phytopathology, 104, 1088–1097. [DOI] [PubMed] [Google Scholar]
- Fits, L. , van der Deakin, E.A. , Hoge, J.H.C. and Memelink, J. (2000) The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium‐mediated plant transformation. Plant Mol. Biol. 43, 495–502. [DOI] [PubMed] [Google Scholar]
- Fonseca, S. , Chini, A. , Hamberg, M. , Adie, B. , Porzel, A. , Kramell, R. , Miersch, O. , Wasternack, C. and Solano, R. (2009) (+)−7‐iso‐Jasmonoyl‐L‐isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Innes, N. (1983) Bacterial blight of cotton. Biol. Rev. 58, 158–176. [Google Scholar]
- Ito, T. , Ng, K.H. , Lim, T.S. , Yu, H. and Meyerowitz, E.M. (2007) The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis . Plant Cell, 19, 3516–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalloul, A. , Montillet, J.L. , Assigbetsé, K. , Agnel, J.P., Delannoy, E., Triantaphylidès, C., Daniel, J.F., Marmey, P., Geiger, J.P. and Nicole, M. (2002) Lipid peroxidation in cotton:Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J. Cell Mol. Biol. 32, 1–12. [DOI] [PubMed] [Google Scholar]
- Jasinski, S. (2002) Comparative molecular and functional analyses of the tobacco cyclin‐dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol. 130, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko, O. , Andersson, M.X. , Hamberg, M. , Brunnström, A. , Göbel, C. , McPhail, K.L. , Gerwick, W.H. , Feussner, I. and Ellerström, M. (2007) Oxo‐phytodienoic acid‐containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol. 145, 1658–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu, A. , Champion, A. , Legrand, J. , Lavenus, J. , Mast, D. , Brunoud, G. , Oh, J. , Guyomarc'h, S. , Pizot, M. , Farmer, E.E. , Turnbull, C. , Vernoux, T. , Bennett, M.J. and Laplaze, L. (2015) A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Commun. 6, 6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert, D. , Schaller, F. and Weiler, E.W. (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase . Planta, 211, 163–165. [DOI] [PubMed] [Google Scholar]
- Lee, D.S. , Nioche, P. , Hamberg, M. and Raman, C.S. (2008) Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature, 455, 363–368. [DOI] [PubMed] [Google Scholar]
- Li, X.B. , Fan, X.P. , Wang, X.L. , Cai, L. and Yang, W.C. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell, 17, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. and Schmittgen, T. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Marmey, P. , Jalloul, A. , Alhamdia, M. , Assigbetse, K. , Cacas, J.L. , Voloudakis, A.E. , Champion, A. , Clerivet, A. , Montillet, J.L. and Nicole, M. (2007) The 9‐lipoxygenase GhLOX1 gene is associated with the hypersensitive reaction of cotton Gossypium hirsutum to Xanthomonas campestris pv malvacearum . Plant Physiol. Biochem. PPB Soc. Fr. Physiol. Végétale, 45, 596–606. [DOI] [PubMed] [Google Scholar]
- Martinez, C. , Montillet, J.L. , Bresson, E. , Agnel, J.P. , Dai, G.H. , Daniel, J.F. , Geiger, J.P. and Nicole, M. (1998) Apoplastic peroxidase generates superoxide anions in cells of cotton cotyledons undergoing the hypersensitive reaction to Xanthomonas campestris pv. malvacearum Race 18. Mol. Plant–Microbe Interact. 11, 1038–1047. [Google Scholar]
- Martinez, C. , Baccou, J.C. , Bresson, E. , Baissac, Y. , Daniel, J.F. , Jalloul, A. , Montillet, J.L. , Geiger, J.P. , Assigbetsé, K. and Nicole, M. (2000) Salicylic acid mediated by the oxidative burst is a key molecule in local and systemic responses of cotton challenged by an avirulent race of Xanthomonas campestris pv malvacearum . Plant Physiol. 122, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T. , Suzuki, K. , Fujimura, T. and Shinshi, H. (2006) Genome‐wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer, K. , Tuominen, H. , Kettunen, R. , Betz, C. , Langebartels, C. , Sandermann, H. and Kangasjärvi, J. (2000) Ozone‐sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide‐dependent cell death. Plant Cell, 12, 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. and Goossens, A. (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell, 23, 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Morreel, K. , De Witte, E. , Lammertyn, F. , Van Montagu, M. , Boerjan, W. , Inzé, D. and Goossens, A. (2008) Mapping methyl jasmonate‐mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc . Natl. Acad. Sci. USA, 105, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Barbero, G.F. , Geerinck, J. , Tilleman, S. , Grunewald, W. , Pérez, A.C. , Chico, J.M. , Bossche, R.V. , Sewell, J. , Gil, E. , García‐Casado, G. , Witters, E. , Inzé, D. , Long, J.A. , De Jaeger, G. , Solano, R. and Goossens, A. (2010) NINJA connects the co‐repressor TOPLESS to jasmonate signalling. Nature, 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Rustérucci, C. , Montillet, J.L. , Agnel, J.P. , Battesti, C. , Alonso, B. , Knoll, A. , Bessoule, J.J. , Etienne, P. , Suty, L. , Blein, J.P. and Triantaphylidès, C. (1999) Involvement of lipoxygenase‐dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 274, 36 446–36 455. [DOI] [PubMed] [Google Scholar]
- Schommer, C. , Palatnik, J.F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E.E. , Nath, U. and Weigel, D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, S. , Okamoto, M. , Seto, H. , Ishizuka, K. , Sano, H. and Ohashi, Y. (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science, 270, 1988–1992. [DOI] [PubMed] [Google Scholar]
- Seo, S. , Sano, H. and Ohashi, Y. (1999) Jasmonate‐based wound signal transduction requires activation of WIPK, a tobacco mitogen‐activated protein kinase. Plant Cell, 11, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I.A. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Stenzel, I. , Hause, B. , Maucher, H. , Pitzschke, A. , Miersch, O. , Ziegler, J. , Ryan, C.A. and Wasternack, C. (2003) Allene oxide cyclase dependence of the wound response and vascular bundle‐specific generation of jasmonates in tomato – amplification in wound signalling. Plant J. Cell Mol. Biol. 33, 577–589. [DOI] [PubMed] [Google Scholar]
- Tuominen, H. , Overmyer, K. , Keinänen, M. , Kollist, H. and Kangasjärvi, J. (2004) Mutual antagonism of ethylene and jasmonic acid regulates ozone‐induced spreading cell death in Arabidopsis . Plant J. Cell Mol. Biol. 39, 59–69. [DOI] [PubMed] [Google Scholar]
- Van der Does, D. , Leon‐Reyes, A. , Koornneef, A. , Van Verk, M.C. , Rodenburg, N. , Pauwels, L. , Goossens, A. , Körbes, A.P. , Memelink, J. , Ritsema, T. , Van Wees, S.C. and Pieterse, C.M. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell, 25, 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley, J.W. , Coughlan, S. , Hudson, M.E. , Covington, M.F. , Kaspi, R. , Banu, G. , Harmer, S.L. and Dehesh, K. (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis‐element. PLoS Genet. 3, 1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2014) Action of jasmonates in plant stress responses and development—applied aspects. Biotechnol. Adv. 32, 31–39. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zarei, A. , Körbes, A.P. , Younessi, P. , Montiel, G. , Champion, A. and Memelink, J. (2011) Two GCC boxes and AP2/ERF‐domain transcription factor ORA59 in jasmonate/ethylene‐mediated activation of the PDF1.2 promoter in Arabidopsis . Plant Mol. Biol. 75, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Genes encoding potential enzymes of jasmonate biosynthesis expressed in response to hormonal treatments in cotton. Expression of GhAOS‐like, GhAOC2‐like and GhACX1a‐like in response to JA and salicylic acid (SA) treatments, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Twelve‐day‐old cotton plants were treated for 2 h with 50 mm JA, 50 mm SA or a combination of the solvents dimethyl sulfoxide (DMSO; 0.1%) and sodium phosphate, pH 7 (0.5 mm) as control (C). Results are averages of three biological replicates; error bars indicate standard deviation (n > 12).
Fig. S2 Expression analysis of GhERF‐IIb3 gene in response to solvent (C, control), jasmonic acid (JA) and salicylic acid (SA) treatments. This experiment was carried out as described in Fig. S1.
Fig. S3 Both transcription factors AtORA47 and GhERF‐IIb3 transactivate the AtAOC2 promoter in cotton protoplasts. Protoplasts were produced and transformed using a protocol adapted from Yoo et al. (2007). β‐Glucuronidase (GUS) and luciferase (LUC) activity tests were performed as described previously (Zarei et al., 2011). Averaged GUS/LUC ratios were expressed relative to the control vector. Three independent experiments ± standard deviation (n = 6).
Fig. S4 Optimization of T‐DNA transfer to cotton cotyledon cells using Agrobacterium tumefaciens LBA1119 virGN54D bacterial vector. (A) Comparison of the transformation efficiency over time in agroinfiltrated cotyledons using two distinct bacterial strains. Agrobacterium tumefaciens strains LBA1119 and LBA1119 virGN54D were both transformed with the pCAMBIA1300::uidA‐intron and further used for the transformation of 10‐day‐old cotton cotyledons, as described in Experimental procedures. The virG‐modified LBA1119 was chosen because it is known to improve the T‐DNA transfer frequency (Fits et al., 2000). β‐Glucuronidase (GUS) activity was followed for 2 days for both strains. For each strain, 24 cotyledons were agroinfiltrated. GUS activities are reported as mean ± standard deviation (SD). Experiments were repeated at least three times with similar results. 4‐methylumbelliferone (MU) (B) Quantitative analysis of GUS activity until 6 days post‐inoculation (dpi) with LBA1119 virGN54D. Twenty‐four cotyledons were agroinfiltrated. GUS activity is reported as mean ± SD. The experiment was repeated twice with similar results. (C) Histochemical analysis of agroinfiltrated cotton cotyledons. The left half of the cotyledon was infiltrated with the empty bacterial strain as control, whereas the right half was infiltrated using the pCAMBIA1300‐uidA‐intron plasmid containing the virG‐modified LBA1119 strain. The photograph (2 dpi) shows the presence of indigo crystals over the whole region that had been transformed by pCAMBIA1300‐GUS‐intron.
Fig. S5 Overexpression of AtORA47 increases the gene expression of the jasmonic acid (JA)‐biosynthesis marker genes and endogenous oxo‐phytodienoic acid (OPDA)/JA levels in cotton. (A) Expression of GhAOS‐like, GhAOC2‐like and GhACX1a‐like genes in cotton cotyledons overexpressing AtORA47 or β‐glucuronidase‐encoding uidA genes. (B) OPDA and JA contents in cotton cotyledons overexpressing AtORA47 or GUS‐encoding uidA genes. Ten‐day‐old Gossypium hirsutum plants were transformed with Agrobacterium tumefaciens carrying either the AtORA47 or uidA construct. At 2 days post‐transformation, gene expression and oxylipin content were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and liquid chromatography‐mass spectrometry (LC‐MS), respectively. Averages ± standard deviation (n = 12) were calculated from biological triplicates.
Fig. S6 Gene expression of jasmonic acid (JA)‐responsive and defence marker genes in transgenic cotton overexpressing GhERF‐IIb3 and inoculated by the virulent Xcm20 strain. Relative transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) at 1 day post‐inoculation with Xcm20 in control tissues expressing GFP6‐ and in GhERF‐IIb3‐overexpressing tissues. Means ± standard deviation (n = 12). Experiments were repeated at least twice with similar results. GFP, green fluorescent protein; Xcm, Xanthomonas citri pv. malvacearum.
Fig. S7 Transient ectopic expression of AtORA47 in cotton cotyledons confers resistance to virulent Xanthomonas citri pv. malvacearum (Xcm) (race 20) bacteria. Cotton cotyledons overexpressing AtORA47 or GFP6 were inoculated at 108 colony‐forming units (cfu)/mL and the bacterial content was determined over time as described in Experimental procedures. Means ± standard deviation (n = 12). Experiments were repeated twice with similar results. GFP, green fluorescent protein.
Table S1 Jasmonic acid (JA) biosynthesis gene candidates ALLENE OXIDE SYNTHASE (AOS), ALLENE OXIDE CYCLASE (AOC) and ACYL‐COENZYME A OXIDASE (ACX) from Gossypium hirsutum share a significant degree of sequence conservation with their Arabidopsis thaliana counterparts.
Table S2 List of primers used in this study.
Table S3 List of APETALA2 (AP2) domains used in phylogenetic analysis.


