Summary
Virus infections in plants cause changes in host gene expression that are common to other environmental stresses. In this work, we found extensive overlap in the transcriptional responses between Arabidopsis thaliana plants infected with Tobacco rattle virus (TRV) and plants undergoing senescence. This is exemplified by the up‐regulation during infection of several senescence‐associated Dark‐inducible (DIN) genes, including AtDIN1 (Senescence 1, SEN1), AtDIN6 (Asparagine synthetase 1, AtASN1) and AtDIN11. DIN1, DIN6 and DIN11 homologues were also activated in Nicotiana benthamiana in response to TRV and Potato virus X (PVX) infection. Reduced TRV levels in RNA interference (RNAi) lines targeting AtDIN11 indicate that DIN11 is an important modulator of susceptibility to TRV in Arabidopsis. Furthermore, low accumulation of TRV in Arabidopsis protoplasts from RNAi lines suggests that AtDIN11 supports virus multiplication in this species. The effect of DIN6 on virus accumulation was negligible in Arabidopsis, perhaps as a result of gene or functional redundancy. However, TRV‐induced silencing of NbASN, the DIN6 homologue in N. benthamiana, compromises TRV and PVX accumulation in systemically infected leaves. Interestingly, NbASN inactivation correlates with the appearance of morphological defects in infected leaves. We found that DIN6 and DIN11 regulate virus multiplication in a step prior to the activation of plant defence responses. We hypothesize on the possible roles of DIN6 and DIN11 during virus infection.
Keywords: Arabidopsis thaliana, Dark‐inducible (DIN) genes, Nicotiana benthamiana, plant viruses, Potato virus X, senescence‐associated genes, Tobacco rattle virus
Introduction
Plants respond to pathogen infection by reprogramming host gene expression, which includes, among others, the induction of a suite of defence‐ and pathogenesis‐related genes (van Loon et al., 2006; Wise et al., 2007). The expression of defence‐related genes is also triggered during senescence, a process of programmed cell death that occurs in response to developmental, physical, physiological and hormonal cues (Buchanan‐Wollaston et al., 2003; Gepstein et al., 2003; Lim and Nam, 2005; Quirino et al., 2000). Interestingly, senescence‐associated genes are activated during pathogen attack, which is suggestive of common events or signalling pathways in the control of gene expression in these two overlapping processes (Lin and Wu, 2004; Quirino et al., 1999). Some senescence‐enhanced genes are detected during the hypersensitive response against incompatible bacteria and fungi and against virulent or avirulent pathogens that cause necrosis (Pontier et al., 1999; Schenk et al., 2005). This observation could lead to the misleading assumption that cell death is the necessary point of convergence between defence and senescence. However, senescence‐related genes are also expressed at elevated levels during compatible interactions between plants and viruses (Espinoza et al., 2007; Whitham et al., 2003). Hence, the execution of the cell death programme seems to be uncoupled from the initial regulation of senescence. Cellular stress is the most likely factor explaining why senescence‐related genes are activated during pathogen‐activated responses. Indeed, many stress‐inducible genes are up‐regulated during senescence, which implies that cells undergoing senescence are subjected to stress conditions (Gepstein et al., 2003; Weaver et al., 1998). In addition, stresses caused by drought, darkness, sugar starvation, wounding or leaf detachment initiate the senescence programme (Lin and Wu, 2004; Weaver et al., 1998). The extent and significance of this overlap and the precise roles of pathogen‐ and senescence‐responsive pathways remain largely unknown.
The production of reactive oxygen species (ROS) is a unifying response to multiple stresses and, consequently, has been proposed as a common signal controlling gene expression in stressed plants (Love et al., 2005; Quirino et al., 2000; Torres and Dangl, 2005). Senescence and pathogen infection are both accompanied by a certain degree of cellular damage caused by oxidative stress and, accordingly, ROS detoxification genes are stimulated in both types of process (Buchanan‐Wollaston et al., 2003; Espinoza et al., 2007; Schippers et al., 2008; Whitham et al., 2003). For instance, glutamate dehydrogenase (GDH), which is activated by both senescence and viral or bacterial infections, is highly expressed under oxidative stress (Pageau et al., 2006). Chemical signal compounds, such as salicylic acid (SA) and methyl jasmonate (MeJA), which have defensive and senescence‐promoting functions, regulate the expression of certain senescence‐ and pathogen‐associated genes (Schenk et al., 2005). Changes in the source‐to‐sink balance, normally associated with pathogen invasions, are also pivotal in regulating leaf senescence (Herbers et al., 2000; Masclaux et al., 2000; Shalitin and Wolf, 2000).
In this study, we have identified a set of genes, collectively named Dark‐inducible (DIN), that actively respond to Tobacco rattle virus (TRV) infection in Arabidopsis thaliana. DIN genes are induced in dark‐adapted and senescing leaves (Fujiki et al., 2001, 2005; Lin and Wu, 2004; Quirino et al., 2000), and inspection of Arabidopsis microarray data using Genevestigator indicates that DIN genes also respond to biotic stimuli. This finding suggests that DIN genes could participate in cellular events that are common to both senescence and pathogen invasion (Zimmermann et al., 2004). However, although some DIN genes, such as DIN1, DIN6 and DIN11, have been used previously as markers to characterize senescence‐associated responses, their regulation and function during compatible interactions between viruses and host plants are largely unknown (Lam et al., 1994; Oh et al., 1996; Schenk et al., 2005). Here, we show that DIN6‐ and DIN11‐encoding enzymatic activities may play critical roles in the modulation of plant susceptibility to virus infection.
Results
Gene expression profiling reveals significant similarities between TRV infection and senescence in Arabidopsis
To discriminate the group of genes whose expression was similarly regulated in response to TRV infection and senescence, we compared microarray data of the Arabidopsis response to TRV with gene expression profiling data during developmental or dark‐induced leaf senescence (Buchanan‐Wollaston et al., 2005; Fernandez‐Calvino et al., 2014; Lin and Wu, 2004). Global transcriptomic changes in TRV‐infected Arabidopsis plants were documented at 8 days after inoculation (dai), when TRV has been shown to exhibit the highest peak of genomic RNA accumulation (Fernandez‐Calvino et al., 2014). TRV‐responsive genes (Bonferroni‐corrected P < 0.01) were compared with the list of genes described above. We found a group of 68 genes that responded similarly to virus infection, dark‐induced senescence and age‐mediated (natural) senescence [Table S1 (Supporting Information) and Fig. 1A]. Interestingly, 61% of the TRV‐responsive genes (724 genes) were altered in the same direction (either induced or repressed) when senescence was artificially induced by darkness or carbon starvation, and 8% (97 genes) were shared between TRV infection and developmental leaf senescence. In both cases, the number of common genes in the intersection between TRV infection and dark‐induced senescence or developmental senescence was higher than that expected in a random distribution (P < 0.0001), indicating significant commonalities in their transcriptional responses (Fig. 1A). Using an independent set of plants, and consistent with microarray data, quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) revealed increased transcript levels of the stress‐inducible Senescence‐associated gene 13 (AtSAG13, At2g29350) at 8 dai (Fig. 1B) (Gan and Amasino, 1997; Weaver et al., 1998).
Figure 1.
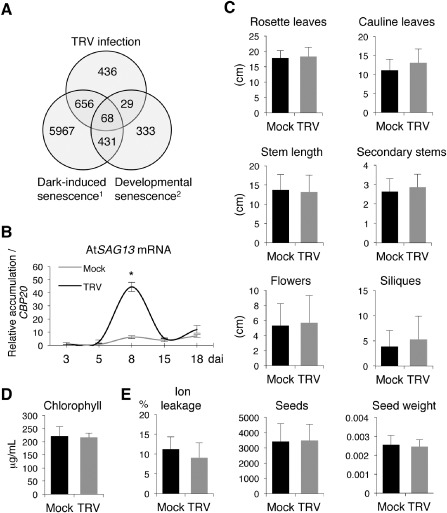
Analysis of senescence‐associated traits during Tobacco rattle virus (TRV) infection in Arabidopsis. (A) Venn diagram to illustrate the number of specific and common misregulated genes in dark‐induced senescence, natural senescence and TRV infection in Arabidopsis. Genes showing significant TRV responsiveness were compared with Affymetrix expression data from the two types of senescence: 1, Lin and Wu (2004); 2, Buchanan‐Wollaston et al. (2005). The numbers of genes within the intersections are higher than those expected by chance. To test whether the significance of matches was higher than explained by random sampling, we assumed a Poisson distribution from a mean value calculated as μ = G1 × (G2/Gt), as any gene in G1 has a G2/Gt possibility of belonging to G2, where G1 and G2 are the subsets of genes to be compared and Gt is the total number of genes represented in the microarray. (B) Time‐course expression of the stress senescence marker AtSAG13 in TRV‐infected plants examined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The means of three measurements ± standard deviation (SD) are shown. Values are relative to those in mock‐inoculated plants at 3 days after inoculation (dai) which were arbitrarily assigned a value of unity after normalization to the AtCBP20 internal control. Asterisks indicate significant difference from the mock‐inoculated control (P < 0.001, Duncan's multiple range test). (C) Effect of TRV infection on the number of rosette and cauline leaves, secondary stems, inflorescences and siliques, and stem length (cm) at 16 dai. Total seed number and weight (g) were used to quantify progeny production. (D) Total chlorophyll content in Arabidopsis leaves at 16 dai. (E) Activation of cell death measured by leakage of ions in mock‐inoculated and TRV‐infected plants at 16 dai. The means and SD of three independent replicates are shown.
Despite the extensive degree of overlap in the transcriptomic responses of TRV‐infected leaves and leaves undergoing senescence (Fig. 1A), TRV infection in Arabidopsis apparently proceeds in the absence of macroscopic alterations that could be diagnostic of accelerated induction of leaf senescence (Lin et al., 2009; Quirino et al., 2000). To accurately determine the effect of virus infection on plant growth and development, we measured a series of morphological traits in vegetative and reproductive organs in mock‐inoculated and TRV‐infected plants at 16 and 29 dai. None of the growth‐related traits tested differed significantly between healthy and TRV‐infected plants (Figs 1C and S1, see Supporting Information). Likewise, equivalent numbers of viable seeds and seed weight were produced by infected and mock‐inoculated plants (Fig. 1C). Chlorophyll breakdown during chloroplast disassembly is a typical visible manifestation of senescence (Quirino et al., 2000). However, chlorophyll content remained at comparable levels in mock‐inoculated and TRV‐infected leaves at 16 and 29 dai (Figs 1D and S1). We also determined electrolyte leakage to assess whether virus infection caused membrane damage. Our results indicated a similar percentage of electrolyte leakage in leaves inoculated with TRV and in upper non‐inoculated TRV‐infected leaves with respect to the corresponding non‐infected leaves, even at later time points after infection (Figs 1E and S1). Collectively, the overlap in the transcriptional responses between TRV infection and both types of senescence suggests a significant interaction between these two stress‐related processes, which, however, did not reflect a premature ongoing senescence‐like phenotype.
TRV stimulates the expression of DIN genes in Arabidopsis
Among the genes that exhibited similar responsiveness to TRV in different Arabidopsis organs (F. J. del Toro and C. Llave, unpublished data), we identified several senescence‐associated genes catalogued as DIN genes. The gene expression data and predicted functions of Arabidopsis DIN genes deduced from their amino acid sequences are shown in Table 1. To validate microarray‐based expression of DIN genes in TRV‐infected Arabidopsis, we conducted qRT‐PCR on the same pooled RNA extracts as used for microarray hybridization, as well as on an independent set of samples (not used for microarrays), to confirm reproducibility in their expression patterns. According to this analysis, AtDIN1 (also Senescence 1, AtSEN1) and Asparagine synthetase 1 (AtASN1), the Arabidopsis homologue of DIN6 (herein referred to as DIN6 for simplicity), transcripts were significantly more abundant in TRV‐infected tissue than in the non‐infected control in the two sets of samples collected from leaves, inflorescences and roots (Fig. 2). These results corroborated the microarray‐based prediction that AtDIN1 and AtDIN6 were strongly and widely activated by TRV. qRT‐PCR demonstrated that AtDIN11, which was not represented in the CATMA (Complete Arabidopsis Transcriptome MicroArray) used, was induced by TRV in all three tissues analysed using independent RNA preparations (Fig. 2). In this study, we focus on AtDIN1, AtDIN6 and AtDIN11, given that these three genes were significantly up‐regulated and exhibited broad responsiveness throughout the plant (Table 1).
Table 1.
Microarray‐based expression changes in Dark‐inducible (DIN) genes in three Arabidopsis organs
| Gene | AGI | Protein description | Fold change† | ||
|---|---|---|---|---|---|
| Leaves | Inflorescences | Roots | |||
| DIN1 | At4g35770 | Senescence‐associated gene 1 (SEN1) | 7.31 *** | 4.75 *** | 3.58 *** |
| DIN2 | At3g60140 | β‐Glucosidase 30, Senescence‐associated gene 2 (SEN2) | 1.04 | −0.98 | 1.04 |
| DIN3 | At3g06850 | Dihydrolipoamide branched chain acyltransferase (BCE2) | 1.06 | 1.44 * | 1.86 *** |
| DIN4 | At3g13450 | Branched chain α‐keto acid dehydrogenase E1 β | 1.91 ** | 2.65 *** | 1.5 ** |
| DIN6 | At3g47340 | Glutamine‐dependent asparagine synthetase 1 (ASN1) | 2.05 *** | 3.01 *** | 1.89 *** |
| DIN9 | At1g67070 | Phosphomannose isomerase 2 (PMI2) | −0.96 | −0.97 | 1.00 |
| DIN10 | At5g20250 | Raffinose synthase 6 (RS6) | −0.82 | 4.5 *** | 1.94 *** |
| DIN11 | At3g49620 | 2‐Oxoacid‐dependent dioxygenase | NDa | ND | ND |
†Bold numbers indicate significant differences in gene expression between mock‐inoculated and Tobacco rattle virus (TRV)‐infected plants. Differences from control values were significant at P < 1 × 10−12 (***), P < 0.01 (**) or P < 0.05 (*) (Bonferroni‐corrected method).
ND, not detected.
Figure 2.
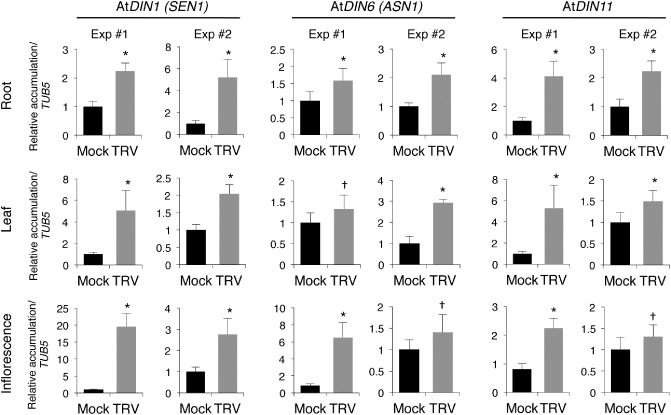
Expression of Dark‐inducible (DIN) genes in response to Tobacco rattle virus (TRV) infection in Arabidopsis. Transcript levels of AtDIN1 (Senescence 1, SEN1), AtDIN6 (Asparagine synthetase 1, ASN1) and AtDIN11 were examined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in two independent experiments (Exp #1 and Exp #2). The means of three measurements ± standard deviation (SD) are shown. Values are given relative to those in mock‐inoculated plants that were arbitrarily assigned a value of unity after AtTUB5 normalization. Samples were collected from roots and non‐inoculated rosette leaves at 8 days after inoculation (dai) and inflorescences at 12 dai. Differences from control values were significant at P < 0.001 (*) or P < 0.05 (†) (Duncan's multiple range test).
Microarray data indicated the deregulation of other dark‐responding genes in the presence of TRV, although changes in their expression patterns were not very reproducible between organs or batches of plants. In general, AtDIN3 and AtDIN10 were activated by TRV in both inflorescences and roots, and their mRNA accumulation patterns were confirmed by qRT‐PCR in independent RNA samples (Fig. S2, see Supporting Information). Although microarray data did not reflect differential changes in AtDIN3 or AtDIN10 expression in leaves, qRT‐PCR showed that AtDIN10 mRNA levels were substantially reduced using independent samples from infected relative to non‐infected leaves, whereas AtDIN3 was either repressed or unaffected by TRV (Fig. S2). We found that AtDIN2 transcripts, which did not exhibit TRV responsiveness on the basis of microarray‐based expression profiling, were significantly more abundant in both sets of samples from roots and leaves when assessed using qRT‐PCR (Fig. S2). Finally, our gene expression tests indicated that AtDIN9 was consistently up‐regulated in inflorescences on TRV challenge, whereas transcript accumulation varied when roots and leaves were analysed (Fig. S2).
AtDIN11 enhances susceptibility to TRV in Arabidopsis
To assess the contribution of DIN1, DIN6 and DIN11 to TRV infection, we used Arabidopsis knockout lines of these genes to monitor virus accumulation. qRT‐PCR revealed that TRV accumulated in single din1 or din6 mutants to the same levels as in the wild‐type background, suggesting that neither AtDIN1 nor AtDIN6 was critical for viral susceptibility in Arabidopsis (Fig. 3A,B). To test AtDIN11 contribution to TRV infection, we obtained transgenic Arabidopsis plants harbouring a hairpin dsRNA‐based RNA interference (RNAi) construct targeted against the AtDIN11 gene. Total RNA from systemically infected rosette leaves of two independent homozygous T3 Arabidopsis lines (L10 and L21) infected with TRV was subjected to qRT‐PCR experiments. In these plants, AtDIN11 transcript levels were reduced by more than 80% compared with wild‐type controls, demonstrating the effective silencing of the targeted gene (Fig. 3C). Silencing of AtDIN11 in both L10 and L21 RNAi lines correlated with a significant two‐fold reduction in TRV accumulation compared with wild‐type plants at 5 and 8 dai (Fig. 3C). Interestingly, TRV levels showed large differences between independent replicates and genetic backgrounds at 16 dai (data not shown), suggesting that the influence of DIN11 on TRV accumulation could be restricted to the early time points of infection during which virus proliferation is particularly dynamic. To test this hypothesis, we isolated protoplasts from Arabidopsis rosette leaves and subjected them to transfection with the infectious pTRV1 vector. qRT‐PCR of total RNA extracted at 24 h post‐transfection demonstrated that viral replication was severely compromised in both L10 and L21 lines (Fig. 3D). This result was highly reproducible, confirming that a functional DIN11 was required to allow virus multiplication. No major visible morphological differences were observed between knockout or RNAi lines and wild‐type plants (data not shown).
Figure 3.
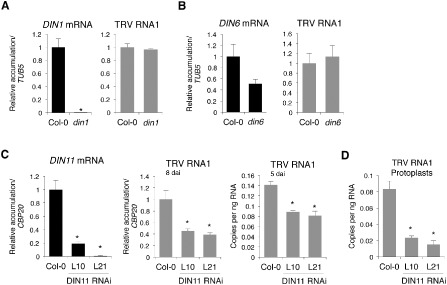
Effect of Dark‐inducible (DIN) genes on Tobacco rattle virus (TRV) susceptibility in Arabidopsis. Transcript accumulation, determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), is given relative to that in wild‐type plants that were assigned a value of unity after normalization. The absolute quantification of TRV RNA1 is given as the number of viral copies per nanogram of total RNA. Means of three measurements ± standard deviation (SD) are shown. Asterisks indicate statistical significance versus the wild‐type control (P < 0.001, Duncan's multiple range test). (A, B) Analysis of AtDIN1 (A) and AtDIN6 (B) expression and accumulation of TRV RNA1 in TRV‐infected Arabidopsis leaves of din1 (A) and din6 (B) mutants at 8 days after inoculation (dai). (C) Analysis of AtDIN11 expression and accumulation of TRV genomic RNA1 in infected leaves of Arabidopsis DIN11 RNAi lines (L10 and L21) at 5 dai (absolute quantification) and 8 dai (relative quantitation). (D) Accumulation of TRV RNA1 in Arabidopsis protoplasts from L10 and L21 lines 24 h after transfection.
TRV and Potato virus X (PVX) promote the expression of NbNRIP1, NbASN and NbDIN11 in infected Nicotiana benthamiana
We were interested in determining whether TRV‐mediated induction of DIN1, DIN6 or DIN11 was unique to Arabidopsis or whether these genes responded to TRV infection in other susceptible hosts. To test this possibility, we first conducted a blast search using Arabidopsis DIN genes as queries to find putative DIN homologue genes in the N. benthamiana expressed sequence tag (EST) database. Our homology‐based search identified chloroplastic N receptor‐interacting protein 1 (NbNRIP1) (EU332891) and NbASN (GQ354808) as genes homologous to Arabidopsis AtDIN1 (At4g35770) and AtDIN6 (At3g47340), respectively. We failed to identify sequences with extensive sequence similarity to Arabidopsis AtDIN11 (At3g49620) through database mining in N. benthamiana (N. benthamiana transcriptome v5 primary transcripts database; http://sydney.edu.au/science/molecular_bioscience/benthamiana/, University of Sydney, Sydney, Australia). However, we were able to amplify an N. benthamiana cDNA product using PCR primers designed from highly conserved sequence domains of Solanum tuberosum (DQ200393.1) and S. lycopersicum ESTs (AK321665.1), that each showed a sequence identity of approximately 67% at the nucleotide level with AtDIN11, suggesting that a DIN11 homologue exists in the N. benthamiana genome.
We then inoculated N. benthamiana plants with TRV, and mRNA accumulation was registered for each DIN gene in the upper non‐inoculated leaves. qRT‐PCR assays indicated that NbNRIP1, NbASN and putative NbDIN11 transcripts accumulated to significantly higher levels in N. benthamiana in the presence of TRV relative to mock‐inoculated plants (Fig. 4). These results were fully reproducible in independent experiments and demonstrated that the DIN homologues NbNRIP1, NbASN and NbDIN11 are induced by TRV in different host species.
Figure 4.
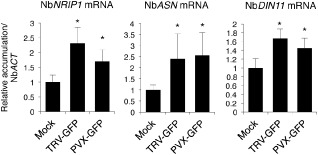
Expression of Dark‐inducible (DIN) homologues in Nicotiana benthamiana in response to Tobacco rattle virus (TRV) and Potato virus X (PVX). Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of NbNRIP1 (N receptor‐interacting protein 1), NbASN (Asparagine synthetase 1) and NbDIN11 expression in upper non‐inoculated leaves of plants infected with TRV‐GFP or PVX‐GFP. Accumulation levels are referenced to the mock‐inoculated controls that were arbitrarily assigned a value of unity after normalization to the NbACT internal control. The means of three measurements ± standard deviation (SD) are shown. Asterisks indicate statistical significance versus the mock‐inoculated control (P < 0.001, Duncan's multiple range test). GFP, green fluorescent protein.
We next wanted to test whether these three DIN genes were activated in N. benthamiana in response to other RNA viruses. To test this, we agroinjected N. benthamiana leaves with a PVX‐green fluorescent protein (PVX‐GFP) clone, and the upper non‐inoculated leaves were analysed for transcript accumulation. qRT‐PCR revealed that NbNRIP1, NbASN and NbDIN11 endogenous transcripts were increased significantly by more than 50% in PVX‐infected plants relative to mock plants agroinfiltrated with an empty vector (Fig. 4). These results demonstrate that DIN1, DIN6 and DIN11 are potential targets of regulation during infections by different viruses in different hosts.
Suppression of NbASN compromises susceptibility to TRV and PVX in N. benthamiana
To investigate the influence of NbNRIP1, NbASN and NbDIN11 genes on TRV susceptibility in N. benthamiana, we tested whether the inactivation of the endogenous DIN genes affected the multiplication and spread of TRV in this species. We took advantage of our TRV infectious clone as a vector for virus‐induced gene silencing (VIGS) to knock down the expression of the target DIN genes. A mixture of Agrobacterium cultures containing TRV1 and a recombinant TRV2 clone that carried a fragment of the target gene was co‐infiltrated on N. benthamiana leaves as described previously (Fig. 5A) (Liu et al., 2002). Expression of the targeted DIN transcripts was monitored alongside TRV accumulation using qRT‐PCR in the upper non‐infiltrated leaves 17 days after agroinjection. A TRV‐GFP clone that carried the GFP coding sequence gene was used as a control. To accurately amplify endogenous NbNRIP1, NbASN and NbDIN11 transcripts, qRT‐PCR primers were designed from regions located outside of those cloned into the corresponding TRV derivatives used for silencing.
Figure 5.
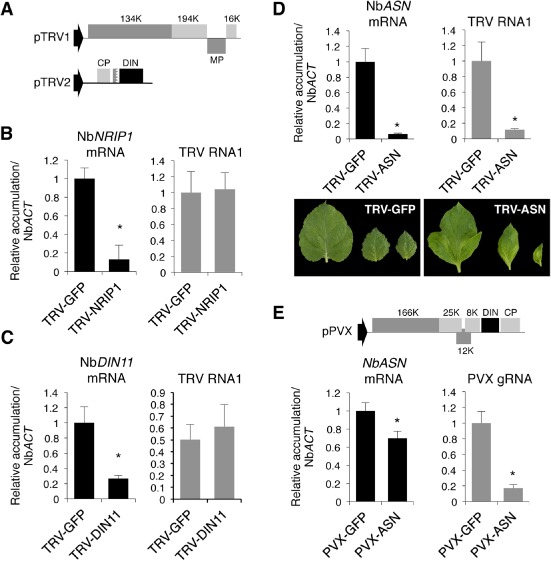
Effect of silencing of Dark‐inducible (DIN) genes on virus accumulation and symptom expression in Nicotiana benthamiana plants. Expression values, determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), are relative to those in plants infected with Tobacco rattle virus‐green fluorescent protein (TRV‐GFP) or Potato virus X‐green fluorescent protein (PVX‐GFP) that were assigned a value of unity after NbACT normalization. The means of three measurements ± standard deviation (SD) are shown. Asterisks indicate significant difference from the TRV‐GFP/PVX‐GFP control (P < 0.001, Duncan's multiple range test). (A) Genome organization of the TRV‐based virus‐induced gene silencing (VIGS) vectors used for silencing of endogenous N. benthamiana DIN homologues. (B, C) Analysis of NbNRIP1 (B) and NbDIN11 (C) expression and TRV RNA1 accumulation in upper leaves of plants infected with TRV‐NbNRIP1 (B) or TRV‐NbDIN11 (C). (D) Analysis of NbASN (Asparagine synthetase 1) expression and accumulation of TRV RNA1 in upper leaves infected with TRV‐NbASN. Symptoms associated with silencing of NbASN in TRV‐infected leaves are shown relative to the asymptomatic infection caused by TRV‐GFP. (E) Analysis of NbASN expression and PVX genomic RNA accumulation in upper leaves infected with PVX‐NbASN and PVX‐GFP. The genome organization of the PVX derivative used to silence NbASN is shown.
When a recombinant TRV‐NbNRIP1 infectious clone that harboured a fragment of the NbNRIP1 gene was introduced into N. benthamiana plants, we observed a five‐fold decrease in endogenous NbNRIP1 transcript levels in the upper leaves relative to plants infected with a TRV‐GFP construct (Fig. 5B). Similarly, infection of N. benthamiana plants with the recombinant TRV‐NbDIN11 clone that carried a fragment of the potato homologue of DIN11 resulted in a two‐ to five‐fold reduction in the relative NbDIN11 transcript levels compared with control plants (Fig. 5C and data not shown). These results demonstrate the robustness of silencing of the target genes and validate our experimental approach. qRT‐PCR also revealed that TRV‐genomic RNA accumulated in these silenced leaves to the same high levels as those in plants infected with TRV‐GFP, suggesting that silencing of NbNRIP1 or NbDIN11 was not sufficient to alter virus accumulation (Fig. 5B,C).
Agrobacterium‐mediated infiltration of TRV‐NbASN, which contained a fragment of the DIN6 Arabidopsis homologue, resulted in approximately 90% inhibition of NbASN expression in systemically infected leaves relative to plants infected with the TRV‐GFP control (Fig. 5D). Strikingly, suppression of NbASN expression in plants infected with TRV‐NbASN was reproducibly accompanied by the appearance of dispersed chlorotic areas and leaf deformation that became conspicuous in the youngest apical leaves at approximately 15 dai (Fig. 5D). In addition, NbASN‐silenced plants were far less susceptible to TRV as they accumulated 60%–90% less viral genomic RNA1 than non‐silenced plants agroinoculated with the control TRV‐GFP construct (Fig. 5D). Reduction of TRV levels occurred only in NbASN‐silenced plants, but not in NbNRIP1‐ or NbDIN11‐silenced plants, which supports the conclusion that low TRV accumulation is caused by knockdown of NbASN.
Given that NbASN was inducible by PVX in N. benthamiana (Fig. 4), we wondered whether silencing of NbASN could also interfere with PVX accumulation in this species. To explore this idea, we adopted a PVX‐based silencing strategy to challenge N. benthamiana plants with an infectious PVX construct containing a fragment of NbASN. qRT‐PCR analysis showed that the endogenous NbASN transcript levels were diminished by about 35% in systemically infected leaves relative to those observed in plants infected with a PVX‐GFP vector (Fig. 5E). Remarkably, partial silencing of NbASN transcripts was sufficient to cause a significant reduction (80%) in PVX RNA levels in the upper infected leaves (Fig. 5E). In these plants, upper silenced leaves infected with PVX‐NbASN did not exhibit a distinguishable phenotype with respect to the non‐silenced leaves infected with PVX‐GFP (data not shown), perhaps because suppression of the endogenous NbASN gene was incomplete in PVX‐NbASN‐infected plants.
Reduced susceptibility to TRV in AtDIN11‐ or NbASN‐silenced plants is not caused by a general activation of defence genes
Our findings that virus accumulation was drastically impaired in Arabidopsis plants in which DIN11 transcripts were silenced or in N. benthamiana leaves with reduced NbASN transcript levels prompted us to investigate whether the inactivation of DIN11 or NbASN stimulated an antiviral plant defence response. To test this possibility, we first conducted expression analyses of Pathogenesis‐related 1 (AtPR1, At2g14610) and Plant defensin 1.2 (AtPDF1.2, At5g44420) genes in the hairpin DIN11 RNAi transgenic L10 and L21 lines. PR1 and PDF1.2 are commonly used to monitor SA‐ and jasmonic acid (JA)‐dependent defence responses (Loake and Grant, 2007). The SA pathway is typically activated during senescence and virus infections, and resistance to certain RNA viruses is SA dependent (Mayers et al., 2005; Whitham et al., 2006). Application of MeJA is known to inhibit the replication of several viruses in compatible interactions (Jameson and Clarke, 2002). Interestingly, we found that AtPDF1.2 expression was compromised in both AtDIN11‐silenced RNAi lines compared with Col‐0, whereas AtPR1 expression somehow differed between the independent RNAi lines tested (Fig. S3, see Supporting Information). qRT‐PCR using RNA preparations from TRV‐infected plants revealed that AtPR1 and AtPDF1.2 transcripts accumulated to comparable levels to those found in non‐infected plants. This finding suggests that lower TRV levels in our DIN11 RNAi lines were not caused by the elevated expression of these two defence markers (Fig. 6A).
Figure 6.
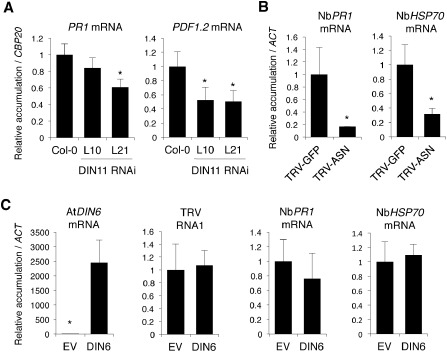
Effect of AtDIN11 (Dark‐inducible 11) and NbASN (Asparagine synthetase 1) on the expression of defence‐related genes in Tobacco rattle virus (TRV)‐infected plants. Expression values, determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), are the means of three measurements ± standard deviation (SD). Asterisks indicate statistical significance versus the controls (P < 0.001, Duncan's multiple range test). (A) Expression analysis of AtPR1 (Pathogenesis‐related 1) and AtPDF1.2 (Plant defensin 1.2) in upper non‐inoculated rosette leaves of Arabidopsis DIN11 RNA interference (RNAi) lines (L10 and L21) systemically infected with TRV. Values in infected Col‐0 plants were assigned a value of unity after AtCBP20 normalization. (B) Expression analysis of NbPR1 and NbHSP70 (Heat‐shock protein 70) in upper N. benthamiana leaves systemically infected with TRV‐NbASN relative to control plants infected with TRV‐GFP which were assigned a value of unity after NbACT normalization. (C) Expression analysis of NbPR1 and NbHSP70 in TRV‐infected N. benthamiana leaves that transiently overexpressed the Arabidopsis AtDIN6 homologue relative to the control leaves in which an empty vector (EV) was agroinoculated (this sample was assigned a value of unity after NbACT normalization).
To determine whether silencing of NbASN in N. benthamiana plants infected with TRV‐NbASN correlated with the activation of defensive genes, the expression of NbPR1 (G1FNL9) and Heat‐shock protein 70 (NbHSP70, GQ354819.1) was examined using the same RNA samples as employed for NbASN and TRV accumulation in transient expression assays. We chose to test NbHSP70 expression because HSPs are associated with plant virus replication (Aranda et al., 1996; Mine et al., 2012). qRT‐PCR revealed that NbPR1 and NbHSP70 transcript levels were reduced by nearly 70%–80% in NbASN‐silenced leaves infected with TRV‐NbASN compared with non‐silenced, TRV‐GFP‐infected controls (Fig. 6B).
We next investigated the expression pattern of NbPR1 and NbHSP70 in TRV‐infected leaves in which a 35S‐driven construct containing the Arabidopsis AtDIN6 homologue gene had been agroinjected 2 days prior to virus inoculation. Transient accumulation of AtDIN6 transcripts was corroborated by qRT‐PCR compared with control plants infiltrated with an empty vector (Fig. 6C). We found that TRV levels in leaves expressing AtDIN6 were similar to those observed in control leaves infiltrated with an empty vector (Fig. 6C). Both NbPR1 and NbHSP70 mRNAs accumulated in the presence of AtDIN6 to the same levels as those observed in control plants infiltrated with empty vector (Fig. 6C). These results suggest that TRV accumulation and induction of NbPR1 and NbHSP70 are unaffected by the ectopic expression of AtDIN6 (Fig. 6C).
Discussion
In our study, we have shown that TRV induces host gene responses that are similar to those activated during senescence, even though TRV infection occurs in the absence of morphological manifestations of disease. This means that the activation of senescence genes during TRV infection does not necessarily imply the initiation of the senescence syndrome. Collectively, DIN genes are activated in leaves undergoing senescence, as well as in leaves subjected to stimuli that normally lead to senescence, such as darkness, phosphate or carbon starvation, wounding or treatment with photosynthesis inhibitors (Fujiki et al., 2001). Microarray‐based analysis of gene expression indicates that the up‐regulation of DIN genes also occurs during responses induced by compatible and incompatible fungal and bacterial pathogens, or on treatment with defence‐inducing chemical signals (Genevestigator data) (Schenk et al., 2005). Therefore, it seems reasonable to anticipate that DIN genes are part of the basal metabolic responses used by cells to cope with stresses that may be common to senescence and pathogen infections. However, like many other stress‐inducible genes, the function of DIN genes remains elusive at this time. In addition, how these genes contribute to the infectious process is an important question that remains unresolved. The main objective of this study was to examine the effect of DIN1, DIN6 and DIN11 on plant susceptibility to virus infection. We found that AtDIN1, AtDIN6 and AtDIN11 were broadly regulated in Arabidopsis in response to TRV, whereas transcripts from NbNRIP1, NbASN and NbDIN11 accumulated to significantly increased levels in N. benthamiana leaves infected with TRV or PVX. This finding indicates that DIN induction probably represents a common response to different viruses in different compatible host species.
Our data indicated that DIN1 was apparently irrelevant for virus infection in both Arabidopsis and N. benthamiana. Similarly, AtDIN1 was shown to be dispensable for disease resistance or tolerance to bacterial and fungal pathogens, although AtDIN1 is strongly elicited during defence responses against these pathogens (Oh et al., 1996; Schenk et al., 2005). However, NbNRIP1, the N. benthamiana homologue of AtDIN1, is required for an effective N‐mediated defence response to Tobacco mosaic virus (TMV) in N. benthamiana (Caplan et al., 2008). VIGS of NbNRIP1 partially abolishes the function of N and allows TMV to spread systemically (Caplan et al., 2008). NbNRIP1 mediates the association of the N immune receptor and the TMV‐encoded 50‐kDa helicase (p50) effector protein to assemble a core recognition complex that allows the plant to detect TMV in infected cells (Caplan et al., 2008). Although TRV accumulation was not altered in NbNRIP1‐silenced plants, it would be interesting to investigate whether NbNRIP1 could also mediate the recognition of putative TRV effector proteins by host immune receptors. The 5′‐upstream sequence of the DIN1 gene contains a TCA motif (TCATCTTCTT) that is highly conserved among genes induced by various stresses (Goldsbrough et al., 1993; Oh et al., 1996). Therefore, this sequence motif may be involved in the induction of DIN1 homologues under the stress caused by TRV or PVX in infected cells.
We found that AtDIN6 was not crucial for TRV infection in Arabidopsis. Likewise, AtDIN6 was unnecessary for basal defence and resistance against bacterial and oomycete infection in Arabidopsis (Hwang et al., 2011). However, inactivation of the AtDIN6 homologue NbASN affected negatively TRV and PVX replication in N. benthamiana. Interestingly, despite the important role of NbASN in viral infection in this species, ectopic expression of AtDIN6 had no significant effects on TRV accumulation. A possible explanation is that DIN6/ASN‐related functions are not affected by elevated expression of DIN6/ASN‐coding genes, as opposed to what is expected from its genetic inactivation. Yet, additional experiments are needed to assess the functionality of the Arabidopsis homologue AtDIN6 in N. benthamiana. We demonstrated that VIGS of NbASN was accompanied by a dramatic reduction in TRV or PVX genomic RNA relative to that in non‐silenced control plants. It is worth noting that TRV‐mediated silencing of the target NbASN transcripts occurs concomitantly with severe morphological defects in the most apical leaves infected with TRV. In contrast, PVX‐NbASN‐infected plants, in which silencing of NbASN was only partial, developed normally with an absence of morphological abnormalities. These observations raise the question of whether symptoms are uniquely caused by disruption of NbASN functions or are also influenced by specific host–virus interactions. Interestingly, VIGS of the DIN6 homologue gene of Capsicum annuum (CaAS1) was found to interfere with pathogen susceptibility by enhancing bacterial infections in pepper (Hwang et al., 2011). The apparent discrepancies between our observations and those reported by Hwang et al. (2011) could be a result of the inherent differences in the experimental approaches and pathogens tested, with different strategies used to infect the plants.
Our results demonstrated that AtDIN11 influences virus susceptibility in Arabidopsis, as manifested by reduced TRV levels in upper non‐inoculated leaves of DIN11 RNAi lines at early stages of infection. Furthermore, our results suggested that AtDIN11 supports TRV replication, as the accumulation of viral genomic RNA was partially inhibited in Arabidopsis protoplasts isolated from knockout lines compared with wild‐type Col‐0. The reduced susceptibility observed in AtDIN11‐ and NbASN‐silenced plants cannot be attributed to a boost in the induction of general defence responses during infection, as none of the defence markers tested in this study showed a changed in expression profile in response to viral infection. This observation is remarkable and supports the idea that AtDIN11 and NbASN have a direct impact on viral susceptibility by modulating virus accumulation upstream and independent of the activation of defence genes. Nevertheless, we cannot exclude the possibility that other components of the defensive reaction respond positively to AtDIN11 or NbASN levels to dampen virus accumulation. Why TRV and PVX are unable to replicate efficiently when AtDIN11 or NbASN activity is compromised is unknown.
DIN6 encodes a glutamine‐dependent asparagine synthetase that regulates the levels of asparagine in the dark or under carbon‐limiting conditions (Lam et al., 1994, 1995, 1998). Induction of DIN6 in virus‐infected plants is presumably linked to the deaminating activity of GDH, a component of senescence and plant defence responses that is up‐regulated in Arabidopsis infected with TRV (Fernandez‐Calvino et al., 2014; Pageau et al., 2006). Glutamate deamination by GDH releases significant amounts of free ammonium that must be rapidly eliminated to avoid deteriorating effects within the cell (Lam et al., 1998; Masclaux‐Daubresse et al., 2006). Given that asparagine is viewed as an ammonia detoxification product, it is sensible to propose that the activation of DIN6 could contribute to counteract the toxic effects of excess ammonium produced during the infection and to help maintain cell integrity, thus allowing effective virus multiplication. Gene or functional redundancy could explain, at least in part, why the genetic inactivation of AtDIN6 in the Arabidopsis din6 mutant has no influence on TRV susceptibility. Indeed, the Arabidopsis genome contains two additional asparagine synthetase homologous genes (AtASN2 and AtASN3), which encode a novel class of asparagine synthetase enzyme with enhanced ammonia‐dependent activity (Lam et al., 1998). DIN11 encodes a protein similar to plant dioxygenases that shows a requirement for 2‐oxoglutarate as a co‐substrate (Fujiki et al., 2001; Prescott and John, 1996). Therefore, like DIN6, the activation of DIN11 in TRV‐infected plants could be coupled to the GDH‐catalysed deamination of glutamate to 2‐oxoglutarate (Masclaux‐Daubresse et al., 2006). However, the precise function of DIN11 remains largely unknown.
To date, very limited information is available concerning the role of host factors in compatible plant–virus interactions. Our work has identified DIN6 and DIN11 as key regulators that support virus infections in systemically infected host plants, and outlines a number of important questions which need to be answered to elucidate the specific mechanisms and molecular pathways involving DIN6 and DIN11 activities during infections in plants.
Experimental Procedures
Plant material, virus inoculation and sampling procedure
All Arabidopsis plants were in the Columbia background (Col‐0), and were grown in controlled environment chambers under 16 h/8 h of light/dark at 19–22 °C. Homozygous plants for T‐DNA insertion lines din1 (NASC code N665464, SALK_020571C) and din6 (NASC code N479505, GK‐829B05) were used. Homozygosity of the mutant alleles was verified by PCR genotyping and by sequencing of the PCR products. din1 contains a T‐DNA insertion in the first intron of the At4g35770 locus (genomic location 16 945 146). din6 contains a T‐DNA insertion in the first exon of the At3g47340 gene (genomic location 17 441 009), 35 nucleotides downstream from the translation initiation site. The primers used for PCR genotyping of homozygous mutants are listed in Table S2 (see Supporting Information).
Nicotiana benthamiana plants were grown in controlled environment chambers under 16 h/8 h of light/dark at 25 °C, and were inoculated at approximately 21 days after germination by infiltration of Agrobacterium tumefaciens cultures containing TRV PpK20 cDNA clones of RNA1 (pTRV1)‐ and RNA2 (pTRV2)‐derived constructs (mixed in a 1:1 ratio), or containing PVX (pgR107)‐recombinant derivatives, as described previously (Liu et al., 2002). Three‐week‐old Arabidopsis plants were inoculated with TRV using extract sap from systemically TRV‐infected N. benthamiana leaves, as described previously (Donaire et al., 2008). Mock inoculation was performed using sap from healthy, non‐infected N. benthamiana leaves. Virus infection was corroborated by RT‐PCR using TRV or PVX sequence‐specific primers (Table S3, see Supporting Information).
Unless otherwise indicated, two or three newly emerging leaves per plant were collected from N. benthamiana infected with TRV or PVX derivatives used for VIGS at ∼16 dai. Two or three upper non‐inoculated rosette leaves per plant were collected from mock‐ and TRV‐inoculated Arabidopsis at ∼8 dai. Each sample consisted of RNA pooled from four to ten plants.
Construction of TRV‐ and PVX‐derived plasmids
The pTRV1, pTRV2 and pgR107‐PVX DNA vectors used in this study have been described previously (Carrington et al., 1999; Liu et al., 2002). To generate the recombinant TRV‐GFP clone, the soluble modified GFP (smGFP) (Chiu et al., 1996) gene cassette under the coat protein promoter of Pea early‐browning tobravirus (PEBV) was inserted into the multiple cloning site (MCS) of pTRV2. To generate the pTRV2 and pgR107 derivatives used for VIGS, cDNA fragments of NbNRIP1 (EU332891.1), AtDIN6/ASN1 (At3g47340) and NbDIN11 transcripts were RT‐PCR amplified from RNA preparations and cloned into pCR2.1 vectors (Invitrogen, Barcelona, Spain) for DNA sequencing. A 303‐bp fragment of N. benthamiana NbNRIP1, which corresponds to nucleotides 58–360, was amplified using NbNRIP1‐F and NbNRIP1‐R primers. A 306‐bp fragment at nucleotide positions 25–330 of Arabidopsis AtDIN6/ASN1, which shares 77% sequence similarity at the nucleotide level with N. benthamiana NbASN (GQ354808/C9DFA8), was amplified using AtDIN6‐F and AtDIN6‐R primers. A 322‐bp fragment, corresponding to nucleotides 462–784 of the DIN11 homologue of S. tuberosum (clone 069B08), was amplified using StDIN11‐F and StDIN11‐R primers. A 606‐bp fragment of the smGFP‐protein coding sequence was amplified using GFP‐F and GFP‐R primers. cDNA fragments were excised from pCR2.1 vectors and inserted into the MCS of TRV2 or pgR107. All primers are listed in Table S3.
Generation of transgenic RNAi lines
The RNAi Gateway® vector pH 7GWIWG2 (II) (Invitrogen) was used to generate an RNAi construct targeted against AtDIN11. The selected target region comprised a 431‐bp fragment from the 5′ terminal region of the Arabidopsis AtDIN11 mRNA that was unique to that particular gene. The primers used are listed in Table S4 (see Supporting Information). All constructs were confirmed by DNA sequencing. The resulting plasmid was transformed into A. tumefaciens strain GV3101 by electroporation. Arabidopsis plants of the Col‐0 ecotype were then transformed using the floral dipping method and selected on the basis of kanamycin resistance (Clough and Bent, 1998). Homozygous T3 transgenic lines were screened for transcript abundance by qRT‐PCR, and only transgenic lines containing reduced levels of DIN11 transcripts relative to the expression in the wild‐type were used for virus infection assays.
RNA preparation and real‐time qRT‐PCR
Details on the minimum information for publication of real‐time qPCR experiments are listed in Tables S5 and S6 (see Supporting Information) (Bustin et al., 2009). To quantify the absolute copy number of TRV RNA1, a standard curve of known concentration of in vitro transcripts was used. Absolute quantification of viral RNA1 was expressed as the number of viral copies per nanogram of total RNA. Sequence‐specific primers are listed in Table S7 (see Supporting Information).
Protoplast preparation and transfection
Arabidopsis protoplasts were isolated from rosette leaves of 3‐week‐old plants and transiently transformed by the polyethylene glycol (PEG) method, as described previously (Yoo et al., 2007), using 30 μg of purified pTRV1 plasmid DNA. Transfected protoplasts were cultured for 24 h in a growth chamber under 16 h of light at 20 °C and 8 h of dark at 22 °C. Protoplasts were harvested by centrifugation, suspended in 500 μL of Trizol reagent and stored at −80 °C for total RNA extraction.
Electrolyte leakage detection and chlorophyll quantification
Electrolyte leakage and chlorophyll a and b levels were measured in inoculated (TRV or mock) and upper non‐inoculated leaves as described previously (De Leon et al., 2002; Inskeep and Bloom, 1985).
Morphological trait analysis and fitness estimation
Measurements were taken at 16 and 29 dai from at least 30 plants per treatment. The weight and number of total seeds produced from healthy and TRV‐infected Arabidopsis plants were measured from at least 30 individual plants per treatment.
Microarray data
CATMAs for gene expression profiling of TRV‐infected Arabidopsis were conducted in our group as described previously (Crowe et al., 2003; Fernandez‐Calvino et al., 2014; Rodrigo et al., 2012), and deposited at the Gene Expression Omnibus (GEO) under accession numbers GSE15557/155562/15558.
Supporting information
Fig. S1 Analysis of senescence‐associated traits during late Tobacco rattle virus (TRV) infection in Arabidopsis.
Fig. S2 Transcript accumulation of Dark‐inducible (DIN) genes in response to Tobacco rattle virus (TRV) infection in Arabidopsis.
Fig. S3 Expression of defence‐related genes in non‐infected DIN11 RNA interference (RNAi) lines.
Table S1 List of genes showing similar responsiveness to Tobacco rattle virus (TRV) infection, dark‐induced senescence and age‐dependent senescence.
Table S2 List of primers used for polymerase chain reaction (PCR) genotyping.
Table S3 List of primers used for virus amplification and in the construction of Tobacco rattle virus (TRV)‐based virus‐induced gene silencing (VIGS) vectors.
Table S4 List of primers used in the construction of Dark‐inducible (DIN)‐overexpressing Arabidopsis transgenic lines.
Table S5 Minimum information required for publication of quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments involving messenger RNAs.
Table S6 Minimum information required for publication of quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments involving absolute quantification of viral RNA.
Table S7 List of primers used in quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR).
Acknowledgements
This work was supported by grants BIO2006‐13107 and BIO2009‐12004 from the Spanish Ministry of Science and Innovation and Fondo Europeo de Desarrollo Regional (FEDER). IG‐B and FJdT were supported by graduate fellowships from MICINN. LF‐C was the recipient of a Junta de Ampliación de Estudios (JAE)‐Doc contract from Consejo Superior de Investigaciones Científicas (CSIC, Spain). VR‐F was the recipient of a Ramon y Cajal contract from the Spanish Ministry of Economy and Competiveness.
The TRV vector was a gift from Dr S. P. Dinesh‐Kumar (Yale University, USA). The PVX vector (pgR107) was kindly provided by Dr David C. Baulcombe (University of Cambridge, UK). We thank Ignacio Hamada (Centro de Investigaciones Biológicas‐CSIC) for technical assistance.
References
- Aranda, M.A. , Escaler, M. , Wang, D. and Maule, A.J. (1996) Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA, 93, 15 289–15 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan‐Wollaston, V. , Earl, S. , Harrison, E. , Mathas, E. , Navabpour, S. , Page, T. and Pink, D. (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol. J. 1, 3–22. [DOI] [PubMed] [Google Scholar]
- Buchanan‐Wollaston, V. , Page, T. , Harrison, E. , Breeze, E. , Lim, P.O. , Nam, H.G. , Lin, J.F. , Wu, S.H. , Swidzinski, J. , Ishizaki, K. and Leaver, C.J. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation‐induced senescence in Arabidopsis. Plant J. 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M.W. , Shipley, G.L. , Vandesompele, J. and Wittwer, C.T. (2009) The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin. Chem. 55, 611–662. [DOI] [PubMed] [Google Scholar]
- Caplan, J.L. , Mamillapalli, P. , Burch‐Smith, T.M. , Czymmek, K. and Dinesh‐Kumar, S.P. (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell, 132, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. , Bisseling, T. , Collmer, A. and Jones, J.D. (1999) Highlights from the Ninth International Congress on Molecular Plant–Microbe Interactions. Plant Cell, 11, 2063–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W. , Niwa, Y. , Zeng, W. , Hirano, T. , Kobayashi, H. and Sheen, J. (1996) Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Crowe, M.L. , Serizet, C. , Thareau, V. , Aubourg, S. , Rouze, P. , Hilson, P. , Beynon, J. , Weisbeek, P. , van Hummelen, P. , Reymond, P. , Paz‐Ares, J. , Nietfeld, W. and Trick, M. (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res. 31, 156–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon, I.P. , Sanz, A. , Hamberg, M. and Castresana, C. (2002) Involvement of the Arabidopsis alpha‐DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 29, 61–62. [DOI] [PubMed] [Google Scholar]
- Donaire, L. , Barajas, D. , Martinez‐Garcia, B. , Martinez‐Priego, L. , Pagan, I. and Llave, C. (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus‐derived small interfering RNAs. J. Virol. 82, 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza, C. , Medina, C. , Somerville, S. and Arce‐Johnson, P. (2007) Senescence‐associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J. Exp. Bot. 58, 3197–3212. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Calvino, L. , Osorio, S. , Hernandez, M.L. , Hamada, I.B. , Del Toro, F.J. , Donaire, L. , Yu, A. , Bustos, R. , Fernie, A.R. , Martínez‐Rivas, J.M. and Llave, C. (2014) Virus‐induced alterations in primary metabolism modulate susceptibility to tobacco rattle virus in Arabidopsis. Plant Physiol. 166, 1821–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y. , Yoshikawa, Y. , Sato, T. , Inada, N. , Ito, M. , Nishida, I. and Watanabe, A. (2001) Dark‐inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111, 345–352. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y. , Nakagawa, Y. , Furumoto, T. , Yoshida, S. , Biswal, B. , Ito, M. , Watanabe, A. and Nishida, I. (2005) Response to darkness of late‐responsive dark‐inducible genes is positively regulated by leaf age and negatively regulated by calmodulin‐antagonist‐sensitive signalling in Arabidopsis thaliana . Plant Cell Physiol. 46, 1741–1746. [DOI] [PubMed] [Google Scholar]
- Gan, S. and Amasino, R.M. (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein, S. , Sabehi, G. , Carp, M.J. , Hajouj, T. , Nesher, M.F. , Yariv, I. , Dor, C. and Bassani, M. (2003) Large‐scale identification of leaf senescence‐associated genes. Plant J. 36, 629–642. [DOI] [PubMed] [Google Scholar]
- Goldsbrough, A.P. , Albrecht, H. and Stratford, R. (1993) Salicylic acid‐inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress‐inducible genes. Plant J. 3, 563–571. [DOI] [PubMed] [Google Scholar]
- Herbers, K. , Takahata, Y. , Melzer, M. , Mock, H.P. , Hajirezaei, M. and Sonnewald, U. (2000) Regulation of carbohydrate partitioning during the interaction of Potato virus Y with tobacco. Mol. Plant Pathol. 1, 51–59. [DOI] [PubMed] [Google Scholar]
- Hwang, I.S. , An, S.H. and Hwang, B.K. (2011) Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67, 749–762. [DOI] [PubMed] [Google Scholar]
- Inskeep, W.P. and Bloom, P.R. (1985) Extinction coefficients of chlorophyll a and b in N,N‐dimethylformamide and 80% acetone. Plant Physiol. 77, 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson, P. and Clarke, S. (2002) Hormone–virus interaction in plants. Crit. Rev. Plant Sci. 21, 205–228. [Google Scholar]
- Lam, H.M. , Peng, S.S. and Coruzzi, G.M. (1994) Metabolic regulation of the gene encoding glutamine‐dependent asparagine synthetase in Arabidopsis thaliana . Plant Physiol. 106, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.M. , Coschigano, K. , Schultz, C. , Melo‐Oliveira, R. , Tjaden, G. , Oliveira, I. , Ngai, N. , Hsieh, M.H. and Coruzzi, G. (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell, 7, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.M. , Hsieh, M.H. and Coruzzi, G. (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana . Plant J. 16, 345–353. [DOI] [PubMed] [Google Scholar]
- Lim, P.O. and Nam, H.G. (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr. Top. Dev. Biol. 67, 49–83. [DOI] [PubMed] [Google Scholar]
- Lin, J.F. and Wu, S.H. (2004) Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628. [DOI] [PubMed] [Google Scholar]
- Lin, S.S. , Wu, H.W. , Elena, S.F. , Chen, K.C. , Niu, Q.W. , Yeh, S.D. , Chen, C.C. and Chua, N.H. (2009) Molecular evolution of a viral non‐coding sequence under the selective pressure of amiRNA‐mediated silencing. PLoS Pathog. 5, e100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. and Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Loake, G. and Grant, M. (2007) Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Love, A.J. , Yun, B.W. , Laval, V. , Loake, G.J. and Milner, J.J. (2005) Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense‐signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol. 139, 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux, C. , Valadier, M.H. , Brugiere, N. , Morot‐Gaudry, J.F. and Hirel, B. (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta, 211, 510–518. [DOI] [PubMed] [Google Scholar]
- Masclaux‐Daubresse, C. , Reisdorf‐Cren, M. , Pageau, K. , Lelandais, M. , Grandjean, O. , Kronenberger, J. , Valadier, M.H. , Feraud, M. , Jouglet, T. and Suzuki, A. (2006) Glutamine synthetase–glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink–source nitrogen cycle in tobacco. Plant Physiol. 140, 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers, C.N. , Lee, K.C. , Moore, C.A. , Wong, S.M. and Carr, J.P. (2005) Salicylic acid‐induced resistance to Cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol. Plant–Microbe Interact. 18, 428–434. [DOI] [PubMed] [Google Scholar]
- Mine, A. , Hyodo, K. , Tajima, Y. , Kusumanegara, K. , Taniguchi, T. , Kaido, M. , Mise, K. , Taniguchi, H. and Okuno, T. (2012) Differential roles of hsp70 and hsp90 in the assembly of the replicase complex of a positive‐strand RNA plant virus. J. Virol. 86, 12 091–12 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.A. , Lee, S.Y. , Chung, I.K. , Lee, C.H. and Nam, H.G. (1996) A senescence‐associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30, 739–754. [DOI] [PubMed] [Google Scholar]
- Pageau, K. , Reisdorf‐Cren, M. , Morot‐Gaudry, J.F. and Masclaux‐Daubresse, C. (2006) The two senescence‐related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. J. Exp. Bot. 57, 547–557. [DOI] [PubMed] [Google Scholar]
- Pontier, D. , Gan, S. , Amasino, R.M. , Roby, D. and Lam, E. (1999) Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39, 1243–1255. [DOI] [PubMed] [Google Scholar]
- Prescott, A.G. and John, P. (1996) DIOXYGENASES: molecular structure and role in plant metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 245–271. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F. , Normanly, J. and Amasino, R.M. (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen‐independent induction of defense‐related genes. Plant Mol. Biol. 40, 267–278. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F. , Noh, Y.S. , Himelblau, E. and Amasino, R.M. (2000) Molecular aspects of leaf senescence. Trends Plant Sci. 5, 278–282. [DOI] [PubMed] [Google Scholar]
- Rodrigo, G. , Carrera, J. , Ruiz‐Ferrer, V. , del Toro, F.J. , Llave, C. , Voinnet, O. and Elena, S.F. (2012) A meta‐analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PLoS ONE, 7, e40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Rusu, A.G. , Manners, J.M. and Maclean, D.J. (2005) The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol. Biochem. 43, 997–1005. [DOI] [PubMed] [Google Scholar]
- Schippers, J.H. , Nunes‐Nesi, A. , Apetrei, R. , Hille, J. , Fernie, A.R. and Dijkwel, P.P. (2008) The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell, 20, 2909–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin, D. and Wolf, S. (2000) Cucumber mosaic virus infection affects sugar transport in melon plants. Plant Physiol. 123, 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. and Dangl, J.L. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Weaver, L.M. , Gan, S. , Quirino, B. and Amasino, R.M. (1998) A comparison of the expression patterns of several senescence‐associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Quan, S. , Chang, H.S. , Cooper, B. , Estes, B. , Zhu, T. , Wang, X. and Hou, Y.M. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Yang, C. and Goodin, M.M. (2006) Global impact: elucidating plant responses to viral infection. Mol. Plant–Microbe Interact. 19, 1207–1215. [DOI] [PubMed] [Google Scholar]
- Wise, R.P. , Moscou, M.J. , Bogdanove, A.J. and Whitham, S.A. (2007) Transcript profiling in host–pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P. , Hirsch‐Hoffmann, M. , Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Analysis of senescence‐associated traits during late Tobacco rattle virus (TRV) infection in Arabidopsis.
Fig. S2 Transcript accumulation of Dark‐inducible (DIN) genes in response to Tobacco rattle virus (TRV) infection in Arabidopsis.
Fig. S3 Expression of defence‐related genes in non‐infected DIN11 RNA interference (RNAi) lines.
Table S1 List of genes showing similar responsiveness to Tobacco rattle virus (TRV) infection, dark‐induced senescence and age‐dependent senescence.
Table S2 List of primers used for polymerase chain reaction (PCR) genotyping.
Table S3 List of primers used for virus amplification and in the construction of Tobacco rattle virus (TRV)‐based virus‐induced gene silencing (VIGS) vectors.
Table S4 List of primers used in the construction of Dark‐inducible (DIN)‐overexpressing Arabidopsis transgenic lines.
Table S5 Minimum information required for publication of quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments involving messenger RNAs.
Table S6 Minimum information required for publication of quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments involving absolute quantification of viral RNA.
Table S7 List of primers used in quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR).


