Summary
A novel virus has been identified by next‐generation sequencing (NGS) in privet (Ligustrum japonicum L.) affected by a graft‐transmissible disease characterized by leaf blotch symptoms resembling infectious variegation, a virus‐like privet disease with an unclear aetiology. This virus, which has been tentatively named ‘privet leaf blotch‐associated virus’ (PrLBaV), was absent in non‐symptomatic privet plants, as revealed by NGS and reverse transcription‐polymerase chain reaction (RT‐PCR). Molecular characterization of PrLBaV showed that it has a segmented genome composed of two positive single‐stranded RNAs, one of which (RNA1) is monocistronic and codes for the viral replicase, whereas the other (RNA2) contains two open reading frames (ORFs), ORF2a and ORF2b, coding for the putative movement (p38) and coat (p30) proteins, respectively. ORF2b is very probably expressed through a subgenomic RNA starting with six nucleotides (AUAUCU) that closely resemble those found in the 5′‐terminal end of genomic RNA1 and RNA2 (AUAUUU and AUAUAU, respectively). The molecular signatures identified in the PrLBaV RNAs and proteins resemble those of Raspberry bushy dwarf virus (RBDV), currently the only member of the genus Idaeovirus. These data, together with phylogenetic analyses, are consistent with the proposal of considering PrLBaV as a representative of the second species in the genus Idaeovirus. Transient expression of a recombinant PrLBaV p38 fused to green fluorescent protein in leaves of Nicotiana benthamiana, coupled with confocal laser scanning microscopy assays, showed that it localizes at cell plasmodesmata, strongly supporting its involvement in viral movement/trafficking and providing the first functional characterization of an idaeovirus encoded protein.
Keywords: chlorosis, idaeovirus, leaf blotch, Ligustrum japonicum, movement protein, privet disease, subcellular localization
Introduction
Infectious chlorosis, variegation of privet, mosaic of ligustrum and yellow mosaic of privet (http://gd.eppo.int/taxon/PYM000) have been considered as synonyms of a virus‐like disease known for more than a century (Baur, 1907). The disease has been transmitted by grafting (Baur, 1907; Schmelzer, 1963), thus differing from the leaf chlorosis or yellowing arising from genetic changes or abiotic causes (Pirone, 1978). Although infectious chlorosis of privet is considered to be widespread in Europe (Cooper, 1993), it is not clear whether the chlorotic symptoms repeatedly reported from different geographical locations refer to the same disease or several disorders caused by different agents, but characterized by similar general symptom manifestation. Moreover, whether some of the viruses already isolated from the affected privet plants—including Prunus necrotic ringspot virus (PNRSV), Cherry leaf roll virus, Tomato bushy stunt virus, Tomato black ring virus (Cooper, 1993; and references therein) and Ligustrum necrotic ringspot virus (LNRSV; Scott and Zimmerman, 2008)—are actually involved in the infectious chlorosis or are just accidental discoveries has not been studied in detail. The first solid proof of the possible involvement of a virus in privet disease has only been published recently. Indeed, a novel ilarvirus, tentatively named ‘privet ringspot virus’ (PrRSV), has been characterized and found to be closely associated with necrotic ringspot disease (NRSD), which is widespread in southeastern USA (Aboughanem‐Sabanadzovic et al., 2016).
The genus Idaeovirus, a monotypic taxon currently not assigned to any family, contains only the species Raspberry bushy dwarf virus (RBDV), a positive single‐stranded RNA virus with a bipartite genome (MacFarlane, 2012). RBDV RNA1 contains a single open reading frame (ORF) encoding a polyprotein with the characteristic motifs of viral RNA methyltransferase, helicase and polymerase, whereas RNA2 is bicistronic. ORF2a encodes a protein resembling cell‐to‐cell movement proteins (MPs) of other viruses, whereas ORF2b codes for the viral coat protein (CP), which is expressed through a subgenomic RNA (sgRNA) (Jones and Baker, 2008; Sabanadzovic and Martin, 2011). Although RBDV has a bipartite genome, it has affinities with viruses in the family Bromoviridae that instead have tripartite genomes (Jones and Baker, 2008). A putative new member in the genus Idaeovirus has been reported from citrus, but its genome has been only partially characterized (Derrick et al., 2006). Finally, a bipartite virus with organization of RNA1 closely resembling RBDV has been characterized from symptomatic ferns in USA and referred to as Japanese holly fern mottle virus (JHFMoV; Valverde and Sabanadzovic, 2009). Despite close resemblance with RBDV in RNA1, the organization of JHFMoV RNA2 is distinct from the typical idaeovirus, as it contains three ORFs, including one coding for umbravirus‐like MPs (Valverde and Sabanadzovic, 2009).
In the present article, we report a novel virus identified in privet plants (Ligustrum japonicum) showing symptoms of leaf blotching. The genome structure of this virus, the molecular features of the encoded proteins and the phylogenetic analyses support the proposal of classifying this virus, tentatively named privet leaf blotch‐associated virus (PrLBaV), as a representative of the second species in the genus Idaeovirus. In addition, this study provides evidence that PrLBaV‐encoded p38 localizes at cell plasmodesmata (PD), thus supporting its active role in viral trafficking.
Results
Symptomatology and graft transmission
Privet (Ligustrum japonicum L., family Oleaceae) plants affected by leaf blotch (LB) disease resembling infectious chlorosis (Baur, 1907) were found in southern Italy (A. Ragozzino, unpublished data). The disease was transmitted by bark grafting to healthy L. japonicum seedlings, suggesting the viral origin. Typical symptoms, consisting of chlorotic, white–cream blotches, sometimes associated with ring spots, appeared on leaves of the inoculated plant (Fig. 1), 1 year after inoculation, suggesting the involvement of viral or virus‐like agent(s) in the disease. These symptoms persisted over the years in the inoculated privet, being more visible at the beginning of the spring season, especially when the plants were exposed to full sun. LBs (Fig. 1B–E), initially yellow (Fig. 1C), became white–cream in a few weeks and sometimes coalesced, thus covering the entire leaf (Fig. 1D). Generally, the expression of symptoms was sectorial and affected only a few branches, whereas other parts of the canopy remained asymptomatic. Occasionally, the central area of the chlorotic spots would turn necrotic in some senescent leaves (Fig. 1E). Attempts to isolate the infectious agent on a range of herbaceous hosts were unsuccessful, thus making the investigation of the putative viral or virus‐like agent of LB disease more difficult.
Figure 1.
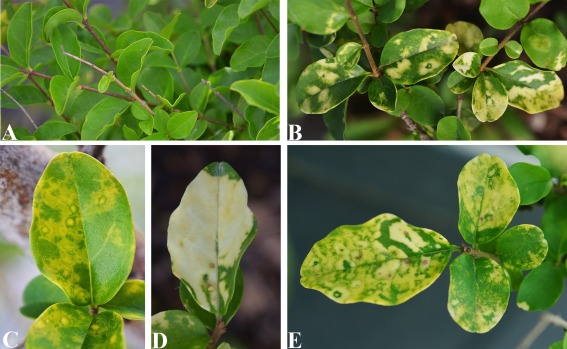
Symptoms of privet leaf blotch disease. (A) Healthy privet. (B–E) Symptomatic leaves from an affected privet: (B) typical leaf blotches; (C) yellow–cream rings; (D) coalesced blotches covering a leaf almost completely; (E) necrotic spots within white–cream blotches.
In the last few years, several novel viruses and viroids have been identified in plants by next‐generation sequencing (NGS) of small RNA (sRNA) libraries (Seguin et al., 2014; Wu et al., 2015). This approach relies on the accumulation of virus‐ and iroid‐derived sRNAs (v‐sRNAs) in the infected tissues that can be sequenced (by NGS) and assembled into larger contigs, which are useful for the identification of the infectious agent by interrogating databases for similar virus and viroid sequences.
Identification of a novel virus by NGS
Two cDNA libraries of sRNAs, generated from one symptomatic and one symptomless privet plant and sequenced by an Illumina Genome Analyzer (HiScan SQ, San Diego, CA, USA), resulted in 5,362,810 and 3,583,927 high‐quality reads (18–26 nucleotides), respectively, which were de novo assembled into larger contigs by Velvet Software (Zerbino and Birney, 2008) adopting a k‐mer of 15–17. blastx search in the GenBank Virus Reference Sequence Database showed that 16 contigs from the symptomatic sample had 35%–86% amino acid identity with proteins encoded by RNA1 or RNA2 of RBDV (Table S1, see Supporting Information). In contrast, no match was obtained when the same analysis was performed using the contigs from the symptomless plant sample, thus suggesting the presence of a putative new virus only in the symptomatic privet. The identified contigs from the symptomatic privet were aligned along the two RNA components of the RBDV genome, thus generating a preliminary genome scaffold of a potential new virus (Fig. S1, see Supporting Information).
The presence of both RNA1 and RNA2 exclusively in the symptomatic privet was further confirmed by reverse transcription‐polymerase chain reaction (RT‐PCR) using primer pairs (Li‐a/Li‐b and Li‐10/Li‐11; Table S2, see Supporting Information) designed on the sequence of different contigs mapping within these RNAs. PCR products with the expected sizes were obtained from the symptomatic sample, whereas no amplicon was generated from the non‐symptomatic sample (Fig. S2, see Supporting Information). Cloning and sequencing of the cDNAs from the symptomatic privet confirmed the partial sequences determined by NGS and allowed the filling of gaps between them. The same RT‐PCR approach was used to test several asymptomatic privet plants, which always tested negative. Altogether, these data strongly support the possibility that a novel virus, taxonomically related to RBDV—the sole member of the genus Idaeovirus to date—could infect the symptomatic privet. Such a possible novel virus was here tentatively designated as ‘privet leaf blotch‐associated virus’ (PrLBaV).
PrLBaV genome organization
The complete nucleotide sequence of the two PrLBaV genomic RNAs was determined and, when they were used to filter the sRNA libraries, the PrLBaV sRNAs (about 35 000 reads, 0.6% of the total reads) were recovered from the library generated from the diseased privet, but not from the healthy control. PrLBaV sRNAs of both polarity strands accumulated at similar levels, with those of 21 and 22 nucleotides being largely prevalent compared with those of the other size classes (data not shown), a result in line with that reported for other plant viruses (Donaire et al., 2009).
PrLBaV RNA1 and RNA2 (GenBank accession numbers LT221868 and LT221869, respectively) shared conserved untranslated regions (UTRs) at the respective 5′ and 3′ ends. In particular, they had an almost identical hexanucleotide sequence (AUAUUU and AUAUAU, respectively) at the 5′ termini and a highly conserved stretch of 22 nucleotides at the 3′ termini, which terminated with four consecutive cytosine residues (Fig. 2). In addition, MFOLD analyses showed that approximately 80 nucleotides at the terminal 3′ UTRs of both genomic RNAs folded into four similar stem‐loop structures, with identical base‐paired nucleotides at several positions within each stem and a 3′ end‐proximal tetracytosine stretch remaining unpaired (Fig. 2). Interestingly, RDBV RNA1 and RNA2 at the 5′ terminus had the same six nucleotides found in PrLBaV genomic RNAs, and at the other terminus adopted similar stem‐loop conformations with a 3′ end‐proximal tetracytosine stretch (Natsuaki et al., 1991).
Figure 2.
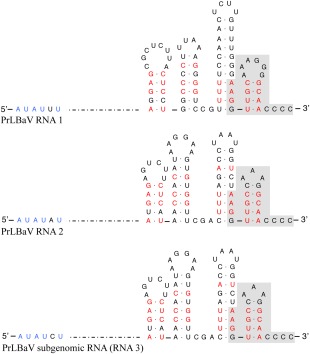
Primary and secondary structures of 5′ and 3′ termini of privet leaf blotch‐associated virus (PrLBaV) RNAs. The genomic RNA1 and RNA2 and the subgenomic RNA3 are reported. Identical nucleotides at the 5′ end are shown in blue; identical sequences at the 3′ termini are shown on a grey background; identical base‐paired nucleotides at several positions within each stem‐loop at the 3′ terminus are denoted in red.
PrLBaV RNA1 (5277 nucleotides) contains a single open reading frame (ORF1), which codes for a long putative polyprotein (p198) of 1738 amino acids (197.8 kDa) (protein_id: CZS63540.1), and is preceded and followed by 5′ and 3′ terminal UTRs of 66 and 94 nucleotides, respectively (Fig. 3). PFAM analyses identified three conserved domains in the predicted protein encoded by ORF1: viral methyltransferase (MTR, amino acids 193–616, Pfam PF01660, E‐value 1.3e‐75), viral helicase (HEL, amino acids 902–1158, Pfam PF01443, E‐value 1.0e‐47) and RNA‐dependent RNA polymerase (RdRp, amino acids 1280–1719, Pfam PF00978, E‐value 1.1e‐115) (Fig. 3). In particular, the three motifs of type 1 methyltransferase, the seven conserved motifs of helicase superfamily 1 and the consensus motifs I–VIII of the tobamovirus lineage of supergroup 3 of the RdRps of positive‐strand RNA viruses (Koonin and Dolja, 1993) were identified. The whole protein shares 48% amino acid identity with the non‐structural protein (NP_620465) encoded by RNA1 (ORF1a) of RBDV, with higher identity values (56%–62%) when the three functional motifs were considered separately (Table 1). Altogether, these data indicate that the putative protein encoded by PrLBaV ORF1 is a viral replicase.
Figure 3.
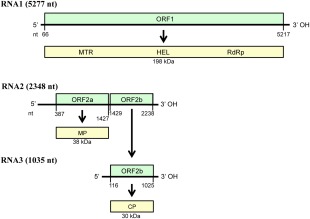
Diagrammatic representation of the genome organization and expression of privet leaf blotch‐associated virus. Horizontal black lines depict RNAs; green boxes show the positions of the open reading frames (ORFs, with nucleotide positions of start and stop codons indicated below); yellow boxes represent the corresponding putative protein products with the molecular mass (kDa) indicated below. RNA3 is a subgenomic RNA generated from the genomic RNA2. CP, coat protein; HEL, helicase; MTR, methyltransferase; MP, movement protein; nt, nucleotide; RdRp, RNA‐dependent RNA polymerase.
Table 1.
Nucleotide and amino acid sequence identity between privet leaf blotch‐associated virus (PrLBaV) and Raspberry bushy dwarf virus (RBDV).
| PrLBaV | RBDV ID | Pairwise identity (%) | blast |
|---|---|---|---|
| Genomic segment | |||
| RNA1 | NC_003739 | 61.46* | 221/328 (67%) ‡ |
| RNA2 | NC_003740 | 55.09* | Not significant |
| Encoded proteins | |||
| p198 (polyprotein) | NP_620465 | 48.41 † | 803/1696 (47%) § |
| Polyprotein domains | |||
| Methyltransferase domain | NP_620465 | 57.52 † | 241/444 (54%) § |
| Helicase domain | NP_620465 | 62.50 † | 197/259 (76%) § |
| RNA‐dependent RNA polymerase domain | NP_620465 | 56.82 † | 250/441 (57%) § |
| p38 (movement protein) | NP_620466 | 25.60 † | 71/249 (28%) § |
| p30 (coat protein) | NP_620467 | 25.66 † | 78/265 (29%) § |
*Nucleotide identity.
†Amino acid identity.
‡ blastn.
§ blastp.
PrLBaV RNA2 (2348 nucleotides) contains two ORFs (ORF2a and ORF2b) separated by an unusual intergenic spacer composed of only one nucleotide. ORF2b is very likely expressed through an sgRNA (RNA3). Rapid amplification of cDNA ends (RACE) experiments mapped the 5′ terminus of RNA3 115 nucleotides upstream of the ORF2b start codon (result supported by six of seven RACE‐derived cDNA clones), corresponding to the nucleotide position 1314 in RNA2 (Fig. 3). Therefore, RNA3 partially overlaps ORF2a (Fig. 3) and starts with six nucleotides (AUAUCU) that closely resemble those found in the 5′‐terminal end of both genomic RNAs. Interestingly, the 5′ most terminal hexanucleotides of RNA1, RNA2 and RNA3 differ from each other only at position 5 (Fig. 2). ORF2a and ORF2b code for a couple of putative proteins of 346 amino acids (38.2 kDa) and 269 amino acids (29.6 kDa), designated p38 and p30 (protein_id: CZS63541.1 and CZS63620.1), respectively (Fig. 3). blastp showed that p38 and p30 had identities of 28% (71/249 amino acids) and 30% (76/265 amino acids) with the putative MP and CP of RBDV, respectively (Table 1). In addition, in PFAM analyses, p30 was identified as related to RBDV CP (Pfam: RBDV_coat, PF06593, e‐value 7.7 e‐22), whereas no match was found for p38 using blastp, PFAM or SMART databases/tools.
RNA‐binding domains (RBDs) in the CP of Alfalfa mosaic virus (AMV, the sole member of the genus Alfamovirus) and in members of the genus Ilarvirus play a role in a phenomenon denoted as ‘genome activation’, which enhances virus infectivity (Bol et al., 1971; for a review, see Jaspars, 1999). Genome activation mediated by CP was also reported in the case of RBDV, although a specific RBD was not conclusively identified (MacFarlane and McGavin, 2009). When PrLBaV p30 protein was analysed with BindN, a few putative RBDs were identified (Fig. 4), including a stretch of residues between amino acid positions 90 and 105, whose RNA‐binding capacity was predicted with very high confidence. Interestingly, a stretch of residues with RNA‐binding properties was also predicted with high confidence at a similar position in the RBDV CP (Fig. 4). These data confirm that p30, encoded by ORF2b, is the CP of PrLBaV. Whether this protein is also involved in PrLBaV genome activation remains to be experimentally addressed and confirmed.
Figure 4.
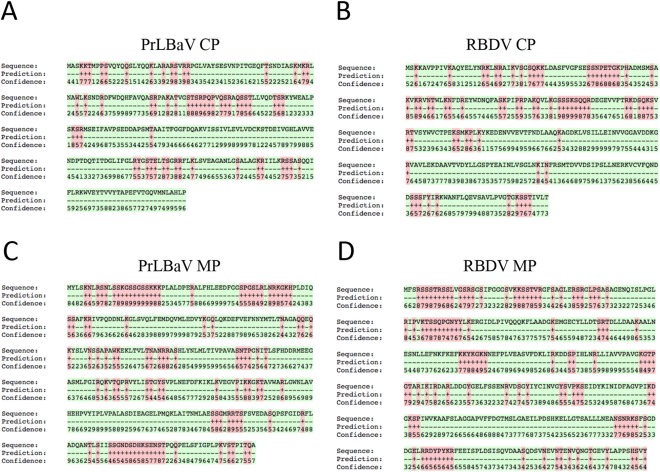
RNA‐binding residues predicted by BindN. Analysis of coat protein (CP) and movement protein (MP) of privet leaf blotch‐associated virus (PrLBaV) (A, C) and Raspberry bushy dwarf virus (RBDV) (B, D). Amino acid sequence, BindN prediction and confidence of the prediction are reported. Binding residues are labelled with ‘+’ and in red; non‐binding residues are labelled with ‘–’ and in green. Confidence: from level 0 (lowest) to level 9 (highest).
p38 of PrLBaV localizes at PD
The PROMALS3D program, which generates multiple protein alignments based on sequences and/or predicted secondary structures (Pei et al., 2008), allowed the identification of the typical signatures of the ‘30K superfamily’ MPs (Melcher, 2000; Mushegian and Elena, 2015) in p38 of PrLBaV, including consecutive β strands, connected to each other by loops with different patterns of sequence conservation, and a nearly invariant aspartic acid residue (the ‘D’ motif) in the third predicted strand (Fig. S3, see Supporting Information). In addition, this protein contains an ‘SIIS’ motif (amino acids 308–311), which recalls the SIS tail found near the C‐termini of most members of the 30K superfamily of virus MPs (Melcher, 2000). Altogether, these features suggest that p38 is an MP belonging to the 30K superfamily.
MPs of the ‘30K superfamily’ also possess nucleic acid‐binding capacity and localize at PD (Waigmann et al., 2004). Accordingly, a unique RBD, located at different positions in the respective MP, has been reported in several representative members in the family Bromoviridae, including AMV, PNRSV (genus Ilarvirus), Brome mosaic virus (BMV, genus Bromovirus) and Cucumber mosaic virus (CMV, genus Cucumovirus) (Fujita et al., 1998; Herranz and Pallás, 2004; Kim et al., 2004; Li and Palukaitis, 1996; Schoumacher et al., 1994). RNA‐binding capability relying on two RBDs (present at the N‐ and C‐termini), instead of one, has been shown recently for the MP of Parietaria mottle ilarvirus (Martínez et al., 2014).
Interestingly, when the amino acid sequence of PrLBaV p38 was analysed by BindN, stretches of residues with predicted RNA‐binding properties (confidence ≥ 6) were found at the N‐ (amino acids 5–23 and 43–55) and C‐termini (amino acids 311–324) of the putative protein (Fig. 4). Whether these predicted RBDs actually play a role in protein functionality remains to be experimentally shown. However, it is worth noting that putative RBDs were also found at similar positions in the presumed MP of RBDV (Fig. 4).
The localization of the putative MP of PrLBaV in the cell was investigated by fusing green fluorescent protein (GFP) to the C‐terminus of PrLBaV p38 (p38:GFP), followed by transient expression by agroinfiltration in Nicotiana benthamiana leaves. Confocal laser scanning microscopy (CLSM) observations revealed that the fused protein emitted fluorescence forming a punctate pattern distributed along the cell wall at 2 days post‐infiltration (Fig. 5A–D), which is in agreement with that observed for other ‘30K’ MPs that have been studied and demonstrated to accumulate at PD (Aparicio et al., 2010; Boyko et al., 2000; Herranz et al., 2005; Martínez et al., 2014; van der Wel et al., 1998). As expected, such a punctate pattern was not observed in cells in which free GFP was transiently expressed (Fig. 5D).
Figure 5.
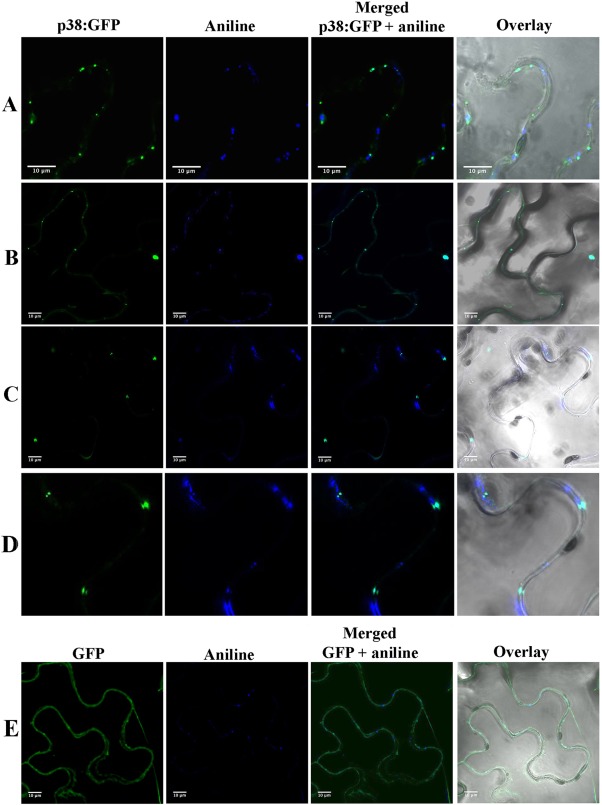
Subcellular localization of privet leaf blotch‐associated virus (PrLBaV) p38 fused to green fluorescent protein (GFP) (p38:GFP). Punctate patterns between neighbouring cells are observed in cells in which p38:GFP is expressed (A–D), with GFP fluorescence co‐localizing with that of aniline, which marks the callose‐rich neck regions of PD (middle and right panels). (D) Closer view of a portion of (C). Co‐localization of GFP and aniline is not observed in negative control cells (E) in which free GFP is transiently expressed.
The accumulation of p38:GFP at PD was further confirmed by the co‐localization of the GFP green punctate signals with the blue fluorescence emitted by aniline blue which, when infiltrated in plant cells, is known to specifically label the callose‐rich neck regions of PD (Martínez et al., 2014). Such co‐localization was absent in the negative controls in which free GFP was transiently expressed (Fig. 5). For the first time, these experiments show that p38 of PrLBaV actually targets PD, providing solid empirical evidence, still lacking in the case of its orthologue in RBDV, for its involvement in virus trafficking.
Phylogenetic relationships of PrLBaV with other viruses
Phylogenetic trees generated using multiple alignments of MTR, HEL and RdRp signatures of PrLBaV, RBDV and several related viruses, including representative members of several genera in the family Bromoviridae and JHFMoV (Valverde and Sabanadzovic, 2009), consistently grouped PrLBaV and RBDV in a clade separated from those formed by members of the other genera in the family Bromoviridae (Fig. 6). A similar tree topography was obtained when the phylogenetic analyses were extended to MPs and CPs of the same representative viruses (Fig. 6). Interestingly, in the case of CP, a third putative idaeovirus, for which only partial sequence data were available at the time of this study (Derrick et al., 2006), clustered together with PrLBaV and RBDV. Summarizing, these analyses highlight the close evolutionary relationships between PrLBaV and RBDV, which are also reflected in the striking similarities in genome organization between the two viruses.
Figure 6.
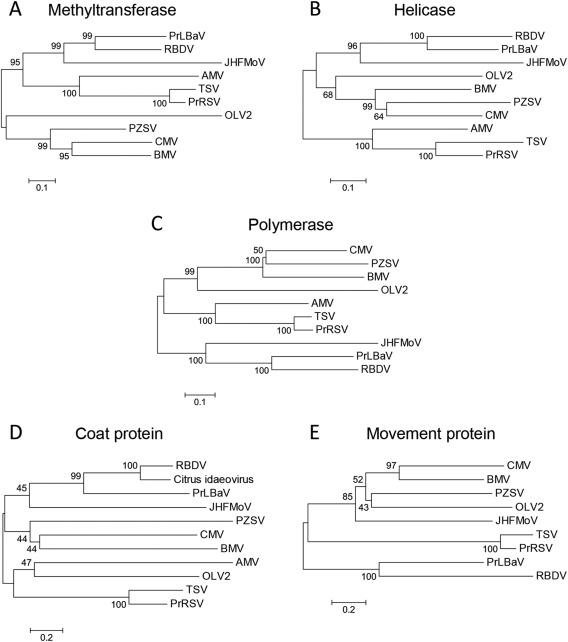
Phylogenetic trees inferred by neighbour‐joining analysis of the methyltransferase (A), helicase (B) and RNA‐dependent RNA polymerase (C) signatures, and of the coat proteins (D) and movement proteins (E) of privet leaf blotch‐associated virus (PrLBaV), RBDV (genus Idaeovirus), JHFMoV and representative viruses in the family Bromoviridae. The number on each branch is the result of bootstrap analysis (1000 replicates). Extended names of the viruses included in the phylogenetic trees are reported below, with the accession numbers of the respective methyltransferase (MTR), helicase (HEL), RNA‐dependent RNA polymerase (RdRp) signatures, and of the coat protein (CP) and movement protein (MP) reported in parentheses. AMV (Alfalfa mosaic virus; MTR and HEL, NP_041192; RdRp, YP_053235; CP, NP_041195; MP, NP_041194), BMV (Brome mosaic virus; MTR and HEL, NP_041196; RdRp, NP_041197; CP, AAA46334; MP, NP_041198), CMV (Cucumber mosaic virus; MTR and HEL, NP_049323; RdRp, NP_049324; CP, NP_040777; MP, NP_040776), JHMoV (Japanese holly fern mottle virus; MTR, HEL and RdRp, YP_003126903; CP, ACT67468; MP, YP_003126905), OLV2 (Olive latent virus 2; MTR and HEL, NP_620042; RdRp, NP_620043; CP, NP_620039; MP, NP_620038), PrRSV (Privet ringspot virus; MTR and HEL, YP_009165996; RdRp, YP_009165997; CP, YP_009166000; MP, YP_009165999), PZSV (Pelargonium zonate spot virus; MTR and HEL, NP_619770; RdRp, NP_619771; CP, NP_619773; MP, NP_619772), RBDV (Raspberry bushy dwarf virus; MTR, HEL and RdRp, NP_620465; CP, NP_620467; MP, NP_620466), TSV (Tobacco streak virus; MTR and HEL, NP_620772; RdRp, NP_620768; CP, NP_620774; MP, NP_620773), Citrus idaeovirus (CP; DQ100358).
Comparison of LB with other similar disorders of privet
None of the eight NRSD‐affected samples of American origin were infected by PrLBaV, and there was no evidence of PrRSV presence in any of the samples affected by LB disease in Italy, suggesting that the two diseases are unrelated.
Discussion
Infectious chlorosis of privet was described at the beginning of the last century (Baur, 1907). Over time, several diseases resembling infectious chlorosis have been reported from different Ligustrum species in various parts of the world, but it is not clear whether or not they are caused by the same infectious agent(s).
Burnett and Youtsey (1962), based on specific symptoms observed in infected plants, clarified that ligustrum necrotic ringspot disease (reported from Florida, USA) and chlorotic spot (originally described from Louisiana, USA) represent the same disease, and proposed the name ‘necrotic ringspot disease’. The same authors doubted that NRSD was related to infectious chlorosis. A novel ilarvirus, referred to as ‘privet necrotic ringspot virus’ (PrRSV), has been identified recently as the possible agent of the former disease (Aboughanem‐Sabanadzovic et al., 2016) as it is found to be consistently associated with NRSD symptoms.
In the present study, we applied NGS to identify a possible causal agent of LB, a graft‐transmissible disease observed in southern Italy and characterized by chlorotic blotches on leaves, resembling infectious chlorosis (Baur, 1907). The characteristic LB symptoms developed on bark‐grafted privet plants at 1 year post‐inoculation. These symptoms are different from those induced by PrRSV, in which chlorotic line patterns and ringspots are observed on young leaves that become necrotic later in the season (Aboughanem‐Sabanadzovic et al., 2016). The sporadic necrotic spots observed in privet affected by LB develop in the middle of white–cream blotches (Fig. 1E) and do not resemble those reported in privet infected by PrRSV (Aboughanem‐Sabanadzovic et al., 2016;), thus suggesting the different nature of the two diseases. Accordingly, PrRSV was not detected in LB‐affected privet by NGS, which is a powerful and unbiased detection method of plant viruses (Massart et al., 2014), especially if the viral genomic sequence is already known. Moreover, none of the other viruses previously reported from privet affected by diseases resembling infectious chlorosis (Cooper, 1993; Scott and Zimmerman, 2008) were detected by NGS in the LB‐affected sample used in this study.
Instead, an apparently novel virus, provisionally named as ‘privet leaf blotch‐associated virus’ (PrLBaV), was identified in LB‐affected privet from Italy. NGS and RT‐PCR assays excluded the presence of PrLBaV in several non‐symptomatic privet controls, thus strengthening the hypothesis of a possible association of this novel virus with the disease. Furthermore, PrLBaV was shown to be absent in several representative samples affected by NRSD in the USA, thus excluding its involvement in that disease. These data also suggest that LB and NRSD are distinct disorders affecting privet.
Sequencing of the complete genome of PrLBaV showed that this virus is closely related to RBDV, with which it shares genomic organization, gene expression strategy and structural elements at both the RNA and protein levels. PrLBaV and RBDV have two genomic RNAs with similar 5′ and 3′ UTRs, consisting of an almost identical hexanucleotide fragment at the 5′ terminus and a stretch of nucleotides assuming a similar secondary structure and terminating with a tetracytidine at the 3′ end. The sizes of RNA1 and RNA2 of PrLBaV are similar to those of the two genomic RNAs of RBDV. Furthermore, the viral replicases encoded by ORF1 in RNA1 of the two viruses share significant amino acid sequence identity to each other. However, the PrLBaV RNA1 is monocistronic, thus lacking the additional putative short ORF present at the 3′ end of RNA1 of RBDV and the related JHFMoV (Ziegler et al., 1992; Valverde and Sabanadzovic, 2009). Interestingly, such an ORF was reported as probably not expressed in RBDV because of the apparent absence of a corresponding sgRNA (Ziegler et al., 1992).
Similar to RBDV, two ORFs (ORF2a and ORF2b coding for MP and CP, respectively) are contained in RNA2 of PrLBaV. The size of the intergenic region (only one nucleotide) in this viral RNA is uncommon and different from that in RBDV RNA2, reported to be more than 40 nucleotides (Mayo et al., 1991). However, the characterization of the 5′ terminus of PrLBaV sgRNA (RNA3) strongly indicates that the gene expression strategy of PrLBaV CP resembles that of RBDV (Jones and Baker, 2008; Sabanadzovic and Martin, 2011). In addition, we showed that PrLBaV RNA3 contains a 5′‐terminal hexanucleotide almost identical to those found in genomic RNA1 and RNA2, indicative of their major role in the modulation of viral gene expression.
Although less conserved than replicase proteins, the putative CP and MP of PrLBaV and RBDV share certain structural features, including putative RBDs (Fig. 4). The RBD in CP could be involved in the phenomenon of genome activation which, similar to AMV and members of the genus Ilarvirus (Jaspars, 1999), has already been reported for RBDV and awaits experimental proof in the case of PrLBaV. The presence of RBDs in p38 of PrLBaV is consistent with its involvement in virus trafficking. In addition, a functional role of p38 as an MP was also supported by the in silico identification of structural elements typical of MPs of the ‘30K superfamily’ in this protein (Fig. S3). Finally, the results of transient expression and confocal microscopy studies carried out in this work provide the ultimate experimental evidence that this protein targets cell PD, a typical feature of plant virus MPs.
The close relationship between PrLBaV and RBDV was further confirmed by phylogenetic analyses which, independent of the protein considered, always grouped these two viruses in the same clade, separated from representative members of genera in the family Bromoviridae. Altogether, these data support the proposal of classifying PrLBaV as the prototype of the second species in the genus Idaeovirus.
Data reported in this study will be useful for further investigation on the epidemiology and spread of PrLBaV in privet and possibly in other hosts. In this context, it is worth noting that PrLBaV most probably also infects ash (Fraxinus excelsior L.). Indeed, a partial sequence of 1323 nucleotides (AN: HM153080), almost identical to PrLBaV RNA1 (96.75% nucleotide identity), has been sequenced from ash and deposited in databases as an RBDV isolate. Pairwise and blast analyses have shown that HM153080 shares only 54% amino acid identity with the MTR domain of several RBDV isolates and 98% amino acid identity with the corresponding region of PrLBaV, indicating that it is an isolate of PrLBaV and not RBDV, as erroneously annotated in GenBank. Interestingly, F. excelsior, like L. japonicum, is a species of the family Oleaceae, thus suggesting that other species in this botanical family could be hosts of PrLBaV.
Experimental Procedures
RNA isolation and sequencing of sRNA libraries
Total RNA was extracted with phenol–chloroform from leaves collected from symptomless and symptomatic privet plants growing in southern Italy, and recovered by ethanol precipitation (Dalmay et al., 1993). sRNAs of 16–30 nucleotides were purified and processed as reported previously (Di Serio et al., 2010) to generate a cDNA library of sRNAs that was subjected to high‐throughput sequencing on a Illumina Genome Analyzer (HiScan SQ apparatus). The raw reads were filtered for quality, reduced to unique reads and de novo assembled into larger contigs using Velvet Software1.2.08 (Zerbino and Birney, 2008) with a k‐mer of 15–17. Following assembly, contigs were screened for sequence homologies using the blastx online resource (http://www.ncbi.nlm.nih.gov/). Partial genomic RNAs of the putative novel virus were reconstructed by manual aligning of contigs with the most closely related viral sequences found in the GenBank database.
Sequencing and analyses of the viral genome
The viral genome was determined by sequencing overlapping cDNA clones generated by RT‐PCR using specific primers designed on selected contigs (Table S2). Total nucleic acids (100 ng), extracted according to the protocol reported by Foissac et al. (2005), were reverse transcribed using random hexamers and Superscript II (Invitrogen‐ThermoFisher Scientific, Waltham, MA, USA) reverse transcriptase, following the manufacturer's instructions. PCR amplifications were performed using Go‐Taq polymerase (Promega, Madison, WI, USA) adopting the following cycling conditions: initial denaturation at 94 °C for 3 min, followed by 30 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min, and a final extension step at 72 °C for 7 min. The 5′ and 3′ ends of viral genomic and sgRNAs were determined using the 5′ RACE strategy with specific reverse primers (Table S2). The PCR products were gel purified, cloned into pGEMT‐Easy vector (Promega) and sequenced.
ORF Finder at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to search for potential ORFs in the virus genomic and anti‐genomic RNAs. SMART analyses (Letunic et al., 2015), which include searches in the PFAM (http://pfam.xfam.org/) database, were used for the identification of conserved protein domains (Finn et al., 2014). The RNAfold web resource (Gruber et al., 2008) was used to predict the secondary structure of the 3′ ends of genomic RNAs. The prediction of potential RNA‐binding residues was performed with BindN (http://bioinfo.ggc.org/bindn/). Alignments for multiple protein sequences and/or structures were performed using PROMALS3D (http://prodata.swmed.edu/promals/promals.php; Pei et al., 2008), which also generated secondary structure prediction for the aligned sequences using the PSIPRED algorithm (Jones, 1999). Amino acid sequence alignments were generated with the ClustalW program (Larkin et al., 2007). Phylogenetic trees were obtained with the neighbour‐joining method (Saitou and Nei, 1987) using 1000 bootstrap replicates in the software package MEGA6 (Tamura et al., 2013).
Cloning of PrLBaV MP fused to GFP and transient expression by Agrobacterium infiltration
PrLBaV ORF2a was amplified by RT‐PCR from total RNA extracted from a symptomatic privet plant using MP‐NcoI‐For (5′‐CCATGGATCTTTCAAAGAATCTTCGTTC‐3′) and MP‐NheI‐Rev (5′‐GCTAGCCGCTTGAGTAATTGGGG‐3′) primers (containing NcoI and NheI sites, respectively; in italic). The PCR product was digested with NcoI and NheI restriction enzymes and inserted into the plasmid pSK35S/GFP (Herranz et al., 2005) linearized with the same enzymes, resulting in a construct (p38:GFP cassette) composed of the cDNA of PrLBaV p38 fused in frame to the 5′ end of GFP cDNA under the control of the 35S Cauliflower mosaic virus (CaMV) promoter. The plasmid obtained was HindIII digested and the p38:GFP cassette was introduced into the plant binary vector pMOG800 (Knoester et al., 1998) to generate pMOG‐p38GFP. Agrobacterium tumefaciens cells (strain C58C5), transformed with pMOG‐GFP and pMOG‐p38GFP, were used for transient expression assays in N. benthamiana leaves. Bacterial cultures were grown in liquid Luria–Bertani medium containing antibiotics at 28 °C for 24 h, resuspended in infiltration buffer [10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.6, 150 µm acetosyringone] at an optical density at 600 nm (OD600) of 0.2 or 0.4, and infiltrated into expanded leaves of 3‐week‐old N. benthamiana plants. Fluorescence was visualized and photographed at 2 days post‐infiltration with a Zeiss LSM 780 AxiObserver (Zeiss, Oberkochen, Germany). Ten minutes before visualization, the leaves were infiltrated with aniline blue solution [0.005% aniline blue (Merck, Darmstadt, Germany) in sodium phosphate buffer, 70 mm, pH 9.0] to specifically label callose‐rich neck regions of PD. Excitation and emission wavelengths were 488 and 508 nm for GFP and 460–535 nm for aniline blue.
Comparison of LB with other similar disorders of privet
In order to clarify whether the LB disorder is related to necrotic ringspot, a disease reported more than 50 years ago from several states in southern USA, we performed RT‐PCR experiments to check for the possible presence of PrRSV in our samples using the primers reported by Aboughanem‐Sabanadzovic et al. (2016). In addition, eight representative samples affected by NRSD collected in Mississippi were checked for the presence of PrLBaV using the primer set Li6‐Li7 (Table S2).
Accession numbers
The GenBank accession numbers LT221868 and LT221869 have been assigned to the sequences of PrLBaV RNA1 and RNA2.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Contigs obtained by assembling small RNAs (sRNAs) from symptomatic leaves of privet sequenced by Illumina technology and showing homology with Raspberry bushy dwarf virus (RBDV).
Table S2 Primers used in this study.
Fig. S1 Schematic representation of the relative positions of privet leaf blotch‐associated virus (PrLBaV) contigs mapped in the genomic RNAs of Raspberry bushy dwarf virus (RBDV) (in blue). Contigs are reported as red lines. Primers designed for the amplification of PrLBaV genomic cDNAs based on this scaffold are indicated by arrows. The sequences of the primers are reported in Table S2.
Fig. S2 Detection by reverse transcription‐polymerase chain reaction (RT‐PCR) of RNA1 (A) and RNA2 (B) in privet plants: 1, non‐symptomatic privet plants; 2, privet plant affected by leaf blotch disease; 3, water control (water instead of RNA was added to the RT‐PCR); 4, DNA molecular weight marker (100‐bp ladder, Geneaid Biotech, Taiwan) with sizes (bp) indicated on the right.
Fig. S3 Multiple sequence alignment of movement proteins from privet leaf blotch‐associated virus (PrLBaV) and plant viruses of the 30K superfamily. Sequences are from representative members of the virus genera in the 30K superfamily. PrLBaV is shown in bold. The GenBank identifier, virus genus and the distance in amino acids from the N‐terminus of the protein are shown on the left side of each sequence. Consensus secondary structure elements, predicted by PSIRED within the PROMALS3D program, are reported at the top with the strands and helices indicated with e and h, respectively. The nearly invariant aspartic acid residue (the D motif) is shown in bold.
Acknowledgements
We wish to thank Dr Federico Aparicio (Instituto de Biologia Molecular y Celular de Plantas UPV‐CSIC, IBMCP, Valencia, Spain) for helpful discussions, Marisol Guascón (IBMCP) for valuable technical assistance and advice with confocal microscopy, Antonia Antonacci for excellent technical assistance, and Professor Sead Sabanadzovic (Mississippi State University) and Professor Ricardo Flores (IBMCP) for critical reading of the manuscript and for suggestions. Work in the laboratories of B.N. and F.D.S. was partially supported by a dedicated grant (CISIA) from the Ministero dell'Economia e Finanze Italiano to the CNR (Legge n. 191/2009).
References
- Aboughanem‐Sabanadzovic, N. , Tzanetakis, I.E. , Lawrence, A. , Stephenson, C.R. and Sabanadzovic, S. (2016) A novel ilarvirus is associated with privet necrotic ringspot disease in the southern USA. Phytopathology, 106, 87–93. [DOI] [PubMed] [Google Scholar]
- Aparicio, F. , Pallás, V. and Sánchez‐Navarro, J.A. (2010) Implication of the C terminus of the prunus necrotic ringspot virus movement protein in cell to cell transport and its interaction with the coat protein. J. Gen. Virol. 91, 1865–1870. [DOI] [PubMed] [Google Scholar]
- Baur, E. (1907) Weitere Mitteilungen über due infektiöse Chlorose der Malvaceen und über einige analoge Erscheinungen bei Ligustum und Laburnum . Ber. Dtsch. Bot. Ges. 24, 416–428. [Google Scholar]
- Bol, J.F. , van Vloten‐Doting, L. and Jaspars, E.M.J. (1971) A functional equivalence of top component a RNA and coat protein in the initiation of infection by alfalfa mosaic virus. Virology, 46, 73–85. [DOI] [PubMed] [Google Scholar]
- Boyko, V. , van der Laak, J. , Ferralli, J. , Suslova, E. , Kwon, M.O. and Heinlein, M. (2000) Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J. Virol. 74, 11 339–11 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett, H.C. and Youtsey, C.O. (1962) Necrotic ringspot, a virus disease of Ligustrum sp. Fla. State Hort. Soc. 75, 472–476. [Google Scholar]
- Cooper, I.J. (1993) Virus Diseases of Trees and Shrubs. London: Chapman & Hall. [Google Scholar]
- Dalmay, T. , Rubino, L. , Burgyán, J. , Kollár, Á and Russo, M. (1993) Functional analysis of cymbidium ringspot virus genome. Virology, 194, 697–704. [DOI] [PubMed] [Google Scholar]
- Derrick, K.S. , Beretta, M.J. and Barthe, G.A. (2006) Detection of an idaeovirus in citrus with implication as to the cause of citrus blight. Proc. Fla. State Hort. Soc. 119, 69–72. [Google Scholar]
- Di Serio, F. , Martínez de Alba, A.E. , Navarro, B. , Gisel, A. and Flores, R. (2010) RNA‐dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a nuclear‐replicating viroid. J. Virol. 84, 2477–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire, L. , Wang, Y. , Gonzalez‐Ibeas, D. , Mayer, K.F. , Aranda, M.A. and Llave, C. (2009) Deep‐sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology, 392, 203–214. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Miller, B.L. , Clements, J. and Bateman, A. (2014) iPfam: a database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res. 42, D364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac, X. , Svanella‐Dumas, L. , Gentit, P. , Dulucq, M.J. , Marais, A. and Candresse, T. (2005) Polyvalent degenerate oligonucleotides reverse transcription–polymerase chain reaction: a polyvalent detection and characterization tool for trichoviruses, capilloviruses, and foveaviruses. Phytopathology, 95, 617–625. [DOI] [PubMed] [Google Scholar]
- Fujita, M. , Mise, K. , Kajiura, Y. , Dohi, K. and Furusawa, I. (1998) Nucleic acid‐binding properties and subcellular localization of the 3a protein of Brome mosaic bromovirus. J. Gen. Virol. 79, 1273–1280. [DOI] [PubMed] [Google Scholar]
- Gruber, A.R. , Lorenz, R. , Bernhart, S.H. , Neuböck, R. and Hofacker, I.L. (2008) The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, M.C. and Pallás, V. (2004) RNA‐binding properties and mapping of the RNA‐binding domain from the movement protein of Prunus necrotic ringspot virus. J. Gen. Virol. 85, 761–768. [DOI] [PubMed] [Google Scholar]
- Herranz, M.C. , Sánchez‐Navarro, J.A. , Saurí, A. , Mingarro, I. and Pallás, V. (2005) Mutational analysis of the RNA‐binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell‐to‐cell movement. Virology, 339, 31–41. [DOI] [PubMed] [Google Scholar]
- Jaspars, E.M. (1999) Genome activation in alfamo‐ and ilarviruses. Arch. Virol. 144, 843–863. [DOI] [PubMed] [Google Scholar]
- Jones, A.T. and Baker, H. (2008) Idaeovirus In: Encyclopedia of Virology, 3rd edn (Mahy B.W.J. and van Regenmortel M.H.V., eds), pp. 37–41. Amsterdam: Academic Press. [Google Scholar]
- Jones, D.T. (1999) Protein secondary structure prediction based on position‐specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Kalinina, N.O. , Andreev, I. , Ryabov, E.V. , Fitzgerald, A.G. , Taliansky, M.E. and Palukaitis, P. (2004) The C‐terminal 33 amino acids of the Cucumber mosaic virus 3a protein affect virus movement, RNA binding and inhibition of infection and translation. J. Gen. Virol. 85, 221–230. [DOI] [PubMed] [Google Scholar]
- Knoester, M. , van Loon, L.C. , van den Heuvel, J. , Hennig, J. , Bol, J.F. and Linthorst, H.J.M. (1998) Ethylene‐insensitive tobacco lacks nonhost resistance against soil‐borne fungi. Proc. Natl. Acad. Sci. USA, 95, 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. and Dolja, V.V. (1993) Evolution and taxonomy of positive‐strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28, 375–430. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. and Palukaitis, P. (1996) Comparison of the nucleic acid‐ and NTP‐binding properties of the movement protein of Cucumber mosaic cucumovirus and Tobacco mosaic tobamovirus. Virology, 216, 71–79. [DOI] [PubMed] [Google Scholar]
- MacFarlane, S.A. 2012. Genus Idaeovirus In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q., Adams M.J., Carstens E.B., and Lefkowitz E.J., eds), pp. 1173–1175. London: Elsevier/Academic Press. [Google Scholar]
- MacFarlane, S.A. and McGavin, W.J. (2009) Genome activation by Raspberry bushy dwarf virus coat protein. J. Gen. Virol. 90, 747–753. [DOI] [PubMed] [Google Scholar]
- Martínez, C. , Coll‐Bonfill, N. , Aramburu, J. , Pallás, V. , Aparicio, F. and Galipienso, L. (2014) Two basic (hydrophilic) regions in the movement protein of Parietaria mottle virus have RNA binding activity and are required for cell‐to‐cell transport. Virus Res. 184, 54–61. [DOI] [PubMed] [Google Scholar]
- Massart, S. , Olmos, A. , Jijakli, H. and Candresse, T. (2014) Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 188, 90–96. [DOI] [PubMed] [Google Scholar]
- Mayo, M.A. , Jolly, C.A. , Murant, A.F. and Raschke, J.H. (1991) Nucleotide sequence of raspberry bushy dwarf virus RNA‐3. J. Gen. Virol. 72, 469–472. [DOI] [PubMed] [Google Scholar]
- Melcher, U. (2000) The ‘30K’ superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R. and Elena, S.F. (2015) Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology, 476, 304–315. [DOI] [PubMed] [Google Scholar]
- Natsuaki, T. , Mayo, M.A. , Jolly, C.A. and Murant, A.F. (1991) Nucleotide sequence of raspberry bushy dwarf virus RNA‐2: a bicistronic component of a bipartite genome. J. Gen. Virol. 72, 2183–2189. [DOI] [PubMed] [Google Scholar]
- Pei, J. , Kim, B.H. and Grishin, N,V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirone, P.P. (1978) Ligustrum diseases In: Diseases and Pests of Ornamental Plants, 5th edn (Pirone P.P., ed.), pp. 340–341. New York: Wiley. [Google Scholar]
- Sabanadzovic, S. and Martin, R.R. (2011). Genus Idaeovirus In Springer Index of Viruses, 2nd edn. (Tidona C. and Darai G., eds), pp. 2005–2009. Heidelberg: Springer. [Google Scholar]
- Saitou, N. and Nei, M. (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schmelzer, K. (1963) Untersuchungen as Viren der Zier und Wildgeholze 2. Mitteilung: virosen as Forsythia, Lonicera, Ligustrum und Laburnum . Phytopathol Z., 46, 105–138. [Google Scholar]
- Schoumacher, F. , Giovane, C. , Maira, M. , Poirson, A. , Godefroy‐Colburn, T. and Berna, A. (1994) Mapping of the RNA‐binding domain of the Alfalfa mosaic virus movement protein. J. Gen. Virol. 75, 3199–3202. [DOI] [PubMed] [Google Scholar]
- Scott, S.W. and Zimmerman, M.T. (2008) The complete sequence of ligustrum necrotic ringspot virus, a novel carlavirus. Arch. Virol. 153, 393–396. [DOI] [PubMed] [Google Scholar]
- Seguin, J. , Rajeswaran, R. , Malpica‐Lopez, N. , Martin, R.R. , Kasschau, K. , Dolja, V.V. , Otten, P. , Farinelli, L. and Pooggin, M.M. (2014) De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS One, 9, e88513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, R.A. and Sabanadzovic, S. (2009) A new plant virus with unique properties infecting Japanese holly fern. J. Gen. Virol. 90, 2542–2549. [DOI] [PubMed] [Google Scholar]
- Waigmann, E. , Ueki, S. , Trutnyeva, K. and Citovsky, V. (2004) The ins and outs of non‐destructive cell‐to‐cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23, 195–250. [Google Scholar]
- van der Wel, N.N. , Goldbach, R.W. and van Lent, J.W. (1998) The movement protein and coat protein of alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology, 244, 322–329. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Ding, S.W. , Zhang, Y. and Zhu, S. (2015) Identification of viruses and viroids by next‐generation sequencing and homology‐dependent and homology‐independent algorithms. Annu. Rev. Phytopathol. 53, 425–444. [DOI] [PubMed] [Google Scholar]
- Zerbino, D.R. and Birney, E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, A. , Natsuaki, T. , Mayo, M.A. , Jolly, C.A. and Murant, A.F. (1992) The nucleotide sequence of RNA‐1 of raspberry bushy dwarf virus. J. Gen. Virol. 73, 3213–3218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Contigs obtained by assembling small RNAs (sRNAs) from symptomatic leaves of privet sequenced by Illumina technology and showing homology with Raspberry bushy dwarf virus (RBDV).
Table S2 Primers used in this study.
Fig. S1 Schematic representation of the relative positions of privet leaf blotch‐associated virus (PrLBaV) contigs mapped in the genomic RNAs of Raspberry bushy dwarf virus (RBDV) (in blue). Contigs are reported as red lines. Primers designed for the amplification of PrLBaV genomic cDNAs based on this scaffold are indicated by arrows. The sequences of the primers are reported in Table S2.
Fig. S2 Detection by reverse transcription‐polymerase chain reaction (RT‐PCR) of RNA1 (A) and RNA2 (B) in privet plants: 1, non‐symptomatic privet plants; 2, privet plant affected by leaf blotch disease; 3, water control (water instead of RNA was added to the RT‐PCR); 4, DNA molecular weight marker (100‐bp ladder, Geneaid Biotech, Taiwan) with sizes (bp) indicated on the right.
Fig. S3 Multiple sequence alignment of movement proteins from privet leaf blotch‐associated virus (PrLBaV) and plant viruses of the 30K superfamily. Sequences are from representative members of the virus genera in the 30K superfamily. PrLBaV is shown in bold. The GenBank identifier, virus genus and the distance in amino acids from the N‐terminus of the protein are shown on the left side of each sequence. Consensus secondary structure elements, predicted by PSIRED within the PROMALS3D program, are reported at the top with the strands and helices indicated with e and h, respectively. The nearly invariant aspartic acid residue (the D motif) is shown in bold.


