Summary
In this article, we describe the presence of genes encoding close homologues of an endogenous plant peptide, rapid alkalinization factor (RALF), within the genomes of 26 species of phytopathogenic fungi. Members of the RALF family are key growth factors in plants, and the sequence of the RALF active region is well conserved between plant and fungal proteins. RALF1‐like sequences were observed in most cases; however, RALF27‐like sequences were present in the Sphaerulina musiva and Septoria populicola genomes. These two species are pathogens of poplar and, interestingly, the closest relative to their respective RALF genes is a poplar RALF27‐like sequence. RALF peptides control cellular expansion during plant development, but were originally defined on the basis of their ability to induce rapid alkalinization in tobacco cell cultures. To test whether the fungal RALF peptides were biologically active in plants, we synthesized RALF peptides corresponding to those encoded by two sequenced genomes of the tomato pathogen Fusarium oxysporum f. sp. lycopersici. One of these peptides inhibited the growth of tomato seedlings and elicited responses in tomato and Nicotiana benthamiana typical of endogenous plant RALF peptides (reactive oxygen species burst, induced alkalinization and mitogen‐activated protein kinase activation). Gene expression analysis confirmed that a RALF‐encoding gene in F. oxysporum f. sp. lycopersici was expressed during infection on tomato. However, a subsequent reverse genetics approach revealed that the RALF peptide was not required by F. oxysporum f. sp. lycopersici for infection on tomato roots. This study has demonstrated the presence of functionally active RALF peptides encoded within phytopathogens that harbour an as yet undetermined role in plant–pathogen interactions.
Keywords: Fusarium, phytopathogen effectors, rapid alkalinization factor (RALF)
Introduction
Fungal phytopathogens are a major threat to all plant ecosystems, both natural and agricultural (Stukenbrock and McDonald, 2008). Therefore, it is crucial to understand how fungal phytopathogens have co‐evolved with their plant hosts to enable better management of crop losses and preserve natural biodiversity. The mechanisms by which pathogenic fungi manipulate their plant hosts are complex and diverse. It is now recognized that pathogen effector molecules (proteins or secondary metabolites) play a significant role in manipulating host plants to facilitate infection (de Jonge et al., 2011). For example, biotrophic fungi produce effectors that allow the fungus to elude or manipulate host defences. Conversely, necrotrophic fungi produce effectors that induce host cell necrosis, thereby producing dead plant tissue for the fungus to colonize. Some effectors are vital for host infection; others are not strictly necessary, but assist in the full exploitation of the host resources (De Wit et al., 2009). Most fungal effector genes remain undiscovered, but the increasing availability of fully sequenced fungal genomes has enabled new methods for the identification of putative effector genes using comparative genomics and candidate gene prediction tools.
The advent of next‐generation sequencing (NGS) has dramatically increased the number of whole‐genome sequences available for organisms from all kingdoms of life. Access to these data has allowed researchers to compare genes from disparate organisms (Thynne et al., 2015b) and has provided significant insights into the genes involved in phytopathogen emergence and niche specialization (Gardiner et al., 2012; Klosterman et al., 2011), including the identification of pathogen genes related to host genes, either through horizontal gene transfer (HGT) or convergent evolution. By studying the molecular evolution of these genes, we can develop hypotheses about their function, including their potential roles as effectors.
Horizontal transfer of pathogenicity genes between fungi is a well‐known phenomenon and an active area of research (Friesen et al., 2006; Gardiner et al., 2012; Klosterman et al., 2011; Richards et al., 2011; Thynne et al., 2015b), although the exact mechanisms of genetic transfer are poorly understood. Recent studies have also demonstrated a role for plant to microorganism gene transfer leading to increased virulence. For example, the secreted proteinaceous Verticillium effector Ave1, which promotes pathogen virulence on the plant host, is hypothesized to have a plant origin (de Jonge et al., 2012). Other examples exist in which plant‐interacting organisms have apparently acquired plant growth and cell structure alteration factors. For example, C‐terminally encoded peptides (CEPs) have been identified in the genome assemblies of phytopathogenic root knot nematodes (Bobay et al., 2013). Similarly, homologues of plant expansins have been identified in the genomes of a range of bacteria and fungi (Nikolaidis et al., 2014). CEPs and expansins are growth regulatory and plant cell‐loosening molecules, respectively (Cosgrove, 2000; Delay et al., 2013). These molecules have been postulated to assist in plant colonization, inhabitation and/or parasitization (Soanes and Richards, 2014). Pathogen genes of plant origin are therefore promising targets for further study as potential virulence factors.
The plant rapid alkalinization factor (RALF) genes encode secreted peptides that were first identified through their ability to trigger a rapid increase in extracellular pH when added to plant cell suspensions (Pearce et al., 2001). RALF peptides have roles in the development and regulation of plant roots, root hairs, legume nodules and pollen tubes (Murphy and De Smet, 2014; Pearce et al., 2010). Subsequent in silico analyses of the Arabidopsis thaliana genome have identified 34 genes within the RALF family (Olsen et al., 2002). These genes were named RALF1 to RALF34, and all homologues since identified in other species have been named according to their similarities to members of the A. thaliana family. Functional studies on a tobacco RALF peptide have revealed that proteolytic cleavage of a larger precursor protein generates a 49‐amino‐acid active peptide (Fig. 1) (Srivastava et al., 2009). Mutational analysis has identified several motifs essential for RALF activity, including a YISY motif and four conserved cysteine residues that are required for disulphide bonding (Pearce et al., 2010). Recently, the Arabidopsis receptor for RALF1 was identified as the plasma membrane receptor kinase FERONIA (FER). Activation of FER results in phosphorylation of plasma membrane H+‐ATPase 2 at serine‐899, causing inhibition of proton transport (Haruta et al., 2014).
Figure 1.

Sequence of the Nicotiania tabacum rapid alkalinization factor (RALF) peptide (NtRALF). The underlined region represents the predicted signal peptide and the arrow indicates the point of cleavage by a subtilisin‐like serine protease (Srivastava et al., 2009). The sequence in red represents the active RALF peptide. Asterisks (*) denote the essential YSIY motif. Cysteine residues required for disulphide bond formation and RALF peptide activity are denoted by hashtags (#) (Pearce et al., 2010).
In this study, we describe the distribution of genes encoding RALF homologues amongst a diverse range of plant‐pathogenic fungal species and RALF‐like hybrid genes in a limited number of bacterial species. Functional testing of synthetic versions of two fungal RALF peptides showed that plants perceive and respond to the fungal peptides. These data suggest that these fungal RALF genes enable pathogenic fungi to manipulate host plant physiology.
Results
Distribution and variation of RALF peptide sequences in the fungal and bacterial kingdoms
The search programs blastp and tblastn were used to screen publicly available fungal genomes for homologues of plant RALF proteins using the full‐length protein sequences of Arabidopsis thaliana plant RALF genes 1–34 (Table 1). Using this approach, 26 different species of fungi were found to possess genes encoding the processed RALF peptide domain (Fig. 2, Table 1). These include the class Pucciniomycetes of the Basidiomycota and the classes Dothideomycetes and Sordariomycetes of the Ascomycota. All of the fungi found to possess RALF homologues are plant pathogens. Both biotrophic and necrotrophic fungi are represented in this list. Phylogenetic analysis of plant and fungal RALF peptide sequences showed that the fungal RALF homologues are interspersed amongst the plant RALFs (Fig. S1, see Supporting Information). Many of the fungal homologues resolved with the plant RALF1 proteins (Fig. S1). The Pseudocercospora fijiensis RALF homologue clustered with RALF homologues of Musa acuminata malaccensis. However, this was only supported with a low posterior probability of 33%. The Septoria populicola and Sphaerulina musiva RALF homologues clustered with the plant RALF27‐like proteins with 100% posterior support (Figs 2 and S1). The best blastp hit to a complete plant protein for both fungal sequences was a RALF27‐like protein from Populus trichocarpa (E‐values of 3e‐14 and 4e‐13); however, this was only weakly supported by the gene tree analysis, with a posterior probability of 50% (Figs 2 and S1). It should be noted that there was no detectable homology between the N‐terminal pro‐peptide domains of the plant and fungal RALF sequences.
Table 1.
Fungal species identified as containing rapid alkalinization factor (RALF) homologues.
| Fungi containing RALF sequences | JGI/NCBI/GenBank/Ensembl/UniProt sequence identifier |
|---|---|
| Ascomycetes | |
| Dothideomycetes | |
| Botryosphaeriaceae | |
| Botryosphaeriaceae spp. | See Table 3 |
| Corynesporascaceae | |
| Corynespora cassiicola | jgi|Corca1|631921 |
| Leptosphaeriaceae | |
| Leptosphaeria biglobosa | GenBank: FO905662.1 |
| Leptosphaeria maculans | NCBI Reference Sequence: XP_003844940.1 |
| Phoma tracheiphila | jgi|Photr1|406253 |
| Mycosphaerellaceae | |
| Pseudocercospora fijiensis | NCBI Reference Sequence: XP_007921540.1 |
| Septoria populicola | jgi|Seppo1|99826 |
| Sphaerulina musiva | GenBank: EMF16709.1 |
| Pleosporaceae | |
| Pyrenophora tritici‐repentis | NCBI Reference Sequence: XP_001937123.1 |
| Pyrenophora teres | jgi|Pyrtt1|194499:17017‐24160 |
| Setosphaeria turcica | jgi|Settu1|scafd_12:1053528‐1053731 |
| Teratosphaeriaceae | |
| Teratosphaeria nubilosa | jgi|Ternu1|214030 |
| Sordariomycetes | |
| Glomerellaceae | |
| Colletotrichum higginsianum | GenBank: CCF44719.1 |
| Plectosphaerellaceae | |
| Verticillium dahliae | GenBank: CP009075.1| |
| Verticillium alfalfae | |
| Nectriaceae | |
| Fusarium spp. | See Table 2 |
| Basidiomycetes | |
| Pucciniomycetes | |
| Melampsoraceae | |
| Melampsora lini | jgi|Melli1|sc_1450:29870‐30703 |
| Melampsora larici‐populina | NCBI Reference Sequence: XP_007408986.1 |
| Cronartiaceae | |
| Cronartium quercuum | jgi|Croqu1|50147 |
Figure 2.
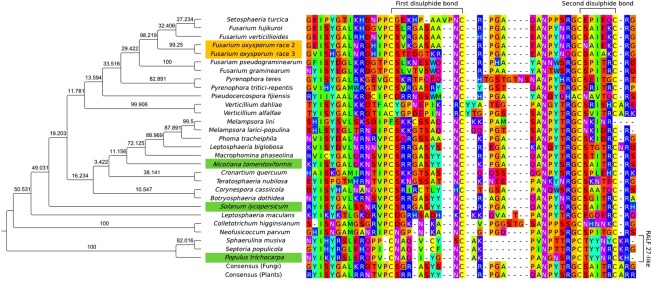
Identification, alignment and analysis of the rapid alkalinization factor (RALF) domain in fungi and selected plants. Aligned RALF peptide domain sequences from five isolates of Fusarium oxysporum, 24 other fungal species and selected plants (highlighted in green) are presented. Cysteine pairs expected to form disulphide bonds are labelled at the top. The Melampsora lini RALF may have an alternative disulphide bond, marked with a dotted line. Support values on each branch refer to the percentage posterior probability of each clade. RALF sequences from two species of fungi, Septoria populicola and Sphaerulina musiva, clustered with plant RALF27‐like genes with 100% posterior support (labelled on the right). The plant RALF27‐like sequences are from Populus trichocarpa (gi:566213100) and Arabidopsis thaliana (gi:15230083). The other plant RALF sequences are from Solanum lycopersicum (gi:460366641), corresponding to the peptide used here in seedling and cell culture assays, and from Nicotiana tomentosiformis (gi:697149758), which is identical to the original RALF sequence reported from N. tabacum and A. thaliana RALF1 (gi:15218637). The divergent RALF sequences of Fusarium oxysporum isolates 4287 and MN25 are highlighted in gold. Individual amino acids are coloured using the Taylor scheme (Taylor, 1997).
Analysis of the Fusarium species showed evidence of four divergent groups of RALF homologues (Fig. 3, Table 2), which were designated as Groups I–IV. Members of Group I are present in all isolates of F. oxysporum (Fox) that have been sequenced to date, although the Group I RALF genes of two Fox isolates encode sequence variations suggesting loss of function (a premature stop codon in the human pathology isolate FOSC 3‐a, and a tyrosine in place of a conserved cysteine in the non‐pathogen Fo47). Group I can be subdivided into two subgroups A and B, with subform IB also present in F. verticillioides, F. fujikuroi and F. circinata (Fig. 3). Interestingly, subform IA distinguishes two tomato pathogens, F. oxysporum f. sp. lycopersici (Fol) isolate MN25 and F. oxysporum f. sp. radicis‐lycopersici (Forl) from a third Fol isolate 4287. The divergent Fol homologues, here designated RALF‐B (from 4287) and RALF‐C (from MN25), were chosen for further study.
Figure 3.
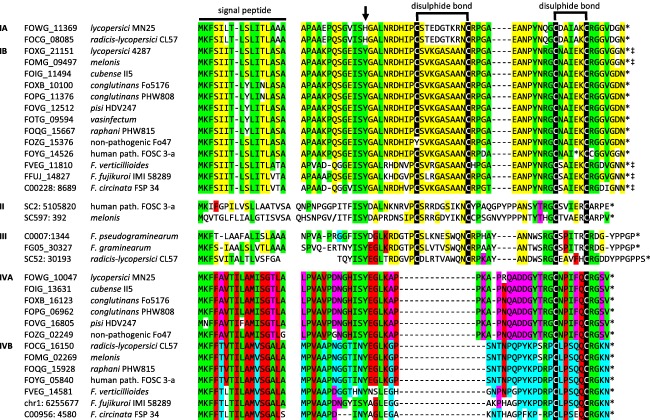
Multiple sequence alignments of Fusarium rapid alkalinization factor (RALF) and RALF‐like homologues. Group I comprises a large family of RALF homologues from Fusarium verticillioides, F. fujikuroi, F. circinata and various formae speciales of F. oxyporum (Fox). It should be noted that the Group IA RALF homologues FOWG_113369 F. oxyporum f. sp. lycopersici 4287 and FOCG_08085 F. oxyporum f. sp. radicis‐lycopersici CL57 are very similar to one another, but divergent from the Group IB RALF sequences at specific points within the predicted mature RALF peptide, notably the sequence between the first two cysteines and a histidine substitution in place of a conserved tyrosine (arrowed). Four cysteine residues involved in disulphide bonding are conserved in all isolates, except FOZG_15376, where the first cysteine is swapped at position 39 to a tyrosine. FOYG_14526 has an early stop codon at site 67, prior to the final cysteine at site 69. Group II comprises a small family of divergent RALF homologues from F. oxyporum f. sp. melonis and a human pathology isolate of Fox. It should be noted that the f. sp. melonis homologue has an apparent frameshift indicated by a slash (/) that could be the consequence of a sequencing error rather than a frameshift mutation. Group III comprises a small family of divergent RALF homologues from F. oxyporum f. sp. radicis‐lycopersici CL57, F. graminearum and F. pseudograminearum. Group IV comprises a large family of RALF homologues that have an internal deletion relative to the RALF homologues shown in Groups I, II and III. This deletion includes the cysteine residues that form the first disulphide bond indicated above the alignments. It should be noted that this family contains two apparently mutually exclusive subforms, as can be seen from the conserved residues highlighted in pink and blue. Conserved cysteines are highlighted in black. Other residues conserved between Groups I and IV, and, to some extent, between Groups II and III are highlighted in green. Residues conserved within Group I and, to some extent, Groups II and III are highlighted in yellow. Residues conserved within Group IV and, to some extent, Groups II and III are highlighted in red. Residues conserved within Group IV, but differing between Groups IVA and IVB, are highlighted in pink and blue. Signal peptide cleavage sites were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Disulphide bond locations are based on Pearce et al. (2001). ‡Transcript and protein sequences predicted using the same splice acceptor site as predicted for all of the other sequences in this group. *Stop codon.
Table 2.
Fusarium isolates and their rapid alkalinization factor (RALF) homologue IDs or DNA sequence origins.
| Fusarium isolate | Protein ID/origin |
|---|---|
| Fusarium fujikuroi IMI 58289 | FFUJ_14827 chromosome 1: 6255677–6255856 |
| Fusarium graminearum PH‐1 (NRRL 31084) | FG05_30327 |
| Fusarium oxysporum f. sp. conglutinans PHW808 (NRRL 54008) |
FOPG_11376 FOPG_06962 |
| Fusarium oxysporum f. sp. conglutinans Fo5176 |
FOXB_10100 FOXB_16123 |
| Fusarium oxysporum f. sp. cubense TR4 II5 (NRRL 54006) |
FOIG_11494 FOIG_13631 |
| Fusarium oxysporum f. sp. lycopersici 4287 (NRRL 34936) | FOXG_21151 |
| Fusarium oxysporum f. sp. lycopersici MN25 (NRRL 54003) |
FOWG_11369 FOWG_10047 |
| Fusarium oxysporum f. sp. melonis (NRRL 26406) |
FOMG_09497 FOMG_02269 Super Contig 597: 392–624 |
| Fusarium oxysporum f. sp. pisi HDV247 (NRRL 37622) |
FOVG_12512 FOVG_16805 |
| Fusarium oxysporum f. sp. radicis‐lycopersici CL57 (NRRL 26381) |
FOCG_08085 FOCG_16150 Super Contig 52: 30193–30411 |
| Fusarium oxysporum f. sp. raphani PHW815 (NRRL 54005) |
FOQG_15667 FOQG_15928 |
| Fusarium oxysporum f. sp. vasinfectum (NRRL 25433) | FOTG_09594 |
| Fusarium oxysporum human path. isolate FOSC 3‐a (NRRL 32931) |
FOYG_14526 FOYG_05840 |
| Fusarium oxysporum non‐pathogenic Fo47 (NRRL 54002) |
FOZG_15376 FOZG_02249 |
| Fusarium pseudograminearum CS3096 | Contig 007: 1344–1371 |
| Fusarium verticillioides 7600 (NRRL 20956) |
FVEG_11810 FVEG_14581 |
| Fusarium circinata FSP34 |
Contig 00228: 8689–8910 Contig 00956: 4580–4747 |
The Group II family of RALF homologues has only two members, one from Fox FOSC 3‐a, which could potentially complement the aberrant FOSC 3‐a Group I homologue, and the other from F. oxysporum f. sp. melonis, which contains an apparent frameshift mutation (Fig. 3). The Group III family of RALF homologues has only three members, one from Forl CL57 and the others from F. graminearum and F. pseudograminearum (Fig. 3).
Genes encoding a fourth family of RALF‐like proteins (Group IV) were also found in Fusarium. These differ from the three families described above by a central deletion encompassing the conserved cysteines that form the first disulphide bond (Fig. 3). Nevertheless, the sequence conservation within this family suggests the retention of function, but whether they possess the same function as the fungal RALF homologues described above requires further investigation. This gene family can also be divided into two sequence‐divergent subgroups that seem to be distributed in a mutually exclusive fashion between different formae speciales of Fox, although two (vasinfectum and Fol 4287) do not possess either form. Interestingly, Group IV genes seem to be absent from plants and other fungal genomes, suggesting that they are unique to Fusarium.
Notably, all four possible combinations of the Group I and IV RALF variants occur in Fox, indicating a polyphyletic origin for at least some of these gene combinations. This is a curious finding given that these genes also occur in F. verticillioides, F. fujikuroi and F. circinata, suggesting that they are located on the core conserved set of non‐mobile chromosomes in Fusarium, rather than the mobile lineage‐specific chromosomes that seem to determine host specificity in Fox (Ma et al., 2010). Indeed, the RALF‐B gene is encoded by core chromosome 12 in Fol 4287.
Subsequent to the preliminary database search, we annotated a number of new RALF sequences within the genomes of members of the Botryosphaeriaceae. Similarly to the Fusarium spp., members of the Botryosphaeriaceae family showed evidence of their own three divergent groups of RALF homologues (Table 3, Fig. S2, see Supporting Information). These three groups were designated BI–BIII. Amino acid variations between these three divergent groups are highlighted in Fig. S2A. BI contains an amino acid string (‘ISNGAM’) apparently rare among both fungal RALFs (only non‐Botryosphaeriaceae RALF is in Colletotrichum higginsianum) and plant RALFs (only observed in Brachypodium distachyon). BII is similar in amino acid composition to the Fusarium Group I. BIII is similar, and perhaps divergent from, BII. Botryosphaeria dothidea contains two RALF homologues (BI and BII). Botryosphaeria dothidea's BI RALF homologue was previously un‐annotated. Diplodia seriata contains one RALF homologue (BIII). Neofusicoccum parvum contains one RALF homologue (BI). Macrophomina phaseolina contains four RALF homologues (BI, two BIIs and BIII). All but one of M. phaseolina's RALFs were previously un‐annotated. With four predicted RALF peptides, M. phaseolina has the highest number of predicted fungal RALF peptides identified in this study. One of the BII RALF peptides shares a conserved RALF peptide domain with the mature RALF peptide from soybean species (Glycine max and Glycine soja). For this region, these species have a 100% amino acid identity. Variation among these species is found in the N‐terminal and C‐terminal regions. In addition, common bean (Phaseolus vulgaris) has a 98% sequence similarity for this conserved region, and mung bean (Vigna radiata) has a 100% sequence similarity for 97% of the query coverage. The only species of Botryosphaeriaceae analysed in this study not to contain RALF peptides are three closely related species: Eutiarosporella darliae, E. pseudodarliae and E. tritici‐australis. These are the causal agents of white grain disorder (WGD) in wheat (Thynne et al., 2015a).
Table 3.
Botryosphaeriaceae isolates and their rapid alkalinization factor (RALF) homologue IDs or DNA sequence origins.
| Botryosphaeriaceae isolate | Protein ID/origin |
|---|---|
| Botryosphaeria dothidea | jgi|Botdo1_1|289944 |
| Node_8012: | |
| (92987–93247) | |
| Diplodia seriata | gi|821070376 |
| Macrophomina phaseolina | gi|407928039 |
| Contig_00502: (91876–92412) | |
| Contig_00151: (16464–16099) | |
| Neofusicoccum parvum | Contig_00254: (124817–124602) |
| gi|615408450 |
We re‐performed the database searches using blastp, this time searching the bacterial portal of the National Center for Biotechnology Information (NCBI) database. We identified certain members of Actinobacteria that possess putative secreted proteins with an incorporated RALF peptide domain motif (Fig. S3, see Supporting Information). In both fungal and plant RALFs, this particular motif is located at the C‐terminus of the mature peptide. Similarly, in these bacterial proteins, the RALF domain motif is at the C‐terminus of the protein. Unlike plant and fungal RALFs, the bacterial RALF domain‐containing proteins also contain a domain homologous to the S1 pertussis toxin subunit. These bacterial species include a number of plant‐pathogenic species, e.g. Streptomyces acidiscabies (a pathogen of potatoes) (Huguet‐Tapia et al., 2012).
Tomato seedling growth and development are severely inhibited by a synthetic RALF‐B peptide
Endogenous RALF peptides arrest the growth and development of plant roots (Pearce et al., 2010). To determine whether fungal RALF peptides share this property, we synthesized RALF peptides based on RALF2 of tomato (Sl‐RALF, NCBI GenBank GI:460366641) and those of Fol race 2 isolate 4287 and race 3 isolate MN25 (RALF‐B and RALF‐C, respectively) (Figs 2 and S4, see Supporting Information). Fol 4287 was the first fully sequenced Fox isolate, and MN25 is a representative of race 3, which appeared in the 1980s and overcame resistance to races 1 and 2 (Bournival et al., 1989). A non‐functional mutant of Sl‐RALF lacking the required YISY motif (Sl‐RALFΔ) was used as a negative control. Germinating tomato seeds were grown in medium containing each RALF peptide at 10 μm (Fig. 4), or with no added peptide. Untreated seedlings remained healthy and showed normal development of roots and shoots. Seedlings grown in medium containing Sl‐RALF developed less biomass than the untreated controls, but this was not significantly different from the Sl‐RALFΔ treatment (Fig. 4A). However, the Sl‐RALF‐treated seedlings were developmentally stunted with minimal root formation compared with the Sl‐RALFΔ‐treated seedlings (Fig. 4B). The growth and root development of seedlings treated with synthetic RALF‐B peptide were severely inhibited relative to mock‐ and Sl‐RALFΔ‐treated seedlings. This was reflected in their significantly lower average weights. In contrast, the synthetic RALF‐C peptide did not inhibit root growth. There was no significant difference in average biomass between seedlings treated with RALF‐C and the mock and Sl‐RALFΔ treatments. Similarly, the seedlings appeared healthy with well‐developed root systems.
Figure 4.
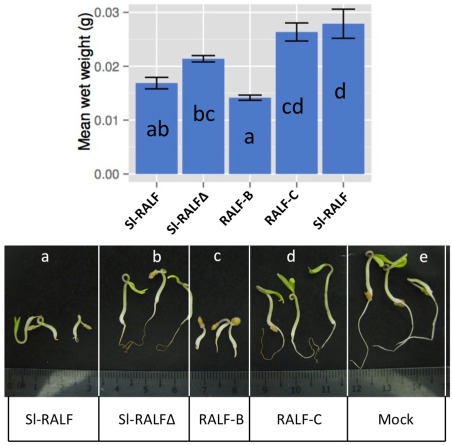
Effect of rapid alkalinization factor (RALF) peptides on germinating tomato seedlings. (A) Mean weights were calculated from whole seedlings grown in medium containing 10 μm synthetic RALF peptides (n = 9). The letter codes a–d indicate statistical significance: treatments which do not share any letter codes are significantly different (P < 0.05, corrected for multiple testing). Error bars correspond to standard error. (B) Digital images of representative samples in (A): a, Sl‐RALF; b, Sl‐RALFΔ; c, RALF‐B; d, RALF‐C; e, mock treatment.
The RALF‐B peptide elicits recognized plant RALF responses
The receptor for the Arabidopsis RALF1 peptide is the CrRLK1L family receptor FER (Haruta et al., 2014). As FER function is associated with the accumulation of extracellular reactive oxygen species (ROS) (Wolf and Höfte, 2014), and this is also a typical consequence of plant defence activation, we asked whether synthetic Fol RALF peptides could also elicit ROS production. Treatment of leaf discs of tomato and another solanaceous species, Nicotiana benthamiana, with Sl‐RALF at 10 μm induced a ROS burst. The ROS burst elicited by Sl‐RALF was stronger in tomato than in N. benthamiana. In contrast, RALF‐B (but not RALF‐C) induced a massive ROS burst in both solanaceous species, only minutes after treatment. RALF‐C was not active in these assays. Neither the mock nor Sl‐RALFΔ treatments elicited a significant ROS response in the leaves of either species.
The dose response for fungal RALF peptides was also determined. As the ROS bursts induced by RALF‐B were comparable between tomato and N. benthamiana, the activity of this peptide was tested in N. benthamiana. RALF‐B was active down to a concentration of 10 nm, with progressive increases in ROS output observed up to 500 nm (Fig. S5, see Supporting Information). In contrast, ROS output subsequent to increasing concentrations of RALF‐C did not exceed control levels, providing further evidence that the peptide does not elicit a typical RALF response in plants.
The ability of RALF‐B to induce alkalinization was also determined. RALF peptides were originally named as such because of their strong and rapid alkalinization of tobacco suspension‐cultured cells (Pearce et al., 2001). To test the ability of RALF‐B to induce alkalinization, the peptide was incubated with cut leaf strips from both tomato and N. benthamiana. For both plants, RALF‐B elicited an alkalinization response not significantly different from that of the tomato RALF peptide (Fig. 5C,D).
Figure 5.
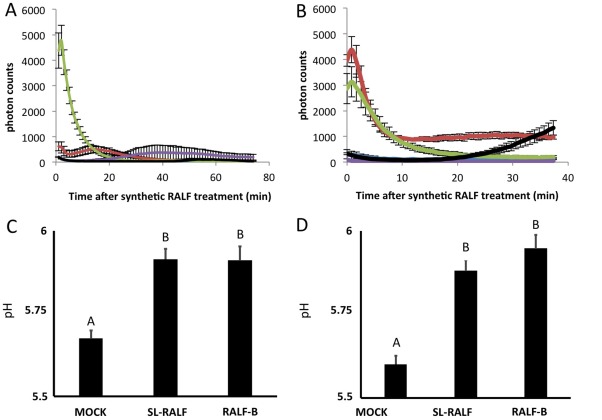
Synthetic rapid alkalinization factor (RALF) peptides induce reactive oxygen species (ROS) production and alkalinization in Nicotiana benthamiana and tomato leaves. The ROS burst over time after treatment with 10 μm of each synthetic RALF peptide in N. benthamiana (A) and tomato (B) leaves. (C, D) Induction of alkalinization in N. benthamiana and tomato, respectively, by RALF‐B and Sl‐RALF. For (A) and (B), line colours are as follows: dark blue, mock treatment; red, Sl‐RALF; green, RALF‐B; purple, RALF‐C; black, Sl‐RALFΔ.
Similarly, the activation of mitogen‐activated protein kinases (MAPKs), a known RALF response (Pearce et al., 2001), was also investigated. Peptide‐induced MAPK activation was measured in N. benthamiana leaf discs as confirmation of the variation of activity in response to the different peptides. Treatments with 10 µm Sl‐RALF or RALF‐C activated MAPK progressively, with greater activity detected at 15 min after peptide addition (Fig. S6, see Supporting Information). Interestingly, the addition of only 100 nm RALF‐B (i.e. 100 times less than that of Sl‐RALF) caused strong activation of MAPK that peaked at 5 min and showed a decline by 15 min. This mirrors the ROS profile arising from RALF‐B treatment. Overall, these data (ROS burst, alkalinization and MAPK activation) prove that plant cells perceive fungal RALF peptides.
The Fol RALF‐B gene is expressed during infection of tomato roots
If RALF‐B plays a role in fungal pathogenicity, it should be expressed during infection of tomato roots by Fol isolates carrying the RALF‐B gene. A reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis was carried out on roots of the susceptible tomato cultivar M82 infected with Fol isolate #1943, which carries the RALF‐B gene, at 3 and 6 days post‐inoculation (dpi). No RALF‐B expression was detected in samples from the roots of mock‐inoculated tomato plants or 5‐day‐old mycelia grown in vitro, but expression was detected in the 3‐ and 6‐dpi samples (Fig. 6). The FEM1 gene (Schoffelmeer et al., 2001) was used as a positive control for fungal gene expression. The RALF‐B primers flanked an intron, thus yielding different‐sized products for cDNA and genomic DNA (Fig. 6). No RALF‐B or FEM1 genomic DNA products were detected in cDNA samples (Fig. 6), and no RALF‐B or FEM1 products were detected in samples prepared without reverse transcriptase (not shown), indicating that the RALF‐B and FEM1 bands observed at 3 and 6 dpi are authentic RT‐PCR products. The absence of RALF‐B expression in mycelia grown in vitro suggests that RALF‐B expression is induced during plant infection, consistent with a potential role as an effector.
Figure 6.
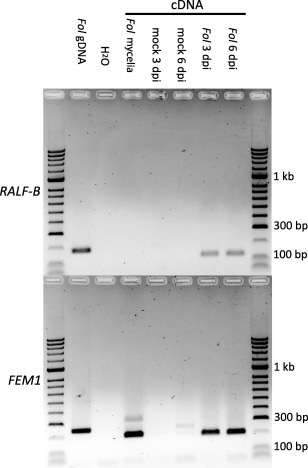
Reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis shows that RALF‐B is expressed during infection of tomato roots by Fusarium oxysporum f. sp. lycopersici (Fol). RT‐PCR analysis of RALF‐B (top gel image) shows bands (expected size, 123 bp) consistent with gene expression in Fol‐infected roots at 3 and 6 days post‐inoculation (dpi), but not in mock‐inoculated tomato plants or 5‐day‐old mycelia grown in vitro. RT‐PCR analysis of FEM1 (bottom gel image) shows bands (expected size, 201 bp) consistent with expression in infected roots at 3 and 6 dpi and in 5‐day‐old mycelia grown in vitro. The FEM1 control shows that cDNA synthesis was successful for the mycelial RNA sample and that the resulting cDNA sample supported PCR. PCR analysis of a Fol genomic DNA sample allowed the detection of the RALF‐B and FEM1 genes in Fol. The RALF‐B and FEM1 genomic DNA and cDNA products differ in size (expected sizes: 179 bp versus 123 bp for RALF‐B and 250 bp versus 201 bp for FEM1), allowing the detection of genomic DNA contamination in cDNA samples. No genomic RALF‐B or FEM1 products were detected in the cDNA samples.
Effect of knocking out the RALF‐B gene in Fol race 3
To test the role of the RALF‐B gene in Fol pathogenicity, knockout (ΔRALF) mutants were obtained by Agrobacterium‐mediated transformation, using a binary T‐DNA vector carrying a hygromycin resistance gene as a selectable marker for gene replacement and a thymidine kinase gene as a counter‐selectable marker against ectopic T‐DNA insertions (Fig. S7A, see Supporting Information). A total of 44 Fol transformants that showed resistance to hygromycin and 5′‐fluoro‐2′‐deoxyuridine were screened by PCR, confirming deletion of the RALF coding sequence in four transformants (named Δ2, Δ23, Δ24 and Δ31) (Fig. S7B).
The four Fol ΔRALF transformants were used to evaluate the effect of the loss of the RALF gene on virulence. The roots of 3‐week‐old plants of the tomato cultivar M82 (susceptible to Fol race 3) were wounded and subsequently inoculated with Fol race 3, a transformant with an ectopic insertion of the T‐DNA (i.e. no deletion of RALF gene) and the two ΔRALF transformants. The disease symptoms were recorded and disease scores were assigned as described by Rep et al. (2004), with scores of 0 = healthy plant to 4 = severely wilted plant.
No consistent significant differences in fungal virulence were found between wild‐type Fol race 3 and any of the transformants (Fig. S7D,E). ΔRALF transformants not the ectopic mutant strain tested. Therefore, using a wound infection assay, we found no strong evidence that the RALF‐B gene is involved in the pathogenicity of Fol race 3 on tomato.
Discussion
Here, we have reported that a number of fungal plant pathogens encode RALF‐like peptides in their genomes, and have provided evidence that at least a subset of these peptides can be perceived by plant cells. RALF peptides have high levels of conservation across a wide range of plant species, and the importance of their roles in the regulation of plant physiology is becoming increasingly apparent (Covey et al., 2010; Murphy and De Smet, 2014; Pearce et al., 2001). The suggestion that fungi deploy RALF peptides as effectors with the assumed consequence of improving fungal interaction with their hosts provides new perspectives both to the understanding of the pathogenic strategies of fungi and the evolutionary mechanisms underlying the acquisition of virulence.
Of the fungal genera carrying RALF homologues, the genus Fusarium has the most diverse array with four groups of RALF homologues (Fig. 3), with two subtypes in Groups I and IV. Each of the four groups has representatives in Fox, making Fox one of the most interesting fungal species for further study of fungal RALF peptides. However, it is possible that this apparent diversity is a result, in part, of the large number of Fusarium species, and Fox formae speciales in particular, whose genomes are publicly available. In bacteria, large numbers of genome sequences for a single species have catalysed the concept of the pan‐genome. The pan‐genome represents the entire genomic coding repertoire of all strains within a species, enabling intraspecific variation to be observed and assessed (Vernikos et al., 2015). This variation can be used to assist in explanations of varied lifestyles between isolates. The idea of the pan‐genome is becoming more common in fungi and the re‐sequencing of fungal genomes has begun to reveal the level of intraspecific variation (Thynne et al., 2015b). With many re‐sequenced Fox genomes and diversity in Fox lifestyles (non‐pathogenic, plant‐pathogenic, insect‐pathogenic and opportunistic human pathogen), the genetic determinants underpinning this diversity can be identified and analysed. Focusing on the RALF sequences identified here, considerable intraspecific variation was observed between different Fox genomes. Intriguingly, the two sequenced Fox isolates that are not reported plant pathogens (isolate FOSC 3‐a, which is a human clinical isolate, and isolate Fo47, which is non‐pathogenic) carry mutations in conserved cysteine residues that should render the RALF peptide biologically inert. Disulphide bonds formed between the two pairs of cysteines in RALF are essential for activity (Pearce et al., 2010). The RALF genes from these isolates appear to be undergoing pseudogenization, perhaps because they are not required outside of the plant‐pathogenic niche. In this study, the Fol RALF‐B variant elicited a strong response in the solanaceous plants tested. In contrast, there was a comparatively negligible response to Fol RALF‐C, with only weak activation of MAPK observed in treated N. benthamiana. Perhaps these differences are related to the swap of a conserved tyrosine with histidine early in the active peptide domain, as this tyrosine is important for RALF activity (Pearce et al., 2010). RALF‐C is found in Forl, which causes crown rot of tomato, and therefore has a different pathogenic lifestyle to Fol. It is possible that the RALF‐C variant plays a different role in Forl than does RALF‐B in Fol, but why then does Fol MN25 carry RALF‐C instead of RALF‐B like Fol 4287? It is possible that Fol MN25 carries other genetic factors, such as the Group IV RALF‐like genes present in Fol MN25, but absent from Fol 4287 (Fig. 3), which compensate for the absence of RALF‐B. These are questions for further study.
Although the RALF variation found within Fox does not reveal the origins of the RALF gene family in fungi, their polyphyletic nature does suggest genetic mobility within Fox. All four pairwise combinations of the two apparently mutually exclusive subtypes of Group I and Group IV RALF homologues (Fig. 3) were found in different formae speciales of Fox, suggesting that one or both of these genes may be mobile, i.e. transmitted horizontally. This opens up yet another area for further investigation.
Using a synthetic RALF‐B peptide, we demonstrated that a fungal RALF peptide can be perceived by plants. This is a highly significant result because it suggests that pathogenic fungi hijack endogenous plant physiological mechanisms to enhance their virulence. RALF was initially defined as a plant peptide able to trigger the alkalinization of plant cell cultures, and subsequently demonstrated to play varied roles in plant development. (Murphy and De Smet, 2014; Pearce et al., 2001). Here, we showed that treatment of plants with both plant and fungal RALF caused seedling growth inhibition, a rapid burst of ROS, MAPK activation and alkalinization, all hallmarks of defence activation by plants. Elicitation of ROS by RALFs has not been reported previously and represents an important addition to the spectrum of responses associated with RALF perception. We tested two representative divergent RALF sequences from the 12 Fox sequences. Only one of these caused consistent activation of plant responses at very low concentrations. Interestingly, RALF‐B was very much more potent than the endogenous Sl‐RALF peptide in treated N. benthamiana, and showed marked differences in both the extent and timing of the plant response. It is interesting to note that Sl‐RALF only induced a strong response in tomato, whereas RALF‐B was able to induce a strong response in both tomato and N. benthamiana. This is consistent with the apparent conservation of RALF peptides across Fox spp. In these species, these peptides are located on the conserved (not lineage‐specific) chromosomes. As such, promiscuity of activity among different potential hosts may be beneficial. At this stage, it is difficult to interpret the differences in activities between peptides, other than to point out that they must be a result of the sequence differences between the peptides. A key objective of this analysis was to demonstrate that a plant could perceive a fungal RALF peptide. This was achieved.
The expression of RALF‐B in Fol during in planta growth was consistent with a potential role in mediating disease. However, under the conditions used for pathogenicity assays in this study, strains of Fol lacking RALF‐B displayed similar disease symptoms to wild‐type Fol, implying that the peptide is not involved in the induction of disease symptoms. These data suggest that RALF‐B plays other as yet undetermined roles in this host–pathogen interaction not trialled in this study. Inoculation was performed with cut roots, dipped in spore solution. This is a routine technique for the assay of Fox infections of tomato. Perhaps the fungal RALFs are involved in the manipulation of the host prior to and/or during colonization. Potential roles for RALF are the focus of ongoing research.
Whilst this article was still under review, Masachis et al. (2016) reported a functional role for RALF‐B in Fol pathogenicity, but clear differences in the infection assays and scoring criteria were apparent when compared with this study. Masachis et al. (2016) grew plants for inoculation in vermiculite with no plant nutrients provided, and it is possible that assays conducted on such plants may have revealed an effect that was not evident in healthy plants grown in potting mix well supplied with nutrients. They also scored plant survival as opposed to disease symptoms and over a much longer period (up to 35 days as opposed to 21 days in our study), and it is interesting to note that their study also showed no significant effects at 21 dpi, i.e. no significant differences in survival. Therefore, we are confident that, under the inoculation conditions and scoring criteria used in this study, we found no significant effect of the RALF‐B knockout on pathogenicity, and can only conclude that any effect of RALF‐B on pathogenicity was too subtle for us to detect under our experimental conditions.
An intriguing observation from the distribution of RALFs among fungi was that the two poplar (P. trichocarpa) pathogens, Sp. musiva (Mycosphaerella populorum) and Se. populicola (Mycosphaerella populicola), encode RALF27‐like homologues. Recent comparisons between the genomes of these two species have uncovered an adaptation event that probably altered the fitness of Sp. musiva to become more pathogenic to poplars in relation to Se. populicola (Dhillon et al., 2015). Similarly, the gain or evolution of this peptide probably assisted the interaction of these fungi with poplars. Unlike most of the other fungal RALF homologues identified, the most similar homologues to the Sp. musiva and Se. populicola RALF27‐like peptides are those of the host they infect. This implies that the fungal peptides have the same role in plants as the endogenous peptides. This is also a potentially interesting model for the study of the function of RALF27. No function has yet been assigned to this member of the RALF family, which is only briefly mentioned in the literature (Lafleur et al., 2015; Marmiroli and Maestri, 2014; Olsen et al., 2002). The characterization of the role of fungal RALF27 homologues during infection may also help to elucidate the function of the host homologue.
Similar to Sp. musiva and Se. populicola, both Pseudocercospora fijiensis and M. phaseolina share RALF homologues similar to their respective hosts. Pseudocercospora fijiensis has a RALF that resolves with banana (Musa acuminata malaccensis), and M. phaseolina has a RALF with sequence similarity to a range of bean species. The RALF peptide from M. phaseolina is particularly interesting as it shares extremely high levels of sequence conservation for the peptide domain with a number of bean species (some of its primary hosts). However, this sequence clearly relates to other fungal RALF peptides, falling with a group of peptides shared among a range of fungal species (e.g. other members of the family Botryosphaeriaceae and the genus Fusarium). This would suggest that the peptide domain of this protein in M. phaseolina has probably evolved to be better attuned with that of its hosts.
The distribution of RALFs in the fungal kingdom is sporadic, with members represented from the Dothideomycete and Sordariomycete classes, as well as in the evolutionarily more distant Pucciniomycetes. Strikingly, the represented species are all economically significant phytopathogens. The strong conservation of the RALF peptide domain, and the in planta transcription and host perception of one such variant, represents important evidence that fungi have co‐opted this endogenous host signalling system to enhance their fitness on plants. The question remains, however, what is the ancestral origin of these conserved genes? The data presented here are consistent with conflicting evolutionary scenarios; HGT or convergent evolution. At this time, we cannot comment with any certainty on which scenario is the most likely.
Although the primary focus of this analysis has been to present the findings that a number of members of the fungal kingdom possess RALF peptides, we also identified a limited number of plant‐pathogenic bacteria which possess genes encoding amino acid sequences with an incorporated RALF motif domain. However, we did not explore any potential functional role for these proteins because of their striking differences from fungal and plant RALFs. The mature peptides of the RALFs discussed in fungi are homologous to the endogenous plant peptides. In contrast, the amino acid sequences in bacteria with a RALF‐like domain appear to be an amalgamation of a C‐terminal RALF domain and the S1 subunit of the pertussis toxin. Pertussis toxin has been studied in detail, particularly its role as a virulence factor in bacterial–animal pathosystems. However, how these two unrelated domains may perform in bacterial–plant interactions is currently unknown.
There remains much to learn about the function of RALF peptides and their role in plant growth and development. Adding to this mystery is how particular plant‐pathogenic organisms, both fungal and bacterial, utilize variants of these peptides to facilitate host infection and/or disease. In this study, we have demonstrated that genomes of specific fungal and bacterial phytopathogens encode RALF peptides, at least some of which are functionally active. We have focused on the interaction between Fol's expression of a particular RALF peptide and infection in tomato. However, there are a number of different RALF homologues utilized in various microorganism–plant pathosystems. We anticipate that this discovery will form the basis of an understanding of how different RALF peptides are utilized by microorganisms to co‐opt plant machinery to facilitate in planta growth.
Experimental Procedures
Identification of RALF homologues in fungal sequence databases
Homologues of RALF were identified with blast searches against the fungal sequence databases in NCBI (http://www.ncbi.nlm.nih.gov) and available via the Mycocosm portal in JGI (http://genome.jgi-psf.org/programs/fungi/index.jsf). The queries used were the Arabidopsis thaliana plant RALF1–RALF34 protein sequences from NCBI (Table S1). Because of the variability in the N‐terminal and C‐terminal regions, the blast search queries were refined where needed to include only the mature RALF peptide region (Pearce et al., 2010). blastp searches were also employed to find RALF homologues using fungal proteins with RALF annotations (Altschul et al., 1990). No e‐value cut‐off was used. Instead, individual blast hits were assessed for the presence of RALF‐associated domains (Murphy and De Smet, 2014), regardless of e‐value. tblastn searches were used to find mRNA sequences and un‐annotated or differently annotated RALF homologues on fungal genome contigs/chromosomes (Condon et al., 2013; Gardiner et al., 2012; Nemri et al., 2014; Ohm et al., 2012). Where annotations were not present or were different, the online Softberry server (http://linux1.softberry.com/berry.phtml) was used to predict mRNA and protein sequences employing the FGNESH+ prediction algorithm (Solovyev et al., 2006). Fungal RALF protein sequences were used as queries against the plant protein database in NCBI to find the top plant blast hits (Altschul et al., 1990).
Alignment and analysis of RALF protein sequences
A representative database of plant RALF proteins was compiled by searching for any plant amino acid sequence in the RefSeq database (Pruitt et al., 2007) labelled with the term ‘RALF’ or ‘Rapid Alkalinization Factor’. To compare RALF protein sequences from fungi and plants, GLAM2 (Frith et al., 2008) was used to identify the conserved region corresponding to the RALF domain from the plant sequences, conserved sites within the RALF domain and to align the conserved sites. Its sister program GLAM2SCAN was then used to align conserved sites from the fungal sequences, which were then added to the plant alignment.
The Bayesian phylogenetics software ExaBayes (Aberer et al., 2014) was then used to infer the posterior distribution of a RALF gene tree based on the aligned sequences. The priors for this analysis were the LG substitution matrix (Le and Gascuel, 2008) (used as a fixed amino acid model prior), and a uniform prior on tree topologies. Eight independent chains were run with different seeds, sampled once every 3000 states. Using Tracer v1.6 (http://beast.bio.ed.ac.uk/Tracer), five chains were observed to converge at the same likelihood after 1.8 million states, and so sampled trees and statistics from the subsequent 3.6 million states of those converged chains were concatenated. The estimated sample size of the concatenated log likelihood was 747, which suggests sufficient sampling of the posterior, as this is much greater than the minimum threshold of 200 (Kuhner, 2009).
A summary tree of 29 fungal RALF sequences and five plant RALF sequences was generated by pruning each tree in the posterior distribution of RALF gene trees to just these 34 tips, and then producing an extended majority‐rule tree from the posterior using PAUP 4.0 (http://paup.csit.fsu.edu /). The resulting summary tree was rooted and ladderized using TreeGraph 2 (Stöver and Müller, 2010). The full summary tree of all sequences was generated using the same method, but without pruning. To produce a combined phylogeny and alignment, the RALF sequences, including non‐conserved residues, were manually aligned, re‐ordered to match the pruned summary tree and individual amino acid residues coloured using JalView (Waterhouse et al., 2009).
Fox RALF homologue sequences were aligned in Geneious 7.1.5 (Biomatters, Auckland, New Zealand) using MUSCLE alignment (Edgar, 2004).
Synthetic peptides
Four synthetic RALF‐like peptides were produced by Mimotopes (Notting Hill, Australia) (Fig. S4). First, synthetic tomato (Solanum lycopersicum) RALF‐like (Sl‐RALF) peptide (9.2 mg with minimum purity of 96%) was chosen as a positive control because of its similar domain structure to the RALF peptides described by Pearce et al. (2010) in a study describing important residues for activity and for its similarity to Fol race 2. The synthetic peptide started at the first amino acids reported to be required for RALF activity (‘YISY’) (Pearce et al., 2010). Second, inactive Sl‐RALF peptide (Sl‐RALFΔ) was used as a negative control peptide (9.4 mg with minimum purity of 82%). This peptide was designed missing the YISY motif required for RALF function (Pearce et al., 2010). We synthesized two peptides of fungal origin: Fol race 2 RALF (RALF‐B) (9.3 mg with minimum purity of 86%) and Fol race 3 RALF (RALF‐C) (9.2 mg with minimum purity of 77%). These two Fox RALF peptide sequences are representative of the two main RALF variants observed in Fox (Fig. 3). All peptides were synthesized in oxidized form, with disulphide bonds between cysteine residues at the indicated positions (Figs 2 and 3).
Root inhibition in response to fungal RALF synthetic peptides
Tomato seeds (Solanum lycopersicum cv. Moneymaker) were surface sterilized for 10 min in 4% sodium hypochlorite–10% ethanol and germinated in 2 mL of 50% Murashige and Skoog (MS) medium on an orbital shaker for 2 days. Three lots of three germinating seeds were placed into 2 mL of fresh 50% MS medium with a final concentration of 10 μm synthetic peptide and incubated on the orbital shaker for 2 days. After this time, seedling weight was measured and comparisons in developmental morphology were made. Tukey's test was used to determine which differences between treatments were significant for all pairwise combinations of treatments, and to correct for multiple testing.
Measurement of ROS, induction of alkalinization and MAPK production in tomato and N. benthamiana in response to synthetic RALF peptides
ROS assays were performed as described previously (Segonzac et al., 2011), except that L‐012 (Wako Chemicals, Cape Charles, USA) was used instead of luminol, and luminescence was measured on a Tecan plate reader, Infinite M200 PRO (Tecan, Männedorf, Switzerland). MAPK activation assays were performed as described previously (Segonzac et al., 2011). For the alkalinization experiment, tomato or N. benthamiana leaves were cut the day before the assay in strips of approximately 1 cm × 0.1 cm, and incubated in double‐distilled H2O (ddH2O) overnight. The next day, 12–15 leaf strips of each were transferred to one well of a 12‐well plate containing 2 mL of sterile ddH2O. Leaves were incubated in the absence or presence of 1 μm RALF peptide for 1 h with shaking at 120 rpm. The pH was measured using a pH electrode until the reading was stable. The graphs show the average of three biological replicates and its estimated standard error. Different letters depict significant differences using Tukey–Kramer honest significant difference (HSD) test with P < 0.01.
RT‐PCR analysis of RALF‐B expression during infection
Two‐week‐old susceptible tomato cv. M82 seedlings were inoculated by dipping their roots in a suspension of 1 × 107 conidia/mL of Fol isolate #1943, or mock inoculated by dipping in water. Fol‐inoculated and mock‐inoculated plants were then grown in a controlled‐environment growth room with a 25 °C, 16‐h day/20 °C, 8‐h night cycle until collection of the samples. Roots of three to four Fol‐infected or mock‐inoculated plants were collected at 3 and 6 dpi, washed with sterile deionized water, pooled in a microcentrifuge tube and frozen in liquid nitrogen ready for RNA extraction. Frozen root samples were ground in liquid nitrogen and total RNA was extracted using a Plant RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Total RNA (2 μg) was treated with 2 μL of RQ1 RNase‐Free DNase (Promega, Madison, WI, USA) in a reaction volume of 20 μL containing 1 × RQ1 DNAse reaction buffer (400 mm Tris‐HCl, pH 8.0, 100 mm MgSO4, 10 mm CaCl2; Promega) and 1 μL of RNasin ribonuclease inhibitor (Promega). The reaction was incubated at 37 °C for 30 min, followed by an inactivation step at 65 °C for 20 min. Treated RNA was reverse transcribed into cDNA using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and an oligo [dT]12–18 primer (Invitrogen) following the manufacturer's instructions. PCR (35 cycles) was carried out using Phire Hot Start II DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) in a reaction volume of 25 μL containing 1 μL of cDNA template according to the manufacturer's instructions. Primers 5′‐GCTGAAGCCAACCCCTATAA‐3′ and 5′‐TTACGATCCGGTTACCAAGC‐3′ were used to amplify RALFFol, and primers 5′‐AGCCTTACACCATCCGCTAC‐3′ and 5′‐CGCTGTAGTTGACCTCACCA‐3′ were used to amplify FEM1 (Fusarium extracellular matrix protein 1) (Schoffelmeer et al., 2001) as a positive control for fungal gene expression. RNA and genomic DNA (50 ng) from non‐inoculated tomato plants and 5‐day‐old cultures of Fol isolates #1943 and 4287 were used as controls. Controls also included reactions in which reverse transcriptase or template DNA was omitted.
Fungal strains and pathogenicity test on tomato
The tomato M82 cultivar susceptible to Fol race 3 was used for pathogenicity tests. Australian Fol race 3 isolate #1943 was used to generate RALF knockout mutants. Pathogenicity tests were performed by the root dip method (Mes et al., 1999; Wellman, 1939). Using this method, roots of 3‐week‐old plants of the tomato cultivar M82 (susceptible to Fol race 3) were wounded and subsequently inoculated with Fol race 3. Disease symptoms were scored based on plant fresh weight and disease index, as described by Rep et al. (2004).
Construction of Fol RALF deletion vectors
Flanking regions of the RALF gene were amplified from Fol race 3 genomic DNA using primers RALF_FR1 and RALF_FR2 for the upstream flanking region and RALF_FR3 and RALF_FR4 for the downstream flanking region (Table S2, see Supporting Information). The upstream flanking region was cloned via KpnI and BsrGI restriction sites into the 5′ end of the hygromycin resistance cassette in the binary vector pPK2HPH (Michielse et al., 2009), and the downstream flanking region was cloned via XbaI and HindIII sites into the 3′ end of the hygromycin resistance cassette to generate pPK2HPH:ΔRALF.
A modified HSVtk gene cassette was generated and used as a second selection marker, as described by Khang et al. (2005). Briefly, the promoter of the trpC gene of Aspergillus nidulans was amplified from the pGpdGFP binary vector (Sexton and Howlett, 2001) using the primers trpC_p‐F and trpC_p‐R (Table S2), and cloned into the pGEMt easy vector (Promega). The HSVtk gene was obtained from DNA of the herpes simplex virus (HSV) isolate UL23 (provided by Professor David Tscharke, The Australian National University, Canberra, Australia) using the primers HSVtk‐F and HSVtk‐R (Table S2), and cloned into the pGEMt:trpC promoter intermediate vector via SpeI and SalI sites. The terminator sequence of the Fol β‐tubulin gene was obtained from Fol genomic DNA using the primers βtub‐F and βtub‐R, and cloned into the pGEMt:trpC promoter:HSVtk coding sequence intermediate vector via SalI and PmeI restriction sites. The entire HSVtk cassette was then transferred to the pPK2HPH:ΔRALF vector and placed next to the RALF downstream flanking region, using HindIII and PmeI sites (Fig. S6A).
Fol transformation was performed using the Agrobacterium tumefaciens strain LBA4404 containing the appropriate binary vector. The transformation protocol was adapted from Mullins et al. (2001) and Takken et al. (2004). The selection medium for the RALF knockout mutants was supplemented with 75 μg/mL augmentin, 50 μg/mL hygromycin and 5 mm 5‐fluoro‐2′‐deoxyuridine (F2dU). Deletion of the RALF gene was confirmed by PCR using the primers scRALF_5′‐F, RALF_int‐R, RALF_int‐F, scRALF_3′‐R and scGDP‐R (Table S2 and Fig. S6B).
Note Added in Proof
Consistent with our findings, Vlaadingerbroek et al. (2016; Mol. Plant Pathol. DOI: 10.1111/mpp.12440) have shown, using the same kind of tomato pathogenicity assay and scoring system we used, that complete loss of Fol chromosome 12, which carries the RALF‐B gene, has no significant effect on Fol race 2 pathogenicity.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Gene tree of all identified rapid alkalinization factor (RALF) sequences from plants and fungi. This tree was inferred using the RALF peptide domain amino acid sequences from 433 plant RALF sequences and 29 fungal RALF sequences (highlighted in bold). Support values on each branch refer to the percentage posterior probability of each clade. Fungal RALF sequences are labelled with the scientific name of the fungal species or isolate. Plant RALF sequences are labelled with their NCBI GI code, binomial name and NCBI RefSeq description. The tree was rooted to split the tree into RALF 27‐like sequences and other RALF sequences, a split with 100% posterior support.
Fig. S2. There are three divergent rapid alkalinization factor (RALF) groups within the Botryosphaeriaceae (A). Particular motifs are unique among these groups (A). In particular, Group BI has a motif shared between few other species (plants and fungi) (B). One of Macrophomina phaseolina's Group BII RALFs shows 100% identity for the mature peptide region with species of soy. These are primary hosts of M. phaseolina.
Fig. S3. A limited number of bacteria encode a protein with a pertussis toxin subunit (S1) and a C‐terminal rapid alkalinization factor (RALF) domain (A). This C‐terminal RALF domain in these bacteria shares homology with plant species RALFs (plant species outlined) (B).
Fig. S4. Amino acid sequences of peptides synthesized for the reactive oxygen species (ROS) burst and mitogen‐activated protein kinase (MAPK) activation assays
Fig. S5. Concentration‐dependent rapid alkalinization factor (RALF)‐mediated production of reactive oxygen species (ROS) in Nicotiana benthamiana leaves. (A) ROS burst over time on treatment with 1, 10, 50, 100 and 500 nm RALF‐B. (B) ROS burst over time on treatment with 100 nm, 1 µm and 10 µm of RALF‐C.
Fig. S6. Activation of mitogen‐activated protein kinases (MAPKs) in Nicotiana benthamiana leaves at 5 and 15 min after treatment with the synthetic rapid alkalinization factor (RALF) peptides as indicated.
Fig. S7. Generation and screening of Fil race 3 transformants containing a knockout of the RALF gene (ΔRALF). (A) Schematic representation of the RALF gene replacement using the hygromycin phosphotransferase (hph) gene as a selectable marker and the thymidine kinase (tk) gene as a counter‐selectable marker. Arrows indicate primers pairs used for screening. (B) Polymerase chain reaction (PCR) detection of RALF gene using scRALF 5′‐F/RALF_int‐R primers (primer set B; left gel image) and RALF_int‐F/scRALF 3′‐R primers (primer set C; right gel image). Both gel images indicate the absence of the RALF coding sequence in the four ΔRALF transformants. Fol, Fusarium oxysporum f. sp. lycopersici DNA. (C) PCR with scRALF 5′‐F/sc_gdp‐R primers (primer set D) indicates the presence of the hph cassette in the ΔRALF transformants. Ect, DNA from a transformant with an ectopic T‐DNA insertion in Fol. (D) Pathogenicity test on M82 plants with Fol wild‐type race 3 (WT), a Fol transformant with an ectopic T‐DNA insertion or Fol ΔRALF transformants Δ23 and Δ24. Photographs were taken at 21 days post‐inoculation (dpi). (E) Distribution of disease scores for the symptoms observed in two experiments for 10 plants for each treatment at 21 dpi. Probability values were obtained using the one‐tailed, non‐parametric, Mann–Whitney test to determine significant differences (P ≤ 0.05) between treatments.
Table S1. Arabidopsis RALF sequences used as BLAST queries.
Table S2. Primers used in this study.
Acknowledgements
The authors would like to thank Dr Markus Albert for technical advice on the alkalinization assay. PSS is an Australian Research Council Future Fellow (FT110100698). BS is supported by an Australian Research Council Discovery Early Career Award (DE150101897). ET is supported by an Australian Postgraduate Award and a Grains Research and Development Corporation Scholarship. ExaBayes was run on a computer cluster managed by the Genome Discovery Unit of the Australian Cancer Research Foundation's Biomolecular Resource Facility.
Contributor Information
David A. Jones, Email: david.jones@anu.edu.au
John P. Rathjen, Email: john.rathjen@anu.edu.au
Peter S. Solomon, Email: peter.solomon@anu.edu.au
References
- Aberer, A. , Kobert, K. and Stamatakis, A. (2014) ExaBayes: massively parallel bayesian tree inference for the whole‐genome era. Mol. Biol. Evol. 31, 2553–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bobay, B.G. , DiGennaro, P. , Scholl, E. , Imin, N. , Djordjevic, M.A. and Bird, D.M. (2013) Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Lett. 587, 3979–3985. [DOI] [PubMed] [Google Scholar]
- Bournival, B. , Scott, J. and Vallejos, C. (1989) An isozyme marker for resistance to race 3 of Fusarium oxysporum f. sp. lycopersici in tomato. Theor. Appl. Genet. 78, 489–494. [DOI] [PubMed] [Google Scholar]
- Condon, B.J. , Leng, Y. , Wu, D. , Bushley, K.E. , Ohm, R.A. , Otillar, R. , Martin, J. , Schackwitz, W. , Grimwood, J. , MohdZainudin, N. , Xue, C. , Wang, R. , Manning, V.A. , Dhillon, B. , Tu, Z.J. , Steffenson, B.J. , Salamov, A. , Sun, H. , Lowry, S. , LaButti, K. , Han, J. , Copeland, A. , Lindquist, E. , Barry, K. , Schmutz, J. , Baker, S.E. , Ciuffetti, L.M. , Grigoriev, I.V. , Zhong, S. and Turgeon, B.G. (2013) Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet. 9, e1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000) Loosening of plant cell walls by expansins. Nature, 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Covey, P.A. , Subbaiah, C.C. , Parsons, R.L. , Pearce, G. , Lay, F.T. , Anderson, M.A. , Ryan, C.A. and Bedinger, P.A. (2010) A pollen‐specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. , Mehrabi, R. , Van Den Burg, H.A. and Stergiopoulos, I. (2009) Fungal effector proteins: past, present and future: review. Mol. Plant Pathol. 10, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay, C. , Imin, N. and Djordjevic, M.A. (2013) CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Dhillon, B. , Feau, N. , Aerts, A.L. , Beauseigle, S. , Bernier, L. , Copeland, A. , Foster, A. , Gill, N. , Henrissat, B. , Herath, P. , LaButti, K.M. , Levasseur, A. , Lindquist, E.A. , Majoor, E. , Ohm, R.A. , Pangilinan, J.L. , Pribowo, A. , Saddler, J.N. , Sakalidis, M.L. , de Vries, R.P. , Grigoriev, I.V. , Goodwin, S.B. , Tanguay, P. and Hamelin, R.C. (2015) Horizontal gene transfer and gene dosage drives adaptation to wood colonization in a tree pathogen. Proc. Natl. Acad. Sci. USA, 112, 3451–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids. Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Frith, M.C. , Saunders, N.F.W. , Kobe, B. and Bailey, T.L. (2008) Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, D.M. , McDonald, M.C. , Covarelli, L. , Solomon, P.S. , Rusu, A.G. , Marshall, M. , Kazan, K. , Chakraborty, S. , McDonald, B.A. and Manners, J.M. (2012) Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8, e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta, M. , Sabat, G. , Stecker, K. , Minkoff, B.B. and Sussman, M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science, 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet‐Tapia, J.C. and Loria, R. (2012) Draft genome sequence of Streptomyces acidiscabies 84–104, an emergent plant pathogen. J. Bacteriol., 194, 1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. and Thomma, B.P.H.J. (2011) How filamentous pathogens co‐opt plants: the ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14, 400–406. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Maruthachalam, K. , Bolton, M.D. , Santhanam, P. , Saber, M.K. , Zhang, Z. , Usami, T. , Lievens, B. , Subbarao, K.V. and Thomma, B.P. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA, 109, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang, C.H. , Park, S.Y. , Lee, Y.H. and Kang, S. (2005) A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum . Fungal Genet. Biol. 42, 483–492. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Subbarao, K.V. , Kang, S. , Veronese, P. , Gold, S.E. , Thomma, B.P.H.J. , Chen, Z. , Henrissat, B. , Lee, Y.‐H. , Park, J. , Garcia‐Pedrajas, M.D. , Barbara, D.J. , Anchieta, A. , de Jonge, R. , Santhanam, P. , Maruthachalam, K. , Atallah, Z. , Amyotte, S.G. , Paz, Z. , Inderbitzin, P. , Hayes, R.J. , Heiman, D.I. , Young, S. , Zeng, Q. , Engels, R. , Galagan, J. , Cuomo, C.A. , Dobinson, K.F. and Ma, L.J. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner, M.K. (2009) Coalescent genealogy samplers: windows into population history. Trends Ecol. Evol. 24, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur, E. , Kapfer, C. , Joly, V. , Liu, Y. , Tebbji, F. , Daigle, C. , Gray‐Mitsumune, M. , Cappadocia, M. , Nantel, A. and Matton, D.P. (2015) The FRK1 mitogen‐activated protein kinase kinase kinase (MAPKKK) from Solanum chacoense is involved in embryo sac and pollen development. J. Exp. Bot. (in press) doi: 10.1093/jxb/eru1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S. and Gascuel, O. (2008) LG: an improved, general amino‐acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Ma, L.J. , Van Der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A.E. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G.J. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmiroli, N. and Maestri, E. (2014) Plant peptides in defense and signaling. Peptides, 56, 30–44. [DOI] [PubMed] [Google Scholar]
- Masachis, S. , Segorbe, D. , Turrà, D. , Leon‐Ruiz, M. , Fürst, U. , El Ghalid, M. , Leonard, G. , López‐Berges, M.S. , Richards, T.A. , Felix, G. and Di Pietro, A. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1, 16043. [DOI] [PubMed] [Google Scholar]
- Mes, J.J. , Weststeijn, E.A. , Herlaar, F. , Lambalk, J.J.M. , Wijbrandi, J. , Haring, M.A. and Cornelissen, B.J.C. (1999) Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology, 89, 156–160. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Cornelissen, B.J.C. and Rep, M. (2009) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. Genome Biol. 10, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, E.D. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D.M. and Kang, S. (2001) Agrobacterium tumefaciens‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Murphy, E. and De Smet, I. (2014) Understanding the RALF family: a tale of many species. Trends Plant Sci. 19, 664–671. [DOI] [PubMed] [Google Scholar]
- Nemri, A. , Saunders, D.G. , Anderson, C. , Upadhyaya, N.M. , Win, J. , Lawrence, G.J. , Jones, D.A. , Kamoun, S. , Ellis, J.G. and Dodds, P.N. (2014) The genome sequence and effector complement of the flax rust pathogen Melampsora lini . Front. Plant Sci. 5, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis, N. , Doran, N. and Cosgrove, D.J. (2014) Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol. Biol. Evol. 31, 376–386. [DOI] [PubMed] [Google Scholar]
- Ohm, R.A. , Feau, N. , Henrissat, B. , Schoch, C.L. , Horwitz, B.A. , Barry, K.W. , Condon, B.J. , Copeland, A.C. , Dhillon, B. , Glaser, F. , Hesse, C.N. , Kosti, I. , LaButti, K. , Lindquist, E.A. , Lucas, S. , Salamov, A.A. , Bradshaw, R.E. , Ciuffetti, L. , Hamelin, R.C. , Kema, G.H. , Lawrence, C. , Scott, J.A. , Spatafora, J.W. , Turgeon, B.G. , de Wit, P.J. , Zhong, S. , Goodwin, S.B. and Grigoriev, I.V. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8, e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N. , Mundy, J. and Skriver, K. (2002) Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol. 2, 441–451. [PubMed] [Google Scholar]
- Pearce, G. , Moura, D.S. , Stratmann, J. and Ryan, C.A. (2001) RALF, a 5‐kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA, 98, 12 843–12 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G. , Yamaguchi, Y. , Munske, G. and Ryan, C.A. (2010) Structure–activity studies of RALF, rapid alkalinization factor, reveal an essential–YISY–motif. Peptides, 31, 1973–1977. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. , Tatusova, T. and Maglott, D.R. (2007) NCBI reference sequences (RefSeq): a curated non‐redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M. , van der Does, H.C. , Meijer, M. , van Wijk, R. , Houterman, P.M. , Dekker, H.L. , de Koster, C.G. and Cornelissen, B.J.C. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Richards, T.A. , Soanes, D.M. , Jones, M.D. , Vasieva, O. , Leonard, G. , Paszkiewicz, K. , Foster, P.G. , Hall, N. and Talbot, N.J. (2011) Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. USA, 108, 15 258–15 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffelmeer, E.A.M. , Vossen, J.H. , Van Doorn, A.A. , Cornelissen, B.J.C. and Haring, M.A. (2001) FEM1, a Fusarium oxysporum glycoprotein that is covalently linked to the cell wall matrix and is conserved in filamentous fungi. Mol. Gen. Genet. 265, 143–152. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, A.C. and Howlett, B.J. (2001) Green fluorescent protein as a reporter in the Brassica–Leptosphaeria maculans interaction. Physiol. Mol. Plant Pathol. 58, 13–21. [Google Scholar]
- Soanes, D. and Richards, T.A. (2014) Horizontal gene transfer in eukaryotic plant pathogens. Annu. Rev. Phytopathol. 52, 583–614. [DOI] [PubMed] [Google Scholar]
- Solovyev, V. , Kosarev, P. , Seledsov, I. and Vorobyev, D. (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, S10–S11. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, R. , Liu, J. , Guo, H. , Yin, Y. and Howell, S. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis . Plant J. 59, 930–939. [DOI] [PubMed] [Google Scholar]
- Stöver, B.C. and Müller, K.F. (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics, 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E.H. and McDonald, B.A. (2008) The origins of plant pathogens in agro‐ecosystems. Annu. Rev. Phytopathol. 46, 75–100. [DOI] [PubMed] [Google Scholar]
- Takken, F.L.W. , Van Wijk, R. , Michielse, C.B. , Houterman, P.M. , Ram, A.F. and Cornelissen, B.J.C. (2004) A one‐step method to convert vectors into binary vectors suited for Agrobacterium‐mediated transformation. Curr. Genet. 45, 242–248. [DOI] [PubMed] [Google Scholar]
- Taylor, W.R. (1997) Residual colours: a proposal for aminochromography. Protein Eng. 10, 743–746. [DOI] [PubMed] [Google Scholar]
- Thynne, E. , McDonald, M.C. , Evans, M. , Wallwork, H. , Neate, S. and Solomon, P.S. (2015a) Re‐classification of the causal agent of white grain disorder on wheat as three separate species of Eutiarosporella . Australas. Plant Pathol. 44, 527–539. [Google Scholar]
- Thynne, E. , McDonald, M.C. and Solomon, P.S. (2015b) Phytopathogen emergence in the genomics era. Trends Plant Sci. 20, 246–255. [DOI] [PubMed] [Google Scholar]
- Vernikos, G. , Medini, D. , Riley, D.R. and Tettelin, H. (2015) Ten years of pan‐genome analyses. Curr. Opin. Microbiol. 23, 148–154. [DOI] [PubMed] [Google Scholar]
- Waterhouse, A.M. , Procter, J.B. , Martin, D.M. , Clamp, M. and Barton, G.J. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics, 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman, F.L. (1939) A technique for studying host resistance and pathogenicity in tomato Fusarium wilt. Phytopathology, 29, 945–956. [Google Scholar]
- Wolf, S. and Höfte, H. (2014) Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell, 26, 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Gene tree of all identified rapid alkalinization factor (RALF) sequences from plants and fungi. This tree was inferred using the RALF peptide domain amino acid sequences from 433 plant RALF sequences and 29 fungal RALF sequences (highlighted in bold). Support values on each branch refer to the percentage posterior probability of each clade. Fungal RALF sequences are labelled with the scientific name of the fungal species or isolate. Plant RALF sequences are labelled with their NCBI GI code, binomial name and NCBI RefSeq description. The tree was rooted to split the tree into RALF 27‐like sequences and other RALF sequences, a split with 100% posterior support.
Fig. S2. There are three divergent rapid alkalinization factor (RALF) groups within the Botryosphaeriaceae (A). Particular motifs are unique among these groups (A). In particular, Group BI has a motif shared between few other species (plants and fungi) (B). One of Macrophomina phaseolina's Group BII RALFs shows 100% identity for the mature peptide region with species of soy. These are primary hosts of M. phaseolina.
Fig. S3. A limited number of bacteria encode a protein with a pertussis toxin subunit (S1) and a C‐terminal rapid alkalinization factor (RALF) domain (A). This C‐terminal RALF domain in these bacteria shares homology with plant species RALFs (plant species outlined) (B).
Fig. S4. Amino acid sequences of peptides synthesized for the reactive oxygen species (ROS) burst and mitogen‐activated protein kinase (MAPK) activation assays
Fig. S5. Concentration‐dependent rapid alkalinization factor (RALF)‐mediated production of reactive oxygen species (ROS) in Nicotiana benthamiana leaves. (A) ROS burst over time on treatment with 1, 10, 50, 100 and 500 nm RALF‐B. (B) ROS burst over time on treatment with 100 nm, 1 µm and 10 µm of RALF‐C.
Fig. S6. Activation of mitogen‐activated protein kinases (MAPKs) in Nicotiana benthamiana leaves at 5 and 15 min after treatment with the synthetic rapid alkalinization factor (RALF) peptides as indicated.
Fig. S7. Generation and screening of Fil race 3 transformants containing a knockout of the RALF gene (ΔRALF). (A) Schematic representation of the RALF gene replacement using the hygromycin phosphotransferase (hph) gene as a selectable marker and the thymidine kinase (tk) gene as a counter‐selectable marker. Arrows indicate primers pairs used for screening. (B) Polymerase chain reaction (PCR) detection of RALF gene using scRALF 5′‐F/RALF_int‐R primers (primer set B; left gel image) and RALF_int‐F/scRALF 3′‐R primers (primer set C; right gel image). Both gel images indicate the absence of the RALF coding sequence in the four ΔRALF transformants. Fol, Fusarium oxysporum f. sp. lycopersici DNA. (C) PCR with scRALF 5′‐F/sc_gdp‐R primers (primer set D) indicates the presence of the hph cassette in the ΔRALF transformants. Ect, DNA from a transformant with an ectopic T‐DNA insertion in Fol. (D) Pathogenicity test on M82 plants with Fol wild‐type race 3 (WT), a Fol transformant with an ectopic T‐DNA insertion or Fol ΔRALF transformants Δ23 and Δ24. Photographs were taken at 21 days post‐inoculation (dpi). (E) Distribution of disease scores for the symptoms observed in two experiments for 10 plants for each treatment at 21 dpi. Probability values were obtained using the one‐tailed, non‐parametric, Mann–Whitney test to determine significant differences (P ≤ 0.05) between treatments.
Table S1. Arabidopsis RALF sequences used as BLAST queries.
Table S2. Primers used in this study.


