Summary
Chitin‐binding lysin motif (LysM) effectors contribute to the virulence of various plant‐pathogenic fungi that are causal agents of foliar diseases. Here, we report the LysM effectors of the soil‐borne fungal vascular wilt pathogen Verticillium dahliae. Comparative genomics revealed three core LysM effectors that are conserved in a collection of V. dahliae strains. Remarkably, and in contrast with the previously studied LysM effectors of other plant pathogens, no expression of core LysM effectors was monitored in planta in a taxonomically diverse panel of host plants. Moreover, targeted deletion of the individual LysM effector genes in V. dahliae strain JR2 did not compromise virulence in infections on Arabidopsis, tomato or Nicotiana benthamiana. Interestingly, an additional lineage‐specific LysM effector is encoded in the genome of V. dahliae strain VdLs17, but not in any other V. dahliae strain sequenced to date. Remarkably, this lineage‐specific effector is expressed in planta and contributes to the virulence of V. dahliae strain VdLs17 on tomato, but not on Arabidopsis or N. benthamiana. Functional analysis revealed that this LysM effector binds chitin, is able to suppress chitin‐induced immune responses and protects fungal hyphae against hydrolysis by plant hydrolytic enzymes. Thus, in contrast with the core LysM effectors of V. dahliae, this lineage‐specific LysM effector of strain VdLs17 contributes to virulence in planta.
Keywords: aggressiveness, chitin, comparative genomics, hyphal protection, lineage‐specific effector, tomato, vascular wilt disease
Introduction
To establish infection, fungal plant pathogens secrete effector molecules that manipulate host physiology, including immune responses that are triggered when plant hosts sense invading pathogens (Cook et al., 2015; Jones and Dangl, 2006; Rovenich et al., 2014). Typically, effectors are small secreted proteins that are species or even lineage‐specific. However, some effectors are more broadly conserved, such as the necrosis‐ and ethylene‐inducing‐like proteins (NLPs) that are produced by bacteria, oomycetes and fungi, and that are particularly known for their phytotoxic activity (Gijzen and Nürnberger, 2006; de Jonge et al., 2011). Another group of conserved fungal effectors are the lysin motif (LysM) effectors, which are found in a wide range of fungal species, including plant and animal pathogens, as well as saprophytes (de Jonge and Thomma, 2009; Kombrink and Thomma, 2013).
LysM effectors are defined as secreted proteins that contain no annotated protein domains apart from a varying number of LysM domains, which are carbohydrate‐binding modules that occur in many prokaryotic and eukaryotic proteins (Buist et al., 2008; de Jonge and Thomma, 2009). The LysM effectors studied to date belong to various fungal plant‐pathogenic species and have all been found to bind chitin, a homopolymer of unbranched β‐1,4‐linked N‐acetylglucosamine (GlcNAc) (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Chitin is a major component of fungal cell walls and plays an important role in the interaction between fungal pathogens and their plant hosts (Bowman and Free, 2006; Kombrink et al., 2011; Rovenich et al., 2016; Sánchez‐Vallet et al., 2015). Plants have evolved to recognize chitin as a ‘non‐self’ molecule and to mount an immune response on chitin perception in order to stop fungal infection (Felix et al., 1993; Shibuya et al., 1993). Several plasma membrane‐localized chitin receptors have been identified in plants that all contain LysM domains (Cao et al., 2014; Kaku et al., 2006; Miya et al., 2007; Wan et al., 2008). LysM effectors of various fungal foliar pathogens, namely the tomato leaf mould fungus Cladosporium fulvum, the Septoria tritici blotch fungus Zymoseptoria tritici (formerly Mycosphaerella graminicola), the rice blast fungus Magnaporthe oryzae and the Brassicaceae anthracnose fungus Colletotrichum higginsianum, have been demonstrated to perturb the activation of chitin‐induced immunity during host colonization and to contribute to virulence (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Recently, the crystal structure of the LysM effector Ecp6, secreted by C. fulvum, has revealed that Ecp6 suppresses chitin‐triggered immunity through two distinct mechanisms (Sánchez‐Vallet et al., 2013). One mechanism involves two LysM domains of a single Ecp6 molecule (LysM1 and LysM3) that cooperatively bind chitin with ultra‐high (pM) affinity and allow Ecp6 to outcompete plant receptors for chitin binding. In addition, the remaining, singular, LysM domain (LysM2) binds chitin and has the capability to suppress chitin‐induced immune responses. However, its relatively low (μM) affinity for chitin probably does not allow this domain to function through the sequestration of chitin fragments, suggesting that LysM2 suppresses chitin‐triggered immunity through another mechanism that potentially involves receptor complex disturbance (Sánchez‐Vallet et al., 2013). In addition, the LysM effectors Mg1LysM and Mg3LysM from Z. tritici have been found to protect fungal hyphae against degradation by plant chitinases (Marshall et al., 2011; Schlumbaum et al., 1986). Although the LysM effectors that have been characterized to date are all secreted by foliar pathogens, they are also found in the genomes of soil‐borne pathogens that infect host plants through their roots, such as Fusarium oxysporum and Verticillium dahliae (de Jonge and Thomma, 2009).
Verticillium dahliae is a soil‐borne fungal pathogen that colonizes the xylem vessels of its host plants, resulting in vascular wilt disease (Fradin and Thomma, 2006; Klimes et al., 2015; Klosterman et al., 2009). Verticillium dahliae infects a wide range of dicotyledonous plant species, including economically important crops, such as cotton, lettuce and tomato. Genetic resistance has been characterized in tomato, and it was shown that the cell surface‐localized immune receptor Ve1 confers resistance against strains of V. dahliae that belong to race 1 (Fradin et al., 2009). However, Ve1 homologues that may recognize V. dahliae are widespread in plants (Song et al., 2016; Zhang et al., 2013). Based on comparative population genomics, it was discovered that Ve1 recognizes the race 1‐specific effector protein Ave1 (for Avirulence on Ve1 tomato) in order to activate effector‐triggered immunity (de Jonge et al., 2012). Furthermore, Ave1 is required for full virulence on tomato genotypes that lack Ve1 (de Jonge et al., 2012). In addition, comparative population genomics revealed that all V. dahliae strains carry lineage‐specific genomic regions that account for 1–5 Mb of their ∼35‐Mb genome and are significantly enriched for in planta expressed genes (Faino et al., 2015, 2016; de Jonge et al., 2013, 2012). Although, in several other plant‐pathogenic species, such as F. oxysporum and Z. tritici, such lineage‐specific regions are found as small dispensable chromosomes (Goodwin et al., 2011; Ma et al., 2010), in V. dahliae these regions are found as islands within the core chromosomes (Faino et al., 2015, 2016; de Jonge et al., 2013).
In this article, we characterize the LysM effector gene family of V. dahliae which consists of three members in the core genome of V. dahliae. Remarkably, one additional LysM effector gene was found in a lineage‐specific region of V. dahliae strain VdLs17.
Results
Three core LysM effectors are encoded in the V. dahliae genome
Initially, LysM effectors were identified in the first available V. dahliae genome sequence, that of strain VdLs17 (Klosterman et al., 2009). However, as this strain has been isolated from the non‐model plant lettuce and is a rather mild pathogen of tomato and Arabidopsis (K. A. Yadeta and B. P. H. J. Thomma, unpublished data), our research focuses on V. dahliae strain JR2 which is particularly aggressive on these plant hosts (de Jonge et al., 2013, 2012; Santhanam et al., 2013, 2016; Santhanam and Thomma, 2013). Recently, we established a gapless whole‐genome sequence of the V. dahliae JR2 strain (Faino et al., 2015; http://fungi.ensembl.org/Verticillium_dahliaejr2/Info/Index). The seven LysM effector genes that were initially identified in strain VdLs17 were also found to occur in JR2, except for the lineage‐specific effector VDAG_05180 (de Jonge et al., 2013; Klosterman et al., 2009). The identification of effector genes is often hampered by erroneous gene annotation (Gibriel et al., 2016). Therefore, for the present study, we revisited the corresponding gene models, revealing that not all of the initially identified LysM effector genes were predicted correctly. First, VdLs17 protein VDAG_03096 (VDAG_JR2_Chr8g05800) contains a single LysM domain that only constitutes a minor portion of the total protein (Fig. S1, see Supporting Information). Furthermore, mapping of RNA sequencing reads from samples of V. dahliae grown in vitro and in planta (Faino et al., 2012; de Jonge et al., 2012) indicated that the initially predicted gene model of the corresponding locus is incorrect, as reads mapped to predicted introns, whereas other parts of the predicted gene, including the LysM domain, were not supported by read mapping (Fig. S1). Thus, VDAG_03096 (VDAG_JR2_Chr8g05800) was disqualified as a bona fide LysM effector. Similarly, deeper analysis of the predicted VdLs17 VDAG_06426 (VDAG_JR2_Chr7g08210) protein sequence by SMART protein domain analysis (http://smart.embl-heidelberg.de) revealed the absence of a signal peptide and the presence of a zinc finger domain (Fig. S1), disqualifying this protein as a bona fide LysM effector. Finally, VdLs17 protein VDAG_08171 (VDAG_JR2_Chr8g01370) was disqualified because of the presence of a glycosylphosphatidylinositol (GPI) anchor, suggesting that it is a membrane‐associated protein, rather than an apoplastic effector protein. The three remaining VdLs17 LysM effectors VDAG_00902, VDAG_04781 and VDAG_06998 are not only encoded by strain JR2 (as VDAG_JR2_Chr1g02480, VDAG_JR2_Chr6g09650 and VDAG_JR2_Chr4g05910, respectively), but also by 18 additional V. dahliae strains that were sequenced (Table S1, see Supporting Information) (de Jonge et al., 2013, 2012; B. P. H. J. Thomma et al., unpublished data), and are further referred to as core VdLysM effectors. As these effectors comprise four, five or six LysM domains, they were named accordingly: Vd4LysM, Vd5LysM and Vd6LysM.
VdLysM sequence analysis in 20 V. dahliae strains
To investigate sequence polymorphisms in VdLysM effector genes within the V. dahliae population, we mapped the sequencing reads of 19 V. dahliae strains (Table S1) to the genome assembly of strain JR2. Subsequently, we analysed single nucleotide polymorphisms (SNPs) in the different V. dahliae strains. This revealed that Vd4LysM is the most conserved LysM effector gene, as only one strain (Vd39) showed seven synonymous SNPs and one non‐synonymous SNP (Figs 1, S2, see Supporting Information). Vd5LysM displays the most SNPs: 14 synonymous and 16 non‐synonymous SNPs were found involving the majority of the strains. For Vd6LysM, seven synonymous and three non‐synonymous SNPs were identified in seven strains in total (Figs 1, S2). Although the sequence variation in Vd5LysM exceeds that of the other VdLysM effector genes, no evidence for negative or positive selection could be found. No SNPs were observed in LysM effector genes of strains VdLs17, 2009‐605, V152, V52, van Dijk and Vd57 when compared with those of JR2, suggesting that these strains are phylogenetically more closely related to JR2 than the remaining 13 strains.
Figure 1.
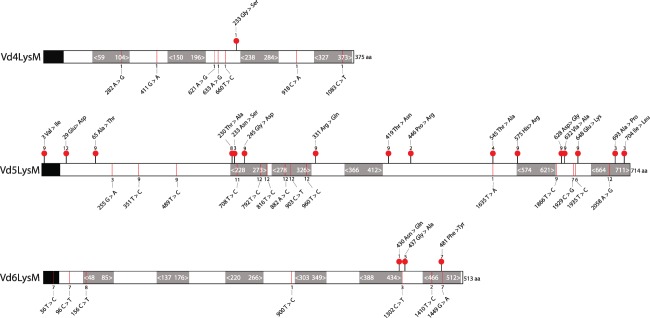
Sequence polymorphisms of core VdLysM effector genes in Verticillium dahliae population. The single nucleotide polymorphisms (SNPs) were compared among 20 V. dahliae strains. The protein models are based on the annotation of VdLysMs in V. dahliae strain JR2. Three core LysM effectors have four to six predicted LysM domains, which are represented by grey squares (amino acid positions are indicated with white font). The N‐terminal black box represents the predicted signal peptide. Sites with synonymous and non‐synonymous nucleotide substitutions are marked by red lines and red dots, respectively. The number above or below the SNP sites indicates the amount of strains that share the same nucleotide polymorphism at that position (detailed information on each individual SNP can be found in Fig. S2).
Core LysM effectors of V. dahliae strain JR2 do not contribute to virulence on tomato
To test whether the core LysM effectors contribute to the virulence of V. dahliae on tomato, gene functional analysis was pursued in V. dahliae strain JR2. Previously, RNA sequencing was performed on samples harvested during a time course of V. dahliae‐inoculated Nicotiana benthamiana plants (Faino et al., 2012; de Jonge et al., 2012). These data were used to assess the expression of VdLysM genes during host invasion. However, no significant VdLysM effector gene expression was observed at any of the time points. In contrast, the expression of the functionally analysed Ave1 effector gene was significantly induced in these samples (de Jonge et al., 2013) (Fig. 2A).
Figure 2.
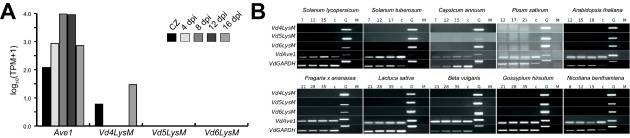
Core VdLysM effector genes of Verticillium dahliae strain JR2 are not expressed in planta. (A) VdLysM effector gene expression and expression of the Ave1 effector gene during growth of strain JR2 on Czapek Dox medium (CZ) and during colonization on Nicotiana benthamiana at 4, 8, 12 and 16 days post‐inoculation (dpi). (B) In planta reverse transcription‐polymerase chain reaction (RT‐PCR) expression analysis of core VdLysM genes. Expression of VdLysM genes could not be detected in 10 different plant species (Solanum lycopersicum, Solanum tuberosum, Capsicum annuum, Pisum sativum, Arabidopsis thaliana, Fragaria × ananassa, Lactuca sativa, Beta vulgaris, Gossypium hirsutum and N. benthamiana) at time points between 7 and 35 dpi. Expression was analysed at three time points per species and referenced to the expression of VdAve1, and VdGAPDH cDNA (c) and genomic DNA (G) from V. dahliae cultured in vitro were used as controls. Primer pairs were designed to span introns to account for amplification from cDNA or DNA templates. Mock (M) treated plants are also shown.
Differential V. dahliae effector gene induction among host plant species has been observed previously, as the NLP2 effector gene was found to be induced in tomato and Arabidopsis, but not in N. benthamiana (Santhanam et al., 2013). Therefore, reverse transcription‐polymerase chain reaction (RT‐PCR) was performed on cDNA generated from samples of tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana) plants inoculated with V. dahliae strain JR2 and harvested at different time points. However, again no expression of any of the VdLysM effector genes was recorded, whereas expression of Ave1 as a positive control was clearly detected (Fig. 2B). In a final attempt to detect in planta expression of core VdLysM effector genes, a taxonomically wide range of plants, including potato (Solanum tuberosum), pepper (Capsicum annuum), Australian tobacco (N. benthamiana), pea (Pisum sativum), strawberry (Fragaria × ananassa), lettuce (Lactuca sativa), beet (Beta vulgaris) and cotton (Gossypium hirsutum), were inoculated with V. dahliae strain JR2 and VdLysM effector gene expression was monitored. However, expression of VdLysM effector genes could not be detected in any of these plant species, whereas the expression of the Ave1 effector gene was observed in all hosts (Fig. 2B).
As it cannot be excluded that VdLysM effector gene expression is induced at low levels only, or only at specific time points or at particular sites, further investigations into the possible contribution of the core VdLysM effector genes to the virulence of V. dahliae strain JR2 were evaluated by targeted deletion. Multiple deletion strains were obtained for the LysM effector genes Vd4LysM, Vd5LysM and Vd6LysM. Importantly, none of the deletion strains obtained showed any phenotypic deviations from the wild‐type JR2 strain on cultivation in vitro (data not shown). Subsequent virulence assays revealed that Arabidopsis, N. benthamiana and tomato plants inoculated with the wild‐type strain or with deletion strains for Vd4LysM, Vd5LysM and Vd6LysM showed similar disease development; the timing and degree of stunting of the plants and wilting of the leaves were similar on inoculation with the various genotypes (Fig. 3). Thus, the lack of VdLysM effector gene expression, together with the observation that VdLysM deletion strains are not affected in virulence on Arabidopsis, N. benthamiana and tomato, strongly suggest that the core VdLysM effectors do not play an important role during host colonization.
Figure 3.

Core VdLysM effector genes do not contribute to the virulence of Verticillium dahliae strain JR2. Infection assay of wild‐type and VdLysM effector gene deletion strains on Arabidopsis, Nicotiana benthamiana and tomato plants showing two deletion strains for each VdLysM effector gene, with the exception of Vd5LysM. At 14 (tomato, N. benthamiana) or 21 days post‐inoculation (Arabidopsis), photographs were taken of one representative plant out of six that were inoculated with the same V. dahliae genotype. The infection assays on Arabidopsis, N. benthamiana and tomato were repeated three times with similar results.
A lineage‐specific LysM effector is required for full virulence on tomato, but not on N. benthamiana or Arabidopsis
It has been demonstrated previously that lineage‐specific effectors of V. dahliae strain JR2 contribute significantly to virulence on tomato (de Jonge et al., 2013), suggesting that lineage‐specific regions of individual V. dahliae genotypes are important for the development of aggressiveness on particular host plants. Intriguingly, one LysM effector gene, VDAG_05180, was originally identified in strain VdLs17, but could not be found in strain JR2 or in any of the other 18 additional V. dahliae strains that were sequenced, revealing that VDAG_05180 is a lineage‐specific effector gene. Interestingly, apart from VdLs17, none of the other sequenced V. dahliae strains carries lineage‐specific LysM effector genes.
It has been shown that the VdLs17 lineage‐specific LysM effector which contains two LysM domains, and is hence designated Vd2LysM, contributes to virulence on tomato (de Jonge et al., 2013). Here, the expression of Vd2LysM during infection of V. dahliae strain VdLs17 on tomato, N. benthamiana and Arabidopsis was investigated using real‐time PCR, and expression was detected at each of the time points monitored (Fig. 4A). To test whether Vd2LysM contributes to virulence, Vd2LysM deletion strains were inoculated on these host plants. Importantly, none of the transformants showed morphological anomalies on growth in vitro when compared with the VdLs17 wild‐type strain (Fig. 4B). As noted previously (de Jonge et al., 2013), the Vd2LysM deletion strains showed significantly reduced virulence on inoculation on tomato when compared with the wild‐type strain VdLs17 (Fig. 4C). The tomato plants that were inoculated with the wild‐type strain showed stronger stunting than plants that were inoculated with two independent ΔVd2LysM strains (Fig. 4C). In accordance with the reduced symptom development, real‐time PCR quantification of the fungal biomass showed that the ΔVd2LysM strains produced significantly less biomass than the wild‐type (Fig. 4E). These results show that the lineage‐specific LysM effector gene Vd2LysM plays a role in the virulence of V. dahliae. To investigate whether the contribution to virulence extends to other plant hosts as well, the Vd2LysM deletion strains were inoculated onto N. benthamiana and Arabidopsis plants. Intriguingly, the virulence of the Vd2LysM deletion strains appeared to be comparable with that of the wild‐type strain on these plant species, as all genotypes induced similar symptomatology on the inoculated plants and real‐time PCR measurements of fungal biomass did not reveal compromised fungal host colonization (Fig. 4D, E). On Arabidopsis, it seems that Vd2LysM deletion leads to enhanced colonization.
Figure 4.
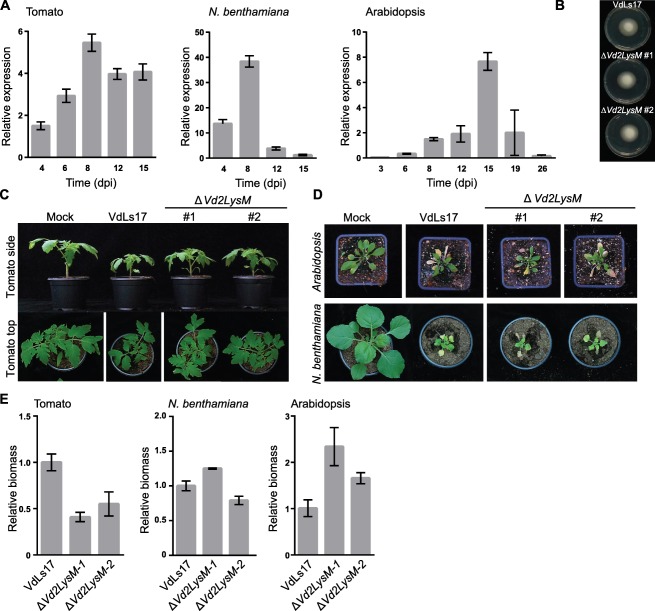
Lineage‐specific LysM effector Vd2LysM contributes to the virulence of Verticillium dahliae strain VdLs17 on tomato. (A) Vd2LysM expression during colonization of V. dahliae strain VdLs17 on tomato, N. benthamiana and Arabidopsis plants at 2–26 days post‐inoculation (dpi). (B) Morphology of wild‐type V. dahliae strain VdLs17 and two Vd2LysM deletion strains after 7 days of incubation on potato dextrose agar (PDA) medium at room temperature. (C) Photographs of representative tomato plants out of eight plants that were either mock‐inoculated or inoculated with wild‐type V. dahliae strain VdLs17 or two deletion strains of Vd2LysM at 14 dpi. The infection assay was repeated three times with similar results. (D) Photographs of representative N. benthamiana and Arabidopsis plants out of eight plants that were either mock‐inoculated or inoculated with wild‐type V. dahliae strain VdLs17 or two deletion strains of Vd2LysM at 14 dpi (N. benthamiana) and 21 dpi (Arabidopsis). The assay was repeated three times with similar results. (E) Fungal biomass accumulation in tomato, N. benthamiana and Arabidopsis plants inoculated with wild‐type strain VdLs17 or Vd2LysM deletion strains at 14 dpi (tomato, N. benthamiana) and 21 dpi (Arabidopsis). Error bars represent the standard error of three replicate experiments.
Vd2LysM binds chitin
Previously characterized LysM effectors of fungal plant pathogens have been demonstrated to bind chitin (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Therefore, the chitin‐binding ability of Vd2LysM was tested. To this end, Vd2LysM was heterologously produced in the yeast Pichia pastoris, which has been used previously for the production of LysM effectors from other fungi (Kombrink, 2012). Subsequently, the purified Vd2LysM protein was used in affinity precipitation assays with the insoluble carbohydrates chitin, chitosan, xylan and cellulose. We observed that Vd2LysM precipitated with all carbohydrates tested (Fig. 5A), which suggests that the protein precipitates by itself rather than binds to any of these carbohydrates. To investigate this further, Vd2LysM was subjected to glycan‐array analysis to test the binding affinity for approximately 600 glycans, taking C. fulvum Ecp6 as a control. As demonstrated previously, Ecp6 specifically binds to the chitin oligosaccharides (GlcNAc)3, (GlcNAc)5 and (GlcNAc)6 that are present on the array (de Jonge et al., 2010). In contrast, Vd2LysM does not bind to any of the glycans on the array (Fig. S3, see Supporting Information). Based on these findings, we conclude that P. pastoris‐produced Vd2LysM probably precipitates spontaneously in the affinity precipitation assay.
Figure 5.
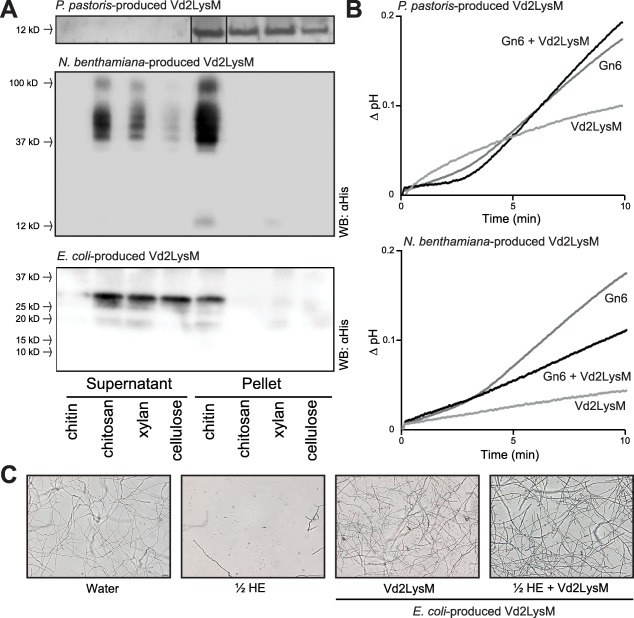
Vd2LysM is a chitin‐binding protein that suppresses chitin‐induced immune responses. (A) Vd2LysM produced in Pichia pastoris, Nicotiana benthamiana and Escherichia coli was used in affinity precipitation experiments with the insoluble polysaccharides chitin, chitosan, xylan and cellulose. After sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and Coomassie brilliant blue staining for P. pastoris‐produced Vd2LysM or western blot analysis for in planta‐ and Escherichia coli‐produced Vd2LysM, the proteins are observed in the insoluble pellet fraction or in the supernatant fraction. Vd2LysM produced in P. pastoris is observed in the insoluble pellet fraction of all polysaccharides. Vd2LysM produced in planta and in E. coli precipitates with chitin, but not with chitosan, xylan or cellulose. (B) pH shift measurements in a tomato cell suspension after the addition of 1 nm chitin or 1 nm chitin that was pre‐incubated with 10 nm Vd2LysM produced in either P. pastoris or in planta. The graphs show a representative experiment out of two to four experiments with similar results. (C) Micrographs of Fusarium oxysporum f.sp. lycopersicum strain 4287 taken 23 h after the addition of water, hydrolytic enzyme mix (HE) or Vd2LysM and pre‐treated with Vd2LysM followed by treatment with hydrolytic enzyme mix. The figure is representative of three independent experiments.
Recently, a crystal structure has been obtained of C. fulvum Ecp6, which was heterologously produced in P. pastoris. The crystal structure of the protein contained chitin, arguably derived from P. pastoris during protein production, in a composite‐binding groove of two LysM domains (Sánchez‐Vallet et al., 2013). To exclude the possibility that potential Vd2LysM substrate‐binding sites were occupied by P. pastoris chitin, the effector was produced in N. benthamiana. The purified protein was subsequently tested for chitin‐binding ability in an affinity precipitation assay using chitin, chitosan, xylan and cellulose. Interestingly, N. benthamiana‐produced Vd2LysM bound to chitin, but not to any of the other carbohydrates (Fig. 5A). Furthermore, we observed that N. benthamiana‐produced Vd2LysM migrated as multiple bands on the gel, which were not visible with P. pastoris‐produced Vd2LysM, which migrated as a single band of the expected 17 kDa. This may indicate that Vd2LysM produced in N. benthamiana forms oligomers. Collectively, these findings suggest that Vd2LysM is a chitin‐binding effector protein, and the chitin that is readily available during heterologous production in P. pastoris occupies the Vd2LysM substrate‐binding sites.
Vd2LysM suppresses chitin‐induced immune responses and protects hyphae against degradation by plant hydrolytic enzymes
Previously, LysM effectors from various fungal plant pathogens have been demonstrated to suppress the chitin‐induced pH shift in a tomato cell suspension, which is indicative of the ability of the effector to perturb chitin‐induced host immune responses (Felix et al., 1993; de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Both in planta‐produced and P. pastoris‐produced Vd2LysM were tested for this capacity, as described previously (de Jonge et al., 2010). Interestingly, P. pastoris‐produced Vd2LysM, which is not able to bind chitin, did not suppress the chitin‐induced pH shift, whereas N. benthamiana‐produced Vd2LysM, which is able to bind chitin, suppressed the chitin‐induced immune response (Fig. 5B). These results suggest that Vd2LysM is able to suppress chitin‐triggered immune responses during V. dahliae colonization of tomato.
As the Z. tritici LysM effectors Mg3LysM and Mg1LysM have been demonstrated previously to protect fungal hyphae against degradation by plant hydrolytic enzymes (Marshall et al., 2011), we also intended to test Vd2LysM for this activity. However, the yield of in planta‐produced Vd2LysM was too low to perform such assays. Therefore, we pursued Vd2LysM production in yet another heterologous system: Escherichia coli. Similar to N. benthamiana‐produced Vd2LysM, E. coli‐produced Vd2LysM was found to bind chitin, but no other insoluble carbohydrates tested (Fig. 5A). Moreover, E. coli‐produced Vd2LysM also showed signs of oligomerization. Interestingly, similar to the Z. tritici LysM effectors Mg3LysM and Mg1LysM, Vd2LysM was also found to protect fungal hyphae against degradation by plant hydrolytic enzymes (Fig. 5C).
LysM domains of Vd2LysM are more similar to those of previously characterized LysM effectors than to those of core VdLysM effectors
Vd2LysM is presently the only LysM effector of V. dahliae for which a role in virulence could be shown, and which is able to perturb chitin‐centred host immune responses, as described previously for Ecp6, Mg1LysM, Mg3LysM, Slp1, ChELP1 and ChELP2 (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Remarkably, the LysM domains of Vd2LysM are more similar to those of Ecp6, Mg1LysM, Mg3LysM, Slp1, ChELP1 and ChELP2 than to those of the core VdLysM effectors (Fig. 6). To further investigate this, we carried out a comparative LysM domain analysis with functionally characterized plant and fungal proteins (Fig. 6). The fungal LysM domains fall into two separate clades (Fig. 6B). One clade was nearly exclusively formed by LysMs of plant receptors and LysMs of fungal effectors with a role in virulence, including Vd2LysM (Bolton et al., 2008; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). This suggests that fungi and plants produce LysM proteins with a conserved motif, and thus possibly originate from a common ancestor. Interestingly, the second clade solely contains the LysMs of the V. dahliae core effectors and the Trichoderma atroviride LysM effector TAL6, which has been shown previously to specifically inhibit the germination of conidia of Trichoderma spp. and has been proposed to act in fungal development rather than in host interactions (Seidl‐Seiboth et al., 2013) (Fig. 6B). Intriguingly, the two clades are characterized by highly divergent consensus motifs for their LysMs, with the ‘fungal‐specific’ LysMs having three highly conserved cysteine residues that are completely lacking from the ‘effector/receptor’ LysMs. Collectively, these findings may suggest that the core LysM effectors of V. dahliae, in contrast with Vd2LysM, do not act in host interactions, but possibly in other physiological processes.
Figure 6.
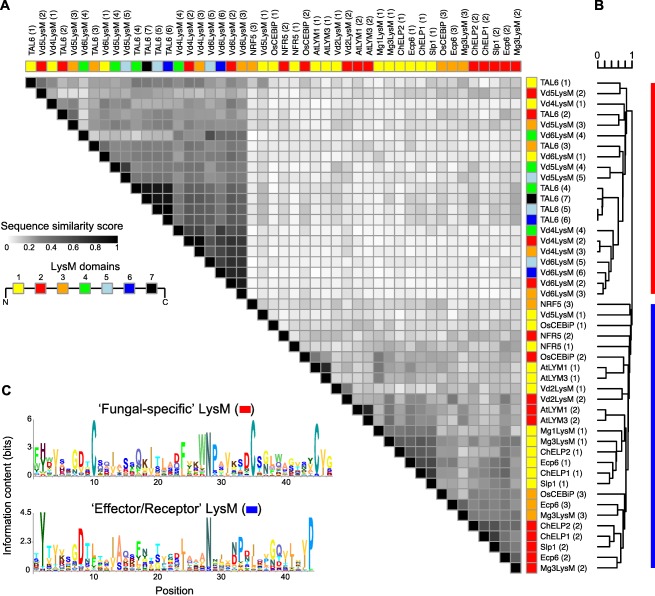
Sequence similarity between LysM domains. (A) Pairwise comparisons of the LysM domains of fungal and plant LysM proteins. Normalized sequence similarities between LysM domains are displayed in the heat map. The colour annotation above the heat map indicates the relative position of each LysM domain within the LysM protein. (B) Hierarchical clustering (UPGMA clustering) based on the (dis)‐similarity matrix of all pairwise LysM domain comparisons is shown above and on the side, and the clustering is used to order the rows and columns of the heat map. (C) Sequence logos of LysM domains identified in fungal effectors as well as in plant receptors (blue) and LysM domains that have been defined previously as ‘fungal specific’.
Discussion
Here, we have described the characterization of the LysM effector catalogue of the soil‐borne vascular wilt pathogen V. dahliae. In‐depth analysis revealed that three VdLysM effector genes occur in the core genome of all strains for which a genome sequence is available. In addition, a lineage‐specific VdLysM effector gene, Vd2LysM, occurs in strain VdLs17 and not in any of the other strains that have been sequenced to date. We initially reasoned that the ubiquitous occurrence of the three core LysM effector genes suggests that these are required for host colonization by V. dahliae. However, a role in virulence cannot be attributed to any of these core LysM effectors. Intriguingly, the lineage‐specific Vd2LysM effector was found to contribute to host‐specific virulence of strain VdLs17.
LysM effectors are found in fungal species with various lifestyles, but, thus far, have only been characterized as virulence factors of plant‐pathogenic fungi (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). Moreover, they have only been studied in foliar pathogens with a (relatively) narrow host range: C. fulvum (Ecp6), Z. tritici (Mg1LysM and Mg3LysM), M. oryzae (Slp1) and C. higginsianum (ChELP1 and ChELP2) (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016). In contrast, V. dahliae is a soil‐borne vascular plant pathogen that infects a broad range of host plants. The expression of the previously characterized LysM effectors is induced during the early stages of infection of the respective pathogens, when evasion of recognition by the host is of particular importance to facilitate tissue colonization. The expression of Vd2LysM peaks at about 1 week after inoculation, which is when V. dahliae biomass is accumulating in the xylem and before wilting and necrosis of the host tissue are visible (Fig. 4A) (Fradin and Thomma, 2006). Ecp6, Mg3LysM, Slp1, ChELP1 and ChELP2 have been demonstrated to function as suppressors of chitin‐induced host immunity and, similar to these proteins, Vd2LysM suppresses chitin‐induced medium alkalinization in a tomato cell suspension. This strongly suggests that Vd2LysM contributes to the virulence of V. dahliae through the perturbation of the activation of chitin‐triggered host immunity. However, in contrast with Ecp6, Slp1, ChELP1 and ChELP2, and similar to Mg3LysM, Vd2LysM is also able to protect fungal hyphae against degradation by plant hydrolytic enzymes. Thus, Vd2LysM plays a broad role in overcoming chitin‐centred host immune responses (Sánchez‐Vallet et al., 2015).
Considering the widespread occurrence of chitin receptor homologues in plant species, and the observation that LysM effectors of diverse plant pathogens have the ability to suppress chitin‐triggered immunity, it seems that this ability is fundamental for fungal plant pathogens to establish infections on their hosts. As most of the sequenced V. dahliae strains are pathogenic on tomato, these strains probably employ molecules other than Vd2LysM to overcome chitin‐centred host immune responses (Sánchez‐Vallet et al., 2015). Alternatively, V. dahliae may employ other mechanisms to prevent the activation of chitin‐triggered immune responses (Rovenich et al., 2016; Sánchez‐Vallet et al., 2015). For example, the α‐1,3‐glucan synthase gene has been demonstrated to be important for the virulence of several fungal pathogens, as the α‐1,3‐glucan layer around hyphae reduces the accessibility of fungal chitin to host chitinases and, consequently, prevents the release of free chitin fragments (Fujikawa et al., 2012). In addition, fungal species have been found to secrete chitin deacetylases that convert chitin into chitosan, which is a poor inducer of immune responses (Gough and Cullimore, 2011).
Structural analysis of the C. fulvum LysM effector Ecp6 revealed that the first and third LysM domains cooperatively bind one chitin molecule with ultra‐high (pM) affinity through intramolecular LysM dimerization, whereas the second, singular LysM domain displays low micromolar affinity (Sánchez‐Vallet et al., 2013). Nevertheless, both binding sites are able to suppress chitin‐triggered immune responses (Sánchez‐Vallet et al., 2013). Intriguingly, all characterized LysM effectors that have the ability to suppress chitin‐triggered immune responses either have three (Ecp6, Mg3LysM) or two (Slp1, ChELP1, ChELP2, Vd2LysM) LysM domains. It remains unknown whether the LysM effectors with two LysM domains contain one composite chitin‐binding site that is composed of the coordinated action of two LysM domains, or two binding sites of separate, singular LysM domain activities. Consequently, the mechanism by which Vd2LysM perturbs the activation of chitin‐triggered immunity requires further investigation.
Recently, evolutionary analyses and consensus pattern profiling of fungal LysM domains have led to their classification into two groups (Akcapinar et al., 2015). The clustering into two phylogenetic clades across taxonomic boundaries is consistent with the presence of a characteristic cysteine residue pattern present in 80% of fungal‐specific LysMs (Akcapinar et al., 2015) (Fig. 6C). Remarkably, the group of fungal LysMs which do not display the conserved cysteine residues contains all of the LysM domains of previously characterized LysM effectors from fungal pathogens (Bolton et al., 2008; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016) (Fig. 6C). It has been noted previously that LysM domains from saprotrophs, mutualists and mycoparasitic fungi are under‐represented in this clade (Akcapinar et al., 2015), pointing to the significantly diverging roles of LysM proteins. To predict the putative roles of the core LysM effectors of Verticillium, we carried out a comparative LysM domain analysis with functionally characterized plant and fungal proteins (Fig. 6). Consistent with previous findings (Akcapinar et al., 2015), the fungal LysM domains analysed here largely fall into two separate clades (Fig. 6B). Interestingly, the V. dahliae core effectors and TAL6 from the mycoparasitic fungus T. atroviride, which selectively inhibits the spore germination of Trichoderma spp. (Seidl‐Seiboth et al., 2013), grouped into the same clade (Akcapinar et al., 2015) (Fig. 6B). Considering the similarity of the core LysM effectors of V. dahliae to TAL6, we hypothesize that they could function in other physiological processes rather than virulence. Although the (single gene) mutants do not display phenotypic deviations from the wild‐type strain, further experimental support is required to confirm this hypothesis. Originally, the LysM domain was identified in bacterial lysozymes that bind and hydrolyse peptidoglycan components of bacterial cell walls (Buist et al., 2008). Therefore, it is possible that fungal LysMs also display peptidoglycan‐specific binding activities, which could help fungi to outcompete bacterial competitors (Kombrink and Thomma, 2013). Similar to the LysM effector functions of plant‐colonizing fungi, LysM effectors of saprotrophs could protect fungal hyphae from hydrolytic enzyme attack secreted by mycoparasites.
In conclusion, we have demonstrated that a lineage‐specific LysM effector of V. dahliae contributes to virulence, whereas three LysM effectors that are present in the core genome do not seem to play a role during host colonization. This finding confirms previous observations that, in particular, those effectors that are encoded in lineage‐specific regions of V. dahliae genomes are important for fungal aggressiveness (de Jonge et al., 2013). It is tempting to speculate that the core LysM effectors play other roles in fungal life. Therefore, it might be worthwhile to investigate whether the core LysM effectors contribute to fungal growth in other stages of the V. dahliae life cycle that were not covered in this study, for example, during saprophytic growth or during survival as resting structures in the soil.
Experimental Procedures
VdLysM effector gene expression analysis
Conidiospores of V. dahliae strain JR2 were inoculated on potato dextrose agar (PDA) plates and grown at room temperature for 7 days before harvest and dilution in 40 mL of potato dextrose broth (PDB) to a concentration of 1 × 106 conidia/mL that was used as inoculum for plant inoculation. Plants of S. lycopersicum, S. tuberosum, C. annuum, P. sativum, A. thaliana, F. × ananassa, L. sativa, B. vulgaris, G. hirsutum and N. benthamiana were grown in the glasshouse at 21°C/19°C during 16‐h/8‐h light/dark photoperiods, respectively, with a relative humidity of ∼75% and 100 W/m2 supplemental light when the light intensity dropped below 150 W/m2. At 10 days post‐germination, the plants were inoculated with V. dahliae or PDB as mock treatment by dipping of the roots of uprooted plants for 6 min before transfer to fresh soil. Three time points were set for each species and five inoculated plants were collected per time point.
A two‐step RT‐PCR was performed to test the expression of VdLysM effectors. RNA was extracted from all sampled plant material using the QuickRNA™ MiniPrep kit (Zymo Research, Irvine, CA, USA) following the protocol provided by the manufacturer. cDNA was synthesized from 1 μg of RNA from each sample by M‐MMLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer's instructions. cDNA and genomic DNA from V. dahliae strain JR2 grown in vitro on PDB were used as controls. Expression analysis was performed by PCR using 2 μL of cDNA as template with the following protocol: 5 min at 95°C, followed by 32 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 1 min. For the strawberry, lettuce, beet and cotton samples, a second PCR was needed to detect the expression of the control genes, owing to the low levels of V. dahliae biomass in infected plants. This second PCR was performed by taking 2 μL of the first PCR as template and run with the same temperature profile, but for only 15 cycles. The primers listed in Table S2 (see Supporting Information) were used to amplify transcripts from Vd4LysM, Vd5LysM and Vd6LysM. VdAVe1 and VdGAPDH were used as controls for effector induction in planta and presence of V. dahliae in the sample, respectively. The primers were designed to span introns to discriminate between amplicons derived from cDNA or DNA templates as a control for DNA contaminations in the synthesized cDNA. Products from all samples were visualized by gel electrophoresis in a 2% agarose gel.
Functional analysis of VdLysM effector genes
VdLysM effector gene deletion strains were generated by amplification of flanking sequences of the coding sequences using the primer sets listed in Table S2. PCR products were subsequently cloned into pRF‐HU2 (Frandsen et al., 2008). Verticillium dahliae transformation and subsequent inoculations on tomato (cv. Motelle and MoneyMaker) plants to assess the virulence of the knock‐out mutants were performed as described by Fradin et al. (2009). In one experiment, six to eight plants were used per inoculation with wild‐type or deletion strains, and the experiment was repeated at least three times. Plants were regularly inspected and representative plants were photographed at 12 and 21 days post‐inoculation (dpi). For biomass quantification, the roots and stem below the cotyledons of four plants per V. dahliae genotype were flash frozen in liquid nitrogen. The samples were ground to powder, and an aliquot was used for DNA isolation (Fulton et al., 1995). Real‐time PCR was conducted with primer sets SlRub‐F1/SlRub‐F2 for tomato RuBisCo and VdGAPDH‐F/VdGAPDH‐R for V. dahliae GAPDH (Table S2). For expression analyses, 3‐week‐old N. benthamiana plants were inoculated with strain VdLs17 as described previously (Fradin et al., 2009), harvested at 4, 6, 8 and 12 dpi, and flash frozen in liquid nitrogen. Total RNA was extracted using the RNeasy Kit (Qiagen, Hilden, Germany) and cDNA was synthesized by SuperScript III (Invitrogen, Carlsbad, CA, USA). Real‐time PCR was conducted with primer sets Q‐VdGAPDH‐F/Q‐VdGAPDH‐R for V. dahliae GAPDH and Q‐Vd2LysM‐F/Q‐Vd2LysM‐R for V. dahliae Vd2LysM (Table S2).
VdLysM sequence analysis and screening of lineage‐specific regions for LysM effectors
The sequencing reads of 19 V. dahliae stains (Table S1) were mapped to the genome assembly of strain JR2 using BWA with default settings (Li and Durbin, 2009). Subsequently, SNPs in genes encoding core LysM proteins were detected using FreeBayes with default settings (Garrison and Marth, 2012). The effects of SNPs were predicted using VEP (McLaren et al., 2010).
We performed pairwise whole‐genome alignments of all V. dahliae strains with NUCmer (using maxmatch option and alignments were filtered by 90% identity), which is part of the mummer package (Kurtz et al., 2004). Subsequently, for each reference strain, the aligned regions were extracted using bedtools (Quinlan and Hall, 2010). Regions in the reference strain that are present in less than 18 strains were considered as lineage‐specific regions. Subsequently, we used all the LysM protein sequences that were compared in this study as queries for a blast search (tblastn; Altschul et al., 1990) against the lineage‐specific regions.
Heterologous Vd2LysM expression in P. pastoris
Vd2LysM was cloned into P. pastoris expression vector pPic9 (Invitrogen) after performing PCR using primers to add the N‐terminal HIS‐ and FLAG‐tag and EcoRI and NotI restriction sites for directional cloning (Table S1). Subsequently, P. pastoris strain GS115 was transformed and a selected clone was cultured in a fermentor (Bioflo 3000) as described previously (Rooney et al., 2005). After removal of cells and concentration of the culture medium, the HIS‐tagged protein was purified using an Ni‐NTA column (Qiagen) according to the manufacturer's protocol. The final protein concentration was determined spectrophotometrically at 280 nm and confirmed using the Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA) with bovine serum albumin (BSA) as a standard.
In planta production of Vd2LysM
PCR was performed to add NheI and SacI restriction sites to Vd2LysM. Using directional cloning, Vd2LysM was cloned into vector pHYG, a modified version of the expression vector pMDC32 (Curtis and Grossniklaus, 2003). The expression vector was subsequently transferred into Agrobacterium tumefaciens strain MOG101. Infiltration of 4–5‐week‐old N. benthamiana with Agrobacterium tumefaciens was performed as described by Westerhof et al. (2012). Four days after infiltration, leaves were harvested and flash frozen in liquid nitrogen. Plant material was ground and homogenized in ice‐cold extraction buffer [1% v/v Tween‐20, 2% w/v immobilized polyvinylpolypyrrolidone (PVPP), 300 mm NaCl, 50 mm NaH2PO4, 10 mm imidazole, 1 mm dithiothreitol (DTT), pH 7.4] using 2 mL/g fresh plant material. After 30 min of homogenization at 4°C, the crude extract was centrifuged at 16 000 g at 4°C. The supernatant was further cleaned using a miracloth filter, and an Ni‐NTA Superflow column (Qiagen) was used to purify the HIS‐tagged protein, according to the manufacturer's protocol. The final protein concentration was determined spectrophotometrically at 280 nm and confirmed using the Pierce BCA Protein Assay Kit (Thermo Scientific) with BSA as a standard.
Escherichia coli production of Vd2LysM
The coding sequence of Vd2LysM was amplified from V. dahliae cDNA and cloned into the pETSUMO (Invitrogen) expression vector according to the manufacturer's instructions prior to E. coli Origami (DE3) transformation. A single transformant was selected and grown in Luria broth medium until an optical density at 600 nm (OD600) of 0.9 was reached. Heterologous production of Vd2LysM was induced with 1 mm Isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) at 26°C during ∼20 h. Cell pellets were lysed using lysozyme from chicken egg (Sigma, St Louis, MO, USA) and Vd2LysM was purified from the soluble protein fraction using an Ni2+‐NTA Superflow column (Qiagen). Purified protein was dialysed against 200 mm NaCl and concentrated over Amicon ultracentrifugal filter units (Molecular weight cut‐off (MWCO) = 3 kDa; Millipore, Billerica, MA, USA).
Sequence similarity between LysM domains
LysM domains within fungal and plant proteins were identified using the SMART database (including PFAM domain and outlier detection) (Schultz et al., 1998). Sequence similarities between LysM domains were approximated using the Needleman–Wunsch algorithm, which has been implemented in the EMBOSS package (Rice et al., 2000). The Needleman–Wunsch score was further normalized to the range 0–1 by NWijnorm = NWij/max(NWii, NWjj). Normalized Needleman–Wunsch scores were displayed in R. Moreover, hierarchical clustering unweighted pair group method with arithmetic mean (UPGMA) based on the (dis)‐similarity matrix of all pairwise LysM domains was performed in R. Sequence logos for effector/receptor LysM domains and other fungal LysM domains were created with Skylign (Wheeler et al., 2014). LysM domain sequences were aligned using mafft (global setting) (Katoh et al., 2002).
Affinity precipitation assays
These assays were performed as described by van den Burg et al. (2016) with 100 μg/mL Vd2LysM. The protein was incubated at room temperature for 1 h with 3 mg of insoluble polysaccharides with gentle rocking.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Disqualification of two previously identified Verticillium dahliae LysM effector genes. Two originally identified VdLysM effector genes are not predicted correctly. The initially predicted gene model of LysM effector gene VDAG_03096 is not supported by the mapping of RNA sequencing reads (in red). Reads map to predicted introns (in yellow), whereas some coding parts of the gene (in green), including the LysM domain that constitutes only a small part of the predicted protein, is not supported by reads. SMART prediction using the amino acid sequence encoded by VDAG_06426 reveals the absence of a signal peptide and the presence of a zinc finger domain. Also in this case, the LysM domain constitutes only a small portion of the predicted protein.
Fig. S2 Sequence polymorphisms of core VdLysM effectors in the Verticillium dahliae population. Synonymous and non‐synonymous polymorphisms and their occurrence in 19 V. dahliae strains are indicated per VdLysM effector gene, and nucleotide and amino acid positions of polymorphisms are indicated, respectively.
Fig. S3 Glycan array analysis of Pichia pastoris‐produced Vd2LysM and Ecp6. Relative fluorescence (RFU, relative fluorescence unit) on scanning of a glycan array that contains probes for 406 glycans after hybridization with Vd2LysM and with Ecp6 as a control. Only Ecp6 hybridizes to the array, and only to 170–172, representing (GlcNAc)6, (GlcNAc)5 and (GlcNAc)3, respectively.
Table S1 Verticillium dahliae strains used in this study.
Table S2 Primers used in this study.
Acknowledgements
M.F.S. and B.P.H.J.T. were supported by a Veni and a Vici grant, respectively, from the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO). The authors declare no conflicts of interest.
References
- Akcapinar, G.B. , Kappel, L. , Sezerman, O.U. and Seidl‐Seiboth, V. (2015) Molecular diversity of LysM carbohydrate‐binding motifs in fungi. Curr. Genet. 61, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , van Esse, H.P. , Vossen, J.H. , de Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J.E. , van den Berg, G.C.M. , Borras‐Hidalgo, O. , Dekker, H.L. , de Koster, C.G. , de Wit, P.J.G.M. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Bowman, M. and Free, S.J. (2006) The structure and synthesis of the fungal cell wall. BioEssays, 28, 799–808. [DOI] [PubMed] [Google Scholar]
- Buist, G. , Steen, A. , Kok, J. and Kuipers, O.P. (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Harrison, S.J. , Joosten, M.H.A.J. , Vervoort, J. and de Wit, P.J.G.M. (2016) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant–Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Liang, Y. , Tanaka, K. , Nguyen, C.T. , Jedrzejczak, R.P. , Joachimiak, A. and Stacey, G. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin‐induced complex with related kinase CERK1. Elife, 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D. , Mesarich, C.H. and Thomma, B.P.H.J. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , de Jonge, R. and Thomma, B.P.H.J. (2012) The transcriptome of Verticillium dahliae‐infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signal. Behav. 7, 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Datema, E. , van den Berg, G.C.M. , Janssen, A. , Wittenberg, A.H.J. and Thomma, B.P.H.J. (2015) Single‐molecule real‐time sequencing combined with optical mapping yields completely finished fungal genome. mBio, 6, e00936–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Shi‐Kunne, X. , Pauper, M. , van den Berg, G.C.M. , Wittenberg, A.H.J. and Thomma, B.P.H.J. (2016) Transposons passively and actively contribute to evolution of the two‐speed genome of a fungal pathogen. Genome Res, 26, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. , Regenass, M. and Boller, T. (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316. [Google Scholar]
- Fradin, E.F. and Thomma, B.P.H.J. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Juarez Ayala, J.C. , Castroverde, C.C.M. , Nazar, R.N. , Robb, J. , Liu, C.‐M. and Thomma, B.P.H.J. (2009) Genetic dissection of Verticillium wilt resistance mediated 1 by tomato Ve1. Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen, R.J.N. , Andersson, J.A. , Kristensen, M.B. and Giese, H. (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa, T. , Sakaguchi, A. , Nishizawa, Y. , Kouzai, Y. , Minami, E. , Yano, S. , Koga, H. , Meshi, T. and Nishimura, M. (2012) Surface α‐1,3‐glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8, e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, T.M. , Chunwongse, J. and Tanksley, S.D. (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13, 207–209. [Google Scholar]
- Garrison, E. and Marth, G. (2012) Haplotype‐based variant detection from short‐read sequencing. Preprint at arXiv:1207.3907v2 [q‐bio.GN].
- Gibriel, H.A. , Thomma, B.P. and Seidl, M.F. (2016) The age of effectors: genome‐based discovery and applications. Phytopathology, 106, 1206–1212. [DOI] [PubMed] [Google Scholar]
- Gijzen, M. and Nürnberger, T. (2006) Nep1‐like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- Gough, C. and Cullimore, J. (2011) Lipo‐chitooligosaccharide signalling in endosymbiotic plant–microbe interactions. Mol. Plant–Microbe Interact. 24, 867–878. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , BenM'Barek, S. , Dhillon, B. , Wittenberg, A.H.J. , Crane, C.F. , Hane, J.K. , Foster, A.J. , Van der Lee, T.A.J. , Grimwood, J. , Aerts, A. , Antoniw, J. , Bailey, A. , Bluhm, B. , Bowler, J. , Bristow, J. , van der Burgt, A. , Canto‐Canché, B. , Churchill, A.C. , Conde‐Ferràez, L. , Cools, H.J. , Coutinho, P.M. , Csukai, M. , Dehal, P. , De Wit, P. , Donzelli, B. , van de Geest, H.C. , van Ham, R.C. , Hammond‐Kosack, K.E. , Henrissat, B. , Kilian, A. , Kobayashi, A.K. , Koopmann, E. , Kourmpetis, Y. , Kuzniar, A. , Lindquist, E. , Lombard, V. , Maliepaard, C. , Martins, N. , Mehrabi, R. , Nap, J.P. , Ponomarenko, A. , Rudd, J.J. , Salamov, A. , Schmutz, J. , Schouten, H.J. , Shapiro, H. , Stergiopoulos, I. , Torriani, S.F. , Tu, H. , de Vries, R.P. , Waalwijk, C. , Ware, S.B. , Wiebenga, A. , Zwiers, L.H. , Oliver, R.P. , Grigoriev, I.V. and Kema, G.H. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. , Kombrink, A. , van den Berg, G.C.M. , Yadeta, K.A. and Thomma, B.P.H.J. (2013) Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. and Thomma, B.P.H.J. (2011) How filamentous pathogens co‐opt plants: the ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14, 400–406. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , Van der Krol, S. , Shibuya, N. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P.H.J. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. and Thomma, B.P.H.J. (2009) Fungal LysM effectors – extinguishers of host immunity? Trends Microbiol. 17, 151–157. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defence signalling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11 086–11 091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kumar, S. and Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimes, A. , Dobinson, K.F. , Thomma B.P.H.J. and Klosterman, S.J. (2015) Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium . Annu. Rev. Phytopathol. 53, 181–198. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Kombrink, A. (2012) Heterologous production of fungal effectors in Pichia pastoris . Methods Mol. Biol. 835, 209–217. [DOI] [PubMed] [Google Scholar]
- Kombrink, A. and Thomma, B.P.H.J. (2013) LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 9, e1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink, A. , Sánchez‐Vallet, A. and Thomma, B.P.H.J. (2011) The role of chitin detection in plant–pathogen interactions. Microbes Infect. 13, 1168–1176. [DOI] [PubMed] [Google Scholar]
- Kurtz, S. , Phillippy, A. , Delcher, A.L. , Smoot, M. , Shumway, M. , Antonescu, C. and Salzberg, S.L. (2004) Versatile and open software for comparing large genomes. Genome Biol. 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.‐J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P.H.J. and Rudd, J. (2011) Analysis of two in planta expressed LysM effector homologues from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant. Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren, W. , Pritchard, B. , Rios, D. , Chen, Y. , Flicek, P. and Cunningham, F. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics, 26, 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlak, T.A. , Kombrink, A. , Shinya, T. , Ryder, L.S. , Otomo, I. , Saitoh, H. , Terauchi, R. , Nishizawa, Y. , Shibuya, N. , Thomma, B.P.H.J. and Talbot, N.J. (2012) Effector‐mediated suppression of chitin‐triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell, 24, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 19 613–19 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A.R. and Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P. , Longden, I. and Bleasby, A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C. , Van't Klooster, J.W. , van der Hoorn, R.A.L. , Joosten, M.H.A.J. , Jones, J.D.G. and de Wit, P.J.G.M. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rovenich, H. , Boshoven, J.C. and Thomma, B.P.H.J. (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103. [DOI] [PubMed] [Google Scholar]
- Rovenich, H. , Zuccaro, A. and Thomma, B.P.H.J. (2016) Convergent evolution of filamentous microbes towards evasion of glycan‐triggered immunity. New Phytol. 212, 896–901. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Saleem‐Batcha, R. , Kombrink, A. , Hansen, G. , Valkenburg, D.‐J. , Thomma, B.P.H.J. and Mesters, J. (2013) Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife, 2, e00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Mesters, J.R. and Thomma, B.P.H.J. (2015) The battle for chitin recognition in plant–microbe interactions. FEMS Microbiol Rev. 39, 171–183. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. and Thomma, B.P.H.J. (2013) Verticillium dahliae Sge1 differentially regulates expression of candidate effector genes. Mol. Plant–Microbe Interact. 26, 249–256. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , van Esse, H.P. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B.P.H.J. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , Boshoven, J.C. , Salas, O. , Bowler, K. , Tohidul Islam, M. , Keykha Saber, M. , van den Berg, G.C.M. , Bar‐Peled, M. and Thomma, B.P.H.J. (2016) Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus Verticillium dahliae . Mol. Plant Pathol. doi: 10.1111/mpp.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumbaum, A. , Mauch, F. , Vogeli, U. and Boller, T. (1986) Plant chitinases are potent inhibitors of fungal growth. Nature, 324, 365–367. [Google Scholar]
- Schultz, J. , Milpetz, F. , Bork, P. and Ponting, C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl‐Seiboth, V. , Zach, S. , Frischmann, A. , Spadiut, O. , Dietsch, C. , Herwig, C. , Ruth, C. , Rodler, A. , Jungbauer, A. and Kubicel, P. (2013) Spore germination of Trichoderma atroviride is inhibited by its LysM protein TAL6. FEBS J. 280, 1226–1236. [DOI] [PubMed] [Google Scholar]
- Shibuya, N. , Kaku, H. , Kuchitsu, K. and Maliarik, M.J. (1993) Identification of a novel high‐affinity binding site for N‐acetylchitooligosaccharide elicitor in the membrane fraction from suspension‐cultured rice cells. FEBS Lett. 329, 75–78. [DOI] [PubMed] [Google Scholar]
- Song, Y. , Zhang, Z. , Seidl, M.F. , Majer, A. , Jakse, J. , Javornik, B. and Thomma, B.P.H.J. (2016) Broad taxonomic characterization of Verticillium wilt resistance genes reveals ancient origin of the tomato Ve1 immune receptor. Mol. Plant Pathol. doi: 10.1111/mpp.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara, H. , Hacquard, S. , Kombrink, A. , Hughes, B. , Halder, V. , Robin, G.P. , Hiruma, K. , Neumann, U. , Shinya, T. , Kombrink, E. , Shibuya, N. , Thomma, B.P.H.J. and O'Connell, R.J. (2016) Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin‐triggered plant immunity. New Phytol. 211, 1323–1337. [DOI] [PubMed] [Google Scholar]
- Wan, J.R. , Zhang, X.C. , Neece, D. , Ramonell, K.M. , Clough, S. , Kim, S.Y. , Stacey, M.G. and Stacey, G. (2008) A LysM receptor‐like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell, 20, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhof, L.B. , Wilbers, R.H.P. , Roosien, J. , van de Velde, J. , Goverse, A. , Bakker, J. and Schots, A. (2012) 3D domain swapping causes extensive multimerisation of human interleukin‐10 when expressed in planta . PLoS One, 7, e46460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, T.J. , Clements, J. and Finn, R.D. (2014) Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics, 15, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Fradin, E.F. , de Jonge, R. , van Esse, H.P. , Smit, P. , Liu, C.M. and Thomma, B.P.H.J. (2013) Optimized agroinfiltration and virus‐induced gene silencing to study Ve1‐mediated Verticillium resistance in tobacco. Mol. Plant–Microbe Interact. 26, 182–190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Disqualification of two previously identified Verticillium dahliae LysM effector genes. Two originally identified VdLysM effector genes are not predicted correctly. The initially predicted gene model of LysM effector gene VDAG_03096 is not supported by the mapping of RNA sequencing reads (in red). Reads map to predicted introns (in yellow), whereas some coding parts of the gene (in green), including the LysM domain that constitutes only a small part of the predicted protein, is not supported by reads. SMART prediction using the amino acid sequence encoded by VDAG_06426 reveals the absence of a signal peptide and the presence of a zinc finger domain. Also in this case, the LysM domain constitutes only a small portion of the predicted protein.
Fig. S2 Sequence polymorphisms of core VdLysM effectors in the Verticillium dahliae population. Synonymous and non‐synonymous polymorphisms and their occurrence in 19 V. dahliae strains are indicated per VdLysM effector gene, and nucleotide and amino acid positions of polymorphisms are indicated, respectively.
Fig. S3 Glycan array analysis of Pichia pastoris‐produced Vd2LysM and Ecp6. Relative fluorescence (RFU, relative fluorescence unit) on scanning of a glycan array that contains probes for 406 glycans after hybridization with Vd2LysM and with Ecp6 as a control. Only Ecp6 hybridizes to the array, and only to 170–172, representing (GlcNAc)6, (GlcNAc)5 and (GlcNAc)3, respectively.
Table S1 Verticillium dahliae strains used in this study.
Table S2 Primers used in this study.


